Abstract
The maturation of [NiFe]-hydrogenase in E. coli is a complex process involving many steps and multiple accessory proteins. The two accessory proteins, HypA and HypB, interact with each other and are thought to cooperate to insert nickel into the active site of the hydrogenase-3 precursor protein. Both of these accessory proteins bind metal individually, but little is known about the metal-binding activities of the proteins once they assemble together into a functional complex. In this study, we investigate how complex formation modulates metal binding to the E. coli proteins HypA and HypB. This work lead to a re-evaluation of the HypA nickel affinity, revealing a KD on the order of 10−8 M. HypA can efficiently remove nickel, but not zinc, from the metal-binding site in the GTPase domain of HypB, a process that is less efficient when complex formation between HypA and HypB is disrupted. Furthermore, nickel release from HypB to HypA is specifically accelerated when HypB is loaded with GDP, but not GTP. These results are consistent with the HypA-HypB complex serving as a transfer step in the relay of nickel from membrane transporter to its final destination in the hydrogenase active site, and suggest that this complex contributes to the metal fidelity of this pathway.
Keywords: Metalloprotein, metal chaperone, nickel metabolism, nickel transfer, [NiFe] hydrogenase, hydrogenase maturation, HypA, HypB
Many enzymes require transition metal ions at their active sites in order to function,1, 2 and access to the appropriate metals is often vital for the survival of the organism. Strikingly, the same metals that are essential can also be toxic when distribution is not properly controlled.3, 4 An intracellular excess of one type of metal may result in competition with other metals required as cofactors for regulatory or enzymatic processes,3, 5 or catalyze the formation of free radicals,6 both detrimental circumstances. For this reason, organisms have intricate systems dedicated to the controlled flow of essential metals throughout the cell.4, 7–9 For example, through the use of metallochaperone proteins metal ions can be directed to where they are needed, minimizing the demand for freely available metals in the cytoplasm.10–13 In many cases, metallochaperones are credited with ensuring that the correct metal ions are delivered to the appropriate metalloenzyme precursor proteins through targeted protein-protein interactions.
Hydrogenases are a group of metalloenzymes that catalyze the reversible oxidation of hydrogen gas to protons and electrons. They contribute to the metabolism of many species, especially under anaerobic conditions.14–16 The active sites of [NiFe]-hydrogenases contain a complex NiFe(CN)2CO bimetallic cofactor and the proper maturation of this metallocenter is a multistep process that requires a team of accessory proteins.14, 17 The isolation of hydrogenase precursor proteins that contain the Fe(CN)2CO cofactor, but no nickel, provided support for two distinct stages of metallocenter assembly in which the nickel ion is delivered after iron insertion.18, 19 Studies of Escherichia coli [NiFe]-hydrogenase-3 suggested that at least seven accessory proteins participate in the maturation process, encoded by the hypABCDEF and slyD genes.14, 20 Nickel insertion requires HypB and HypA,17, 21 the latter protein is replaced by the homologous HybF protein in the production of hydrogenases-1 and -2.22 Cells lacking either of these accessory proteins produce immature, inactive hydrogenase that can be at least partially restored by growing the mutant bacteria in media supplemented with extra nickel,22–24 suggesting that these factors are responsible for shepherding nickel to the hydrogenase enzyme under nickel limited conditions. SlyD also enhances the ability of E. coli to produce functioning hydrogenase by contributing to nickel delivery,25 although it is not absolutely required.
E. coli HypB is a GTPase that contains two metal-binding sites.23, 26 The first site, essential for hydrogenase production,27 is located in the GTPase domain (G-domain) and binds nickel or zinc with micromolar affinities (KD of 1 HM vs 12 μM for zinc and nickel, respectively).26 Binding of either metal to the G-domain inhibits the GTPase activity, with zinc exerting a stronger impact than nickel.28 Although the molecular role of the GTPase cycle in hydrogenase biosynthesis has not yet been defined, the impact of metal binding on GTP hydrolysis suggested that this metal site has a regulatory function. E. coli HypB also contains a second, high-affinity metal-binding site, located at the N-terminal CXXCGC motif.26, 29 This site binds nickel with a sub-picomolar affinity,26 and is also known to bind other transition metals.30 Despite being absent in some HypB homologs (notably, HypB from Helicobacter pylori), the high-affinity metal-binding site of HypB is also essential for hydrogenase production in E. coli.27
The other nickel accessory protein required for hydrogenase-3 biosynthesis, HypA, forms a complex with HypB and is also capable of binding two equivalents of metal.31, 32 The first site binds zinc tightly through a Cys4 coordination and is thought to act as a structural motif,31, 33, 34 although spectroscopic analysis of H. pylori HypA suggested that it may contribute to a switch in HypA function in this organism.35 The second HypA metal-binding site is capable of binding nickel with a reported KD in the mid-micromolar range.31, 32, 36 Although the nickel coordination site of HypA remains to be completely defined, it includes His2 at the N-terminus,32, 34, 36 and mutation of this residue in homologous proteins abrogates hydrogenase production.32, 36
HypA and HypB can interact with each other in E. coli in the absence of the hydrogenase precursor protein, suggesting that they preassemble before reaching HycE.37, 38 Furthermore, deletion of the hypA gene prevents HypB from forming a complex with HycE, suggesting that HypA serves as a scaffold that docks the other nickel accessory proteins onto the hydrogenase precursor protein.37 There is also evidence that HypA is only capable of associating with the hydrogenase enzyme after it has been loaded with iron,37 consistent with the model of two distinct stages of metal delivery to the enzyme active site.
While HypA and HypB have been studied extensively in isolation, it is unclear how they behave once they assemble during production of the metallocenter of the [NiFe]-hydrogenase-3. In this study, we examined metal binding within the HypA-HypB complex, and discovered that nickel relocates from the G-domain site of HypB to HypA. Disruption of the protein-protein interaction slows metal transfer. Furthermore, this process is modulated by the nucleotide-loaded state of HypB, and is not observed for zinc. These results shed light on the role of the HypA-HypB complex, including regulation by the GTPase cycle, during nickel delivery to the [NiFe]-hydrogenase precursor protein.
Materials and Methods
Materials
Restriction enzymes, Pfu DNA polymerase, and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). Kanamycin, IPTG (isopropyl β-D-1-thiogalactopyranoside), TCEP (tris(2-carboxyethyl)phosphine), Tris (tris(hydroxymethyl)aminomethane), and PMSF (phenylmethanesulfonylfluoride) were purchased from BioShop (Toronto, ON) and all chromatography media were purchased from GE Healthcare, with the exception of the UnoQ column, which was purchased from Bio-Rad Canada (Mississauga, ON) and the Streptactin-sepharose resin, which was purchased from IBA Life Sciences (Goettingen, Germany). PMB (para-mercury-benzenesulfonic acid), PAR (4-(2-pyridylazo)resorcinol) and DTNB (5,5′-dithiobis(2-nitrobenzoic acid)) were purchased from Sigma-Aldrich, as were NiSO4 and ZnSO4 (>99.99% purity).
Plasmid Construction
Plasmids bearing the genes for the WT proteins were previously described,26,39,37 and used without modification. The L78A, V80A mutation was introduced into hypB-containing plasmids using the Phusion mutagenesis technique (Thermo Scientific). The plasmid was amplified using the 5′-phosphorylated primers (Integrated DNA Technologies) 5′-GCGGAAATTGACGTGCTGGAC-3′ (forward) and 5′-TTCCGCCATCCGACGCTG-3′ (reverse), where the mutated bases are underlined. The resulting product was digested with DpnI for two hours to remove any template DNA, then ligated with T4 DNA ligase prior to transformation. The K24E, R25E mutant was introduced in hypA-containing plasmids using the QuikChange protocol (Agilent Technologies). The plasmid was amplified using the forward primer 5′-CGCAAAACACGGCGCAGAAGAAGTAACTGGGGTCTGGCTC-3′ and the reverse primer 5′-GAGCCAGACCCCAGTTACTTCTTCTGCGCCGTGTTTTGCG-3′. The resulting PCR product was digested with DpnI to remove any template DNA.
All plasmids were transformed into NEB Turbo cells for amplification, and purified using a Qiagen Miniprep Kit. The sequences of the mutated genes were verified by dideoxy sequencing (ACGT, Toronto).
Protein Expression and Purification
Wild-type and mutant E. coli HypAStr were purified using the Strep-II purification system (IBA Life Sciences). The strA-pET24b plasmid was transformed into E. coli BL21 Star (DE3) (Invitrogen). Cells were grown at 37 °C in LB media supplemented with 50 μg/mL kanamycin, until the culture reached an OD 0.6 – 0.8. At this point, 1 μM ZnSO4 was added and overexpression of HypAStr was induced by addition of 700 μM IPTG, followed by overnight growth at 15 °C and then centrifugation at 4 °C. The resulting cell pellets were resuspended in lysis buffer (20 mM Tris, pH 7.6, 100 mM NaCl). The cells were lysed by using sonication in the presence of 5 mM PMSF and 5 mM TCEP, and then spun at 25000 g for 45 min at 4 °C to remove cell debris. The lysate was passed through a Streptactin-Sepharose column, followed by washing with 10 volumes of high salt wash buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 10% glycerol). The protein was then eluted from the column using 2.5 mM desthiobiotin (Sigma) in the same high salt wash buffer. Fractions were screened on a 15% SDS-polyacrylamide gel. HypAStr-containing fractions were dialyzed into Buffer AG (20 mM Tris, pH 7.5, 10% glycerol), and further purified through an UnoQ anionic exchange column by using a NaCl gradient in Buffer AG. HypAStr typically eluted at 400 mM NaCl. HypAStr concentration was determined by using the extinction coefficient of 18350 M−1cm−1 at 280 nm.40 The molecular mass of purified, denatured HypAStr was observed by electrospray ionization mass spectrometry (ESI-MS, Department of Chemistry, University of Toronto) to be 14208 Da (calculated mass: 14208.3 Da). Similarly, the molecular mass of the K24E, R25E HypAStr was observed to be 14181 Da (calculated: 14182.1 Da).
Wild-type and mutant E. coli HypB were overexpressed and purified as previously described.26 The oxidation state of proteins was determined by DTNB assay. Briefly, free thiols in the protein were quantified in the presence of 4 M GuHCl and 1 mM EDTA. The product of the reaction of DTNB with free thiols, 5-mercapto-2-nitrobenzoic acid, was quantified at 412 nm, and compared to a standard curve prepared with β-mercaptoethanol. Proteins were judged to be fully reduced when >95% of the cysteine residues in a protein were reactive with DTNB (data not shown).
Both HypB and HypAStr were co-purified with bound metals. HypB was typically purified with one equivalent of nickel, likely bound to the N-terminal high-affinity site. HypAStr co-purified with one equivalent of zinc. The presence of these metals after purification was considered to be an indicator of proper protein folding and oxidation state, and no steps were taken to remove them. Bound metal was quantified in purified proteins by using a PAR assay. Proteins were incubated with 4 M GuHCl and 1 mM PMB. After one hour of incubation, 100 μM PAR was added to the sample, and the amount of metal present was quantified by using the signal at 494 nm, which corresponds to the (PAR)2Me(II) complex. HypAStr samples were compared to a standard curve of ZnSO4, while HypB samples were instead compared to a standard curve of NiSO4. Proteins were deemed fit for further experiments when > 90% metal loading was observed (data not shown). More details regarding the metal-loaded states of each protein and the corresponding nomenclature can be found in Supplemental Table 1.
Determination of HypAStr Nickel Affinity
To determine the KD of HypAStr for Ni(II), as-purified HypAStr protein was dialyzed into working buffer (25 mM HEPES, pH 7.5, 100 mM KCl) in an anaerobic glovebox (95% N2 and 5% H2). Competition experiments using mag-fura-2 (MF2) (Invitrogen) were performed by incubating 20 HM HypA and 20 HM MF2 with 0–60 HM of NiSO4 overnight at 4 °C in an anaerobic glovebox. The absorbance of apo-MF2 was monitored at 369 nm, then converted to fractional saturation using the equation θ = (A369 – A369i)/(A369f – A369i), where θ is the fractional saturation, A369 is the absorbance of the sample at 369 nm, A369i is the absorbance of the sample in the absence of metal, and A369f is the absorbance of the sample under metal saturation conditions. The data were fitted in the program DYNAFIT41 using a custom script (Supplemental Table 2).
Electrospray Mass Spectrometry
Protein samples were buffer exchanged into MS buffer (10 mM ammonium acetate, pH 7.5) using two consecutive PD-10 desalting columns (GE Healthcare). For time-resolved MS experiments, the indicated amounts of NiSO4 were added to HypAStr immediately prior to rapid infusion into the MS, which was sufficient time for nickel binding. For time-resolved nickel transfer experiments, HypB was incubated with ten equivalents of nickel sulfate at 4 °C overnight in an anaerobic glovebox, and then desalted through a PD-10 column into MS buffer to remove unbound nickel. HypAStr was added immediately prior to rapid infusion into the mass spectrometry. Experiments were monitored for 10 min, and the relative abundance of protein species were extracted from the mass spectra based on the height of the reconstructed peak.
The mass spectrometry data were recorded in positive ion mode using an AB Sciex QStar XL mass spectrometer equipped with a hot source-induced desolvation (HSID) interface (Ionics Mass Spectrometry Group Inc.). Ions were scanned from 800–3000 m/z with 1 s accumulation and no interscan delay. Instrument parameters were as follows: ion source temperature, 200 °C; ion source gas, 50 psi; curtain gas, 50 psi; ion spray voltage, 5000 V; declustering potential, 60 V; focusing potential, 60 V; collision gas, 3; MCP detection, 2200 V. Mass spectra were reconstructed by using the Bayesian protein program contained within Analyst QS (v1.1) software.
Benzyl Viologen Hydrogenase Assays
Cultures were grown in modified TYEP media, containing 10 g/L of tryptone, 5 g/L of yeast extract, 69 mM K2HPO4 and 22 mM KH2PO4, 25 supplemented with 1 μM sodium molybdate, 1 μM sodium selenite, 30 mM sodium formate, 0.8% glycerol, 100 μM arabinose, and 100 mg/L ampicillin. After inoculation with 1% (v/v) of an overnight aerobically-grown culture, the cultures were grown anaerobically in a sealed flask at 37 °C for either 6 h or 18 h. The MC4100 strain was used as a wild-type control, while DHBP cells (MC4100 ΔhypB) were either analyzed on their own or following transformation with a pBAD24-hypB plasmid. The cells were harvested by centrifugation, then washed with cold 50 mM potassium phosphate, pH 7.6, and resuspended in the same buffer supplemented with 200 μM PMSF. The cells were sonicated on ice, and the lysate separate from cell debris by centrifuging for 20 minutes at 21000 g. When cell lysate was not analyzed immediately, it was stored at −80 °C.
Total hydrogenase activity of crude cell lysates was monitored by measuring the hydrogen-dependent reduction of benzyl viologen.42, 43 Samples were prepared inside an anaerobic glovebox (95% N2 and 5% H2) and contained within a septum-sealed cuvette during the reaction. Activity was measured in units/mg of total protein, where one unit of activity corresponds to 1 Hmol of benzyl viologen reduced/min. The amount of reduced benzyl viologen was quantified by electronic absorption spectroscopy and an extinction coefficient of 7400 M−1cm−1 at 600 nm. BCA protein assays (Pierce) were used to determine total protein concentration.
Western Blot Analysis
Proteins were resolved on either 12.5% or 15% SDS- polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore) after electrophoresis. The blots were probed with the anti-HypB polyclonal rabbit antibody (Cedarlane Labs, Burlington, Canada), raised against purified HypB protein, at a 1:1000 dilution and then 2° goat anti-rabbit (Bio-Rad) antibodies conjugated to horseradish peroxidase, at a dilution of 1:30000. Enhanced chemiluminescence (SuperSignal West Pico Chemiluminscence, Pierce) was used for detection.
Metal Transfer Experiments
Metal transfer from 50 μM HypB was monitored by electronic absorption spectroscopy. Samples with a final volume of 200 μL were mixed in a quartz cuvette, and the absorbance was measured from 190 – 800 nm with an Agilient 8452 spectrophotometer. Nickel loading in the G-domain of HypB was monitored by the absorbance at 340 nm, and its extinction coefficient determined experimentally (ε = 2660 M−1cm−1 for the 1:1 HypB-nickel complex). Nickel binding to HypAStr produces no significant change in absorbance at this wavelength. Kinetic experiments were performed by first measuring the spectrum of the solution, followed by addition of the metal acceptor (either HypAStr at 70–220 μM, or a small molecule chelator at 1 mM). The absorbance at 340 nm was measured every 2 s for 5 min, then the samples were removed from the cuvette and set aside. After two hours, the solution was measured again, to determine the end point of the reaction. When nucleotides were to be included in the experiment, GDP (guanosine diphosphate) (Sigma-Aldrich, >96% purity) or GppCp (guanosine-5′-[(β,γ)-methyleno]triphosphate) (Sigma-Aldrich, >98% purity) were used at concentrations of 100 μM. All kinetic experiments were performed at room temperature in the presence of 1 mM TCEP to prevent protein oxidation. The half-lives of metal transfer reactions were calculated empirically, by noting the time at which the absorbance of the HypB-nickel absorbance band reached an intensity that is halfway between the starting point and the end point.
Results
Metal binding to HypAStr
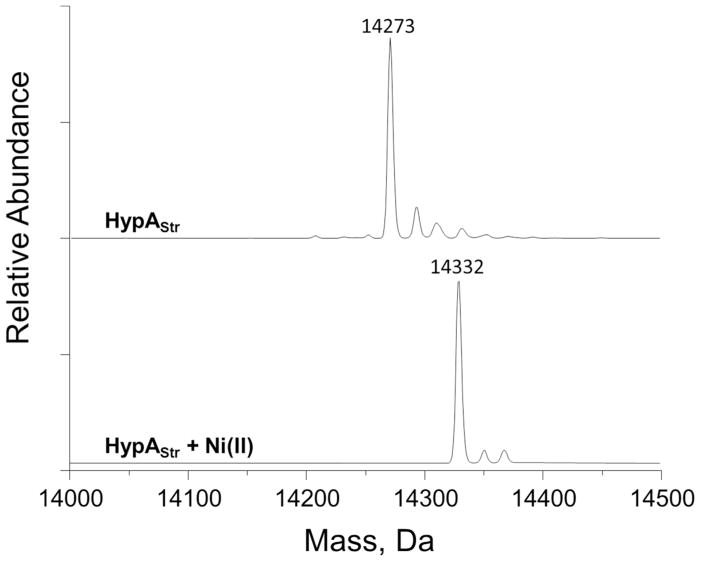
In order to simplify the purification of E. coli HypA, a Strep-tag II was appended to the C-terminus of the protein (referred to as HypAStr onwards), affording a two-step purification process of the protein.44 Previous work demonstrated that the HypAStr construct is functional in vivo.37 HypAStr co-purified with a single equivalent of zinc, likely bound to the previously characterized Cys4 metal-binding site.31 The identity of the metal ion was elucidated by mass spectrometry (Figure 1), and the stoichiometry (0.98 ± 0.05 equivalents) was confirmed by using an assay with the metallochromic indicator PAR.
Figure 1.
Mass spectrometry of Ni(II) binding to HypA. (Top) HypA was copurified with one equivalent of zinc bound to the protein (Calculated mass: 14273 Da). (Bottom) Upon addition of one equivalent of nickel, the mass of the protein shifts by 59 Da, corresponding to nickel binding (Calculated mass: 14332 Da). In addition to the expected protein peaks, small peaks at approximately +23 Da and +39 Da were observed, corresponding to sodium and potassium adducts.
Nickel binding to HypAStr was also monitored by using mass spectrometry (Figure 1). When as-purified HypAStr was mixed with one equivalent of nickel sulfate, the reconstructed mass of the protein shifted by 59 Da, corresponding to the mass of a nickel ion. Metal binding occured rapidly, and reached completion within minutes (data not shown). This result demonstrates that HypAStr is capable of binding a nickel and zinc ion simultaneously in two separate metal-binding sites, consistent with previous studies of wild-type HypA and its homologs.31, 32, 34–36 In contrast to nickel, upon the addition of supplemental zinc a HypA-Zn(II)2 species could not be detected by mass spectrometry, despite numerous attempts (data not shown), suggesting that if zinc can bind to some or all of the nickel ligands of HypA, it is with weak affinity.
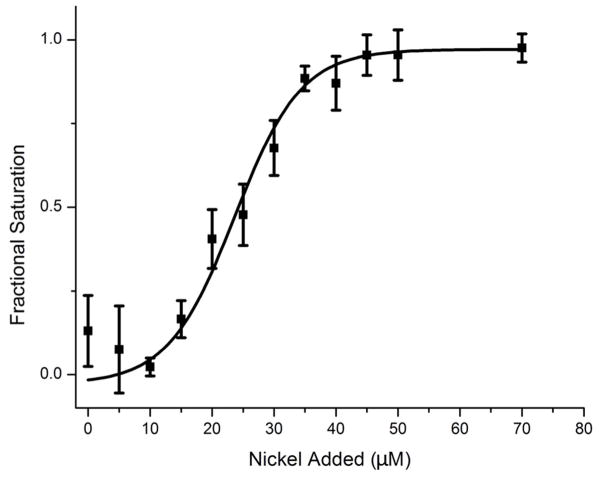
In a prior study, we reported a KD for the E. coli HypA nickel complex in the range of 10−5 M.31 This value was determined indirectly by monitoring a decrease in intrinsic tryptophan fluorescence that occurs upon titration with nickel. However, mass spectrometry revealed that HypAStr exhibited quantitative loading of nickel even at low micromolar concentrations of protein, suggesting a much tighter KD than expected. In order to resolve this issue, the affinity of the HypAStr nickel-binding site was measured in competition experiments with the metal-sensitive dye mag-fura-2 (MF2). Even though MF2 exhibits an apparent KD of 150 nM, (A. Sydor, D. B. Z., personal communication) competition for nickel between MF2 and HypAStr was observed (Figure 2). Fitting the data revealed that under these experimental conditions HypAStr bound nickel with a calculated KD of 75 ± 46 nM.
Figure 2.
Nickel-binding competition between mag-fura-2 and E. coli HypAStr. The electronic absorbance of 20 HM mag-fura-2 was measured at 369 nm in the presence of 20 HM HypAStr and converted to fractional saturation. Competition between mag-fura-2 and HypAStr suggests similar affinities for nickel. The data were fit to an apparent KD of 75 ± 46 nM for the HypAStr-nickel complex. The data are the average of three independent trials, and the error bars represent one standard deviation.
Metal binding in the HypAStr-HypB complex
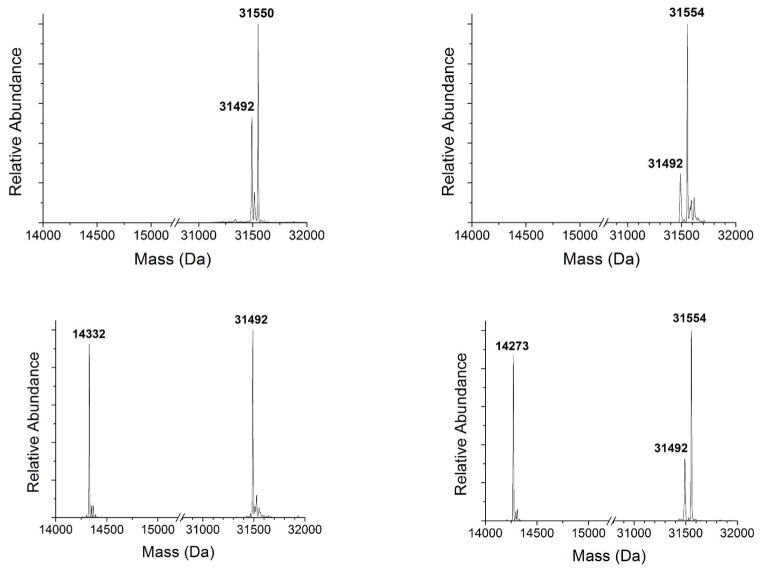
To examine the metallation states of HypA and HypB mass spectrometry was used to concurrently monitor both proteins after mixing together. These experiments were performed with as-purified HypB, which has nickel in the N-terminal high-affinity site.26 In these and in all subsequent experiments, no impact was detected on the zinc bound to as-purified HypAStr or the high-affinity nickel loading of HypB. If HypB was loaded with a second nickel ion in the G-domain site and mixed with as-purified HypAStr, HypB lost a nickel ion (Figure 3, top left, detected as a loss of 59 Da), while HypAStr gained the nickel ion (Figure 3, bottom left), indicating nickel transfer between the two proteins. In contrast, if HypB was loaded with zinc in the G-domain (Figure 3, right) the addition of HypAStr did not produce a change in the masses of either protein, indicating that only nickel can be transferred from HypB to HypAStr.
Figure 3.
Mass spectrometry reveals nickel transfer between HypB and HypAStr. (Top) Mass spectra of as-purified HypB incubated with one equivalent of either NiSO4 (left, 31550 Da) or ZnSO4 (right, 31554 Da). Under the experimental conditions used (5 μM HypB), one equivalent of metal does not yield quantitative metal loading of the HypB G-domain site. (Bottom) The addition of 5 HM HypAStr (14273 Da) to nickel-loaded HypB (left) results in the complete transfer of a nickel ion from HypB to HypAStr after five minutes of incubation at room temperature. However, the same experiment with zinc-loaded HypB (right) demonstrates that HypAStr does not affect the zinc bound to the low-affinity site of HypB. The masses of each protein in the different metal-loaded states are listed in Supplemental Table 2.
To further explore metal transfer between HypB and HypAStr, electronic absorption spectroscopy was used. Upon binding nickel in the G-domain site, HypB exhibits an absorbance band at 340 nm (Figure 4),26 corresponding to a cysteine-to-nickel ligand-to-metal charge transfer (LMCT) band. Nickel binding to HypAStr does not result in a significant change in the electronic absorption spectrum of the protein, so metal binding to HypB can be monitored at 340 nM in solutions containing both proteins. The addition of 50 μM nickel sulfate to 50 HM HypB resulted in approximately 70% loading of the G-domain metal site with nickel, consistent with the previously reported affinity of this site for nickel.26 When HypB loaded with nickel is mixed with 70 HM HypAStr, the band at 340 nm rapidly diminishes, reaching baseline within a few minutes (Figure 4). This observation is consistent with the nickel transfer from HypB to HypAStr that was observed in the mass spectrometry experiments. Furthermore, increasing the concentration of HypAStr in solution did not result in any further increase in the rate of nickel release from the HypB G-domain site (Figure 5). Finally, when HypB was incubated with nickel-loaded HypAStr, the low-affinity site absorption band at 340 nm was not observed, suggesting that nickel cannot be transferred in the reverse direction to HypB from HypAStr.
Figure 4.
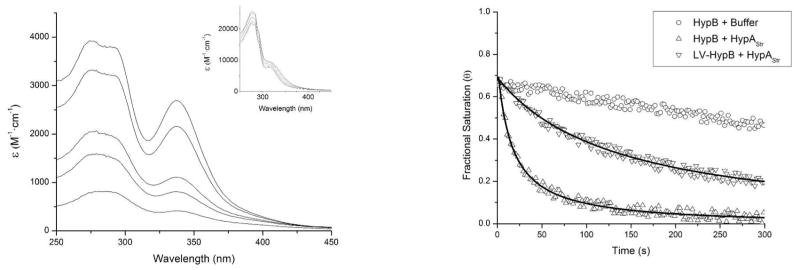
Nickel loss from the HypB G-domain site monitored by electronic absorption spectroscopy. (Left) Difference spectra and observed spectra (inset) of nickel-loaded HypB mixed with HypA and the change in the spectra over time. Initially, a peak at 340 nm was observed, corresponding to nickel binding in the G-domain of HypB. As time progresses, this peak decreased, suggesting that nickel was transferred to the spectroscopically silent nickel site in HypAStr. (Right) HypB or LV-HypB (50 μM) was mixed with one equivalent of nickel, followed by addition of 70 μM HypAStr. The 340 nm band of HypB decreased much faster in the presence of HypAStr (triangles) than in the absence of HypAStr (circles) corresponding to the transfer of nickel from the G-domain site of HypB to HypAStr. When WT HypB was used as the nickel source, nickel transfer occurred with a half-life of approximately 12 ± 3 seconds. When LV-HypB was used (upside-down triangles), the transfer occurred with a slower rate (t1/2 = 72 ± 10 s). Both transfer reactions could be fit to second order decays.
Figure 5.
Rate dependence of nickel loss from WT or mutant HypB on HypAStr concentrations. HypB loaded with nickel in the G-domain site was mixed with varying concentrations of HypAStr and the absorbance at 340 nm was monitored. The half-life of the nickel transfer was independant of acceptor concentration when 50 HM WT HypB was used as the donor. However, when 50 HM LV-HypB was used as the donor, the half-life of nickel transfer decreased with increasing concentrations of HypAStr. Concentrations of HypAStr higher than 250 μM resulted in protein precipitation, preventing accurate rate measurements. Points are the average values from three separate trials, and error bars represent one standard deviation.
Role of the HypA-HypB complex in nickel transfer
Previous in vitro analysis demonstrated that purified HypB and HypA form a complex,31, 32 and there is also evidence that this interaction occurs in vivo.37, 45 In order to explore the role of complex formation during metal transfer between the two proteins, mutations were sought out that would disrupt it. Initially, residues at the interface of the HypA-HypB complex were identified via crosslinking experiments. HypAStr and HypB were covalently trapped together by using EDC (Supplemental Figure 5), as previously reported,31 and Glu93 of HypB and Lys24 of HypAStr were identified by LC-MS/MS as the crosslinker-reactive residues, suggesting an electrostatic interaction between the two proteins. To confirm this finding, Lys24 and Arg25 of HypAStr were replaced with glutamate residues. EDC trapping of a complex between HypB and the mutant HypAStr was not detected (Supplemental Figure 5), indicating that Lys24 is a key component of the reaction with the carbodiimide. However, complex formation between HypB and the HypAStr mutant was still detectable by a pull-down assay (Supplemental Figure 6), demonstrating that reversing the charge at that location on HypA is not sufficient to disrupt the interaction, and that other parts of HypA also contribute to complex formation with HypB.
While this work was in progress, it was reported that Leu78 and Val80 of HypB are important for complex formation with HypA,45 so these two residues were replaced with alanine and the mutant protein was analyzed in vitro (L78A, V80A, or “LV-HypB”). LV-HypB retains many of the characteristics of WT HypB. It still binds nickel in the G-domain (Supplemental Figure 3) and it binds GDP with a similar affinity as WT HypB (Supplemental Figure 4). However, when complex formation with HypAStr was monitored by pull-down assay, the LV-HypB-HypAStr interaction appears to be significantly weaker than that of WT-HypB (Supplemental Figure 1), although some LV-HypB is still observed to elute with HypAStr, indicating that complex formation is not completely disrupted. Furthermore, expression of LV-HypB from an inducible plasmid is only able to partially restore hydrogenase production in a ΔhypB strain of E. coli (Supplemental Figure 2), while WT HypB restores almost 100% of the activity. These data are in agreement with the conclusion45 that Leu78 and Val80 of HypB contribute to the formation of the HypA-HypB complex and that this interaction is important during [NiFe]-hydrogenase maturation.
Next, nickel transfer from LV-HypB to HypAStr was examined. Although nickel release to HypAStr was observed, it was approximately 6-fold slower than when WT-HypB was used (Figure 4). Furthermore, in contrast to the reaction with wild-type HypB, the addition of increasing concentrations of HypAStr caused nickel release from LV-HypB to speed up (Figure 5). Altogether, these results suggest that protein-protein interactions between HypB and HypA are an important component of nickel transfer between the two proteins.
In addition to the metal site in the G-domain, E. coli HypB also binds nickel with high affinity to a site located at the N-terminus.26, 29 To determine if HypA can modulate nickel binding to the N-terminal site of HypB, nickel loss from as-purified HypB to EDTA in the presence of HypAStr was examined. Unlike the other nickel accessory protein SlyD, which accelerates nickel release from the tight site of HypB,39 HypA had no effect on the rate of nickel release from the HypB high-affinity site (Supplemental Figure 7), indicating that the impact of HypA on HypB is constrained to the metal site of the G-domain.
Nucleotide loading of HypB affects the rate of nickel loss
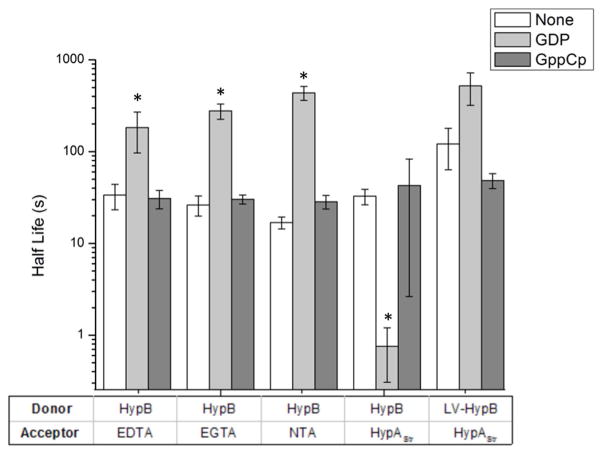
HypB is a GTPase,23 and the metal-binding ligands of the G-domain site of HypB are embedded within the GTPase motifs of the protein. To investigate how nucleotide loading of HypB affects nickel transfer to HypAStr, the rate of nickel loss from wild-type HypB was monitored in the presence of 100 μM GDP or the non-hydrolyzable GTP analogue guanosine-5′-[(β,γ)-methyleno]triphosphate (GppCp) and 5 mM MgSO4. These nucleotide concentrations should be sufficient to load the G-domain of HypB, given the reported micromolar GDP binding constant.28,23 The rate of nickel transfer when GppCp was bound to HypB was comparable to the rate in the absence of nucleotide (Figure 6). In contrast, when GDP was added the rate of nickel transfer to HypAStr increases significantly, with the reaction reaching completion in seconds (Figure 6). The same experiments were performed using LV-HypB, and in this case GDP loading does not increase the rate of metal release, suggesting that the faster nickel transfer induced by GDP is dependent on formation of the HypA-HypB complex. To test this model further, metal transfer experiments were performed by using several different small molecule chelators as the metal acceptor instead of HypAStr. These experiments were performed with EDTA, EGTA, or NTA, and they were present in considerable excess (1 mM) to ensure that the nickel released from HypB would be trapped by the strong chelators. In the absence of nucleotide, the rates of metal loss from HypB to the chelators were similar to that observed with HypAStr as the acceptor. Likewise, loading HypB with GppCp did not affect the rates of nickel release. However, when GDP was bound to HypB, nickel transfer to small molecule chelators was significantly slowed, the opposite effect compared to that observed on nickel transfer to HypAStr.
Figure 6.
Half-lives of nickel transfer from the G-domain of HypB or LV-HypB to an excess of metal acceptor. Small molecule acceptors (EDTA, EGTA, NTA) were added to 50 μM HypB with a final concentration of 1 mM, while HypA was added to a final concentration of 70 μM. Note that the y-axis is a log scale. Each bar represents the average of at least three separate experiments, while the error bars represent one standard deviation. Astericks indicate that nickel loss from GDP-loaded HypB is significantly different than in the absence of nucleotide (p < 0.05).
Discussion
HypA and HypB are required for the biosynthesis of the [NiFe]-hydrogenase metallocenter, and these proteins have been assigned a role in the nickel delivery stage of this process. Both individual proteins bind nickel, but as we started to examine metal binding in the context of the protein-protein complex, it became clear that HypAStr was binding nickel more readily than expected. In our previous study E. coli HypA,31 a change in intrinsic tryptophan fluorescence upon addition of nickel to apo-protein was used to calculate an apparent KD of 60 μM, a number consistent with the nickel affinities of homologous proteins.32, 36 However, mass spectrometry revealed quantitative nickel binding by low micromolar concentrations of HypA, leading us to revisit the HypA-nickel affinity. Competition experiments with a metallochromic indicator revealed an affinity of 75 nM, consistent with the HypA mass spectrometry data as well as the rapid nickel transfer observed from HypB to HypA. It is possible that the Strep-II tag added to the C-terminus affects nickel binding to HypA, but this construct is functional in hydrogenase biosynthesis in vivo,37 and all of the residues implicated in nickel binding are at the other end of the primary sequence.32, 34, 36 Furthermore, preliminary experiments with HypAStr revealed a similar change in intrinsic fluorescence in response to nickel as previously observed with untagged protein (data not shown), suggesting that this signal is not reporting on specific nickel binding to HypA and the basis of the fluorescence change remains to be determined.
There is significant evidence that HypA and HypB assemble together during hydrogenase biosynthesis,37, 45 but the role of this complex has not been defined. This interaction does not dramatically impact the tightest metal sites of each of the proteins. Zinc binding to HypA was not disturbed by HypB under any of the conditions examined, consistent with the role of this metal as a structural element, although changes in coordination cannot be ruled out. Similarly, HypA does not affect nickel binding to the N-terminal high-affinity site of HypB. This latter result is in contrast to the impact of another hydrogenase nickel accessory protein, SlyD, which accelerates nickel release from HypB.39, 46 Instead, HypA mediates nickel transfer from the G-domain of HypB to the binding site on HypA. This metal transfer from HypB (KD = 12 μM) to HypA (KD = 75 nM) is thermodynamically favorable given that the relative affinities of the two sites differ by more than two orders of magnitude.
Complex formation between HypB and HypA is key to the rapid nickel transfer, because disrupting this protein-protein interaction by mutating Leu78 and Val80 of HypB to alanine slows the process down considerably. Titrating increasing amounts of HypAStr into LV-HypB allows nickel transfer to occur more efficiently, consistent with the observation that this mutation in HypB only weakens the interaction between the two proteins without blocking it completely. In contrast, under the conditions used (50 μM HypB and 70 μM HypAStr), adding more HypAStr to wild-type HypB does not promote the rate of transfer any further, suggesting that HypB is saturated with HypAStr. If the rate of nickel transfer is proportional to the amount of HypA-HypB complex being formed, we can estimate the KD for WT-HypB-HypAStr to be < 10−5 M, and on the order of 10−4 M for the LV-HypB-HypAStr complex.
One possible mechanism for nickel transfer is that complex formation between the two proteins results in positioning the HypA nickel site in close proximity to the HypB G-domain, such that HypA functions as a nickel sink and metal transfer is simply controlled by thermodynamics. The decreased rate of transfer to HypAStr when LV-HypB is used as the nickel donor may be due to the mutation decreasing the local concentration of HypA, a result of the weakened affinity of the HypA-HypB complex. In support of this model, nickel loss from nucleotide-free or GppCp-loaded HypB occurs under the same timescale regardless of the identity of the nickel acceptor. Similar rates were observed with HypA as with small molecule acceptors that have a range of nickel affinities (from 10−9 M to 10−18 M), suggesting that the process is governed by an intrinsic property of HypB, such as the off rate of nickel dissociation from the protein.
The situation changes dramatically once HypB is in the GDP-loaded state. Nickel transfer from GDP-loaded HypB to any of the small molecule chelators is substantially slower compared to that observed in the absence of nucleotide or with the GTP analog. This decrease in the off rate of nickel from HypB could be caused by a change in the overall conformation of HypB to make the nickel less solvent accessible, or a rearrangement of the nickel ligands in the G-domain of HypB. The latter case has been observed with H. pylori HypB, for which nucleotide loading affects the coordination sphere of nickel bound to the G-domain site of the protein (A. Sydor, D. B. Z., personal communication).
In contrast, upon loading HypB with GDP, the nickel transfer to HypA becomes faster by several orders of magnitude, such that the process goes to completion within seconds. This accelerated transfer is not observed when LV-HypB is used as the nickel donor, suggesting that it is dependent on the formation of a complex between the two partner proteins. Furthermore, the fact that the metal loss is only faster when HypA is the acceptor suggests that GDP loading of HypB induces a “readied state” in the metal-binding site, resulting in directed nickel transfer to HypA. Whether this GDP-induced state of HypB results in a change in the nickel coordination and/or modifies the interaction with HypA remains to be determined.
The role of the conserved metal-binding site in the G-domain of HypB is not clear. The observation that metal binding modulates the GTPase activity in both the E. coli and H. pylori HypB proteins suggested that the metal may have a regulatory role.28, 47, 48 However, the results reported here are also consistent with a model in which this site serves as a source of nickel for the hydrogenase pathway, particularly in conjunction with HypA. The fact that nickel transfer to HypA is fastest in the post-hydrolysis state of HypB appears to be in conflict with the observation that nickel inhibits the GTPase activity, but it is likely that other components of the hydrogenase biosynthetic pathway will impact the activities of HypB and HypA. One obvious candidate for such a role is SlyD, which increases the kcat of the HypB GTPase activity by several fold.46 However, SlyD can also pull nickel out of the G-domain site of HypB,46 so whether there would be competition with HypA is not clear. There is evidence for a SlyD-HypB-HypA tertiary complex in vivo,37, 49 but such a complex has yet to be isolated and studied in vitro. Furthermore, HypA acts as a “docking” protein between the immature HycE and HypB,37 and it is possible that in the context of a functional HycE-HypA-HypB complex that GTP hydrolysis is accelerated and nickel delivery is allowed to proceed.
At this point, it is unclear which metal is the “cognate metal” in the G-domain of HypB. Zinc also binds to this site with an affinity tighter than that of nickel,26 and zinc is more effective at inhibiting GTP hydrolysis by HypB.28, 47, 48 However, the concentration of available zinc in healthy E. coli is kept at extremely low levels,50, 51 so perhaps under normal growth conditions there is insufficient zinc to fill the HypB G-domain site. If zinc does reach this site, such as in a case of improper metal regulation, the GTPase cycle of HypB would be repressed and the process would come to a halt, as zinc is not passed on to HypA. In this manner, the HypB G-domain metal site may act as a mechanism for maintaining the metal fidelity of the process, and blocking hydrogenase maturation until intracellular metal concentrations can be normalized.
Whether the molecular details of metal delivery are completely conserved remains to be established. Both nickel and zinc bind more tightly to H. pylori HypB than to the E. coli homolog,26,48 whereas the H. pylori HypA has a weaker nickel affinity than that reported here,32, 35 so nickel transfer would be thermodynamically driven from HypA to HypB, instead of the reverse. Furthermore, the strength of the H. pylori HypA-HypB complex is weaker than that estimated indirectly for the E. coli proteins in the metal transfer experiments.38 It is possible that the accessory proteins have adapted to the distinct metal metabolism of different organisms. For example, the H. pylori HypB does not have the N-terminal high-affinity site that is required for hydrogenase biosynthesis in E. coli,27 and the H. pylori proteins play multiple roles because they also contribute to urease biosynthesis in that organism.52
The work discussed here presents a significant step in understanding the route that nickel takes in its journey from nickel importer to the active site of the [NiFe]-hydrogenase enzymes. This allows for an updated working model of HypA-HypB complex formation. If the G-domain site of HypB is filled with nickel, such as in nickel-replete cellular conditions, then GTP hydrolysis is inhibited. Upon docking with the iron-loaded HycE, it is possible that GTP hydrolysis is accelerated, activating rapid nickel transfer from HypB to HypA. Whether this nickel ion is ultimately delivered to the hydrogenase precursor protein, how the metal-binding affinities are modulated in the context of the ternary complex, and how the high-affinity nickel site of HypB contributes to this pathway are issues that need to be resolved in future work.
Supplementary Material
Table 1.
Plasmids used in this study
| Plasmid | Gene | Parent Plasmid | Reference |
|---|---|---|---|
| strA-pET24b | E. coli hypA with C-terminal Strep-tag II | pET-24b | 37 |
| mstrA-pET24b | E. coli hypA K24E, R25E with C-terminal Strep-tag II | pET-24b | This work |
| hypB-pET24b | E. coli hypB | pET-24b | 26 |
| hypB-pBAD24 | E. coli hypB | pBAD2453 | 39 |
| lvhypB-pET24b | E. coli hypB, L78A, V80A | pET-24b | This work |
| lvhypB-pBAD24 | E. coli hypB, L78A, V80A | pBAD24 53 | This work |
Acknowledgments
We thank Dr. Matthew Forbes for assistance with mass spectrometry, and Michael Lam for assistance with preparation of proteins. We also thank Prof. A. Böck for the generous donation of the MC4100 strains of E. coli, A. Sydor for critical reading of this manuscript, and members of the Zamble laboratory for helpful discussions.
Funding Sources
This work was supported by funding from the Canadian Institutes of Health Research as well as a graduate fellowship from the Natural Sciences and Engineering Research Council of Canada (to C.D.D.).
ABBREVIATIONS
- E. coli
Escherichia coli
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid
- EDTA
ethylenediaminetetraacetate
- EGTA
ethyleneglycol tetraacetate
- ESI-MS
electrospray ionization mass spectrometry
- GDP
Guanosine-5′-diphosphate
- GTP
Guanosine-5′-triphosphate
- GuHCl
guanidine hydrochloride
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HypAStr
HypA with C-terminal Strep II affinity tag
- KD
dissociation constant
- LMCT
ligand-to-metal charge transfer
- LV-HypB
L78A, V80A HypB mutant
- MF2
mag-fura-2
- NTA
nitrilotriacetic acid
- PAR
4-(2-pyridylazo)resorcinol
- PMB
para-mercury-benzenesulfonic acid
- PMSF
phenylmethanesulfonylfluoride
- Tris
tris(hydroxymethyl)aminomethane
Footnotes
This work was supported by funding from the Canadian Institutes of Health Research as well as graduate fellowships from the Natural Sciences and Engineering Research Council of Canada (to C.D.D. and H. K.).
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Control experiments of mutant variants of HypB, scripts for fitting, crosslinking experiments, and predicted masses of metal-protein species are contained within the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Bioinorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Gray HB. Biological inorganic chemistry at the beginning of the 21st century. Proc Natl Acad Sci U S A. 2003;100:3563–3568. doi: 10.1073/pnas.0730378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macomber L, Elsey SP, Hausinger RP. Fructose-1,6-bisphosphate aldolase (class II) is the primary site of nickel toxicity in Escherichia coli. Mol Microbiol. 2011;82:1291–1300. doi: 10.1111/j.1365-2958.2011.07891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bleackley MR, Macgillivray RT. Transition metal homeostasis: from yeast to human disease. BioMetals. 2011;24:785–809. doi: 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- 5.Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol. 2012;78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imlay J, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 7.Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- 8.Ma Z, Jacobsen FE, Giedroc DP. Coordination chemistry of bacterial metal transport and sensing. Chem Rev (Washington, DC, U S) 2009;109:4644–4681. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sydor A, Zamble D. Nickel Metallomics: General Themes Guiding Nickel Homeostasis. In: Banci L, editor. Metallomics and the Cell. Springer Netherlands; 2013. pp. 375–416. [DOI] [PubMed] [Google Scholar]
- 10.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenzweig AC. Metallochaperones: Bind and deliver. Chem Biol. 2002;9:673–677. doi: 10.1016/s1074-5521(02)00156-4. [DOI] [PubMed] [Google Scholar]
- 12.O’Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 13.Kuchar J, Hausinger RP. Biosynthesis of metal sites. Chem Rev. 2004;104:509–526. doi: 10.1021/cr020613p. [DOI] [PubMed] [Google Scholar]
- 14.Böck A, King PW, Blokesch M, Posewitz MC. Maturation of Hydrogenases. In: Robert KP, editor. Adv Microb Physiol. Academic Press; 2006. pp. 1–225. [DOI] [PubMed] [Google Scholar]
- 15.Vignais PM, Billoud B. Occurance, classification, and biological function of hydrogenases: an overview. Chem Rev (Washington, DC, U S) 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 16.Maier RJ. Use of molecular hydrogen as an energy source substrate by human pathogenic bacteria. Biochem Soc Trans. 2005;33:83–85. doi: 10.1042/BST0330083. [DOI] [PubMed] [Google Scholar]
- 17.Forzi L, Sawers R. Maturation of [NiFe]-hydrogenases in Escherichia coli. BioMetals. 2007;20:565–578. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- 18.Löscher S, Zebger I, Andersen LK, Hildebrandt P, Meyer-Klaucke W, Haumann M. The structure of the Ni-Fe site in the isolated HoxC subunit of the hydrogen-sensing hydrogenase from Ralstonia eutropha. FEBS Lett. 2005;579:4287–4291. doi: 10.1016/j.febslet.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 19.Winter G, Buhrke T, Lenz O, Jones AK, Forgber M, Friedrich B. A model system for [NiFe] hydrogenase maturation studies: Purification of an active site-containing hydrogenase large subunit without small subunit. FEBS Lett. 2005;579:4292–4296. doi: 10.1016/j.febslet.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 20.Leach M, Zamble D. Metallocenter assembly of the hydrogenase enzymes. Curr Opin Chem Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Kaluarachchi H, Chan Chung KC, Zamble DB. Microbial nickel proteins. Nat Prod Rep. 2010;27:681–694. doi: 10.1039/b906688h. [DOI] [PubMed] [Google Scholar]
- 22.Hube M, Blokesch M, Bock A. Network of hydrogenase maturation in Escherichia coli: Role of accessory proteins HypA and HybF. J Bacteriol. 2002;184:3879–3885. doi: 10.1128/JB.184.14.3879-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maier T, Jacobi A, Sauter M, Bock A. The product of the hypB gene, which is required for nickel incorporation into hydrogenases, is a novel guanine nucleotide-binding protein. J Bacteriol. 1993;175:630–635. doi: 10.1128/jb.175.3.630-635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobi A, Rossmann R, Böck A. The hyp operon gene products are required for the maturation of catalytically active hydrogenase isoenzymes in Escherichia coli. Arch Microbiol. 1992;158:444–451. doi: 10.1007/BF00276307. [DOI] [PubMed] [Google Scholar]
- 25.Zhang JW, Butland G, Greenblatt JF, Emili A, Zamble DB. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J Biol Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- 26.Leach M, Sandal S, Sun H, Zamble D. Metal binding activity of the Escherichia coli hydrogenase maturation factor HypB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- 27.Dias AV, Mulvihill CM, Leach MR, Pickering IJ, George GN, Zamble DB. Structural and biological analysis of the metal sites of Escherichia coli hydrogenase accessory protein HypB. Biochemistry. 2008;47:11981–11991. doi: 10.1021/bi801337x. [DOI] [PubMed] [Google Scholar]
- 28.Cai F, Ngu T, Kaluarachchi H, Zamble D. Relationship between the GTPase, metal-binding, and dimerization activities of E. coli HypB. J Biol Inorg Chem. 2011;16:857–868. doi: 10.1007/s00775-011-0782-y. [DOI] [PubMed] [Google Scholar]
- 29.Chan Chung KC, Cao L, Dias AV, Pickering IJ, George GN, Zamble DB. A high-affinity metal-binding peptide from Escherichia coli HypB. J Am Chem Soc. 2008;130:14056–14057. doi: 10.1021/ja8055003. [DOI] [PubMed] [Google Scholar]
- 30.Douglas CD, Dias AV, Zamble DB. The metal selectivity of a short peptide maquette imitating the high-affinity metal-binding site of E. coli HypB. Dalton Trans. 2012;41:7876–7878. doi: 10.1039/c2dt30132f. [DOI] [PubMed] [Google Scholar]
- 31.Atanassova A, Zamble DB. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watanabe S, Arai T, Matsumi R, Atomi H, Imanaka T, Miki K. Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. J Mol Biol. 2009;394:448–459. doi: 10.1016/j.jmb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 34.Xia W, Li H, Sze K-H, Sun H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. J Am Chem Soc. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 35.Herbst RW, Perovic I, Martin-Diaconescu V, O’Brien K, Chivers PT, Pochapsky SS, Pochapsky TC, Maroney MJ. Communication between the zinc and nickel sites in dimeric HypA: metal recognition and pH sensing. J Am Chem Soc. 2010;132:10338–10351. doi: 10.1021/ja1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blokesch M, Rohrmoser M, Rode S, Böck A. HybF, a zinc-containing protein involved in NiFe hydrogenase maturation. J Bacteriol. 2004;186:2603–2611. doi: 10.1128/JB.186.9.2603-2611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan Chung KC, Zamble DB. Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase. J Biol Chem. 2011;286:43081–43090. doi: 10.1074/jbc.M111.290726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia W, Li H, Yang X, Wong K-B, Sun H. Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. J Biol Chem. 2012;287:6753–6763. doi: 10.1074/jbc.M111.287581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leach M, Zhang J, Zamble D. The role of complex formation between the Escherichia coli hydrogenase accessory factors HypB and SlyD. J Biol Chem. 2007;282:16177–16186. doi: 10.1074/jbc.M610834200. [DOI] [PubMed] [Google Scholar]
- 40.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petr K. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 42.Peck HD, Gest H. A new procedure for assay of bacterial hydrogenases. J Bacteriol. 1956;71:70–80. doi: 10.1128/jb.71.1.70-80.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballantine SP, Boxer DH. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol. 1985;163:454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt TGM, Skerra A. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protocols. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 45.Chan K-H, Lee K-M, Wong K-B. Interaction between hydrogenase maturation factors HypA and HypB is required for [NiFe]-hydrogenase maturation. PLoS One. 2012;7:e32592. doi: 10.1371/journal.pone.0032592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaluarachchi H, Zhang JW, Zamble DB. Escherichia coli SlyD, more than a Ni(II) reservoir. Biochemistry. 2011;50:10761–10763. doi: 10.1021/bi201590d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng T, Li H, Yang X, Xia W, Sun H. Interaction of SlyD with HypB of Helicobacter pylori facilitates nickel trafficking. Metallomics. 2013 doi: 10.1039/c3mt00014a. [DOI] [PubMed] [Google Scholar]
- 48.Sydor AM, Liu J, Zamble DB. Effects of metal on the biochemical properties of Helicobacter pylori HypB, a maturation factor of [NiFe]-hydrogenase and urease. J Bacteriol. 2011;193:1359–1368. doi: 10.1128/JB.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan Chung KC, Zamble DB. The Escherichia coli metal-binding chaperone SlyD interacts with the large subunit of [NiFe]-hydrogenase 3. FEBS Lett. 2011;585:291–294. doi: 10.1016/j.febslet.2010.12.024. [DOI] [PubMed] [Google Scholar]
- 50.Outten CE, O’Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 51.Wang D, Hosteen O, Fierke CA. ZntR-mediated transcription of zntA responds to nanomolar intracellular free zinc. J Inorg Biochem. 2012;111:173–181. doi: 10.1016/j.jinorgbio.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 53.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.