Abstract
Interactions of the hydrophobic anticancer drug tamoxifen (TAM) with lipid model membranes were studied using calcein-encapsulated vesicle leakage, attenuated total reflection Fourier transform infrared (FTIR) spectroscopy, small-angle neutron scattering (SANS), atomic force microscopy (AFM) based force spectroscopy, and all-atom molecular dynamics (MD) simulations. The addition of TAM enhances membrane permeability, inducing calcein to translocate from the interior to the exterior of lipid vesicles. A large decrease in the FTIR absorption band’s magnitude was observed in the hydrocarbon chain region, suggesting suppressed bond vibrational dynamics. Bilayer thickening was determined from SANS data. Force spectroscopy measurements indicate that the lipid bilayer area compressibility modulus KA is increased by a large amount after the incorporation of TAM. MD simulations show that TAM decreases the lipid area and increases chain order parameters. Moreover, orientational and positional analyses show that TAM exhibits a highly dynamic conformation within the lipid bilayer. Our detailed experimental and computational studies of TAM interacting with model lipid membranes shed new light on membrane modulation by TAM.
Introduction
Tamoxifen (TAM) is an important anticancer drug that is commonly used in the prevention and treatment of breast cancer (1), and also exhibits antioxidant and cardioprotective effects (2). One mechanism by which TAM inhibits cancer cell growth is competitive blocking of estrogen receptors (3). However, TAM also inhibits the growth of estrogen-receptor-negative breast cancer cells (4). This implies the presence of additional mechanisms that are not related to estrogen receptor mediation. TAM was also found to antagonize protein kinase C without interacting with the enzyme’s active site (5). Other studies have shown that TAM can cause liver toxicity by affecting mitochondria functions (6). Since TAM is highly lipophilic (7), modulation of cell membrane structural and mechanical properties may be a reason for its anticancer activity (2) and associated adverse effects (8).
Model lipid bilayers composed of one or a few lipid species are suitable systems for investigating the interactions of membrane active compounds with cell membranes. Previous studies of TAM and lipid bilayers mainly used fluorescence anisotropy, differential scanning calorimetry (DSC), and Fourier transform infrared (FTIR) spectroscopy. In particular, anisotropy and DSC measurements have shown that TAM decreases lipid-phase transition temperatures (9–14). Studies using anisotropy (9) and FTIR (15) measurements reported that TAM disorders gel-phase lipid organization, although another study found no discernible anisotropy change (11). Other studies using FTIR have reported mixed effects on gel-phase chain packing as a function of TAM concentration (12–14,16). Similar discrepancies have been reported for fluid-phase lipid chain organization. Chain ordering (9,11,15,17), disordering (10,18,19), and concentration-dependent effects (12–14,16) have all been reported using anisotropy and FTIR measurements (on saturated phospholipids). These mixed reports are complicated by the fact that both fluorescence anisotropy and FTIR are not very sensitive to small changes in lipid bilayer properties. For example, we observed no band shift in our FTIR spectra, whereas our other techniques clearly showed membrane modulation exerted by TAM (see Results section). Similarly, we have unpublished data showing that fluorescence anisotropy is not a sensitive technique when only small changes take place in a fluid phase bilayer. Moreover, fluorescence anisotropy relies on membrane-solubilized dye molecules, which can modify lipid bilayer properties (20). Finally, many of the above-mentioned studies used bilayers composed of lipids with two saturated chains, mainly because this facilitates access to the gel and fluid phases. Although these lipids are experimentally appealing, lipids with one saturated and one unsaturated chain are better mimics of biological membranes.
Here, we examined the effects of TAM on a lipid bilayer composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC). Since biological membranes often contain anionic lipids, we also added 10 mol % of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) to the POPC bilayer. For simplicity, we will refer to this mixture as a POPC/POPG bilayer. We first used a fluorescence leakage assay to elucidate the perturbation effect of TAM on POPC/POPG bilayer integrity. We also used attenuated total reflection FTIR spectroscopy to explore lipid bond vibrational dynamics (21). Due to the differences in neutron scattering length between the lipid bilayer’s hydrocarbon chains and the D2O solvent, small-angle neutron scattering (SANS) is widely used to determine the lipid bilayer hydrophobic thickness, as well as other important structural parameters (22). We collected SANS data under three external contrast conditions for POPC/POPG bilayers, with and without TAM, and determined measurable changes in the lipid bilayer thickness. We investigated the effects of TAM on lipid bilayer mechanical properties using atomic force microscopy (AFM)-based force spectroscopy measurements. We then used the force-distance curves obtained as the AFM tip approached a solid supported planar bilayer with different TAM concentrations to extract the lipid bilayer area compressibility moduli KA. Finally, we performed all-atom molecular dynamics (MD) simulations to gain a mechanistic understanding of TAM interactions with neighboring lipids. We carried out detailed analyses pertaining to TAM orientation and its impact on lipid bilayer properties, such as the average lipid area, hydrocarbon chain thickness, and chain order parameters.
Materials and Methods
Materials
Synthetic POPC and POPG, in powder form, were purchased from Avanti Polar Lipids (Alabaster, AL). TAM and calcein were purchased from Sigma Aldrich (St. Louis, MO). All other chemicals were of HPLC grade.
POPC, POPG, and TAM stock solutions were prepared using organic solvents (chloroform for POPC and TAM, and chloroform/methanol 3:1 by volume for POPG). Lipid mixtures were prepared by mixing appropriate ratios of stock solutions in glass test tubes. Organic solvents were removed by a gentle stream of argon gas and then placed under vacuum overnight. The resultant lipid dry films were used for experimental measurements.
Dye leakage
Calcein-encapsulated unilamellar vesicles (ULVs) were prepared using the method described in the Supporting Material. Fluorescence intensity at 25°C was measured using an FP-8300 fluorometer (Jasco, Easton, MD). To study the degree of dye leakage as a function of the TAM/lipid ratio, r, we added different amounts of TAM solubilized in DMSO to microcentrifuge tubes containing 1 ml of HEPES buffer B (Supporting Material), and added 15 μl of calcein-encapsulated ULVs at the end. To assess the effect of DMSO on calcein leakage, we performed control experiments with DMSO alone. No discernible dye leakage was observed for DMSO concentrations of <3%. Based on these data, all calcein leakage experiments were carried out with <1.5% of DMSO.
A time-course measurement was first carried out to establish the equilibration time needed for TAM to induce maximum calcein leakage (Fig. S13). TAM caused a rapid increase in fluorescence intensity during the first 9 min, followed by a gradual increase. The final equilibrium state was reached in ∼8 h. To eliminate complications associated with incubation time, all TAM/lipid samples were measured after they were incubated overnight in the dark under gentle shaking.
After incubation, TAM/lipid/calcein solutions were excited by a wavelength of 494 nm. The emission spectrum was recorded from 498 nm to 600 nm. Both the excitation and emission bandwidths were 2.5 nm. The emission maximum near 514 nm was taken as the fluorescence intensity I for each sample. The fluorescence intensity without any quenching, Imax, was determined by completely breaking up the ULVs using 100 mM Triton X-100 (after taking dilution into account). The percentage of calcein leakage is defined as
| (1) |
where I0 is the fluorescence intensity from calcein-encapsulated ULVs without TAM.
FTIR spectroscopy
A Vertex 70 Bio ATRII spectrometer (Bruker, Billerica, MA) was used for attenuated total reflection FTIR measurements. All samples were maintained at 25°C using a Huber Ministat water circulator. POPC lipid (8.0 mg) mixed with different amounts of TAM was hydrated with 0.25 ml of HEPES buffer A (Supporting Material). A multilamellar vesicle (MLV) solution (30 μl) was loaded into a sample cell. Infrared (IR) absorption spectra were collected at a range of wavenumbers from 400 to 4000 cm−1 with a resolution of 2 cm−1 (1500 scans). Each spectrum was corrected for background absorption arising from the water buffer. The final spectrum was normalized by the lipid concentration.
SANS experiments
Lipid dry films, with and without 10 mol % TAM, were hydrated with D2O buffer. ULVs were obtained by an extrusion procedure similar to that used for the leakage experiments. The extruded ULVs in 100% D2O were aliquoted into three microcentrifuge tubes and diluted to the desired external contrast conditions (i.e., 100%, 75%, and 50% D2O) using appropriate ratios of D2O and H2O buffers. SANS data were obtained at 25°C on the CG-3 Bio-SANS instrument located at Oak Ridge National Laboratory (ORNL). The beam wavelength was 6.0 Å with a dispersion of 12%. The collected scattering images were corrected for dark current, solid angle, detector sensitivity, and sample transmission. One-dimensional scattering curves were obtained by powder averaging after correcting for water background. All data reduction was performed using the Mantid software provided by ORNL (http://dx.doi.org/10.5286/SOFTWARE/MANTID).
AFM experiments
AFM experiments were performed using a Multimode 8 atomic force microscope (Bruker, Santa Barbara, CA) operated in a mode referred to as PeakForce quantitative nanomechanics (QNM). This mode allows simultaneous mapping of a sample’s nanomechnical and topographical properties. Small unilamellar vesicles (SUVs) were used for AFM measurements. Briefly, lipid films with different TAM concentrations were hydrated with 10 mM of HEPES buffer. SUVs were prepared using an ultrasonicator operated at 40 W for a total duration of 15 min. The obtained SUVs were centrifuged to remove metal particles that detached from the probe. Dynamic light scattering measurements showed that the ultrasonication-prepared SUVs had an average diameter of 60 nm. The SUVs were injected into a liquid cell via a syringe pump. After incubation for 1 h, a mica supported bilayer was formed. HEPES buffer was injected to flush out excess vesicles. We scanned hydrated bilayers using a Si3N4 cantilever with a spring constant of 0.1 N/m, which was determined using the thermal noise method (23). After obtaining a topographic image, we collected multiple force-distance curves at different locations on the bilayer. To extract the lipid bilayer area compressibility modulus, we determined the AFM tip radius using a tip-quantification function provided by the Nanoscope analysis software. The tip radius determined for this study was 11.0 nm.
MD simulations
We generated the initial coordinates for two POPC/POPG bilayer models made up of 200 lipids (i.e., 180 POPC and 20 POPG) using the CHARMM-GUI Membrane Builder (24) for one model and Packmol (25) for the other. Despite their different initial structural properties, the two model bilayers yielded similar results, with comparable average lipid area and chain thickness (after reaching equilibrium). This highlights that our simulations do not depend on initial conditions, thus supporting the convergence of our simulated bilayers. We generated the initial coordinates for the two POPC/POPG/TAM lipid bilayer systems (i.e., 162 POPC, 18 POPG, and 20 TAM) using Packmol with different random seeds and packing protocols. All hydrogen atoms were explicitly included (all-atom model) in addition to ∼10,800 water molecules and counterions to neutralize the system. MD simulations were performed using NAMD 2.9 (26) and the CHARMM 36 lipid force field (27,28) for lipid molecules, and the CHARMM general force field for TAM was generated using the ParamChem server (http://www.paramchem.org) (29). Periodic boundary conditions were applied. For each system, we first minimized the atomic coordinates using the conjugated gradient algorithm for 5000 steps, followed by 20 ns of equilibration in a constant particle NPT ensemble. The z axis was allowed to expand and contract independently of the x-y plane (semi-isotropic pressure coupling). Subsequently, the four simulations (two POPC/POPG binary mixtures and two POPC/POPG mixed with 10 mol % TAM) were each run for ∼150 ns in the NPT ensemble using the Langevin piston method (target pressure: 1 atm; oscillation period: 200 fs; and damping timescale: 100 fs) and Langevin dynamics to control the temperature (298 K; damping coefficient: 2.0) during the entire simulation (30,31). In all simulations, the van der Waals (vdW) interactions were truncated via a force-based switching function. Starting from a switching distance of 8 Å, the vdW force was brought smoothly to zero at the cutoff distance of 12 Å. Electrostatic interactions were treated using the particle-mesh Ewald (PME) method with a 1.0 Å grid spacing (32,33). The r-RESPA multiple-time-step method (34) was employed with a 2 fs time step for bonded interactions, and 2 and 4 fs time steps for short-range nonbonded and long-range electrostatic interactions, respectively. The bonds between hydrogen and other atoms were constrained using the SHAKE algorithm (35). All simulations were conducted on the Hopper supercomputer located at the National Energy Research Scientific Computing Center (NERSC).
Only the last 100 ns of all simulations were used for structural analyses. The orientational order parameter −2SCD for each carbon of POPC’s palmitoyl and oleoyl chains was calculated from the angle θ of the C-D bond relative to the bilayer normal (36):
| (2) |
Results
Calcein leakage and bilayer integrity
When TAM is introduced to calcein-encapsulated ULVs, its large hydrophobicity prompts it to bind to surrounding ULVs. The resulting membrane perturbation causes encapsulated calcein to leak out. By monitoring the change of fluorescence intensity, one can estimate the amount of calcein that is released. Fluorescence spectra of POPC/POPG ULVs incubated with different amounts of TAM are shown in Fig. S13. The emission maximum is located near 514 nm and is independent of TAM concentration. The calculated percentage of calcein leakage is shown in Fig. 1. It is clear that the leakage percentage increases rapidly as the TAM/lipid ratio, r, increases. Maximum leakage is reached near r = 1.0.
Figure 1.
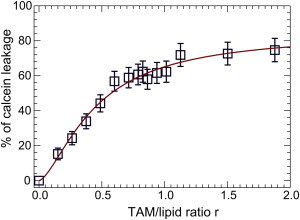
Leakage of calcein-encapsulated ULVs induced by TAM. The solid line is a fit using the empirical Hill equation. To see this figure in color, go online.
We use a Hill equation to qualitatively describe calcein leakage:
| (3) |
where Gmax is the maximum leakage, r is the TAM/lipid ratio, r0 is the ratio at which the half-maximum of calcein leakage occurs, and n is the Hill coefficient. We note that the use of Eq. 3 is purely empirical. A nonlinear least-square fit gives rise to a maximum leakage Gmax of 85%, a value smaller than unity. This indicates that even at maximum leakage, there is still a portion of calcein encapsulated within the ULVs. Partial maximum leakage has also been reported for flavanoid genistein and its derivatives (37), and fibril-forming amyloid peptides (38). The fit to the data also results in an r0 of 0.46 at half-maximum leakage, and a Hill coefficient n of 1.5. Overall, our fluorescence data highlight that TAM impairs bilayer integrity and enhances the permeability of calcein across the bilayer. The fact that the maximum leakage is less than 100% suggests that TAM only modifies the structural properties of the lipid bilayer. This is different from surfactants (39) and antimicrobial peptides (40), which solubilize or completely permeabilize lipid vesicles at high concentrations.
FTIR spectroscopy and lipid bond vibrational dynamics
To study the effect of TAM on lipid vibrational dynamics, we performed attenuated total reflection FTIR measurements on POPC MLVs mixed with different concentrations of TAM. Fig. 2 illustrates the absorption bands corresponding to three chemical groups: the hydrocarbon chain region (CH2, Fig. 2 A), the backbone ester carbonyl (C=O; Fig. 2 B), and the water-solvated headgroup (PO2−; Fig. 2 C). The spectrum for each sample is normalized by its lipid concentration. This is corroborated by the nearly identical absorption peaks located around 1229 and 1087 cm−1, which correspond to PO2−’s antisymmetric and symmetric vibrations, respectively (Fig. 2 C). No discernible band shift is observed for the two bands corresponding to the hydrocarbon chain CH2’s antisymmetric (2923 cm−1) and symmetric (2853 cm−1) vibrations as TAM concentration increases (Fig. 2 A). The absence of band shift is also noted for the ester carbonyl C=O (Fig. 2 B). These results contrast with the FTIR measurements obtained for TAM in fully saturated lipids (10,13,14,16).
Figure 2.

Attenuated total reflection FTIR spectrum for POPC MLVs containing different concentrations of TAM. (A) Lipid IR absorption bands located near 2923 and 2853 cm−1 correspond to hydrocarbon chain CH2 groups undergoing antisymmetric and symmetric stretching vibrations, respectively. (B) The absorption band of the backbone ester carbonyl C=O occurs near 1733 cm−1. (C) Antisymmetric and symmetric absorption bands corresponding to the phosphate PO2− are observed near 1229 and 1087 cm−1, respectively. The width of each panel is proportional to the span of its wavenumber. To see this figure in color, go online.
In contrast to the band shift, the band magnitudes for CH2 and C=O decrease with increasing TAM concentration (Fig. 2, A and B). Since the absorption spectrum is normalized by the lipid concentration, the band magnitude is associated with a change in the bond length (or dipole moment) during bond vibration. Therefore, a reduction in the band magnitude implies that TAM dampens the bond-length change for CH2 and C=O groups. This implies a more rigid configuration in the hydrocarbon chains and carbonyl backbone region. In contrast to CH2 and C=O, the vibrational bands for PO2− remain unaltered after addition of TAM (Fig. 2 C), suggesting that TAM does not interact with POPC’s phosphate headgroup.
SANS data and bilayer thickness
Hydrogen and its isotope, deuterium, possess very different neutron scattering lengths. This results in a large difference in neutron scattering length density (NSLD) between the lipid bilayer’s protiated hydrocarbon chains and the D2O water solvent. In this study, we collected SANS data under three external contrast conditions (i.e., 100%, 75%, and 50% D2O) for POPC/POPG ULVs, with and without 10 mol % TAM. We used a three-strip model (Supporting Material) (41) to simultaneously analyze the three sets of SANS data (Fig. 3). The corresponding structural parameters are listed in Table S1. At each D2O concentration, good overlap is observed between the model and the experimental data. The resulting hydrocarbon chain thickness of 24.6 Å is smaller by 4 Å compared with previously determined POPC (42) and POPG (43) bilayers. For the headgroup thickness, an overestimation is observed. This is consistent with a previous finding that multi-strip models tend to overestimate the polar headgroup thickness (41), possibly due to an oversimplification of the heterogeneous NSLD distribution within the headgroup region.
Figure 3.
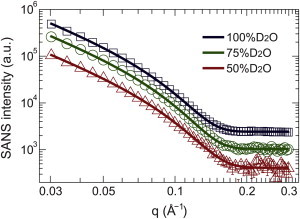
Simultaneous three-strip model fits (solid lines) to SANS data (open symbols) for POPC/POPG ULVs at three external contrast conditions. Intensities were shifted vertically for better visualization. Error bars at small q are less than the dimensions of the symbols. To see this figure in color, go online.
Addition of TAM increases the hydrocarbon chain thickness of the lipid bilayer by 2.8 Å. The observed chain thickening is consistent with TAM’s rigidifying effect on the lipid chain vibrational dynamics. Similar to what has been reported for cholesterol (44,45), TAM’s triple-ring structure could restrict the dynamics of nearby hydrocarbon chains, forcing them to adopt a more linear conformation and thus yielding a thicker bilayer. Note that our SANS data reveal an overall change in the lipid bilayer structure. An increase in hydrocarbon chain thickness could simply be due to the addition of TAM without altering the lipid properties. However, this was not the case, as was revealed by our MD simulations (discussed further below).
AFM
We used AFM-based force spectroscopy to explore lipid bilayer mechanical properties. Based on the height profile of a partially formed POPC/POPG bilayer (Supporting Material), the bilayer surface protrudes by 6 nm above the mica substrate. After a large bilayer patch is formed, as shown by the topographic image, we collected force-distance curves by monitoring the cantilever deflection d and the piezoelectric distance z as the AFM tip approached and retracted from the lipid bilayer. For the lipid bilayer, a jump near 5 nN was observed in the approaching curve (Supporting Material). When the tip is far from the bilayer, no resistance force exists. As the tip moves in the vicinity of the bilayer, vdW and electrostatic interactions emerge. In the intermediate regime where the tip makes contact but does not puncture the bilayer, bilayer deformation becomes the predominant source contributing to the tip force. In this regime, tip indentation causes nearby lipid molecules to adopt a stretched conformation. The free energy associated with the increased lipid area under the curved tip can be described by the area compressibility modulus KA (46). The relation between the tip force F and the tip indentation z0 can be approximated as
| (4) |
where R is the tip radius and h is the intact bilayer height. To obtain the tip indentation, we convert the force-distance curves into force-separation curves. The separation refers to the distance between the tip apex and the mica substrate. A force-separation curve is shown in Fig. 4 A. It is clear that the bilayer deformation mainly takes place when the separation changes from 6 to 3 nm. It is interesting to note that there is an ∼3-nm-thick space between the substrate and the point where the bilayer is punctured. This region corresponds to a water layer cushioning the bilayer. Within the bilayer elastic deformation regime, the tip indentation z0 is the difference between the bilayer height and the tip-substrate separation. The bilayer KA is obtained by fitting the force-separation data using Eq. 4. An example of the data fitting is shown in Fig. 4 B. Good agreement is obtained between the theoretical predication and the experimental data. Note that we only include data points with separations of <6 nm. Inclusion of data points at larger separations only slightly changes KA. Another consideration is that Eq. 4 is only valid in the bilayer deformation regime (i.e., the separation should be no larger than the bilayer height). After an analysis of >300 force curves, the averaged KA for the pure lipid bilayer is 278 mN/m with an uncertainty of ∼15%. Our force-spectroscopy-derived KA is in good agreement with results obtained from micropipette aspiration measurements (47).
Figure 4.
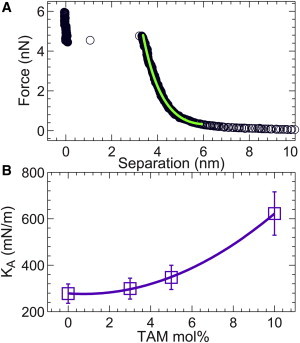
Lipid bilayer area compressibility modulus KA obtained from force-separation curves. (A) A typical force-separation curve for a solid supported lipid bilayer. The increased force from 6 to 3 nm as the tip approaches the substrate is predominantly contributed by tip indentation into the lipid bilayer. The green line is a theoretical fit based on the bilayer deformation model outlined in the main text. (B) KA at different TAM concentrations. The KA values for 0, 3, 5, and 10 mol % TAM are 278, 299, 348, and 622 mN/m, respectively. The uncertainty of each KA is ∼15%. The solid line is a quadratic fit. To see this figure in color, go online.
The same analyses were performed for force curves obtained from bilayers with different amounts of TAM. The results are shown in Fig. 4 B. It is clear that KA increases with TAM concentration. At 10 mol % TAM, the KA value is doubled compared with that obtained for the pure lipid bilayer. Similar to our findings with TAM, cholesterol was previously observed to increase the KA of a POPC bilayer (48). However, 30 mol % of cholesterol is needed to double the KA. In contrast to the fused-ring system of cholesterol, TAM contains an extended triple-ring structure, which may contribute to the larger resistance under lateral compression.
MD simulations
We performed all-atom MD simulations of a POPC/POPG bilayer before and after adding 10 mol % TAM. To assess the effect of the initial configurations of TAM molecules within the lipid bilayer, we built two TAM-lipid systems. The initial tilt angles of the 20 TAM molecules are populated within a range of 7–38° in one leaflet, and 142–172° in the opposite leaflet (Supporting Material). Tilt angle is defined by the vector O-C11 connecting the oxygen and C11 atoms of TAM with respect to the bilayer normal (Fig. 5 A). The time trajectories for the 20 TAM molecules indicate that the individual tilt angles oscillate between 30° and 150°, with a periodicity of ∼20 ns (Supporting Material), highlighting a highly dynamic conformation of TAM within the lipid bilayer. To test the convergence of our simulations, we calculated time-block averaged TAM orientational and positional distributions. We found that the 120 ns block (from 80 ns to 120 ns) overlapped very well with the 160 ns block (120–160 ns) with regard to both orientation and position. Moreover, the two independent sets of TAM-lipid simulations show similar distributions and time-averaged structural parameters (Supporting Material), supporting the convergence of our simulations.
Figure 5.
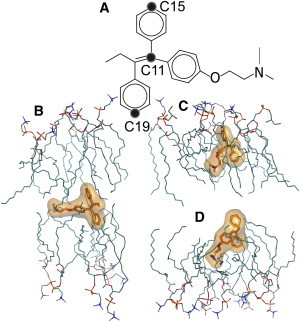
TAM orientation within the POPC/POPG bilayer. (A) Schematic of a TAM molecule and the atoms used for orientation analysis. Two vectors, C15-C19 and O-C11, are defined by the lines connecting the corresponding atoms. (B–D) Snapshots of TAM molecules exhibiting different orientations. To see this figure in color, go online.
A comparison between equilibrated bilayers, with and without TAM, reveals that the addition of 10 mol % TAM increases the hydrocarbon chain thickness 2DC by 0.8 Å (Supporting Material). This value is smaller than that determined from SANS data. However, the differences between our simulation and SANS results are not surprising considering the simple model we used to analyze the SANS data. Moreover, the 2DC values from our simulations match very well with the actual bilayer hydrocarbon chain thicknesses for POPC and POPG bilayers (22,49). Applying the principle that the total volume probability along the bilayer normal equals unity (50), we determined the hydrocarbon chain volume Vchain from our simulations. Using the box relation that lipid area equals Vchain/DC, this gives rise to a lipid area of 64.8 and 63.2 Å2 for bilayers without and with TAM, respectively. Overall, our simulation data indicate that addition of TAM increases the lipid bilayer hydrocarbon chain thickness (by 0.8 Å) and consequently decreases the lipid area (by 1.6 Å2). These results are in qualitative agreement with our SANS data.
The two vectors connecting the C15 and C19 atoms (C15-C19), and the oxygen and C11 atoms (O-C11) of TAM (Fig. 5 A) are used to describe TAM orientation in the lipid bilayer. As mentioned above, the time trajectories show fast fluctuation dynamics of the TAM molecules (Supporting Material). Snapshots of TAM molecules exhibiting different conformations are shown in Fig. 5, B–D. TAM can reside at the center of the bilayer or upright in the hydrocarbon chain region. The time-averaged distributions of the two vectors, with respect to the bilayer normal, are shown in Fig. S10. Both vectors exhibit a broad distribution of a nearly equal probability between ∼30° and 150°.
To visualize where TAM is located within the lipid bilayer, we calculated atom-number averaged number density distributions for the six lipid components (Fig. 6 A) and the tail nitrogen atom of TAM (N-TAM). The results are shown in Fig. 6 B. Overall, the N-TAM atom resides within the hydrocarbon chain region, with maximum probabilities located at the bilayer center and near the glycerol-carbonyl backbone. This observation is consistent with the polar nature (basicity) of the tertiary amine group. Similar to what has been observed for the hydroxyl group of cholesterol, one favorable location for the hydrophilic nitrogen atom is near the backbone region. The water-amine interactions are largely due to hydrogen bonding between the proton in the water molecule and the lone electron pair on the nitrogen atom. However, the hydrophilicity of the tertiary amine group is not strong enough to keep TAM in the backbone region, as revealed by time trajectories. The second most favorable region for the nitrogen atom to reside is at the bilayer center, where TAM preferentially assumes a nearly horizontal orientation to minimize the interruption of local lipid chain packing.
Figure 6.
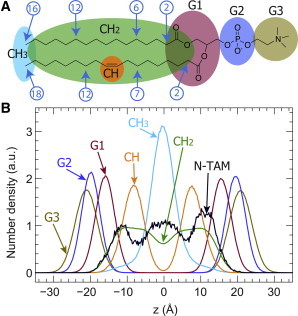
Number density distributions for lipid components and the tail nitrogen atom of TAM (N-TAM). (A) A POPC lipid molecule is parsed into six components: G1, G2, and G3 for the headgroup, and CH, CH2, and CH3 for the hydrocarbon chains. Selected carbon indices used for order parameter calculations (Fig. 7) are indicated. (B) Atom-number averaged number density distributions for the six lipid components depicted in A. The number density distribution of N-TAM was normalized to match the integral of lipid components. To see this figure in color, go online.
We also calculated 2D probabilities of the position and orientation of TAM within the lipid bilayer. The position and orientation of TAM are represented by the z axis position of the C11 atom and the tilt angle of the vector O-C11 (with respect to the bilayer normal), respectively. A clear correlation between the two parameters is observed (Supporting Material). Specifically, for the z position ranging from 5 to 10 Å, the most probable tilt angles are between 20° and 90°. As the C11 atom moves toward the bilayer center, the most probable angles are confined to a region near 90°. This observation echoes the notion that TAM exhibits different orientations depending on where it is located.
We also calculated the orientational order parameter −2SCD for each carbon atom in POPC chains (51). The results are shown in Fig. 7. The carbon indices for each chain are indicated in Fig. 6 A. For the palmitoyl chain, a slight increase is observed for carbon numbers 4–10. A similar increase is observed for the monounsaturated oleoyl chain, mainly for carbon numbers 4–9. For carbon atoms located near the bilayer center, no ordering is observed. The averaged order parameters for POPC chain carbons are 0.141 (without TAM) and 0.144 (with TAM)—an overall 2% increase. This is a rather small change compared with that induced by cholesterol, which orients upright and increases lipid chain order parameters by a significant amount (52). The small increase of the lipid chain order parameters by TAM is consistent with the small increase in 2DC.
Figure 7.
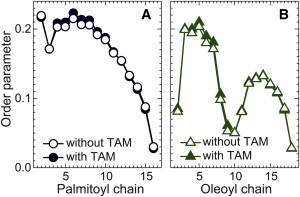
Order parameters −2SCD for POPC chains obtained from MD simulations of a POPC/POPG bilayer before (open symbols) and after (solid symbols) the addition of 10 mol % TAM. (A) Palmitoyl chain. (B) Oleoyl chain. To see this figure in color, go online.
Discussion
Cell membranes are important targets for many drugs (53). Elucidating the molecular interactions that occur between lipid model membranes and administered drugs will aid in the development of more effective drugs. Here, we studied the interactions of TAM with a model lipid membrane. Time-course fluorescence measurements revealed a two-phase leakage behavior. This can be explained by TAM first binding to the outer leaflet of ULVs. The subsequent asymmetric distribution of TAM between the inner and outer leaflets stresses the bilayer, causing calcein to leak out at a fast rate from the vesicle interior. After some time, TAM gradually transverses into the inner leaflet through a slow process, such as flip-flop (54). This accounts for the slow leakage phase. Engelk et al. (11) also observed TAM-induced dye leakage using a lipid mixture composed of egg-PC, cholesterol, and α-tocopherol, but did not identify any two-phase leakage behavior. Moreover, the authors observed that 5 mol % TAM induced 80% dye leakage after only 50 min of incubation. These differences may highlight the role of lipids in modulating TAM activity.
High-resolution AFM has emerged as an invaluable tool for exploring membrane nanomechanics. Our AFM-based force spectroscopy measurements indicate that the KA is doubled in the presence of 10 mol % TAM. It is known that membrane proteins are sensitive to the force from lipids (55). Modulating the mechanical properties of the membrane may be a means by which TAM controls protein functions. One problem in cancer treatment is drug efflux caused by multidrug resistant transporters. TAM has been found to effectively reverse multidrug resistance in several cancer cell lines (56). Altering the membrane's mechanical properties offers a new way to explore transporter inhibition by TAM.
We examined atomic-level interactions between TAM and lipid bilayers in MD simulations. In particular, the addition of TAM increased hydrocarbon chain thickness by 0.8 Å and decreased the average lipid area by 1.6 Å2. The small changes of these two parameters are consistent with lipid chain order parameter calculations. The structural parameters obtained from our simulations are in qualitative agreement with the bilayer thickening shown by SANS data and the reduced lipid bond vibrational dynamics revealed by FTIR measurements. It is worth noting that both our SANS measurements and MD simulations indicate that TAM increases lipid bilayer thickness. However, no discernible band shift was observed in the FTIR spectra. This suggests that band shift is not a good criterion for detecting small changes in bilayer thickness.
Our orientational order parameter calculations indicate that lipid chain carbons located near the backbone are more susceptible to ordering by TAM. The reduced influence on lipid chain order toward the bilayer center has also been observed for a voltage sensor toxin (57). This observation can be explained by considering the orientation of TAM’s triple-ring structure. Our 2D probability calculations indicate that the ring structure has a tilt angle range of 20–90° when it is near the backbone and 80–120° when it is near the bilayer center (Supporting Material). Since the largest ordering effect occurs when the ring structure is parallel to the chain direction, the upper chain carbons are more susceptible to ordering by the ring structure of TAM than those near the bilayer center. In addition, the central carbons are likely to be disordered by the bulky ring structure, which adopts a nearly parallel orientation when residing at the bilayer center.
Our simulation data show that TAM does not have a preference for associating with either POPC or POPG (Supporting Material). This is understandable because TAM molecules are located in the hydrocarbon chain region, and POPC and POPG have the same chain composition. More importantly, we find that TAM can reside either at the bilayer center or upright in the hydrocarbon chain region. The main TAM axis displays a broad angular distribution. Similarly, the nitrogen atom is broadly distributed within the hydrocarbon chain region, with its maximum probability located at the bilayer center or near the backbone. TAM’s fast conformational dynamics within the lipid bilayer may be beneficial when rapid recognition of TAM by the binding pockets of estrogen receptors is required.
It is well known that the permeability of small solutes is inversely related to the hydrocarbon chain thickness and proportionally related to the free fraction of the lipid area (58). Indeed, an increasing cholesterol content leads to a thicker bilayer with a lower water permeability (59). This is in drastic contrast to TAM, which thickens the lipid bilayer but enhances the calcein permeability. Therefore, factors other than the lipid bilayer thickness should be responsible for increasing the solute permeability in the presence of TAM. It is conceivable that the highly dynamic conformation of TAM could induce transient defects to allow solutes to translocate across the bilayer.
Conclusions
We studied the effects of the anticancer drug TAM on a model membrane’s permeability, bond vibrational dynamics, bilayer thickness, and bilayer nanomechanics, as well as its interactions with surrounding lipids, using a number of experimental and computational techniques, including fluorescence dye leakage, FTIR spectroscopy, SANS measurements, AFM-based force spectroscopy, and all-atom MD simulations. We found that TAM impairs lipid bilayer integrity, causing calcein molecules to leak from the interior of lipid vesicles. FTIR spectroscopy shows that TAM suppresses bond vibrational dynamics located in the hydrocarbon chain region. TAM also increases bilayer hydrocarbon chain thickness, as revealed by our SANS data and MD simulations. AFM-based force spectroscopy measurements show that TAM increases the lipid bilayer area compressibility modulus by a large amount. Detailed analyses of our MD simulation data show that: 1) the orientation of TAM oscillates between 30° and 150° with respect to the bilayer normal; 2) TAM can reside at any depth within the hydrocarbon chain region; 3) TAM’s orientation varies as a function of its depth within the bilayer; and 4) TAM’s hydrophilic tertiary amine group preferentially locates either at the bilayer center or at the headgroup-chain interface. The various experimental and computational results obtained from our studies should prove useful for illustrating TAM’s effects on lipid membranes.
Author Contributions
J.P. and X.C. designed the project. N.K.K., C.S.H., and J.P. performed the experiments and analyzed the experimental data. X.C. performed the MD simulations and analyzed the simulation data. X.C., J.K., and J.P. wrote the manuscript.
Acknowledgments
We thank Dr. Frederick A. Heberle for help with sample preparation and data collection for the SANS experiments. We also thank Dr. Martin Muschol and Ms Tatiana Miti for help with osmotic pressure measurements.
J.P. is supported through a startup package from the University of South Florida. X.C. is partially supported by the Laboratory Directed R&D (LDRD) program at Oak Ridge National Laboratory. J.K. is supported through the Department of Energy (DOE) Scientific User Facilities Division, Office of Basic Energy Sciences. This work received additional support from the DOE Office of Biological and Environmental Research for the BioSANS instrument at the ORNL Center for Structural Molecular Biology. This research used resources of the National Energy Research Scientific Computing Center, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under contract No. DE-AC02-05CH11231.
Supporting Material
References
- 1.Clarke M., Collins R., Grp E.B.C.T.C. Tamoxifen for early breast cancer: an overview of the randomised trials. Early Breast Cancer Trialists’ Collaborative Group. Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 2.Wiseman H. Tamoxifen: new membrane-mediated mechanisms of action and therapeutic advances. Trends Pharmacol. Sci. 1994;15:83–89. doi: 10.1016/0165-6147(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 3.Shiau A.K., Barstad D., Greene G.L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 4.Yang L.H., Tseng H.S., Chen D.R. Survival benefit of tamoxifen in estrogen receptor-negative and progesterone receptor-positive low grade breast cancer patients. J. Breast Cancer. 2012;15:288–295. doi: 10.4048/jbc.2012.15.3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Brian C.A., Liskamp R.M., Weinstein I.B. Inhibition of protein kinase C by tamoxifen. Cancer Res. 1985;45:2462–2465. [PubMed] [Google Scholar]
- 6.Ribeiro M.P.C., Santos A.E., Custódio J.B.A. Mitochondria: the gateway for tamoxifen-induced liver injury. Toxicology. 2014;323:10–18. doi: 10.1016/j.tox.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Custódio J.B.A., Almeida L.M., Madeira V.M.C. A reliable and rapid procedure to estimate drug partitioning in biomembranes. Biochem. Biophys. Res. Commun. 1991;176:1079–1085. doi: 10.1016/0006-291x(91)90394-m. [DOI] [PubMed] [Google Scholar]
- 8.Baum M. Has tamoxifen had its day? Breast Cancer Res. 2002;4:213–217. doi: 10.1186/bcr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Custódio J.B.A., Almeida L.M., Madeira V.M.C. The anticancer drug tamoxifen induces changes in the physical properties of model and native membranes. Biochim. Biophys. Acta. 1993;1150:123–129. doi: 10.1016/0005-2736(93)90080-j. [DOI] [PubMed] [Google Scholar]
- 10.Dicko A., Morissette M., Di Paolo T. Effect of estradiol and tamoxifen on brain membranes: investigation by infrared and fluorescence spectroscopy. Brain Res. Bull. 1999;49:401–405. doi: 10.1016/s0361-9230(99)00066-0. [DOI] [PubMed] [Google Scholar]
- 11.Engelk M., Bojarski P., Diehl H. Tamoxifen perturbs lipid bilayer order and permeability: comparison of DSC, fluorescence anisotropy, laurdan generalized polarization and carboxyfluorescein leakage studies. Biophys. Chem. 2001;90:157–173. doi: 10.1016/s0301-4622(01)00139-9. [DOI] [PubMed] [Google Scholar]
- 12.Kazanci N., Severcan F. Concentration dependent different action of tamoxifen on membrane fluidity. Biosci. Rep. 2007;27:247–255. doi: 10.1007/s10540-007-9050-3. [DOI] [PubMed] [Google Scholar]
- 13.Bilge D., Sahin I., Severcan F. Interactions of tamoxifen with distearoyl phosphatidylcholine multilamellar vesicles: FTIR and DSC studies. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014;130:250–256. doi: 10.1016/j.saa.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 14.Bilge D., Kazanci N., Severcan F. Acyl chain length and charge effect on tamoxifen-lipid model membrane interactions. J. Mol. Struct. 2013;1040:75–82. [Google Scholar]
- 15.Boyar H., Severcan F. Tamoxifen-model membrane interactions: an FT-IR study. J. Mol. Struct. 1997;408:265–268. doi: 10.1042/bst025493s. [DOI] [PubMed] [Google Scholar]
- 16.Severcan F., Kazanci N., Zorlu F. Tamoxifen increases membrane fluidity at high concentrations. Biosci. Rep. 2000;20:177–184. doi: 10.1023/a:1005515502954. [DOI] [PubMed] [Google Scholar]
- 17.Wiseman H., Quinn P., Halliwell B. Tamoxifen and related compounds decrease membrane fluidity in liposomes. Mechanism for the antioxidant action of tamoxifen and relevance to its anticancer and cardioprotective actions? FEBS Lett. 1993;330:53–56. doi: 10.1016/0014-5793(93)80918-k. [DOI] [PubMed] [Google Scholar]
- 18.Luxo C., Jurado A.S., Madeira V.M. Use of Bacillus stearothermophilus as a model to study tamoxifen-membrane interactions. Toxicol. In Vitro. 1996;10:463–471. doi: 10.1016/0887-2333(96)00019-7. [DOI] [PubMed] [Google Scholar]
- 19.Suwalsky M., Hernández P., Sotomayor C.P. Interaction of the anticancer drug tamoxifen with the human erythrocyte membrane and molecular models. Z. Naturforsch. C. 1998;53:182–190. doi: 10.1515/znc-1998-3-407. [DOI] [PubMed] [Google Scholar]
- 20.Ayuyan A.G., Cohen F.S. Lipid peroxides promote large rafts: effects of excitation of probes in fluorescence microscopy and electrochemical reactions during vesicle formation. Biophys. J. 2006;91:2172–2183. doi: 10.1529/biophysj.106.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mannock D.A., Lewis R.N.A.H., McElhaney R.N. A calorimetric and spectroscopic comparison of the effects of ergosterol and cholesterol on the thermotropic phase behavior and organization of dipalmitoylphosphatidylcholine bilayer membranes. Biochim. Biophys. Acta. 2010;1798:376–388. doi: 10.1016/j.bbamem.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Kučerka N., Nagle J.F., Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 24.Jo S., Lim J.B., Im W. CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martínez L., Andrade R., Martínez J.M. PACKMOL: a package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009;30:2157–2164. doi: 10.1002/jcc.21224. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feller S.E., Yin D., MacKerell A.D., Jr. Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys. J. 1997;73:2269–2279. doi: 10.1016/S0006-3495(97)78259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanommeslaeghe K., Hatcher E., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martyna G.J., Tobias D.J., Klein M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 31.Feller S.E., Zhang Y.H., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 32.Darden T., York D., Pedersen L. Particle mesh Ewald: an N.Log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 33.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 34.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular-dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 35.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 36.Seelig A., Seelig J. The dynamic structure of fatty acyl chains in a phospholipid bilayer measured by deuterium magnetic resonance. Biochemistry. 1974;13:4839–4845. doi: 10.1021/bi00720a024. [DOI] [PubMed] [Google Scholar]
- 37.Sroda K., Michalak K., Hendrich A.B. Genistein derivatives decrease liposome membrane integrity—calcein release and molecular modeling study. Biophys. Chem. 2008;138:78–82. doi: 10.1016/j.bpc.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Sheynis T., Friediger A., Jelinek R. Aggregation modulators interfere with membrane interactions of β2-microglobulin fibrils. Biophys. J. 2013;105:745–755. doi: 10.1016/j.bpj.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heerklotz H., Seelig J. Leakage and lysis of lipid membranes induced by the lipopeptide surfactin. Eur. Biophys. J. 2007;36:305–314. doi: 10.1007/s00249-006-0091-5. [DOI] [PubMed] [Google Scholar]
- 40.Ladokhin A.S., Wimley W.C., White S.H. Leakage of membrane vesicle contents: determination of mechanism using fluorescence requenching. Biophys. J. 1995;69:1964–1971. doi: 10.1016/S0006-3495(95)80066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pencer J., Krueger S., Katsaras J. Method of separated form factors for polydisperse vesicles. J. Appl. Cryst. 2006;39:293–303. [Google Scholar]
- 42.Kučerka N., Nieh M.P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 43.Kučerka N., Holland B.W., Katsaras J. Scattering density profile model of POPG bilayers as determined by molecular dynamics simulations and small-angle neutron and X-ray scattering experiments. J. Phys. Chem. B. 2012;116:232–239. doi: 10.1021/jp208920h. [DOI] [PubMed] [Google Scholar]
- 44.Hung W.C., Lee M.T., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan J., Mills T.T., Nagle J.F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. Lett. 2008;100:198103. doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das C., Sheikh K.H., Connell S.D. Nanoscale mechanical probing of supported lipid bilayers with atomic force microscopy. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:041920. doi: 10.1103/PhysRevE.82.041920. [DOI] [PubMed] [Google Scholar]
- 47.Rawicz W., Olbrich K.C., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Evans E., Rawicz W., Smith B.A. Back to the future: mechanics and thermodynamics of lipid biomembranes. Faraday Discuss. 2013;161:591–611. doi: 10.1039/c2fd20127e. [DOI] [PubMed] [Google Scholar]
- 49.Pan J., Marquardt D., Katsaras J. Revisiting the bilayer structures of fluid phase phosphatidylglycerol lipids: Accounting for exchangeable hydrogens. Biochim. Biophys. Acta. 2014;1838:2966–2969. doi: 10.1016/j.bbamem.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Petrache H.I., Feller S.E., Nagle J.F. Determination of component volumes of lipid bilayers from simulations. Biophys. J. 1997;72:2237–2242. doi: 10.1016/S0006-3495(97)78867-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sodt A.J., Sandar M.L., Lyman E. The molecular structure of the liquid-ordered phase of lipid bilayers. J. Am. Chem. Soc. 2014;136:725–732. doi: 10.1021/ja4105667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendrich A.B., Michalak K. Lipids as a target for drugs modulating multidrug resistance of cancer cells. Curr. Drug Targets. 2003;4:23–30. doi: 10.2174/1389450033347172. [DOI] [PubMed] [Google Scholar]
- 54.Kornberg R.D., McConnell H.M. Inside-outside transitions of phospholipids in vesicle membranes. Biochemistry. 1971;10:1111–1120. doi: 10.1021/bi00783a003. [DOI] [PubMed] [Google Scholar]
- 55.McIntosh T.J., Simon S.A. Roles of bilayer material properties in function and distribution of membrane proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 56.Ramu A., Glaubiger D., Fuks Z. Reversal of acquired resistance to doxorubicin in P388 murine leukemia cells by tamoxifen and other triparanol analogues. Cancer Res. 1984;44:4392–4395. [PubMed] [Google Scholar]
- 57.Mihailescu M., Krepkiy D., White S. Structural interactions of a voltage sensor toxin with lipid membranes. Proc. Natl. Acad. Sci. USA. 2014;111:E5463–E5470. doi: 10.1073/pnas.1415324111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagle J.F., Mathai J.C., Tristram-Nagle S. Theory of passive permeability through lipid bilayers. J. Gen. Physiol. 2008;131:77–85. doi: 10.1085/jgp.200709849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawicz W., Smith B.A., Evans E. Elasticity, strength, and water permeability of bilayers that contain raft microdomain-forming lipids. Biophys. J. 2008;94:4725–4736. doi: 10.1529/biophysj.107.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


