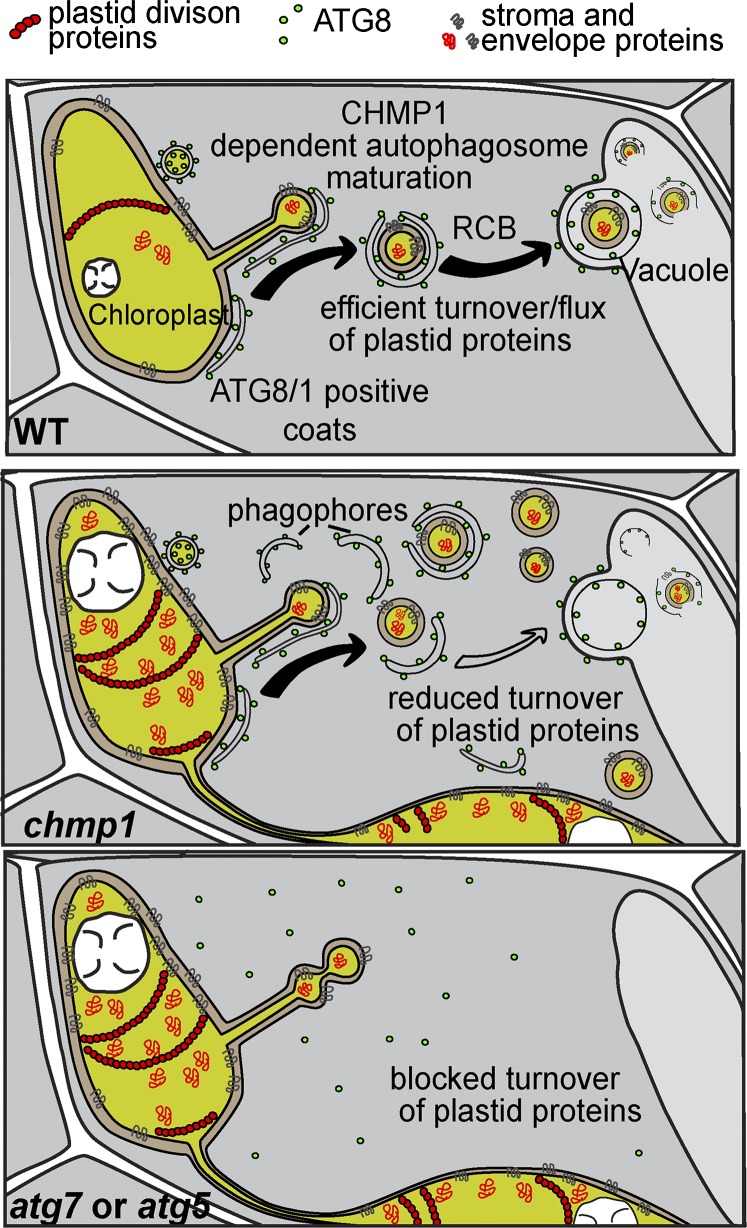
Analysis of mutant plants points to a direct role for CHMP1 in plastid autophagy, demonstrating a connection between the ESCRT machinery and the autophagic clearance of plant plastids.
Abstract
Endosomal Sorting Complex Required for Transport (ESCRT)-III proteins mediate membrane remodeling and the release of endosomal intraluminal vesicles into multivesicular bodies. Here, we show that the ESCRT-III subunit paralogs CHARGED MULTIVESICULAR BODY PROTEIN1 (CHMP1A) and CHMP1B are required for autophagic degradation of plastid proteins in Arabidopsis thaliana. Similar to autophagy mutants, chmp1a chmp1b (chmp1) plants hyperaccumulated plastid components, including proteins involved in plastid division. The autophagy machinery directed the release of bodies containing plastid material into the cytoplasm, whereas CHMP1A and B were required for delivery of these bodies to the vacuole. Autophagy was upregulated in chmp1 as indicated by an increase in vacuolar green fluorescent protein (GFP) cleavage from the autophagic reporter GFP-ATG8. However, autophagic degradation of the stromal cargo RECA-GFP was drastically reduced in the chmp1 plants upon starvation, suggesting that CHMP1 mediates the efficient delivery of autophagic plastid cargo to the vacuole. Consistent with the compromised degradation of plastid proteins, chmp1 plastids show severe morphological defects and aberrant division. We propose that CHMP1 plays a direct role in the autophagic turnover of plastid constituents.
INTRODUCTION
Protein transport and sorting are important cellular processes for all organisms. Eukaryotic cells in particular have evolved complex systems for protein and membrane trafficking of cellular components to appropriate destinations for further processing, proper function, and/or degradation. Two compartments that mediate vacuolar delivery of proteins are autophagosomes and multivesicular bodies (MVBs) (Klionsky, 2007; Hanson and Cashikar, 2012). Autophagosomes arise from a cup-shaped phagophore membrane structure that expands to encircle cytoplasmic material. Phagophore closure generates the double-membrane-bound autophagosome, the outer membrane of which fuses with the vacuolar membrane to deposit its cargo encapsulated by the inner membrane into the vacuolar lumen. The released vesicle, also called an autophagic body, then undergoes rapid breakdown by vacuolar hydrolases, thus completing a degradative process called macroautophagy (hereafter referred to as autophagy).
Autophagy-related (ATG) proteins, the core machinery that controls autophagy, are largely conserved across eukaryotes (Li and Vierstra, 2012). Central to the formation of the autophagosome are the ubiquitin-fold proteins ATG8 and ATG12. Via an ATP-dependent cascade initiated by ATG7, ATG12 becomes attached to ATG5, and the ATG12-ATG5 conjugate then directs the ligation of the lipid phosphatidylethanolamine (PE) to ATG8. The ATG8-PE adduct decorates the enveloping phagophore and helps with vesicle closure, cargo recruitment, and fusion of the resulting autophagosome with the lysosomes/vacuole (Slobodkin and Elazar, 2013). In mouse (Mus musculus), mutations blocking ATG8 and ATG12 function lead to compromised embryogenesis (Kuma et al., 2004), whereas comparable atg mutants in Arabidopsis thaliana are hypersensitive to nutrient deprivation and senesce prematurely (Thompson et al., 2005; Phillips et al., 2008).
Autophagosomes were initially thought to be dedicated to the bulk removal of cytosolic components during starvation but are now known to also remove specific cargo using dedicated autophagy receptors (Klionsky, 2007; Noda et al., 2010; Johansen and Lamark, 2014; Okamoto, 2014). Through these receptors, autophagosomes selectively engulf peroxisomes (pexophagy), mitochondria (mitophagy), endoplasmic reticulum (reticulophagy), RNAs (RNautophagy), ribosomes (ribophagy), and other cellular components. Chloroplast dismantling during senescence also involves the delivery of chloroplastic constituents to vacuoles for degradation (Chiba et al., 2003; Ishida et al., 2008; Martínez et al., 2008; Ono et al., 2013). ATG8-decorated bodies containing Rubisco and other stromal proteins accumulate in the vacuolar lumen of wild-type Arabidopsis plants but not in autophagy mutants (Ishida et al., 2008; Wada et al., 2009). However, the precise mechanism(s) by which autophagy transfers plastid proteins to the vacuole are unclear. Recently, a novel autophagic structure devoid of Rubisco and decorated with ATG8-Interacting Protein 1 (AT1-PS bodies) has been postulated to mediate the vacuolar degradation of some stroma, envelope, and thylakoid proteins (Michaeli et al., 2014).
MVBs regulate the sorting and vacuolar delivery of plasma membrane proteins for degradation. The ESCRT (Endosomal Sorting Complex Required for Transport) machinery, which comprises five distinct complexes and accessory proteins, sorts ubiquitylated membrane proteins into the intraluminal vesicles of MVBs for degradation in vacuoles/lysosomes (Hanson and Cashikar, 2012). One of these complexes, ESCRT-III, associates directly with endosomal membranes and is thought to mediate changes in the membrane architecture that ultimately lead to intraluminal vesicle scission (Schuh and Audhya, 2014). Studies with Drosophila melanogaster, Caenorhabditis elegans, plants, and mammalian cells showing that ESCRT mutants hyperaccumulate autophagosomes (Roudier et al., 2005; Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2007; Rusten and Stenmark, 2009; Djeddi et al., 2012; Katsiarimpa et al., 2013) implied that the ESCRT machinery plays a role in autophagy. Given that the scission of nascent intraluminal vesicles at endosomes and the closure of phagophores into sealed autophagosomes are topologically similar, ESCRT proteins could participate directly in phagophore closure. However, whether the ESCRT machinery has an intimate role in autophagosome dynamics is under debate (Manil-Segalén et al., 2012).
Here, we report a connection between the ESCRT machinery and the autophagic clearance of plant plastids. Whereas ATG5 and ATG7 are necessary for the formation of cytoplasmic bodies containing plastid material, the ESCRT subunit paralogs CHARGED MULTIVESICULAR BODY PROTEIN1 (CHMP1A) and CHMP1B are necessary for phagophore maturation and the efficient delivery of autophagic plastid bodies to the vacuole. However, autophagic output as measured by the vacuolar green fluorescent protein (GFP) cleavage of the autophagy tracer GFP-ATG8 is increased in the double homozygous chmp1a chmp1b (hereafter referred to as chmp1) mutant, suggesting that CHMP1 promotes the efficient sequestration of cargo from plastids into autophagosomes.
RESULTS
Loss of CHMP1 Causes Plastid Clustering and Division Defects
The ESCRT protein CHMP1 is required for proper endosomal sorting of plasma membrane proteins (Howard et al., 2001). A transcript-null mutant for the two functionally redundant Arabidopsis CHMP1 genes, CHMP1A and CHMP1B, mis-sorts crucial plasma membrane proteins required for development and exhibits embryo or early seedling lethality (Spitzer et al., 2009). chmp1 mutants develop into mature but sterile plants when grown on low-strength Murashige and Skoog (MS) medium, thus providing a method to analyze CHMP1 function during subsequent stages of Arabidopsis growth and development.
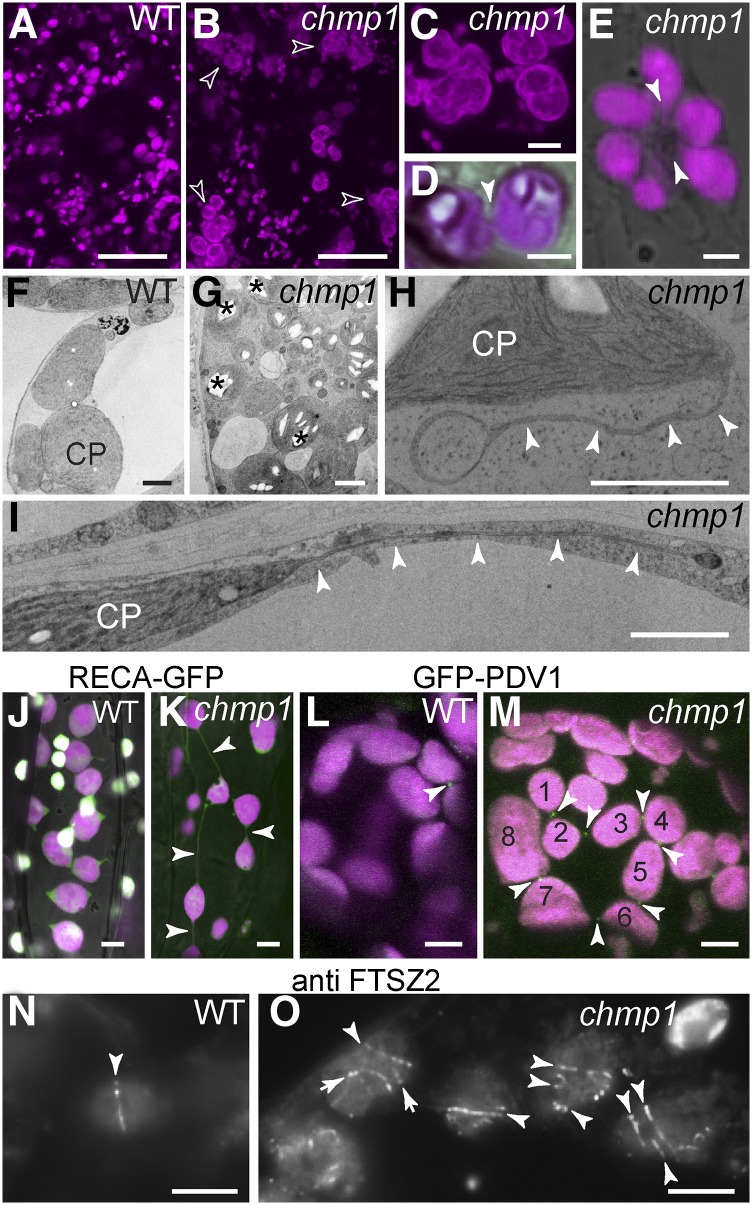
Plastids in 2-week-old chmp1 seedlings contained large starch granules and were frequently found in complex clusters with long extensions/stromules and interconnecting bridges (Figures 1A to 1I; Supplemental Figures 1A to 1H and 2A to 2E). We expressed the stromal marker RECA-GFP (transit peptide of Arabidopsis RECA, a protein involved in plastid DNA repair, fused to GFP) (Köhler et al., 1997) in CHMP1A/chmp1a chmp1b/chmp1b plants and analyzed seedlings from the progeny, which were either double homozygous for chmp1a/b or wild-type-looking siblings containing at least one wild-type CHMP1A allele. We found an unusually high frequency of plastid connections in chmp1 seedlings (Figures 1J and 1K). Plastid clustering and abnormal stromules have been reported in mutants blocked in plastid division, a process mediated by large ring-shaped protein complexes that direct constriction and fission. The plastid division proteins FTSZ1/2 and ARC3 are localized to the stroma, ARC6 and PARC6 to the inner envelope, and PLASTID DIVISION1 (PDV1), PDV2, and DRP5B to the outer envelope (Yang et al., 2008). We localized PDV1-GFP (Miyagishima et al., 2006) and the endogenous FTSZ2 protein (Vitha et al., 2001) in the chmp1 mutant by fluorescence microscopy. In both wild-type and chmp1 cells, PDV1-GFP was readily detected at the constriction of dividing plastids (Figures 1L and 1M), but in mutant cells, PDV1-GFP rings remained at the connections between the chain-like plastid clusters (Figure 1M). Whereas we detected only one FTSZ2 ring per plastid in wild-type cells, the chmp1 mutant plastids retained multiple FTSZ2 rings that could reflect aborted fission events or remnants of past constrictions (Figure 1N to 1O; Supplemental Figures 1I to 1L). Taken together, these results indicate that plastid division is abnormal without CHMP1.
Figure 1.
Plastid Defects in the Arabidopsis chmp1 Mutant.
(A) to (E) Chlorophyll autofluorescence (magenta) in wild-type (A) and chmp1 ([B] to [E]) cells. Large plastid clusters (hollow arrowheads) and connections between plastids in chmp1 hypocotyl cells in (D) and (E) (solid arrowheads) are indicated.
(F) to (I) Transmission electron micrographs of wild-type and chmp1 mutant chloroplasts (CP) in cotyledons of 15-d-old seedlings. Mutant chloroplast clusters with starch granules ([G]; asterisks). Plastid extensions/bridges in mutant plastids are indicated by arrowheads in (H) and (I).
(J) and (K) RECA-GFP (green) and chlorophyll autofluorescence (magenta) in wild-type and chmp1 cells. Connecting bridges between mutant plastids are indicated by arrowheads. PDV1-GFP-positive rings ([L] and [M]; white arrows) in dividing plastids of wild-type and chmp1 mutant cells. Note that PDV1-GFP rings are assembled on constricted bridges in a cluster of at least eight interconnected plastids (M).
(N) and (O) Immunofluorescence detection of FTSZ2 rings (arrowheads) in plastids.
Bars = 50 μm in (A) and (B), 10 μm in (C), 2 μm in (D) to (I), and 5 μm in (J) to (O).
The chmp1 Mutant Accumulates Stromal and Plastid Envelope Proteins
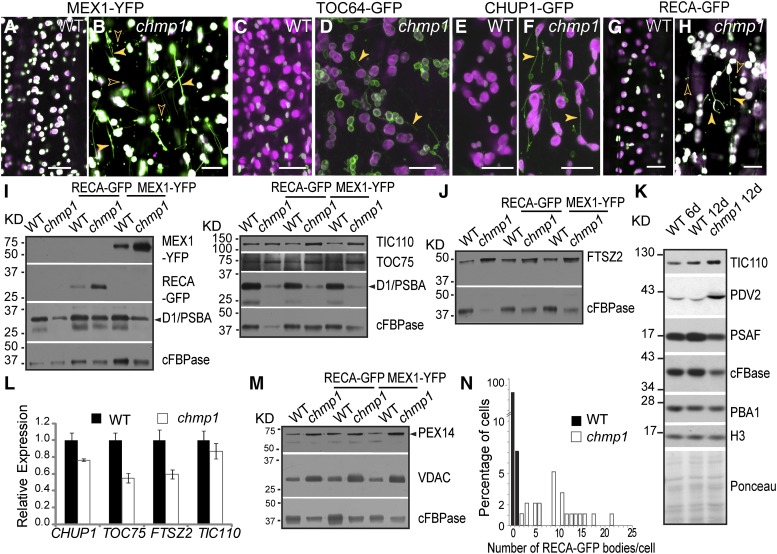
To analyze the structure and dynamics of plastids, we expressed several fluorescently labeled plastid proteins in chmp1 plants and wild-type-looking siblings. Analyses of the inner envelope marker MEX1-YFP (yellow fluorescent protein) (Niittylä et al., 2004) and the outer envelope markers CHUP1-GFP (Oikawa et al., 2008) and TOC64-GFP (Breuers et al., 2012) confirmed the presence of plastid clusters and bridges (Figures 2A to 2F) and demonstrated that the observed plastid defects were not limited to adult green tissues but could also be found in embryos (Supplemental Figures 3A to 3J) and root hairs (Supplemental Figures 3K to 3R). Imaging of RECA-GFP, MEX1-YFP, CHUP1-GFP, and TOC64-GFP also revealed a strong increase in overall fluorescent signal in chmp1 plastids, some of which was detected in stromules/bridges and cytoplasmic vesicles (Figures 2A to 2H; Supplemental Figure 3).
Figure 2.
Plastid Stromal and Envelope Proteins in chmp1.
(A) to (H) Confocal microscopy imaging of fluorescently tagged plastid proteins in hypocotyl cells. Plastid extensions/bridges (solid arrowheads) and cytoplasmic bodies containing plastid proteins (hollow arrowheads) are observed in chmp1 cells. GFP/YFP and chlorophyll autofluorescence are in green and magenta, respectively.
(I) and (J) Immunoblot of MEX1-YFP, RECA-GFP, TIC110, TOC75, FSTZ2, and D1/PBSA. GFP/YFP-tagged proteins were detected with anti-GFP antibodies. Cytoplasmic FBPase (cFBPase) was used as loading control.
(K) Immunoblot detection of TIC110, PDV2, and PSAF in protein extracts from 6-d-old wild-type and 12-d-old wild-type and chmp1 seedlings. cFBPase, PBA1, and H3 were used as internal controls. A Ponceau-stained membrane is shown to illustrate amounts of loaded protein from each sample.
(L) Quantitative PCR of CHUP1, TOC75, FTSZ2, and TIC110 transcripts. Expression was normalized to UBC9. Four biological replicates were analyzed. Bars represent se.
(M) Immunoblot detection of the peroxisomal PEX14 and the mitochondrial VDAC proteins using specific antibodies. cFBPase was used as loading control.
(N) Quantification of RECA-GFP-positive bodies (n = 7 RECA-GFP bodies in 95 wild-type cells and 218 RECA-GFP bodies in 21 chmp1 cells).
The densitometric quantifications of the immunoblots are shown in Supplemental Table 1. Bars = 20 μm in (A) to (H).
To test whether these plastid proteins were more abundant in mutant cells, we quantified their levels by immunoblotting total protein extracts, using cytoplasmic FBPase as a loading control. Consistent with the observed increase in fluorescent signals, higher levels of the RECA-GFP (stroma) and MEX1-YFP (inner envelope) markers, as well as the native proteins FTSZ2 (stroma), TIC110 (inner envelope), and TOC75 (outer envelope), were evident in the chmp1 plants compared with the wild type, suggesting a stabilization of plastid constituents (Figures 2I and 2J; Supplemental Table 1). Because the chmp1 seedlings develop much slower than their wild-type counterparts (Supplemental Figure 2A), as an additional control, we extracted total proteins from 12-d-old wild-type and chmp1 seedlings (same chronological age) and 6-d-old wild-type seedlings (developmentally more similar to the 12-d-old chmp1 seedlings). The 12-d-old chmp1 seedlings contained more TIC110 and PDV2, a plastid division protein localized to the outer envelope, than either 6-d-old or 12-d-old wild-type seedlings, relative to not only cFBPase, but also to PROTEASOME SUBUNIT β1 (PBA1) and HISTONE3 (H3) (Figure 2K; Supplemental Table 1).
Because expression of RECA-GFP, MEX1-YFP, CHUP1-GFP, and TOC64-GFP was driven by the non-native CaMV35S promoter, we tested whether protein accumulation was due to transcriptional upregulation in the chmp1 background by checking GFP abundance in both wild-type and chmp1 plants expressing CaMV35S:GFP. No difference in GFP abundance was detected (Supplemental Figure 4F and Supplemental Table 1). Likewise, levels of the FTSZ2, TIC110, TOC75, and CHUP1 transcripts were not elevated in chmp1 seedlings (Figure 2L), indicating that the increases in mRNA abundance was not the underlying cause for plastid protein hyperaccumulation. However, the content of not all plastid proteins was elevated in chmp1 plants. The abundance of the thylakoid proteins D1/PsbA and PHOTOSYSTEM I SUBUNIT F (PSAF) relative to cytoplasmic and nuclear proteins used as internal controls was unchanged (Figures 2I and 2K; Supplemental Table 1). This distinction might reflect the fact that the several pathways participate in the degradation of plastid proteins (Ling et al., 2012; Lee et al., 2013; Michaeli et al., 2014). Whereas stromal components are thought to be degraded in the vacuole and envelope proteins by the proteasome (Ling et al., 2012; Jarvis and López-Juez, 2013) and the vacuole (Michaeli et al., 2014), D1/PSBA is removed internally by stroma-localized proteases (Haussühl et al., 2001).
Taken together, our results indicate that impaired CHMP1 function leads to the accumulation of stromal and plastid envelope proteins through a posttranscriptional mechanism, most likely protein degradation. Also of note, we detected elevated amounts of the peroxisomal PEX14 protein and outer mitochondrial membrane protein VOLTAGE-DEPENDENT ANION CHANNEL (VDAC) in chmp1 (Figure 2M; Supplemental Table 1), suggesting that the turnover of peroxisomal and mitochondrial constituents is also affected by CHMP1A/B.
The chmp1 Mutant Accumulates Cytoplasmic Bodies with Plastid Proteins
Besides abnormal plastids, chmp1 cells contained an unusually high number of cytoplasmic bodies (0.81 μm in diameter ± 0.35 μm; sd, n = 19 cells) labeled with RECA-GFP and MEX1-YFP (Figures 2B, 2H, and 2N). Many membrane-bound carriers that deliver plastid contents to the vacuole for degradation during leaf senescence have been characterized. The RECA-GFP bodies seen in chmp1 cells morphologically resemble Rubisco-containing bodies (RCBs) and senescence-associated vacuoles previously documented to arise during leaf senescence and proposed to help transport stromal contents like Rubisco and RECA-GFP to the vacuole for breakdown (Chiba et al., 2003; Ishida et al., 2008; Martínez et al., 2008), but appear distinct from the much smaller ATI1-PS bodies (50 to 100 nm) (Michaeli et al., 2014). Our results indicate that not only stromal but also envelope proteins such as MEX1-YFP become incorporated into these bodies. Based on the imaging of RECA-GFP, we found a 140-fold increase in the number of cytoplasmic RCB-like structures in the mutant cells compared with the wild type (Figure 2N).
RCBs are part of a degradation pathway for stromal, but not for thylakoid components, and their degradation in vacuoles of senescing tissues depends on the autophagy components ATG5 and ATG7 (Ishida et al., 2008; Lee et al., 2013). RCBs are thought to form by tip-shedding of stromules, but experimental evidence for their formation and transport is limited (Ishida and Yoshimoto, 2008; Ishida et al., 2008). Our observations that stromal and plastid envelope proteins together with RCB-like vesicles accumulate in a CHMP1-deficient mutant supports a model whereby the ESCRT machinery promotes the autophagic delivery of RCB to the vacuole, but not RCB formation.
Autophagy Proteins Associate with Chloroplasts and Are Required for Releasing RECA-GFP-Containing Bodies
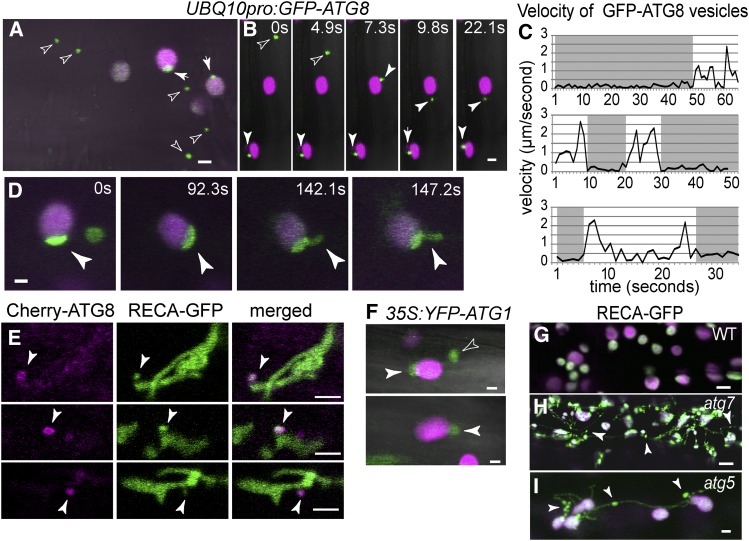
To reveal the underlying autophagic mechanism responsible for plastid protein degradation, we analyzed in more detail the dynamics of the autophagic tracer GFP-ATG8 (Thompson et al., 2005; Kuma et al., 2007) in wild-type seedlings. ATG8 is recruited to phagophores and decorates the autophagosome upon membrane closure. From the analysis of root cells by confocal microscopy, GFP-ATG8 was localized to the limiting membrane of compartments 0.89 μm ± 0.34 μm (sd, n = 16 cells) in diameter that likely represent autophagosomes (Figures 3A and 3B). These autophagosomes moved freely in the cytoplasm, but upon encountering plastids they often stopped and associated with the plastid surface from a few seconds up to 1 min (Figures 3B and 3C; Supplemental Movie 1). In some cases, individual GFP-ATG8-positive autophagosomes could be seen visiting multiple plastids in succession (Supplemental Movie 1). In addition, we identified GFP-ATG8-decorated coats associating with the stromules/plastid extensions that transiently protruded from individual plastids (Figure 3D; Supplemental Movie 2).
Figure 3.
Dynamics of Autophagic Membranes and Plastids in Wild-Type Arabidopsis.
(A) and (B) GFP-ATG8-positive autophagosomes transiently associated with plastids. Chlorophyll autofluorescence and GFP-ATG8 in magenta and green, respectively. Free (hollow arrowheads) and plastid-associated (solid arrowheads) GFP-ATG8-decorated autophagosomes in representative control hypocotyl cell (A) and in still images extracted from Supplemental Movie 1 (B).
(C) Graphs depicting the measured velocities of GFP-ATG8 autophagosomes. Autophagosomes moved freely within the cytoplasm (white regions) but stopped upon association with chloroplasts (gray regions).
(D) Still images extracted from Supplemental Movie 2 showing distinct GFP-ATG8-positive coats assembled on plastids and on plastid extensions.
(E) Epidermal cells at the root expansion zone expressing mCherry-ATG8 and RECA-GFP. Arrowheads show mCherry-ATG8-decorated membranes completely surrounding RECA-GFP-filled plastid extensions.
(F) Association of YFP-ATG1A-decorated autophagosomes with plastids.
(G) to (I) RECA-GFP imaging in wild-type and autophagy mutants. Note plastid bridges and extensions with unreleased RECA-GFP bodies/RCB (arrowheads) in the atg mutants. Bars = 2 μm.
To further investigate this ATG8/plastid interaction, we analyzed by confocal microscopy wild-type root cells stably expressing both RECA-GFP and mCherry-ATG8. We captured multiple events where the mCherry-ATG8 signal surrounded RECA-GFP-positive plastid extensions (Figure 3E). We imaged a second autophagic reporter YFP-ATG1A, which also localizes to autophagic membranes (Suttangkakul et al., 2011). Similar to GFP-ATG8, the YFP-ATG1A signal transiently associated with plastids (Figure 3F).
Previous studies showed that Arabidopsis plants deficient in ATG5 more frequently generate plastid extensions and fail to accumulate RCBs within vacuoles (Ishida et al., 2008). Our observations of RECA-GFP in the null atg5-1 and atg7-2 mutant seedlings confirmed these observations and also showed that, similar to chmp1, some plastids in atg5-1 and atg7-2 mutants abort scission during division and remain connected by long bridges (Figures 3G to 3I). Moreover, dark-treated atg5-1 and atg7-2 seedlings failed to accumulate RECA-GFP-positive bodies in the cytoplasm. The plastid extensions in atg5-1 and atg7-2 developed a rosary-like profile, consistent with the impaired release of RCBs. Collectively, our data indicate that autophagic vesicles physically associate with plastids and suggest that they form a dynamic coat that accompanies the release of plastid material into the cytoplasm. CHMP1 acts subsequently, possibly by promoting the delivery of the resulting RCBs to the vacuole.
Impaired Autophagosome Formation and Autophagic Cargo Transport in chmp1
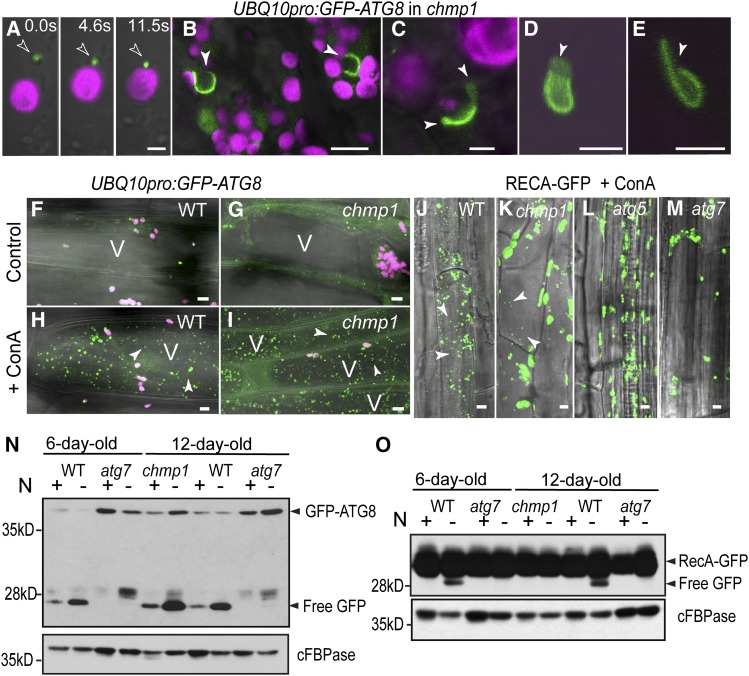
To define the relationship between ESCRT and autophagy, we also analyzed the GFP-ATG8 reporter in the chmp1 background. Similar to those in wild-type cells, GFP-ATG8-decorated membranes transiently interacted with chmp1 plastids (Figure 4A), suggesting that GFP-ATG8/plastid association does not require CHMP1. However, chmp1 cells accumulated aberrant GFP-ATG8 structures possibly with a double-membrane, open configuration that strongly resembled enlarged phagophores (Figures 4B to 4E). Phagophores are short-lived structures and are not commonly detected in normal cells (Kirisako et al., 1999; He et al., 2008; Yen et al., 2010; Le Bars et al., 2014). In agreement, we unambiguously identified 29 GFP-ATG8-positive, phagophore-type structures in 153 randomly selected mutant root cells, but found none in 133 wild-type cells.
Figure 4.
GFP-ATG8-Decorated Autophagosomes and Coats Associated with Chloroplasts in chmp1 Hypocotyl Cells.
(A) GFP-ATG8-decorated autophagosome (arrowhead) associated with a chmp1 plastid at different time points. Chlorophyll autofluorescence and GFP-ATG8 in magenta and green, respectively.
(B) to (E) GFP-ATG8-positive phagophore-like structures in chmp1 hypocotyl cells. The open end of the putative phagophores is indicated by the arrowheads.
(F) to (I) Dark-induced autophagy in wild-type and chmp1 hypocotyl cells expressing GFP-ATG8 and treated with ConA. Note the presence of autophagic bodies (arrowheads) inside the vacuole (V) in (H) and (I).
(J) to (M) Dark-induced autophagy in seedlings expressing RECA-GFP and treated with ConA. RECA-GFP bodies inside the vacuoles are indicated by arrowheads.
(N) and (O) Detection of cleaved GFP from GFP-ATG8 (N) and RECA-GFP (O) in Columbia-0 (WT) and mutant seedlings grown with or without N.
The densitometric quantifications of the immunoblots in (N) are shown in Supplemental Table 2. Bars = 2 μm in (A), 5 μm in (B) and (J) to (M), 1 μm in (C) to (E), and 20 μm in (F) to (I).
To determine whether the lack of CHMP1 affects autophagy progression, we induced autophagy in GFP-ATG8- and RECA-GFP-expressing wild-type and chmp1 seedlings by either dark treatment or nitrogen (N) starvation followed by exposure to concanamycin A (ConA), a drug that attenuates the vacuolar breakdown of autophagic bodies. Under these conditions, the accumulation of autophagic bodies can be used as a readout of autophagy completion (Li et al., 2014). We detected GFP-ATG8- and RECA-GFP-decorated autophagic bodies in both chmp1 and wild-type vacuoles (Figures 4F to 4K), indicating that autophagy is not completely abolished in the absence of CHMP1. However, whereas a comparable density of GFP-ATG8-decorated autophagic bodies was found in the two genotypes (Figures 4H and 4I), there was a drastic reduction in the subset of RECA-GFP-decorated autophagic bodies in the chmp1 background (Figures 4J and 4K). As expected and reported previously by Ishida et al. (2008), under the same conditions, no RECA-GFP-containing autophagic bodies were detected inside the vacuoles of atg5 and atg7 mutant seedlings (Figures 4L and 4M).
Prior studies demonstrated that when autophagic reporters labeled with GFP are delivered to the vacuole, the GFP moiety is cleaved and accumulates in a free form due to its stability inside the vacuole (Djeddi et al., 2012; Li et al., 2014). In this way, the ratio of free GFP to GFP-tagged protein can be used as a semiquantitative measure of autophagic flux and whether this delivery changes with environmental or genetic perturbations. For example, whereas the release and accumulation of GFP from GFP-ATG8 is readily detectable in wild-type Arabidopsis starved for N, it is completely absent in atg7-2 (Li et al., 2014) (Figure 4N). Autophagic flux as measured by the abundance of vacuolar cleaved GFP relative to GFP-ATG8 appeared not to be compromised in chmp1 plants both under normal and starvation conditions (Figure 4N; Supplemental Table 2). However, overall autophagy was elevated in the chmp1 background as evidenced by the increased levels of both GFP-ATG8 and its cleavage product (Figure 4N; Supplemental Table 2). We also used a similar assay to confirm that the RECA-GFP reporter was indeed delivered to the vacuole via a CHMP1-dependent autophagic route. When chmp1 plants were assayed for the release of free GFP from RECA-GFP, a drastic reduction of free GFP was detected similar to that observed with atg7-2 plants (Figure 4O), indicating that CHMP1 does affect the efficient autophagic delivery of chloroplast cargo to the vacuole.
To further dissect genetically the interaction between the ESCRT and autophagy pathways, we analyzed triple mutant combinations of chmp1a and chmp1b with either the atg5-1 or the atg7-2 alleles. From a screen of 1414 seeds derived from six selfed plants heterozygous for chmp1a and homozygous mutant for both the chmp1b and atg5-1 alleles, and 941 seeds from four selfed plants heterozygous for chmp1a and homozygous mutant for both the chmp1b and atg7-2 alleles, we failed to recover triple homozygous individuals. From analysis of seeds, we observed that 37 and 38% of the embryos carrying the atg7-2 and atg5-1 mutations, respectively, had aborted (Table 1). Arrested embryos with the morphological defects frequently seen in chmp1a/b homozygous mutants were common within these aborted seeds (Figure 5), indicating that the combination of chmp1a chmp1b with either atg5-1 or atg7-2 leads to early embryo lethality. The fact that chmp1 seeds cannot germinate without ATG7 or ATG5 suggests that autophagy partially compensates for the lack of ESCRT function.
Table 1. Percentage of Aborted Seeds in Various Genotypes Carrying atg5-1, atg7-2, chmp1a, and chmp1b Mutant Alleles.
| Genotype of Mother Plant | Percentage of Aborted Seeds | Total No. of Seeds |
|---|---|---|
| atg7-2/atg7-2 | 15% | 233 |
| atg5-1/atg5-1 | 26% | 283 |
| CHMP1A/chmp1a chmp1b/chmp1b | 6% | 272 |
| CHMP1A/chmp1a chmp1b/chmp1b atg7-2/atg7-2 | 37% | 319 |
| CHMP1A/chmp1a chmp1b/chmp1b atg5-1/atg5-1 | 38% | 207 |
| CHMP1A/chmp1a CHMP1B/chmp1b ATG5/atg5-1 | 4% | 307 |
| Columbia-0 | 1% | 283 |
Figure 5.

Arrested Seed Development in Plants Segregating atg5 chmp1a chmp1b and atg7 chmp1a chmp1b Mutants.
Normal and arrested seeds produced by a CHMP1A/chmp1a chmp1b/chmp1b atg5/atg5 plant were cleared for analysis. Arrested embryos are indicated by asterisks. Bar = 5 μm.
DISCUSSION
A Mechanistic Connection between ESCRT Proteins and Autophagy
Previous studies have linked autophagy and ESCRT functions based on the increased number of autophagosomes in ESCRT mutants (Rusten and Stenmark, 2009; Hurley and Hanson, 2010; Katsiarimpa et al., 2013). However, whether the increase in autophagosome frequency reflects a direct role for ESCRT in the formation/maturation of autophagosomes or autophagy induction remained controversial. The chmp1 mutant shares many phenotypic defects with atg mutants, including reduced degradation of some plastid proteins, aberrant plastid division, and a hyperaccumulation of starch (Lee et al., 2013; Wang et al., 2013). These phenomena, together with the presence of arrested phagophores and the reduced autophagy-mediated degradation of the plastid marker RECA-GFP, clearly point to a direct role for CHMP1 in plastid autophagy. Like the Arabidopsis chmp1 mutant, C. elegans ESCRT mutants (Djeddi et al., 2012) show increased autophagy as measured by GFP cleavage from the autophagic reporter GFP-ATG8. This observation led to the conclusion that the ESCRT machinery is not required for autophagosome maturation and that the accumulation of autophagosomes in ESCRT mutants is due to the upregulation of the autophagy pathway (Djeddi et al., 2012). However, in Arabidopsis, the autophagic degradation of plastid components is greatly reduced in the absence of CHMP1 even though more GFP-ATG8 is delivered to the vacuole. This indicates that ESCRT mutants exhibit both an upregulation of autophagy and a reduction in the degradation of autophagic cargo, likely due to the inefficient sequestration of cargo within autophagosomes as indicated by the abundant abnormal phagophores in the chmp1 mutant. It is noteworthy that the chmp1 mutant also hyperaccumulates peroxisomal and mitochondrial proteins, suggesting that such an ESCRT-mediated autophagic route represents a common mechanism for recycling material from organelles in plants and potentially other eukaryotes.
The degradation of plastid components requires autophagy-dependent and -independent pathways. Stroma contents are exported from naturally or artificially senescing chloroplasts to the vacuole via RCBs, senescence-associated vacuoles, and the smaller ATI1-PS bodies (Chiba et al., 2003; Otegui et al., 2005; Ishida et al., 2008; Martínez et al., 2008; Michaeli et al., 2014). Whereas the vacuolar degradation of RCB and ATI1-PS bodies depends on autophagy (Ishida et al., 2014; Michaeli et al., 2014), the formation of senescence-associated vacuoles seems to be autophagy independent (Otegui et al., 2005). The degradation of plastid envelope proteins depends on the proteasome (Ling et al., 2012) and at least for some envelope proteins, on ATI1-PS bodies (Michaeli et al., 2014). The degradation of the thylakoid D1/PSBA protein, by contrast, is mediated by the stroma-localized serine endopeptidase DEGP2 (Haussühl et al., 2001) and the metalloprotease FTSH (Lindahl et al., 2000).
Although our work does not explore all possible pathways for plastid turnover, it clearly demonstrates a connection between the ESCRT machinery and autophagy-dependent turnover of plastid constituents. It also shows that the autophagy machinery is required for the release of RCBs into the cytoplasm, in addition to their delivery to the vacuole. Such a mechanism implies a sorting process that sequesters unwanted plastid material into domains of stromules/extensions for subsequent encapsulation into cytoplasmic RBCs. CHMP1 does not participate in the release of RCB from plastids but in their delivery to the vacuole. Possibly by promoting phagophore maturation/closure, CHMP1 helps efficiently sequester autophagic plastid cargo into autophagosomes. In the absence of CHMP1, phagophore closure is delayed, thus allowing RCBs to escape and accumulate in the cytoplasm (Figure 6).
Figure 6.
Model of the Functions of CHMP1 and ATG Proteins in the Autophagy-Dependent Degradation of Plastid Material.
Autophagic components mediate the release of RCBs containing plastid cargo into the cytoplasm. Impaired sequestration of this cargo into autophagosomes in the absence of CHMP1 causes the accumulation of phagophore-like structures and cytoplasmic RCBs and compromises the autophagic turnover of stromal and envelope proteins. Abnormally high levels of plastid division proteins such as FTSZ2 and PDV1 interfere with plastid division, leading to plastid clustering.
Plastid Division Defects in ESCRT and Autophagy Mutants
Based on our observations, plastids do not seem to complete abscission after division in chmp1. Plastid division involves constriction of four concentric rings, including the internal, stroma-localized FTSZ ring composed of FTSZ1 and FTSZ2, and the cytoplasmic dynamin-like DRP5B ring. These ring complexes are connected across the two envelope membranes through interactions with membrane-anchored envelope proteins (Osteryoung and Pyke, 2014). In Arabidopsis, overexpression of FTSZ1 or FTSZ2 in the wild-type background results in dose-dependent plastid division defects, with plants having fewer, larger chloroplasts than in wild-type, heterogeneous chloroplast morphology, randomly organized FSTZ filaments, and at least in the case of FTSZ1, long interconnecting bridges (Vitha et al., 2001; McAndrew et al., 2001; Raynaud et al., 2004). Impaired division is observed in plants overexpressing FTSZ1 by <3-fold (Stokes et al., 2000; Schmitz et al., 2009). Overexpression of several other plastid division proteins also causes related but distinct abnormalities in chloroplast division, FTSZ filament organization, and chloroplast morphology (Vitha et al., 2003; Maple et al., 2007; Zhang et al., 2009, 2013). By contrast, overexpression of PDV1 and/or PDV2 has the opposite effect in that it results in an increased plastid division rate and smaller and more numerous plastids per cell (Okazaki et al., 2009).
Here, we detected accumulation of both FTSZ2 and PDV2 together with other stroma and envelope proteins in chmp1 plants, suggesting that the abundance of other plastid division proteins is also misregulated. We detected a 7-fold increase in the amount of FTSZ2 in our immunoblots of chmp1. Much lower increases in either FTSZ1 or FTSZ2 (<3- or 4-fold) are sufficient to cause division defects in wild-type plants. However, the plastid division defects in chmp1 only partly resemble those caused by FTSZ1 overexpression, namely, multiple and disorganized FTSZ filaments and plastids with long interconnecting bridges (Vitha et al., 2001), and they are generally distinct from those caused by overexpression of other division proteins. It is hard to predict or even experimentally mimic the effect of overaccumulation of multiple plastid division proteins considering that overexpression has different, sometimes opposite effects. However, it is likely that the tightly regulated abundance of these factors is critical for normal plastid division and morphology such that even small changes in their levels could affect division activity (Vitha et al., 2001; Okazaki et al., 2009). Therefore, we postulate that the plastid division defects in chmp1 are a secondary effect of impaired degradation of plastid division constituents. The fact that similar plastid division defects were observed in atg mutants (Figures 3H and 3I) further supports this hypothesis. Based on our solid evidence indicating that autophagosome maturation and vacuolar delivery of autophagic plastid cargo is partially impaired, we propose that the plastid defects in chmp1 are due to abnormal autophagy-mediated turnover of the plastid division machinery.
It is noteworthy that the defects in plastid morphology seem to be more severe in chmp1 than in atg mutants. Several possibilities could explain this observation. First, in addition to its function in autophagy, CHMP1 could have a direct role in plastid division. Unfortunately, appending fluorescent tags to ESCRT-III proteins renders them nonfunctional, thus preventing us from testing if CHMP1 specifically associates with dividing plastids. However, we consider this scenario unlikely given that ESCRT-III proteins have high affinity for negatively curved membranes, such as the neck of a forming endosomal vesicle, the constriction zone of a diving animal cell, the neck of a retrovirus-induced vesicle at the plasma membrane, or a closing autophagosome (Fyfe et al., 2011; Henne et al., 2013), but not for positively curved membranes such as the cytoplasmic face of the constricted division site of a plastid. Second, CHMP1 could also affect an autophagy-independent pathway for plastid turnover. We have not found any evidence of a role for the ESCRT-dependent endosomal pathway in plastid degradation, though a connection between CHMP1 and some of the autophagy-independent pathways for plastid turnover cannot be completely ruled out at this point. Third and most likely, the drastic accumulation of starch, together with its severe developmental and growth defects due to the general mislocalization of plasma membrane proteins in chmp1 (Spitzer et al., 2009), could in turn enhance the defects in plastid morphology induced by impaired autophagy.
The confocal images of RECA-GFP and other fluorescently tagged plastid proteins (Figure 2G; Supplemental Figure 4) showed increased accumulation of these proteins inside plastids in chmp1 plants compared with wild-type controls. This seems in contradiction to the observation that the chmp1 mutant is able to remove RCBs from plastids. However, a defect in any step of a trafficking pathway could affect the overall flux of its components. For example, as a typical ESCRT mutant, chmp1 is impaired in the endosomal-mediated degradation of plasma membrane proteins internalized by endocytosis (Spitzer et al., 2009). Nonetheless, both the polarization and abundance of the auxin efflux carriers PIN1 and PIN2 at the plasma membrane, which depend on trafficking processes not directly related to ESCRT-III function (e.g., polar exocytosis, endocytosis, and early endosomal recycling; Luschnig and Vert, 2014), are strongly misregulated in the chmp1 mutant. Presumably, impaired degradation generates an excess of PIN1 and PIN2, which leads to misregulation of their abundance through all steps of their trafficking pathways (Spitzer et al., 2009). In the same way, the inability to degrade RCBs could feedback in protein accumulation in plastids just as a defect in endosomal sorting results in accumulation of plasma membrane proteins at the cell surface.
METHODS
Plant Material
The following lines were used in this study: chmp1a/CHMP1A chmp1b/chmp1b (Spitzer et al., 2009), ProUBQ10:GFP-ATG8a (Kim et al., 2013), CaMV35S:RecA-GFP (Köhler et al., 1997), ProPVD1:PDV1-GFP (Miyagishima et al., 2006); CaMV35S:MEX1-YFP (Niittylä et al., 2004), CaMV35S:CHUP1-GFP (Oikawa et al., 2008); atg5-1 and atg7-2 (Thompson et al., 2005; Chung et al., 2010), ProUBQ10:mCherry-ATG8a, and CaMV35S:YFP-ATG1a (Suttangkakul et al., 2011). Fluorescent protein expression cassettes were introgressed into CHMP1A/chmp1a chmp1b/chmp1b, atg5-1, and atg7-2 plants by crossing.
Plants expressing CaMV35S:TOC64-III-GFP were generated by Agrobacterium tumefaciens-mediated transformation of the corresponding plasmid (Breuers et al., 2012) into CHMP1A/chmp1a chmp1b/chmp1b plants. Seeds were grown on agar plates with 0.25× MS, stratified for 4 d, and grown in 16/8-h light/dark cycle at 22 to 25°C. From the progeny of CHMP1A/chmp1a chmp1b/chmp1b plants, homozygous chmp1 mutant embryos and seedlings and wild-type-looking siblings, which contain at least one wild-type copy of CHMP1A (Spitzer et al., 2009), were analyzed.
Confocal Microscopy
Control and mutant seedlings were grown until the emergence of the second true leaf in control seedlings (∼2-week-old seedlings) and mounted in water between a Gold Seal glass slide (Thermo Scientific) and a cover slip. Imaging was performed with a Zeiss LSM 510 META using a Plan-Apochromat 63×/NA1.4 oil differential interference contrast objective. The following filters/settings were used: for chlorophyll autofluorescence, excitation with krypton/argon laser line 488 nm, MBS: HFT 488 and band-pass 650 to 710 IR filter; for chlorophyll autofluorescence and GFP/YFP, excitation with krypton/argon laser line 488 nm, MBS: HFT 488 and band-pass 500 to 530 IR and band-pass 650 to 710 IR filters; for GFP and mCherry, excitation with krypton/argon laser line 458 nm and helium/neon laser line 543, MBS: HFT KP 700/543, metadetector 602 to 698 nm and 505 to 570 nm; for propidium iodide, excitation with krypton/argon laser line 488 nm, MBS: HFT 488, and LP560 filter.
ConA Treatment
Carbon starvation was initiated by transferring seedlings to liquid media containing 0.2 g/L MS. Seedlings were incubated under gentle agitation for 24 h in darkness after which ConA in DMSO was added to a final concentration of 1 μM. As control, 1% DMSO was used. After 36 h in the dark with gentle agitation, seedlings were mounted for analysis.
For the GFP cleavage assays, either 6- or 12-d-old seedlings expressing GFP-ATG8a or RECA-GFP grown on 0.5× MS plate with 1% sucrose under long-day conditions (16 h light/8 h dark) were transferred to nitrogen-deficient (-N) liquid medium containing 1% sucrose and incubated under gentle agitation for 24 h in darkness. Total protein extracted from treated seedlings was performed as described previously (Suttangkakul et al., 2011). Clarified protein extracts were then subjected to SDS-PAGE and immunoblot analysis with anti-GFP antibodies (Roche) with dilution 1:5000.
RNA Extraction and Quantitative PCR
Four independent biological samples were analyzed for each control and chmp1a chmp1b mutant seedlings. Approximately 10 15-d-old seedlings (20 to 35 mg fresh weight) per biological sample were harvested. RNA was extracted using Trizol (Invitrogen), treated with DNase I (Invitrogen) and RNaseOUT (Invitrogen) according to the manufacturer’s recommendations, and quantified with a Nanodrop 2000 UV spectrophotometer (Thermo Scientific). cDNA was synthesized from 1 µg RNA with SuperScript III (Invitrogen) and used as template for PCR with MAXIMA SYBR Green/ROX qPCR Master Mix (Thermo Scientific) according to manufacturer’s recommendations. Reactions of all eight samples (four mutant and four control) were run with technical triplicates on a Stratagene MX3000P qPCR system and analyzed with LinRegPCR (version 2013.0, http://www.hartfaalcentrum.nl/index.php?main=files&sub=LinRegPCR). Amplification of UBC9 (UBIQUITIN CONJUGATING ENZYME9) was used as reference. Primers used for quantitative PCR were designed with Quantprime (http://www.quantprime.de/main.php): 5′-TCCTACTTCATGTAGCGCAGGAC-3′ and 5′-TCCTCCAGAATAAGGGCTATCCG-3′ for UBC9 (At4g27960); 5′-CGTCGTAGGAAAGATCAAGC-3′ and 5′-CGCTCACTGAATCTAGCTC-3′ for CHUP1 (At3g25690); 5′-TCTGTCCGTGGCTACAACATG-3′ and 5′-ATTCTGATCTCAGCACCGACC-3′ for TOC75 (At3g46740); 5′-TTGTAGATCCAGCCCTCAGC-3′ and TTGTAGCTCCAACTGACGCAG-3′ for FTSZ2 (At2g36250); and 5′-TCCAAGCCGTGGCATTACTCA-3′ and GCAAATCATTCAGCGACAAGACC for TIC110 (At1g06950).
Immunofluorescence of FTSZ2-1
Tissue samples from 2-week-old seedlings were fixed, embedded, and sectioned as described previously (Vitha and Osteryoung, 2011). Slides were incubated in blocking buffer (2% nonfat dry milk and 0.05% Tween 20 in PBS, pH 7.4) for 1 h at room temperature, followed by overnight incubation with anti-FTSZ2-1 antibody (1:500; Stokes et al., 2000) in buffer (2% normal goat serum in blocking buffer). After four washes of 10 min in PBST buffer (PBS containing 0.05% Tween), slides were incubated for 3 h with Alexa Fluor 488 goat anti-rabbit IgG (1:500; Invitrogen). Finally, washed slides were prepared for observation with mounting medium (0.1% p-phenylendiamine and 90% glycerol in PBS, pH 8.0). For epifluorescence, fluorescein isothiocyanate (excitation 455 to 495 nm; emission 512 to 575 nm) or Texas red (excitation 535 to 585 nm; emission 607 to 682 nm) filter sets were used and images acquired using a Leica DMRA2 microscope equipped with Q-Capture camera control software (Q-imaging).
Electron Microscopy
Control and chmp1a chmp1b seedlings were high-pressure frozen in a Baltec HPM 010 and freeze-substituted in 2% OsO4 in acetone for 4 d. Samples were embedded in Eponate 12, sectioned, and stained with 2% uranyl acetate in 70% methanol and lead citrate (2.6% lead nitrate and 3.5% sodium citrate, pH 12).
Protein Blot Analysis
Control (plants with genotypes chmp1a/CHMP1A chmp1b/chmp1b and CHMP1A/CHMP1A chmp1b/chmp1b) and chmp1a chmp1b double homozygous mutant seedlings of same age were harvested not later than the time of appearance of the third and fourth leaves in control seedlings. Between 5 and 9 mg fresh tissue was ground in 100 μL lysis buffer (50 mM Tris/HCl, pH 6.8, 50 mM DTT, 2% SDS, 1 mM EDTA, 10% glycerol, and Roche Complete protease inhibitor) and centrifuged at 10,000g for 30 s. Protein content in the supernatant was quantified (Bio-Rad) and ∼20 μg total protein was loaded and resolved on SDS-PAGE gels using Bio-Rad Mini Protean 3 gel system and blotted on nitrocellulose membranes (Amersham). Nitrocellulose membranes were blocked 1 h at room temperature in 5% low-fat milk, and antibody incubation was performed in PBS (0.1% Tween 20) with 5% low-fat milk. Membranes were incubated in primary antibodies at 4°C overnight and in secondary antibodies for 90 min at room temperature. Protein amounts between control and mutant samples were adjusted with the cytosolic protein cFBPase. The following rabbit polyclonal anti-PsbA/D1 (Agrisera AS05084), anti-cFBPase (Agrisera AS04043), anti-TIC110 (Lübeck et al., 1996), anti-Toc75 (Nielsen et al., 1997), anti-PBA1 (Smalle et al., 2002), anti-PDV2 (Glynn et al., 2008), anti-PSAF (Agrisera AS06104), anti-H3 (Abcam ab1791), anti-FtsZ2 (Stokes et al., 2000), and anti-PEX14 (Agrisera) antibodies and the mouse monoclonal anti-GFP (Roche 11814460001) and anti-VDAC (Subbaiah et al., 2006) antibodies were used in this study.
Goat anti-rabbit serum coupled to horseradish peroxidase (Santa Cruz Biotechnology) was used for detection of rabbit polyclonal primary antibodies, and goat anti-mouse serum coupled to horse radish peroxidase (Santa Cruz) was used for detection of mouse monoclonal anti-GFP and anti-VDAC antibodies. Densitometric analyses were performed using ImageJ as described (http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATG1A (At3g61960), ATG5 (At5g17290), ATG7 (At5g45900), ATG8A (At4g21980), CHMP1A (At1g73030), CHMP1B (At1g17730), CHUP1 (At3g25690), FTSZ2 (At2g36250), MEX1 (At5g17520), PBA1 (At4g31300), PDV1 (At5g53280), PDV2 (At2g16070), RECA (At1g79050), PSAF (At1g31330), TOC64-III (At3g17970), and UBC9 (At4g27960).
Supplemental Data
Supplemental Figure 1. Plastid Defects in chmp1a chmp1b Mutant Seedlings.
Supplemental Figure 2. Seedling Morphology and Chloroplast Ultrastructure.
Supplemental Figure 3. Abnormal Plastid Morphology in chmp1 Embryos and Root Cells.
Supplemental Figure 4. Protein Expression in Wild-Type and chmp1 seedlings.
Supplemental Table 1. Densitometric Quantification of Immunoblots in Figure 2 and Supplemental Figure 4.
Supplemental Table 2. Densitometric Quantification of Immunoblot in Figure 4N.
Supplemental Movie 1. Association of GFP-ATG8-Decorated Autophagosomes with Chloroplasts in Wild-Type Cells.
Supplemental Movie 2. Dynamics of GFP-ATG8 Coats on Wild-Type Chloroplasts that Extend and Surround Stromules.
Supplementary Material
Acknowledgments
We thank Thomas Elthon (University of Nebraska) for providing the anti-VDAC antibody, John E. Froehlich (Michigan State University) for the anti-TIC110 and anti-TOC75 antibodies, David Kramer (Michigan State University) for the anti-PSAF antibodies, and all contributors of marker lines as described in Methods. This work was supported by National Science Foundation Grants MCB1157824 to M.S.O. and IOS 1339325 to R.D.V. and M.S.O. FTSZ localization was supported by Grant DE-FG02-06ER15808 to K.W.O. by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy.
AUTHOR CONTRIBUTIONS
C.S. and M.S.O. designed experiments. C.S. generated plant lines, performed imaging analysis and immunoblots of plastid proteins, and assembled figures and videos. F.L. performed the immunodetection of free GFP, plastid, and nuclear proteins. H.R. isolated higher order mutants and analyzed segregation of mutant alleles. R.B. and C.S. performed quantitative PCR analysis. M.Z. performed localization of FTSZ2. K.W.O., T.C., and R.D.V. contributed materials integral to the research and analyzed data. C.S. and M.S.O. wrote the article with advice from the other authors.
Glossary
- MVB
multivesicular body
- MS
Murashige and Skoog
- RCB
Rubisco-containing body
- ConA
concanamycin A
References
- Breuers F.K., Bräutigam A., Geimer S., Welzel U.Y., Stefano G., Renna L., Brandizzi F., Weber A.P.M. (2012). Dynamic remodeling of the plastid envelope membranes – a tool for chloroplast envelope in vivo localizations. Front. Plant Sci. 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba A., Ishida H., Nishizawa N.K., Makino A., Mae T. (2003). Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol. 44: 914–921. [DOI] [PubMed] [Google Scholar]
- Chung T., Phillips A.R., Vierstra R.D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 62: 483–493. [DOI] [PubMed] [Google Scholar]
- Djeddi A., Michelet X., Culetto E., Alberti A., Barois N., Legouis R. (2012). Induction of autophagy in ESCRT mutants is an adaptive response for cell survival in C. elegans. J. Cell Sci. 125: 685–694. [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007). Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179: 485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe I., Schuh A.L., Edwardson J.M., Audhya A. (2011). Association of ESCRT-II with VPS20 generates a curvature sensitive protein complex capable of nucleating filaments of ESCRT-III. J. Biol. Chem. 286: 34262–34270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn J.M., Froehlich J.E., Osteryoung K.W. (2008). Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20: 2460–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Cashikar A. (2012). Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28: 337–362. [DOI] [PubMed] [Google Scholar]
- Haussühl K., Andersson B., Adamska I. (2001). A chloroplast DegP2 protease performs the primary cleavage of the photodamaged D1 protein in plant photosystem II. EMBO J. 20: 713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Baba M., Cao Y., Klionsky D.J. (2008). Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell 19: 5506–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Stenmark H., Emr S.D. (2013). Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 5: a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard T.L., Stauffer D.R., Degnin C.R., Hollenberg S.M. (2001). CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114: 2395–2404. [DOI] [PubMed] [Google Scholar]
- Hurley J.H., Hanson P.I. (2010). Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat. Rev. Mol. Cell Biol. 11: 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K. (2008). Chloroplasts are partially mobilized to the vacuole by autophagy. Autophagy 4: 961–962. [DOI] [PubMed] [Google Scholar]
- Ishida H., Izumi M., Wada S., Makino A. (2014). Roles of autophagy in chloroplast recycling. Biochim. Biophys. Acta 1837: 512–521. [DOI] [PubMed] [Google Scholar]
- Ishida H., Yoshimoto K., Izumi M., Reisen D., Yano Y., Makino A., Ohsumi Y., Hanson M.R., Mae T. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148: 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P., López-Juez E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14: 787–802. [DOI] [PubMed] [Google Scholar]
- Johansen T., Lamark T. (2014). Selective autophagy goes exclusive. Nat. Cell Biol. 16: 395–397. [DOI] [PubMed] [Google Scholar]
- Katsiarimpa A., Kalinowska K., Anzenberger F., Weis C., Ostertag M., Tsutsumi C., Schwechheimer C., Brunner F., Hückelhoven R., Isono E. (2013). The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25: 2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee H., Lee H.N., Kim S.-H., Shin K.D., Chung T. (2013). Autophagy-related proteins are required for degradation of peroxisomes in Arabidopsis hypocotyls during seedling growth. Plant Cell 25: 4956–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8: 931–937. [DOI] [PubMed] [Google Scholar]
- Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042. [DOI] [PubMed] [Google Scholar]
- Kuma A., Matsui M., Mizushima N. (2007). LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3: 323–328. [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004). The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Le Bars R., Marion J., Le Borgne R., Satiat-Jeunemaitre B., Bianchi M.W. (2014). ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5: 4121. [DOI] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. (2007). ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17: 1561–1567. [DOI] [PubMed] [Google Scholar]
- Lee T.A., Vande Wetering S.W., Brusslan J.A. (2013). Stromal protein degradation is incomplete in Arabidopsis thaliana autophagy mutants undergoing natural senescence. BMC Res. Notes 6: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Vierstra R.D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Li F., Chung T., Vierstra R.D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26: 788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M., Spetea C., Hundal T., Oppenheim A.B., Adam Z., Andersson B. (2000). The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q., Huang W., Baldwin A., Jarvis P. (2012). Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659. [DOI] [PubMed] [Google Scholar]
- Lübeck J., Soll J., Akita M., Nielsen E., Keegstra K. (1996). Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO J. 15: 4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Vert G. (2014). The dynamics of plant plasma membrane proteins: PINs and beyond. Development 141: 2924–2938. [DOI] [PubMed] [Google Scholar]
- Manil-Segalén M., Lefebvre C., Culetto E., Legouis R. (2012). Need an ESCRT for autophagosomal maturation? Commun. Integr. Biol. 5: 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J., Vojta L., Soll J., Møller S.G. (2007). ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep. 8: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez D.E., Costa M.L., Gomez F.M., Otegui M.S., Guiamet J.J. (2008). ‘Senescence-associated vacuoles’ are involved in the degradation of chloroplast proteins in tobacco leaves. Plant J. 56: 196–206. [DOI] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. (2001). Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127: 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- Michaeli S., Honig A., Levanony H., Peled-Zehavi H., Galili G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26: 4084–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima S.Y., Froehlich J.E., Osteryoung K.W. (2006). PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18: 2517–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E., Akita M., Davila-Aponte J., Keegstra K. (1997). Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16: 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niittylä T., Messerli G., Trevisan M., Chen J., Smith A.M., Zeeman S.C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89. [DOI] [PubMed] [Google Scholar]
- Noda N.N., Ohsumi Y., Inagaki F. (2010). Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584: 1379–1385. [DOI] [PubMed] [Google Scholar]
- Oikawa K., Yamasato A., Kong S.G., Kasahara M., Nakai M., Takahashi F., Ogura Y., Kagawa T., Wada M. (2008). Chloroplast outer envelope protein CHUP1 is essential for chloroplast anchorage to the plasma membrane and chloroplast movement. Plant Physiol. 148: 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K. (2014). Organellophagy: eliminating cellular building blocks via selective autophagy. J. Cell Biol. 205: 435–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K., Kabeya Y., Suzuki K., Mori T., Ichikawa T., Matsui M., Nakanishi H., Miyagishima S.Y. (2009). The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation. Plant Cell 21: 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Wada S., Izumi M., Makino A., Ishida H. (2013). Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36: 1147–1159. [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Pyke K.A. (2014). Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 65: 443–472. [DOI] [PubMed] [Google Scholar]
- Otegui M.S., Noh Y.S., Martínez D.E., Vila Petroff M.G., Staehelin L.A., Amasino R.M., Guiamet J.J. (2005). Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J. 41: 831–844. [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C., Cassier-Chauvat C., Perennes C., Bergounioux C. (2004). An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16: 1801–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier N., Lefebvre C., Legouis R. (2005). CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic 6: 695–705. [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Stenmark H. (2009). How do ESCRT proteins control autophagy? J. Cell Sci. 122: 2179–2183. [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., Stenmark H. (2007). ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17: 1817–1825. [DOI] [PubMed] [Google Scholar]
- Schmitz A.J., Glynn J.M., Olson B.J., Stokes K.D., Osteryoung K.W. (2009). Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2: 1211–1222. [DOI] [PubMed] [Google Scholar]
- Schuh A.L., Audhya A. (2014). The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit. Rev. Biochem. Mol. Biol. 49: 242–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodkin M.R., Elazar Z. (2013). The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 55: 51–64. [DOI] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Babiychuk E., Kushnir S., Durski A., Vierstra R.D. (2002). Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Reyes F.C., Buono R., Sliwinski M.K., Haas T.J., Otegui M.S. (2009). The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 21: 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes K.D., McAndrew R.S., Figueroa R., Vitha S., Osteryoung K.W. (2000). Chloroplast division and morphology are differentially affected by overexpression of FtsZ1 and FtsZ2 genes in Arabidopsis. Plant Physiol. 124: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Palaniappan A., Duncan K., Rhoads D.M., Huber S.C., Sachs M.M. (2006). Mitochondrial localization and putative signaling function of sucrose synthase in maize. J. Biol. Chem. 281: 15625–15635. [DOI] [PubMed] [Google Scholar]
- Suttangkakul A., Li F., Chung T., Vierstra R.D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23: 3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.R., Doelling J.H., Suttangkakul A., Vierstra R.D. (2005). Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138: 2097–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Osteryoung K. (2011). Immunofluorescence microscopy for localization of Arabidopsis chloroplast proteins. In Chloroplast Research in Arabidopsis, Jarvis R.P., ed (New York: Springer: ), pp. 33–58. [DOI] [PubMed] [Google Scholar]
- Vitha S., McAndrew R.S., Osteryoung K.W. (2001). FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 153: 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S., Froehlich J.E., Koksharova O., Pyke K.A., van Erp H., Osteryoung K.W. (2003). ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15: 1918–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Ishida H., Izumi M., Yoshimoto K., Ohsumi Y., Mae T., Makino A. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149: 885–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yu B., Zhao J., Guo J., Li Y., Han S., Huang L., Du Y., Hong Y., Tang D., Liu Y. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Glynn J.M., Olson B.J., Schmitz A.J., Osteryoung K.W. (2008). Plastid division: across time and space. Curr. Opin. Plant Biol. 11: 577–584. [DOI] [PubMed] [Google Scholar]
- Yen W.L., Shintani T., Nair U., Cao Y., Richardson B.C., Li Z., Hughson F.M., Baba M., Klionsky D.J. (2010). The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 188: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Hu Y., Jia J., Li D., Zhang R., Gao H., He Y. (2009). CDP1, a novel component of chloroplast division site positioning system in Arabidopsis. Cell Res. 19: 877–886. [DOI] [PubMed] [Google Scholar]
- Zhang M., Schmitz A.J., Kadirjan-Kalbach D.K., Terbush A.D., Osteryoung K.W. (2013). Chloroplast division protein ARC3 regulates chloroplast FtsZ-ring assembly and positioning in Arabidopsis through interaction with FtsZ2. Plant Cell 25: 1787–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.