Abstract
The 2009 H1N1 influenza virus outbreak is the first pandemic of the twenty-first century. Epidemiological data reveal that of all the people afflicted with H1N1 virus, <5% are over 51 y of age. Interestingly, in the uninfected population, 33% of those >60 y old have pre-existing neutralizing Abs against the 2009 H1N1 virus. This finding suggests that influenza strains that circulated 50–60 y ago might provide cross-protection against the swine-origin 2009 H1N1 influenza virus. To test this, we determined the ability of representative H1N1 influenza viruses that circulated in the human population from 1930 to 2000, to induce cross-reactivity to and cross-protection against the pandemic swine-origin H1N1 virus, A/California/04/09. We show that exposure of mice to the 1947 virus, A/FM/1/47, or the 1934 virus, A/PR/8/34, induced robust cross-protective immune responses and these mice were protected against a lethal challenge with mouse-adapted A/California/04/09 H1N1 virus. Conversely, we observed that mice exposed to the 2009 H1N1 virus were protected against a lethal challenge with mouse-adapted 1947 or 1934 H1N1 viruses. In addition, exposure to the 2009 H1N1 virus induced broad cross-reactivity against H1N1 as well as H3N2 influenza viruses. Finally, we show that vaccination with the older H1N1 viruses, particularly A/FM/1/47, confers protective immunity against the 2009 pandemic H1N1 virus. Taken together, our data provide an explanation for the decreased susceptibility of the elderly to the 2009 H1N1 outbreak and demonstrate that vaccination with the pre-1950 influenza strains can cross-protect against the pandemic swine-origin 2009 H1N1 influenza virus.
Influenza virus is lipid enveloped, with a segmented negative sense RNA genome. The envelope of the virion contains two types of surface glycoproteins, which play essential roles in viral infection. The hemagglutinin (HA) protein is responsible for attachment of the virus to sialic acid-containing glycan receptors on the host cell surface (1, 2), whereas the neuraminidase (NA) is a receptor-destroying enzyme, which has important functions in viral release and cell-to-cell spread (3, 4). There are three distinct serotypes of influenza viruses, designated A, B, and C, with types A and B viruses playing the major role in human infection. Influenza A viruses also occur in birds, pigs, and other species, whereas types B and C are found primarily in humans. Human influenza viruses are continuously evolving owing to mutations in the viral genome RNA, resulting in variants with surface glycoproteins that have distinct antigenic properties. These mutations are responsible for seasonal epidemics that occur with both influenza A and B viruses. Less frequently, influenza A viruses occur with novel HA proteins that are unrelated to pre-existing human strains with respect to antigenic properties. These major antigenic shifts result in novel antigenic subtypes of the HA and sometimes the NA glycoproteins, which can spread rapidly, causing global disease pandemics (5–8).
The first known swine H1N1 influenza virus was isolated in 1930 (9). This virus was shown to exhibit similarities in sequence to the 1918 H1N1 virus that was recently reconstructed from preserved patient specimens (10, 11). The first human influenza virus isolates were also of the H1N1 serotype, which persisted in the human population until the appearance of the H2N2 virus in 1957 (12). In 1977, the H1N1 virus reappeared and has been cocirculating with H3N2 viruses until the present time. In April 2009, a distinct H1N1 virus of swine origin was identified in North America, and it has since spread rapidly in multiple geographic regions, resulting in the declaration of a new pandemic by the World Health Organization in June 2009. It is a quadruple reassortant virus containing a unique combination of gene segments derived from the classical swine, North American avian, human (H3N2), and Eurasian avian-like swine influenza viruses (13). Although most human infections with the 2009 swine-origin H1N1 viruses have been mild, resembling typical seasonal influenza infections, >700 deaths and numerous hospitalizations have been reported, suggesting that the new virus is more pathogenic in mammalian hosts than are seasonal H1N1 viruses that circulated in recent years. Typically, during seasonal influenza outbreaks, the elderly, persons with underlying chronic diseases, infants, and young children who have not been previously exposed to the virus manifest the most severe disease symptoms. This pattern does not seem to hold completely true for the 2009 pandemic H1N1 virus; those >50 y old seem to be spared. Preliminary analysis of patients afflicted with the H1N1 virus showed that in >700 confirmed cases in the United States the majority were young adults and only 5% were >51 y old (14). This observation raises the possibility of pre-existing immunity to the 2009 pandemic H1N1 virus in the population. In addition, Katz and colleagues (15) recently showed that seasonal influenza vaccines from 2005 to 2009 did not induce cross-reactive Abs against the 2009 H1N1 virus. Interestingly, they found that ~33% of those >60 y old in their study had pre-existing cross-reactive Abs against the 2009 H1N1 virus (15). As a result, the frequency of hospitalization has been highest in individuals from 24 to 60 y of age, and very low in those >60 y old.
In this study, we determined the extent to which pre-existing immunity to representative H1N1 viruses that appeared in the human population from 1930 to 2000 protects against the 2009 pandemic H1N1 virus. Our findings suggest that exposure to pre-1950 H1N1 influenza strains can offer protection against the 2009 H1N1 virus, limiting disease severity in this population. We also find that exposure to the 2009 swine-origin H1N1 virus induces broad cross-reactivity against H1N1 and H3N2 influenza viruses and this is dependent on both Abs and CD8 T cells.
Materials and Methods
Mice, viruses, and immunization
BALB/c mice, purchased from Charles River Laboratories (Wilmington, MA), were housed under specific pathogen-free conditions at the Emory Vaccine Center vivarium. The wild-type A/California/04/09 virus was a kind gift from Dr. Richard Webby, St. Jude Hospital, Memphis, TN. All the H1N1 (A/PR/8/34, A/FM/1/47, A/Denver/1/57, A/California/10/78, A/Chile/1/83, and A/New Caledonia/20/99) and H3N2 (A/Aichi/2/68, A/Udorn/307/72, A/Victoria/3/75, and A/Arizona/2/80) influenza viruses were propagated in embryonated eggs, as described (16). All the mouse-adapted strains were generated by eight serial passages in BALB/c mice with the exception of A/California/04/09 virus, which was passaged five times. We then determined LD50 for mouse-adapted virus using the Reed–Muench equation (17). For the challenge and infection studies, mice were anesthetized with isoflurane and then infected with virus in solution by intranasal instillation. The animals were either infected with 0.1 × LD50 of the old strains (1930–2000), followed by challenge with 3 × LD50 the swine-origin A/California/04/09, or they were infected with 0.5 × LD50 A/California/04/09 and challenged with 3 × LD50 of the mouse-adapted A/PR/8/34 (H1N1), A/FM/1/47 (H1N1), A/Aichi/2/68 (H3N2), or A/Victoria/3/75 (H3N2) virus.
Challenge of mice with influenza virus
To determine postchallenge survival rates and immune responses, mice were challenged 1 mo after either primary immunization or infection by intranasal instillation of 30 μl virus with 3 × LD50 live mouse-adapted virus and monitored for 14 d. As a negative control group, we included naive mice. A weight loss exceeding 25% was used as the experimental end point, and mice reaching this end point were euthanized according to Institutional Animal Care and Use Committee (IACUC) guidelines. The challenged mice were monitored daily for signs of morbidity (body weight changes, fever, and hunched posture) and mortality.
Evaluation of humoral immune responses
Sera were collected from individual mice, and anti-influenza–specific Ab levels were determined quantitatively by ELISA, as described (16). The reagents for ELISA were purchased from Southern Biotechnology Associates (Birmingham, AL).
We determined the hemagglutination inhibition (HAI) titers based on the World Health Organization protocol, as described previously (18). Sera were treated with receptor-destroying NA (Roche Diagnostics, Indianapolis, IN) overnight at 37°C, heat inactivated for 30 min at 56°C, and incubated with packed chicken or turkey RBCs for 1 h at 4°C to remove any cryoglobulins interfering with the agglutination reaction. They were then serially diluted and preincubated at room temperature with 4 HA Units/50 μl the panel of 1930–2009 viruses, for 30 min. An equal volume of 0.5% chicken or turkey RBCs was then added to each well for 30 min incubation at room temperature.
Depletion of CD8 T cells in vivo
To deplete CD8 T cells in vivo, we used a rat IgG2a anti-mouse CD8 mAb produced from the hybridoma, YTS169.4 (19). We administered the mAb i.p. twice, at a dose of 500 μg 3 d and 1 d prior to viral challenge. We tested the effectiveness of the mAb to deplete the CD8 population by flow cytometry using fluorochrome-labeled rat anti-mouse CD8 mAb (clone 53-6.7; BD Biosciences, San Jose, CA). The anti-CD8 mAb treatment depleted CD8 T cells completely (data not shown).
Influenza HA cloning from allantoic fluid or mouse lung homogenate
We cloned and sequenced the HA gene segments from all the influenza viruses used in this study. Briefly, viral RNA was extracted from cell-free allantoic fluid derived from infected eggs or infected mouse lung homogenate, using the QIAmp Viral RNA Mini Kit (Qiagen, Valencia, CA). RT-PCR was performed using the Qiagen One-Step RT-PCR Kit and primers complementary to the 5′ and 3′ untranslated regions specific to the HA RNA segment (primer sequences). Amplified HA segments were cloned using the TOPO TA Cloning Kit (Invitrogen; Carlsbad, CA). Plasmids were first confirmed by restriction mapping and then sequenced (Operon-MWG; Huntsville, AL). Protein sequences were deduced from DNA sequences, using the standard genetic code.
Influenza hemagglutinin multiple sequence alignments and homology modeling
Multiple sequence alignments were performed using the MegAlign software (Lasergene, v7.0) and the BioEdit software (Ibis Biosciences, Carlsbad, CA) using the ClustalW alignment algorithm. Homology modeling of mouse- and egg-adapted influenza hemagglutinin proteins was performed using SWISS-MODEL (http://swissmodel.expasy.org/) and analyzed using Chimera (University of California, San Francisco, version 1.3).
Statistics
The statistical significance of the difference was calculated by two-tailed unpaired Student t test and one-way or two-way ANOVA (including the Bonferroni multiple comparison test). Values were considered significant for p ≤ 0.05. Unless otherwise stated, data were pooled from at least two independent experiments.
Results
Sera from mice sublethally infected with pre-1950 influenza strains cross-react with the swine-origin H1N1 influenza virus, A/CA/04/2009
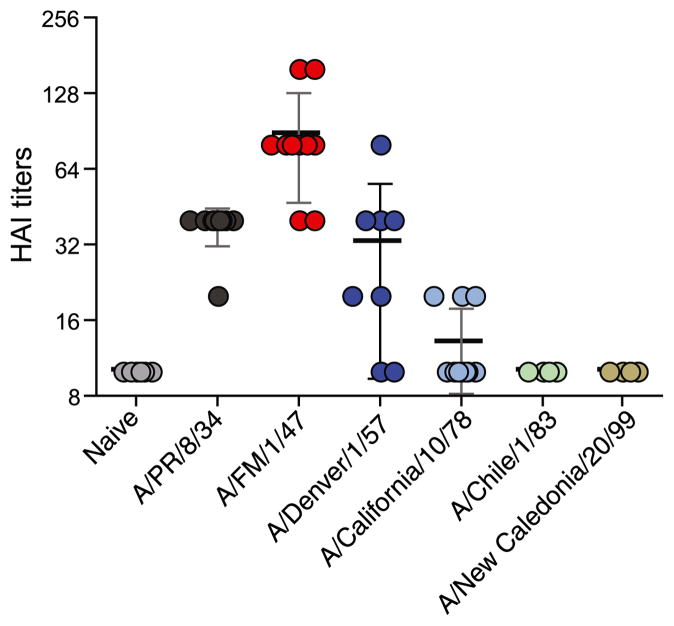
To evaluate the degree of cross-reactivity of the older influenza strains to the new swine-origin H1N1 virus, A/California/04/09, we sublethally infected cohorts of BALB/c mice with live mouse-adapted H1N1 viruses, A/PR/8/34, A/FM/1/47, A/Denver/1/57, A/Califor-nia/10/78, A/Chile/1/83, and A/New Caledonia/20/99. These are representative of H1N1 viruses that circulated in the human population from 1930 to 2000. One month later, we collected sera and tested their ability to cross-react with the novel 2009 H1N1 virus by determining their ability to inhibit hemagglutination of turkey RBCs by A/California/04/09 virus in a HAI (Fig. 1). Immune sera from mice infected with A/FM/1/47virus exhibited the highest HAI titers, and this was followed by A/PR/8/34. In the group of mice infected with the 1957 H1N1 virus, A/Denver/1/57, half of them exhibited mean HAI titers >32.5. A/PR/8/34-immune mice had a mean HAI titer of 38, and most interestingly, A/FM/1/47-immune mice had significantly higher HAI titers than all the other groups (p < 0.001). In contrast, mice exposed to H1N1 viruses from 1957 to 2000 exhibited minimal cross-reactivity; the mean HAI titers for these groups were <13. This finding was not due to a lack of Abs against the homologous viruses; all sublethally infected mice exhibited HAI titers >640 against the homologous viruses. Taken together, these data demonstrate that mice immune to pre-1950 H1N1 viruses, especially A/FM/1/47 and A/PR/8/34, had cross-reactive Abs against A/California/04/09 virus.
FIGURE 1.
Sera from mice sublethally infected with pre-1950 influenza strains cross-react with the swine-origin H1N1 influenza virus, A/CA/04/2009. Cohorts of BALB/c mice were infected intranasally with 0.1 × LD50 of live mouse-adapted influenza viruses spanning from 1930 to 2000 (A/PR/8/34, A/FM/1/47, A/Denver/1/57, A/California/10/78, A/Chile/1/83, and A/New Caledonia/20/99). A month following infection, we collected their sera and tested their ability to inhibit hemagglutination of turkey RBCs by the 2009 swine-origin A/California/04/09 virus. The HAI titers were read as the reciprocal of the highest dilution of serum that conferred inhibition of hemagglutination. The values are expressed as mean ± SD. Each data point represents an individual animal.
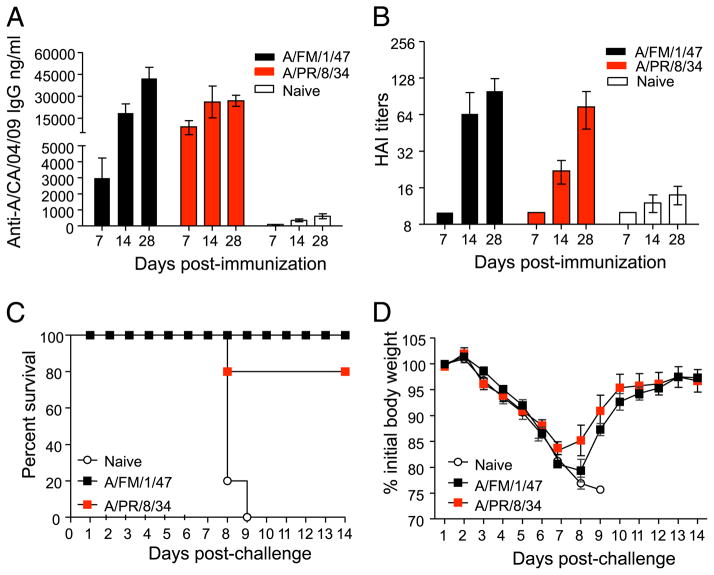
Mice exposed to 1934 and 1947 H1N1 viruses are protected against lethal challenge with mouse-adapted swine-origin H1N1 influenza A/California/04/09 virus
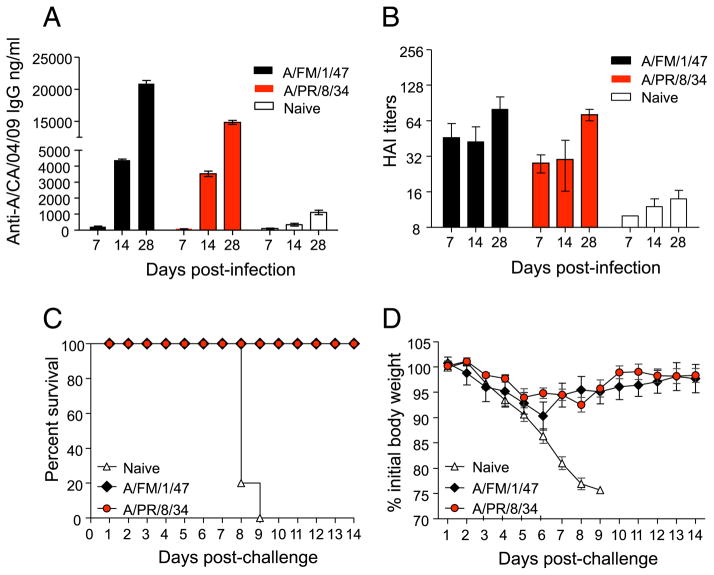
Because the A/PR/8/34 and A/FM/1/47 viruses induced significantly higher cross-reactive HAI titers than all the other groups, we determined the kinetics of their cross-reactive responses. Briefly, we infected cohorts of BALB/c mice intranasally with sublethal doses (0.1 × LD50) of live mouse-adapted A/FM/1/47 and A/PR/8/34 viruses. Both viruses induced robust immune responses; 28 d postexposure, the mean (±SEM) HAI titers against A/PR/8/34 and A/FM/1/47 were 896 ± 156 and 640, respectively. Mock infected mice served as naive controls. At days 7, 14, and 28 postinfection, we collected their sera and tested their ability to cross-react with A/California/04/09, using ELISA (Fig. 2A) and HAI assays (Fig. 2B). The data show that cross-reactive IgG Abs specific for A/California/04/09 virus could be detected by ELISA as early as day 7 and significant levels were induced by day 28 (Fig. 2A). Exposure to the older viruses also induced significant levels of HAI activity against the A/CA/04/09 virus (Fig. 2B). To determine the extent to which immunity to A/FM/1/47 or A/PR/8/34 could cross-protect against lethal challenge with A/California/04/09, we lethally challenged the A/FM/1/47 and A/PR/8/34 immune mice (shown in Fig. 2A, 2B) with 3 × LD50 of live mouse-adapted A/California/04/09 virus and monitored them for survival (Fig. 2C) and morbidity (Fig. 2D) for the next 14 d. Of the mice previously exposed to A/PR/8/34 or A/FM/1/47, 100% survived lethal challenge with the 2009 pandemic H1N1 strain. In contrast, all of the naive control mice succumbed to the mouse-adapted A/California/04/09 virus challenge and either died or lost >25% of their initial body weight and had to be euthanized per IACUC guidelines by days 8–9 postchallenge. Following challenge, their morbidity, as measured by weight loss, was minimal in the A/FM/1/47- and A/PR/8/34-immune groups. These mice lost ~8–10% of their body weight by day 6 and exhibited no outward signs of morbidity, such as hunched posture, ruffled fur, or failure to thrive. All of these mice regained their weights by 2 wk (Fig. 2D). Taken together, our data show that previous exposure to A/FM/1/47 or A/PR/8/34 virus gives protective immunity against lethal challenge with A/California/04/09 virus.
FIGURE 2.
Exposure to A/PR/8/34 and A/FM/1/47 induces protective immunity against A/California/04/09. Cohorts of BALB/c mice were sublethally infected intranasally with 0.1 × LD50 live mouse-adapted A/PR/8/34 and A/FM/1/47 influenza viruses. We collected sera from these mice at days 7, 14, and 28 and determined their A/California/04/09-specific IgG titers (A) and HAI titers (B). One month later, these mice were challenged with 3 × LD50 live mouse-adapted A/California/04/09 virus and monitored for survival up to 14 d postchallenge (C). Body weight changes, indicative of morbidity, were also monitored up to 14 d (D). The graphs represent the mean ± SEM.
Abs produced in response to the 2009 H1N1 A/California/04/09 virus cross-react with a large spectrum of H1N1 influenza viruses
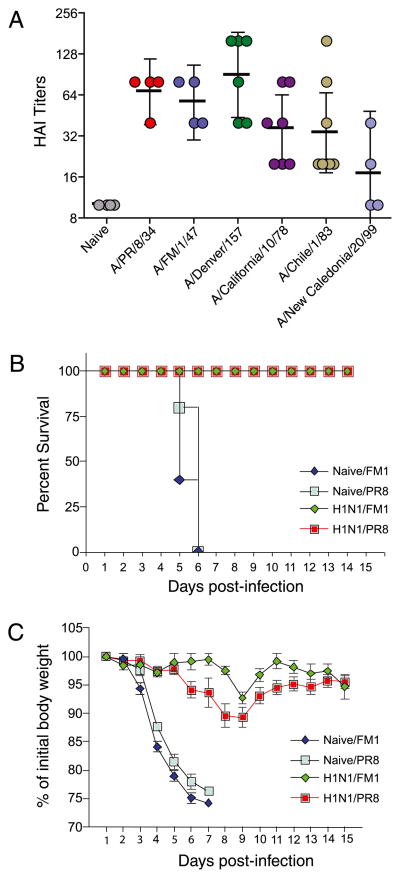
We also tested whether exposure to A/California/04/09 could induce responses that cross-react with older H1N1 influenza strains. We infected cohorts of BALB/c mice with 0.5 × LD50 live mouse-adapted A/California/04/09 virus. A month later, we bled them and tested the ability of sera to cross-react with A/PR/8/34, A/FM/1/47, A/Denver/1/57, A/California/10/78, A/Chile/1/83, and A/New Caledonia/20/99, using HAI assay (Fig. 3A). Serum samples exhibited high HAI titers with all the older H1N1 strains tested, except for the A/New Caledonia/20/1999 strain. The cross-reactivity was highest for A/Denver/1/57, A/California/10/78, A/FM/1/47, and A/PR/8/34, and, to a lesser extent, for A/Chile/1/83. The HAI titers were minimal against A/New Caledonia/20/99. We next tested whether A/California/04/09-immune mice could be protected against lethal challenge with 3 × LD50 mouse-adapted A/PR/8/34 and A//FM/1/47 virus; the survival was 100% for both these viruses (Fig. 3B). In contrast, 100% of the naive control mice challenged with either 3 × LD50 A/PR/8/34 or A/FM/1/47 succumbed to the infection and either died or had to be euthanized by day 6 owing to excessive weight loss. In both the A/PR/8/34- and A/FM/1/47-challenged mice, the weight loss was only ~8–10% at its peak (day 8) (Fig. 3C). Taken together, these data show that mice exposed to A/California/04/09 are protected against lethal challenge with either A/PR/8/34 or A/FM/1/47 viruses.
FIGURE 3.
Abs produced in response to A/California/04/09 cross-react with a large spectrum of older H1N1 influenza viruses. We infected cohorts of BALB/c/mice sublethally with live mouse-adapted swine-origin A/CA/04/2009, collected their sera 1 mo postinfection, and tested their ability to cross-react with a panel of 1930–2000 H1N1 viruses, using HAI assay (A). We then challenged these animals with 3 × LD50 mouse-adapted A/PR/8/34 or A/FM/1/47 strains and monitored for survival (B) and signs of morbidity (C), as reflected in the body weight changes up to 14 d postchallenge. The graphs represent the mean ± SD.
Abs produced in response to the 2009 H1N1 A/California/04/09 virus cross-react with H3N2 influenza viruses
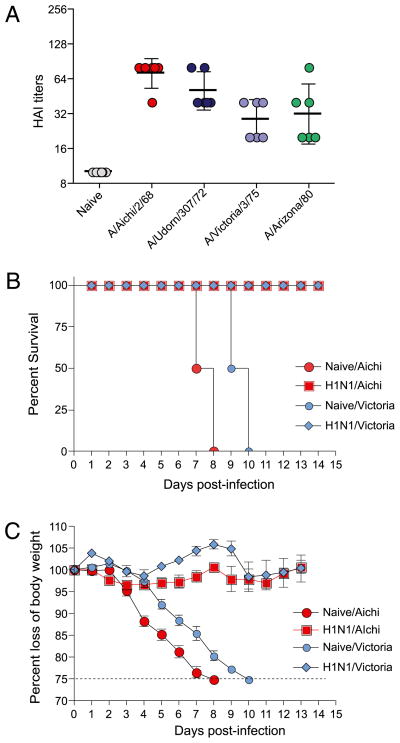
Because Abs produced in response to the 2009 H1N1 virus cross-reacted with a large spectrum of H1N1 viruses, we also tested whether these Abs would cross-react against H3N2 influenza viruses, A/Achi/2/68, A/Udorn/307/72, A/Victoria/3/75, and A/Arizona/2/80. Interestingly, we observed cross-reactivity against all four H3N2 viruses (Fig. 4A). Next we determined whether this heterosubtypic immunity could induce protection from a lethal challenge with mouse-adapted H3N2 viruses. We infected cohorts of BALB/c mice with 0.5 × LD50 live mouse-adapted A/California/04/09 virus. A month later, we lethally challenged these animals with 3 × LD50 of mouse-adapted H3N2 viruses, A/Aichi/2/68 or A/Victoria/3/75, and monitored them for survival (Fig. 4B) and morbidity (Fig. 4C). Of the mice previously exposed to A/California/04/2009, 100% survived lethal challenge with A/Aichi/2/68 or A/Victoria/3/75. In contrast, all naive control mice that were lethally challenged with the two H3N2 viruses succumbed to the infection and, by days 8–11, either died or lost >25% of their body weight and were euthanized per IACUC guidelines. Morbidity, as measured by weight loss (Fig. 4C), hunched posture, ruffled fur, and failure to thrive, was minimal in mice previously exposed to A/California/04/2009 virus. These mice lost <5% of their body weight following lethal challenge with H3N2 viruses. Taken together, our data show that previous exposure to A/California/04/09 virus gives heterosubtypic protective immunity against lethal challenge with H3N2 influenza viruses.
FIGURE 4.
Exposure to A/California/04/09 induces heterosubtypic cross-reactivity and protective immunity against H3N2 influenza viruses. We infected cohorts of BALB/c/mice sublethally with live mouse-adapted swine-origin A/CA/04/2009, collected their sera 1 mo postinfection, and tested their ability to cross-react with a panel of H3N2 viruses, using HAI assay (A). We then challenged these animals with 3 × LD50 mouse-adapted H3N2 viruses, A/Aichi/2/68 or A/Victoria/3/75, and monitored them for survival (B) and signs of morbidity (C), as reflected in the body weight changes up to 14 d postchallenge. The graphs represent the mean ± SD of five mice per group.
Vaccination with older H1N1 viruses, especially A/FM/1/47, protects against lethal challenge with mouse-adapted swine-origin H1N1 influenza A/California/04/09 virus
To determine whether vaccination with older H1N1 viruses—A/PR/8/34 or A/FM/1/47 viruses—could induce protective immunity against the 2009 H1N1 virus, we immunized cohorts of mice with 10 μg (1800 HA units) of formalin-inactivated A/FM/1/47 or A/PR/8/34 influenza viruses. The immunizations induced HAI titers (mean ± SEM) of 960 ± 149 and 1180 ± 67 against homologous viruses, A/PR/8/34 and A/FM/1/47, respectively, at day 28 postvaccination. We measured the cross-reactive serum IgG levels (Fig. 5A) and HAI (Fig. 5B) against A/California/04/09 virus at days 7, 14, and 28 postimmunization. At day 28, both A/FM/1/47- and A/PR/8/34-vaccinated mice had significantly higher titers of cross-reactive IgG Abs than did control mice (p < 0.001 for A/FM/1/47 and p < 0.05 for A/PR/8/34). Similarly, immunization with A/FM/1/47 or A/PR/8/34 induced significantly higher cross-reactive HAI titers against A/CA/04/2009. Next, we sought to determine whether these cross-reactive Abs would protect immunized mice against lethal challenge. One month following immunization with inactivated A/FM/1/47 or A/PR/8/34 virus, we challenged these mice with 3 × LD50 live mouse-adapted A/California/04/09 virus and monitored their survival (Fig. 5C) and morbidity (Fig. 5D). All of the naive control naive mice either died or were euthanized by day 8–9 because they lost >25% of their initial body weight. Mice immunized with A/FM/1/47 virus fared better than those immunized with A/PR/8/34. The survival rates were 100% for mice immunized with A/FM/1/47 virus and 80% for mice vaccinated with A/PR/8/34. We also assessed morbidity by monitoring their body weights daily (Fig. 5D). Mice immunized with A/FM/1/47 lost less weight than did the A/PR/8/34-immunized group. Maximal weight loss of 17% occurred at day 7; by day 14 they regained, on average, 96% of their initial body weight. Mice immunized with A/PR/8/34 exhibited maximal weight loss of 21% at day 8, and by day 14 they regained their weights. Taken together, these data demonstrate that immunization with inactivated A/FM/1/47 and A/PR/8/34 viruses induces protective immunity against A/California/04/09 and that the A/FM/1/47 virus is more effective than A/PR/8/34 in inducing protective immunity.
FIGURE 5.
Vaccination with inactivated A/FM/1/47 or A/PR/8/34 virus induces robust Ab responses and protective immunity against A/CA/04/2009. We immunized cohorts of BALB/c mice with 10 μg (1800 HA units) of formalin-inactivated A/FM/1/47 or A/PR/8/34 influenza virus. We collected their sera at days 7, 14, and 28 postimmunization and determined the A/California/04/09-specific ELISA (A) and HAI titers (B). Naive, unimmunized mice served as negative controls. The results are the average of two independent experiments, and error bars represent SEM. One month following immunization, we challenged them by intranasal infection with 3 × LD50 of live mouse-adapted A/California/04/09 virus. Postchallenge, the survival rates (C) and body weight changes (D), indicative of morbidity, were monitored for 14 d. The graphs represent the mean ± SEM of five mice per group at each time point.
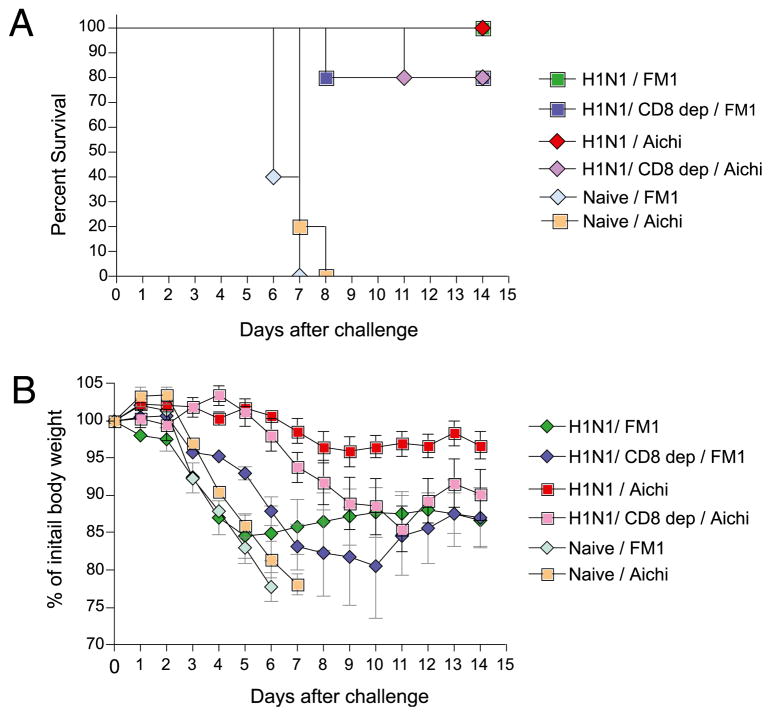
CD8+ T cells are critical for cross-protective immunity
On the basis of the data presented (Figs. 3, 4), it is clear that exposure to 2009 H1N1 virus induces cross-protection against H1N1 strains as well as H3N2 strains. Although it is evident from cross-reactive HAI titers that Abs play a critical role in cross-protection, the extent to which cross-reactive CD8 T cells participate in this is unknown. To address this issue, we did in vivo CD8 depletion studies. Briefly, we administered rat anti-mouse CD8 mAb (clone YTS169.4) to cohorts of mice previously immune to A/California/04/09. Control immune mice received normal rat IgG. The CD8 depletion was complete, as determined by flow cytometry using fluorochrome-labeled rat anti-mouse CD8 mAb (clone 53-6.7) (data not shown). Cohorts of A/California/04/09-immune mice, either CD8 T cell depleted or nondepleted, were challenged intranasally with live mouse-adapted 3 × LD50 A/FM/1/47 (H1N1) or A/Aichi/2/68 (H3N2) influenza viruses. The animals were monitored for survival (Fig. 6A) and body weight changes (Fig. 6B) for 14 d. As expected, 100% of the A/California/04/09-immune mice whose CD8 T cells were not depleted survived lethal challenge with A/FM/1/47 or A/Aichi/2/68 viruses. In contrast, 80% of the CD8-depleted mice survived lethal challenge (Fig. 6A). All of the naive control mice died or lost >25% of their initial body weight and were euthanized per IACUC guidelines by day 6 following lethal challenge. In addition, the morbidity, as measured by body weight loss, was more severe in the CD8-depleted group (Fig. 6B). The maximal weight loss in the nondepleted group was ~5%, compared with 15% in the CD8-depleted cohort. Altogether, these data show that CD8 T cells also play a critical part in cross-protection and suggest that the 2009 H1N1 virus shares CD8 T cell epitopes with the A/FM/1/47 or A/Aichi/2/68 virus.
FIGURE 6.
CD8 T cells are critical for cross-protection. Cohorts of BALB/c mice sublethally infected with 0.1 × LD50 live mouse-adapted swine origin A/CA/04/2009 and allowed to proceed to memory phase were depleted of CD8 T cells with anti-CD8 mAbs. Following CD8 depletion, these mice were challenged intranasally with 3 × LD50 mouse-adapted A/FM/1/47 or A/Aichi/2/68 strains and monitored for survival (A) and body weight changes (B) up to 14 d postchallenge. Immune yet non–CD8-depleted mice and naive mice served as controls. The body weight changes of the survivors are plotted as mean ± SEM.
Discussion
Our data provide a basis for the recent reports that people >50 y of age are more resistant to the new H1N1 pandemic virus. Using mouse-adapted influenza viruses, we demonstrate that of the animals exposed to 1930–2000 influenza viruses only those infected with strains that appeared prior to 1957 cross-react successfully with the 2009 swine-origin influenza virus. Moreover we found a reciprocal phenomenon; sera from mice infected with the A/California/04/09 strain cross-reacted with the older H1N1 strains. In addition to the in vitro ELISA and HAI studies of cross-reactivity, we further showed that exposure to A/FM1/47 or A/PR/8/34 protected against lethal challenge by A/California/04/09 and that animals previously exposed to A/California/04/09 survived the lethal challenge with A/PR/8/34 or A/FM/1/47 viruses. Thus our data show that in mice, previous exposure to old H1N1 strains can provide cross-protection against the 2009 H1N1 strain.
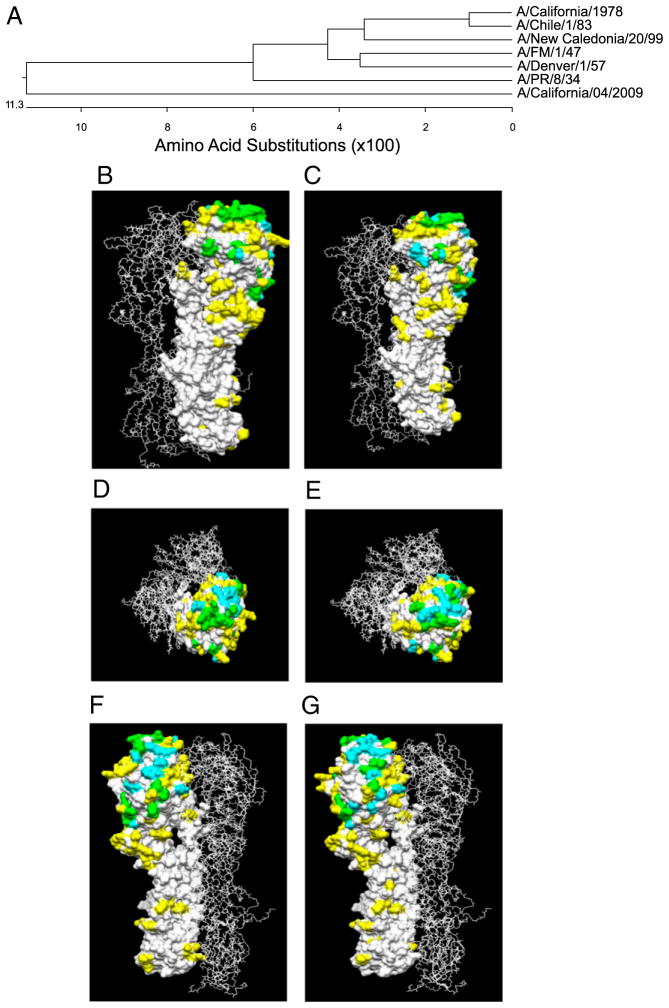
To determine a plausible explanation for the observed cross-protection between 2009 H1N1 and other H1N1 viruses, we compared the amino acid sequences, as well as the immunodominant antigenic epitopes, between the strains. First, we aligned the H1 HA amino acid sequence of A/California/04/09 with the HA amino acid sequences from A/PR/8/34, A/FM/1/47, A/Denver/1/57, A/California/10/1978, A/Chile/1/83, and A/New Caledonia/20/99. Overall, the HA protein from A/California/04/09 shared <90% sequence similarity with the 1930–2000 isolates (Fig. 7A). The HA molecule comprises a membrane-distal globular head (HA1) and a membrane-proximal stalk region (HA2). The HA1 chain of A/California/04/09 exhibits the highest sequence identity to A/PR/8/34 (75.2%), followed by A/Denver/1/57 (74.2%) and A/FM/1/47 (73.7%), and the lowest identity is to the chronologically more distant strain A/New Caledonia/20/99 (72.1%). In contrast, the HA2 chain of the 1930–2000 strains shares sequence similarity to A/California/04/09, ranging from 91–93%. Overall, the HAs of A/PR/8/34 and A/FM/1/47 are 92% identical, and these two HAs are 80% identical to A/California/04/09.
FIGURE 7.
A, Phylogenetic analysis of the 1930–2000 H1N1 influenza virus strains. The H1 HA amino acid sequence from A/California/04/09 was aligned to the HA amino acid sequences from A/Puerto Rico/8/34, A/Fort Monmouth/1/47, A/Denver/1/57, A/California/10/1978, A/Chile/1/83, and A/New Caledonia/20/99. The multisequence alignment and the phylogenetic analysis showed the sequence identity of the tested isolates to the swine-origin H1N1 and their relatedness. Comparative modeling of A/PR/8/34, A/FM/1/47, and A/California/04/09 are shown in B–G. The HA proteins were superimposed onto the crystal structure of the A/PR/8/34 HA trimer. The differences in primary amino acid sequence between the A/California/04/09 HA and the A/FM/1/47 (B, D, F) and the A/California/04/09 HA and A/PR/8/34 (C, E, G) are highlighted in yellow. Antigenic sites determined by mapping on the A/PR/8/34 HA protein (22) are highlighted in cyan, and regions of overlap are highlighted in green.
The crystal structure of the A/PR/8/34 HA molecule has been solved (20). In addition, antigenic mapping of this HA has defined five immunodominant B cell epitopes: Sa, Sb, Ca1, Ca2, and Cb (21). Because we observed the highest cross-reactive HAI titers against A/California/04/09 in mice exposed to A/FM/1/47 and A/PR/8/34 strains, we determined whether the immunodominant B cell epitopes are shared between these viruses by comparing the conservation of these B cell epitopes. We did this by superimposing pairwise the HAs of A/California/04/09 with A/FM/1/47 (Fig. 7B, 7D, 7F) or A/PR/8/34 (Fig. 7C, 7E, 7G) onto the crystal structure of the A/PR/8/34 HA trimer (PDB 1HG1) (22). We observed that the Cb, Ca1, and Ca2 antigenic sites are 100%, 75%, and 80% conserved between A/California/04/2009 and A/FM/1/47 or A/PR8/34, respectively. The Sa antigenic site in A/California/04/2009 was conserved 100% in A/FM/1/47 and 66% in AP/PR/834, and the Sb site was conserved 66% in A/FM/1/47 and 50% in A/PR/8/34, respectively, as compared with A/California/04/2009. Taken together, there is a high degree of conservation in the predicted immunodominant B cell epitopes between these three viruses; this could plausibly explain the observed cross-reactivity.
Our data also show that exposure to the 2009 H1N1 virus can cross-protect against H3N2 viruses. Such heterosubtypic cross-protection is not unique to mice (23, 24); it has been demonstrated in chickens (25), ferrets (26), pigs (27, 28), and cotton rats (29). The effect of heterosubtypic immunity is still debated in humans (30–34). However, there is indirect evidence in humans of cross-protection against influenza strains due to cross-reactive T cells (35) and even Abs (36).
A critical role for CD8 T cells in heterosubtypic cross-protection has been proposed in the past (37, 38). Our studies agree with this idea; depletion of CD8 T cells in mice immune to the 2009 H1N1 virus, followed by lethal challenge with 3 × LD50 mouse-adapted A/Aichi/2/68, led to significant decrease in the survival rate. This result suggests that cross-reactive CD8 T cells may perhaps function in heterosubtypic immunity. We also found that CD8 T cells are important for cross-reactivity against the 1947 H1N1 virus, A/FM/1/47. Altogether, our data suggest that CD8 T cell epitopes are conserved between the 2009 H1N1 as well as the 1968 H3N2 and 1947 H1N1 viruses.
Our results also show that vaccination with older strains, particularly A/FM/1/47, gives protection against lethal challenge with 3 × LD50 mouse-adapted A/California/04/2009. This observation is important because it suggests that pre-existing vaccine strains can be used for inducing protective immunity against the 2009 H1N1 virus. In addition, the cross-reactivity was greater in the reverse order: mice exposed to A/California/04/2009 exhibited cross-reactive HAI titers to H1N1 viruses, A/Denver/1/57, A/California/10/78, A/FM/1/47, and A/PR/8/34, and, to a lesser extent, A/Chile/1/83. Abs produced against A/California/04/2009 also cross-reacted with the four H3N2 influenza viruses that we tested. This finding suggests that HAI Abs produced in response to A/California/04/2009 virus can cross-react with B cell epitopes that are found in a variety of H1N1 and H3N2 influenza viruses. It is intriguing that the reverse is not true (i.e., recent H1N1 strains [A/California//10/78, A/Chile/1/83, and A/New Caledonia/20/99] do not induce cross-reactive Abs against the 2009 H1N1 virus). A plausible explanation could presumably be that cross-reactive B cell epitopes on these viruses are not immunogenic or immunodominant because they are immunologically inaccessible owing to mutation or glycosylation in the neighboring residues. The key to better understanding this enigma lies in the identification of these cross-reactive epitopes. Nonetheless, the fact that the 2009 H1N1 virus can induce such cross-reactive Abs raises the intriguing possibility that viruses such as A/California/04/2009 can be used for vaccines to induce broadly cross-reactive humoral immune responses against influenza viruses. Identifying the mechanism behind this broad reactivity may enable us to design broadly cross-reactive universal influenza vaccines.
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Contract HHSN266 200700006C. J.J. is a research scholar of the American Cancer Society.
We thank Dr. Peter Palese (Mt. Sinai Medical Center, New York, NY) and members of the Jacob laboratory for helpful discussions, as well as Leela Thomas for excellent mouse colony management and Dianne Agapito for technical assistance.
Abbreviations used in this paper
- HA
hemagglutinin
- HAI
hemagglutination inhibition
- IACUC
Institutional Animal Care and Use Committee
- NA
neuraminidase
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Gottschalk A, Graham ER. 6-alpha-D-Sialyl-N-acetyl-galactosamine: the neuraminidase-susceptible prosthetic group of bovine salivary mucoprotein. Biochim Biophys Acta. 1959;34:380–391. doi: 10.1016/0006-3002(59)90290-2. [DOI] [PubMed] [Google Scholar]
- 2.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 3.Palese P, Schulman JL. Mapping of the influenza virus genome: identification of the hemagglutinin and the neuraminidase genes. Proc Natl Acad Sci USA. 1976;73:2142–2146. doi: 10.1073/pnas.73.6.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 6.Kawaoka Y, Bean WJ, Webster RG. Evolution of the hemagglutinin of equine H3 influenza viruses. Virology. 1989;169:283–292. doi: 10.1016/0042-6822(89)90153-0. [DOI] [PubMed] [Google Scholar]
- 7.Schäfer JR, Kawaoka Y, Bean WJ, Süss J, Senne D, Webster RG. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194:781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz B, Gellin B. Vaccination strategies for an influenza pandemic. J Infect Dis. 2005;191:1207–1209. doi: 10.1086/428952. [DOI] [PubMed] [Google Scholar]
- 9.Dowdle WR, Hattwick MA. Swine influenza virus infections in humans. J Infect Dis. 1977;136(Suppl):S386–S389. doi: 10.1093/infdis/136.supplement_3.s386. [DOI] [PubMed] [Google Scholar]
- 10.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solórzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, García-Sastre A. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 11.Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir Ther (Lond) 2007;12(4 Pt B):581–591. [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210. doi: 10.1016/s0168-1702(02)00027-8. [DOI] [PubMed] [Google Scholar]
- 13.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–2615. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- 15.Katz J, Hancock K, Veguilla V, Zhong W, Lu WH, Sun H, Butler E, Dong L, Liu F, Zin ZN, DeVos J, Gargiullo P. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep. 2009;58:521–524. [PubMed] [Google Scholar]
- 16.Skountzou I, Quan FS, Jacob J, Compans RW, Kang SM. Transcutaneous immunization with inactivated influenza virus induces protective immune responses. Vaccine. 2006;24:6110–6119. doi: 10.1016/j.vaccine.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Reed LJaMH. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 18.WHO/CDS/CSR/NCS. WHO Manual of Animal Influenza Diagnosis and Surveillance. Department of Communicable Disease Surveillance and Response; 2002. [Google Scholar]
- 19.Noort WA, Benner R, Savelkoul HF. Differential effectiveness of anti-CD8 treatment on ongoing graft-versus-host reactions in mice. Transpl Immunol. 1996;4:198–202. doi: 10.1016/s0966-3274(96)80017-7. [DOI] [PubMed] [Google Scholar]
- 20.Russell RJ, Gamblin SJ, Haire LF, Stevens DJ, Xiao B, Ha Y, Skehel JJ. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology. 2004;325:287–296. doi: 10.1016/j.virol.2004.04.040. [DOI] [PubMed] [Google Scholar]
- 21.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 22.Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 23.Benton KA, Misplon JA, Lo CY, Brutkiewicz RR, Prasad SA, Epstein SL. Heterosubtypic immunity to influenza A virus in mice lacking IgA, all Ig, NKT cells, or gamma delta T cells. J Immunol. 2001;166:7437–7445. doi: 10.4049/jimmunol.166.12.7437. [DOI] [PubMed] [Google Scholar]
- 24.Kreijtz JH, Bodewes R, van Amerongen G, Kuiken T, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. Primary influenza A virus infection induces cross-protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine. 2007;25:612–620. doi: 10.1016/j.vaccine.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 25.Imai K, Nakamura K, Mase M, Tsukamoto K, Imada T, Yamaguchi S. Partial protection against challenge with the highly pathogenic H5N1 influenza virus isolated in Japan in chickens infected with the H9N2 influenza virus. Arch Virol. 2007;152:1395–1400. doi: 10.1007/s00705-007-0953-x. [DOI] [PubMed] [Google Scholar]
- 26.Suguitan AL, Jr, McAuliffe J, Mills KL, Jin H, Duke G, Lu B, Luke CJ, Murphy B, Swayne DE, Kemble G, Subbarao K. Live, attenuated influenza A H5N1 candidate vaccines provide broad cross-protection in mice and ferrets. PLoS Med. 2006;3:e360. doi: 10.1371/journal.pmed.0030360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinen PP, de Boer-Luijtze EA, Bianchi AT. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J Gen Virol. 2001;82:2697–2707. doi: 10.1099/0022-1317-82-11-2697. [DOI] [PubMed] [Google Scholar]
- 28.Reeth KV, Brown I, Essen S, Pensaert M. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 2004;103:115–124. doi: 10.1016/j.virusres.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Straight TM, Ottolini MG, Prince GA, Eichelberger MC. Evidence of a cross-protective immune response to influenza A in the cotton rat model. Vaccine. 2006;24:6264–6271. doi: 10.1016/j.vaccine.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 30.Jameson J, Cruz J, Terajima M, Ennis FA. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J Immunol. 1999;162:7578–7583. [PubMed] [Google Scholar]
- 31.Steinhoff MC, Fries LF, Karron RA, Clements ML, Murphy BR. Effect of heterosubtypic immunity on infection with attenuated influenza A virus vaccines in young children. J Clin Microbiol. 1993;31:836–838. doi: 10.1128/jcm.31.4.836-838.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. Cross-subtype protection in humans during sequential, overlapping, and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. J Infect Dis. 1985;151:81–88. doi: 10.1093/infdis/151.1.81. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien RJ, Noble GR, Easterday BC, Kendal AP, Shasby DM, Nelson DB, Hattwick MA, Dowdle WR. Swine-like influenza virus infection in a Wisconsin farm family. J Infect Dis. 1977;136(Suppl):S390–S396. doi: 10.1093/infdis/136.supplement_3.s390. [DOI] [PubMed] [Google Scholar]
- 34.Gaydos JC, Hodder RA, Top FH, Jr, Soden VJ, Allen RG, Bartley JD, Zabkar JH, Nowosiwsky T, Russell PK. Swine influenza A at Fort Dix, New Jersey (January–February 1976). I. Case finding and clinical study of cases. J Infect Dis. 1977;136(Suppl):S356–S362. doi: 10.1093/infdis/136.supplement_3.s356. [DOI] [PubMed] [Google Scholar]
- 35.Boon AC, de Mutsert G, van Baarle D, Smith DJ, Lapedes AS, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. Recognition of homo- and heterosubtypic variants of influenza A viruses by human CD8+ T lymphocytes. J Immunol. 2004;172:2453–2460. doi: 10.4049/jimmunol.172.4.2453. [DOI] [PubMed] [Google Scholar]
- 36.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villán E, Palese P, Basler CF, García-Sastre A. Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 2010;6:e1000745. doi: 10.1371/journal.ppat.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura S, Tanimoto T, Kurata T. Mechanisms of broad cross-protection provided by influenza virus infection and their application to vaccines. Jpn J Infect Dis. 2005;58:195–207. [PubMed] [Google Scholar]
- 38.Selin LK, Brehm MA, Naumov YN, Cornberg M, Kim SK, Clute SC, Welsh RM. Memory of mice and men: CD8+ T-cell cross-reactivity and heterologous immunity. Immunol Rev. 2006;211:164–181. doi: 10.1111/j.0105-2896.2006.00394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]