Abstract
“Far-West” Africa is known to be a secondary contact zone between the two major malaria vectors Anopheles coluzzii and A. gambiae. We investigated gene-flow and potentially adaptive introgression between these species along a west-to-east transect in Guinea Bissau, the putative core of this hybrid zone. To evaluate the extent and direction of gene flow, we genotyped site 702 in Intron-1 of the para Voltage-Gated Sodium Channel gene, a species-diagnostic nucleotide position throughout most of A. coluzzii and A. gambiae sympatric range. We also analyzed polymorphism in the thioester-binding domain (TED) of the innate immunity-linked thioester-containing protein 1 (TEP1) to investigate whether elevated hybridization might facilitate the exchange of variants linked to adaptive immunity and Plasmodium refractoriness. Our results confirm asymmetric introgression of genetic material from A. coluzzii to A. gambiae and disruption of linkage between the centromeric "genomic islands" of inter-specific divergence. We report that A. gambiae from the Guinean hybrid zone possesses an introgressed TEP1 resistant allelic class, found exclusively in A. coluzzii elsewhere and apparently swept to fixation in West Africa (i.e. Mali and Burkina Faso). However, no detectable fixation of this allele was found in Guinea Bissau, which may suggest that ecological pressures driving segregation between the two species in larval habitats in this region may be different from those experienced in northern and more arid parts of the species’ range. Finally, our results also suggest a genetic subdivision between coastal and inland A. gambiae Guinean populations and provide clues on the importance of ecological factors in intra-specific differentiation processes.
Introduction
Anopheles gambiae (Giles) and A. coluzzii (Coetzee & Wilkerson sp. n.) (formerly defined as A. gambiae s.s. M and S molecular forms based on X-linked SNPs in ribosomal DNA [1] are arguably the most important cryptic species of mosquitoes transmitting human malaria in sub-Saharan Africa. Restricted gene flow between A. gambiae and A. coluzzii in natural populations from sympatric areas of West and Central Africa is attributed to pre-mating mechanisms of reproductive isolation, selection against hybrids and ecologically-driven divergent selection [2, 3, 4, 5, 6]. Variation in larval habitats strongly influences species segregation: A. gambiae is associated with small ephemeral puddles, whereas A. coluzzii breeds in larger and more stable ponds, often created by agriculture, urbanization, or other human activities [7, 8, 9].
Genetic divergence between A. gambiae and A. coluzzii appears to be concentrated in "genomic islands of divergence" located in peri-centromeric regions of chromosome-X, -2 and -3 [10, 11], but it is also detectable in other smaller areas across the genome, some outside of centromeres [12, 13, 14, 15, 16]. A comparative genome-wide scan identified a significant area of inter-specific divergence on chromosome-3L, including five known or suspected immune response genes [17]. Of these, the thioester-containing protein 1 (TEP1) encodes a complement-like opsonin, binding of which triggers killing of gram-negative bacteria and protozoa via phagocytosis [18]. TEP1 is highly polymorphic [19] and shows amino acid substitutions in the thioester-binding domain (TED) associated with pathogen resistance phenotypes [18]. In fact, experimental infections demonstrated that laboratory-reared A. gambiae individuals homozygous or heterozygous for TEP1*R1 [18] and TEP1r B [17] alleles are significantly more resistant to Plasmodium and bacterial infections than mosquitoes carrying other TEP1 alleles. In contrast individuals carrying TEP1*R2 [18] and TEP1r A [17] alleles show less resistant phenotypes. TEP1 genotyping of natural populations indicates that TEP1r B is absent or very rare in A. gambiae, but is fixed in A. coluzzii from West Africa (i.e. Mali and Burkina Faso) [17]. Given the relatively low rates and intensities of natural Plasmodium infection in both mosquito species, it was speculated that the most likely source of pathogen-mediated selection for resistance came from larval habitat [17]. Specifically, the longer-lasting and more biotically diverse aquatic milieu exploited by A. coluzzii, presumably harboring richer pathogen populations than temporary breeding sites exploited by A. gambiae, would exert higher selective pressures on the immune system of the former species [17, 20].
Although recent analyses suggest that hybridization between A. gambiae and A. coluzzii may be more dynamic than previously appreciated [6], the “Far-West” African region likely represents the most stable hybridization zone. High frequencies of A. gambiae x A. coluzzii hybrids have been repeatedly recorded in The Gambia (up to 7%) [21] and Guinea Bissau (>20%) [22, 23] leading to the hypothesis that these “Far-West” areas of the species’ range may represent a secondary contact zone in which local ecological settings have significantly disrupted reproductive barriers [21].
Hybrid zones offer an excellent opportunity to examine the outcome of genetic exchange of traits responsible for species segregation and to identify possible changes in ecological conditions inducing relaxation of the reproductive isolation found elsewhere in the sympatric range of their distribution [24]. Data collected so far on the genetic background of parental and hybrid individuals from the A. gambiae/A. coluzzii secondary contact zone indicate a preferential acquisition by A. gambiae of A. coluzzii alleles suggesting asymmetric introgression from A. coluzzii to A. gambiae [6, 23, 25].
In this paper we surveyed parental and hybrid mosquitoes from a West-to-East transect in Guinea Bissau to investigate gene-flow and potentially adaptive introgression between A. coluzzii and A. gambiae in their secondary contact zone. First, we evaluated the extent and direction of gene flow, using the species-informative site 702 in Intron-1 (Int-1702) of the Voltage-Gated Sodium Channel (VGSC) gene, located within the chromosome-2L "genomic island". This site is characterized by species-specific alleles in West and Central Africa (A. coluzzii = Int-1C; A. gambiae = Int-1T), which define species-specific Intron-1 haplogroups in strong linkage disequilibrium with the species-diagnostic X-linked rDNA SNPs [26, 27]. Second, we investigated the effect of hybridization on the exchange of adaptive alleles in the secondary contact zone by analyzing polymorphism in the catalytic TED domain of the TEP1 gene on chromosome-3.
Materials and Methods
Field collected samples and species identification
Anopheles gambiae s.l. adult females were selected from a larger sample collected in the rainy season of 2010 (October) in five villages located along a West-to-East geographical transect in Guinea Bissau: Antula (11° 53’ 49” N—15° 35’ 29” W), Safim (11° 57’ 00” N—15° 39’ 00” W), Mansoa (12° 04' 00'' N—15° 19' 00'' W), Ga-Mbana (12° 03' 00'' N—14° 55' 00'' W) and Leibala (11° 52' 53.96'' N—15° 37' 4.06'' W) (Fig 1). Field collections (not conducted in protected areas, nor involving endangered or protected species) were approved by and carried out under the guidance of the Guinea Bissau National Institute of Public Health (INASA). Indoor sampling was performed in private houses after permission by owners (informed on research aims) with CDC light traps in Antula and Ga-Mbana and with mechanical aspirators (Indoor Resting Collection, IRC) in Safim, Mansoa and Leibala.
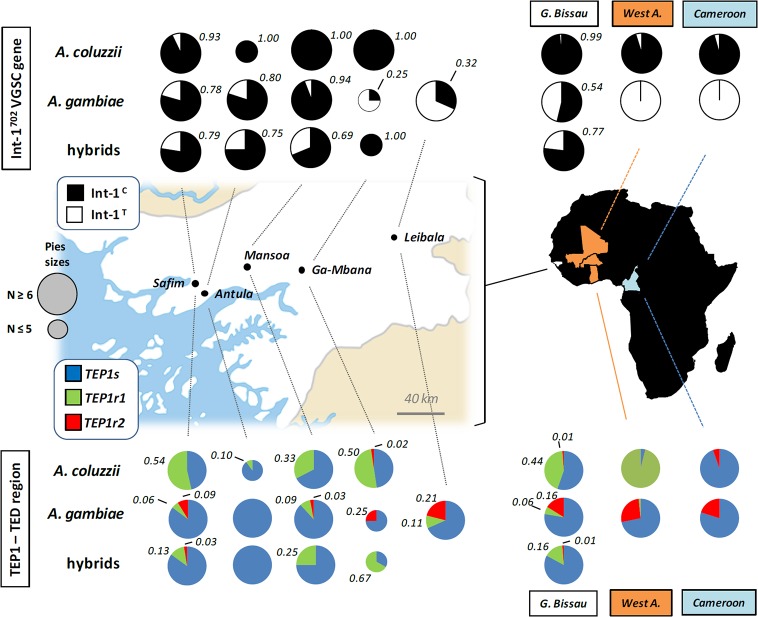
Fig 1. Int-1702 and TEP1 allele frequencies along a west-to-east geographic transect in Guinea Bissau.
Relative frequencies of Int-1702 VGSC (above) and TEP1 (below) alleles are reported overall and in each of the five sampled villages for A. coluzzii, A. gambiae and hybrids. Numbers refer to relative frequencies of Int-1C and of TEP1r1 and TEP1r2 alleles in Guinean sample. Overall allele frequencies in Guinea Bissau for both markers (this study) and from West-Africa and Cameroon (data from Gentile et al., 2004 and White et al., 2011) are reported on the right for comparison. GPS coordinates (UTM) of sampled Guinean villages are as follows: Antula (11° 53’ 49” N—15° 35’ 29” W), Safim (11° 57’ 00” N—15° 39’ 00” W), Mansoa (12° 04' 00'' N—15° 19' 00'' W), Ga-Mbana (12° 03' 00'' N—14° 55' 00'' W) and Leibala (11° 52' 53.96'' N—15° 37' 4.06'' W). Geographic map modified from Guinea Bissau sm03.png (Wikipedia image in public domain)
Morphological identification was performed using the available taxonomic keys [28, 29, 30]. Specimens were stored in silica gel-filled tubes until DNA extraction using DNAzol (Life Technologies) or DNeasy Blood & Tissue Kit (Qiagen) was carried out. Identification of A. gambiae s.s. and A. coluzzii was carried out using two methods: SINE-PCR based on SINE insertion [31] and IMP-PCR based on IGS mutations [32]. We chose samples to be genotyped from within each sample site to increase the number of specimens of the less-frequent species and of the hybrid category. The latter includes all individuals heterozygous for both diagnostic markers (i.e. MSSINE/MSIGS, N = 31) and specimens showing discordant SINE and IGS PCR patterns (i.e. MMSINE/MSIGS, N = 4; SSSINE/MSIGS, N = 4; MSSINE/MMIGS, N = 2; from Safim and Mansoa), interpreted as being advanced generation hybrids (see [6, 11, 33, 34] for further details on species and hybrid identification in the secondary contact area).
Genotyping of Int-1702 SNP
We genotyped Int-1702 using a primer-introduced restriction analysis assay (PIRA-PCR) designed on available Int-1 alignments [26, 27]. A forward primer, INTeco-f (5'-ATTATGCTCTTTACAATGCCAACGgAAT-3'), was designed to incorporate a C-to-G mismatch at the 4th base from the 3'-end. In the presence of a ‘T’ at site 702 and of a fixed ‘C’ at site 703 (as observed in West Africa [26, 27 35]), the -3’ gAAT sequence of INTeco-f creates a recognition site for the EcoRI restriction enzyme (i.e. G’AATTC) within the PCR product amplified with INTeco-f and reverse primer INTa-r (5’-GGAATCTATCCACATTATCTG-3’). The restriction produces a 265 bp or a 240 bp band for Int-1C/C and Int-1T/T homozygotes, respectively; heterozygotes display both bands. PCR reactions were carried out in a 10 μl reaction which contained 1x Buffer, 1 pmol of each primer, 0.2 mM of each dNTP, 1.5 mM MgCl2, 2.5 U Taq DNA polymerase, and 8–10 ng of template DNA extracted from a single mosquito. Thermocycler conditions were 94°C for 10 min followed by thirty-five cycles of 94°C for 30 sec, 54°C for 30 sec and 72°C for 1 min, with a final elongation at 72°C for 10 min. Five microliters of each PCR product were digested with 10 U of EcoRI enzyme (New England Biolabs, UK) with 1× buffer in a final volume of 15 μl incubated at 37°C for 1 hour. The restriction products were run on 3% agarose gels stained with ethidium bromide. To obtain stronger bands on templates that proved difficult, a semi-nested PCR protocol was employed: on a first round of amplification, PCR products were obtained using INTeco-f and INTb-r (5’-ATCTTGGCAGATCATGGTCGG-3’), then diluted 1:100 and used as template for a second round of amplification under the PCR conditions described above.
TEP1 amplification and sequencing
A 456 bp fragment of the TEP1-TED domain was amplified with primers OB2996F (5'-CACGGTCATCAAGAACCTGGAC-3') [19] and EMTep1R (5’-TCCAGCAATGCCATCAACACAT-3'), the latter specifically designed for the aim of this work in order to avoid co-amplification of other TEP-related paralogs, which were instead pervasively co-amplified with other primer couples available in literature. Amplifications were performed in a 15 μl reaction-mix using 0.5–1.0 μl of template DNA using the High Fidelity AccuPrime Taq DNA Polymerase kit (Life Technologies) following manufacturer's guidelines. Thermocycler conditions were as follows: initial denaturation at 94°C for 2 min followed by 35 cycles of 94°C for 30 sec, 54°C for 30 sec, 68°C for 1 min, with a final elongation at 68°C for 7 min. The resulting products were analysed on 1–2% agarose gels stained with GelRed (Biotium), purified with the SureClean Kit (Bioline) and sequenced at the BMR Genomics s.r.l. (Padua, Italy). Sequences are available in GenBank under Accession Numbers KR091079—KR091309.
Sequence and population genetic analysis
TEP1 chromatograms were edited and trimmed to remove low-quality base calls with Staden Package ver. 2003.1.6 [36]. A final 387 bp alignment of genotype sequences was produced using MAFFT ver. 7 [37] and alleles phased using the PHASE algorithm [38]. Resulting TEP1 alleles were identified as susceptible or resistant based on key residues in the catalytic loop and pre-α- loop found in the TED portion [39]. The sequenced TED fragment did not allow us to discriminate among previously described variants within susceptible and resistant TEP1 allelic classes, because such distinction is also based on residues outside the catalytic and pre-α- loop region [17, 18]. So, for the purpose of this study, we chose to name allelic classes as follows: TEP1s = 'susceptible' class, comparable to TEP1*S1, TEP1*S2, TEP1*S3 [18] and TEP1s [17]; TEP1r1 = 'resistant' allele comparable to TEP1*R1 [18] and TEP1r B [17]; TEP1r2 = TEP1 'resistant' allele plausibly corresponding to TEP1*R2 [18] and TEP1r A [17]. A median-joining network was built for TEP1s and TEP1r allelic classes with NETWORK ver. 4.510 [40]. TEP1 genotyping data obtained by B. White and collaborators [17] were used for comparison.
DnaSP v5.10.1 [41] was used to estimate genetic polymorphism and to perform neutrality tests, i.e. Tajima's D, Fu & Li D* and Wall's Q statistics. F-statistics (F ST and F IS) [42], departures from Hardy-Weinberg Equilibrium (HWE) and linkage disequilibrium (LD) were estimated for TEP1 and Int-1702 genotyping data using ARLEQUIN 3.5 [43] and GENEPOP ‘007 [44].
Results and Discussion
The westernmost extreme of the sympatric range of Anopheles coluzzii and A. gambiae is believed to be the core of a secondary contact region where disruption of genetic association is observed among "genomic islands" of divergence on centromeres of chromosome-X, -2 and -3, and a preferential transfer of genetic material from A. coluzzii to A. gambiae occurs (i.e., asymmetric introgression) [21, 23, 25, 34]. These phenomena are confirmed by our results from the genotyping of the Int-1702 SNP of the VGSC gene in Guinean populations (Fig 1). Results show that, as in the rest of the species range, Int-1C allele is almost fixed in Guinean A. coluzzii. However, in contrast to other areas, it is also found at high frequencies (up to 94%) in sympatric A. gambiae populations from the coastal/cropland areas of Antula, Safim and Mansoa. Thus, the association between Int-1702 and species-specific markers on chromosome-X (i.e. IGS, SINE) observed elsewhere [26, 27] is lost in these populations (S1 Table). It is worth noting that the very low frequency of kdr-associated mutations in these three A. gambiae sample sites (Vicente JL, personal communication) precludes the explanation that this unprecedented Int-1702 pattern might result from hitchhiking driven by insecticide selective pressure acting on kdr locus, as shown to occur in other African regions [35, 45].
Previously, no data were available from the “Far-West” secondary contact region on functional polymorphisms of potential adaptive significance (such as in immune-related genes) that differentiate A. coluzzii and A. gambiae in the rest of their sympatric range. Our genetic analysis of TEP1-TED—whose TEP1r1 allele confers high resistance to pathogens and is confined to A. coluzzii in West and Central Africa [17] (Fig 1)—provides the first insights on the exchange and polymorphism of this potentially adaptive protein in the “Far-West” region. First, susceptible (TEP1s) and resistant (TEP1r1 and TEP1r2) alleles are shown to be shared by the two species (Fig 1). Indeed, the occurrence of the TEP1r1 allele in A. gambiae at frequencies up to 11% contrasts with the virtual absence of this allele in the rest of the species range. Lack of TEP1r1 private haplotypes in Guinean A. gambiae suggest that this allele was acquired from A. coluzzii (Fig 2).
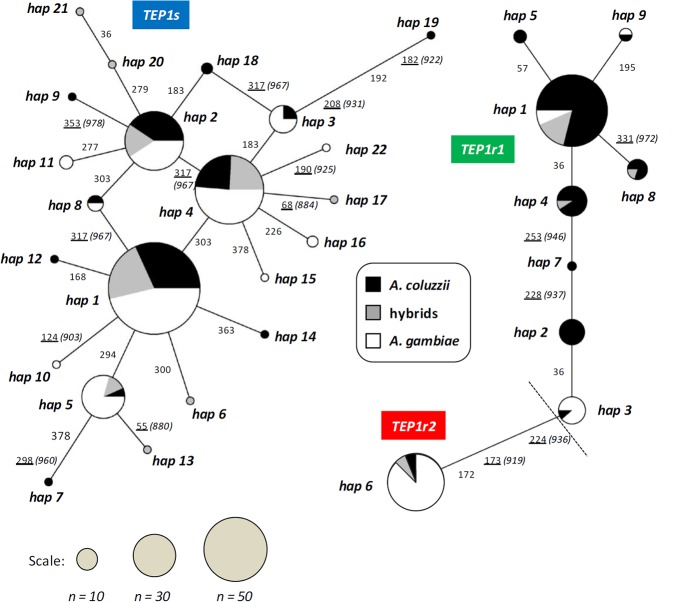
Fig 2. TEP1 median-joining networks depicting relationships among TEP1s (left) and TEP1r (right) related haplotypes.
Haplotype frequencies are proportional to pie sizes. Site numbering of synonymous and non-synonymous (underlined) mutations along branches follows our 387 bp TEP1 alignment, those of replaced amino acids (in brackets) follows Blandin et al., 2009. TEP1s and TEP1r networks are separated by a total of 32 fixed mutations, 12 at synonymous and 20 non-synonymous sites, respectively.
Yet, despite evidence of inter-specific gene-flow, significant genetic differentiation between A. coluzzii and A. gambiae remains (F ST = 0.19, S2 Table), probably reflecting past segregation of TEP1 resistant alleles in the hybridizing parental species. TEP1r1 is more frequent in A. coluzzii than in A. gambiae (χ 2 = 46.9; p<0.001), whereas the opposite is observed for TEP1r2 (χ 2 = 20.8; p<0.001) (Fig 1, Table 1). Second, our data highlights that TEP1r1 allele frequency varies from 0.1% to 0.5% in Guinean A. coluzzii but never reach fixation (Table 2, Fig 3) as it occurs in co-specific populations from northern savanna areas of West Africa [17].
Table 1. Int-1702 and TEP1 genotype frequencies and F IS.
| Int-1 702 genotype frequencies (%) | TEP1 genotype frequencies (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Int-1 C/C | Int-1 C/T | Int-1 T/T | F IS | s/s | s/r1 | r1/r1 | s/r2 | r2/r2 | F IS | ||
| Overall | A. coluzzii | 85 | 97.7 | 2.3 | - | -0.01 | 28.2 | 51.8 | 17.7 | 2.3 | - | -0.07 |
| hybrids | 41 | 65.8 | 22.0 | 12.2 | 0.39 * | 68.3 | 26.9 | 2.4 | 2.4 | - | -0.01 | |
| A. gambiae | 105 | 43.8 | 20.0 | 36.2 | 0.60 ** | 59.0 | 12.4 | - | 24.8 | 3.8 | -0.01 | |
| Total | 231 | 67.5 | 13.9 | 18.6 | 0.64 ** | 49.3 | 29.4 | 6.9 | 12.6 | 1.7 | 0.07 ** | |
| Antula | A. coluzzii | 5 | 100.0 | - | - | n.a. | 80.0 | 20.0 | - | - | - | n.a. |
| hybrids | 10 | 70.0 | 10.0 | 20.0 | 0.76 * | 100.0 | - | - | - | - | n.a. | |
| A. gambiae | 10 | 60.0 | 40.0 | - | -0.20 | 100.0 | - | - | - | - | n.a. | |
| Safim | A. coluzzii | 14 | 85.7 | 14.3 | - | -0.04 | 21.4 | 50.0 | 28.6 | - | - | -0.04 |
| hybrids | 20 | 60.0 | 35.0 | 5.0 | 0.02 | 70.0 | 25.0 | - | 5.0 | - | -0.12 | |
| A. gambiae | 17 | 64.7 | 29.4 | 5.9 | 0.13 | 70.6 | 11.8 | - | 17.6 | - | -0.10 | |
| Mansoa | A. coluzzii | 23 | 100.0 | 0.0 | - | n.a. | 34.8 | 65.2 | - | - | - | -0.47 * |
| hybrids | 8 | 62.5 | 12.5 | 25.0 | 0.74 | 50.0 | 50.0 | - | - | - | -0.27 | |
| A. gambiae | 17 | 94.1 | - | 5.9 | 1.00 ** | 76.5 | 17.6 | - | 5.9 | - | -0.08 | |
| Ga-Mbana | A. coluzzii | 43 | 100.0 | - | - | n.a. | 20.9 | 48.8 | 25.6 | 4.6 | - | -0.01 |
| hybrids | 3 | 100.0 | - | - | n.a. | - | 66.7 | 33.3 | - | - | -0.33 | |
| A. gambiae | 4 | 25.0 | - | 75.0 | 1.00 | 50.0 | - | - | 50.0 | - | -0.20 | |
| Leibala | A. gambiae | 57 | 21.1 | 21.1 | 57.8 | 0.52 ** | 43.9 | 14.0 | - | 35.1 | 7.0 | -0.04 |
Genotype frequencies of Int-1702 and TEP1 in A. gambiae s.s. (i.e. SSSINE/SSIGS), A. coluzzii (i.e. MMSINE/MMIGS) and hybrids (i.e. 31 MSSINE/MSIGS, 4 MMSINE/MSIGS, 4 SSSINE/MSIGS and 2 MSSINE/MMIGS) are reported as percentages (%). N = sample size, F IS = inbreeding coefficient. Significant F IS indicating deviations from HWE are reported in bold:
*p< 0.05
**p<0.001.
Table 2. TEP1 polymorphism and neutrality tests.
| n | H | Hd | S | Eta | π | θ | N-SYN | SYN | πA | πS | Dsyn | D* | Q | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| By allelic classes (all) | TEP1s | 325 | 22 | 0.75 | 21 | 21 | 0.3 | 0.9 | 9 | 12 | 0.1 | 1.0 | -1.27 | -5.16 * | 0.09 |
| TEP1r1 | 100 | 8 | 0.59 | 6 | 6 | 0.3 | 0.0 | 3 | 3 | 0.2 | 0.5 | -0.55 | 1.12 | 0.00 | |
| TEP1r2 | 37 | 1 | - | - | - | - | - | - | - | - | - | n.a. | n.a. | n.a. | |
| TEP1s (by groups) | A. coluzzii | 94 | 12 | 0.73 | 12 | 12 | 0.3 | 0.6 | 5 | 7 | 0.2 | 0.9 | -1.03 | -3.49 * | 0.17 |
| A. gambiae | 163 | 11 | 0.77 | 9 | 9 | 0.3 | 0.4 | 3 | 6 | 0.1 | 1.1 | -0.27 | -1.13 | 0.00 | |
| hybrids | 68 | 9 | 0.72 | 8 | 8 | 0.3 | 0.4 | 3 | 5 | 0.1 | 0.9 | -0.62 | -1.75 | 0.00 | |
| TEP1r1 (by groups) | A. coluzzii | 74 | 8 | 0.56 | 6 | 6 | 0.3 | 0.3 | 3 | 3 | 0.2 | 0.5 | -0.50 | 0.23 | 0.00 |
| A. gambiae | 13 | 3 | 0.56 | 3 | 3 | 0.3 | 0.2 | 2 | 1 | 0.3 | 0.2 | -1.15 | 0.05 | 0.67 | |
| hybrids | 13 | 3 | 0.41 | 2 | 2 | 0.1 | 0.2 | 1 | 1 | 0.1 | 0.3 | -0.27 | -0.41 | 0.00 | |
| In each locality (all allelic classes) | Antula | 50 | 8 | 0.69 | 46 | 46 | 0.7 | 2.7 | n.a. | n.a. | 0.4 | 1.7 | -2.15 * | -5.51 * | 0.76 *** |
| Safim | 102 | 16 | 0.85 | 52 | 52 | 4.4 | 2.6 | n.a. | n.a. | 3.3 | 8.3 | 2.34 * | 1.11 | 0.50 *** | |
| Mansoa | 96 | 16 | 0.79 | 50 | 51 | 4.3 | 2.5 | n.a. | n.a. | 3.3 | 7.8 | 1.67 | 1.06 | 0.53 ** | |
| Ga-Mbana | 100 | 14 | 0.80 | 48 | 48 | 5.7 | 5.7 | n.a. | n.a. | 4.3 | 10.2 | 3.49 *** | 1.69 * | 0.58 *** | |
| Leibala | 114 | 10 | 0.83 | 45 | 45 | 4.5 | 4.5 | n.a. | n.a. | 3.2 | 9.0 | 3.09 ** | 1.91 * | 0.67 *** | |
| Overall | 462 | 31 | 0.85 | 60 | 61 | 4.7 | 2.3 | n.a. | n.a. | 3.5 | 8.8 | 2.61 * | -0.58 | 0.34 *** |
Summary statistics for TEP1 sequence polymorphism are reported a) within each TEP1 allelic class in the whole sample, b) within TEP1s and TEP1r1 in A. gambiae, A. coluzzii and hybrids, c) for the whole Guinean sample and within each locality (all TEP1 allelic classes). n = n° of alleles, H = n° of haplotypes, Hd = haplotype diversity, S = n° of segregating sites, Eta = total n° of mutations, π = nucleotide diversity (at non-synonymous = πa or at synonymous = πs sites), θ = Watterson's estimator, N-SYN = n° of non-synonymous mutations, SYN = n° synonymous mutations, Tajima Dsyn = Tajima's D test based on synonymous substitutions only. n.a. = not applicable. Deviations from neutrality tests are in bold:
* p< 0.05
** p< 0.01
*** p< 0.001.
Fig 3. Comparison of TEP1 genotype distribution in West/Central Africa and Guinea Bissau.
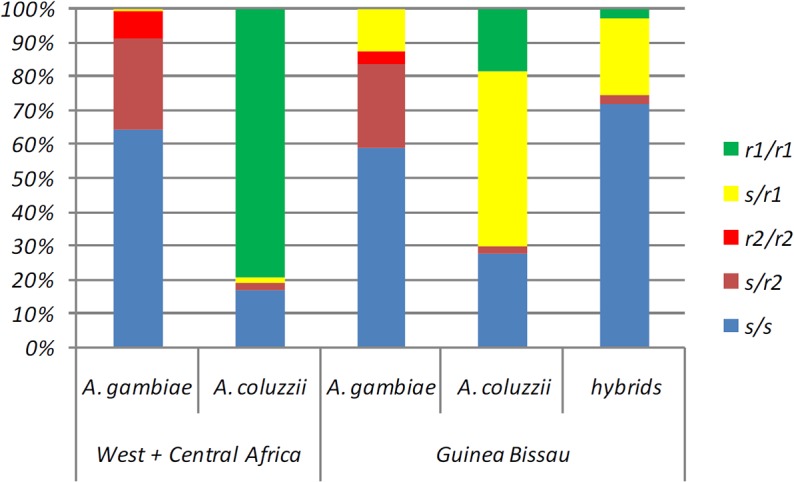
Relative frequencies of TEP1 genotypes in Guinea Bissau are reported in Table 1 and those from West and Central Africa are from White et al. (2011).
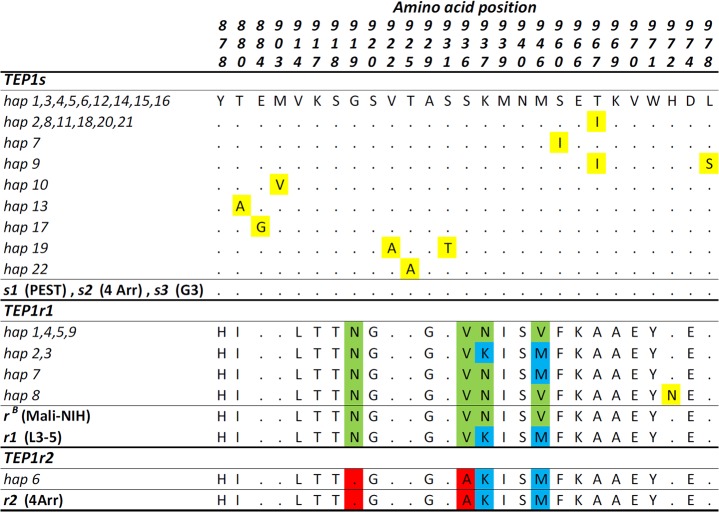
Polymorphism analysis shows that the TEP1-TED portion analyzed is well-conserved within each allelic class (TEP1s: πS = 1.0, πA = 0.1; TEP1r1: πS = 0.5, πA = 0.2; Table 2), likely retaining functionally-relevant residues known to confer resistance or susceptibility to pathogens [18, 39] (Fig 4). The four most common TEP1s haplotypes (-hap1,-hap2,-hap4 and-hap5) are those found in the rest of Africa [17, 19] and are shared between species (Fig 2). Of the resistant alleles (Figs 2 and 4), the most frequent TEP1r1 haplotype found is related to a functional variant found exclusively in West-Africa (i.e. TEP1r B) [17], and a single TEP1r2 haplotype is observed along the entire transect, corresponding to Tep1*R2 [18] and TEP1r A, which has a broad distribution [17]. Some novel TEP1r1 protein variants (i.e.-hap7,-hap8) are also observed; the phenotypic consequences of these are currently unknown.
Fig 4. Amino acid variability in TEP1-TED region in Guinea Bissau.
Protein alignment showing variable positions in the 387 bp of TED region of TEP1 analyzed is shown. Positions are numbered following Blandin et al., 2009. Shading highlights amino acid differences within and between TEP1s, TEP1r1 and TEP1r2 allelic classes. TEP1 allele designations refer to sequences from laboratory strains (Mali L3-5, Mali-NIH, PEST, 4Arr, and G3). TEP1 haplotypes (based on nucleotide sequences and named as in Fig 2) reported in the left side of each row code for the same protein variant.
Finally, our analysis suggests intra-specific genetic subdivision within A. gambiae from coastal areas of Guinea Bissau eastwards (Fig 1). Firstly, A. gambiae populations from Western coastal/flooded cropland areas (i.e. Safim, Antula and Mansoa) show higher frequencies of Int-1C (mean = 85.2%) than those from drier savanna-like areas in the eastern part of the geographical transect (i.e. Ga-Mbana and Leibala; mean Int-1C frequency = 29.2%) (χ 2 = 20.1; p<<0.001). These coastal A. gambiae populations are also significantly differentiated from the easternmost population examined for the TEP1 locus (i.e. Leibala; F ST = 0.34–0.45, p<0.05; S2 Table). Moreover, TEP1r2 frequency does not exceed 9.0% in the A. gambiae coastal populations, but is significantly higher (21%) in Leibala (χ 2 = 6.855; p<0.01) (Fig 1, Table 1). In addition, A. gambiae from Leibala also differs from coastal populations via near-exclusivity of the TEP1r1-hap3 haplotype (shared with only one A. coluzzii from Ga-Mbana, Fig 2), whose coding sequence contains two residues ('K' and 'M') related to the phenotypically-resistant TEP1*R1 [18], a possible recombinant TEP1r B/*R2 allele [17] (Fig 4). Notably, this West-to-East genetic discontinuity is also revealed by the analysis of chromosome-3 microsatellite polymorphisms along the same geographic transect in Guinea Bissau (Pinto J., personal communication). This pattern may be due either to a different origin of “coastal” and “inland” A. gambiae populations, or to natural selection promoting niche partitioning and, hence, genetic splitting at a local scale within this species. It is worth noting that intra-specific genetic divergence between geographically close populations of A. gambiae belonging to different ecological settings has already been reported from the “Far-West” region (i.e. from The Gambia), where a potential role of introgressive hybridization in triggering this divergence at a meso-geographical scale has been hypothesized [46].
Conclusions
The introgression of the TEP1r1 allele from A. coluzzii to A. gambiae in Guinea Bissau (Fig 1) shows that hybridization can promote the transfer of potentially adaptive immune-resistant alleles from a 'donor' (A. coluzzii) to a 'recipient' (A. gambiae) vector species. Introgressive hybridization may favor the rapid acquisition of advantageous traits from one species to another, but the adaptive significance (or fitness effects) of the genetic variant entering the 'recipient' species should be ascertained [47]. It is tempting to speculate that the observed absence of fixation and/or relatively low frequency of the ‘novel’ TEP1r1 acquired by A. gambiae in Guinea Bissau could be due to a lower adaptive benefit of this allele to the 'recipient' species in this region, where hybridization between A. gambiae and A. coluzzii is highest and stable. It would be interesting to monitor TEP1 allele exchange also in other African regions where assortative mating was shown to occasionally break down [6] and to assess whether introgressed resistant-alleles increase in frequency in A. gambiae: this could enhance the immune responsiveness of this species and, thus, its ability to compete with A. coluzzii in permanent larval sites, with potential repercussions on vector ecology, distribution and, eventually, malaria transmission especially in those areas where larval habitats are strongly segregated. In fact, it has been hypothesized that the more biotically diverse aquatic milieu and the higher bacterial load in larval sites typical of A. coluzzii in dry savannah areas are the most likely selective forces driving fixation of TEP1r1 in these northern populations [17, 20]. The observed lack of fixation of TEP1r1 in Guinea Bissau might be related to a greater availability of water resources due to a relatively higher annual rainfall regime in this westernmost region when compared to northern savanna areas [48]. The more humid ecological context of Guinea Bissau is likely to cause frequent replenishment of A. coluzzii larval habitats with clean-water all-year round, thus diluting the bacterial load in permanent ponds and possibly reducing immune stress for mosquito larvae. Indeed, mean annual precipitation in Guinea Bissau (e.g. ~1500 mm/yr in Leibala to 2000 mm/yr in Antula) is considerably higher than in northern regions (e.g., Mali: Bamako, 1100 mm/yr; Burkina Faso: Ouagadougo, 800 mm/yr; Bobo Dioulasso, 1100 mm/yr) [25, 49]. The present data on TEP1 allele distributions in Guinea Bissau could indirectly support the role of pathogens in potentially driving and maintaining fixation of TEP1r1 in A. coluzzii larvae in Mali and Burkina Faso, where a lower and seasonal precipitation regime may increase water stagnation and concentration of organic matter in permanent breeding sites, thereby increasing pathogen density and diversity. This might also explain the lower frequency or absence of the TEP1r1 allele in A. coluzzii collected close to coastal areas of Ghana and Cameroon [17]. Further studies testing correlations among TEP1 genotype frequencies, chromosomal inversion polymorphisms (known to be highly differentiated between coastal Guinea Bissau and inland/northern savannah areas [50]), climatic/ecological conditions and pathogen loads in breeding sites are needed to confirm these hypotheses.
Supporting Information
LnLHood LD = likelihood of linkage disequilibrium; LnLHood LE = likelihood of linkage equilibrium; p (LD) = probabilities from likelihood ratio tests (significant LD are in bold).
(DOC)
Pairwise comparisons of FST between species (co. = A. coluzzii, ga = A. gambiae) and hybrids (= hyb.) are reported either overall in Guinea Bissau, or within/among populations. Significance of FST was assessed by performing 500 replicates with a non-parametric permutation test; significant p<0.05 are in bold.
(DOC)
Acknowledgments
We are grateful to E. Levashina and to the EU-MALVECBLOK Consortium for having stimulated our interest on genetics of innate-immunity in Anopheles gambiae. We thank M. Calzetta, V. Pichler, P. Serini, and F. Polin for helping in optimization of TEP1 and Int-1 genotyping. This work was supported by: European Union’s INFRAVEC project (Grant agreement no. 228421, under FP7 program) to AdT and JP; FCT Portugal/FEDER (through program COMPETE) co-funds (PTDC/BIA-EVF/120407/2010); AWARD 2013 grant by 'Sapienza' Università di Roma to AdT, MIUR-FIRB “Futuro in Ricerca 2010” grant to BC (Grant N° RBFR106NTE).
Data Availability
All sequence files are available from the GenBank database (accession numbers: KR091079 - KR091309).
Funding Statement
This work was supported by the European Union’s INFRAVEC project (Grant agreement no. 228421, under FP7 program) to AdT and JP; FCT Portugal/FEDER (through program COMPETE) co-funds (PTDC/BIA-EVF/120407/2010); AWARD 2013 grant by 'Sapienza' Università di Roma to AdT, MIUR-FIRB “Futuro in Ricerca 2010” grant to BC (Grant N° RBFR106NTE).
References
- 1. della Torre A, Fanello C, Akogbeto M, Dossou-yovo J, Favia G, Petrarca V. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10: 9–18. [DOI] [PubMed] [Google Scholar]
- 2. della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35: 755–769. [DOI] [PubMed] [Google Scholar]
- 3. Diabaté A, Dao A, Yaro AS, Adamou A, Gonzalez R, Manoukis NC, et al. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae . Proc. Biol Sci. 2009;276: 4215–4222. 10.1098/rspb.2009.1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pennetier C, Warren B, Dabiré KR, Russell IJ, Gibson G. ‘‘Singing on the wing” as a mechanism for species recognition in the malarial mosquito Anopheles gambiae . Curr Biol. 2010;20: 131–136. 10.1016/j.cub.2009.11.040 [DOI] [PubMed] [Google Scholar]
- 5. Coetzee M, Hunt RH, Wilkerson R, della Torre A, Coulibaly MB, Besansky NJ. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 2013;3619: 246–274. [PubMed] [Google Scholar]
- 6. Lee Y, Marsden CD, Norris LC, Collier TC, Main BJ, Fofana A, et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae . Proc Natl Acad Sci USA 2013;110: 19854–19859. 10.1073/pnas.1316851110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehmann T, Diabaté A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet and Evol. 2008;8: 737–746. 10.1016/j.meegid.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gimonneau G, Pombi M, Choisy M, Morand S, Dabiré RK, Simard F. Larval habitat segregation between the molecular forms of the mosquito Anopheles gambiae in a rice field area of Burkina Faso, West Africa. Med Vet Entomol. 2012;26: 9–17. 10.1111/j.1365-2915.2011.00957.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kamdem C, Fouet C, Etouna J, Etoa F, Simard F, Besansky NJ, et al. Spatially explicit analyses of anopheline mosquitoes indoor resting density: implications for malaria control. PLoS One 2012;7: e31843 10.1371/journal.pone.0031843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae . PLoS Biology 2005;3: e285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae . Mol Ecol. 2010;19: 925–939. 10.1111/j.1365-294X.2010.04531.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lawniczak MKN, Emrich SJ, Holloway AK, Regier AP, Olson M, White B, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 2010;330: 512–514. 10.1126/science.1195755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neafsey DE, Lawniczak MKN, Park DJ, Redmond SN, Coulibaly MB, Traoré SF, et al. SNP genotyping defines complex gene-flow boundaries among african malaria vector mosquitoes. Science 2010;330: 514–517. 10.1126/science.1193036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ. Gene flow–dependent genomic divergence between Anopheles gambiae M and S forms. Mol Biol Evol. 2012;29: 279–291. 10.1093/molbev/msr199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reidenbach KR, Neafsey DE, Costantini C, Sagnon N, Simard F, Ragland GJ, et al. Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biol Evol. 2012;4: 1202–1212. 10.1093/gbe/evs095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clarkson CS, Weetman D, Essandoh J, Yawson AE, Maslen G, Manske M, et al. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 2014;5: 4248 10.1038/ncomms5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. White BJ, Lawniczak MKN, Cheng C, Coulibaly MB, Wilson MD, Sagnon N, et al. Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium . Proc Natl Acad Sci USA. 2011;108: 244–249. 10.1073/pnas.1013648108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, et al. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae . Science 2009;326: 147–150. 10.1126/science.1175241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Obbard DJ, Callister DM, Jiggins FM, Soares DC, Yan G, Little TJ. The evolution of TEP1, an exceptionally polymorphic immunity gene in Anopheles gambiae . BMC Evol Biol. 2008;8: 274 10.1186/1471-2148-8-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cassone BJ, Kamdem C, Cheng C, Tan JC, Hahn MW, Costantini C, et al. Gene expression divergence between malaria vector sibling species Anopheles gambiae and An. coluzzii from rural and urban Yaoundé Cameroon. Mol Ecol. 2014;23: 2242–2259. 10.1111/mec.12733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Caputo B, Santolamazza F, Vicente JL, Nwakanma DC, Jawara M, Palsson K, et al. The “Far-West” of Anopheles gambiae molecular forms. PLoS ONE 2011;6: e16415 10.1371/journal.pone.0016415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oliveira E, Salgueiro P, Palsson K, Vicente JL, Arez AP, Jaenson TG, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45: 1057–1063. [DOI] [PubMed] [Google Scholar]
- 23. Marsden CD, Lee Y, Nieman CC, Sanford MR, Dinis J, Martins C, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20: 4983–4994. 10.1111/j.1365-294X.2011.05339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, et al. Hybridization and speciation. J Evol Biol. 2005;26: 229–246. [DOI] [PubMed] [Google Scholar]
- 25. Nwakanma DC, Neafsey DE, Jawara M, Adiamoh M, Lund E, Rodrigues A, et al. Breakdown in the process of incipient speciation in Anopheles gambiae . Genetics 2013;193: 1221–1231. 10.1534/genetics.112.148718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gentile G, Santolamazza F, Fanello C, Petrarca V, Caccone A, della Torre A. Variation in an intron sequence of the voltage-gated sodium channel gene correlates with genetic differentiation between Anopheles gambiae s.s. molecular forms. Insect Mol Biol. 2004;13: 371–377. [DOI] [PubMed] [Google Scholar]
- 27. Santolamazza F, Caputo B, Nwakanma DC, Fanello C, Petrarca V, Conway DJ, et al. Remarkable diversity of intron-1 of the para voltage-gated sodium channel gene in an Anopheles gambiae/Anopheles coluzzii hybrid zone. Malaria J. 2015;14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gillies MT, de Meillon B. The Anophelinae of Africa south of the Sahara (Ethiopian Zoogeographical Region). 2nd edn. South African Institute for Medical Research, Johannesburg. Publications of the South African Institute for Medical Research no. 54, 1968.
- 29.Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara (Afrotropical Region). South African Institute for Medical Research, Johannesburg. Publications of the South African Institute for Medical Research no. 55, 1987.
- 30. Hervy JP, Le Goff G, Geoffroy B, Hervé JP, Manga L, Brunhes J. Les anophèles de la région Afro-tropicale. Logiciel d’identification et d’enseignement ORSTOM édition, série Didactiques; Paris, France, 1998. [Google Scholar]
- 31. Santolamazza F, Mancini E, Simard F, Qi Y, Tu Z, della Torre A. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7: 163 10.1186/1475-2875-7-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilkins EE, Howell PI, Benedict MQ. IMP PCR primers detect single nucleotide polymorphisms for Anopheles gambiae species identification, Mopti and Savanna rDNA types, and resistance to dieldrin in Anopheles arabiensis . Malar. J. 2006;19: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santolamazza F, Caputo B, Calzetta M, Vicente JL, Mancini E, Petrarca V, et al. Comparative analyses reveal discrepancies among results of commonly used methods for Anopheles gambiae molecular form identification. Malar J. 2010;10: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee Y, Marsden CD, Nieman C, Lanzaro CG. A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae . Mol Ecol Resour. 2014;14: 297–305. 10.1111/1755-0998.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinto J, Lynd A, Vicente JL, Santolamazza F, Randle NP, Gentile G, et al. Multiple origins of knockdown resistance mutations in the Afrotropical mosquito vector Anopheles gambiae . PLoS One 2007;2: e1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132: 115–130. [DOI] [PubMed] [Google Scholar]
- 37. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le BV, Williams M, Logarajah S, Baxter RH. Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles. PLoS Pathog. 2012;8: e1002958 10.1371/journal.ppat.1002958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16: 37–48. [DOI] [PubMed] [Google Scholar]
- 41. Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009;25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 42. Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution 1984;38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 43. Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2007;1: 47–50. [PMC free article] [PubMed] [Google Scholar]
- 44. Rousset F. Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008;8: 103–106. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 45. Etang J, Vicente JL, Nwane P, Chouaibou M, Morlais I, Do Rosario VE, et al. Polymorphism of intron-1 in the voltage-gated sodium channel gene of Anopheles gambiae s.s. populations from Cameroon with emphasis on insecticide knockdown resistance mutations. Mol Ecol. 2009;18: 3076–3086. 10.1111/j.1365-294X.2009.04256.x [DOI] [PubMed] [Google Scholar]
- 46. Caputo B, Nwakanma D, Caputo FP, Jawara M, Oriero EC, Hamid-Adiamoh M, et al. Prominent intraspecific genetic divergence within Anopheles gambiae sibling species triggered by habitat discontinuities across a riverine landscape. Mol Ecol. 2014;23: 4574–4589. 10.1111/mec.12866 [DOI] [PubMed] [Google Scholar]
- 47. Hedrick PW. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol Ecol. 2013;22: 4606–4618. 10.1111/mec.12415 [DOI] [PubMed] [Google Scholar]
- 48. Liebmann B, Bladé I, Kiladis GN, Carvalho LMV, Senay GB, Allured D, et al. Seasonality of African Precipitation from 1996 to 2009. J Climate 2012;25: 4304–4322. [Google Scholar]
- 49. Kangah PAD. Rainfall and agriculture in Central West Africa since 1930: impact on socioeconomic development LAP LAMBERT academic publisher, Saarbrücken, 2010. [Google Scholar]
- 50. Petrarca V, Carrara GC, Di Deco MA, Petrangeli G. The Anopheles gambiae complex in Guinea Bissau. Parassitologia 1983;25: 29–39. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
LnLHood LD = likelihood of linkage disequilibrium; LnLHood LE = likelihood of linkage equilibrium; p (LD) = probabilities from likelihood ratio tests (significant LD are in bold).
(DOC)
Pairwise comparisons of FST between species (co. = A. coluzzii, ga = A. gambiae) and hybrids (= hyb.) are reported either overall in Guinea Bissau, or within/among populations. Significance of FST was assessed by performing 500 replicates with a non-parametric permutation test; significant p<0.05 are in bold.
(DOC)
Data Availability Statement
All sequence files are available from the GenBank database (accession numbers: KR091079 - KR091309).