Abstract
Objective Social media is becoming increasingly popular as a platform for sharing personal health-related information. This information can be utilized for public health monitoring tasks, particularly for pharmacovigilance, via the use of natural language processing (NLP) techniques. However, the language in social media is highly informal, and user-expressed medical concepts are often nontechnical, descriptive, and challenging to extract. There has been limited progress in addressing these challenges, and thus far, advanced machine learning-based NLP techniques have been underutilized. Our objective is to design a machine learning-based approach to extract mentions of adverse drug reactions (ADRs) from highly informal text in social media.
Methods We introduce ADRMine, a machine learning-based concept extraction system that uses conditional random fields (CRFs). ADRMine utilizes a variety of features, including a novel feature for modeling words’ semantic similarities. The similarities are modeled by clustering words based on unsupervised, pretrained word representation vectors (embeddings) generated from unlabeled user posts in social media using a deep learning technique.
Results ADRMine outperforms several strong baseline systems in the ADR extraction task by achieving an F-measure of 0.82. Feature analysis demonstrates that the proposed word cluster features significantly improve extraction performance.
Conclusion It is possible to extract complex medical concepts, with relatively high performance, from informal, user-generated content. Our approach is particularly scalable, suitable for social media mining, as it relies on large volumes of unlabeled data, thus diminishing the need for large, annotated training data sets.
Keywords: adverse drug reaction, ADR, social media mining, pharmacovigilance, natural language processing, machine learning, deep learning word embeddings
INTRODUCTION
Adverse drug reactions (ADRs) are a major public health concern and are among the top causes of morbidity and mortality.1 Clinical drug trials have limited ability to detect all ADRs due to factors such as small sample sizes, relatively short duration, and the lack of diversity among study participants.2 Postmarket drug safety surveillance is therefore required to identify potential adverse reactions in the larger population to minimize unnecessary, and sometimes fatal, harm to patients. Spontaneous reporting systems (SRSs) are surveillance mechanisms supported by regulatory agencies such as the Food and Drug Administration in the United States, which enable healthcare providers and patients to directly submit reports of suspected ADRs. When compared to reports from other healthcare providers, patients’ reports have been found to contain different drug-ADR pairs, contain more detailed and temporal information, increase statistical signals used to detect ADRs, and increase the discovery of previously unknown ADRs.3–6 However, under-reporting limits the effectiveness of SRSs. It is estimated that more than 90% of ADRs are under-reported.7 To augment the current systems, there are new ways to conduct pharmacovigilance using expanded data sources—including data available on social media sites, such as Twitter,8,9 or health-related social networks, such as DailyStrength (DS).10 While a few individuals’ experiences may not be clinically useful, thousands of drug-related posts can potentially reveal serious and unknown ADRs. Figure 1 shows examples of ADR-relevant user postings from Twitter (a) and DS (b), with labeled mentions.
Figure 1.

Examples of user-posted drug reviews in Twitter (a) and DailyStrength (b).
Our prior research analyzed user postings in DS, and was the first study which demonstrated that natural language processing (NLP) techniques can be used for the extraction of valuable drug-safety information from social media.11 Other publications further explored the topic,12–14 relying primarily on string comparison techniques over existing or custom built ADR lexicons. However, there are important challenges that make a pure lexicon-based approach to the problem suboptimal, namely:
Consumers do not normally use technical terms found in medical lexicons. Instead, they use creative phrases, descriptive symptom explanations, and idiomatic expressions. For example the phrase “messed up my sleeping patterns” was used to report “sleep disturbances.”
Even when correctly identified, matched terms are not necessarily adverse effects. The terms used to describe ADRs can also be used for indications (reason to use the drug; e.g., “infection” in Figure 1b), beneficial effects, or other mention types.
User postings are informal, and deviate from grammatical rules. They include misspellings, abbreviations, and phrase construction irregularities that make extraction more difficult compared to other corpora (such as news or biomedical literature).
In this work, we introduce ADRMine, a machine learning sequence tagger for concept extraction from social media. We explore the effectiveness of various contextual, lexicon-based, grammatical, and semantic features. The semantic features are based on word clusters generated from pretrained word representation vectors (also referred to as word embeddings15), which are learned from more than one million unlabeled user posts, using a deep learning technique. Deep learning, a new class of machine learning methods based on nonlinear information processing, typically uses neural networks (NNs) for automatic feature extraction, pattern analysis, and classification.16 Deep learning methods have shown promising results in NLP tasks, including sentence chunking and named entity recognition, in well-formatted domains such as news or Wikipedia content.15,17 However, to the best of our knowledge, these methods have not previously been explored for medical concept extraction from social media data.
In this study, we hypothesized that ADRMine would address many of the abovementioned challenges associated with social media data, and would accurately identify most of the ADR mentions, including the consumer expressions that are not observed in the training data or in the standard ADR lexicons. Furthermore, we hypothesized that incorporating the “embedding cluster features” would diminish the need for large amounts of labeled data, which are generally required to train supervised machine learning classifiers.
RELATED WORK
Various resources have been studied for extraction of postmarketing drug safety information, including electronic health records,18,19 biomedical literature,20–22 and SRSs.23,24 However, the surfeit of user-posted online health information has recently encouraged researchers to explore other resources for drug-safety information extraction including various health social networking sites, such as DS,11,25,26 PatientsLikeMe,27,28 and MedHelp;29 generic social networks such as Twitter;8,9,30 and users’ web search logs.31
Given that there are standard and extensive ADR lexicons, such as Side Effect Resource (SIDER, containing known ADRs),32,33 Consumer Health Vocabulary (CHV, containing consumer alternatives for medical concepts),34 Medical Dictionary for Regulatory Activities (MedDRA),35 and Coding Symbols for a Thesaurus of Adverse Reaction Terms (COSTART), most prior studies11,12,14,36–38 focused on exploring existing or customized/expanded lexicons to find ADR mentions in user posts. To address some of the limitations of the lexicon-based extraction methods in our own previous work,11 we applied association rule mining, a data mining technique, to learn the language patterns that were used by patients or their caregivers to report ADRs.25 The technique generated extraction patterns based on the immediate local context of the ADR mentions in the training sentences. The patterns could successfully extract a subset of the ADRs. However, performance of such an approach is highly dependent on training set size, making it difficult to locate concepts expressed in less frequent and more complex sentences.
Overall, there has been limited progress on automated medical concept extraction approaches from social media, and advanced machine learning-based NLP techniques have been underutilized for the task. Specifically, there has been less effort in addressing the introduced challenges, such as finding “creative” consumer expressions, handling misspellings, distinguishing ADRs from other semantic types (e.g., indications), and mapping such creative expressions to the standard medical terminologies.
METHODS
Data collection and annotation
We collected user posts about drugs from two different social media resources: DS and Twitter. In this study, 81 drugs were used (the drug list is available for download at: http://diego.asu.edu/downloads/publications/ADRMine/drug_names.txt). A pharmacology expert selected the drugs mainly based on widespread use in the US market. The set also includes relatively newer drugs that were released between 2007 and 2010; this provides a time cushion for market growth and helps to ensure that we can find patient discussions on social media. For more information about the data and the collection process please refer to prior publications using Twitter data or DS.8,9
A team of two expert annotators independently annotated the user posts under the supervision of the expert pharmacologist. The annotations include mentions of medical signs and symptoms with the following semantic types:
adverse drug reaction – a drug reaction that the user considered negative;
beneficial effect – an unexpected positive reaction to the drug;
indication – the condition for which the patient is taking the drug; and
other – any other mention of signs or symptoms.
Every annotation includes the span of the mention (start/end position offsets), the semantic type, the related drug name, and the corresponding UMLS (Unified Medical Language System) concept ID—assigned by manually selecting concepts in the ADR lexicon (see “ADR lexicon” Section). To measure the inter-annotator agreement, we used Cohen’s kappa approach.39 The calculated kappa value for approximate matching of the concepts is 0.85 for DS and 0.81 for Twitter, which can be considered high agreement.40 Finally, the gold standard was generated by including only the reviews with complete inter-annotator agreement. From the DS corpus, 4720 reviews are randomly selected for training (DS train set) and 1559 for testing (DS test set). The Twitter corpus contains 1340 tweets for training (Twitter train set) and 444 test tweets (Twitter test set). The Twitter annotated corpus is available for download from http://diego.asu.edu/downloads/publications/ADRMine/download_tweets.zip.
For unsupervised learning, we collected an additional 313 833 DS user reviews, associated with the most-reviewed drugs in DS, and 397 729 drug related tweets, for a total of 711 562 postings. This unlabeled set (Unlabeled_DS_Twitter set), excluding the sentences in DS test and Twitter test sets, consists of more than one million sentences.
ADR lexicon
We compiled an exhaustive list of ADR concepts and their corresponding UMLS IDs. The lexicon, expanded from our earlier work,11 currently includes concepts from COSTART, SIDER, and a subset of CHV. In order to compile a list of only ADRs, we filtered the CHV phrases by excluding the concepts with UMLS IDs that were not listed in SIDER. For example, we did not add “West Nile virus” since the related UMLS ID (C0043125) was not listed in SIDER. The final lexicon contains over 13 591 phrases, with 7432 unique UMLS IDs. In addition, we compiled a list of 136 ADRs frequently tagged by the annotators in the training data. This additional list was not used during annotation and only is used in our automatic extraction techniques. The ADR lexicon has been made publicly available at http://diego.asu.edu/downloads/publications/ADRMine/ADR_lexicon.tsv.
Concept extraction approach: sequence labeling with CRF
A supervised sequence labeling CRF classifier is used in ADRMine to extract the ADR concepts from user sentences. CRF is a well-established, high performing classifier for sequence labeling tasks.15,41,42 We used CRFsuite, the implementation provided by Okazaki,43 as it is fast and provides a simple interface for training/modifying the input features.15,43 Generating the input CRFsuite train and test files with calculated features for 88 565 tokens in DS train/test sets takes about 40 min, while building the CRF model and assigning labels for test sentences takes about 2 min on a PC with a dual core CPU and 10 GB of RAM running the Ubuntu operating system.
The CRF classifier is trained on annotated mentions of ADRs and indications, and it attempts to classify individual tokens in sentences. Although the focus is to identify the ADR mentions, our preliminary empirical results show that including indication labels in the model improves the performance of ADR extraction. We also consider the mentions of beneficial effects as indications, since there are very limited number of annotated beneficial effects and they are similar to indications. For encoding the concepts’ boundaries, ADRMine uses the inside, outside, beginning (IOB) scheme—where every token can be the beginning, inside, or outside of a semantic type. Therefore, it learns to distinguish five different labels: B-ADR, I-ADR, B-Indication, I-Indication, and Out.
CRF features
To represent the classification candidates (i.e., individual tokens), we explored the effectiveness of the following feature sets:
Context features: Context is defined with seven features including the current token (ti), the three preceding (ti−3, ti−2, ti−1), and three following tokens (ti+1, ti+2, ti+3) in the sentence. The preprocessed token strings are values of these features. Preprocessing includes spelling correction and lemmatization. For spelling correction, we utilize the Apache Lucene44 spell checker library, which suggests the correct spelling based on an index of English words. The index is generated using the ADR lexicon, described in the previous section, and a list of common English words from Spell Checker Oriented Word Lists.45 For lemmatization, we used the Dragon toolkit46 lemmatizer, which returns the WordNet47 root of the input word.
ADR Lexicon: A binary feature that shows whether or not the current token exists in the ADR lexicon.
POS: Part of speech of the token, which was generated using Stanford parser.48
Negation: This feature indicates whether or not the token is negated. The negations are identified based on syntactic dependency rules between lexical cues (e.g., no, not, cannot) and the token.49,50
Learning word embeddings
One potential problem with the abovementioned features is that the classifier may struggle with unseen or rarely occurring tokens. To address this issue, we incorporated a set of semantic similarity-based features. We model the similarity between words by utilizing more than one million unlabeled user sentences (Unlabeled_DS_Twitter set) about drugs to generate the word embeddings. The embeddings are meaningful real-valued vectors of configurable dimension (usually, 150–500 dimensions). We generate 150-dimensional vectors using the word2vec tool,51 which learns the vectors from an input text corpus. We split the sentences in each user post, lemmatize all the tokens, and lowercase them for generalization. Word2vec first constructs a vocabulary from the input corpus, and then learns word representations by training a NN language model. The NN learns a word’s embedding based on the word’s contexts in different sentences. As a result, the words that occur in similar contexts are mapped into close vectors. More information about generating the embeddings can be found in the related papers.15,52,53
Embedding cluster features
We compute word clusters with the word2vec tool, which performs K-means clustering on the word embeddings. The words in the corpus are grouped into n (=150) different clusters, where n is a configurable integer number. Examples of generated clusters with a subset of words in each cluster are shown in Table 1. Seven features are then defined based on the generated clusters. The features include the cluster number for the current token, three preceding and three following tokens. These features add a higher level abstraction to the feature space by assigning the same cluster number to similar tokens. For instance, as Table 1 illustrates, the drug names “Abilify” and “Adderall” are assigned to the same cluster, which includes only drug names. The value of n and the embedding vectors’ dimension were selected based on preliminary experiments targeted at optimizing CRF performance for values of n between 50 and 500. Generating the embedding clusters from raw input texts is very fast and takes around 30 s. The generated word embeddings and clusters are made available for download http://diego.asu.edu/Publications/ADRMine.html.
Table 1.
Examples of the unsupervised learned clusters with the subsets of the words in each cluster; ci is an integer between 0 and 149
| Cluster# | Semantic category | Examples of clustered words |
|---|---|---|
| c1 | Drug | Abilify, Adderall, Ambien, Ativan, aspirin, citalopram, Effexor, Paxil, … |
| c2 | Signs/Symptoms | hangover, headache, rash, hive, … |
| c3 | Signs/Symptoms | anxiety, depression, disorder, ocd, mania, stabilizer, … |
| c4 | Drug dosage | 1000 mg, 100 mg, .10, 10 mg, 600 mg, 0.25, .05, … |
| c5 | Treatment | anti-depressant, antidepressant, drug, med, medication, medicine, treat, … |
| c6 | Family member | brother, dad, daughter, father, husband, mom, mother, son, wife, … |
| c7 | Date | 1992, 2011, 2012, 23rd, 8th, April, Aug, August, December, … |
The “Semantic category” titles are manually assigned and are not used in the system.
Figure 2 shows the calculated features for three CRF classification instances.
Figure 2.

Calculated features representing three CRF classification instances.
Baseline extraction techniques
We aimed to analyze the performance of ADRMine relative to four baseline techniques: two simple baselines based on MetaMap,54 a lexicon-based approach for extraction of candidate ADR phrases based on our ADR lexicon, and an SVM classifier that identifies the semantic type of the extracted phrases.
Lexicon-based candidate phrase extraction
To locate the ADR lexicon concepts in user sentences, we use an information retrieval approach based on Lucene, which is similar to those applied for ontology mapping55,56 and entity normalization.57 A Lucene index is built from the ADR lexicon entries. For each concept in the lexicon, the content and the associated UMLS IDs are added to the index. Before indexing, the concepts are preprocessed by removing the stop words and lemmatization.
To find the concepts presented in a given sentence, we generate a Lucene search query after preprocessing and tokenizing the sentence. The retrieval engine returns a ranked list of all the lexicon concepts that contain a subset of the tokens presented in the input query. We consider a retrieved concept present in the sentence if all of the concept’s tokens are present in the sentence. The span of the concepts in the sentence are then identified by using string comparison via regular expressions. This technique is flexible enough to identify both single and multi-token concepts, regardless of the order or the presence of other tokens in between them. For example, the sentence “… I gained an excessive amount of weight during six months.” is correctly matched with the lexicon concept “weight gain.” We apply two constraints before accepting the presence of a retrieved lexicon concept: the distance between the first and the last included token should be equal or less than a configurable size (=5), and there should not be any punctuation or connectors like “but” or “and” in between the tokens.
SVM semantic type classifier
Since not all mentions that match with the lexicon are adverse reactions, we train a multiclass SVM classifier to identify the semantic types of the candidate phrases. Every SVM classification candidate is a phrase (may include more than one token) that is already matched with the ADR lexicon. The possible semantic types for a candidate phrase are ADR, Indication or Other. SVM was chosen because it has been shown to perform very well in text classification problems.58 We used SVMlight59 to build the SVM model. The SVM features for representing the candidate phrases are similar to CRF features and include: the phrase tokens, three preceding and three following tokens around the phrase (neighbor tokens), the negation feature, and the embedding cluster number for the phrase tokens and the neighbor tokens.
MetaMap baselines
We use MetaMap to identify the UMLS concept IDs and semantic types in the user reviews, and add two baselines to evaluate the performance of MetaMap on this type of data. In the first baseline (MetaMapADR_LEXICON), all identified mentions by MetaMap that their assigned UMLS IDs are in our lexicon are considered to be ADRs. In the second baseline (MetaMapSEMANTIC_TYPE), all concepts belonging to specific UMLS semantic types are considered to be ADRs. The selected semantic types include: injury or poisoning, pathologic function, cell or molecular dysfunction, disease or syndrome, experimental model of disease, finding, mental or behavioral dysfunction, neoplastic process, signs or symptoms, mental process.
RESULTS
We evaluate the performance of the extraction techniques using precision (p), recall (r), and F-measure (f):
True positives (tp), false positives (fp), and false negatives (fn) are calculated by comparing the systems’ extracted concepts with the annotated ADRs in the gold standard via approximate matching.60 The effectiveness of the proposed techniques is evaluated using DS and Twitter corpora independently. Table 2 shows the details about the sentences and the number of annotated concepts in each corpus.
Table 2.
Number of user reviews and annotation details in train/test sets for DailyStrength (DS) and Twitter corpora
| Dataset | No. of user posts | No. of sentences | No. of tokens | No. of ADR mentions | No. of indication mentions |
|---|---|---|---|---|---|
| DS train set | 4720 | 6676 | 66 728 | 2193 | 1532 |
| DS test set | 1559 | 2166 | 22 147 | 750 | 454 |
| Twitter train set | 1340 | 2434 | 28 706 | 845 | 117 |
| Twitter test set | 444 | 813 | 9526 | 277 | 41 |
The performance of ADRMine is compared with the baseline techniques in Table 3. We found that ADRMine significantly outperforms all baseline approaches (p < 0.05). Furthermore the utility of different techniques in concept extraction is consistent between the two tested corpora. We compute the statistical significance (p-value) by using the model proposed by Yeh61 and implemented by Pado.62
Table 3.
Comparison of ADR classification precision (P), recall (R), and F-measure (F) of ADRMine with embedding cluster features (ADRMineWITH_CLUSTER) and the baselines systems on two different corpora: DS and Twitter
| Method | DS | |||||
|---|---|---|---|---|---|---|
| P | R | F | P | R | F | |
| MetaMapADR_LEXICON | 0.470 | 0.392 | 0.428 | 0.394 | 0.309 | 0.347 |
| MetaMapSEMANTIC_TYPE | 0.289 | 0.484 | 0.362 | 0.230 | 0.403 | 0.293 |
| Lexicon-based | 0.577 | 0.724 | 0.642 | 0.561 | 0.610 | 0.585 |
| SVM | 0.869 | 0.671 | 0.760 | 0.778 | 0.495 | 0.605 |
| ADRMineWITHOUT_CLUSTER | 0.874 | 0.723 | 0.791 | 0.788 | 0.549 | 0.647 |
| ADRMineWITH_CLUSTER | 0.860 | 0.784 | 0.821 | 0.765 | 0.682 | 0.721 |
The highest values in each column are highlighted in bold.
To investigate the contribution of each feature set in the CRF model, we performed leave-one-out feature experiments (Table 4). We found that the most contributing groups of features are the context and the embedding clusters. The combination of both is sufficient to achieve the highest result for DS.
Table 4.
The effectiveness of different CRF feature groups; all feature set (All) includes: context, lexicon, POS, negation, and embedding clusters (cluster)
| CRF Features | DS | |||||
|---|---|---|---|---|---|---|
| P | R | F | P | R | F | |
| All | 0.856 | 0.776 | 0.814 | 0.765 | 0.682 | 0.721 |
| All – lexicon | 0.852 | 0.781 | 0.815 | 0.765 | 0.646 | 0.701 |
| All – POS | 0.853 | 0.776 | 0.812 | 0.754 | 0.653 | 0.700 |
| All – negation | 0.854 | 0.769 | 0.810 | 0.752 | 0.646 | 0.695* |
| All – context | 0.811 | 0.665 | 0.731* | 0.624 | 0.498 | 0.554* |
| All – cluster | 0.851 | 0.745 | 0.794* | 0.788 | 0.549 | 0.647* |
| Context + cluster | 0.860 | 0.784 | 0.821* | 0.746 | 0.628 | 0.682* |
Statistically significant changes (p < 0.05), when compared with All feature set, are marked with asterisks.
The highest values in each column are highlighted in bold.
To further investigate the power of the embedding clusters, we performed several experiments to compare them with context features. These experiments were only performed on DS as we had a relatively larger set of training data available. The size of the training data was varied while the test set remained unchanged. Starting with 20% (944 reviews) of the original DS training set, we increased its size by 20% each time via random sampling without replacement. Figure 3 shows that adding the cluster features (context + clusters) constantly improves F-measure (Figure 3b), gives significant rise to the recall (Figure 3a), but slightly decreases the precision.
Figure 3.
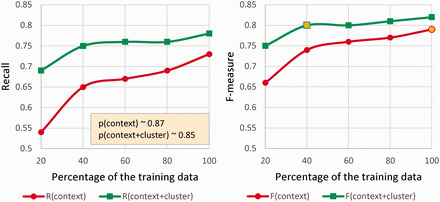
The impact of embedding clusters on precision, recall (a), and F-measure (b), when training the CRF on variable training set sizes and testing on the same test set.
DISCUSSION
Twitter vs. DailyStrength corpus
As shown in Table 3, the extraction performance for DS is much higher than Twitter. This is partially related to the fact that there was less annotated data available for Twitter. In general, however, compared to DS extracting ADR information from Twitter poses a more challenging problem. Whereas DS is a health-focused site that fosters discussion from patients about their personal experiences with a drug, Twitter is a general networking site where users may be inclined to mention a particular drug and its side effects for any number of reasons. Some may include personal experiences, but others may tweet about side effects they heard about, be sharing of a news report, or a sarcastic remark. These nuances may be difficult for even annotators to detect as the limited length of the tweets can make it more challenging for the annotator to ascertain the context of the mention. For instance in this tweet: “Hey not sleeping. #hotflashes #menopause #effexor,” it is difficult to determine whether the patient is taking the drug for their problem or if they are reporting ADRs.
Comparison of the concept extraction methods
Evaluation of the baseline lexicon-based technique (Table 3) demonstrated that it can extract ADR mentions with relatively high recall but very low precision. The recall is anticipated to even further improve in future by augmenting the ADR lexicon with a larger subset of MedDRA entries and a more comprehensive list of common consumer expressions for ADRs. This high recall indicates that the utilized lexicon-based techniques were effective in handling term variability in the user sentences. However, the low precision was mainly due to the matched mentions with semantic types other than ADRs. When we used SVM to distinguish the semantic types, the precision markedly increased, while the recall decreased but the overall extraction performance improved (Table 3). Both MetaMap baselines performed poorly, showing the vulnerability of MetaMap when applied to informal text in social media. ADRMine significantly outperformed all the baseline approaches. Figure 4 illustrates examples of ADRs that could only be extracted by ADRMine.
Figure 4.

Examples of successfully extracted concepts using ADRMine.
The effectiveness of classification features
Feature evaluations (Table 4) indicated that lexicon, POS, and negation features added no significant contribution to the results when CRF was trained on comparatively larger number of training instances (DS train set), while they could still make small contributions to the performance when less data was available (Twitter train set or DS with less number of training instances). However, for both corpora, the context features were fundamental for achieving both high precision and recall; and the embedding cluster features were critical in improving the recall which resulted in a significant boost in F-measure. Examples of ADRs that were extracted after adding the cluster features are starred (asterisks) in Figure 5.
Figure 5.

Examples of concepts that could only be extracted after adding the embedding cluster features to ADRMine. These concepts are starred and other extracted concepts are highlighted.
Interestingly, as Figure 3b illustrates, the F-measure of the CRF with cluster features and 40% training data is higher than the F-measure without cluster features and 100% training data. Therefore, these features can be advantageous in situations where less annotated data is available. As shown in Table 3, the contribution of the cluster features in ADRMine was substantially higher for the Twitter corpus, which also confirms this finding.
Error analysis
For error analysis, we randomly selected 50 fp and 50 fn ADR mentions from DS test set and categorized the likely sources of errors. A summary of this is depicted in Figure 6, with example fp/fn concepts shown within brackets. The majority of fp errors were caused by mentions that were confused with indications or non-ADR clinical mentions. We believe that incorporating more context (e.g., a longer window), and sentiment analysis features, which model the positivity/negativity of the context, will diminish such errors in future.
Figure 6.
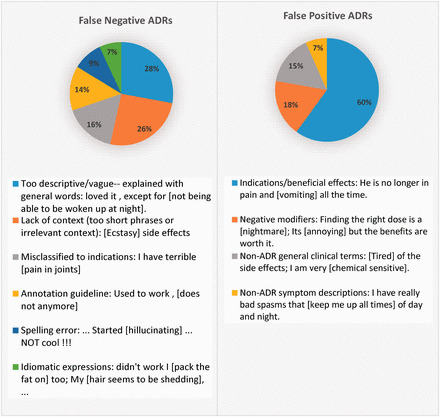
Analysis of false positive and false negatives produced by the ADR extraction approach.
Twenty-eight percent of fn ADRs were expressed in long, descriptive phrases, which rarely included any technical terms. Sentence simplification techniques might be effective in extracting such fns.63 Irrelevant immediate context or the lack of context in too short, incomplete sentences, made it difficult for the CRF to generalize, and contributed to 26% of fns. Other fns were related to specific rules in the annotation guideline, mentions expressed with complex idiomatic expressions or uncorrected spelling errors. Future research is needed to identify an optimized set of features that could potentially minimize these errors.
CONCLUSION
In this study, we proposed ADRMine, a machine learning-based sequence tagger for automatic extraction of ADR mentions from user posts in social media. ADRMine achieved an F-measure of 0.82 for DS, and 0.72 for Twitter corpus, outperforming all baseline techniques. We explored the effectiveness of various classification features in training the CRF model, and found that context and embedding clusters were the most contributing features. Furthermore, we utilized a large volume of unlabeled user posts for unsupervised learning of the embedding clusters, which enabled similarity modeling between the tokens, and gave a significant rise to the recall. Considering the rapid increasing volume of user posts in social media that contain new and creative consumer expressions for medical terms, and the fact that we generally have a comparatively small number of annotated sentences, we showed that using the embedding clusters diminished the dependency on large numbers of annotated data.
Considering the challenges of task-specific feature design, and given the success of the deep learning techniques in generating the word embeddings, our future work will involve exploring the effectiveness of training a deep learning NN, instead of the CRF, for simultaneous learning of both classification features and the labels. In addition, in this study we focused on concept extraction, but future research should examine normalization techniques. It is needed to map an extracted mention to the corresponding concept in standard ontologies, such as UMLS and MedDRA. Moreover, we believe that the proposed features and extraction techniques may prove applicable for extraction of other medical concepts from social media and similar contents.
FUNDING
This work was supported by National Institutes of Health (NIH) National Library of Medicine (NLM) grant number NIH NLM 5R01LM011176. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM or NIH.
COMPETING INTERESTS
None.
CONTRIBUTORS
This study was conducted under the supervision of G.G. and she provided critical review and substantial edits to the manuscript. A.N. designed the study, performed experiments, analyzed the data, and drafted and revised the manuscript. A.S. performed the MetaMap experiments, contributed to data preparation, and helped to edit the manuscript. K.O. made contributions in drafting the literature review and part of the discussions. R.G. contributed to details about the data collection. K.O. and R.G. annotated the corpora and provided significant edits to the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Dr Karen L. Smith for supervising the annotation process and Pranoti Pimpalkhute, Swetha Jayaraman, and Tejaswi Upadhyaya for their technical support.
REFERENCES
- 1. Pirmohamed M, James S, Meakin S, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sultana J, Cutroneo P, Trifirò G. Clinical and economic burden of adverse drug reactions. J Pharmacol Pharmacother. 2013;4:S73–S77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aagaard L, Nielsen LH, Hansen EH. Consumer reporting of adverse drug reactions: a retrospective analysis of the Danish adverse drug reaction database from 2004 to 2006. Drug Saf. 2009;32:1067–1074. [DOI] [PubMed] [Google Scholar]
- 4. Avery AJ, Anderson C, Bond CM, et al. Evaluation of patient reporting of adverse drug reactions to the UK “Yellow Card Scheme”: literature review, descriptive and qualitative analyses, and questionnaire surveys. Southampton: NIHR HTA; 2011. doi:10.3310/hta15200. [DOI] [PubMed] [Google Scholar]
- 5. Van Geffen ECG, van der Wal SW, van Hulten R, et al. Evaluation of patients’ experiences with antidepressants reported by means of a medicine reporting system. Eur J Clin Pharmacol. 2007;63:1193–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vilhelmsson A, Svensson T, Meeuwisse A, et al. What can we learn from consumer reports on psychiatric adverse drug reactions with antidepressant medication? Experiences from reports to a consumer association. BMC Clin Pharmacol. 2011;11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hazell L, Shakir SAW. Under-reporting of adverse drug reactions. Drug Saf. 2006;29:385–396. [DOI] [PubMed] [Google Scholar]
- 8. Ginn R, Pimpalkhute P, Nikfarjam A, et al. Mining Twitter for adverse drug reaction mentions: a corpus and classification benchmark. In: proceedings of the Fourth Workshop on Building and Evaluating Resources for Health and Biomedical Text Processing (BioTxtM). Reykjavik, Iceland; May 2014. [Google Scholar]
- 9. O’Connor K, Nikfarjam A, Ginn R, et al. Pharmacovigilance on Twitter? Mining Tweets for adverse drug reactions. In: American Medical Informatics Association (AMIA) Annual Symposium. November, 2014. [PMC free article] [PubMed] [Google Scholar]
- 10. DailyStrength. http://www.dailystrength.org/. Accessed June, 2014.
- 11. Leaman R, Wojtulewicz L, Sullivan R, et al. Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. In: Proceedings of the 2010 Workshop on Biomedical Natural Language Processing; July, 2010:117–125. [Google Scholar]
- 12. Yates A, Goharian N. ADRTrace: detecting expected and unexpected adverse drug reactions from user reviews on social media sites. Adv Inf Retr. 2013;7814 LNCS:816–819. [Google Scholar]
- 13. Yang C, Jiang L, Yang H, et al. Detecting signals of adverse drug reactions from health consumer contributed content in social media. In: Proceedings of ACM SIGKDD Workshop on Health Informatics; Beijing, August, 2012. [Google Scholar]
- 14. Benton A, Ungar L, Hill S, et al. Identifying potential adverse effects using the web: a new approach to medical hypothesis generation. J Biomed Inform. 2011;44:989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turian J, Ratinov L, Bengio Y. Word representations: a simple and general method for semi-supervised learning. In: Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics. July 2010:384–394. [Google Scholar]
- 16. Deng L, Yu D. Deep Learning: Methods and Applications, Foundations and Trends in Signal Processing. 2014;7:197–387. http://dx.doi.org/10.1561/2000000039. [Google Scholar]
- 17. Collobert R, Weston J, Bottou L, et al. Natural language processing (almost) from scratch. J Mach Learn Res. 2011;1:2493–2537. [Google Scholar]
- 18. Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Heal Technol Inf. 2010;160:739–743. [PubMed] [Google Scholar]
- 19. Friedman C. Discovering novel adverse drug events using natural language processing and mining of the electronic health record. In: AIME ’09 Proceedings of the 12th Conference on Artificial Intelligence in Medicine: Artificial Intelligence in Medicine. Verona, Italy, July 2009. [Google Scholar]
- 20. Wang W, Haerian K, Salmasian H, et al. A drug-adverse event extraction algorithm to support pharmacovigilance knowledge mining from PubMed citations. AMIA Annu Symp Proc. 2011;2011:1464–1470. [PMC free article] [PubMed] [Google Scholar]
- 21. Gurulingappa H, Rajput A, Toldo L. Extraction of adverse drug effects from medical case reports. J Biomed Semantics. 2012;3:15 doi:10.1186/2041-1480-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Toldo L, Bhattacharya S, Gurulingappa H. Automated identification of adverse events from case reports using machine learning. In: Proceedings XXIV Conference of the European Federation for Medical Informatics. Workshop on Computational Methods in Pharmacovigilance. August 2012;26–29. [Google Scholar]
- 23. Harpaz R, DuMouchel W, Shah NH, et al. Novel data-mining methodologies for adverse drug event discovery and analysis. Clin Pharmacol Ther. 2012;91:1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Polepalli Ramesh B, Belknap SM, Li Z, et al. Automatically recognizing medication and adverse event information from food and drug administration’s adverse event reporting system narratives. JMIR Med Informatics. 2014;2:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikfarjam A, Gonzalez G. Pattern mining for extraction of mentions of adverse drug reactions from user comments. AMIA Annu Symp Proc. 2011;2011:1019–1026. [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X, Chen H. AZDrugMiner: an information extraction system for mining patient-reported adverse drug events. In: Proceedings of the 2013 international conference on Smart Health. August 2013;134–150. [Google Scholar]
- 27. Chee BW, Berlin R, Schatz B. Predicting adverse drug events from personal health messages. AMIA Annu Symp Proc. 2011;2011:217–226. [PMC free article] [PubMed] [Google Scholar]
- 28. Wicks P, Vaughan TE, Massagli MP, et al. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol. 2011;29:411–414. [DOI] [PubMed] [Google Scholar]
- 29. Yang CC, Yang H, Jiang L, et al. Social media mining for drug safety signal detection. In: Proceedings of the 2012 international workshop on Smart health and wellbeing. New York, USA: ACM Press; October, 2012:33–40. [Google Scholar]
- 30. Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. Journal of biomedical informatics. 2014; In Press. doi:10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. White RW, Tatonetti NP, Shah NH, et al. Web-scale pharmacovigilance: listening to signals from the crowd. J Am Med Inform Assoc. 2013;20:404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. SIDER 2 — Side Effect Resource. http://sideeffects.embl.de/. Accessed September, 2014.
- 34. Zeng-Treitler Q, Goryachev S, Tse T, et al. Estimating consumer familiarity with health terminology: a context-based approach. J Am Med Informatics Assoc. 2008;15:349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mozzicato P. MedDRA: an overview of the medical dictionary for regulatory activities. Pharmaceut Med. 2009;23:65–75. [Google Scholar]
- 36. Liu X, Liu J, Chen H. Identifying adverse drug events from health social media: a case study on heart disease discussion. In: International Conference on Smart Health. July 2014:25–36. [Google Scholar]
- 37. Gurulingappa H, Rajput AM, Roberts A, et al. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J Biomed Inform. 2012;45:885–892. [DOI] [PubMed] [Google Scholar]
- 38. Jiang L, Yang C, Li J. Discovering consumer health expressions from consumer-contributed content. In: Proceedings of International Conference on Social Computing, Behavioral-Cultural Modeling, and Prediction. Washington, D.C., April 2013:164–174. [Google Scholar]
- 39. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- 40. Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med. 2005;37:360–363. [PubMed] [Google Scholar]
- 41. Ritter A, Clark S, Etzioni O. Named entity recognition in tweets: an experimental study. In: Proceedings of the Conference on Empirical Methods in Natural Language Processing. July 2011:1524–1534. [Google Scholar]
- 42. Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pacific Symp Biocomput. 2008;13:652–663. [PubMed] [Google Scholar]
- 43. Okazaki N. CRFsuite: a fast implementation of Conditional Random Fields (CRFs). 2007. http://www.chokkan.org/software/crfsuite/. Accessed July, 2014. [Google Scholar]
- 44. Apache Lucene. http://lucene.apache.org/. Accessed November, 2014.
- 45. Atkinson K. SCOWL (Spell Checker Oriented Word Lists). http://wordlist.aspell.net/. Accessed November, 2014. [Google Scholar]
- 46. Zhou X, Zhang X, Hu X. Dragon Toolkit: incorporating auto-learned semantic knowledge into large-scale text retrieval and mining. In: proceedings of the 19th IEEE International Conference on Tools with Artificial Intelligence (ICTAI). 2007;2:197–201. [Google Scholar]
- 47. Miller GA. WordNet: a lexical database for English. Commun ACM. 1995;38:39–41. [Google Scholar]
- 48. Manning CD, Klein D. Accurate unlexicalized parsing. In: Proceedings of the 41st Meeting of the Association for Computational Linguistics. July 2003:423–430. doi:10.3115/1075096.1075150. [Google Scholar]
- 49. Kilicoglu H, Bergler S. Syntactic dependency based heuristics for biological event extraction. In: Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task. 2009:119–127. [Google Scholar]
- 50. Nikfarjam A, Emadzadeh E, Gonzalez G. A hybrid system for emotion extraction from suicide notes. Biomed Inform Insights. 2012;5:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. word2vec. https://code.google.com/p/word2vec/. Accessed June, 2014.
- 52. Bengio Y, Ducharme R, Vincent P, Janvin C. A neural probabilistic language model. J Mach Learn Res. 2003;3:1137–1155. [Google Scholar]
- 53. Mikolov T, Chen K, Corrado G, et al. Efficient estimation of word representations in vector space. In: Proceedings of International Conference on Learning Representations, Scottsdale, Arizona, May 2013. [Google Scholar]
- 54. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. In: Proc AMIA Symp. November 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 55. Lopez V, Sabou M, Motta E. PowerMap: mapping the real semantic web on the fly. In: Proceedings of the 5th International Semantic Web Conference. November 2006:414–427. [Google Scholar]
- 56. Emadzadeh E, Nikfarjam A, Ginn R, et al. Unsupervised Gene Function Extraction using Semantic Vectors. Database. 2014; 2014 doi: 10.1093/database/bau084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang M, Liu J, Zhu X. GeneTUKit: a software for document-level gene normalization. Bioinformatics. 2011;27:1032–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Joachims T. Text categorization with support vector machines: learning with many relevant features. Mach Learn ECML-98. 1998;1398:137–142. [Google Scholar]
- 59. Joachims T. Making large scale SVM learning practical. In: Schölkopf B, Burges C, Smola A.Advances in kernel methods - support vector learning .Cambridge, MA, MIT Press; 1999: 169–184. [Google Scholar]
- 60. Tsai RT-H, Wu S-H, Chou W-C, et al. Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinformatics. 2006;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yeh A. More accurate tests for the statistical significance of result differences. In: Proceedings of the 18th Conference on Computational linguistics. Saarbrueken, Germany, July 2000:947–953. [Google Scholar]
- 62. Pado S. User’s guide to sigf: significance testing by approximate randomisation. 2006. http://www.nlpado.de/~sebastian/software/sigf.shtml. Accessed November, 2014. [Google Scholar]
- 63. Jonnalagadda S, Gonzalez G. Sentence simplification aids protein-protein interaction extraction. In Proceedings of the 3rd International Symposium on Languages in Biology and Medicine, November 2009:109–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


