Summary
Over the past several years, there has been an increasing research effort focused on inhibition of protein-protein interactions (PPIs) to develop novel therapeutic approaches for cancer, including hematologic malignancies. These efforts have led to development of small molecule inhibitors of PPIs, some of which already advanced to the stage of clinical trials while others are at different stages of pre-clinical optimization, emphasizing PPIs as an emerging and attractive class of drug targets. Here, we review several examples of recently developed inhibitors of protein-protein interactions highly relevant to hematologic cancers. We address the existing skepticism about feasibility of targeting PPIs and emphasize potential therapeutic benefit from blocking PPIs in hematologic malignancies. We then use these examples to discuss the approaches for successful identification of PPI inhibitors and provide analysis of the protein-protein interfaces, with the goal to address ‘druggability’ of new PPIs relevant to hematology. We discuss lessons learned to improve the success of targeting new protein-protein interactions and evaluate prospects and limits of the research in this field. We conclude that not all PPIs are equally tractable for blocking by small molecules, and detailed analysis of PPI interfaces is critical for selection of those with the highest chance of success. Together, our analysis uncovers patterns that should help to advance drug discovery in hematologic malignancies by successful targeting of new protein-protein interactions.
Keywords: protein-protein interactions, drug discovery, hematologic malignancies
Introduction
Limited success and toxicity of conventional chemotherapy in the treatment of hematologic malignancies emphasize the need for development of targeted therapies, which are expected to be more effective and less toxic than chemotherapy agents. Targeted agents, exemplified by the number of monoclonal antibodies and small molecule inhibitors currently in clinical use or in clinical trials, have a history of success in the treatment of hematologic malignancies, either as single agents or in combination with chemotherapy agents (1–3). The approval of imatinib (Gleevec) in 2001 for the treatment of chronic myeloid leukemia (CML) and its high success rate (~90% five-year survival) support the extensive efforts to develop novel molecularly targeted therapies for hematologic cancers (4). The majority of small molecule targeted agents developed for hematologic malignancies block activity of protein kinases, including FLT3, Aurora kinase, JAK1/2, Akt, and mTOR (2, 3). Additional examples include proteasome inhibitors (5) and epigenetic/demethylating agents, such as inhibitors of histone deacetylases and DNA or histone methyltransferases (2, 3, 6–8). These agents, however, are still under active investigation at different stages of clinical trials to validate their clinical applicability (1–3).
Studies on the mechanistic basis of tumorigenesis allow understanding how genetic and epigenetic modifications lead to different subtypes of cancers (9–11). In many hematologic cancers genetic profiles are well defined (12). The ability to develop new effective drugs relies on understanding biological mechanisms connecting specific genetic abnormality with disease progression. Likewise, analysis of the molecular basis of tumorigenesis reveals proteins that play a critical role in oncogenesis and provides novel molecular targets for therapeutic intervention. Among molecular targets critical in pathogenesis of different types of cancer, the protein-protein interactions (PPIs) play a very important role (13, 14). Under normal physiological conditions PPIs hold together multi-protein complexes in cells to control essentially all cellular processes (15, 16). Therefore it is not surprising that many PPIs have been recognized as an emerging class of molecular targets in different diseases, including cancer (13, 16, 17). Protein-protein interactions were, however, considered as either very challenging or even ‘undruggable’ targets. The main challenges in targeting PPIs were linked to the flexibility, large size, and complex topology of PPI interfaces. These features, together with poor compatibility of PPI interfaces with small molecules available in screening libraries limited the applicability of classical drug screening approaches for identification of PPI inhibitors (18). Despite these challenges and limitations, a number of PPIs were successfully explored for drug discovery purposes, both in academia and industry, leading to small molecules that have already entered clinical trials in oncology (18–20) (Table 1). These successful examples have changed the general concept of targeting PPIs, which are no longer considered uniformly ‘undruggable’.
Table 1.
Small molecule inhibitors of protein-protein interactions currently in clinical trials for hematologic malignancies
| Drug candidate | Target protein | Indication | Status | Reference |
|---|---|---|---|---|
| RO5045337 | MDM2-p53 | Hematologic neoplasms (leukemia) | Phase 1 | (194) |
| PRI-724 | CBP/beta-catenin | AML, CML | Phase 1/2 | (195) |
| TL32711 | SMAC mimetic | AML, ALL, MDS | Phase 1/2 | (196) |
| LCL161 | SMAC mimetic | Multiple myeloma | Phase 2 | (197) |
| ABT-263 | Bcl-2-BH3 | Hematological cancers (lymphoma, leukemia) | Phase 1/2 | (198) |
| GX15-070 | Bcl-2 family – BH3 | Hematological cancers (lymphoma, leukemia) | Phase 1/2 | (199) |
| GSK525762 | BET bromodomains | Hematologic Malignancies | Phase 1 | (88) |
| OTX015 | BET bromodomains | Hematologic Malignancies | Phase 1 | NCT01713582a |
| CPI-0610 | BET bromodomains | Lymphoma | Phase 1 | NCT01949883a |
Clinical trial identifier.
The limited success of conventional chemotherapy currently used for treatments in hematologic cancers clearly supports the need for development of new treatment options (1–3, 21). Improved understanding of PPI networks and success in targeting PPIs by small molecules (19, 22) has encouraged a number of research groups to pursue similar studies in hematologic cancers. In this review we discuss examples of recently developed small molecule inhibitors of PPIs that are highly relevant to hematologic cancers (Fig. 1), to address the existing skepticism about feasibility of targeting PPIs and emphasize the potential therapeutic benefit from blocking PPIs in hematology. These examples are particularly instructive, as they represent different types of structurally characterized PPIs, providing the opportunity to analyze which PPIs are most tractable as drug targets. We then use these examples to discuss the approaches for successful identification of PPI inhibitors, deliberate on the role of structural biology in this process, and propose when lead optimization should convert into a drug discovery project. This analysis will assess lessons learned to improve the success of targeting new protein-protein interfaces and will evaluate prospects and limits of the research in this field, where inhibitors are still difficult to identify. Together, our analysis uncovers patterns that should help to advance drug discovery in hematologic malignancies by successful targeting of new protein-protein interfaces.
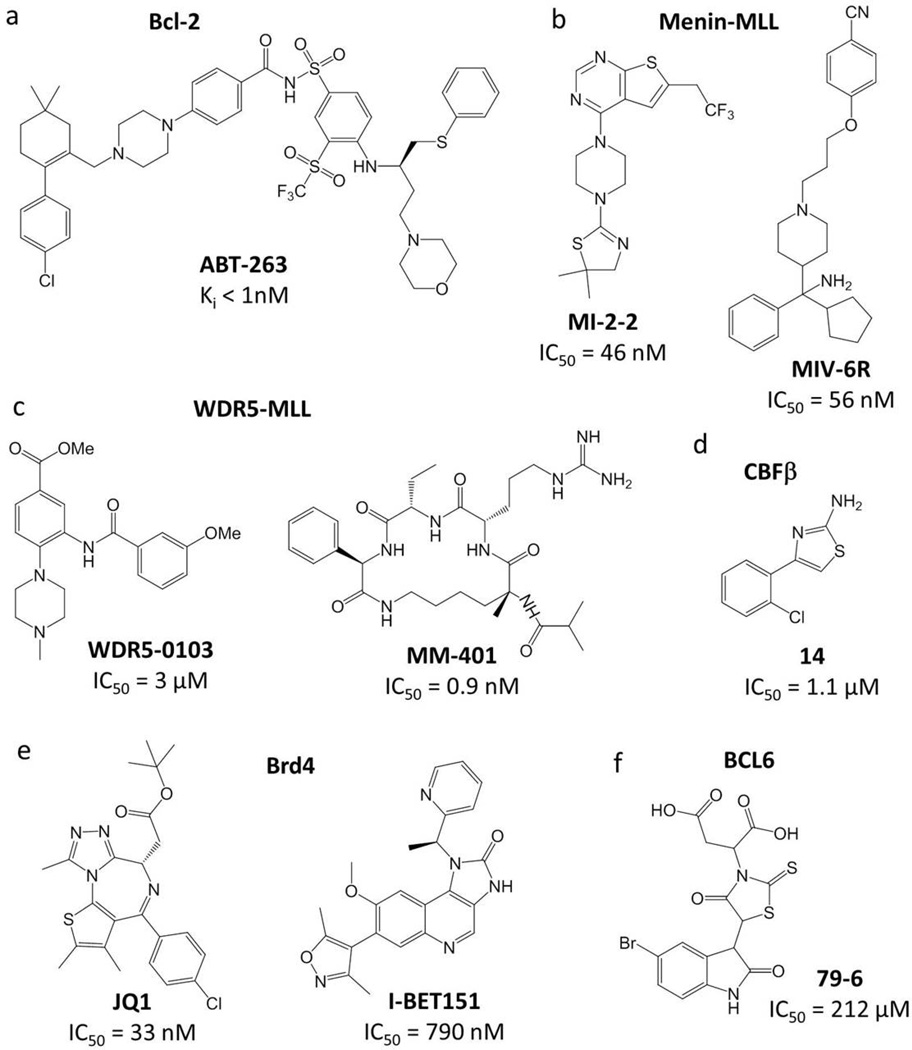
Fig. 1. Chemical structures and inhibitory activity of PPI inhibitors developed for hematology-related protein targets.
(A) ABT-263 targeting Bcl-2 family of proteins. (B) Small molecule inhibitors of the menin-MLL interaction: MI-2-2 and MIV-6R. (C) Inhibitors of WDR5-MLL interaction: small molecule WDR5-0103 and peptidomimetic MM-401. (D) Small molecule inhibitor of CBFβ-Runx1 interaction: cpd 14. (E) Inhibitors of BET bromodomains: JQ1 and I-BET151. (F) Small molecule inhibitor of BCL6: 79-6.
This review article starts with the discussion on how attractive PPIs are as drug targets, followed by addressing major challenges in targeting protein-protein interfaces with small molecules. We then provide several successful examples of small molecules blocking protein-protein interactions relevant to hematology to demonstrate that PPIs are tractable drug targets. This is followed by lessons learned from blocking PPIs, including a description of methods and approaches used for identification of PPI inhibitors, the importance of crystal structures, discussion on druggability of PPI interfaces, as well as the role of academic laboratories in this process. This article concludes with a discussion about what has changed over time in targeting PPIs, which questions still remain to be answered, and what is the future of targeting PPIs for development of novel therapeutics.
Are PPIs attractive drug targets?
Targeting PPIs with small molecules is considered much more challenging than inhibiting classical drug targets such as enzymes, receptors, or ion channels (23–25). Therefore, the key questions are whether such efforts will pay off and whether inhibitors of PPIs will offer significant benefits. PPIs play critical roles in many biological processes, under both physiological and pathological conditions. It has been estimated that 130,000 to 650,000 PPIs may occur in human cells (26, 27), and therefore the number of potential PPIs as drug targets is large and significantly exceeds the druggable human kinome (28). Targeting PPIs also provides an attractive opportunity to directly target proteins that drive disease development. For example, oncogenic activity of mixed lineage leukemia (MLL) fusion proteins in leukemia is dependent of their interactions with menin (29). Hence, direct inhibition of the menin-MLL protein-protein interaction with small molecules may represent a more attractive strategy to reverse oncogenic activity of MLL fusion proteins than targeting downstream signaling pathways.
As discussed previously, targeting PPIs may offer advantages over enzyme inhibition (25). Development of selective inhibitors to block enzymatic activity of kinases (e.g. ATP mimetics) or methyltransferases (e.g. S-adenosylmethionine mimetics) represents a challenge due to high conservation of binding sites across family members. Since PPI interfaces vary significantly between different protein complexes, inhibitors targeting PPIs may be more selective than kinase inhibitors competing for relatively conserved ATP-binding sites. Furthermore, activity of kinases is regulated via multiple PPIs, and therefore targeting these interactions may allow achievement of higher level of selectivity or different biological responses as compared to blocking kinase activity by the ATP competitive inhibitors (25). Availability of PPI inhibitors might allow for combinatorial treatment with kinase inhibitors to more efficiently block signaling pathways.
One of the major concerns about targeted therapy is development of resistance mechanism and loss of drug efficacy, which may arise from mutations in the target protein leading to the loss of drug binding. For example, mutations in the catalytic domain of BCR-ABL lead to resistance to imatinib (Gleevec) in CML patients (30, 31). Furthermore, the C481S mutation in the catalytic domain of the Bruton’s tyrosine kinase (BTK) was recently reported to confer resistance to ibrutinib, an inhibitor of BTK that has been approved in 2014 for the treatment of CLL (32). Mutations of other mechanisms, such as amplification of the receptor kinase or activation of an independent signaling pathway, may result in resistance to the kinase inhibitor drugs (33). In contrast, there is an attractive opportunity that drugs targeting PPIs will be less prone to the resistance mechanism, particularly when resistance is due to mutations in the protein target. Residues at the PPI interfaces are highly evolutionarily conserved and they evolve slower than residues at non-binding surfaces (34, 35). Furthermore, the PPI interfaces have relatively tight packing (36), and mutations predominantly weaken PPIs (37). Therefore, spontaneous mutations at PPI interfaces would have high likelihood of disrupting or weakening the interactions between natural protein partners. Obviously, the same mutations may impair small molecule binding. However, small molecules typically utilize significantly smaller binding interfaces (Figs 2, 3), and therefore there is a higher probability that mutations at PPI interfaces will disrupt or weaken protein-protein interactions rather than small molecule binding to the target protein. This has been exemplified by the recent studies by Jung et al., in which mutations in BRD4 bromodomain were evaluated for their impact on interactions with either a natural binding partner, an acetylated histone 4 (ac-H4) derived peptide, or a potent small molecule inhibitor JQ1 (38). The majority of mutations decreased binding of both, ac-H4 peptide and JQ1, although binding of ac-H4 was impaired more pronouncedly. There was only one mutation (P82A), which resulted in significantly stronger reduction of JQ1 binding to BRD4 as compared to ac-H4. This example demonstrates that drug resistant mutations in BRD4 are possible; however, a great majority of mutations would also impair binding of the protein partner.
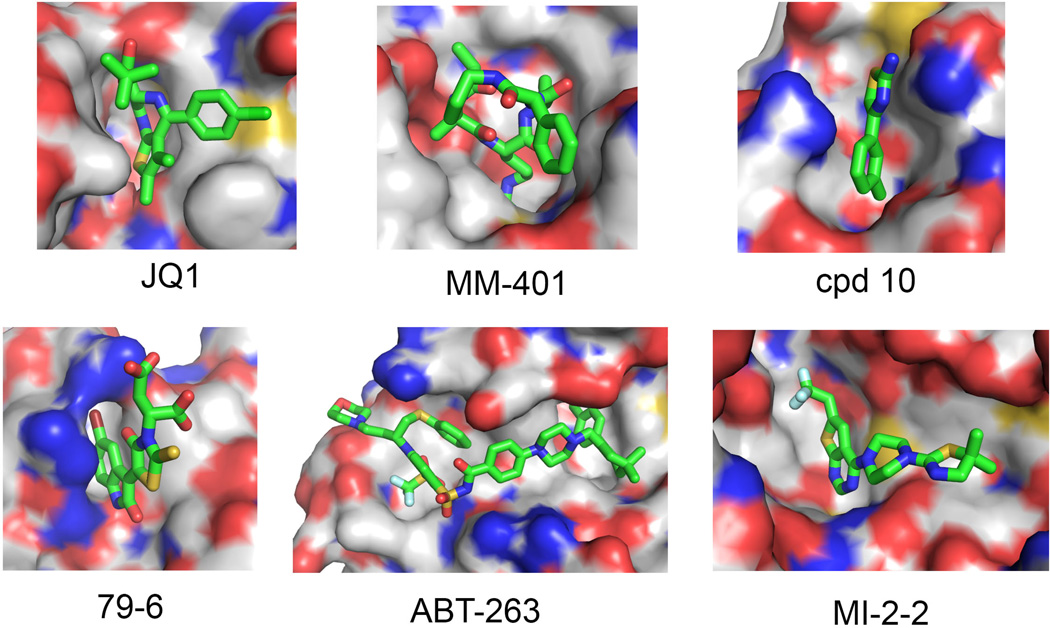
Fig. 2. Binding modes of selected PPI inhibitors to protein targets.
JQ1:Brd4 (PDB code 3MXF), ABT-263:Bcl-2 (4LVT), MM-104:WDR5 (4GM9), 79-6:BCL6 (3LBZ), MI-2-2-menin (4GQ4), and cpd 10-CBFβ (43), demonstrating how small molecule inhibitors bind to the surface pockets at PPI interfaces. Small molecule inhibitors are shown in stick representation with carbons in green, oxygens in red, nitrogens in blue, sulfur in yellow, and fluorines in cyan. Protein is shown in surface representation with white carbons, blue nitrogen, red oxygen, and yellow sulfur atoms.
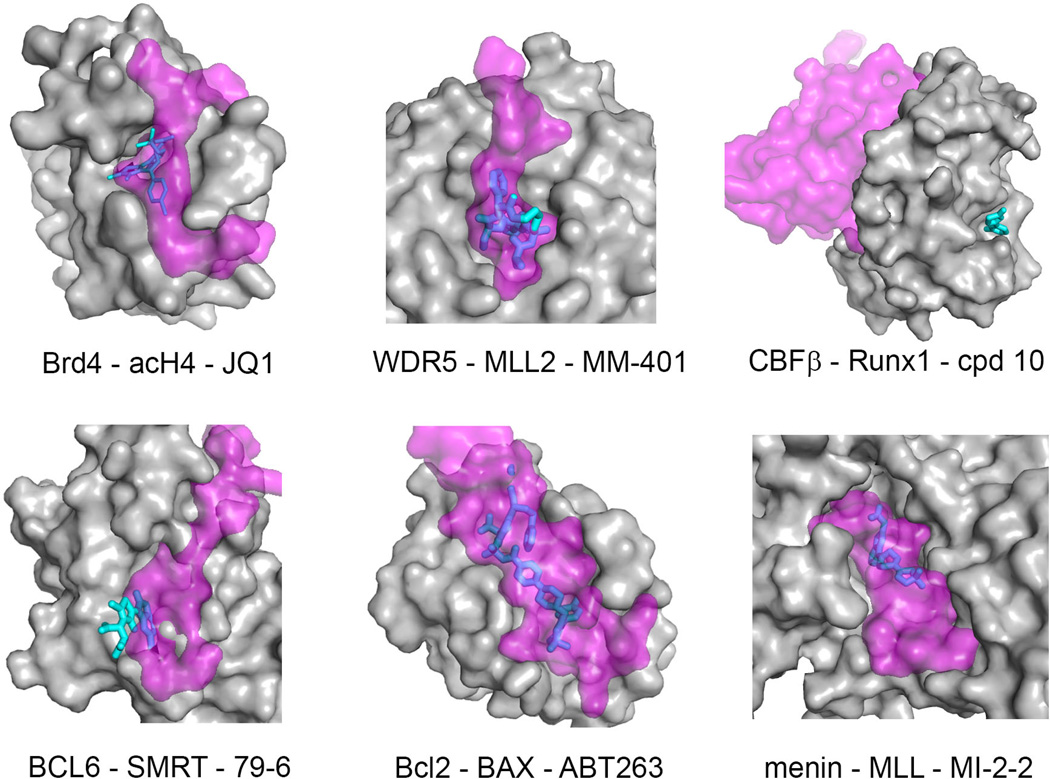
Fig. 3. Comparison of protein-small molecule contacts with protein-protein (or protein-peptide) interaction interfaces.
Target protein is shown in surface representation (gray), protein (or peptide) binding partner is shown in semi-transparent surface (magenta) and inhibitors are shown as sticks (cyan/blue). Blue color corresponds to the region of the ligand molecule that overlaps with binding of the protein (peptide) partner, while cyan color corresponds to the ligand portion that does not overlap with binding of the protein (peptide) partner. PDB codes for PPI complexes are as follows: Brd4-acH4 (3UVW), WDR5-MLL2 (3UVK), BCL6-SMRT (1R2B), Bcl-2-BAX (2×A0); menin-MLL (3U88); CBFβ-Runx1 (1H9D). The PDB structures for protein-inhibitor complexes are the same as shown in Fig. 2.
Several agents targeting PPIs are currently being evaluated in clinical trials in hematological cancers (Table 1), and to our knowledge, no naturally occurring mutations that would impair drug binding to the target protein have been described. These compounds, however, are still at relatively early stages of clinical evaluation (Table 1), and the hypothesis that targeting PPIs would lead to fewer resistance mutations remains to be robustly tested. Incoming data from clinical trials will address this important issue and provide a better understanding of resistance mechanism to PPI inhibitors.
Challenges in targeting protein-protein interactions
Successes and failures in targeting PPIs allow better understanding of the features at PPI interfaces that limit development of small molecule inhibitors. The main challenges in targeting PPIs arise from the complexity and flexibility of PPI interfaces and the relatively poor compatibility of binding pockets at the interfaces with compounds currently available in screening libraries.
Complexity of PPI interfaces
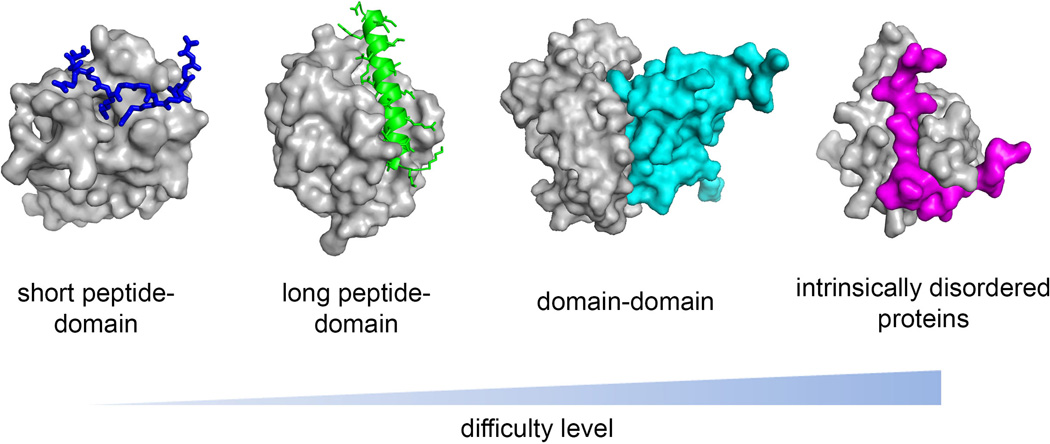
PPI interfaces are complex and vary significantly between different types of protein-protein complexes. Three different types of protein-protein interfaces can be distinguished: (i) globular domain-peptide interactions (Fig. 4A,B), (ii) interactions involving folded globular domains (Fig. 4C), and (iii) interactions of intrinsically unstructured proteins. The majority of the best characterized PPIs fall into the class of complexes involving globular domains. These interactions usually have noncontiguous nature and involve relatively flat and large surface areas (1200 to 3000 Å2) (39, 40). They often lack surface pockets, which make them difficult for disruption by orthosteric inhibitors that bind to the PPI interfaces. Development of allosteric inhibitors might represent an alternative approach to target such PPIs (41–43). The feasibility of developing allosteric inhibitors is, however, difficult to predict until such molecules are identified in screens and validated for binding to the allosteric sites on proteins using structural biology or mutagenesis studies.
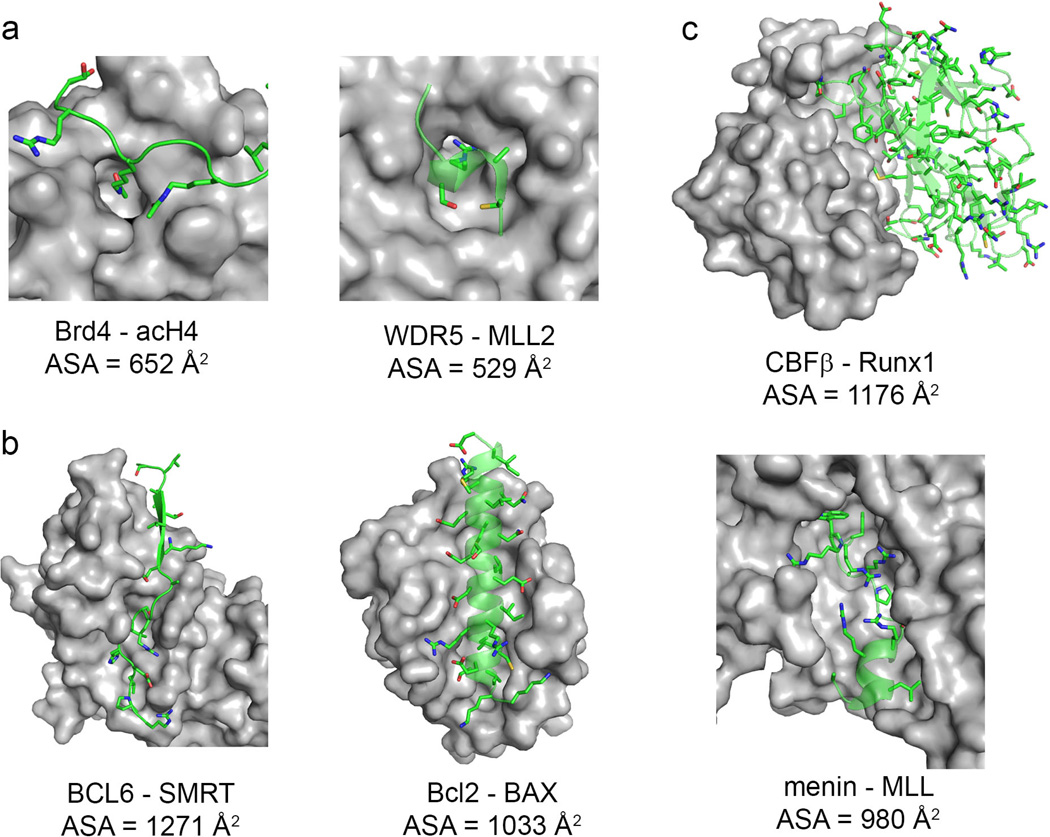
Fig. 4. Types of ‘druggable’ PPI interfaces.
(A) Short peptide - domain interaction. (B) Long peptide – domain interaction. (C) Domain-domain interaction. Target proteins are shown in surface representation (gray), and binding partners are shown in ribbon and stick representations (carbon atoms in green, oxygen in red, nitrogen in blue, sulfur in yellow). Accessible solvent area (ASA) buried in complex formation has been calculated with 2P2I inspector software (200) using the same PDB structures as in Fig. 3.
More attractive opportunities for small molecule development represent PPIs where globular domain engages a peptide-like fragment of a protein partner. These protein-peptide type interactions account for up to 40% of all PPIs (44). The average surface areas at the protein-peptide interfaces are much smaller, and reach about 500 A2 (45). Importantly, these interfaces frequently contain well-defined binding pockets and have been recognized as more susceptible to targeting by small molecule inhibitors.
The most complex and least understood are PPIs involving intrinsically disordered proteins (IDPs) (46), which remain unfolded in the absence of protein partners and fold into a globular structure upon binding (47, 48). Formation of IDP complexes typically results in large contact areas and complex PPI interfaces. Intrinsically disordered proteins have been identified as hubs in the PPI networks (49), and are potentially valuable drug targets. However, whether such interactions can be disrupted by small molecules still remains to be determined.
Despite large size and complexity of PPI interfaces, it has been demonstrated that relatively few residues at the interfaces are essential for high affinity interactions (50, 51). These residues contribute to the majority of binding energy and are called ‘hot spots’ (50–53). ‘Hot spot’ residues constitute less than half of the binding surface and are usually found in the center of the contact interface (54). The ‘hot spot’ amino acids are frequently conserved across different protein families and became buried upon complex formation (35). Hydrophobic residues, such as tryptophan, tyrosine, phenylalanine, methionine and arginine, are the most frequent ‘hot spots’ identified at PPI interfaces (51, 52). Identification of ‘hot spot’ residues can be achieved via alanine scanning mutagenesis (50). Localization of ‘hot spots’ at PPI interface is valuable to guide ligand development, and it has been found that PPI ‘hot spots’ largely correlate with the sites where small molecules bind (55).
Pockets at PPI interfaces are not compatible to bind small molecules
Structural analysis of protein complexes demonstrates that despite a common belief about the lack of pockets at PPI interfaces, such pockets are indeed frequently present. However, the topology of these pockets differs from those found in classical drug targets, such as enzymes, ion channels and receptors (56–58). It has been noted that high affinity PPIs involving fairly localized contacts are most amenable to inhibition, while weaker interactions with large contact areas are much more difficult to disrupt using small molecules (59). Pockets at PPI interfaces are also fairly hydrophobic and enriched in aromatic residues (52, 60). These features make PPIs not fully compatible with physicochemical properties of known small molecule drugs (61).
Flexibility and dynamics of residues at PPI interfaces
Availability of protein structures is critically important for development of PPI inhibitors. However, PPI interfaces are frequently dynamic, and unbound proteins may exist in multiple conformational states (62). In fact, flexibility at PPI interfaces is an intrinsic property of many proteins and might be functionally important. For example, conformational plasticity in the Bcl-2 family of proteins allows for binding to multiple distinct BH3-only proteins (63).
Flexibility and dynamics at PPI interfaces represents a challenge for designing small molecule inhibitors, particularly when structure-based drug design methods are applied. On the other hand, dynamics of PPI interfaces may create opportunities for inhibitor discovery. For example, pockets might not be present in the crystal structures of uncomplexed proteins, while large pockets suitable to bind small molecules may exist in solution (64). Small molecules are also capable of causing conformational changes upon binding to the orthosteric or allosteric sites and induce formation of new pockets (65).
Structural information on protein-protein complexes is typically obtained from analysis of the crystal structures, which provide a static picture of PPI interfaces. Protein conformation in the crystal state represents a selected low energy conformation, while multiple conformations may exist in solution. In addition, crystal structures may be distorted due to crystal packing forces. NMR spectroscopy provides a valuable alternative method for protein structure determination in solution. However, structural studies on protein-ligand complexes by NMR still represent a challenge (66). Another approach to account for conformational dynamics of proteins is molecular dynamics (MD). Multiple snapshots of protein conformations can be taken from MD simulations to be subsequently used as a basis for ligand docking (67–69). Overall, flexibility at PPI interfaces complicates structure-based design of small molecule inhibitors but simultaneously offers an attractive opportunity for identification of new unexpected binding sites and allows for targeting of challenging PPIs.
PPIs are tractable drug targets in hematologic malignancies: case studies
Successful identification and development of small molecules to block PPIs in cancer have been demonstrated for a number of protein targets (19, 22, 23, 70). These examples include inhibitors of the anti-apoptotic Bcl-2 family of proteins implicated in different hematologic malignancies (71–74), with selected compounds (e.g. ABT-263) (Fig. 1A) currently in clinical trials for lymphoid malignancies (22, 72, 73). Inhibition of the MDM2-p53 interaction with small molecules represents another example of PPIs where very potent drug-like molecules were developed (75, 76), and are being evaluated in clinic for different types of cancers, including AML and lymphoma (22, 77). More recently, there has been extensive progress in development of small molecule inhibitors targeting other protein-protein interactions implicated in hematologic cancers. Successful examples include development of inhibitors targeting PPIs critical to oncogenic activity of MLL fusion proteins in MLL rearranged leukemias (78–82), compounds blocking the Core Binding Factor beta (CBFβ) in acute leukemia (43, 83), and inhibitors of the BET family of bromodomains, which demonstrated activity in AML and multiple myeloma (84–86) (Fig. 1B–E). Furthermore, small molecules targeting the protein-protein interface on Bcl-6 have also been developed as a potential therapeutic strategy for B-cell lymphoma (87) (Fig. 1F). Many of these PPI inhibitors have been developed within the last five years, primarily in academic laboratories, and are currently at different stages of pre-clinical optimization, with BET bromodomain inhibitors already advanced to clinical trials (Table 1) (88), as discussed below in details. These examples represent different types of PPIs and are accompanied by detailed structural characterization of the protein-ligand complexes, providing the opportunity to analyze which PPIs are most tractable as drug targets to find common features for improving the success of targeting new protein-protein interfaces relevant to human diseases.
Small molecule inhibitors of the menin-MLL interaction
Chromosomal translocations that affect the MLL (mixed lineage leukemia) gene (in this review MLL uniformly refers to the MLL1 gene) occur in about 5–10% of acute leukemias in adults (89) and ~70% of acute leukemias in infants (90). Translocations of MLL result in expression of chimeric MLL fusion proteins, which retain the N-terminal MLL fragment of approximately 1400 amino acids fused with one out of over 60 fusion partners (91–94). Patients with MLL leukemias are refractory to currently available treatments (91, 95, 96), emphasizing the urgent need for development of novel therapies. Indeed, different novel therapeutic strategies are being explored, including small molecule inhibition of the Dot1L histone methyltransferase (8, 97), Flt3 receptor tyrosine kinase (98), GSK3 kinase (99), and cyclin dependent kinase 6 (CDK6) (100), all of which rely on inhibition of the enzymatic activity of proteins implicated in pathogenesis of MLL leukemia.
The chromosomal rearrangements of the MLL gene affect only one allele, while the second allele almost always remains intact (101). MLL is a member of the mixed lineage leukemia family of histone methyltransferases (HMTs), which catalyzes methylation of histone H3 on K4 through the SET domain located at the C-terminus of MLL (102, 103). Thiel et al. (104) demonstrated that histone methyltransferase activity of the wildtype MLL is required to cooperate with MLL-AF9 in leukemogenic transformation. MLL requires other proteins, including WDR5, ASH2L, and RbBP5, to assemble a catalytically active complex (105), and the interaction between MLL and WDR5 is critical for the integrity of this complex and for its methyltransferase activity, representing a potential drug target in MLL leukemias (see below) (105, 106).
Our own efforts to block oncogenic MLL fusion proteins have been focused on applying a different approach, specifically on inhibiting the protein-protein interaction between MLL fusion proteins and menin, which has been well validated to play a critical role in development and progression of MLL leukemias (29, 107, 108). Menin is a highly specific binding partner of MLL fusion proteins, and this interaction is essential for their leukemogenic activity (29). Therefore, disruption of the PPI between menin and MLL fusion proteins represents a very attractive therapeutic strategy to develop new targeted drugs for MLL leukemias.
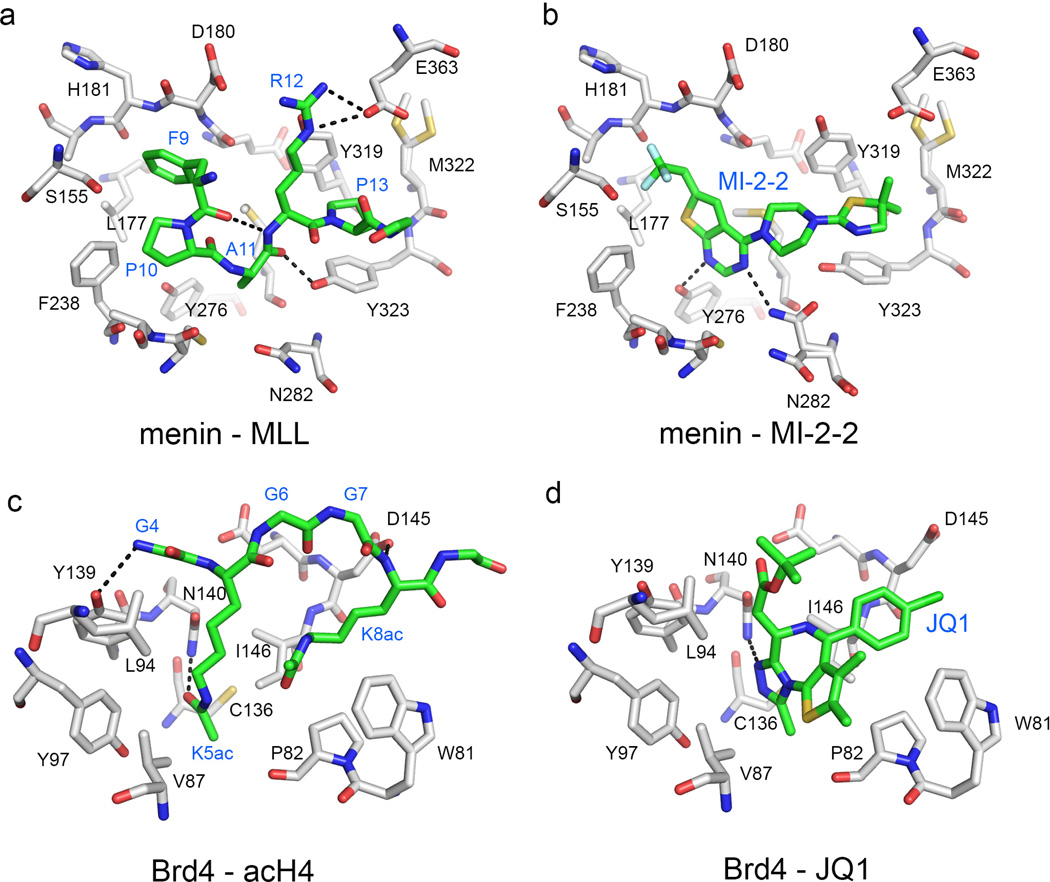
Menin interacts with two N-terminal fragments of MLL, namely MBM1 and MBM2 (menin-binding motif 1 and 2), with MBM1 (MLL4–15) representing high affinity binding motif that binds to menin with Kd of 56 nM (109). Studies from our group (79, 110) and by others (111) resulted in determination of the crystal structure of menin and the menin-MLL complex, demonstrating that MLL binds to the large central cavity on menin (Fig. 4B). Detailed analysis of the crystal structure and alanine scanning mutagenesis revealed that three hydrophobic residues of MLL, F9, P10 and P13 that bind to well-defined hydrophobic pockets on menin, contribute most to the binding affinity (79, 109, 111)(Fig. 5A).
Fig. 5. Comparison of binding modes of natural protein partners and small molecule inhibitors of PPIs.
Details of the interaction of MLL derived peptide (A) and MI-2-2 (B) with menin, demonstrating that MI-2-2 occupies the same region of the binding site and closely mimics key interactions of MLL (in particular residues F9 and P13) with menin (PDB codes: 4GQ6 and 4GQ4, respectively). Comparison of the binding mode of acetylated H4 peptide (D) and JQ1 (D) to Brd4 (PDB codes: 3UVW and 3MXF, respectively). Protein residues, peptides and small molecules are shown in stick representations, with carbon atoms in gray (proteins) or green (peptides and small molecules). Color coding for other heavy atoms remains the same for all complexes: oxygens in red, nitrogens in blue, sulfur in yellow and fluorines in cyan. Dashed lines correspond to hydrogen bonds.
We reported development of two classes of small molecule inhibitors of the menin-MLL interaction (78–80), in addition to recently reported peptidomimetics (112). By applying high throughput screening (HTS) of 49,000 small molecules we identified a thienopyrimidine class of menin-MLL inhibitors, with the most potent being MI-1, which directly binds to menin and inhibits the menin-MLL interaction with IC50 of 1.9 µM (78). Our initial medicinal chemistry efforts performed in the absence of structural information on the menin-ligand complexes resulted in MI-2 (IC50 = 0.46 µM). Further optimization of this class of menin-MLL inhibitors was possible when MI-2 was co-crystalized with menin (79). Using structure-based design we developed MI-2-2 with ~10-fold increased in vitro inhibitory activity (IC50 = 46 nM, Kd = 22nM)(79)(Figs 1B, 5B). Interestingly, MI-2-2 has a similar binding affinity to menin as the 12 amino acid MBM1 MLL derived peptide, despite almost fivefold smaller molecular weight. Strong potency of MI-2-2 is attributed to the fact that it binds to the MLL binding site on menin (Figs 2,3) and closely mimics key interactions of MLL with menin, in particular the interactions involving F9 and P13 residues of MLL (79)(Fig. 5A,B). This demonstrates that small molecule inhibitors of PPIs can achieve strong potency by mimicking the interactions identified for the natural protein partner. When tested in MLL leukemia cells, both MI-2 and MI-2-2 selectively blocked proliferation, induced apoptosis and differentiation and reversed the MLL fusion protein mediated leukemic transformation by downregulating MLL fusion protein target genes, including Hoxa9 and Meis1 (78, 79). Furthermore, both compounds also depleted the MLL-AF9 complex from the Hoxa9 locus and reduced H3K4me3 and H3K79me2 methylation level (78, authors’ unpublished data), validating their specific mechanism of action. The cellular effects in MLL leukemia cells are more pronounced for MI-2-2, correlating with its stronger in vitro inhibition of the menin-MLL interaction (79).
We have also reported another class of menin-MLL inhibitors, the methyl-piperidine compounds, identified by HTS of ~280,000 compounds at the NIH MLPCN (Molecular Libraries Probe Production Centers Network, https://mli.nih.gov/mli) (80). The initial HTS hit, MIV-1, showed only modest inhibitory activity (IC50 = 12 µM) and was further optimized using structure-based design approach to develop MIV-6R (IC50 = 56 nM, Kd = 85nM) (80)(Fig. 1B). Interestingly, the methyl-piperidine menin-MLL inhibitors more closely mimic the MLL binding mode to menin than thienopyrimidine compounds, as they occupy all three pockets on menin required for high affinity binding of MLL (F9, P10, P13). MIV-6R demonstrated specific growth arrest and differentiation in MLL leukemia cells, accompanied by downregulation of MLL fusion protein target genes, demonstrating a specific mechanism of action (80). Overall, the two classes of small molecule inhibitors of the menin-MLL interaction described here validate that pharmacologic inhibition of this PPI is feasible and can reverse the MLL-fusion protein mediated oncogenic transformation. These compounds are currently under development in our laboratory to further improve their potency and other drug-like properties to develop compounds for therapeutic applications.
The MLL-derived macrocyclic peptidomimetics were reported recently as potent in vitro inhibitors of the menin-MLL interaction (Ki = 4.7 nM for MCP-1) (112). However, cellular activity of these compounds was not reported, suggesting that optimization of their drug-like properties is likely required to identify therapeutically useful compounds. Nevertheless, the success with developing different classes of small molecules and peptidomimetics demonstrates that the menin-MLL interaction represents a druggable target for therapeutic intervention. Because this PPI is essential for the MLL-fusion mediated leukemogenesis, we believe that disruption of this interaction with small molecules will directly inactivate MLL fusion proteins and will represent an optimal approach for therapeutic intervention in MLL leukemias.
Inhibitors of the WDR5-MLL interaction
The WDR5-MLL interaction represents another example of PPIs relevant to the MLL fusion protein oncogenic transformation (see above) (81) and an attractive molecular target for small molecule intervention in MLL rearranged leukemias, WDR5 binds to the catalytic subunit of MLL with nanomolar affinity (Kd = 120 nM) using a 12 amino acid fragment called ‘WIN motif’ (113). Further studies identified a three amino acid fragment of MLL, Ac-ARA-NH2, which binds to WDR5 with the same potency as the catalytic subunit of MLL (114). The crystal structure of the MLL derived peptide in complex with WDR5 revealed that the Arg3765 side chain of MLL binds to a deep pocket on WDR5 and is involved in an extensive network of hydrogen bonds (113), providing the structural basis for targeting this PPI.
Two groups reported development of small molecule and peptidomimetic inhibitors of the WDR5-MLL interaction (81, 82, 115, 116). Small molecule inhibitors of WDR5-MLL interactions were identified by HTS of 16,000 diverse small molecules, resulting in WDR5-0101 (82). Exploration of commercially available analogs yielded WDR5-0103, with about 10-fold higher potency than the parent compound (Kd = 0.45 µM, IC50 = 3 µM) (82) (Fig. 1C). Structural studies of WDR5 with a close analog of WDR5-0103 showed that the piperazine moiety of the ligand fits into the pocket on WDR5 that accommodates the arginine residue from the ‘WIN motif’ (82). Importantly, WDR5-0103 was shown to inhibit catalytic activity of MLL, albeit with moderate affinity (IC50 = 83 µM), by antagonizing the interaction of WDR5 with MLL. More potent analogues, however, are required to fully assess the effect of small molecule inhibitors of WDR5 in cancer cells.
A potent class of peptidomimetics has been developed to block the WDR5-MLL protein-protein interaction (81, 116). The linear peptidomimetics, including MM-101 and MI-102 with very potent binding affinity to WDR5 (Kɨ < 1nM, IC50 < 3 nM), were developed first (116) and used for structure-based design of cyclic compound MM-401 (81)(Fig. 1C). The MM-401maintained high binding affinity to WDR5 (Kd < 1 nM) and very strong inhibition of the WDR5-MLL interaction (IC50 = 0.9 nM) due to a restricted conformation and additional hydrophobic contacts over MM-102, as validated by the crystal structure of the complex (81) (Figs 2–3). MM-401 was effective and selective in inhibiting the MLL1 histone methyltransferase activity in vitro, although with modest affinity (IC50 = 0.32 µM) (81). Importantly, MM-401 was shown to specifically inhibit the MLL-dependent H3K4 methylation at Hoxa9 loci in murine MLL-AF9 transformed cells and reduce expression of Hoxa9 and Hoxa10 genes (81). Furthermore, this compound selectively inhibited growth of MLL leukemia cells, induced apoptosis and differentiation of these cells, albeit relatively high concentration of MM-401 (>10 µM) was required to demonstrate cellular activity (81). Limited drug-like properties of peptidomimetics imply that MM-401would need to be re-designed to improve its cellular activity and validate the effect of WDR5-MLL inhibitors in in vivo models of MLL leukemia. Nevertheless, successful development of both small molecule and peptidomimetic inhibitors of the WDR5-MLL interaction imply that this PPI represents a druggable target and inhibition of this interaction can modulate enzymatic activity of MLL in hematologic malignancies.
The two examples of PPIs presented above demonstrate that multiple interactions of MLL with its protein partners (e.g. with menin or WDR5) could be disrupted by small molecules for potential therapeutic applications. This is a consequence of complexity of MLL and MLL fusion protein complexes, with numerous domains playing a role in leukemogenesis. Similar strategies have been proposed for targeting other multidomain proteins relevant in oncology, including EZH2 histone methyltransferase, which can be blocked by either inhibition of the enzymatic activity (117, 118) or by disruption of the EZH2-EED protein-protein interaction (119).
Inhibitors targeting core binding factor β
Development of small molecules targeting core binding factor β (CBFβ) in acute leukemias represents a successful example of disrupting PPIs involving two globular domains (43). The two subunits of the heterodimeric transcription factor CBF, core binding factor β (CBFβ) and Runx1 (CBF), are frequent targets of chromosomal translocations found in acute leukemias (120, 121). The CBFβ is the target of a common chromosomal translocation, inv(16), found in ~15% of AML cases (120), which results in a fusion of the N-terminal 165 amino acid fragment of CBFβ to the smooth muscle myosin heavy chain (SMMHC), leading to the expression of CBFβ-SMMHC fusion protein. CBFβ-SMMHC causes dysregulation of CBF function, and binding of Runx1 to CBFβ is required for dysregulation associated with the fusion protein (121, 122). Therefore, the protein-protein interaction between CBFβ and Runx1represents a valuable target for inhibition as a potential therapeutic strategy for acute leukemias associated with CBF rearrangements.
The interaction between CBFβ and Runx1 has been characterized in detail, including structural characterization of the heterodimer (Figs 3, 4C), its complex with DNA and structures of individual monomers (123, 124). The Runx1-CBFβ represents a high affinity interaction (Kd= 54 nM) (122), which involves two globular domains with a relatively large and flat binding interface devoid of well-defined pockets. The energetically important ‘hot spot’ residues have been mapped on this PPI interface using alanine scanning mutagenesis (125).
Efforts by Bushweller and colleagues (126) led to development of small molecule inhibitors targeting CBFβ to block its interaction with the Runt domain. Initial compounds were identified employing virtual screening based on the NMR structure of CBFβ, followed by experimental evaluation of selected hits by NMR (43). These efforts resulted in an aminothiazole class of compounds, which were validated for binding to CBFβ using NMR and demonstrated inhibition of the CBFβ-Runx1 interaction as measured by FRET-based and ELISA biochemical assays (43). Subsequent chemistry optimization yielded analogues with improved inhibitory activity, with the most potent compounds inhibiting the CBFβ-Runx1 interaction with low micromolar affinities (43)(Fig. 1D). Combination of NMR chemical shift perturbations and computational approaches was used to assess the binding mode of these compounds to CBFβ (43). Strikingly, these compounds bind to a novel allosteric site on CBFβ, offset from the CBFβ-Runx1 interface (Figs 2,3). Because these compounds do not bind directly at the PPI interface, their inhibitory effect occurs by means of an allosteric mechanism, likely via inducing conformational changes, which are transmitted through the protein to the residues at the CBFβ-Runx1 interface.
The allosteric inhibitors of CBFβ were shown to inhibit the CBFβ-Runx1 interaction in mammalian cells and inhibited cell proliferation, induced apoptosis and differentiation in the ME-1 human leukemia cell line expressing CBFβ-SMMHC (43). Finally, these compounds reduced Runx1 binding to DNA in cells, the interaction that is known to be enhanced by CBFβ. Overall, successful development of these compounds and their effect in functional assays suggest that CBFβ-Runx1 inhibitors might have a potential therapeutic value, although their further development is required to identify compounds suitable for in vivo studies in animal models of inv(16) leukemia. From the prospective of successful targeting of protein-protein interactions, this case provides an important example where allosteric inhibition might represent a feasible approach to target large and flat interfaces. More recently, another inhibitor of the CBFβ-SMMHC oncoprotein, AI-10-49, that disrupts its binding to Runx1 has been described in the abstract form (126). This compound has been reported to demonstrate both in vitro and in vivo efficacy in inv(16) leukemia models, but detailed information about this molecule is not available at this time.
Inhibitors of BET bromodomains
Small molecule inhibitors of the BET family of bromodomains represent a highly successful example of PPI inhibitors with strong therapeutic potential in hematologic malignancies, particularly in AML and multiple myeloma (84–86). The members of BET family of bromodomains, which include Brd2, Brd3, Brd4 and Brdt, bind chromatin to induce transcriptional activation (127, 128). The shRNA screening for different chromatin regulators identified Brd4 as a target protein in AML (84).
Bromodomains are known chromatin binding modules that recognize short peptides with acetylated lysine residues (129). Structural studies of Brd4 complex with the acetylated histone derived peptide revealed that the acetylated lysine binds to the relatively small and deep binding site on Brd4 (130, 131)(Figs 4A, 5C). To date, multiple potent inhibitors of the BET bromodomain family have been identified (129). Among these, JQ1, reported by Bradner and colleagues (128), is one of the most broadly used bromodomain inhibitors (Fig. 1E). JQ1 binds to the first and second bromodomain of Brd4 with Kd of 50 and 90 nM, respectively, and inhibits the Brd4 interaction with the acetylated histone H4-derived peptide with IC50 of 77 nM and 33 nM (128).The crystal structure of JQ1 in complex with the Brd4 bromodomains revealed that JQ1 binds to the acetylated lysine pocket and shows remarkable shape complementarity with this pocket (128)(Figs 2, 3, 5D). Furthermore, JQ1 mimics the interactions of the acetylated lysine from the acH4 peptide with Brd4, including the hydrogen bond with Asn140 (Fig. 5C,D). Pharmacologic activity of JQ1 was evaluated in multiple AML cell lines and AML patient samples, demonstrating strong anti-proliferative effect, with the GI50 values better than 500 nM (84). In addition, treatment with JQ1 resulted in myeloid differentiation of MLL-AF9 leukemia cells (84). Importantly, JQ1 delayed progression of MLL leukemia in vivo and significantly extended survival of MLL-AF9 leukemia mice. Furthermore, activity of JQ1 was demonstrated in experimental models of multiple myeloma in vivo, leading to a significant improvement in survival (86).
Inhibitors of BET bromodomains have also been actively developed by GlaxoSmithKline (85, 88, 132). Dawson et al. (85) have tested whether displacement of BET proteins from chromatin by a small molecule inhibitor may have a therapeutic role in MLL leukemias. They used I-BET-151 dimethylisoxazole small molecule inhibitor (Fig. 1E), which potently binds to and inhibits BET bromodomains (Kd of 20 nM and 100 nM for Brd3 and Brd4, respectively, and IC50 < 0.7 µM for both proteins), and has optimized in vivo pharmacokinetics (85). I-BET-151 showed potent and selective anti-proliferative effect in a panel of MLL leukemia cells, with the GI50 values below 200 nM. In vivo studies with I-BET151 in the MLL-AF4 and MLL-AF9 leukemia models showed strongly reduced leukemia progression and marked survival benefit. In the MLL primary patient samples, I-BET151 accelerated apoptosis and abrogated clonogenic efficiency (85). Taken together, bromodomain inhibitors demonstrate very promising activity in multiple hematological malignancies, demonstrating a therapeutic rationale for inhibiting this PPI. This exemplifies a great potential in targeting PPIs, and several BET bromodomain inhibitors are currently being evaluated in clinical trials (Table 1).
Small molecule inhibitors of BCL6
The BCL6 transcriptional repressor is the most frequent oncogene involved in diffuse large B-cell lymphoma (DLBCL) that is constitutively expressed in the majority of patients with aggressive B-cell lymphomas (133). Delivery of shRNA or peptide inhibitors of BCL6 kills DLBCL cells, validating BCL6 as an attractive therapeutic target (133–135). BCL6 is a member of the BTB/POZ family of transcription factors, which recruits SMRT, N-CoR, and BCOR co-repressors (135). Several crystal structures of BCL6 BTB domains bound with approximately 17 amino acid long co-repressor derived peptides were reported (136, 137). Co-repressors bind to the lateral groove formed at the interface of the BTB domain dimer, and this binding occurs with low micromolar affinities, which is relatively weak for PPIs (136, 137).
Melnick and colleagues (87) applied computational methods to identify small molecules that bind to the BCL6 BTB domain at the co-repressor interaction site and inhibit this protein-protein interaction. The step-wise procedure, including virtual screening searches of a library of ~1 million compounds followed by experimental evaluation of 100 best scored hits, was applied to identify initial lead compounds. One class of small molecules, with the most potent compound 79-6 (Fig. 1F), was rigorously characterized, using both in vitro and functional assays. Direct binding of 79-6 to the co-repressor interaction site on BCL6 was validated using both NMR spectroscopy and X-ray crystallography (87)(Fig. 1). The indazoline ring anchors 79-6 in the binding groove on BCL6 BTB, while the carboxyl tail becomes solvent exposed (Figs 2,3). The NMR studies revealed relatively weak binding affinity of 79-6 to BCL6 (Kd = 138 µM), further supported by the fluorescence polarization data, demonstrating IC50 value of 212 µM for inhibition of the BCL6-SMRT derived peptide interaction (87). Despite weak in vitro activity, 79-6 was shown to disrupt recruitment of N-CoR or SMRT to BCL6 at the atr promoter in DLBCL cells, thus affecting BCL6 transcriptional complexes, and altered expression of BCL6 target genes in DLBCL cell lines dependent on BCL6. Interestingly, 79-6 was found to accumulate in cancer cells and selectively kill BCL6-dependent DLBCL cells, with the GI50 values at middle to high micromolar range (87). The 79-6 compound showed favorable pharmacokinetic profile (PK) and was tested in the xenograft models of DLBCL, demonstrating significant inhibition of tumor growth in the BCL6 dependent but no effect in the BCL6 independent xenografts. Taken together, the protein-protein interaction between BCL6 and co-repressors represents a druggable target, and targeting this PPI with small molecules might lead to an effective anti-lymphoma strategy. The 79-6 compound would require, however, further optimization to develop compounds with optimized potency and improved drug-like properties for more advanced pre-clinical and potentially clinical evaluation.
Targeting protein-protein interactions: lessons learned
How to identify inhibitors of protein-protein interactions?
Successful identification of the initial lead molecule that can bind to the protein at the PPI interface represents a major difficulty in the drug discovery campaign. In contrast to enzymes, protein-protein interactions do not have a natural small molecule-like ligand to substitute with a low molecular weight compound. Nevertheless, the success in identifying small molecule inhibitors of PPIs over the last decade highlights different approaches to identify PPI modulators, which can be classified as: (i) HTS of synthetic chemical libraries and natural products, (ii) peptidomimetic approaches, (iii) biophysical and structural biology methods (NMR and X-ray crystallography), and (iv) computational structure-based drug design approaches (18). The first two approaches have proven most successful in identification of PPI modulators (18), possibly due to limited structural information available for PPIs (16). Multiple reviews have described these approaches in detail, providing specific examples (24, 138–141). Here, we intend to highlight the methods used for identification of PPI inhibitors targeting hematology-related protein targets described above and refer to the key aspects that are likely to guide discovery of new PPI modulators.
High throughput screening
High throughput screening of synthetic compounds and natural product libraries represents a traditional approach in drug discovery, which has been successfully applied to the discovery of lead compounds for enzymatic protein targets. A growing number of examples demonstrates that HTS also represents an effective approach for lead discovery of PPIs (18, 78, 80, 142). This approach is particularly valuable when structural information on the protein target is not available, limiting application of structure-based design or peptidomimetic approaches (18). The success of HTS in identification of PPI inhibitors depends on multiple factors. First, the ability of a particular PPI interface to be disrupted by a small molecule (druggability), a property difficult to predict until such compounds are identified or detailed structural analysis of the PPI interface is performed. Second, small molecule compound libraries with a high degree of molecular diversity and complexity are more desired to block PPI interfaces than simply large size compound libraries typically used for HTS (18). Finally, the choice of biochemical assay for identification of PPI modulators, which is in most cases a competitive binding assay, is important for successful HTS, with fluorescence-based screening assays playing a prevalent role (78, 80, 143–145).
While for the majority of PPIs more than one biochemical assay can be developed, providing a choice for HTS, selection of the appropriate compound library represents a much bigger limitation. HTS screening collections are typically limited to commercial libraries, particularly in academic drug discovery centers, which might not sample the appropriate chemical space for non-classical drug targets, such as PPIs (58, 146, 147). The majority of current HTS libraries represent a relatively small number of chemical scaffolds and include molecules resembling existing drugs (146). Inhibitors of PPIs, however, tend to be more complex (e.g. higher molecular weight, high number of stereocenters, macrocycles) than enzyme inhibitors (59). It has been postulated that diversity-oriented synthesis (DOS), which results in compound libraries with increased scaffold complexity (148, 149), and natural product libraries (58, 150, 151), might be more suitable for identification of PPI modulators.
Despite these limitations, HTS has been successfully applied to develop multiple PPI inhibitors, including our own efforts to identify small molecule inhibitors of the menin-MLL interaction (Fig. 1B), as described above (78, 80). The crystal structure of menin was not available when HTS was performed, and therefore druggability of this PPI interface could not be evaluated at that time. Biophysical and biochemical characterization of the menin-MLL interaction demonstrated that this is a protein-peptide type of interface (109). We performed two independent HTS screens, with total ~330,000 compounds tested to identify menin-MLL inhibitors (78, 80, 152). The striking result from both screens was a very low hit rate (< 0.08%) as compared to the typical range of 0.1%–5% hit rate in HTS for classical drug targets. Therefore, despite a relatively large size of the screening collection and non-stringent criteria applied for hit selection (IC50 < 200 µM), only ~20 small molecule inhibitors of the menin-MLL interaction were identified from HTS (78, 80).
Another example of successful HTS for PPIs in hematologic malignancies is identification of small molecule inhibitors of the WDR5-MLL interaction (82). Biochemical screening of a library of 16,000 diverse small molecules resulted in only one compound with IC50 better than 60 µM (WDR5-0101), demonstrating again a very low hit rate as compared to the HTS with classical drug targets. Overall, these examples demonstrate that HTS can be successfully applied to identify new PPI inhibitors, although a low number of hits is expected. Results will vary, however, depending on the nature of the PPI interface to block and libraries applied for screening, as discussed above.
Peptidomimetic approaches
Peptides and peptidomimetics derived from PPI interfaces have potential to serve as leads for development of PPI modulators (18). On the other hand, peptides and peptidomimetics might have drawbacks in drug discovery, due to large molecular weight, conformational flexibility, limited cellular permeability and proteolytic liability. Regardless of these obstacles, a number of successful examples of peptide derivatives and peptidomimetics have been reported (18, 153, 154). Inhibitors of the WDR5-MLL interaction represent a successful example of peptidemimetic approaches applied for PPI relevant to acute leukemia, with both linear and cyclic peptidomimetics developed using structure-based design (81, 116) (Fig 1C), as described above in detail. Despite a very potent in vitro activity (e.g. IC50 < 1nM for MI-401), modest cellular activity of this compound (>10 µM concentration required for activity in MLL leukemia cells) demonstrates limited applicability and/or the need for extensive optimization of peptidomimetics in drug discovery projects.
Computational structure-based drug design
Computational approaches can assist different stages of drug discovery process (18, 155). In the lead identification process, virtual screening of large databases of compounds based on the crystal or NMR structures of proteins have proven successful in reducing the size of compound collection for experimental evaluation (18, 156). Relatively limited success of computational approaches to lead identification for PPIs often arises from limited structural information available for PPI interfaces (16), flexibility of amino acids at the interfaces, and the need for more accurate computational methodologies. Despite these challenges, successful examples of applying computational methods in lead identification for PPIs have been reported (157, 158). One of the examples supporting the utility of this approach is identification of small molecule inhibitors of the BCL6 protein identified by virtual screening searches at the co-repressor binding interface (87). One class of compounds, represented by 79-6 (Fig. 1F), was validated for binding to BCL6 (Fig. 2), and demonstrated activity in multiple in vitro and in vivo models as described above. This example validates that computer-aided drug design methods can be successfully used for lead identification, albeit the activity of resulting hits might not be very high, as demonstrated for BCL6 modulators (IC50 > 100 µM) (87).
Biophysical methods in lead discovery
Biophysical and structural biology methods play a major role in identifying molecules that bind to proteins via direct measurement of the binding event. Different biophysical methods have been applied to lead identification, including NMR, isothermal titration calorimetry (ITC), and surface plasmid resonance (SPR) (58, 159). NMR has been particularly valuable for detection of direct ligand binding to the protein as it provides a possibility of mapping the ligand binding site on the protein structure (58). This has been exemplified by discovery of small molecule inhibitors of CBFβ, which were found to bind to the new allosteric site on the protein, as assessed by mapping the NMR chemical shift perturbations on the CBFβ structure (43). Therefore, NMR is an extremely valuable method not only for identification of new ligands but also for mapping ligand binding sites on proteins and de novo binding site identification in targets where allosteric site is desired.
Fragment-based drug discovery
Fragment-based drug discovery (FBDD) is a relatively novel approach used both in the pharmaceutical industry and academia for identification of new leads for previously intractable biological targets, including PPIs (22, 160, 161). FBDD relies on identification of small molecular weight ligands (< 250 Da) that directly bind to the protein target by screening of relatively small compound libraries (up to few thousand compounds). Fragment screening libraries sample chemical space much more effectively than HTS libraries, as hit rates typically observed in FBDD are 10–1,000 times higher than in conventional HTS screens (160, 162). The fragment-based approach requires sensitive screening methods, such as NMR spectroscopy, SPR or thermal shift assay, which allow identification of hits with weak affinities (high micromolar to milimolar range) (163). Fragment hits are subsequently extensively modified using medicinal chemistry guided by structural biology to improve their potency and pharmacological properties. FBDD has been validated as a valuable drug discovery approach, and Vemurafenib, a small molecule inhibitor of BRAF kinase, was the first approved drug developed using this approach (164).
One of the most impressive stories in applying FBDD for lead identification and optimization for protein-protein interactions is discovery of Bcl-2 family inhibitors: ABT-737 and ABT-263 (Fig. 1). By applying a ‘SAR by NMR’ strategy and screening few thousand fragment-like compounds, Abbot identified very weak (Kd > 300 µM) fragments that bind to the adjacent sites on the Bcl-xL protein (165). Extensive medicinal chemistry efforts supported by structural biology studies resulted in ABT-737 and ABT-263, with Ki < 1nM (166, 167), both of which are currently in clinical trials (Table 1). More recently, a fragment-based approach has been successfully applied to lead identification for other PPIs, including Mcl-1 (168) and XIAP (169), which illustrates a strong potential of FBDD to access new regions of chemical space required for PPI inhibitors (170).
Is a crystal structure required for successful targeting of protein-protein interactions?
Availability of structural information on a protein target is invaluable even before a small molecule development program is initiated, and it is even more critical at the stage of lead identification and optimization. In an ideal situation, the structure of a target protein alone and in complex with the protein partner is desired to analyze the PPI interface and account for potential conformational changes to address protein dynamics at the interface. Detailed analysis of the protein-protein interfaces facilitates a druggability assessment to evaluate the potential for identification of small molecule modulators of the PPI before screening is initiated. Such studies are typically focused on analysis of the PPI interface with respect to the size and presence of binding pockets, which could possibly be occupied by small molecules.
Availability of the crystal structure of a protein target is also very beneficial during lead identification process, in particular when NMR is used as a screening method. The NMR chemical shift perturbations observed upon ligand binding can be mapped on the protein structure to validate direct binding and identify ligand binding site, as demonstrated for compounds targeting CBFβ, where an unexpected allosteric site was identified (43). Furthermore, hits identified using other screening methods, e.g. HTS, can be validated by NMR for their direct binding to the target protein and their binding sites can be mapped onto the protein structure.
The crystal structures of the protein and ideally protein-ligand complexes are essential for successful lead optimization to efficiently direct medicinal chemistry efforts and develop potent inhibitors with optimized drug-like properties. This requirement is even stronger in the context of inhibitors targeting PPIs due to the lack of a natural small molecule modulator that binds to the same site on the protein and could be mimicked by a small molecule inhibitor. In our own studies availability of the crystal structures for the menin-inhibitor complexes enabled rapid optimization of two classes of compounds, resulting in development of compounds with nanomolar binding affinities(79, 80)(Figs 1B,5B). Similarly, for peptidomimetic inhibitors of the WDR5-MLL interaction, extensive structure-based design led to identification of very potent compounds (Ki < 1nM) (81, 116)(Fig. 1C). Other examples include inhibitors of Bcl-2 family of proteins, ABT-737 and ABT-263 (166, 167) (Fig. 1A). It is important to point out that structural data for protein-ligand complexes is critical for both optimization of potency and other drug-like properties (e.g. solubility, PK profile) to recognize which sites on the ligand molecule can be modified or substituted to modulate these properties.
Which PPI interfaces are druggable?
Despite an increasing number of successful examples of PPI inhibitors there is an emerging picture that not all interactions are amenable to disruption by small molecules. Analysis of the successful examples for hematological targets reveals that PPI most amenable to inhibition by small molecules are protein-peptide type of interactions (Fig. 4A,B). This type of PPIs is typically characterized by a presence of well-defined concave binding sites at the PPI interface and binding pockets with hydrophobic amino acids (51, 52)(Fig. 5). Protein-peptide interactions are more amenable to disruption by orthosteric inhibitors that bind to the peptide binding site on the target protein (Fig. 6). Significantly more challenging are PPIs involving globular domains (Figs 4C,6), and the most unapproachable are PPIs composed of intrinsically disordered proteins, which undergo coupled folding upon binding to a protein partner (Fig. 6). Below we briefly summarize features of different PPIs and provide successful examples of their targeting by small molecule inhibitors.
Fig. 6. Difficulty level in targeting PPIs.
Short peptide-domain complex is represented by Brd4-acH4 complex (3UVW), long peptide- domain complex is represented by Bcl-2-BAX (2×A0), domain-domain complex is represented by CBFβ-Runx1 (1H9D), and complex of intrinsically disordered proteins is exemplified by AF9-AF4 (2LM0). Target proteins are shown in gray, and binding partners are shown in different colors.
Protein-peptide interactions: short peptide fragment binds to a small and deep pocket
In this type of PPIs, short fragment of a protein partner encompassing 6–8 amino acids binds to well-defined and deep pocket located on a globular domain of a target protein. Examples include WDR5-MLL interaction (113) and binding of acetylated histone peptides with BRD4 bromodomain (131)(Figs 3, 4A, 5A). The contact surface formed between peptide and binding site is relatively small in this type of PPIs, approximately 1200–1400 Å2 (Fig. 4A), and there is typically a key residue within the peptide essential for high affinity interaction, which inserts into a main binding pocket and mediates key contacts with the protein partner. For example, an arginine side chain of MLL plays such a critical role to form a stable complex with WRD5 (113). Contacts with the backbone are less extensive in such complexes, and it has been shown that three amino acid long peptide derived from MLL is sufficient to maintain a high affinity interaction with WDR5 (114). Bromodomains are also known to bind short (~8 amino acid long) peptides and recognize one or two acetylated lysine residues (171)(Fig. 5C). Capability to bind short peptides with a molecular weight of 600–800 Da suggests that such proteins could also bind effectively small molecules (Fig. 6). Indeed, this type of PPIs has been recognized as a particularly amenable target for inhibition by small molecules (129). Domains that recognize short peptides are abundant in chromatin binding proteins (e.g. PWWP, MBT, tudor, PHD domains), strongly suggesting a wealth of ‘druggable’ PPI interfaces in the human genome.
Protein-peptide interactions: long peptide fragment binds to the large site on globular domain
In this type of PPI a much longer peptide, typically encompassing 12–30 residues, binds to a globular domain. Within examples described above this type of PPI is represented by complexes: menin-MLL (109), Bcl-2 with BH3-domains (172) and BCL6 with SMRT, BCOR co-repressors (136, 137)(Fig. 4B). In contrast to protein-short peptide interactions, here the binding site is significantly larger and spreads over a large part of a protein target, with multiple pockets involved in the interactions. The interface is more complex and buries a large surface area, typically exceeding 1000 Å2 (Fig. 4B). The peptide motif in one of the binding partners is unstructured when free in solution and undergoes folding upon complex formation, frequently adapting -helical or β-strand conformation (45). PPIs involving larger surface contacts are more prone to induce conformational changes and form new pockets, as observed for the Bcl-2 family of proteins (63). Development of small molecule inhibitors targeting these interactions benefits from accurate mapping of ‘hot-spot’ residues in order to identify key sites, which could be occupied by a small molecule inhibitor (55)(Fig. 6). Examples include menin-MLL inhibitor MI-2-2, which closely mimics the key interactions of MLL with menin (79)(Fig. A,B). High affinity synthetic inhibitors targeting these interfaces are typically complex molecules with elongated shape, capable of filling out multiple pockets, and with a larger molecular weight than classical drugs (61), as exemplified here by inhibitors targeting Bcl-2 (Figs 1A, 2,3).
Interfaces involving globular domains
Much less progress has been made in targeting interactions involving globular domains. This type of PPIs is usually characterized by a large and flat interface and lack of well-defined pockets, making it very challenging to disrupt by small molecules. The absence of pockets at PPI interfaces, however, does not discriminate from identification of small molecules as conformational dynamics of proteins may induce formation of transient pockets at the interfaces (65). Furthermore, cryptic pockets not present in the crystal structures of the apo protein may occur at allosteric sites (64). In the context of hematology related targets, the heterodimeric transcription factor CBFβ-Runx1 (Figs 3,4) represents a successful example, where small molecule inhibitors that bind to the allosteric site on CBFβ were identified (43)(Figs 1D,3,4). These compounds bind, however, to a relatively small pocket on CBFβ (Figs 2,3) and have moderate activity (IC50 ~ 1µM). It remains to be shown whether more potent inhibitors of this interaction can be developed. Overall, the absence of deep pockets does not exclude PPIs from initiating small molecule inhibitor development, it does, however, indicate a greater challenge to successfully identify such compounds (Fig. 6).
PPIs involving intrinsically disordered proteins
Intrinsically disordered proteins (IDPs), which undergo coupled folding upon binding to protein partner, belong to one of the most challenging targets for drug discovery. Approximately 25% of mammalian proteins are predicted to be fully disordered, and half of all proteins contain disordered regions longer than 30 amino acids (173). IDPs participate widely in the PPI network (49) and represent attractive targets for inhibitor development. However, it has been recognized that designing drugs targeting IDPs remains a big puzzle (174). These proteins represent a particular challenge because there are no available computational methods to predict binding of small molecule ligand to a disordered protein (174).
A well characterized example of PPIs involving IDPs in hematological malignancies involves AF4 and AF9 proteins, which are among the most common translocation partners of MLL found in patients with acute leukemias (175). It has been demonstrated that disruption of the AF4–AF9 protein-protein interaction by a short AF4-mimetic peptide in cells harboring MLL translocations results in cell death (176). Therefore, inhibition of the AF4–AF9 interaction may represent an attractive therapeutic target for MLL leukemias. Recent structural studies showed that the AHD domain of AF9 and an interacting fragment of AF4 are intrinsically disordered when free in solution and adapt a well–defined structure as an AF4–AF9 complex (48) (Fig. 6). Such coupled folding results in burying an extensive hydrophobic interface between the two proteins and leads to formation of a very stable complex. A small molecule inhibitor capable of disrupting such a complex would need to bind to the unstructured AHD domain of AF9, and either form a stable complex with AF9 or stabilize the disordered protein to prevent binding of AF4. Development of small molecules targeting such PPIs represents a very difficult task (Fig. 6), and feasibility of identifying such compounds would need to be demonstrated. One possibility to block this PPI would be to develop peptidomimetics derived from the AF4 structure.
When to move from lead identification to a drug discovery program?
Validation of hits identified by screening compound libraries for their direct binding to the target protein represents the first step in selection of lead compounds for further optimization. This step is critical as many screening methods, particularly HTS, provide a large number of false positives (143, 177, 178). Multiple biophysical methods are applied for validation of direct binding of screening hits to proteins (159, 179–181). Typically less than 30% of HTS hits identified for PPIs are confirmed for their direct binding to the target protein. Once validated, the most promising lead compounds are selected for medicinal chemistry optimization, based on their activity, chemical tractability and physicochemical properties. Structural information on the protein-ligand complex is critical in lead optimization (see above).
Validated screening hits targeting PPIs typically have in vitro activity at the micromolar range and, therefore, substantial optimization of these compounds is required to initiate cell-based studies. For example, for the menin-MLL inhibitors at least 50-fold improvement in the in vitro activity of the HTS hits was required to achieve IC50 ~ 50nM, before these compounds demonstrated relatively potent activity and selectivity in cell-based assays (79, 80). In addition to in vitro potency, optimization of physicochemical properties, including solubility and cellular permeability, are equally important. For example, very potent (Ki < 1nM) peptidomimetics were developed for the WDR5-MLL1 interaction (81, 116), however their effect in MLL leukemia cells is rather modest likely due to limited cellular permeability. This demonstrates that very potent in vitro inhibition of PPIs by small molecules does not guarantee a strong effect in biological experiments, and optimization of drug-like properties is typically required before compounds demonstrate activity in cells and animal models of diseases.
The question to answer in a lead optimization process is when to move from lead identification to a drug discovery program focused on development of compounds with in vivo efficacy and eventually a clinical candidate? Comprehensive validation of the mechanism of action of a new PPI inhibitor, both in vitro and in relevant cellular assays, are required to consider investing substantial resources into further optimization of a particular lead class within a drug discovery project. If supported, the next step typically includes efficacy evaluation of compounds in the appropriate animal models. To reach this stage, potent cellular activity (GI50 at submicromolar level or better), favorable pharmacokinetic profile (PK) and low toxicity are required. In many drug discovery programs extensive medicinal chemistry efforts are necessary to develop compounds with the desired drug-like properties and satisfactory in vivo effect in animal models, as exemplified for PPI inhibitors of Bcl-2 family of proteins (166, 167), BET bromodomains (88) and in our own efforts with optimization of the menin-MLL inhibitors (not shown). An exception is the Bcl-6 inhibitor, 79-6, which demonstrates significant in vivo efficacy in the Bcl-6 dependent xenografts of lymphoma, despite relatively weak cellular activity (GI50 at middle to high micromolar level), likely due to accumulation of the compound in the tumor samples (87).
How far we can pursue drug discovery programs in academia?
Over the past decade the contribution of academic research to drug discovery has been significantly increased, as recently reviewed in a number of articles (182–185). The role of academic laboratories is particularly valuable in drug discovery for rare and neglected diseases (186), which receive less industrial attention. Furthermore, novel and more risky drug targets not yet validated clinically, such as the majority of PPIs, are also primarily explored in academic laboratories. Academic researchers can contribute to different stages of the pre-clinical drug discovery process (184, 186). Historically, academic laboratories have a great record of success with target identification and validation. More recently, substantial investment has been provided to academic institutions to set-up early stage drug discovery platforms, such as HTS screening capabilities, both at NIH (MLPCN) (https://mli.nih.gov/mli/mlpcn/) and at individual academic institutions (152). Furthermore, some academic centers have created core facilities for medicinal chemistry, PK and animal studies, all of which are critical to successfully pursue drug discovery projects (183). Our own efforts with developing PPI inhibitors strongly support the model of utilizing different core facilities available to academic laboratories. Collaboration with the HTS screening center and other drug discovery oriented core facilities available at the University of Michigan (152) was essential to the successful development of menin-MLL inhibitors (78–80).
Academic drug discovery research is, however, limited by multiple factors. First, such projects require substantial resources, especially at more advanced pre-clinical stages where extensive animal studies and outsourced assays to assess drug-like properties of compounds need to be conducted. Classical research grant funding might not be sufficient to efficiently pursue such studies to identify candidates for advanced pre-clinical or clinical work. This limitation can sometimes be overcome by additional drug discovery supporting mechanisms available from the National Institutes of Health (NIH), such as the NEXT (NCI Experimental Therapeutics Program) program (http://next.cancer.gov/), which allows Investigators to advance potential therapies by accessing resources within NIH to facilitate drug development. In the context of hematologic malignancies, the role of the Leukemia and Lymphoma Society Therapy Acceleration Program (www.lls.org) is invaluable to help with advancing very promising drug discovery projects to clinical stages.
Another limitation in academic drug discovery projects might be a limited access to specific drug discovery resources, such as appropriate screening libraries for HTS (discussed above). Again, the support from NIH, such as access to the NIH MLPCN screening collection of over 300,000 compounds, represents an alternative to partly address this problem. Indeed, by screening the NIH compound collection we identified several novel chemical scaffolds as menin-MLL inhibitors (80). An additional limitation in academic drug discovery is the need for very extensive medicinal chemistry optimization of lead compounds to optimize their drug-like properties and develop compounds appropriate for efficacy studies in animals, with optimized PK and ADME (Adsorption, Distribution, Metabolism, Excretion) properties. To overcome these limitations and also to successfully move towards more advanced pre-clinical stages and eventually to the clinic, a partnership with pharmaceutical or biotech companies might be required.
Taken together, drug discovery projects can be successfully pursued in academic institutions, especially at the earlier stages of pre-clinical development. For novel and more challenging drug targets, such as the majority of PPIs in oncology and hematology, comprehensive target validation studies and in vivo proof-of-principle studies might be required to attract industrial attention. Despite all difficulties and limitations, we expect that academic drug discovery research will play a very significant role, particularly in the area of challenging new targets like PPIs.
What has changed in targeting PPIs?
Drug-like small molecule inhibitors of protein-protein interactions have been, in general, considered very difficult to develop (23–25, 187). Until recently, the potential of successful development of such compounds was viewed with much skepticism. It was believed that targeting PPI interfaces is too difficult due to: (i) unfavorable topology of PPI interfaces, (ii) lack of natural small molecules that bind to PPI interfaces, (iii) lack of appropriate screening libraries (23, 187, 188). However, the last several years have resulted in multiple examples of small molecule inhibitors targeting new PPIs (18, 22, 25, 156, 189). Importantly, several PPI inhibitors have already reached clinical trial stages (22) (Table 1), revealing that drug-like properties of these compounds can be optimized despite the larger molecular weight and more hydrophobic nature of these compounds as compared to existing drugs. The progress in targeting PPIs with small molecules is in part associated with better understanding of ‘druggable’ PPI interfaces (see above). Structural studies on PPI interfaces and co-crystal structures of small molecule inhibitors in complex with target proteins provided important guidelines which PPIs are more tractable for inhibition (Fig. 6). Repertoire of methods used in drug discovery has been expanded by fragment based approach, which is particularly suitable for targeting more difficult PPIs, as exemplified by development of inhibitors of Bcl-2 family that are currently evaluated in clinical trials (161). Besides, discovery of new druggable classes of PPIs with a strong link to human diseases, such as bromodomains, increases optimism about feasibility of successful targeting of PPIs.
The overall conclusion from the studies performed within the last 10–15 years in the field of PPI inhibition proved that they can be blocked by small molecules but are in general more challenging than classical drug targets. Moreover, not all PPIs are equal in this regard, as some are more tractable for inhibition by small molecules than others, with inhibitors already in clinical trials (Table 1) or at advanced pre-clinical stages (see above), providing a big opportunity for future therapies. Nevertheless, successful development of these compounds provides a substantial promise to hematology patients and strong foundation to the researchers to continue development of PPI inhibitors. Detailed characterization and analysis of the interfaces at the molecular level is crucial for selection of PPIs with the highest likelihood of success for a new drug discovery program.
PPI inhibition: questions to answer
Despite recent progress in targeting PPIs, multiple questions still remain to be answered. One of the critical questions is whether PPI inhibitors can be advanced to achieve the level of potency and selectivity, as well as other properties, including cellular permeability and pharmacokinetic profile, required for drugs. Some of the examples discussed above, including Brd4, Bcl-2 family, and MDM2 inhibitors that have reached the stage of clinical trials (Table 1), suggest that this goal can be achieved. Other PPI inhibitors described here (Fig. 1) are still at the stage of pre-clinical optimization, and more efforts are required to address this question. The success in advancing PPI inhibitors to drug molecules might depend on both druggability of a particular PPI interface and the chemical nature of compounds selected for optimization. In general, small molecules are more amenable to optimization of drug-like properties than peptides and peptidomimetics, although the success depends on the chemical scaffold.
Another important question is what chemical space corresponds to the effective blocking of PPIs? With the increasing number of successful examples of PPI inhibitors, especially those that have entered the clinic, it became evident that the molecular weight of PPI inhibitors is much larger (Mw > 500 Da) and they are more hydrophobic (clogP > 4.0) than classical drugs (20), and therefore most of them do not fit into the Lipinski’s ‘Rule-of-Five’ for orally bioavailable drugs (190). Despite these differences, successful optimization of these compounds to a clinically acceptable profile was possible, demonstrating that new rules might apply to PPI inhibitors with regard to their size and physicochemical properties (20).
An additional question is whether inhibiting PPIs would present a therapeutically useful potential and advantage over classical drugs such as enzyme inhibitors. Promising pre-clinical and early stage clinical data for compounds described above, as well as clinical use of PPI inhibitors blocking tubulin polymerization and the mTOR-FKBP12 interaction (22), demonstrate that PPI inhibition can be therapeutically beneficial. However, more clinical examples are required to prove the therapeutic value of this approach, and the next several years should provide such data for PPI inhibitors currently in clinical trials (22).
The remaining question associated with targeting PPIs is how to predict druggability of new protein-protein interfaces. Structural analysis of PPI interfaces can be particularly helpful in druggability assessment (20, 25), as discussed above in details, for development of orthosteric inhibitors. It is also important to consider allosteric inhibition as an alternative solution to overcome limitations of PPI interfaces less predisposed to inhibition by small molecules.
Future of targeting PPIs for therapeutic applications
The increasing number of new PPI inhibitors being discovered and success with advancing these compounds into the clinic, implies that small molecule inhibitors of protein-protein interactions will remain an attractive opportunity for developing new therapeutics in the next decade. Given the size of the human interactome with the estimated number of PPIs reaching ~650,000 (27), many of which are already known to play a critical role in human diseases (13), it becomes evident that many new PPIs will be explored for targeting by small molecules. Due to challenges and higher risk associated with targeting new PPIs, academic research will likely play an important role in drug discovery for protein-protein interfaces. In fact, the majority of PPI inhibitors discussed here were developed in academia. Once the critical proof of concept regarding druggability of a new PPI and its essential role in the diseases progression are established, attention from pharmaceutical industry will become more apparent.
The growing number of successful examples of small molecule inhibitors of PPIs and an increase in structural data for disease relevant PPIs will be very helpful in predicting, which interfaces are more tractable for drug discovery. The success in targeting PPIs within the next several years will also depend on advancement in protein structure determination and in silico screening methods. Broader application of biophysical methods to eliminate false positives from screening assays (18, 179, 180, 191) will also have to grow. The impact of more recent methodologies, such as fragment-based screening applied to PPIs (168, 192, 193), is expected to increase in the future with new libraries developed and advancement in methods used for compound screening. Design and preparation of appropriate compound libraries, both for HTS and fragment-based screening, will also need to be addressed based on the growing number of PPI inhibitors to incorporate structural features present in these compounds.
In lead optimization for PPI targets we will have to diverge from the drug-like properties reflected by the ‘Rule-of-Five’ (190), as already exemplified for inhibitors of the Bcl-2 family (Table 1). The lead optimization process will likely be fairly challenging to properly manage the pharmacokinetic and ADME properties of larger compounds such as PPI inhibitors for advancing them to the clinic (18). In summary, despite challenges highlighted here, a combination of existing knowledge on successful PPI inhibitors and advances in technologies applied to develop such compounds within the past decade should facilitate discovery of new PPI modulators for treatment of cancer and other diseases, including hematologic malignancies. The fact that a number of small molecule inhibitors of PPIs are currently in clinical trials provides hope that more compounds will advance to this stage and might eventually be approved for clinical use.
Acknowledgements
This work was funded by the National Institutes of Health grant R01 (1R01CA160467) to J.G., a Leukemia and Lymphoma Society (LLS) TRP grants (6116-12 to J.G. and 6111-14 to T.C.), LLS Scholar (1215-14) to J.G., American Cancer Society grants (RSG-11-082-01-DMC to T.C. and RSG-13-130-01-CDD to J.G.), and the Department of Pathology, University of Michigan. The authors thank members of the Grembecka’s and Cierpicki’s groups for critical reading of this manuscript.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Moore AS, Kearns PR, Knapper S, Pearson AD, Zwaan CM. Novel therapies for children with acute myeloid leukaemia. Leukemia. 2013;27:1451–1460. doi: 10.1038/leu.2013.106. [DOI] [PubMed] [Google Scholar]
- 2.Napper AD, Watson VG. Targeted drug discovery for pediatric leukemia. Front Oncol. 2013;3:170. doi: 10.3389/fonc.2013.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tasian SK, Pollard JA, Aplenc R. Molecular Therapeutic Approaches for Pediatric Acute Myeloid Leukemia. Front Oncol. 2014;4:55. doi: 10.3389/fonc.2014.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown P, Hunger SP, Smith FO, Carroll WL, Reaman GH. Novel targeted drug therapies for the treatment of childhood acute leukemia. Expert Rev Hematol. 2009;2:145. doi: 10.1586/ehm.09.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niewerth D, Dingjan I, Cloos J, Jansen G, Kaspers G. Proteasome inhibitors in acute leukemia. Expert Rev Anticancer Ther. 2013;13:327–337. doi: 10.1586/era.13.4. [DOI] [PubMed] [Google Scholar]
- 6.Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–2085. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- 7.Giannini G, Cabri W, Fattorusso C, Rodriquez M. Histone deacetylase inhibitors in the treatment of cancer: overview and perspectives. Future Med Chem. 2012;4:1439–1460. doi: 10.4155/fmc.12.80. [DOI] [PubMed] [Google Scholar]
- 8.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 11.Meldi KM, Figueroa ME. Epigenetic deregulation in myeloid malignancies. Transl Res. 2014 doi: 10.1016/j.trsl.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Ofran Y, Rowe JM. Genetic profiling in acute myeloid leukaemia--where are we and what is its role in patient management. Br J Haematol. 2013;160:303–320. doi: 10.1111/bjh.12135. [DOI] [PubMed] [Google Scholar]
- 13.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 16.Kar G, Gursoy A, Keskin O. Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol. 2009;5:e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryan DP, Matthews JM. Protein-protein interactions in human disease. Curr Opin Struct Biol. 2005;15:441–446. doi: 10.1016/j.sbi.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Zinzalla G, Thurston DE. Targeting protein-protein interactions for therapeutic intervention: a challenge for the future. Future Med Chem. 2009;1:65–93. doi: 10.4155/fmc.09.12. [DOI] [PubMed] [Google Scholar]
- 19.Arkin M. Protein-protein interactions and cancer: small molecules going in for the kill. Curr Opin Chem Biol. 2005;9:317–324. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Morelli X, Bourgeas R, Roche P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I) Curr Opin Chem Biol. 2011;15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- 21.Peloquin GL, Chen YB, Fathi AT. The evolving landscape in the therapy of acute myeloid leukemia. Protein Cell. 2013;4:735–746. doi: 10.1007/s13238-013-3057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nero TL, Morton CJ, Holien JK, Wielens J, Parker MW. Oncogenic protein interfaces: small molecules, big challenges. Nat Rev Cancer. 2014;14:248–262. doi: 10.1038/nrc3690. [DOI] [PubMed] [Google Scholar]
- 23.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 24.Fry DC, Vassilev LT. Targeting protein-protein interactions for cancer therapy. J Mol Med (Berl) 2005;83:955–963. doi: 10.1007/s00109-005-0705-x. [DOI] [PubMed] [Google Scholar]
- 25.Arkin MR, Whitty A. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr Opin Chem Biol. 2009;13:284–290. doi: 10.1016/j.cbpa.2009.05.125. [DOI] [PubMed] [Google Scholar]
- 26.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stumpf MP, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Daly RJ. Targeting the human kinome for cancer therapy: current perspectives. Crit Rev Oncog. 2012;17:233–246. doi: 10.1615/critrevoncog.v17.i2.70. [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123:207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 30.Gambacorti-Passerini CB, Gunby RH, Piazza R, Galietta A, Rostagno R, Scapozza L. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol. 2003;4:75–85. doi: 10.1016/s1470-2045(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 31.O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations, drug resistance, and the road to a cure for chronic myeloid leukemia. Blood. 2007;110:2242–2249. doi: 10.1182/blood-2007-03-066936. [DOI] [PubMed] [Google Scholar]
- 32.Furman RR, et al. Ibrutinib resistance in chronic lymphocytic leukemia. N Engl J Med. 2014;370:2352–2354. doi: 10.1056/NEJMc1402716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SH. Molecular basis of drug resistance: epidermal growth factor receptor tyrosine kinase inhibitors and anaplastic lymphoma kinase inhibitors. Tuberc Respir Dis (Seoul) 2013;75:188–198. doi: 10.4046/trd.2013.75.5.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma B, Elkayam T, Wolfson H, Nussinov R. Protein-protein interactions: structurally conserved residues distinguish between binding sites and exposed protein surfaces. Proc Natl Acad Sci U S A. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Keskin O, Ma B, Nussinov R, Liang J. Protein-protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J Mol Biol. 2004;344:781–795. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 36.Keskin O, Ma B, Nussinov R. Hot regions in protein--protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 37.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 38.Jung M, et al. Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1. J Biol Chem. 2014;289:9304–9319. doi: 10.1074/jbc.M113.523019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 41.Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 43.Gorczynski MJ, et al. Allosteric inhibition of the protein-protein interaction between the leukemia-associated proteins Runx1 and CBFbeta. Chem Biol. 2007;14:1186–1197. doi: 10.1016/j.chembiol.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Petsalaki E, Russell RB. Peptide-mediated interactions in biological systems: new discoveries and applications. Curr Opin Biotechnol. 2008;19:344–350. doi: 10.1016/j.copbio.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 45.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Demarest SJ, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 48.Leach BI, Kuntimaddi A, Schmidt CR, Cierpicki T, Johnson SA, Bushweller JH. Leukemia fusion target AF9 is an intrinsically disordered transcriptional regulator that recruits multiple partners via coupled folding and binding. Structure. 2013;21:176–183. doi: 10.1016/j.str.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 50.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 51.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 52.Ma B, Nussinov R. Trp/Met/Phe hot spots in protein-protein interactions: potential targets in drug design. Curr Top Med Chem. 2007;7:999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- 53.DeLano WL. Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol. 2002;12:14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 54.Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 55.Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D. Relationship between hot spot residues and ligand binding hot spots in protein-protein interfaces. J Chem Inf Model. 2012;52:2236–2244. doi: 10.1021/ci300175u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci U S A. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108:13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makley LN, Gestwicki JE. Expanding the number of 'druggable' targets: non-enzymes and protein-protein interactions. Chem Biol Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith MC, Gestwicki JE. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expert Rev Mol Med. 2012;14:e16. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lanzarotti E, Biekofsky RR, Estrin DA, Marti MA, Turjanski AG. Aromatic-aromatic interactions in proteins: beyond the dimer. J Chem Inf Model. 2011;51:1623–1633. doi: 10.1021/ci200062e. [DOI] [PubMed] [Google Scholar]
- 61.Villoutreix BO, Labbe CM, Lagorce D, Laconde G, Sperandio O. A leap into the chemical space of protein-protein interaction inhibitors. Curr Pharm Des. 2012;18:4648–4667. doi: 10.2174/138161212802651571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr Opin Struct Biol. 2004;14:104–109. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 63.Lee EF, et al. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J Biol Chem. 2009;284:30508–30517. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eyrisch S, Helms V. Transient pockets on protein surfaces involved in protein-protein interaction. J Med Chem. 2007;50:3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 65.Arkin MR, et al. Binding of small molecules to an adaptive protein-protein interface. Proc Natl Acad Sci U S A. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Widmer H, Jahnke W. Protein NMR in biomedical research. Cell Mol Life Sci. 2004;61:580–599. doi: 10.1007/s00018-003-3382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sivanesan D, Rajnarayanan RV, Doherty J, Pattabiraman N. In-silico screening using flexible ligand binding pockets: a molecular dynamics-based approach. J Comput Aided Mol Des. 2005;19:213–228. doi: 10.1007/s10822-005-4788-9. [DOI] [PubMed] [Google Scholar]
- 68.Caballero J, Alzate-Morales JH. Molecular dynamics of protein kinase-inhibitor complexes: a valid structural information. Curr Pharm Des. 2012;18:2946–2963. doi: 10.2174/138161212800672705. [DOI] [PubMed] [Google Scholar]
- 69.Lexa KW, Carlson HA. Full protein flexibility is essential for proper hot-spot mapping. J Am Chem Soc. 2011;133:200–202. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White AW, Westwell AD, Brahemi G. Protein-protein interactions as targets for small-molecule therapeutics in cancer. Expert Rev Mol Med. 2008;10:e8. doi: 10.1017/S1462399408000641. [DOI] [PubMed] [Google Scholar]
- 71.Vitagliano O, Addeo R, D'Angelo V, Indolfi C, Indolfi P, Casale F. The Bcl-2/Bax and Ras/Raf/MEK/ERK signaling pathways: implications in pediatric leukemia pathogenesis and new prospects for therapeutic approaches. Expert Rev Hematol. 2013;6:587–597. doi: 10.1586/17474086.2013.827415. [DOI] [PubMed] [Google Scholar]
- 72.Brinkmann K, Kashkar H. Targeting the mitochondrial apoptotic pathway: a preferred approach in hematologic malignancies? Cell Death Dis. 2014;5:e1098. doi: 10.1038/cddis.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goard CA, Schimmer AD. An evidence-based review of obatoclax mesylate in the treatment of hematological malignancies. Core Evid. 2013;8:15–26. doi: 10.2147/CE.S42568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogler M, Dinsdale D, Dyer MJ, Cohen GM. Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 2009;16:360–367. doi: 10.1038/cdd.2008.137. [DOI] [PubMed] [Google Scholar]
- 75.Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. 2009;49:223–241. doi: 10.1146/annurev.pharmtox.48.113006.094723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun D, et al. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J Med Chem. 2014;57:1454–1472. doi: 10.1021/jm401753e. [DOI] [PubMed] [Google Scholar]
- 77.Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 78.Grembecka J, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8:277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi A, et al. Structural insights into inhibition of the bivalent menin-MLL interaction by small molecules in leukemia. Blood. 2012;120:4461–4469. doi: 10.1182/blood-2012-05-429274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He S, et al. High-affinity small-molecule inhibitors of the menin-mixed lineage leukemia (MLL) interaction closely mimic a natural protein-protein interaction. J Med Chem. 2014;57:1543–1556. doi: 10.1021/jm401868d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cao F, et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell. 2014;53:247–261. doi: 10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Senisterra G, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem J. 2013;449:151–159. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cunningham L, et al. Identification of benzodiazepine Ro5-3335 as an inhibitor of CBF leukemia through quantitative high throughput screen against RUNX1-CBFbeta interaction. Proc Natl Acad Sci U S A. 2012;109:14592–14597. doi: 10.1073/pnas.1200037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cerchietti LC, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mirguet O, et al. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56:7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 89.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haematol. 2011;152:141–154. doi: 10.1111/j.1365-2141.2010.08459.x. [DOI] [PubMed] [Google Scholar]
- 90.Tomizawa D, et al. Outcome of risk-based therapy for infant acute lymphoblastic leukemia with or without an MLL gene rearrangement, with emphasis on late effects: a final report of two consecutive studies, MLL96 and MLL98, of the Japan Infant Leukemia Study Group. Leukemia. 2007;21:2258–2263. doi: 10.1038/sj.leu.2404903. [DOI] [PubMed] [Google Scholar]
- 91.Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- 92.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trends Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 93.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 94.Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94:984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Slany RK. When epigenetics kills: MLL fusion proteins in leukemia. Hematol Oncol. 2005;23:1–9. doi: 10.1002/hon.739. [DOI] [PubMed] [Google Scholar]
- 96.Dimartino JF, Cleary ML. Mll rearrangements in haematological malignancies: lessons from clinical and biological studies. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 97.Daigle SR, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knapper S. FLT3 inhibition in acute myeloid leukaemia. Br J Haematol. 2007;138:687–699. doi: 10.1111/j.1365-2141.2007.06700.x. [DOI] [PubMed] [Google Scholar]
- 99.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Placke T, et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood. 2014 doi: 10.1182/blood-2014-02-558114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 102.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 103.Milne TA, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 104.Thiel AT, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17:148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 106.Song JJ, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J Biol Chem. 2008;283:35258–35264. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14:36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cierpicki T, Grembecka J. Challenges and opportunities in targeting the menin-MLL interaction. Future Med Chem. 2014;6:447–462. doi: 10.4155/fmc.13.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grembecka J, Belcher AM, Hartley T, Cierpicki T. Molecular basis of the mixed lineage leukemia-menin interaction: implications for targeting mixed lineage leukemias. J Biol Chem. 2010;285:40690–40698. doi: 10.1074/jbc.M110.172783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286:31742–31748. doi: 10.1074/jbc.M111.258186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang J, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482:542–546. doi: 10.1038/nature10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou H, et al. Structure-based design of high-affinity macrocyclic peptidomimetics to block the menin-mixed lineage leukemia 1 (MLL1) protein-protein interaction. J Med Chem. 2013;56:1113–1123. doi: 10.1021/jm3015298. [DOI] [PubMed] [Google Scholar]
- 113.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J Biol Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 114.Karatas H, Townsend EC, Bernard D, Dou Y, Wang S. Analysis of the binding of mixed lineage leukemia 1 (MLL1) and histone 3 peptides to WD repeat domain 5 (WDR5) for the design of inhibitors of the MLL1-WDR5 interaction. J Med Chem. 2010;53:5179–5185. doi: 10.1021/jm100139b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bolshan Y, et al. Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction. ACS Med Chem Lett. 2013;4:353–357. doi: 10.1021/ml300467n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karatas H, et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein-protein interaction. J Am Chem Soc. 2013;135:669–682. doi: 10.1021/ja306028q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 118.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 119.Kim W, et al. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9:643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu P, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 121.Castilla LH, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- 122.Lukasik SM, et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat Struct Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- 123.Warren AJ, Bravo J, Williams RL, Rabbitts TH. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tahirov TH, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 125.Tang YY, et al. Energetic and functional contribution of residues in the core binding factor beta (CBFbeta) subunit to heterodimerization with CBFalpha. J Biol Chem. 2000;275:39579–39588. doi: 10.1074/jbc.M007350200. [DOI] [PubMed] [Google Scholar]
- 126.Bushweller JH, et al. A small molecule inhibitor of the CBFb-SMMHC/RUNX interaction attenuates inv(16) leukemia in vivo. Blood. 2012;120:286. [Google Scholar]
- 127.Marushige K. Activation of chromatin by acetylation of histone side chains. Proc Natl Acad Sci U S A. 1976;73:3937–3941. doi: 10.1073/pnas.73.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 130.Liu Y, et al. Structural basis and binding properties of the second bromodomain of Brd4 with acetylated histone tails. Biochemistry. 2008;47:6403–6417. doi: 10.1021/bi8001659. [DOI] [PubMed] [Google Scholar]
- 131.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ye BH. BCL-6 in the pathogenesis of non-Hodgkin's lymphoma. Cancer Invest. 2000;18:356–365. doi: 10.3109/07357900009012179. [DOI] [PubMed] [Google Scholar]
- 134.Cerchietti LC, et al. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 2009;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Polo JM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 136.Ghetu AF, Corcoran CM, Cerchietti L, Bardwell VJ, Melnick A, Prive GG. Structure of a BCOR corepressor peptide in complex with the BCL6 BTB domain dimer. Mol Cell. 2008;29:384–391. doi: 10.1016/j.molcel.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ahmad KF, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 138.Fry DC. Protein-protein interactions as targets for small molecule drug discovery. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- 139.Pagliaro L, et al. Emerging classes of protein-protein interaction inhibitors and new tools for their development. Curr Opin Chem Biol. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 140.Toogood PL. Inhibition of protein-protein association by small molecules: approaches and progress. J Med Chem. 2002;45:1543–1558. doi: 10.1021/jm010468s. [DOI] [PubMed] [Google Scholar]
- 141.Loregian A, Palu G. Disruption of protein-protein interactions: towards new targets for chemotherapy. J Cell Physiol. 2005;204:750–762. doi: 10.1002/jcp.20356. [DOI] [PubMed] [Google Scholar]
- 142.Moerke NJ, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–267. doi: 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 143.Inglese J, et al. High-throughput screening assays for the identification of chemical probes. Nat Chem Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 144.Gotoh Y, Nagata H, Kase H, Shimonishi M, Ido M. A homogeneous time-resolved fluorescence-based high-throughput screening system for discovery of inhibitors of IKKbeta-NEMO interaction. Anal Biochem. 2010;405:19–27. doi: 10.1016/j.ab.2010.05.028. [DOI] [PubMed] [Google Scholar]
- 145.Glickman JF, et al. A comparison of ALPHAScreen, TR-FRET, and TRF as assay methods for FXR nuclear receptors. J Biomol Screen. 2002;7:3–10. doi: 10.1177/108705710200700102. [DOI] [PubMed] [Google Scholar]
- 146.Bauer RA, Wurst JM, Tan DS. Expanding the range of 'druggable' targets with natural product-based libraries: an academic perspective. Curr Opin Chem Biol. 2010;14:308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dandapani S, Marcaurelle LA. Grand challenge commentary: Accessing new chemical space for 'undruggable' targets. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 148.Galloway WR, Isidro-Llobet A, Spring DR. Diversity-oriented synthesis as a tool for the discovery of novel biologically active small molecules. Nat Commun. 2010;1:80. doi: 10.1038/ncomms1081. [DOI] [PubMed] [Google Scholar]
- 149.Burke MD, Schreiber SL. A planning strategy for diversity-oriented synthesis. Angew Chem Int Ed Engl. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 150.Boldi AM. Libraries from natural product-like scaffolds. Curr Opin Chem Biol. 2004;8:281–286. doi: 10.1016/j.cbpa.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 151.Koehn FE, Carter GT. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- 152.Larsen MJ, et al. The role of HTS in drug discovery at the University of Michigan. Comb Chem High Throughput Screen. 2014;17:210–230. doi: 10.2174/1386207317666140109121546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.De Clercq E. The design of drugs for HIV and HCV. Nat Rev Drug Discov. 2007;6:1001–1018. doi: 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- 154.Sakurai K, Chung HS, Kahne D. Use of a retroinverso p53 peptide as an inhibitor of MDM2. J Am Chem Soc. 2004;126:16288–16289. doi: 10.1021/ja044883w. [DOI] [PubMed] [Google Scholar]
- 155.Zhong S, Macias AT, MacKerell AD., Jr Computational identification of inhibitors of protein-protein interactions. Curr Top Med Chem. 2007;7:63–82. doi: 10.2174/156802607779318334. [DOI] [PubMed] [Google Scholar]
- 156.Falchi F, Caporuscio F, Recanatini M. Structure-based design of small-molecule protein-protein interaction modulators: the story so far. Future Med Chem. 2014;6:343–357. doi: 10.4155/fmc.13.204. [DOI] [PubMed] [Google Scholar]
- 157.Siddiquee K, et al. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci U S A. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Song H, Wang R, Wang S, Lin J. A low-molecular-weight compound discovered through virtual database screening inhibits Stat3 function in breast cancer cells. Proc Natl Acad Sci U S A. 2005;102:4700–4705. doi: 10.1073/pnas.0409894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Renaud JP, Delsuc MA. Biophysical techniques for ligand screening and drug design. Curr Opin Pharmacol. 2009;9:622–628. doi: 10.1016/j.coph.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 160.Murray CW, Rees DC. The rise of fragment-based drug discovery. Nat Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 161.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 162.Schuffenhauer A, et al. Library design for fragment based screening. Curr Top Med Chem. 2005;5:751–762. doi: 10.2174/1568026054637700. [DOI] [PubMed] [Google Scholar]
- 163.Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 164.Kumar A, Voet A, Zhang KY. Fragment based drug design: from experimental to computational approaches. Curr Med Chem. 2012;19:5128–5147. doi: 10.2174/092986712803530467. [DOI] [PubMed] [Google Scholar]
- 165.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 166.Wendt MD, et al. Discovery and structure-activity relationship of antagonists of B-cell lymphoma 2 family proteins with chemopotentiation activity in vitro and in vivo. J Med Chem. 2006;49:1165–1181. doi: 10.1021/jm050754u. [DOI] [PubMed] [Google Scholar]
- 167.Park CM, et al. Discovery of an orally bioavailable small molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51:6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 168.Friberg A, et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J Med Chem. 2013;56:15–30. doi: 10.1021/jm301448p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Huang JW, et al. Fragment-based design of small molecule X-linked inhibitor of apoptosis protein inhibitors. J Med Chem. 2008;51:7111–7118. doi: 10.1021/jm8006992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Leach AR, Hann MM. Molecular complexity and fragment-based drug discovery: ten years on. Curr Opin Chem Biol. 2011;15:489–496. doi: 10.1016/j.cbpa.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 171.Moriniere J, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 172.Ku B, Liang C, Jung JU, Oh BH. Evidence that inhibition of BAX activation by BCL-2 involves its tight and preferential interaction with the BH3 domain of BAX. Cell Res. 2011;21:627–641. doi: 10.1038/cr.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 174.Chen CY, Tou WI. How to design a drug for the disordered proteins? Drug Discov Today. 2013;18:910–915. doi: 10.1016/j.drudis.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 175.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu Rev Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Palermo CM, Bennett CA, Winters AC, Hemenway CS. The AF4-mimetic peptide, PFWT, induces necrotic cell death in MV4-11 leukemia cells. Leuk Res. 2008;32:633–642. doi: 10.1016/j.leukres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Feng BY, Shelat A, Doman TN, Guy RK, Shoichet BK. High-throughput assays for promiscuous inhibitors. Nat Chem Biol. 2005;1:146–148. doi: 10.1038/nchembio718. [DOI] [PubMed] [Google Scholar]
- 178.Shoichet BK. Screening in a spirit haunted world. Drug Discov Today. 2006;11:607–615. doi: 10.1016/j.drudis.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pellecchia M, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discov. 2008;7:738–745. doi: 10.1038/nrd2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chaires JB. Calorimetry and thermodynamics in drug design. Annu Rev Biophys. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- 181.Cooper MA. Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002;1:515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 182.Stevens AJ, Jensen JJ, Wyller K, Kilgore PC, Chatterjee S, Rohrbaugh ML. The role of public-sector research in the discovery of drugs and vaccines. N Engl J Med. 2011;364:535–541. doi: 10.1056/NEJMsa1008268. [DOI] [PubMed] [Google Scholar]
- 183.Frye S, Crosby M, Edwards T, Juliano R. US academic drug discovery. Nat Rev Drug Discov. 2011;10:409–410. doi: 10.1038/nrd3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fishburn CS. Translational research: the changing landscape of drug discovery. Drug Discov Today. 2013;18:487–494. doi: 10.1016/j.drudis.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 185.Schultz-Kirkegaard H, Valentin F. Academic drug discovery centres: the economic and organisational sustainability of an emerging model. Drug Discov Today. 2014 doi: 10.1016/j.drudis.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 186.Coles LD, Cloyd JC. The role of academic institutions in the development of drugs for rare and neglected diseases. Clin Pharmacol Ther. 2012;92:193–202. doi: 10.1038/clpt.2012.83. [DOI] [PubMed] [Google Scholar]
- 187.Whitty A, Kumaravel G. Between a rock and a hard place? Nat Chem Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 188.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 189.Berg T. Small-molecule inhibitors of protein-protein interactions. Curr Opin Drug Discov Devel. 2008;11:666–674. [PubMed] [Google Scholar]
- 190.Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 191.Huber W, Mueller F. Biomolecular interaction analysis in drug discovery using surface plasmon resonance technology. Curr Pharm Des. 2006;12:3999–4021. doi: 10.2174/138161206778743600. [DOI] [PubMed] [Google Scholar]
- 192.Frank AO, et al. Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach. J Med Chem. 2013;56:9242–9250. doi: 10.1021/jm401333u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vu B, et al. Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development. ACS Med Chem Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Emami KH, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci U S A. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Condon SM, et al. Birinapant, a smac-mimetic with improved tolerability for the treatment of solid tumors and hematological malignancies. J Med Chem. 2014;57:3666–3677. doi: 10.1021/jm500176w. [DOI] [PubMed] [Google Scholar]
- 197.Houghton PJ, et al. Initial testing (stage 1) of LCL161, a SMAC mimetic, by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2012;58:636–639. doi: 10.1002/pbc.23167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Gandhi L, et al. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol. 2011;29:909–916. doi: 10.1200/JCO.2010.31.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Schimmer AD, et al. A phase I study of the pan bcl-2 family inhibitor obatoclax mesylate in patients with advanced hematologic malignancies. Clin Cancer Res. 2008;14:8295–8301. doi: 10.1158/1078-0432.CCR-08-0999. [DOI] [PubMed] [Google Scholar]
- 200.Basse MJ, et al. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013;41:D824–D827. doi: 10.1093/nar/gks1002. [DOI] [PMC free article] [PubMed] [Google Scholar]