Abstract
Background
Securing negative surgical margins is a critical goal for head and neck surgery. Local recurrence occurs even in some cases with histologically negative surgical margins. Minimal residual tumor cells may lead to locoregional recurrence despite clear histologic margins reported at the time of resection of head and neck squamous cell carcinoma (HNSCC). In order to identify subclinical residual disease, we analyzed deep margin imprint samples collected on one layer nitrocellulose sheets.
Methods
Bisulfite treated DNA from 73 eligible cases was amplified by quantitative methylation-specific polymerase chain reaction (QMSP) targeting 6 genes (DCC, EDNRB, HOXA9, KIF1A, NID2, NR2B). QMSP values were dichotomized as positive or negative. Associations between the QMSP status of deep margin samples and clinical outcomes were evaluated.
Results
Two gene methylation combinations among DCC, EDNRB and HOXA9 were associated with decreased locoregional recurrence-free survival (LRFS), recurrence-free survival (RFS) and overall survival (OS). The methylated gene combination EDNRB and HOXA9 in margin imprints was the most powerful predictor of poor LRFS (HR=3.31, 95% CI=1.30-8.46; P=0.012), independent of standard histological factors. In addition, methylation of EDNRB/HOXA9 showed a trend toward reduced RFS (HR=2.74, 95% CI=0.90-8.33; P=0.075) and OS (HR=5.78, 95% CI=0.75-44.7; P=0.093) by multivariable analysis.
Conclusion
A panel of gene methylation targets in deep surgical margin imprints provides a potential predictive marker of post-operative locoregional recurrence. Intra-operative use of molecular margin-imprint analysis might assist surgeons to obtain rigorously negative surgical margins and improve the outcome of head and neck surgery.
Keywords: head and neck cancer, SCC, surgical margin, methylation, locoregional recurrence
Introduction
Squamous cell carcinoma of the upper aerodigestive tract (head and neck squamous cell carcinoma [HNSCC]) is one of the major malignancies affecting males1. HNSCC is usually diagnosed at an advanced stage. Over one-third of tumors are classified at the Union for International Cancer Control’s (UICC) TNM Stage IV, which has a recurrence rate of over 50% during the first two years after surgery2. Patients with poor pathologic prognostic factors after surgery are recommended to receive postoperative therapy (radiation +/- chemotherapy). These factors include close or involved surgical margins, extranodal spread of tumor in metastatic nodes, multiple lymph node metastases and poorly differentiated histology with perineural or perivascular spread3. Hence, securing negative surgical margins is an important therapeutic goal. In a series of 352 HNSCC cases, 59% of cases with positive margins went on to develop locoregional recurrence4. However, a high recurrence rate (over 40%) was also found in the 303 margin negative cases. This observation indicates that there must have been residual cancer cells or partially transformed precancerous cells in the resection bed which were not detected by histological margin examination.
Huang et al. demonstrated that deep surgical margin status was more strongly correlated with tumor recurrence than mucosal margins5. In deep margins, histologically undetected residual cancer cells may be left behind due to the ‘tumor budding’ phenomenon, defined as a single cancer cell or a cluster of <5 cancer cells protruding into the stroma beyond the invasive front6-8. This phenomenon is thought to be due to loss of cellular cohesion and active invasive movement6, 9. These papers showed that the existence of tumor budding is one of the significant prognostic markers of SCC. Intra-tumoral proliferation in lymphatic vessels and blood vessels has also been seen in HNSCC 10. Some of the vessels around a tumor contain detached cancer cells. These are hard to detect in intra-operative frozen samples.
Molecular margin analysis offers several advantages over light microscopy for detecting small numbers of cancer cells. Tumor specific molecular alterations may be more sensitive and specific than detection of cellular morphologic change characterized as atypia. In addition, molecular analysis allows for evaluation of the entire sample harvested from the surgical margin, and processed with high-throughput techniques. Analysis can be automated and standardized, eliminating the need for subjective expertise reliant on training and experience.
Twenty years ago, Brennan, et al, examined the TP53 mutation status of histologically negative surgical margins of HNSCCs11. The probability of locoregional recurrence was significantly correlated with positive molecular margins. Remarkably, no locoregional recurrence was found in cases with TP53 mutation negative margins during 2 years after the surgery. However, to date, one of the limitations of mutation analysis is the lack of universal targets. Although TP53 mutation is one of the most frequent molecular targets in HNSCCs, it can be found in only about 50% of tumors12, 13. Epigenetic alteration of margin samples was first examined in our laboratory in 2004 by Goldenberg, et al, using quantitative methylation-specific polymerase chain reaction (QMSP) 14. In some histologically negative margins, tumor-specific methylation of P16 and MGMT genes was detected. The presence of methylation markers including P16, CCNA1, DCC15, DAPK, ECAD16 and TMEFF 17 in margin specimens has been associated with clinical outcomes. However, these papers are based on small retrospective patient cohorts, and do not take into account the tumor methylation status. It is logical to assume that the relevant molecular markers for margin analysis are those present in the tumor.
In the current study, we made two innovations in molecular margin analysis. First, we prospectively collected tumor samples and matched margin samples from 101 consecutive head and neck cancer patients. This approach avoids potential bias in sample selection since QMSP measurement was completed before patient outcome was available. Secondly, we focused on deep margins where head and neck surgical specimens are surrounded by muscle or other connective tissues. In order to effectively collect margin specimens from the wider area of an irregular deep margin surface, we employed an innovative margin imprint procedure using nitrocellulose membrane to harvest DNA. Gaston, et al., first reported this approach for prostate specimens and extracted RNA from the cells collected on the membrane surface 18. Our group adapted the procedure to collect DNA from imprint samples19.
The primary outcome of this study was the prognostic value of a panel of methylation markers detected in deep margins for predicting recurrence free survival, and locoregional recurrence free survival. The secondary outcome was to determine whether the deep margin imprint procedure is equal or superior to deep margin tissue sample analysis for prognostic evaluation.
Materials and Methods
Surgical cases
The study cohort was prospectively collected from Johns Hopkins Hospital between May, 2009 and December, 2013. Surgical specimens were collected from 101 sequential head and neck cancer patients from whom written informed consent was obtained. The protocol was approved by the Johns Hopkins Hospital Institutional Review Board. Twenty-eight cases were excluded; 19 cases had been enrolled at the time of a second surgical resection attempt which may have complicated deep margin analysis due to prior scarring, 5 others had histologically positive final surgical margins, 3 were found not to be squamous cell carcinoma and 1 had only dysplasia. Thus, 73 cases were eligible for analysis in this study. The average time elapsed from surgery to assessment of outcome was 2.3 years. Locoregional recurrence includes local recurrences and regional lymph node metastasis.
Sample collection procedure
At the time of surgical resection, frozen section samples were sent from the mucosal margins and deep wound bed per routine practice. Only when these margins were read as negative, was the case processed for molecular margin analysis. After removal of the tumor specimen with a rim of apparently normal margin, free liquid on the surface of the specimen was removed by blotting with cloth towels on a back table. Then, margin imprints were collected by pressing 3×3 cm Hybond-C Extra nitrocellulose membranes (GE Healthcare, Little Chalfont, UK) directly on the specimen for 10 seconds18. The membranes were placed into a coded 50 ml tube with 10 mL 1% SDS-PK solution. Three repeat membrane samples were taken from each facet of the specimen. Thereafter, matched margin tissue samples (approximately 4×4 mm) were sharply collected, placed in a coded tube and stored in liquid nitrogen. In order to avoid possible float-on contamination of cells from the tumor, tumor imprints and tissues were collected last using the same method as for margin sampling. A fragment of normal muscle and/or normal mucosa was also collected from an area more than 5 cm distant from the tumor as a negative control.
Target gene selection
Candidate genes were selected for this study based on work previously done in our laboratory to develop a panel for HNSCC detection and surveillance in body fluids including aberrant promoter hypermethylation of DCC20, EDNRB21, HOXA922, KIF1A21, NID222 (detected in HNSCC tumor tissues), and NR2B (detected in esophageal SCC cell lines) 23.
DNA extraction and Bisulfite treatment
For frozen tissues, 50 micron thick sections were harvested, microdissected and digested with 1% SDS (Sigma-Aldrich, St. Louis, MO) and 50μg/ml proteinase K (Invitrogen, Carlsbad, CA). The presence of cancer in each tumor tissue sample was histologically confirmed in slides taken before and after sample harvesting. After four rounds of proteinase K exposure during two overnight periods, phenol/chloroform extraction and ethanol precipitation was performed as previously described 24. DNA from margin imprints were immediately eluted, digested and extracted by the same procedure. DNA (200 ng) extracted from each tissue or imprint sample was subjected to bisulfite treatment using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA).
Quantitative methylation-specific PCR
The bisulfite-modified DNA was used as a template for fluorescence-based real-time PCR as described 25. Primers and probes sequences are available on Supplementary Table S1. Real-time methylation-specific PCR reaction was performed in triplicate using the 7900HT Sequence Detector System (Applied Biosystems, Foster City, CA). Thermal cycling was initiated with a denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Each plate included patient DNA samples, positive standards (Bisulfite Converted Universal Methylated Human DNA standard (Zymo Research, Irvine, CA)), and multiple water blanks as no-template controls. The average of triplicate samples was used for analysis. The relative level of methylated DNA (relative QMSP) for each gene was determined as a ratio of its QMSP value to ACTB gene’s QMSP value × 100.
Statistical analysis
Surgical margin imprint and tissue data were analyzed separately. Patient clinicopathological characteristics were compared using the Fisher’s exact test for categorical variables. For each individual gene, methylation status was dichotomized at zero (positive versus negative). Molecular margin analysis was considered to be positive if target gene methylation was detected in both tumor and at least one matched margin, and negative if it was detected in tumor but not in any margin. If no methylation was detected in tumor, the gene was not used to evaluate the margin samples. Genes were first analyzed individually and then in combination for association with clinical outcomes (LRFS, RFS and OS). Evaluation of gene combinations was performed in the subset of margin samples that had both target genes methylated in main tumor. Panels of 2-gene combinations were considered as molecular margin positive if at least one methylated gene was detected. LRFS was defined as the time from surgery to the time of first documentation of locoregional disease recurrence. Distant recurrence without prior locoregional recurrence was considered as a competing-risk event. Those who remained alive without disease recurrence were censored at the time of their last clinic assessment. RFS was defined as the time from surgery to the time of first documentation of any disease recurrence. OS was defined as the time from surgery to the time of death from any cause. Those who remained alive were censored at the last date the subject was known to be alive. For LRFS, calculation, testing, and regression modeling with hazard ratios and the corresponding 95% confidence intervals of subdistribution functions in competing risks were performed using the Fine and Gray method 26. Association of gene methylation in margins with RFS and OS was evaluated using the Cox proportional hazards model with hazard ratios and 95% confidence intervals estimated. Combinations of all paired genes were initially evaluated in univariate regression models. Promising combinations with P<0.05 in the univariate analysis were selected and further evaluated in a multivariate regression model adjusting for potential confounders (e.g., differentiation, TNM stage). All tests were two-sided and considered statistically significant and clinically promising at P<0.05. Adjustment for multiple comparisons were not performed as the subsets for each marker pair were not identical, and thus the analysis must be considered exploratory. Statistical analyses were carried out using SAS (version 9.3, SAS Institute, Cary, NC) and R statistical software (version 2.15.2).
Results
Patient clinicopathological characteristics
Advanced HNSCCs were dominant in this cohort. The 73 eligible HNSCC cases are characterized in Supplementary Table S2. Among 73 cases, margin imprints were collected from 65 cases, whereas margin tissues were collected from 63 cases. There were 55 cases with both imprints and tissues available, 10 cases with only imprints and 8 cases with only tissues. There was no significant difference in clinicopathological features between margin imprint collected cases (n=65) and margin tissue collected cases (n=63).
Relative QMSP value between tumor and normal mucosa or normal muscle
Although each of the 6 candidate genes had already been established as good methylation markers in HNSCCs 20-23, we confirmed the tumor-specificity of these genes in our cohort (Supplementary Figure S1, Supplementary Table S3). As many deep surgical margins consist of muscle tissues, we collected both normal mucosa and normal muscle from the surgical field far away from the main tumor. Almost all of the 6 genes showed significant tumor-specificity. Only KIF1A had relative QMSP values that were not significantly different between tumor and normal mucosa (P=0.143). However, the difference between tumor and normal muscle was significant (P=0.004). We also checked 11 serum samples from the cohort, and no tumor-specific methylation of serum DNA was detected (data not shown).
Association of clinicopathological factors and QMSP status in tumor with clinical outcome
As an initial step, univariate analyses of the association of clinicopathological and molecular factors with clinical outcomes were performed. The result of margin imprint collected cases (n=65) is shown in Supplementary Table S4, and margin tissue collected cases (n=63) in Supplementary Table S5. In both subsets, smoking history, tumor differentiation and TNM stage were significantly associated with RFS and OS. However, as previously reported27, the presence of single methylated genes in the tumor was not found to be associated with clinical outcome.
Relative QMSP values in histologically positive margins
Before using the QMSP assay for surgical margin analysis, the methylation detection technique was piloted using 6 HNSCC surgical margin tissues which had been previously collected and found to be cancer-positive by light microscopy (positive control) (Supplementary Table S6). DCC, HOXA9 and NID2 genes showed methylation signal in all 6 margins, while EDNRB, KIF1A and NR2B showed no methylation in 2 out of 6 margins. The methylation status of the main tumors in these cases was not available. If no methylation of a gene is present in the main tumor, it is unlikely to be detected even in histologically positive margins. This observation highlights the need for a number of highly sensitive methylation markers to assemble a panel of genes covering all cases.
Relative QMSP value analysis in margin samples
Positive QMSP rates of main tumors in 65 imprint cases and 63 tissue cases were similar (Supplementary Table S7). On the other hand, positive QMSP rates of surgical margins in 65 imprint cases were much higher than those in 63 tissue cases (Supplementary Table S7). This may reflect the advantage of the margin imprint technique which can collect cells from the entire cut specimen surface.
Univariate analysis of individual 6 genes in association with clinical outcomes
The association of the presence of methylation of individual genes in margins with clinical outcomes (LRFS, RFS and OS) is shown in Supplementary Table S8. The presence of methylation of no single gene in tumor was able to predict the clinical outcome.
Univariate and multivariable analysis of gene combinations in association with clinical outcomes
Next, we looked for an association between clinical outcome and methylated gene combinations in margins. The number of cases with concomitantly positive QMSP for two genes in margin samples from the imprint cohort (n=65, upper left white area) and tissue cohort (n=63, lower right grey area) is listed in Table 1. Coverage percentages (the proportion of cases displaying both markers within the tumor) were quite similar for each cohort. Gene combinations showing promise for association with LRFS were selected in Table 2 from all possible pairs listed in Supplementary Table S9. In the margin tissue cohort, no combination of methylation markers was associated with clinical outcomes. On the other hand, in the margin imprint cohort, the presence of EDNRB and HOXA9 combination was significantly predictive of poor LRFS (HR=3.10, 95%CI: 1.19-8.06, P=0.020) and RFS (HR=3.31, 95%CI: 1.11-9.84, P=0.031). This marker pair also was associated with OS (HR=7.62, 95%CI: 0.99-58.6, P=0.051). The cumulative incidence curves for LRFS indicated that patients with molecular margins negative for EDNRB and HOXA9 had no locoregional recurrence during 25 months after surgical treatment (Figure 1a, P=0.033, Gray’s test). This marker combination was also a strong predictor of RFS (Figure 1b). In multivariable analysis (Table 3), this gene combination remained significantly associated with poor LRFS (HR:3.31, 95%CI: 1.30-8.46, P=0.012), after adjusting for differentiation and pathological TNM stage. Distribution of clinicopathological parameters was comparable between the marker combination positive and negative cases (Supplementary Table S10).
Table 1.
Numbers of both genes QMSP positive cases in the main tumor
| Number | DCC | EDNRB | HOXA9 | KIF1A | NID2 | NR2B |
|---|---|---|---|---|---|---|
| DCC | *** | 44 (67.7%) | 50 (76.9%) | 27 (41.5%) | 53 (81.5%) | 31 (47.7%) |
| EDNRB | 39 (61.9%) | *** | 43 (66.2%) | 24 (36.9%) | 45 (69.2%) | 26 (40.0%) |
| HOXA9 | 48 (76.2%) | 41 (65.1%) | *** | 27 (41.5%) | 56 (86.2%) | 30 (46.2%) |
| KIF1A | 39 (61.9%) | 33 (52.4%) | 42 (66.7%) | *** | 29 (44.6%) | 18 (27.7%) |
| NID2 | 48 (76.2%) | 41 (65.1%) | 52 (82.5%) | 40 (63.5%) | *** | 32 (49.2%) |
| NR2B | 27 (42.9%) | 23 (36.5%) | 32 (50.8%) | 23 (36.5%) | 30 (47.6%) | *** |
Number of cases with concomitantly positive QMSP for two genes among each imprint cohort (n=65, upper left white area) and tissue cohort (n=63, lower right grey area) were listed. Coverage percentage was calculated as a ratio of the number of the cases with a both positive main tumor to the total number of each cohort.
Table 2.
Univariate analysis for QMSP gene combinations in association with clinical outcomes
| Factors | LRFS | RFS | OS | ||||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| DCC/HOXA9 | Imprint | 2.99(0.96, 9.27) | 0.058 | 3.93(0.91, 16.91) | 0.067 | 4.06(0.53, 31.25) | 0.178 |
| EDNRB/HOXA9 | Imprint | 3.10(1.19, 8.06) | 0.020 | 3.31(1.11, 9.84) | 0.031 | 7.62(0.99, 58.6) | 0.051 |
Combination of other genes resulted in P values >0.1.
Figure 1.
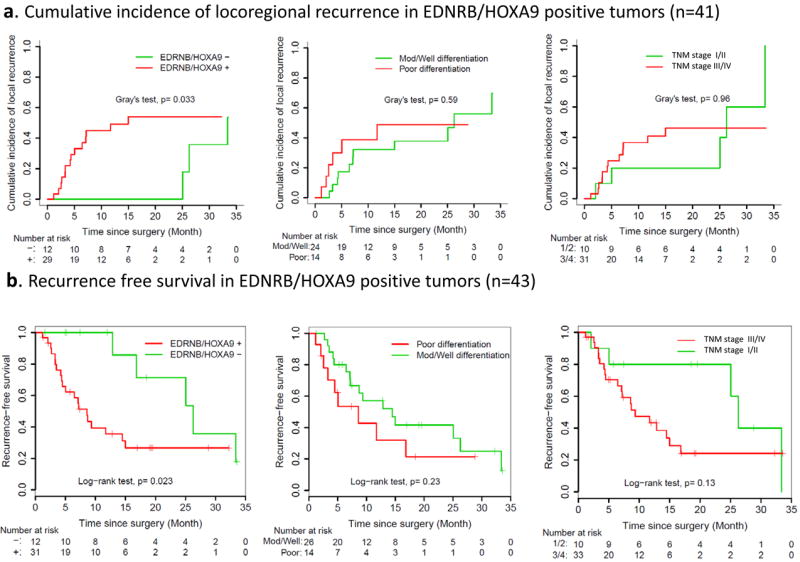
Cumulative incidence curves of locoregional recurrence (Figure 1a) and Kaplan-Meier curves of recurrence free survival (RFS) (Figure 1b). Note: Figure 1a pertains to 41 cases for which the site of recurrence was well documented. Figure 1b includes two others that recurred without precise information on site of recurrence (n=43) In both curves, the molecular marker was a more powerful predictor than histological findings (differentiation and pathological TNM stage). Remarkably, no recurrence was found in the molecular marker negative group during the first 2 years in Figure 1a.
Table 3.
Multivariable analysis for QMSP gene combinations in association with clinical outcomes
| Factors | LRFS | RFS | OS | ||||
|---|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| EDNRB/HOXA9 imprint | Positive vs negative | 3.31(1.30, 8.46) | 0.012 | 2.74(0.90, 8.33) | 0.075 | 5.78(0.75,44.7) | 0.093 |
| Differentiation | Poor vs moderate, well | 1.04(0.36, 3.00) | 0.95 | 1.93(0.81, 4.57) | 0.14 | 3.68(1.15, 11.7) | 0.028 |
| pTNM Stage | III, IV vs I, II | 0.72(0.24, 2.21) | 0.57 | 2.12(0.73, 6.18) | 0.17 | 4.72(0.91, 24.6) | 0.065 |
Discussion
Surgery remains a cornerstone in the treatment of head and neck cancer. The goal of obtaining negative margins is critical to the effectiveness of surgical extirpation. Although histologic diagnosis using frozen samples has long been the gold standard for intraoperative margin assessment, histologically negative surgical margins do not guarantee recurrence-free survival4. For evaluating surgical margins more effectively, detection of cancer-specific molecular alterations within the operative time-frame can be considered as a next-generation strategy. Quantitative methylation specific PCR (QMSP) is a well-established application for detecting low levels of tumor DNA in various body fluids such as saliva28, 29 and urine30. Adaptation of this approach to DNA harvested by imprint began by using promising tumor-specific methylation markers of HNSCC19. One of the practical challenges for the application of this approach is that margin analysis ideally should be performed before the completion of the surgical procedure so that the results may influence the extent of the surgical resection. Although the QMSP assay is a time consuming procedure consisting of DNA extraction, bisulfite treatment and QMSP, we have already streamlined the process such that it can be accomplished within 3 hours by one person without compromising the quality of the results or increasing the cost31. While the process still exceeds the time needed for frozen section analysis, if the primary tumor is resected at the beginning of the operation and margin samples harvested, margin analysis may be finished during subsequent neck dissection(s) and/or the reconstructive phase of the operation, allowing surgeons to resect additional margins if the QMSP margin is positive.
Our results indicate that neither methylation of EDNRB or HOXA9 in HNSCC tumor is associated with clinical outcome. Yet detection of either of these tumor-specific markers in deep margin imprint DNA is associated with a higher risk of recurrence. These findings suggests that positive tumor-specific signal in the margin is indicative of persistent tumor cells in general rather than of a particularly virulent tumor phenotype. The gene combination of EDNRB and HOXA9 was previously investigated in a prospectively collected cohort of 10 consecutive surgical margin samples 31. In that small pilot study, this combination was present in one case that later recurred. The HOXA9 gene encodes a DNA-binding transcription factor that regulates gene expression, morphogenesis, and differentiation. The HMGA2/TET/HOXA9 signaling pathway regulates breast cancer growth and metastasis32. Frequent promoter methylation of HOXA9 has been detected in lung cancer, bladder cancer, and head and neck cancer 22, 33-35. A study from our laboratory showed HOXA9 promoter methylation had 85% sensitivity and 97% specificity for histologic diagnosis of HNSCC 22. The EDNRB gene encodes a protein called endothelin receptor type B, which is located on the surface of cells and functions as a regulator of several critical biological processes, including the development of blood vessels and the stimulation of cell growth and division. Altering the endothelin pathway is reported to be associated with tumorgenesis36. Frequent promoter methylation of EDNRB has been detected in gastric cancer, prostate cancer, and head and neck cancer 27, 37-39. A study from our laboratory indicated this gene was methylated in 97% of primary HNSCC tissues and 6.6% of normal control salivary rinses 21. Besides extremely high sensitivity and specificity, the comparatively high relative QMSP values in HNSCC combine to make these excellent tumor-specific markers for margin analysis. It is possible that other methylation marker combinations with similar tumor population coverage, sensitivity and specificity would match or exceed HOXA9/EDNRB in utility for molecular margin imprint analysis and prediction of risk of recurrence.
In the imprint margin cohort, the cumulative incidence curves for LRFS (Figure 1a) indicated that cases lacking both of the combination marker (EDNRB and HOXA9) had no locoregional recurrence during 25 months after surgical treatment. Interestingly, the tails of these curves were converging at the end of the follow-up period. Upon further scrutiny of the clinical records, this observation was found to be due to the late identification of neck lymph node metastasis in the imprint margin negative group. The size of this cohort does not permit more exhaustive exploration of the underlying clinical or biological basis for this observation.
Postoperative radiation therapy was given to patients in the cohort according to standard clinical practice. Among the 65 imprint margin cases, 27 patients received post-operative radiotherapy (pTNM stage I:2, II:1, III:4, IV:20), and 38 patients did not receive any additional therapy (pTNM stage 0:1, I:11, II:4, III:7, IV:15). [Late stage cases did not receive radiation for a variety of reasons including: distant past history of head and neck radiation, complete removal with clear margins and no nodal metastases (total laryngectomy or laryngopharyngectomy), patient refusal, early postoperative recognition of distant disease.]. Since postoperative adjuvant radiation is intended to control minimal residual disease, its application could profoundly affect outcome and obscure the predictive power of molecular margin detection. Indeed, the difference between molecular margin negative and positive cases was more remarkable in patients who did not receive post-operative radiotherapy after surgery (Supplementary Figure S2 a). In order to elucidate the influence of post-operative radiation on the molecular margin analysis, we compared the clinical outcome of the 10 cases which received post-operative radiation and the 19 which did not among the 29 EDNRB and HOXA9 imprint margin positive cases (Supplementary Figure S2 b). As expected, radiated cases had significantly lower cumulative incidence of local recurrence (P=0.006, Gray’s test). On the other hand, there was no significant difference in outcome in molecular margin negative cases between those who received radiation and those who did not. (Supplementary Figure S2 c). This result not only clearly suggests the usefulness of this marker pair (EDNRB and HOXA9) for the selection of radiation candidates, but also the real presence of subclinical cancer cells in the methylation positive surgical bed.
There are some assay limitations when using QMSP for evaluating margin samples. There is no established QMSP threshold to score surgical margin samples as positive or negative. QMSP cut-off points have been established for a number of tumors after microdissection to ensure over 70-80% cancer cells compared to matched normal tissues. However, margin samples are expected to contain only very rare cancer cells. For this reason, a simple binary scoring paradigm was employed with the presence of any signal being read as positive. Secondly, in order to achieve maximum signal specificity, it is necessary to avoid cancer cell contamination when collecting the margin samples, especially for imprint margins. If detached cancer cells from tumor are deposited on the surrounding surface of the specimen and collected by margin tissue or imprint procedure, a false positive result would be scored. In the present study, we collected margin samples from multiple facets of the surgical specimen. When methylation signal was present, it was limited to one or two squares of nitrocellulose in most cases. Fully negative samples within the set can be considered as internal negative controls and support the contention that a positive signal from other quadrants indicates the true presence of tumor cells.
There is room for improvement of our molecular surgical margin analysis. In this study, some candidate methylation genes including p16 were excluded because of the low methylation level in the 73 tumors. Also, the most promising pair (EDNRB and HOXA9) covered only 43 of 65 imprint cases (66.2%) (Table 3). As a future study, we will continue to test other novel candidate methylated genes. Our ultimate goal is to establish a panel of several methylated genes that covers almost all HNSCC. Moving forward, we will obtain methylated gene information from the pre-operative tumor biopsy in order to minimize the number of candidate genes that need to be examined for intraoperative margin analysis, and focus only on high quality targets present within the individual’s cancer, (made-to-order methylated gene panel). As we have already developed an innovative rapid QMSP pipeline31, molecular margin analysis is ready for application in the operating room. Another future direction is to use Digital PCR technology as a highly sensitive process for the detection of small amounts of tumor-specific methylation40.
In conclusion, our methylation gene panel analysis of margin imprints successfully identified HNSCC patients at increased risk for locoregional recurrence after surgical treatment. In multivariable analysis, the combination of EDNRB and HOXA9 methylation analysis of margin imprints was a more powerful predictor of locoregional recurrence (HR=3.31, 95% CI=1.30-8.46, P=0.012) than standard histological findings (differentiation and pathological TNM stage). Although these findings will need to be externally validated in an independent cohort, we have demonstrated that securing not only a histologic, but also a molecularly negative surgical margin may ultimately lead to improved patient outcomes.
Supplementary Material
Acknowledgments
Grant support
This work is supported by grants from National Institute of Health and the National Institute of Dental and Craniofacial Research (R01 DE013152-11).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Iro H, Waldfahrer F. Evaluation of the newly updated TNM classification of head and neck carcinoma with data from 3247 patients. Cancer. 1998;83:2201–2207. [PubMed] [Google Scholar]
- 3.Sanderson RJ, Ironside JA. Squamous cell carcinomas of the head and neck. BMJ. 2002;325:822–827. doi: 10.1136/bmj.325.7368.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones AS, Bin Hanafi Z, Nadapalan V, Roland NJ, Kinsella A, Helliwell TR. Do positive resection margins after ablative surgery for head and neck cancer adversely affect prognosis? A study of 352 patients with recurrent carcinoma following radiotherapy treated by salvage surgery. Br J Cancer. 1996;74:128–132. doi: 10.1038/bjc.1996.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang X, Pateromichelakis S, Hills A, et al. p53 mutations in deep tissues are more strongly associated with recurrence than mutation-positive mucosal margins. Clin Cancer Res. 2007;13:6099–6106. doi: 10.1158/1078-0432.CCR-07-1369. [DOI] [PubMed] [Google Scholar]
- 6.Almangush A, Bello IO, Keski-Santti H, et al. Depth of invasion, tumor budding, and worst pattern of invasion: Prognostic indicators in early-stage oral tongue cancer. Head Neck. 2013 doi: 10.1002/hed.23380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarioglu S, Acara C, Akman FC, et al. Tumor budding as a prognostic marker in laryngeal carcinoma. Pathol Res Pract. 2010;206:88–92. doi: 10.1016/j.prp.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Koike M, Kodera Y, Itoh Y, et al. Multivariate analysis of the pathologic features of esophageal squamous cell cancer: tumor budding is a significant independent prognostic factor. Ann Surg Oncol. 2008;15:1977–1982. doi: 10.1245/s10434-008-9901-6. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Huang H, Huang Z, et al. Tumor budding correlates with poor prognosis and epithelial-mesenchymal transition in tongue squamous cell carcinoma. J Oral Pathol Med. 2011;40:545–551. doi: 10.1111/j.1600-0714.2011.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beasley NJ, Prevo R, Banerji S, et al. Intratumoral lymphangiogenesis and lymph node metastasis in head and neck cancer. Cancer Res. 2002;62:1315–1320. [PubMed] [Google Scholar]
- 11.Brennan JA, Mao L, Hruban RH, et al. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995;332:429–435. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 12.Boyle JO, Hakim J, Koch W, et al. The incidence of p53 mutations increases with progression of head and neck cancer. Cancer Res. 1993;53:4477–4480. [PubMed] [Google Scholar]
- 13.Maestro R, Dolcetti R, Gasparotto D, et al. High frequency of p53 gene alterations associated with protein overexpression in human squamous cell carcinoma of the larynx. Oncogene. 1992;7:1159–1166. [PubMed] [Google Scholar]
- 14.Goldenberg D, Harden S, Masayesva BG, et al. Intraoperative molecular margin analysis in head and neck cancer. Arch Otolaryngol Head Neck Surg. 2004;130:39–44. doi: 10.1001/archotol.130.1.39. [DOI] [PubMed] [Google Scholar]
- 15.Tan HK, Saulnier P, Auperin A, et al. Quantitative methylation analyses of resection margins predict local recurrences and disease-specific deaths in patients with head and neck squamous cell carcinomas. Br J Cancer. 2008;99:357–363. doi: 10.1038/sj.bjc.6604478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Supic G, Kozomara R, Jovic N, Zeljic K, Magic Z. Prognostic significance of tumor-related genes hypermethylation detected in cancer-free surgical margins of oral squamous cell carcinomas. Oral Oncol. 2011;47:702–708. doi: 10.1016/j.oraloncology.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Shaw RJ, Hobkirk AJ, Nikolaidis G, et al. Molecular staging of surgical margins in oral squamous cell carcinoma using promoter methylation of p16(INK4A), cytoglobin, E-cadherin, and TMEFF2. Ann Surg Oncol. 2013;20:2796–2802. doi: 10.1245/s10434-012-2713-8. [DOI] [PubMed] [Google Scholar]
- 18.Gaston SM, Soares MA, Siddiqui MM, et al. Tissue-print and print-phoresis as platform technologies for the molecular analysis of human surgical specimens: mapping tumor invasion of the prostate capsule. Nat Med. 2005;11:95–101. doi: 10.1038/nm1169. [DOI] [PubMed] [Google Scholar]
- 19.Roh JL, Westra WH, Califano JA, Sidransky D, Koch WM. Tissue imprint for molecular mapping of deep surgical margins in patients with head and neck squamous cell carcinoma. Head Neck. 2012;34:1529–1536. doi: 10.1002/hed.21982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carvalho AL, Chuang A, Jiang WW, et al. Deleted in colorectal cancer is a putative conditional tumor-suppressor gene inactivated by promoter hypermethylation in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9401–9407. doi: 10.1158/0008-5472.CAN-06-1073. [DOI] [PubMed] [Google Scholar]
- 21.Demokan S, Chang X, Chuang A, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127:2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guerrero-Preston R, Soudry E, Acero J, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4:1061–1072. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim MS, Yamashita K, Baek JH, et al. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–3418. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 24.Shao C, Tan M, Bishop JA, et al. Suprabasin is hypomethylated and associated with metastasis in salivary adenoid cystic carcinoma. PLoS One. 2012;7:e48582. doi: 10.1371/journal.pone.0048582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harden SV, Tokumaru Y, Westra WH, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003;9:1370–1375. [PubMed] [Google Scholar]
- 26.Fine JP, G R. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 27.Roh JL, Wang XV, Manola J, Sidransky D, Forastiere AA, Koch WM. Clinical correlates of promoter hypermethylation of four target genes in head and neck cancer: a cooperative group correlative study. Clin Cancer Res. 2013;19:2528–2540. doi: 10.1158/1078-0432.CCR-12-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Righini CA, de Fraipont F, Timsit JF, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007;13:1179–1185. doi: 10.1158/1078-0432.CCR-06-2027. [DOI] [PubMed] [Google Scholar]
- 29.Schussel J, Zhou XC, Zhang Z, et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin Cancer Res. 2013;19:3268–3275. doi: 10.1158/1078-0432.CCR-12-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi M, Guerrero-Preston R, Okamura J, et al. Innovative Rapid Gene Methylation Analysis of Surgical Margin Tissues in Head and Neck Cancer. Ann Surg Oncol. 2014 doi: 10.1245/s10434-014-3661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun M, Song CX, Huang H, et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang SH, Kim KU, Kim JE, et al. Detection of HOXA9 gene methylation in tumor tissues and induced sputum samples from primary lung cancer patients. Clin Chem Lab Med. 2011;49:699–704. doi: 10.1515/CCLM.2011.108. [DOI] [PubMed] [Google Scholar]
- 34.Kim YJ, Yoon HY, Kim JS, et al. HOXA9, ISL1 and ALDH1A3 methylation patterns as prognostic markers for nonmuscle invasive bladder cancer: array-based DNA methylation and expression profiling. Int J Cancer. 2013;133:1135–1142. doi: 10.1002/ijc.28121. [DOI] [PubMed] [Google Scholar]
- 35.Reinert T, Borre M, Christiansen A, Hermann GG, Ørntoft TF, Dyrskjøt L. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One. 2012;7:e46297. doi: 10.1371/journal.pone.0046297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welch AK, Jacobs ME, Wingo CS, Cain BD. Early progress in epigenetic regulation of endothelin pathway genes. Br J Pharmacol. 2013;168:327–334. doi: 10.1111/j.1476-5381.2012.01826.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011;2:123–150. doi: 10.1007/s13148-011-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phé V, Cussenot O, Rouprêt M. Interest of methylated genes as biomarkers in urothelial cell carcinomas of the urinary tract. BJU Int. 2009;104:896–901. doi: 10.1111/j.1464-410X.2009.08696.x. [DOI] [PubMed] [Google Scholar]
- 39.Tao K, Wu C, Wu K, et al. Quantitative analysis of promoter methylation of the EDNRB gene in gastric cancer. Med Oncol. 2012;29:107–112. doi: 10.1007/s12032-010-9805-8. [DOI] [PubMed] [Google Scholar]
- 40.Muraoka T, Soh J, Toyooka S, et al. The degree of microRNA-34b/c methylation in serum-circulating DNA is associated with malignant pleural mesothelioma. Lung Cancer. 2013;82:485–490. doi: 10.1016/j.lungcan.2013.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


