Expression of Gle1 variants with modified InsP6 binding pockets rescues mRNA export defects of the ipk1 InsP6-deficient mutant, suggesting a critical role for Gle1 in mediating InsP6 function.
Abstract
Myo-inositol-1,2,3,4,5,6-hexakisphosphate (InsP6), also known as phytic acid, accumulates in large quantities in plant seeds, serving as a phosphorus reservoir, but is an animal antinutrient and an important source of water pollution. Here, we report that Gle1 (GLFG lethal 1) in conjunction with InsP6 functions as an activator of the ATPase/RNA helicase LOS4 (low expression of osmotically responsive genes 4), which is involved in mRNA export in plants, supporting the Gle1-InsP6-Dbp5 (LOS4 homolog) paradigm proposed in yeast. Interestingly, plant Gle1 proteins have modifications in several key residues of the InsP6 binding pocket, which reduce the basicity of the surface charge. Arabidopsis thaliana Gle1 variants containing mutations that increase the basic charge of the InsP6 binding surface show increased sensitivity to InsP6 concentrations for the stimulation of LOS4 ATPase activity in vitro. Expression of the Gle1 variants with enhanced InsP6 sensitivity rescues the mRNA export defect of the ipk1 (inositol 1,3,4,5,6-pentakisphosphate 2-kinase) InsP6-deficient mutant and, furthermore, significantly improves vegetative growth, seed yield, and seed performance of the mutant. These results suggest that Gle1 is an important factor responsible for mediating InsP6 functions in plant growth and reproduction and that Gle1 variants with increased InsP6 sensitivity may be useful for engineering high-yielding low-phytate crops.
INTRODUCTION
Phytic acid (myo-inositol-1,2,3,4,5,6-hexakisphosphate [InsP6]) is the major form of phosphorus in plant seeds, where it serves as a reservoir for phosphate, minerals, and inositol to support seed germination and seedling growth (Raboy, 2003, 2009; Munnik and Nielsen, 2011). The high abundance of phytic acid in cereal grains and legumes, however, is known to cause nutritional and environmental problems (Raboy, 2001, 2009; Brinch-Pedersen et al., 2006; Beardsley, 2011). Phytic acid is a strong chelator of mineral cations such as iron, zinc, and calcium, forming stable salts called phytins. Phytins are indigestible to humans and nonruminant animals and are mostly excreted, contributing to mineral deficiency. The unavailability of phytic acid phosphorus for absorption increases feed costs for swine, poultry, and fish due to the need to supplement phosphorus for optimal growth. Furthermore, the excretion of undigested phytic acid in animal waste is an important source of water pollution. Thus, seed phytic acid has many negative impacts on both human/animal nutrition and the environment.
InsP6 is ubiquitous in eukaryotic cells and plays a role in diverse cellular processes. In yeast and mammals, InsP6 has been linked to mRNA export, translational control, RNA editing, and DNA repair (Hanakahi and West, 2002; Macbeth et al., 2005; Bolger et al., 2008; Montpetit et al., 2011). In plants, InsP6 has been associated with hormonal and signaling processes in addition to its function as a storage form (phytic acid) in seeds. InsP6 stimulates Ca2+ release in guard cells in response to abscisic acid, inducing stomatal closure (Lemtiri-Chlieh et al., 2003). InsP6 was identified in the x-ray crystal structure of the auxin receptor TIR (transport inhibitor response), possibly as a structural cofactor (Tan et al., 2007). InsP6 is also involved in plant defense reactions (Murphy et al., 2008). However, the detailed function of InsP6 and the significance of InsP6 in plant development remain to be elucidated.
We previously reported that the nuclear pore protein Rae1 (yeast Gle2p) plays a dual role in plants in mRNA export in interphase and in spindle assembly in mitosis (Lee et al., 2009). The result is consistent with recent findings that nuclear pore complex (NPC) proteins perform functions aside from their roles as structural components of the NPC (Blower et al., 2005; Jeganathan et al., 2005; Orjalo et al., 2006; Franks and Hetzer, 2013; Vollmer and Antonin, 2014). To identify plant NPC proteins with noncanonical functions, we evaluated the phenotypes of Nicotiana benthamiana NPC genes using virus-induced gene silencing (VIGS), using N. benthamiana Rae1 and Nup96 (involved in auxin signaling) as controls, and identified Gle1 as a candidate. Gle1 is an essential multifunctional protein that is highly conserved from yeast to humans. In yeast, Gle1 and its cofactor InsP6 activate the DEAD-box ATPase Dbp5 for mRNA export at the NPC (Alcázar-Román et al., 2006, 2010; Dossani et al., 2009; Montpetit et al., 2011). Gle1 is also found in the cytosol and plays a role in translation initiation and termination in Dbp5-independent and -dependent manners, respectively (Bolger et al., 2008; Kutay and Panse, 2008). In plants, the cellular functions of Gle1 are unknown except that the T-DNA insertion mutation of Arabidopsis thaliana Gle1 causes an embryo lethal phenotype (Braud et al., 2012). Here, we reveal the cellular function of Gle1 in plants in relation to LOS4 and InsP6 and show that modified Gle1 with increased InsP6 sensitivity complements the ipk1 low-phytate mutation.
RESULTS
Silencing of Gle1 Results in Growth Retardation and mRNA Export Defects in Arabidopsis and N. benthamiana
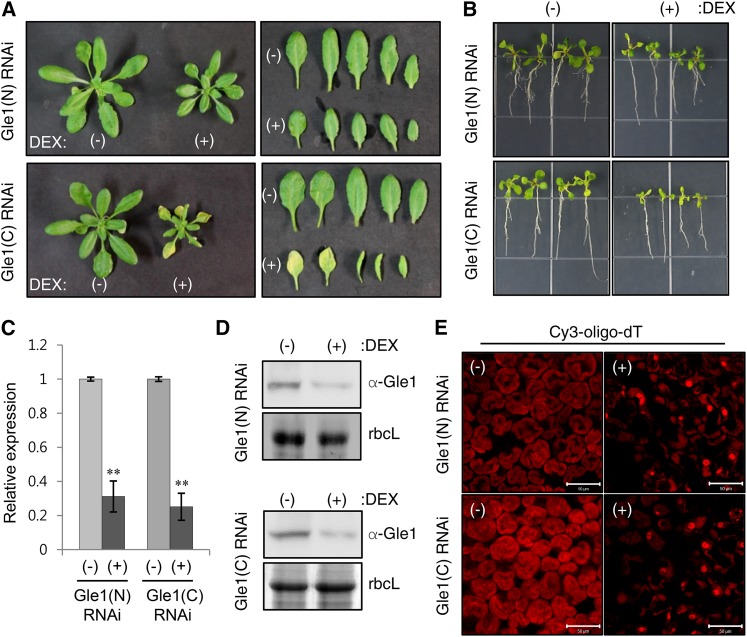
Multispecies sequence alignment revealed that Gle1 is universally found in eukaryotes and conserved from yeast to humans and plants, particularly in the Gle1 domain (Supplemental Figures 1 and 2 and Supplemental Data Set 1). To determine the in vivo effects of Gle1 deficiency in Arabidopsis and N. benthamiana, we employed dexamethasone (DEX)-inducible RNA interference (RNAi) and VIGS. Transgenic Arabidopsis plants (Columbia-0 ecotype) carried RNAi constructs containing an inverted repeat of a 356-bp N-terminal or a 330-bp C-terminal region of the Arabidopsis Gle1 cDNA under the control of the DEX-inducible transcription system and were designated Gle1(N) and Gle1(C) RNAi, respectively. DEX-inducible Gle1 RNAi plants were grown in soil and sprayed with either ethanol (−DEX) or 30 μM DEX. Upon DEX spraying, both Gle1(N) and Gle1(C) RNAi plants showed growth retardation (Figure 1A). When grown on Murashige and Skoog (MS) medium containing 10 μM DEX, both Gle1(N) and Gle1(C) RNAi seedlings exhibited retarded shoot and root development (Figure 1B). Real-time quantitative RT-PCR and immunoblotting with anti-Gle1 antibodies revealed reduced Gle1 mRNA and protein levels in seedlings grown on (+)DEX medium compared with the (−)DEX control, suggesting DEX-induced Gle1 silencing (Figures 1C and 1D). Since Gle1 is an NPC component, we examined whether Gle1 deficiency causes poly(A) RNA export defects using in situ hybridization (Figure 1E). After hybridization with a 45-nucleotide oligo(dT) probe end-labeled with Cy3 (Cy3-oligo-dT), poly(A) RNA signals were broadly distributed in the cytosol and nuclei in (−)DEX leaf cells. By contrast, (+)DEX leaves accumulated much stronger poly(A) RNA signals in the nuclei, suggesting that mRNA export from the nucleus to the cytosol was disturbed by Gle1 deficiency (Figure 1E).
Figure 1.
Analyses of Gle1-Silencing Phenotypes Using DEX-Inducible RNAi in Arabidopsis.
(A) Plant phenotypes of two different Arabidopsis DEX-inducible Gle1 RNAi lines [Gle1(N) and Gle1(C) RNAi] upon DEX treatment. Plants were grown in soil for 14 d and then sprayed with either ethanol (−) or 30 μM DEX (+) for 7 d.
(B) Seedling phenotypes of the two Gle1 RNAi lines that were grown for 10 d on MS medium containing either ethanol (−) or 10 μM DEX (+).
(C) Real-time quantitative RT-PCR analysis to determine Gle1 transcript levels. Transcript levels in (+)DEX samples are expressed relative to those in (−)DEX samples. UBC10 mRNA levels were used as a control. Values represent means ± sd of three replicates per experiment. Asterisks denote statistical significance of the differences between (−)DEX and (+)DEX samples based on two-tailed Student’s t tests: *P ≤ 0.05 and **P ≤ 0.01.
(D) Immunoblotting with anti-Gle1 antibodies to determine the endogenous Gle1 protein levels. Coomassie blue-stained rbcL (Rubisco large subunit) was used as a control.
(E) In situ hybridization of leaves of the two Gle1 RNAi lines after spraying with ethanol (−) or 30 μM DEX (+) for 7 d using the Cy3-oligo-dT probe for confocal microscopy. Bars = 50 μm.
VIGS was performed in N. benthamiana with two constructs, NbGle1(N) and NbGle1(C), containing a 615-bp N-terminal or a 570-bp C-terminal region of the NbGle1 cDNA, respectively (Supplemental Figures 3A to 3D). VIGS of NbGle1 using either construct resulted in a similar phenotype of growth retardation and abnormal leaf development compared with TRV control plants. Immunoblotting using anti-Gle1 antibodies revealed that endogenous Gle1 protein levels were reduced in both NbGle1(N) and NbGle1(C) VIGS leaves compared with the TRV control. After in situ hybridization, NbGle1(N) leaves exhibited mRNA export defects as observed in Arabidopsis Gle1 RNAi plants.
Gle1 Is Localized to the Nuclear Envelope and the Cytosol
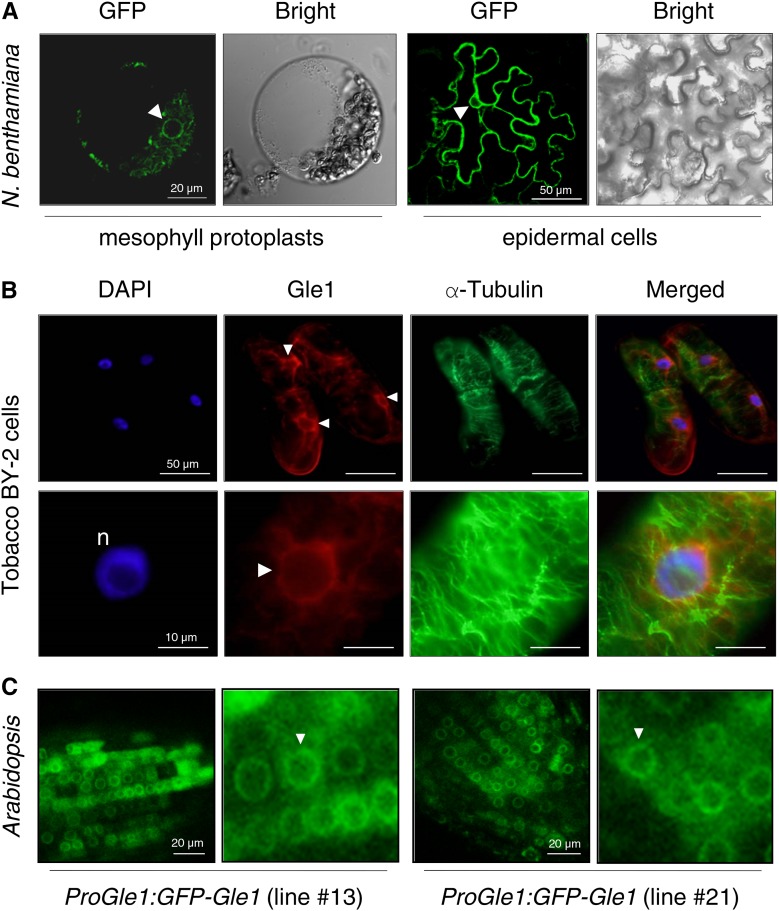
We examined the subcellular localization of Gle1 by expressing a GFP fusion protein of Arabidopsis Gle1 (GFP-Gle1) in N. benthamiana leaves via agroinfiltration. Confocal laser scanning microscopy of mesophyll protoplasts and leaf epidermal cells revealed that Gle1 is enriched around the nuclear envelope and in the cytosol (Figure 2A; Supplemental Figure 4). Next, immunolabeling of tobacco BY-2 cells with anti-Gle1 antibodies revealed that endogenous Gle1 localized in the nuclear periphery and cytosol of BY-2 cells, whereas 4′,6-diamidino-2-phenylindole (DAPI) staining and anti-α-tubulin antibodies labeled the nuclei (n) and the cortical microtubules, respectively (Figure 2B). Finally, root cells of transgenic Arabidopsis plants carrying the GFP-Gle1 construct fused to the endogenous Gle1 promoter (1944 bp upstream of the start codon) were observed by confocal microscopy. Green fluorescent signals of GFP-Gle1 were mainly detected in the nuclear envelope and cytosol of root cells (Figure 2C).
Figure 2.
Subcellular Localization of Gle1.
The GFP signal in the nuclear envelope is marked with arrowheads.
(A) A DNA construct encoding GFP-Gle1 under the control of the cauliflower mosaic virus 35S promoter was expressed in N. benthamiana leaves via agroinfiltration. GFP fluorescence was observed by confocal microscopy.
(B) Tobacco BY-2 cells were fixed and doubled-labeled with anti-Gle1 antibodies (red) and anti-α-tubulin antibodies (green) and stained with DAPI for confocal microscopy.
(C) GFP fluorescence in root cells of the Arabidopsis transgenic plants designated ProGle1:GFP-Gle1, which express GFP-Gle1 under the endogenous Gle1 promoter, was observed by confocal microscopy. Two independent transgenic lines (lines #13 and #21) were examined for this analysis.
Gle1 Interacts with the DEAD-Box ATPase LOS4
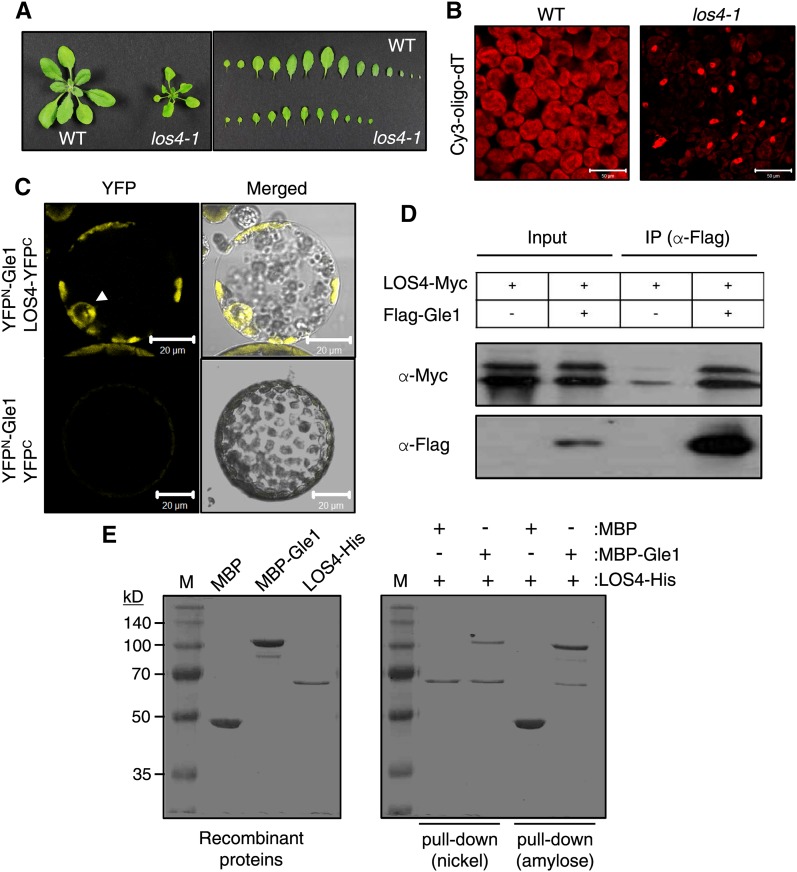
It has been reported that Gle1 associates with the DEAD-box ATPase Dbp5 and Nup159 to form the mRNA-exporting module in yeast (Montpetit et al., 2011). The Arabidopsis homolog of yeast Dbp5 is LOS4 (low expression of osmotically responsive genes 4), which plays a critical role in cold-responsive gene expression and chilling and freezing tolerance of plants (Gong et al., 2002, 2005). The los4-1 mutant plants exhibited growth retardation under normal growth conditions (Figure 3A). In situ hybridization revealed that the los4-1 mutation severely disrupted mRNA export (Figure 3B), as reported (Gong et al., 2005). To determine whether Arabidopsis Gle1 and LOS4 interact with each other, we first used bimolecular fluorescence complementation (BiFC). Coexpression of YFPN-Gle1 and LOS4-YFPC resulted in YFP fluorescence in the nuclear periphery and cytosol (Figure 3C). No fluorescence was detected between YFPN-Gle1 and YFPC, despite the expression of the proteins, indicating a lack of protein interaction (Figure 3C; Supplemental Figure 5). Next, we performed coimmunoprecipitation assays (Figure 3D). Expression of Flag-fused Gle1 (Flag-Gle1) and Myc-fused LOS4 (LOS4-Myc) was detected by immunoblotting with anti-Flag and anti-Myc antibodies, respectively (input). When expressed in N. benthamiana leaves, two forms of LOS4-Myc proteins were consistently detected in immunoblots. Flag-Gle1 was immunoprecipitated from leaf extracts using anti-Flag antibodies (immunoprecipitation), and then immunoblotting with anti-Myc antibodies allowed the detection of LOS4-Myc as a coimmunoprecipitant, suggesting in vivo interactions between Gle1 and LOS4. A small amount of LOS4-Myc was detected in the control experiment due to nonspecific interactions. For in vitro binding assays, maltose binding protein (MBP)-fused Gle1 (MBP-Gle1), 6x-histidine-fused LOS4 (LOS4-His), and MBP were purified (Figure 3E, left). LOS4-His in combination with MBP-Gle1 or MBP were incubated and bound to nickel resin (for His-tag) or amylose resin (for MBP-tag). After extensive washing of the resins, resin-bound proteins were eluted and subjected to Coomassie blue staining (Figure 3E, right). Nickel resin-bound LOS4-His could pull down MBP-Gle1, but not MBP, whereas amylose resin-bound MBP-Gle1, but not MBP alone, could pull down LOS4-His, suggesting a direct interaction between Gle1 and LOS4 in vitro. Collectively, these results suggest that Gle1 and LOS4 interact with each other at the nuclear rim and in the cytosol.
Figure 3.
Gle1 Interacts with LOS4.
(A) Phenotype and leaf series of the los4-1 mutant grown for 3 weeks in soil compared with the wild type.
(B) In situ hybridization of los4-1 mutant leaves with the Cy3-oligo-dT probe. Signal was detected by confocal microscopy. Bars = 50 μm.
(C) BiFC to visualize the interaction between Gle1 and LOS4. YFPN- and YFPC-fusion proteins were expressed together in N. benthamiana leaves by agroinfiltration. YFP fluorescence in mesophyll protoplasts prepared from the infiltrated leaves was examined by confocal microscopy. Arrowhead indicates the nucleus. Bars = 20 μm.
(D) Coimmunoprecipitation of Gle1 and LOS4. After agroinfiltration to coexpress Flag-Gle1 and LOS4-Myc proteins, total leaf proteins were immunoprecipitated with anti-Flag antibodies, and the coimmunoprecipitate was detected by anti-Myc antibodies. IP, immunoprecipitation.
(E) Pull-down assays showing direct interaction between Gle1 and LOS4. Purified recombinant proteins were stained with Coomassie blue (left). A mixture of proteins was bound to nickel resin or amylose resin, and resin-bound proteins were eluted and stained with Coomassie blue (right).
Gle1 Stimulates the ATPase Activity of LOS4
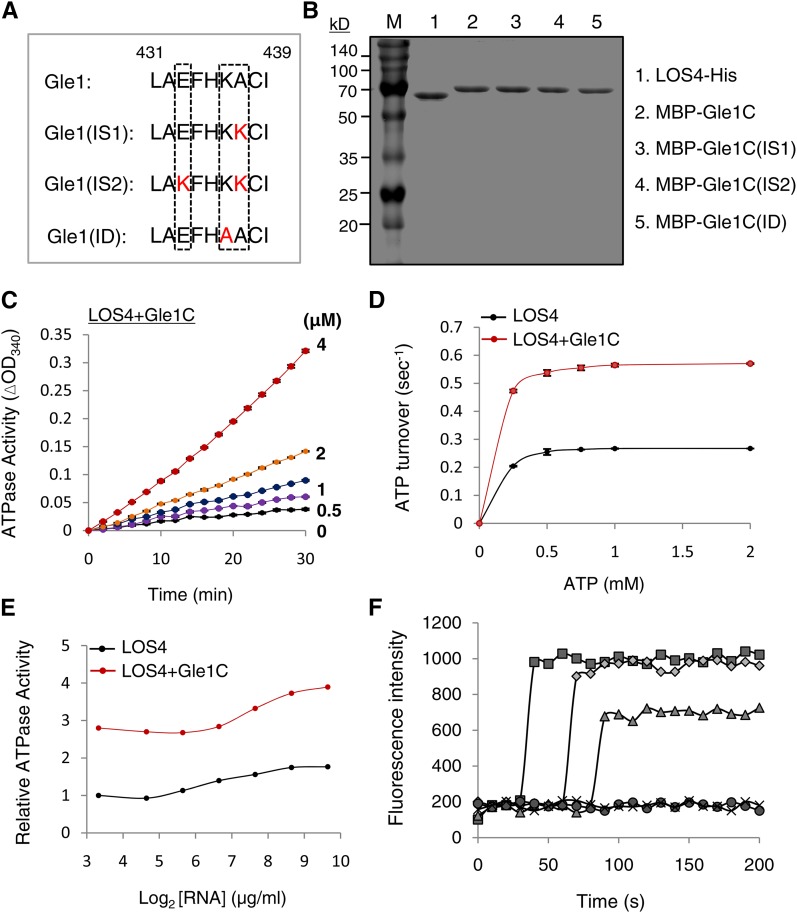
To explore the functional relationship between Gle1 and LOS4, we measured LOS4 ATPase activity in the presence or absence of Gle1. First, we purified LOS-His, MBP-Gle1, and MBP fusion protein of the C-terminal domain of Gle1 (MBP-Gle1C; 244 to 611 amino acid residues) (Figures 3E and 4B). The ATPase activity of LOS4-His was measured by a coupled steady state spectrophotometric assay as described in the Methods. The addition of increasing amounts of MBP-Gle1C proteins (0 to 4 μM) activated LOS4-His ATPase activity (1 μM) in a concentration-dependent manner (Figure 4C). MBP-Gle1C alone did not show intrinsic ATPase activity, and both MBP-Gle1 and MBP-Gle1C exhibited similar abilities to stimulate the ATPase activity of LOS4-His (Supplemental Figure 6). The addition of MBP-Gle1C (2 μM) to LOS4-His (1 μM) led to a substantial increase in ATP turnover (Figure 4D), and the apparent kcat values for LOS4-His alone and LOS4-His with MBP-Gle1C were calculated to be 0.267 and 0.565 s−1, respectively, resulting in an approximate twofold increase in the overall catalytic rate of LOS4 by Gle1. When LOS4-His and MBP-Gle1C were used at a 1:2 ratio for the ATPase assay, the addition of RNA (polyadenylic acid) further increased the LOS4-stimulating activity of Gle1C over a broad range of RNA concentrations, resulting in maximally about threefold activation (Figure 4E).
Figure 4.
Stimulation of LOS4 Activity by Gle1.
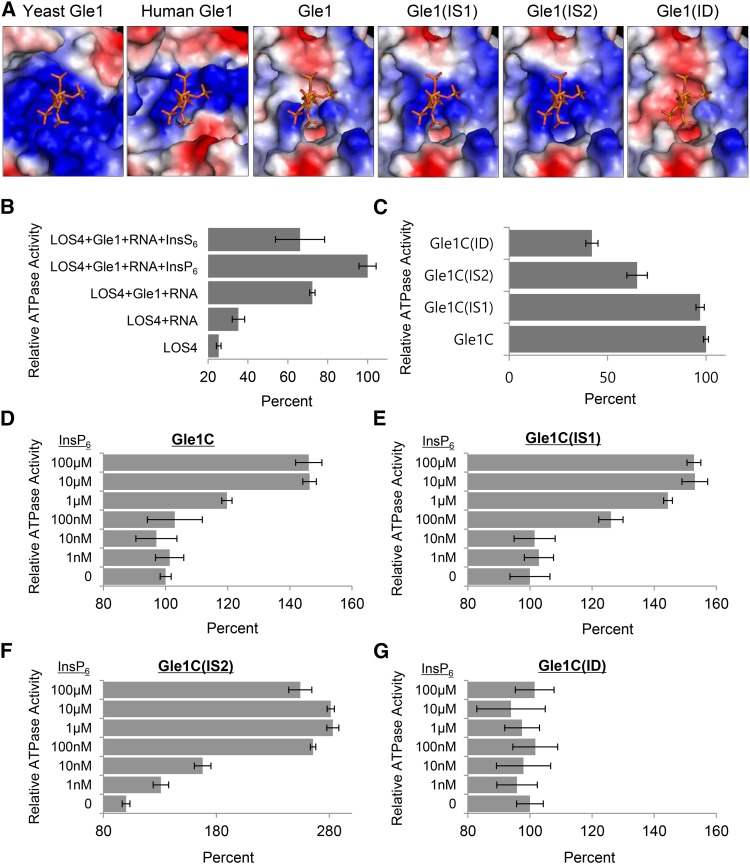
(A) Sequence alignment of key residues of the InsP6 binding pocket of Gle1 and its variants (IS1, IS2, and ID). The key residues are boxed. Modified residues in the variants are shown in red.
(B) Purified recombinant proteins were stained with Coomassie blue. Gle1C indicates the C-terminal region of Gle1 (244 to 611 amino acid residues).
(C) Steady state ATPase assays were performed with 1 μM LOS4-His, 2 mM ATP, and 50 μg/μL polyadenylic acid (RNA) in the presence of 0, 0.5, 1, 2, and 4 μM MBP-Gle1C.
(D) Stimulation of LOS4 ATPase activity by Gle1C. Steady state ATPase assays were performed with 1 μM LOS4-His and 2 μM MBP-Gle1C in the presence of 0 to 2 mM ATP. Data points represent means ± sd of three replicates per experiment.
(E) Dependence of LOS4 ATPase activity on RNA concentration. ATPase assays were performed with 1 μM LOS4-His and 2 mM ATP in the presence or absence of 2 μM MBP-Gle1C with different RNA concentrations.
(F) In vitro nucleic acid-melting assays using 78-nucleotide-long, 9-bp-containing, hairpin-shaped molecular beacons with different ratios of LOS4-His and MBP-Gle1 (full-length) proteins (squares, LOS4:Gle1 [1:4]; diamonds, LOS4:Gle1 [1:2]; triangles, LOS4:Gle1 [1:1]; circles, LOS4 only; and x, control). Beacon fluorescence was measured by fluorescence spectrophotometry.
LOS4 belongs to the DEAD-box RNA helicase family in Arabidopsis, and its yeast homolog Dbp5 possesses an ATP-dependent RNA helicase activity (Tseng et al., 1998; Gong et al., 2005). To test whether Gle1 activates the RNA helicase activity of LOS4, we performed in vitro nucleic acid-melting assays with LOS4-His and MBP-Gle1 recombinant proteins using 78-nucleotide-long, 9-bp-containing, hairpin-shaped molecular beacons (Kim et al., 2007, 2010). In the presence of ATP, the beacon itself (control) or the addition of LOS4-His did not increase beacon fluorescence (Figure 4F). However, the addition of LOS4-His and MBP-Gle1 at 1:1, 1:2, and 1:4 ratios caused a significant increase in fluorescence in a dose-dependent manner, implying a role for Gle1 as an activator of LOS4 RNA helicase activity. The inability of LOS4-His to induce fluorescence by itself suggests that this technique may not be sensitive enough to detect the low basal activity of LOS4. Taken together, these results suggest that Gle1 functions as an activator of LOS4.
Plant Gle1 Proteins Have Modifications in Several Key Residues in the InsP6 Binding Pocket
Recently, the structure of the yeast Dbp5-InsP6-Gle1 complex was resolved by protein crystallography, which suggests that InsP6 stabilizes the interaction between Gle1 and Dbp5 by acting as a small-molecule tether (Montpetit et al., 2011). InsP6 binds to a positively charged pocket at the interface between Gle1 and the C-terminal domain of Dbp5; residues Lys-264, Lys-333, His-337, Arg-374, Lys-377, and Lys-378 of Gle1, and Lys-477 and Lys-481 of Dbp5 are involved in the interaction with the phosphate groups of InsP6 (Montpetit et al., 2011). Particularly, the two residues Lys-377 and Lys-378 of yeast Gle1 were identified as critical residues for InsP6 binding according to site-directed mutagenesis (Alcázar-Román et al., 2010). Computational modeling predicted the surface potential of the Arabidopsis and human Gle1 domain based on that of the yeast Gle1 domain (Supplemental Figure 7). The InsP6 binding pocket present on the surface of yeast Gle1 exclusively comprises basic amino acid residues to accommodate the negatively charged phosphate groups of InsP6 (Figure 5A). Human Gle1 was also predicted to have a similar feature in the InsP6 binding pocket. However, the surface charge of the InsP6 binding site in Arabidopsis Gle1 was predicted to be only partially basic, indicating modifications of the pocket (Figure 5A).
Figure 5.
InsP6 Sensitivity of Gle1 and Gle1 Variants for Stimulation of LOS4 ATPase Activity.
(A) InsP6 and InsP6 binding regions of human Gle1 and Arabidopsis Gle1 and its variants (IS1, IS2, and ID), which were predicted based on the structure of the yeast Gle1 domain. The electrostatic surface potential is shown: acidic, basic, and neutral residues are shown in red, blue, and white, respectively.
(B) Relative ATPase activity of LOS4 with different combinations of cofactors. ATPase assays were performed with 1 μM LOS4-His and 2 mM ATP in the presence or absence of the following cofactors: 2 μM MBP-Gle1 (full-length), 50 μg/μL polyadenylic acid (RNA), 10 μM InsP6, and 10 μM InsS6. Values in (B) to (G) represent means ± sd of three replicates per experiment.
(C) Stimulation of LOS4 ATPase activity by Gle1 and Gle1 variants in the absence of InsP6. ATPase assays were performed with 1 μM LOS4-His, 2 mM ATP, and 50 μg/μL RNA in the presence of MBP-Gle1C or MBP-Gle1C variants at a concentration of 2 μM.
(D) to (G) InsP6 sensitivity of Gle1 and Gle1 variants for LOS4 stimulation. ATPase assays were performed as described in (C) but in the presence of different InsP6 concentrations.
Arabidopsis Gle1 residues corresponding to the Arg-374, Lys-377, and Lys-378 residues of yeast Gle1 could be unambiguously identified due to the high homology around the residues (Supplemental Figure 8). Interestingly, Gle1 proteins of higher plants (dicot, monocot, and gymnosperm) that were examined commonly had a Glu residue instead of Arg-374, and a neutral residue instead of Lys-378 of yeast Gle1, while they maintained a Lys/Arg residue corresponding to Lys-377. Vertebrates (human, mouse, cow, and zebra fish) had His, Lys, and Lys residues corresponding to Arg-374, Lys-377, and Lys-378 of yeast Gle1, maintaining the basic charge. We thus investigated how mutations of these two residues affect the predicted surface charge of the InsP6 binding pocket of Arabidopsis Gle1 (Figures 4A and 5A). A mutation from Ala-437 to Lys was designated IS1 (InsP6-Sensitive 1); a double mutation of Glu-433 to Lys and Ala-437 to Lys was designated IS2; and a mutation from Lys-436 to Ala was designated ID (InsP6-Dead). Computational modeling suggests that IS1 and IS2 mutations progressively increased the basic charge of the InsP6 binding surface, while the ID mutation abrogated the basic charge (Figure 5A).
Gle1(IS1) and Gle1(IS2) Variants Show Increased Sensitivity to InsP6 Concentrations for the Stimulation of LOS4 ATPase Activity
We first determined whether the Gle1-dependent stimulation of LOS4 ATPase activity is affected by the presence of InsP6 (Figure 5B). ATPase assays were performed with LOS4-His and ATP in the presence or absence of cofactors, including RNA (polyadenylic acid), MBP-Gle1, and InsP6. The addition of RNA moderately stimulated LOS4 ATPase activity, whereas the addition of both RNA and Gle1 resulted in an approximate 3-fold induction of ATPase activity. The addition of InsP6, but not of its analog InsS6, to RNA and MBP-Gle1 further stimulated ATPase activity, causing more than a 4-fold increase, suggesting that InsP6 stimulates LOS4 ATPase activity in conjunction with Gle1 and RNA (Figure 5B). Interestingly, addition of InsP5, but not of InsP3 or InsP4, substantially stimulated LOS4 ATPase activity in vitro (Supplemental Figure 9), as similarly observed in yeast (Weirich et al., 2006).
We next tested whether the Gle1 variants with a modified InsP6 binding pocket have different sensitivity to InsP6 in the stimulation of LOS4 ATPase activity. The Gle1 variants Gle1C(IS1), Gle1C(IS2), and Gle1C(ID) were generated by site-directed mutagenesis and purified as MBP fusion proteins (Figure 4B). First, in the absence of InsP6, when incubated with LOS4-His and RNA, MBP-Gle1C and MBP-Gle1C(IS1) activated LOS4 ATPase activity to similar degrees, whereas MBP-Gle1C(IS2) and MBP-Gle1C(ID) showed reduced abilities to stimulate LOS4 (Figure 5C). Particularly, the K436A mutation in the MBP-Gle1C(ID) variant resulted in more than a 2-fold reduction in stimulating activity. We next tested the LOS4-stimulating activity of Gle1C and its variants in the presence of RNA and variable concentrations of InsP6 (0 to 100 μM). MBP-Gle1C maintained basal stimulating activity with 1 to 100 nM InsP6 but increased activity with 1 μM to reach maximal activity with 10 μM InsP6 (Figure 5D). MBP-Gle1C(IS1) could increase basal activity in the presence of 100 nM InsP6, and with 1 μM, almost reached maximal stimulation activity (Figure 5E). MBP-Gle1C(IS2), which shows the highest similarity to yeast Gle1 in the InsP6 binding pocket among the variants, was responsive to 1 nM InsP6 and was able to fully stimulate LOS4 ATPase activity in the presence of 100 nM InsP6 (Figure 5F). Thus, IS1 and IS2 mutations conferred Arabidopsis Gle1 with increased InsP6 sensitivity to LOS4 stimulation in vitro. By contrast, MBP-Gle1C(ID) was unaffected by InsP6 regardless of its concentration, suggesting that the Lys-436 residue is critical for InsP6 binding to Gle1 (Figure 5G).
Expression of InsP6-Sensitive Gle1 Variants Improves Vegetative Growth of the ipk1 InsP6 Biosynthetic Mutant
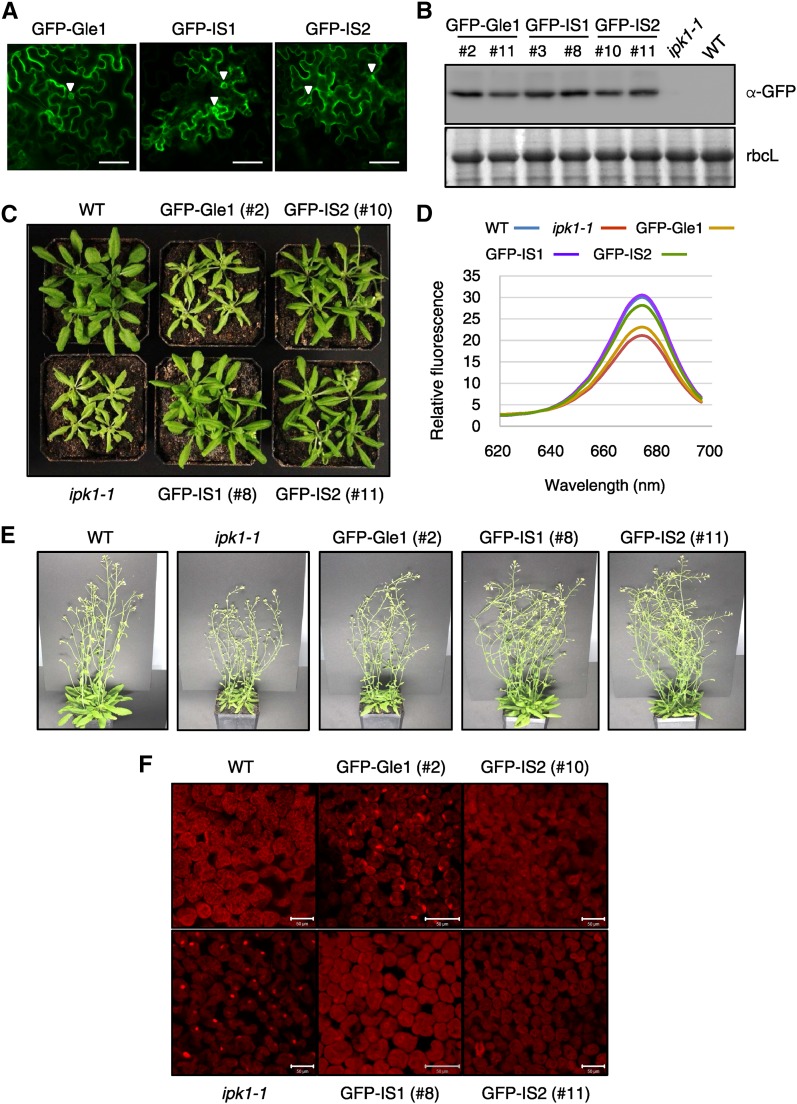
We next investigated whether the Gle1(IS1) and Gle1(IS2) variants function better in a low InsP6 background in vivo than wild-type Gle1 by testing their abilities to complement the ipk1-1 mutation in Arabidopsis. IPK1 encodes inositol 1,3,4,5,6-pentakisphosphate 2-kinase, an enzyme that catalyzes the final step of InsP6 biosynthesis, the conversion of InsP5 to InsP6 (Stevenson-Paulik et al., 2005; Monserrate and York, 2010; Munnik and Nielsen, 2011). The ipk1-1 T-DNA insertion mutant of Arabidopsis had significantly reduced InsP6 levels, ∼17% and 7.5% of wild-type levels in seeds and seedlings, respectively, and exhibited growth defects that were aggravated by nutrient-rich conditions (Stevenson-Paulik et al., 2005). Using Agrobacterium tumefaciens-mediated transformation, we introduced GFP fusion constructs of wild-type Gle1 (GFP-Gle1), Gle1(IS1) [GFP-IS1], and Gle1(IS2) [GFP-IS2] into the ipk1-1 mutant under the control of a cauliflower mosaic virus 35S promoter. Confocal microscopy showed that GFP-IS1 and GFP-IS2 are localized in the nuclear envelope and cytosol, as is GFP-Gle1 (Figure 6A). Immunoblotting with anti-GFP antibodies using leaf extracts from independent transgenic lines confirmed expression of GFP-fused Gle1 and Gle1 variants (Figure 6B). To assess growth, the plants were grown in soil inside a controlled growth chamber. The ipk1-1 mutant plants were significantly smaller and yellower than the wild type (Columbia-0) under the growth conditions examined and exhibited abaxial curling of the rosette leaves, which became more prominent upon aging (Figures 6C and 6D; Supplemental Figure 10). After 4 to 5 weeks, ipk1-1 mutants frequently developed necrosis at the leaf margins. Compared with the parental ipk1-1 mutant, the transgenic GFP-IS1 and GFP-IS2 lines showed significant improvements in vegetative growth with an increase in leaf size and leaf greening, and a lack of necrotic margins, although the leaf curling phenotype still remained (Figures 6C and 6D; Supplemental Figure 10). By contrast, GFP-Gle1 lines showed only marginal improvements in plant growth. Leaf chlorophyll contents of GFP-IS1 and GFP-IS2 plants were higher than those of the ipk1-1 mutant and GFP-Gle1 plants, suggesting increased photosynthetic capacity (Figure 6D). When GFP-IS1 and GFP-IS2 plants were about to bolt, their sizes were comparable to those of wild-type plants (Figure 6C). However, the ipk1-1 mutant and all of the transgenic lines showed early flowering along with early inhibition of rosette leaf growth and developed increased numbers of inflorescence stems compared with wild-type plants (Figure 6E). It is noteworthy that the los4-2/cryophyte site-specific mutation strongly induced early flowering (Gong et al., 2005).
Figure 6.
Phenotypes of Transgenic Arabidopsis Plants that Express Gle1 and Gle1 Variants in an ipk1-1 Background.
(A) Subcellular localization of GFP-Gle1, GFP-IS1, and GFP-IS2 in N. benthamiana leaf epidermal cells. The GFP signal in the nuclear envelope is marked with arrowheads. Bars = 50 μm.
(B) Immunoblotting with anti-GFP antibodies to determine expression levels of GFP-Gle1, GFP-IS1, and GFP-IS2 in independent transgenic lines of the T3 generation. Coomassie blue-stained rbcL was used as a control.
(C) Enhanced vegetative growth of the ipk1-1 mutant by expression of the Gle1 variants. The plants were grown in soil for 3 weeks.
(D) A fluorescence emission spectrum for chlorophyll measurement. Spectrofluorometry of chlorophylls was performed with a fluorescence spectrophotometer.
(E) Phenotypes of the plants grown for 6 weeks in soil.
(F) In situ hybridization was performed with leaves of the plants using the Cy3-oligo-dT probe. Bars = 50 μm.
In situ hybridization revealed that the ipk1-1 mutant leaf cells accumulated the poly(A) RNA signal inside the nucleus (Figure 6F; Supplemental Figure 11), suggesting that an mRNA export defect contributed to the abnormal growth of the mutant. However, GFP-IS1 and GFP-IS2 leaves exhibited a normal distribution of the poly(A) RNA signal in the cytosol and nucleus as observed in wild-type leaves, suggesting that expression of the Gle1 variants can rescue the mRNA export defect of the ipk1 mutant. By contrast, expression of GFP-Gle1 only slightly reduced nuclear accumulation of poly(A) RNA.
Expression of InsP6-Sensitive Gle1 Variants Improves Seed Yield and Seed Performance of the ipk1 Mutant
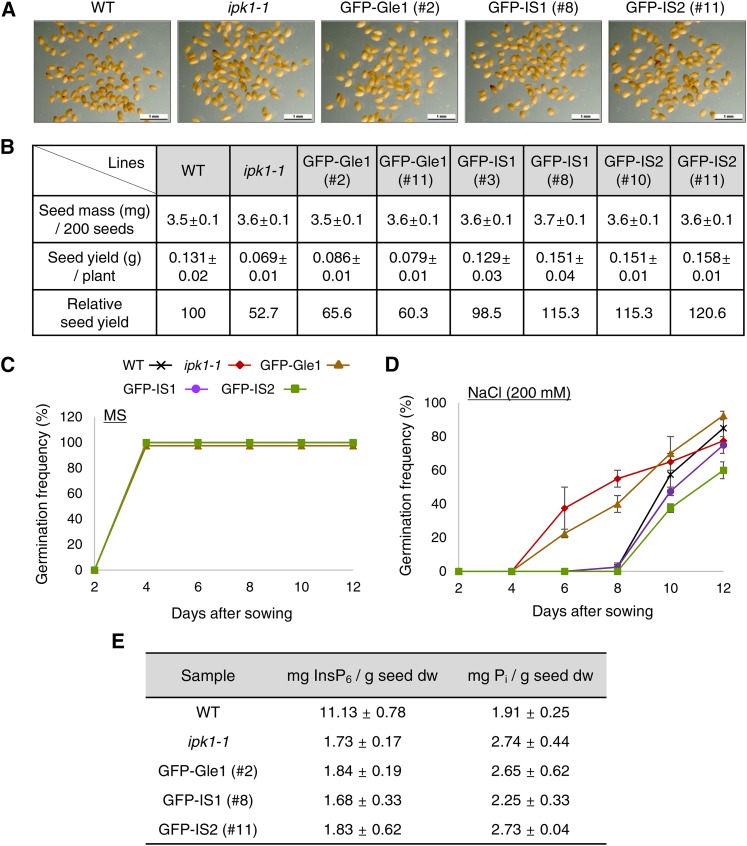
The transgenic lines expressing the Gle1 variants were assessed for seed weight, seed yield, and seed germination rate and compared with the wild type and parental ipk1-1 mutant. There were no apparent differences in seed morphology or seed weight among these lines (Figures 7A and 7B). However, seed yield of the ipk1 mutant was only ∼52% of the wild-type level because many siliques of the mutant plant contained aborted seeds (Figure 7B). By contrast, GFP-IS1 and GFP-IS2 plants had seed yields that were comparable to or even higher than the wild-type level, whereas GFP-Gle1 plants had only slightly increased seed yield compared with the ipk1-1 mutant (Figure 7B). Seed germination rates of the mutant, wild-type, and transgenic lines were very similar on MS medium (Figure 7C). However, in response to 200 mM NaCl, the ipk1-1 seeds germinated significantly earlier than wild-type seeds with green open cotyledons, and soon perished (Figure 7D). This reduced sensitivity to salt stress suggests disrupted stress signaling in the mutant seeds. GFP-IS1 and GFP-IS2 seeds, intriguingly, retained normal sensitivity to salt stress, while GFP-Gle1 seeds behaved similar to the mutant seeds, suggesting that Gle1 function is critical for plant responses to salt stress. In response to other abiotic stresses, such as sucrose and mannitol, no significant differences in germination rates were observed among these seeds (Supplemental Figure 12). These results demonstrate that expression of Gle1(IS1) and Gle1(IS2) variants can largely rescue the defects of InsP6 deficiency in seeds as well as in vegetative tissues.
Figure 7.
Seed Phenotypes of Transgenic Plants Expressing Gle1 and Gle1 Variants.
(A) Morphology of mature dry seeds. Bars = 1 mm.
(B) Seed mass and seed yield. Values represent means ± sd from 12 independent plants for each line.
(C) Seed germination rates on MS medium (n = 200).
(D) Seed germination rates on MS medium containing 200 mM NaCl (n = 200).
(E) Seed phytic acid (InsP6) and free phosphate (Pi) contents based on HPIC analyses. Values represent means ± sd of three replicates per experiment.
We performed high-performance ion chromatography (HPIC) analyses to measure seed phytate content in the transgenic plants (Figure 7E; Supplemental Figure 13). Seed phytate levels were reduced to ∼15% of wild-type levels in all of the transgenic lines and the ipk1-1 mutant, whereas inorganic phosphate levels in the seeds only moderately increased (Figure 7E). Instead, the transgenic lines and the mutant accumulated high levels of InsP5 in seeds (Supplemental Figure 13). These results agreed well with those from a previous report (Stevenson-Paulik et al., 2005) and revealed that expression of the Gle1 variants did not alter cellular InsP6 levels in the transgenic plants. Furthermore, the highly accumulated InsP5 could not rescue the defective mRNA export in the ipk1 mutant (Figure 6F), suggesting that InsP6 specifically functions as a cofactor of LOS4/Gle1-mediated mRNA export in vivo.
IPK1 Interacts with LOS4, but Not with Gle1, in the Nuclear Envelope and Cytosol
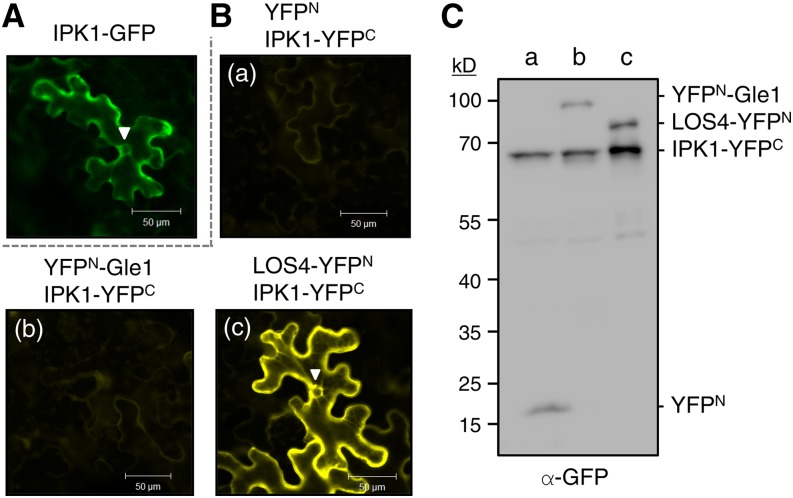
In mature seeds, phytic acid accumulates in protein storage vacuoles as stable salts (phytins) by binding to mineral cations (Lott et al., 1995). It was proposed that phytic acid is synthesized in association with the endoplasmic reticulum, deposited into the endoplasmic reticulum lumen, and then transported in vesicles to the protein storage vacuoles in developing seeds (Otegui et al., 2002). Recently, Nagy et al. (2009) reported that the Arabidopsis ATP binding cassette protein MRP5 (Multidrug resistance-related proteins 5) localized in the vacuolar membrane functions as a high-affinity InsP6 transporter for phytate storage. We examined the subcellular localization of Arabidopsis IPK1 by GFP fusion. Confocal microscopy detected IPK1-GFP fluorescence mainly in the cytosol and around the nuclear envelope (Figure 8A). Furthermore, BiFC suggested that IPK1 interacts with LOS4 in the nuclear envelope and cytosol, but not with Gle1 or control YFPN, despite normal expression of the proteins (Figures 8B and 8C). The close proximity of IPK1 to LOS4 and Gle1 may provide local enrichment of InsP6 to support mRNA export and possibly other LOS4/Gle1-mediated processes, which take place in the nuclear envelope or cytosol.
Figure 8.
Interaction between IPK1 and LOS4.
(A) IPK1-GFP fusion protein was expressed in N. benthamiana leaves by agroinfiltration, and the leaves were examined by confocal microscopy. The GFP signal in the nuclear envelope is marked with an arrowhead. Bars = 50 μm.
(B) BiFC-mediated visualization of the IPK1-LOS4 interaction. YFPN and IPK1-YFPC (A), YFPN-Gle1 and IPK1-YFPC (B), and LOS4-YFPN and IPK1-YFPC (C) were coexpressed in N. benthamiana leaves. The YFP signal in the nuclear envelope is marked with an arrowhead. Bars = 50 μm.
(C) Protein expression in BiFC analyses shown in (B). Expression of YFPN- and YFPC-fused proteins in the infiltrated N. benthamiana leaves was determined by immunoblotting with anti-GFP antibodies.
DISCUSSION
In yeast, plants, and mammals, Gle1 is a component of the NPC but is also localized to the cytosol. In this study, we investigated the nuclear function of Gle1 in plants. Plant Gle1 is involved in nuclear mRNA export by interacting with LOS4 to stimulate LOS4 ATPase/RNA helicase activity. Functions of Gle1 are essential for embryogenesis (Braud et al., 2012) and critical for the postembryonic growth of plants (Figure 1), which is reminiscent of essential Gle1 functions in both yeast and mammals (Murphy and Wente, 1996; Nousiainen et al., 2008). Montpetit et al. (2011) recently proposed the molecular mechanism of Dbp5/Gle1-driven mRNA export in yeast. In the model, Gle1 induces conformational changes in Dbp5 to stimulate RNA release, which is known to be a rate-limiting step in the hydrolytic cycle of DEAD-box RNA helicases. RNA release subsequently causes Nup159 binding to Dbp5 to prevent rebinding of the RNA and allow enzyme recycling. Furthermore, InsP6 bridges and stabilizes the Gle1-Dbp5 interaction by binding to a pocket at the interface between Gle1 and Dbp5 (Montpetit et al., 2011).
Although detailed mechanisms of Gle1 and LOS4 action in the mRNA export pathway in plants remain to be determined, the results of this study suggest that LOS4 is activated by Gle1 in a similar manner as described for yeast Dbp5. However, the requirement of Gle1 for coactivator InsP6 appears to differ between yeast and plants. In yeast, the addition of InsP6 (100 nM) to Gle1 and RNA causes an additional 3- to 4-fold increase in Dbp5 ATPase activity (Dossani et al., 2009; Montpetit et al., 2011), whereas it results in only a 30% additional increase in LOS4 activity in plants even at higher concentrations of InsP6 (10 μM) (Figure 5B). Since the InsP6-interacting Lys-477 and Lys-481 residues of Dbp5 are conserved in LOS4, the relative inefficiency of InsP6 as a Gle1 cofactor for LOS4 activation is likely caused by the modification of the key residues of plant Gle1 proteins, which reduces the basicity of the InsP6 binding pocket. Despite the seemingly imperfect structure of the pocket in plant Gle1, the LOS4/Gle1-mediated mRNA export pathway is functional in plants, unless cellular InsP6 levels are substantially reduced, as in the ipk1 mutant (Figure 6F). The finding that the expression of Gle1 variants containing a more basic InsP6 binding pocket fully restores mRNA export in the ipk1 mutant strongly suggests that InsP6 is an important element in LOS4/Gle1-mediated mRNA export in plants. InsP6 may be required to fine-tune the interaction strength between Gle1 and LOS4 in order to stimulate LOS4 activity above a certain threshold level required for its normal function.
InsP6 has been shown to possess a diverse set of cellular functions in yeast, mammals, and plants (Lemtiri-Chlieh et al., 2000; Hanakahi and West, 2002; Macbeth et al., 2005; Tan et al., 2007; Bolger et al., 2008; Murphy et al., 2008; Montpetit et al., 2011). The ∆ipk1 mutation in yeast abolished InsP6 accumulation and caused synthetic lethality combined with alleles of Gle1, Dbp5, and Nup159 (Miller et al., 2004; Weirich et al., 2004, 2006). IPK1−/− mutant mice died during early embryogenesis, suggesting the importance of InsP6 for early development of mammals, but their heterozygous littermates developed normally with normal cellular InsP6 levels and elevated InsP5 levels (Verbsky et al., 2005). The ipk1-1 mutation in Arabidopsis, which caused >70% decrease in IPK1 mRNA levels, caused stunted growth, reduced seed yields, and abnormal seed germination (Figures 6 and 7). Our finding that expression of the Gle1 variants significantly restores the growth and yield of the ipk1 mutant suggests that Gle1 plays a critical role in mediating InsP6 functions in plant growth and reproduction (Figures 6 and 7). It would be interesting to examine whether plant Gle1 and InsP6 are involved in translational control in the cytosol as reported in yeast (Bolger et al., 2008; Kutay and Panse, 2008), in addition to their NPC-related function. The observed incomplete complementation of the ipk1 phenotypes by the Gle1 variants is likely caused by other functions of InsP6 that are not mediated by Gle1 (Figure 6). Interestingly, expression of the Gle1 variants rescued the salt-insensitive germination of the ipk1-1 seeds (Figure 7D). Thus, restoration of mRNA export in the ipk1-1 seeds might have led to restoration of normal stress signaling for salt adaptation. In yeast, the intact NPC is essential for cell survival at high osmolarity, and the HOG1 stress-activated kinase phosphorylates nucleoporins to facilitate mRNA export upon osmostress (Regot et al., 2013). Arabidopsis los4-1 and los4-2 mutations are linked to chilling and/or heat stresses by causing defective mRNA export under stress conditions (Gong et al., 2002, 2005). These results suggest that LOS4-InsP6-Gle1-mediated mRNA export is involved in plant responses to environmental stresses.
To solve the nutritional and environmental problems related to dietary seed phytic acid, low-phytic-acid (lpa) mutants of maize (Zea mays), barley (Hordeum vulgare), wheat (Triticum aestivum), rice (Oryza sativa), and soybean (Glycine max) have been isolated (Raboy et al., 2000; Meis et al., 2003; Shi et al., 2003, 2005, 2007; Bregitzer and Raboy, 2006; Raboy, 2009). Some of the genes affected in the lpa mutations include the myo-inositol-3-phosphate synthase, myo-inositol kinase, inositol polyphosphate kinase (IPK), and multidrug resistance-associated (MRP) ATP binding cassette transporter genes. However, when these mutations were expressed in a tissue-general manner, they frequently resulted in undesirable agronomic traits, such as stunted vegetative growth, reduced seed weight, and poor seed germination. Thus, more recently, seed-targeted low-phytate engineering has been attempted using seed-specific promoters. Shi et al. (2007) reported that downregulation of MRP4 under embryo-specific promoters resulted in low-phytate maize and soybean without significant effects on seed dry weight and germination rate. Ali et al. (2013) also developed low-phytate rice with no apparent developmental defects by seed-specific silencing of IPK1. However, defects in seed weight, germination frequency, and seedling emergence were observed in several other trials, reflecting a role for InsP6 in seed development (Bilyeu et al., 2008; Kuwano et al., 2009; Li et al., 2014). In this study, we have shown that expression of InsP6-sensitive Gle1 variants enhanced plant growth, seed yield, and seed performance of the phytate-deficient ipk1 mutant, suggesting a novel strategy for improving lpa plants. Testing whether the Gle1 variants enhance agricultural traits of diverse lpa mutants, particularly in crops, is crucial before this technology can be used to engineer high-yielding low-phytate seed crops.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia-0) plants were grown in a growth chamber at 22°C and 150 μmol m−2 s−1 under a 16-h-light/8-h-dark cycle. The ipk1-1 (SALK_065337) and los4-1 (CS24938) mutants were obtained from Salk and ABRC, respectively. Nicotiana benthamiana plants were grown in a growth room at 22°C and 80 μmol m−2 s−1 under a 16-h-light/8-h-dark cycle.
Seed Germination Assays
Seeds were sterilized and sown on medium containing MS salts including vitamins and 0.8% phytoagar with or without the addition of NaCl (100 and 200 mM), sucrose (150 and 250 mM), or mannitol (200 and 400 mM). Before sowing, seeds were imbibed for 3 d at 4°C. Seeds were incubated at 4°C for 1 d after sowing on the medium and then transferred to a growth chamber (22°C, continuous light conditions). Seed germination was scored when cotyledon emergence was visible.
Generation of Arabidopsis DEX-Inducible Gle1 RNAi Lines
For Gle1(N) RNAi lines, a 356-bp Gle1 cDNA fragment was amplified by PCR using 5′-ATGGGGATTGTTTTGGAAC-3′ and 5′-GGTTCATGATCAAACTCTTCAT-3′ primers containing XhoI and HindIII sites for the sense construct, and SpeI and EcoRI sites for the antisense construct. For Gle1(C) RNAi lines, a 330-bp Gle1 cDNA fragment was amplified by PCR using 5′-CACAAAGCTTGCATTTACACT-3′ and 5′-ATGCTCTCTCACAACATTCAC-3′ primers containing XhoI and ClaI sites for the sense construct, and SpeI and BamHI sites for the antisense construct. Using these constructs, DEX-inducible Gle1 RNAi Arabidopsis lines were generated as described (Ahn et al., 2011). For induction of RNAi, the transgenic seedlings were grown on medium containing 10 μM DEX in ethanol (0.033%). Alternatively, the RNAi seedlings were sprayed with 30 μM DEX in ethanol (0.033%) and Tween 20 (0.01% [w/v]).
VIGS
VIGS was performed in N. benthamiana as described (Lee et al., 2009; Ahn et al., 2011; Cho et al., 2013).
Agrobacterium tumefaciens-Mediated Transient Expression
Agroinfiltration was performed as described (Ahn et al., 2011; Cho et al., 2013).
In Situ Hybridization of Poly(A) RNA
In situ hybridization of poly(A) RNA was performed using a 45-nucleotide oligo(dT) probe end-labeled with Cy3 as described (Lee et al., 2009). Cy3 fluorescence was detected by confocal laser scanning microscopy (Zeiss LSM510).
Real-Time Quantitative RT-PCR
Real-time quantitative RT-PCR was performed as described (Ahn et al., 2011; Cho et al., 2013) using the following primers; 5′-CATGGATGGGCTTGGTTAGC-3′ and 5′-TGTCGCAGTGGCTCTGTTG-3′ to detect Gle1 transcripts in the RNAi-Gle1(N) lines, 5′-TCAGCCAATTACTAACACAACCTT-3′ and 5′-GACATGCATTACAAATCCTCCA-3′ to detect Gle1 transcripts in the RNAi-Gle1(C) lines, and 5′-ATGGGTCCTTCAGAGAGTCCT-3′ and 5′-TGGAACACCTTGGTCCTAAAG-3′ for UBC10 transcripts.
Measurement of Chlorophyll Contents
Chlorophylls were extracted from Arabidopsis plants as described (Terry and Kendrick, 1999). Spectrofluorometry was performed using a fluorescence spectrophotometer (Hitachi F-2000) at an excitation wavelength of 440 nm and an emission wavelength of 600 to 700 nm as described (Terry and Kendrick, 1999).
Immunoblotting
Anti-Gle1 antibodies were generated in rabbits against two oligopeptides, EEARRKERAHQEEK and MRLYGALVQT, which correspond to amino acid residues 228 to 241 and 465 to 474 of Arabidopsis Gle1, respectively, using the antibody production services of Cosmogenetech. Immunoblotting was performed with mouse monoclonal antibodies against the Myc tag (1:5000; ABM) or the Flag tag (1:10,000; Sigma-Aldrich), rabbit polyclonal antibody against Gle1 (1:1000; Cosmogenetech), or goat polyclonal antibody against GFP (1:5000; ABM). Next, the membranes were treated with horseradish peroxidase-conjugated goat anti-mouse IgG antibody (1:5000; Invitrogen), goat anti-rabbit IgG antibody (1:10,000; Invitrogen), or donkey anti-goat antibodies (1:10,000; Santa Cruz Biotechnology). Signals were detected by Imagequant LAS 4000 (GE Healthcare Life Sciences).
ATPase Assays
Steady state ATPase assays were performed as described (Alcázar-Román et al., 2006) with minor modifications. ATPase assays were performed with LOS4-His proteins in a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 3 mM MgCl2, 1 mM DTT, 2 mM ATP, 6 mM phosphoenolpyruvate, 1.2 mM NADH, 1 mg/mL BSA, and 2% (v/v) pyruvate kinase/lactate dehydrogenase (Sigma-Aldrich) in a total volume of 100 μL in a 96-well plate. Polyadenylic acid was added to 50 μg/mL or other concentrations as described. Absorbance at 340 nm was measured using a VersaMax Absorbance Microplate Reader (Molecular Devices), and the data were analyzed using SoftMax Pro software (Molecular Devices).
Nucleic Acid-Melting Assays
Nucleic acid-melting assays were performed as described (Kim et al., 2007) using 78-nucleotide-long, 9-bp-containing, hairpin-shaped molecular beacons conjugated with fluorophore (tetramethylrhodamine) and quencher (Dabcyl). Spectrofluorometry was performed using a fluorescence spectrophotometer (Hitachi F-2000) at an excitation wavelength of 555 nm and an emission wavelength of 575 nm.
Immunolabeling of Tobacco BY-2 Cells
Immunocytochemistry of BY-2 cells was performed as described (Lee et al., 2009). For double-labeling of Gle1 and α-tubulin, BY-2 cells were fixed, permeabilized, and immunolabeled with anti-Gle1 (rabbit polyclonal) antibodies (1:200; Cosmogenetech) and anti-α-tubulin (mouse monoclonal) antibodies (1:1000; Sigma-Aldrich). Then, the cells were incubated with Alexa Fluor 563-conjugated anti-rabbit IgG antibodies (1:1000; Invitrogen) and Alexa Fluor 488-conjugated anti-mouse IgG antibodies (1:1000; Invitrogen). After brief staining with DAPI, the BY-2 cells were observed by confocal laser scanning microscopy (Zeiss LSM510).
Coimmunoprecipitation
Flag-Gle1 and LOS4-Myc fusion proteins were coexpressed in N. benthamiana leaves by agroinfiltration. Coimmunoprecipitation was performed following the manufacturer’s instructions using an ANTI-FLAG M2 Affinity Gel (Sigma-Aldrich). After elution with 3X FLAG Peptide (F4799; Sigma-Aldrich), proteins were subjected to SDS-PAGE and immunoblotting.
Purification of Recombinant Proteins
Gle1 and Gle1C (244 to 611 amino acid residues) were cloned into pMAL C2X vector (New England Biolabs) for MBP fusion, and LOS4 was cloned into pET-29a vector (Novagen) for His fusion. MBP-Gle1 and MBP-Gle1C proteins were expressed in BL21 (DE3) strain, and LOS4-His protein was expressed in Rosseta (DE3) strain of Escherichia coli. Cells were grown at 37°C to an A600 of 0.4, shifted to 16°C, and then induced by 0.25 mM isopropyl β-d-1-thiogalactopyranoside for 16 h. The MBP- and His-fused proteins were purified following the manufacturer’s instructions using MBP Excellose (Bioprogen) and His60 Ni Superflow Resin (Clontech), respectively.
In Vitro Pull-Down Assay
MBP and MBP-Gle1 proteins immobilized on MBP Excellose (Bioprogen) were incubated with LOS4-His proteins for 2 h at room temperature. Similarly, LOS4-His proteins immobilized on His60 Ni Superflow resin (Clontech) were incubated with MBP or MBP-Gle1 for 2 h at room temperature. After extensive washing of the resins, bound proteins were eluted with 2× SDS sample buffer, and eluted proteins were visualized by Coomassie Brilliant Blue staining.
HPIC
Seed extracts were prepared from mature desiccated seeds as described (Stevenson-Paulik et al., 2005) with minor modifications. Approximately 15 mg of seeds and 15 mg of acid-washed glass beads (425 to 600 mm; Sigma-Aldrich) were mixed with 20 volumes of 0.4 M HCl. Samples were pulverized using a Mini-BeadBeater 16 (BioSpec Products) for 5 min and then boiled for 5 min. Samples were pulverized again for 5 min and seed extracts were collected by centrifugation for 10 min at 15,000g. The extracts were passed through filters (PTFE, 0.2 μm; Whatman) and analyzed by HPIC (ICS-3000; Dionex) as described (Kim and Tai, 2011) with minor modifications. An IonPac AS11 anion exchange column (4 × 250 mm; Dionex) was eluted with a linear gradient of NaOH from 5 to 80 mM under a flow rate of 1 mL/min for 70 min at 35°C. A conductivity detector was used with an electrolytically regenerated suppressor (ERS 500; Dionex) operated with the external water mode at a current of 300 mA. Inositol phosphate was purchased from Sigma-Aldrich, and a standard curve was established for the quantification. All the measurements were performed in triplicate and expressed as an average with sd.
Statistical Analyses
Two-tailed Student’s t tests were performed using the Minitab 16 program to determine the statistical differences between the samples.
Accession Numbers
The sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: Gle1, At1g13120; LOS4, At3g53110; and IPK1, At5g42810.
Supplemental Data
Supplemental Figure 1. Phylogenetic Tree of Gle1.
Supplemental Figure 2. Protein Structure and Sequence Alignment of Gle1.
Supplemental Figure 3. Analyses of Gle1-Silencing Phenotypes Using VIGS in N. benthamiana.
Supplemental Figure 4. Subcellular Localization of Gle1.
Supplemental Figure 5. Protein Expression in BiFC Analyses.
Supplemental Figure 6. Control Experiments for the ATPase Assay.
Supplemental Figure 7. Computational Modeling of the Gle1 Domain.
Supplemental Figure 8. Alignment of Amino Acid Residues Surrounding the Key Residues of the InsP6 Binding Pocket of Gle1.
Supplemental Figure 9. Efficiency of InsP3, InsP4, and InsP5 in Stimulating LOS4 ATPase Activity.
Supplemental Figure 10. Leaf Series of the Plants Showing the Six Largest Leaves.
Supplemental Figure 11. In Situ Hybridization to Visualize Poly(A) RNA Export Defects.
Supplemental Figure 12. Seed Germination Rates in Response to Diverse Abiotic Stresses.
Supplemental Figure 13. Analysis of Seed Phytate (InsP6) Contents Using High-Performance Ion Chromatography.
Supplemental Data Set 1. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Hunseung Kang (Chonnam National University, Korea) for providing molecular beacons, Ji-Hyun Lee and Hee-Kyung Ahn (Yonsei University, Korea) for helpful discussions on the article, and Yerim Lee (Yonsei University, Korea) for propagating transgenic Arabidopsis lines. This research was supported by the Cooperative Research Program for Agriculture Science and Technology Development (project numbers PJ011147 [PMBC] and PJ011189 [SSAC]) from the Rural Development Administration of Republic of Korea.
AUTHOR CONTRIBUTIONS
H.-S.L. performed most of the experiments and analyzed the results together with D.-H.L. and H.K.C. S.H.K. and J.H.A. performed HPIC. H.-S.P. designed the experiments and wrote the article. All authors discussed the results and commented on the article.
Glossary
- NPC
nuclear pore complex
- VIGS
virus-induced gene silencing
- DEX
dexamethasone
- RNAi
RNA interference
- MS
Murashige and Skoog
- DAPI
4′,6-diamidino-2-phenylindole
- BiFC
bimolecular fluorescence complementation
- HPIC
high-performance ion chromatography
- InsP6
myo-inositol-1,2,3,4,5,6-hexakisphosphate
References
- Ahn C.S., Han J.A., Lee H.S., Lee S., Pai H.S. (2011). The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell 23: 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar-Román A.R., Bolger T.A., Wente S.R. (2010). Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 285: 16683–16692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar-Román A.R., Tran E.J., Guo S., Wente S.R. (2006). Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 8: 711–716. [DOI] [PubMed] [Google Scholar]
- Ali N., Paul S., Gayen D., Sarkar S.N., Datta K., Datta S.K. (2013). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene (IPK1). PLoS ONE 8: e68161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardsley T.M. (2011). Peak phosphorus. Bioscience 61: 91. [Google Scholar]
- Bilyeu K.D., Zeng P., Coello P., Zhang Z.J., Krishnan H.B., Bailey A., Beuselinck P.R., Polacco J.C. (2008). Quantitative conversion of phytate to inorganic phosphorus in soybean seeds expressing a bacterial phytase. Plant Physiol. 146: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Nachury M., Heald R., Weis K. (2005). A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234. [DOI] [PubMed] [Google Scholar]
- Bolger T.A., Folkmann A.W., Tran E.J., Wente S.R. (2008). The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braud C., Zheng W., Xiao W. (2012). LONO1 encoding a nucleoporin is required for embryogenesis and seed viability in Arabidopsis. Plant Physiol. 160: 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregitzer P., Raboy V. (2006). Effects of four independent low-phytate mutations on barley agronomic performance. Crop Sci. 46: 1318–1322. [Google Scholar]
- Brinch-Pedersen H., Hatzack F., Stöger E., Arcalis E., Pontopidan K., Holm P.B. (2006). Heat-stable phytases in transgenic wheat (Triticum aestivum L.): deposition pattern, thermostability, and phytate hydrolysis. J. Agric. Food Chem. 54: 4624–4632. [DOI] [PubMed] [Google Scholar]
- Cho H.K., Ahn C.S., Lee H.S., Kim J.K., Pai H.S. (2013). Pescadillo plays an essential role in plant cell growth and survival by modulating ribosome biogenesis. Plant J. 76: 393–405. [DOI] [PubMed] [Google Scholar]
- Dossani Z.Y., Weirich C.S., Erzberger J.P., Berger J.M., Weis K. (2009). Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1. Proc. Natl. Acad. Sci. USA 106: 16251–16256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., Hetzer M.W. (2013). The role of Nup98 in transcription regulation in healthy and diseased cells. Trends Cell Biol. 23: 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Lee H., Xiong L., Jagendorf A., Stevenson B., Zhu J.K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99: 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Dong C.H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanakahi L.A., West S.C. (2002). Specific interaction of IP6 with human Ku70/80, the DNA-binding subunit of DNA-PK. EMBO J. 21: 2038–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K.B., Malureanu L., van Deursen J.M. (2005). The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438: 1036–1039. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Park S.J., Kwak K.J., Kim Y.O., Kim J.Y., Song J., Jang B., Jung C.H., Kang H. (2007). Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 35: 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.I., Tai T.H. (2011). Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genomics 286: 119–133. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Jung H.J., Kwak K.J., Kim M.K., Oh S.H., Han Y.S., Kang H. (2010). The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell 22: 3951–3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Panse V.G. (2008). Gle1 does double duty. Cell 134: 564–566. [DOI] [PubMed] [Google Scholar]
- Kuwano M., Mimura T., Takaiwa F., Yoshida K.T. (2009). Generation of stable ‘low phytic acid’ transgenic rice through antisense repression of the 1D-myo-inositol 3-phosphate synthase gene (RINO1) using the 18-kDa oleosin promoter. Plant Biotechnol. J. 7: 96–105. [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Lee H.S., Wi S.J., Park K.Y., Schmit A.C., Pai H.S. (2009). Dual functions of Nicotiana benthamiana Rae1 in interphase and mitosis. Plant J. 59: 278–291. [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A.C., Brearley C.A. (2000). Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc. Natl. Acad. Sci. USA 97: 8687–8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F., MacRobbie E.A.C., Webb A.A.R., Manison N.F., Brownlee C., Skepper J.N., Chen J., Prestwich G.D., Brearley C.A. (2003). Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc. Natl. Acad. Sci. USA 100: 10091–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.X., Zhao H.J., Pang W.Q., Cui H.R., Poirier Y., Shu Q.Y. (2014). Seed-specific silencing of OsMRP5 reduces seed phytic acid and weight in rice. Transgenic Res. 23: 585–599. [DOI] [PubMed] [Google Scholar]
- Lott J.N.A., Greenwood J.S., Batten G.D. (1995). Mechanisms and regulation of mineral nutrient storage during seed development. In Seed Development and Germination, Kigei J. and G. Galli, eds (New York: Marcel Dekker; ), pp. 215–235. [Google Scholar]
- Macbeth M.R., Schubert H.L., Vandemark A.P., Lingam A.T., Hill C.P., Bass B.L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meis S.J., Fehr W.R., Schnebly S.R. (2003). Seed source effect on field emergence of soybean lines with reduced phytate and raffinose saccharides. Crop Sci. 43: 1336–1339. [Google Scholar]
- Miller A.L., Suntharalingam M., Johnson S.L., Audhya A., Emr S.D., Wente S.R. (2004). Cytoplasmic inositol hexakisphosphate production is sufficient for mediating the Gle1-mRNA export pathway. J. Biol. Chem. 279: 51022–51032. [DOI] [PubMed] [Google Scholar]
- Monserrate J.P., York J.D. (2010). Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 22: 365–373. [DOI] [PubMed] [Google Scholar]
- Montpetit B., Thomsen N.D., Helmke K.J., Seeliger M.A., Berger J.M., Weis K. (2011). A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472: 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Nielsen E. (2011). Green light for polyphosphoinositide signals in plants. Curr. Opin. Plant Biol. 14: 489–497. [DOI] [PubMed] [Google Scholar]
- Murphy A.M., Otto B., Brearley C.A., Carr J.P., Hanke D.E. (2008). A role for inositol hexakisphosphate in the maintenance of basal resistance to plant pathogens. Plant J. 56: 638–652. [DOI] [PubMed] [Google Scholar]
- Murphy R., Wente S.R. (1996). An RNA-export mediator with an essential nuclear export signal. Nature 383: 357–360. [DOI] [PubMed] [Google Scholar]
- Nagy R., Grob H., Weder B., Green P., Klein M., Frelet-Barrand A., Schjoerring J.K., Brearley C., Martinoia E. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284: 33614–33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen H.O., Kestilä M., Pakkasjärvi N., Honkala H., Kuure S., Tallila J., Vuopala K., Ignatius J., Herva R., Peltonen L. (2008). Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat. Genet. 40: 155–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo A.V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S.G., Fontoura B., Briggs S., Dasso M., Forbes D.J. (2006). The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell 17: 3806–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui M.S., Capp R., Staehelin L.A. (2002). Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14: 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboy V. (2001). Seeds for a better future: ‘low phytate’ grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 6: 458–462. [DOI] [PubMed] [Google Scholar]
- Raboy V. (2003). myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64: 1033–1043. [DOI] [PubMed] [Google Scholar]
- Raboy V. (2009). Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 177: 281–296. [Google Scholar]
- Raboy V., Gerbasi P.F., Young K.A., Stoneberg S.D., Pickett S.G., Bauman A.T., Murthy P.P.N., Sheridan W.F., Ertl D.S. (2000). Origin and seed phenotype of maize low phytic acid 1-1 and low phytic acid 2-1. Plant Physiol. 124: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regot S., de Nadal E., Rodríguez-Navarro S., González-Novo A., Pérez-Fernandez J., Gadal O., Seisenbacher G., Ammerer G., Posas F. (2013). The Hog1 stress-activated protein kinase targets nucleoporins to control mRNA export upon stress. J. Biol. Chem. 288: 17384–17398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang H., Hazebroek J., Ertl D.S., Harp T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42: 708–719. [DOI] [PubMed] [Google Scholar]
- Shi J., Wang H., Wu Y., Hazebroek J., Meeley R.B., Ertl D.S. (2003). The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 131: 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wang H., Schellin K., Li B., Faller M., Stoop J.M., Meeley R.B., Ertl D.S., Ranch J.P., Glassman K. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25: 930–937. [DOI] [PubMed] [Google Scholar]
- Stevenson-Paulik J., Bastidas R.J., Chiou S.T., Frye R.A., York J.D. (2005). Generation of phytate-free seeds in Arabidopsis through disruption of inositol polyphosphate kinases. Proc. Natl. Acad. Sci. USA 102: 12612–12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645. [DOI] [PubMed] [Google Scholar]
- Terry M.J., Kendrick R.E. (1999). Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 119: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.S., Weaver P.L., Liu Y., Hitomi M., Tartakoff A.M., Chang T.H. (1998). Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17: 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbsky J., Lavine K., Majerus P.W. (2005). Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc. Natl. Acad. Sci. USA 102: 8448–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B., Antonin W. (2014). The diverse roles of the Nup93/Nic96 complex proteins - structural scaffolds of the nuclear pore complex with additional cellular functions. Biol. Chem. 395: 515–528. [DOI] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., Berger J.M., Weis K. (2004). The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore. Mol. Cell 16: 749–760. [DOI] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., Flick J.S., Berger J.M., Thorner J., Weis K. (2006). Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8: 668–676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.