Abstract
Autoimmune thyroid diseases (AITD) and Type 1 diabetes (T1D) frequently occur in the same individual pointing to a strong shared genetic susceptibility. Indeed, the cooccurrence of T1D and AITD in the same individual is classified as a variant of the autoimmune polyglandular syndrome type 3 (designated APS3v). Our aim was to identify new genes and mechanisms causing the co-occurrence of T1D+AITD (APS3v) in the same individual using a genome-wide approach. For our discovery set we analyzed 346 Caucasian APS3v patients and 727 gender and ethnicity matched healthy controls. Genotyping was performed using the Illumina Human660W-Quad.v1. The replication set included 185 APS3v patients and 340 controls. Association analyses were performed using the PLINK program, and pathway analyses were performed using the MAGENTA software. We identified multiple signals within the HLA region and conditioning studies suggested that a few of them contributed independently to the strong association of the HLA locus with APS3v. Outside the HLA region, variants in GPR103, a gene not suggested by previous studies of APS3v, T1D, or AITD, showed genome-wide significance (p<5×10−8). In addition, a locus on 1p13 containing the PTPN22 gene showed genome-wide significant associations. Pathway analysis demonstrated that cell cycle, B-cell development, CD40, and CTLA-4 signaling were the major pathways contributing to the pathogenesis of APS3v. These findings suggest that complex mechanisms involving T-cell and B-cell pathways are involved in the strong genetic association between AITD and T1D.
Keywords: Type 1 diabetes, Graves’ disease, Hashimoto’s thyroiditis, Gene, HLA
1. INTRODUCTION
The most common autoimmune endocrine disorders are type 1 (autoimmune) diabetes (T1D) and autoimmune thyroid diseases (AITD). T1D and AITD are both characterized by T-cell infiltration and production of autoantibodies directed at the target organs (pancreatic islets and thyroid, respectively), resulting in their dysfunction or destruction [1]. Epidemiological data have shown that T1D and AITD frequently occur together in the same family and in the same individual, suggesting a strong shared genetic susceptibility [1]. In different studies, up to 44% of T1D patients were positive for thyroid antibodies (TAb) (thyroid peroxidase [TPO] and/or thyroglobulin [Tg] antibodies) [2;3]. Similarly, 2.3% of children with AITD have islet cell antibodies compared with 0% of controls [4]. Indeed, the co-occurrence of T1D and AITD in the same individual is classified as one of the variants of autoimmune polyglandular syndrome type 3 (APS3) [5] (since the phenotype of T1D+AITD in the same individual is a known as a variant of APS3 we refer to it as APS3v in this manuscript).
Family studies also support a strong shared genetic susceptibility to T1D and AITD. One of the largest family studies of T1D and AITD in the US [6;7], showed that among female diabetic probands Hashimoto’s thyroiditis (HT) was diagnosed in 54 – 75% of cases, and among female relatives, the frequency of HT was 22 – 44%. Two other studies, one from the UK [8] and one from Colombia [9] showed similar results. Thus, epidemiological data support a significant shared genetic susceptibility to T1D and AITD. However, while much has been learned about the genetics of T1D and AITD individually, less is known about the joint genetic etiology of these two diseases.
In view of the strong evidence for shared susceptibility for T1D and AITD we have previously mapped joint susceptibility genes for T1D and AITD using linkage studies in a large cohort of multiplex families in which T1D and AITD clustered. We used both the candidate gene approach [10;11], and whole genome linkage approach [12]. A striking finding of these studies was that the phenotype of T1D+AITD in the same individual (APS3v) was a unique phenotype with a genetic predisposition distinct from that of T1D or AITD alone [10;12]. The most significant contribution to T1D+AITD (APS3v) genetic susceptibility came from a sequence variant in HLA-DR [13]. In addition to the HLA class II susceptibility contribution we mapped three non-MHC loci showing evidence for linkage - CTLA-4, PTPN22, and FOXP3 [10–12]. These genes as well as other genes, such as IL-2α/CD25 and TNFα have been reported by other groups studying APS3v [14]. The aim of the present study was to identify genes unique for APS3v using the robust genome wide association study (GWAS) approach in order to identify novel shared mechanisms and pathways for T1D and AITD.
2. PARTICIPANTS AND METHODS
2.1 Study participants
The project was approved by the Icahn School of Medicine Institutional Review Board. SEARCH for Diabetes in the Youth study participants ≥ 18 years old or a parent/guardian of participants < 18 years provided written informed consent for data collection including DNA. We performed a two-stage GWAS using a discovery set and an independent replication set.
Discovery set (Table 1A)
Table 1.
| A: Discovery Set: Non-Hispanic White Participants with Type 1 Diabetes (N=346) from the SEARCH for Diabetes in Youth Study, Clinical Characteristics and Family History of diabetes | ||||
|---|---|---|---|---|
| All n=346 |
Females n=216 (62%) |
Males n=130 (38%) |
p-Value* | |
| Age at diagnosis (y) [Mean (SD)] | 8.5 (4.2) | 8.5 (3.9) | 8.4 (4.7) | 0.7 8 |
| Age at study visit (y) [Mean (SD)] | 13.9 (4.0) | 13.8 (4.1) | 14.1 (4.0) | 0.66 |
| Disease duration (y) [Mean (SD)] | 5.0 (4.5) | 4.9 (4.5) | 5.1 (4.4) | 0.43 |
| TPO-Ab (IU/L) [Mean (SD)] | 15.9 (10.0) | 16.0 (10.1) | 15.6 (9.3) | 0.75 |
| Additional diagnosis [n (%)] | ||||
| Hypothyroidism | 55/346 (16%) | 39/216 (18%) | 16/129 (12%) | 0.18 |
| Hyperthyroidism | 24/345 (7%) | 17/216 (8%) | 7/129 (5%) | 0.51 |
| Vitiligo | 6/343 (1.7%) | 3/215 (1.4%) | 3/128 (2.3%) | 0.68 |
| Addison disease | 1/345 (0.3%) | 1/216 (0.5%) | 0 | 1 |
| Celiac disease | 0 | 0 | 0 | - |
| Family History of DM [n (%)] | ||||
| Mother | 21/344 (6%) | 9/215 (4%) | 12/129 (9%) | 0.065 |
| Father | 20/340 (6%) | 12/214 (6%) | 8/126 (6%) | 1.00 |
| Maternal grandfather | 52/324 (16%) | 33/198 (17%) | 18/128 (14%) | 0.64 |
| Maternal grandmother | 58/335 (17%) | 34/208 (16%) | 24/127 (19%) | 0.40 |
| Paternal grandfather | 55/318 (17%) | 33/199 (17%) | 22/119 (18%) | 0.76 |
| Paternal grandmother | 53/318 (16%) | 29/207 (14%) | 24/121 (20%) | 0.21 |
| Age at the time of Diagnosis DM (years, Mean (SD)] | ||||
| Mother | 24.7 (11.8) | 25.3 (11.2) | 24.4 (12.5) | 0.99 |
| Father | 31.7 (15.9) | 31.5 (15.3) | 32.0 (18.1) | 0.95 |
| Maternal grandfather | 55.1 (15.8) | 57.5 (13.5) | 51.0 (18.8) | 0.55 |
| Maternal grandmother | 54.6 (14.3) | 52.5 (16.5) | 58.9 (6.0) | 0.78 |
| Paternal grandfather | 56.2 (16.5) | 57.9 (16.0) | 54.3 (17.5) | 0.44 |
| Paternal grandmother | 51.0 (21.6) | 52.4 (23.0) | 49.4 (20.5) | 0.52 |
| B: Replication set Patients Characteristics | ||||
|---|---|---|---|---|
| All n=185 |
Females n=128 (70%) |
Males n=57 (30%) |
||
| Age of diagnosis (y) [Mean (SD)] | 19.7 (14.3) | 20.5 (15.4) | 17.9 (11.5) | |
| Age of inclusion (y) [Mean (SD)] | 43.5 (14.5) | 46.1 (14.5) | 37.8 (12.6) | |
| Additional diagnosis [n (%)] | ||||
| Hashimotos’ Thyroiditis | 146/185 (79%) | 108/128 (84%) | 38/57 (67%) | |
| Graves’ Disease | 39/185 (21%) | 20/128 (16%) | 19/57 (33%) | |
n, Number, y, Years, SD, Standard Deviation. TPO-Ab, Thyroid Peroxydase Antibodies, DM, Diabetes Mellitus (any type)
p-Values are for comparing Females vs. Males
n, Number, y, Years, SD, Standard Deviation.
The discovery set included 346 non-Hispanic White (NHW) patients with T1D who were also confirmed to be positive for TPO antibodies. These patients represent a subset of the SEARCH for Diabetes in Youth study prevalent and incident cohorts (for a full description of the SEARCH cohort see [15]). T1D was based on physicians’ diagnosis of T1D and confirmed at the time of the SEARCH study visit based on a positive autoantibody titer for GAD65 or IA-2, consistent with the ADA criteria for type 1A (autoimmune) diabetes [16]. Controls included 727 healthy Caucasians (NHW) that were obtained from the Illumina iControlDB database (http://genomeinformaticsalliance.org/science/icontroldb.ilmn) [17]. Patients and controls were gender and ethnicity matched. The breakdown of the discovery set recruitment is detailed in Supplemental Table G.
Replication set (Table 1B)
The replication set included a separately enrolled dataset of 185 Caucasian (NHW) patients with T1D+AITD (consisting of a T1D+AITD cohort previously described [13] with additional patients enrolled since 2010); 340 healthy Caucasians (NHW) were used as replication controls. The breakdown of the replication set recruitment is detailed in Supplemental Table G.
2.2 Genotyping and principal component analysis (PCA)
Genotyping of the cases and controls was performed at the Cincinnati Children’s Hospital Medical Center DNA core. DNA samples were genotyped using the Illumina Human660W-Quad v1 BeadChips assays [18]. This chip provides 87% genomic coverage in Caucasians at r2 > 0.8. Quality control (QC) testing (including testing for Hardy-Weinberg equilibrium, identity by descent, missing rate < 5%, and minor allele frequency [MAF] > 0.01) was performed using PLINK (http://pngu.mgh.harvard.edu/purcell/plink/). 514,008 single nucleotide polymorphisms (SNPs) passed the QC testing and were used in the association analysis. To ensure that our cases and controls were appropriately matched for ethnicity, avoiding effects of population stratification, we performed Principal Component Analysis (PCA). For the PCA we used 100,000 representative SNPs from the GWAS panel and analyzed them using 2 PC’s [19]. Cases and controls were matched on their principal component scores. The program EIGENSOFT was used to perform the PCA [19].
2.3 Association analyses in the discovery set
After QC testing, association analysis was performed using PLINK for PCA matched samples (346 patients and 727 controls) via logistic regression while using the first 2 principal components as covariates to adjust for the effect of population stratification on the results. SNPs with a p-value < 5×10−8 were considered to have genome-wide level of significance, while SNPs with a p-value ≤ 1×10−5 were considered showing suggestive evidence for association [18].
2.4 Replication set analysis
SNPs were selected for analysis in the replication set based on the following criteria: (1) To reduce the likelihood of false positive results there had to be at least 2 associated SNPs (p ≤ 1×10−5) within the same gene in the discovery set for the gene to be analyzed in the replication set; (2) we did not analyze in the replication set SNPs within HLA class I & II genes (since these have already been shown to be strongly associated with APS3v [11;13]), as well as SNPs within multiple MHC genes that are in tight Linkage Disequilibrium (LD) with each other. Based on these criteria we selected for the replication set 116 SNPs showing suggestive evidence for association (p ≤ 1×10−5). To these we added 28 SNPs that showed borderline significance (p< 5×10−4), but were located within genes already selected for replication based on the above 2 criteria to improve coverage of these genes. The list of 24 genes (containing 144 SNPs) with suggestive evidence for association that were tested in the replication set is shown in Supplemental Table A. These 144 SNPs were genotyped in the replication set using the Illumina custom-made Goldengate assays. PCA was not performed in the replication set since only 144 SNPs were genotyped in that set and they did not include ancestry informative markers. In order to test the effect the PCA might have had on the replication set we redid the association analysis on the 144 SNPs identified in the discovery set without adjusting the results by PCA. The analysis showed that for all the SNPs except one (rs4589926) the results with and without adjusting by PCA were comparable (Supplemental Table B). These results confirmed that the lack of adjustment for PCA in the replication set should have had minimal if any effect on the results.
2.5 Meta-analysis combining the discovery and replication sets
The PLINK association output including odds ratio and standard error values were generated from the discovery and replication datasets, respectively. Meta-analysis was then performed on these two association outputs using PLINK to derive meta-p-values with Mantel-Haenszel correction.
2.6 Linkage disequilibrium (LD) analysis
Linkage disequilibrium (LD) analysis was performed for the MHC and 1p13 loci since several genes in these loci were found to be associated with APS3v. For the LD analysis we used the Haploview program and analyzed the discovery set data [20].
2.7 Logistic regression analysis (Conditional haplotype-based testing)
MHC locus (6p)
To test whether the non-HLA genes within the MHC locus that showed genome-wide significant associations had independent effects or were associated due to strong LD with HLA class I and II genes (known to be associated with APS3v) we ran logistic regression models and performed a Likelihood Ratio- (LR) based omnibus association test on the discovery set. This analysis enabled us to test whether the overall haplotype variation of the SNPs within these genes influenced the presence of the APS3v phenotype. We then performed conditional LR-based tests in order to (1) test for an independent effect of each SNP (independent of the haplotypic effects formed by the remaining SNPs); and (2) test for any marginal omnibus significant association controlling for each SNP (i.e., we tested if for SNPs that had independent effects, the remaining SNPs in the haplotype jointly contributed association information beyond the tested SNP alone).
For this analysis we used 23 SNPs in the MHC region that showed genome wide significance. They included the 4 highest scoring SNPs within the HLA class II genes, 3 highest scoring SNPs within the HLA class I genes, and 16 highest scoring SNPs within the non-HLA MHC genes that showed significant association (Supplemental Table C1). This analysis was performed using PLINK v1.07.
Chromosome 1p13 locus
Logistic regression analysis was also performed on the 4 most associated SNPs within 4 genes in this locus, MAGI3, PHTF1, PTPN22, BCL2L15, in order to determine whether any of these genes had independent effects.
Chromosome 4q27 locus
3 SNPs within this locus showed significant associations with APS3v and we performed logistic regression analysis using allelic, as well as dominant and recessive models to determine which model is most likely contributing to the association with APS3v.
2.8 Pathway analysis
Pathway analysis was performed using the Meta-Analysis Gene-set Enrichment of variant Associations (MAGENTA) program (http://www.broadinstitute.org/mpg/magenta/). For the pathway analysis we used only the discovery dataset. The 1000 SNPs that had the lowest p-values were used in this analysis. The probability that SNPs for a pathway/gene-set were significantly enriched when compared to randomly selected SNP-sets was computed. A p-value < 0.05 was considered significant.
3. RESULTS
3.1 Association analysis in the discovery set
Detailed characteristics of the discovery set are provided in Table 1. The discovery set included 346 Caucasian (NHW) APS3v patients; of them 216 (62%) were females and 130 (38%) were males resulting in a female to male (F:M) ratio of 1.6:1. This ratio is significantly higher than the reported F:M ratio in T1D which is close to 1.0 [21], but is consistent with the female preponderance of AITD. The control discovery set consisted of 727 North American Caucasians. There were 451 (62%) females and 276 (38%) males in the control group (F:M ratio 1.6:1). In order to control for population stratification we performed a PCA in the discovery set. PCA demonstrated that our APS3v patients and controls were ethnically matched North American Caucasians of European ancestry along the axes of statistically significant principal components. The lambda gc (genomics inflation factor) of the PCA was 1.016 demonstrating a very close ethnicity matching of patients and controls.
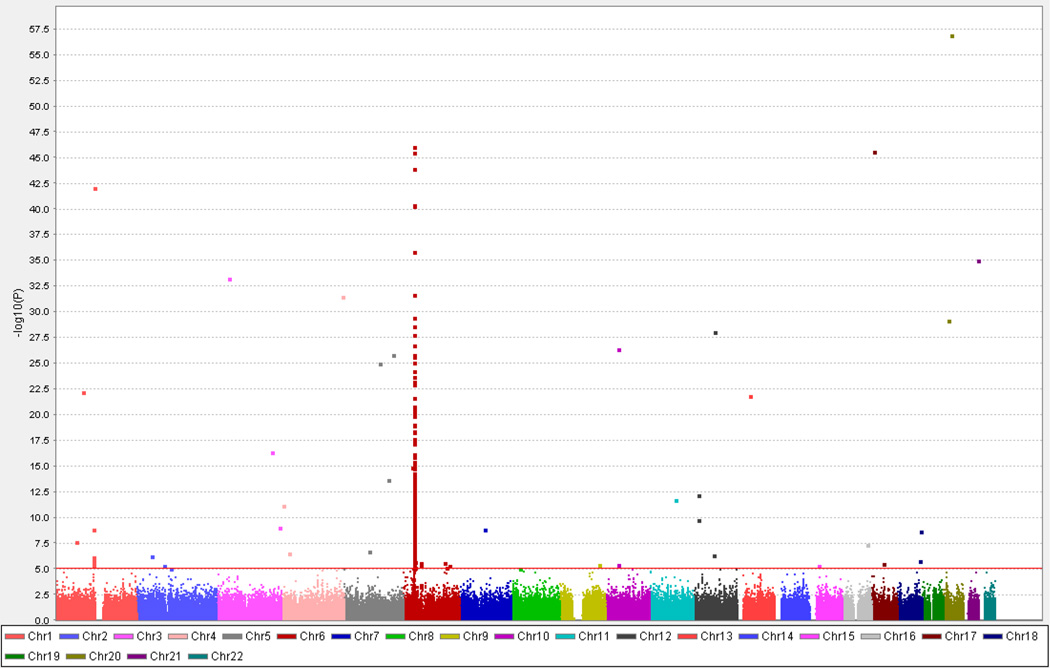
The quantile-quantile (QQ) plot, which illustrates results of the distribution of association analyses for all the tested SNPs, showed an excess of SNPs in several loci that were significantly associated with APS3v at the tail of the distribution (Supplemental Figure A, blue line). To examine the effects of HLA genes (known to be strongly associated with APS3v) on the QQ plot we re-analyzed the QQ plot after removal of SNPs within HLA class I and class II genes (Supplemental Figure A, red line). The QQ plot without HLA genes showed a substantial reduction in the number of SNPs that are significantly associated with APS3v, confirming the major role of HLA genes in the etiology of APS3v. The results of the association analysis using the Human660W-Quad v1 SNPs in APS3v patients compared to controls are summarized in the Manhattan plot shown in Figure 1. The strongest association was seen at the MHC (6p) locus with many SNPs in this locus showing genome wide significance (p < 5 × 10−8); however, other non-MHC loci also showed significant associations (Figure 1). 307 SNPs showed suggestive evidence of association (p ≤ 1 × 10−5) (Supplemental Tables D1 and D2).
Figure 1.
Manhattan plot showing the results of the association analysis in the Discovery set. The X-axis shows the chromosomal location of the analyzed SNPs and the Y-axis shows the – log(p-value). As expected the strongest association was seen on chromosome 6 at the MHC (HLA) locus with many SNPs in this locus showing genome wide significance (p < 5×10−8); however, other non-MHC loci also showed significant associations.
3.2 Association analysis in the replication set
For the replication analysis we tested 144 selected SNPs. These SNPs were chosen because they were located within or near 24 genes-loci showing suggestive evidence of association (p ≤ 1×10−5) in at least 2 SNPs, excluding HLA class I and II genes. After QC, 11 SNPs were removed leaving a total of 133 that were analyzed. The replication analysis identified 107 SNPs within 22 genes-loci that showed a nominal p-value of less than 0.05 (Supplemental Table E). Of these, 20 genes-loci contained one or more SNPs that showed genome wide significance (p < 5×10−8) in the combined sets (Table 2) and one gene (MAGI3) had a SNP giving a combined p-value close to genome wide significance (p = 4.18×10−7). Of these 21 replicated genes-loci, 16 are non-HLA genes located within the MHC locus on chromosome 6p (Figure 2), and 5 genes (MAGI3, PHTF1, PTPN22, BCL2L15 and GPR103) are located outside the MHC locus. Interestingly 4 of the 5 non-MHC genes (MAGI3, PHTF1, PTPN22, and BCL2L15) are located on chromosome 1p13.
Table 2.
Combined association analysis in the Discovery and Replication sets in 22 genes that showed nominal association (p<0.05) in at least one SNP for the Replication set
| Discovery Set | Replication Set | Meta P | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | CHR | GENE | BP | Risk Allele |
P | OR | P | OR | P | OR |
| rs1270942 | 6 | CFB | 32,026,839 | C | 9.02 × 10−11 | 2.371 | 4.13 × 10−18 | 4.923 | 4.65 × 10−25 | 3.0494 |
| rs1150752 | 6 | TNXB | 32,172,704 | G | 2.03 × 10−13 | 2.804 | 1.81 × 10−23 | 59.57 | 2.27 × 10−24 | 3.8666 |
| rs2857595 | 6 | NCR3 | 31,676,448 | T | 1.17 × 10−06 | 1.734 | 2.20 × 10−22 | 3.717 | 1.87 × 10−23 | 2.3765 |
| rs2251396 | 6 | MICA | 31,472,686 | T | 7.37 × 10−11 | 1.92 | 1.11 × 10−14 | 2.893 | 9.87 × 10−23 | 2.2132 |
| rs1015166 | 6 | TAP2 | 32,906,709 | T | 1.88 × 10−11 | 1.925 | 5.57 × 10−13 | 2.62 | 3.84 × 10−22 | 2.1428 |
| rs1980493 | 6 | BTNL2 | 32,471,193 | G | 1.83 × 10−10 | 2.211 | 3.61 × 10−12 | 3.093 | 1.57 × 10−20 | 2.5034 |
| rs3099844 | 6 | HCP5 | 31,556,955 | A | 5.70 × 10−10 | 2.229 | 1.33 × 10−12 | 3.502 | 3.94 × 10−20 | 2.6093 |
| rs2476601 | 1 | PTPN22 | 114,179,091 | A | 2.88 × 10−11 | 2.384 | 8.63 × 10−11 | 3.637 | 7.35 × 10−20 | 2.7073 |
| rs3131296 | 6 | NOTCH4 | 32,280,971 | A | 4.48 × 10−08 | 1.983 | 5.72 × 10−13 | 3.325 | 3.27 × 10−18 | 2.389 |
| rs3115663 | 6 | BAT2 | 31,709,822 | G | 3.70 × 10−10 | 2.027 | 2.69 × 10−09 | 2.413 | 8.72 × 10−18 | 2.1609 |
| rs2227956 | 6 | HSPA1L | 31,886,251 | T | 6.32 × 10−15 | 0.2047 | 2.09 × 10−3 | 0.4768 | 1.99 × 10−15 | 0.2912 |
| rs886424 | 6 | IER3 | 30,889,981 | A | 5.96 × 10−07 | 1.894 | 1.04 × 10−09 | 3.006 | 2.89 × 10−14 | 2.2107 |
| rs3130564 | 6 | PSORS1C1 | 31,209,653 | T | 2.04 × 10−05 | 1.601 | 1.23 × 10−10 | 2.708 | 5.75 × 10−13 | 1.9116 |
| rs2844657 | 6 | DDR1 | 30,937,501 | C | 1.19 × 10−05 | 1.618 | 3.35 × 10−09 | 2.534 | 3.02 × 10−12 | 1.8746 |
| rs2358994 | 1 | BCL2L15 | 114,230,984 | A | 7.41 × 10−08 | 1.844 | 4.74 × 10−05 | 1.919 | 1.56 × 10−11 | 1.8688 |
| rs2071591 | 6 | NFKBIL1 | 31,623,778 | T | 6.28 × 10−06 | 1.529 | 1.72 × 10−06 | 1.895 | 1.14 × 10−10 | 1.6416 |
| rs1041981 | 6 | LTA | 31,648,763 | A | 1.30 × 10−05 | 1.507 | 8.01 × 10−07 | 1.935 | 1.50 × 10−10 | 1.637 |
| rs7679475 | 4 | GPR103 | 122,533,490 | A | 1.28 × 10−07 | 1.651 | 6.64 × 10−4 | 1.562 | 3.51 × 10−10 | 1.6198 |
| rs1111695 | 1 | PHTF1 | 114,045,422 | G | 6.95 × 10−07 | 1.586 | 5.72 × 10−3 | 1.434 | 1.65 × 10−08 | 1.5331 |
| rs2523989 | 6 | TRIM31 | 30,186,254 | A | 1.41 × 10−4 | 1.589 | 1.28 × 10−05 | 2.126 | 1.86 × 10−08 | 1.7499 |
| rs2153977 | 1 | MAGI3 | 113,881,594 | T | 2.44 × 10−4 | 1.431 | 3.74 × 10−4 | 1.611 | 4.18 × 10−07 | 1.491 |
| rs6924102 | 6 | PSMB8 | 32,919,361 | G | 7.27 × 10−07 | 1.614 | 4.64 × 10−08 | 0.4888 | 0.4578 | 1.0594 |
SNP, single nucleotide polymorphism, CHR, chromosome, BP, location on chromosome in base pairs, P, p-value, OR, odds ratio, MetaP, p-value for Meta-analysis
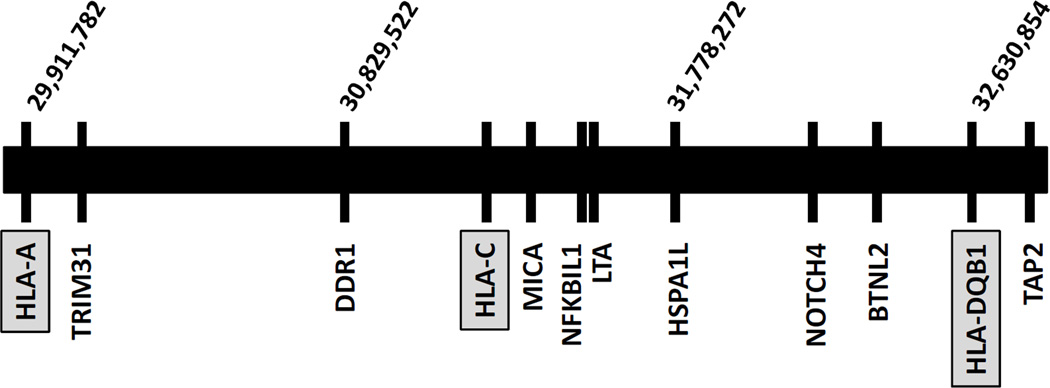
Figure 2.
Schematic diagram of the MHC region showing non-HLA genes within the MHC locus found to be associated with T1D+AITD (APS3v). Of 21 replicated genes 16 were located within the MHC locus on chromosome 6p but are not HLA genes. Shown are HLA-A, HLA-C, and HLA-DQB1 (gray boxes), as well as 9 of the 16 genes within the MHC region that were replicated. Gene names are indicated below the line representing chromosome 6p and their chromosomal location in bp is indicated above the line.
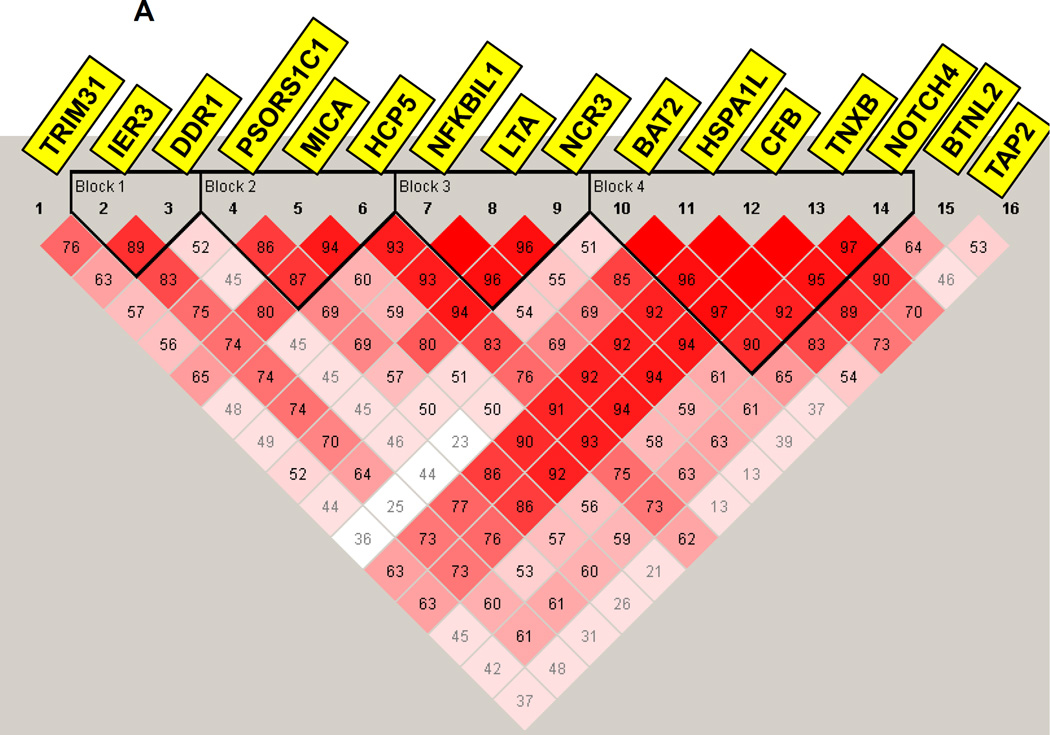
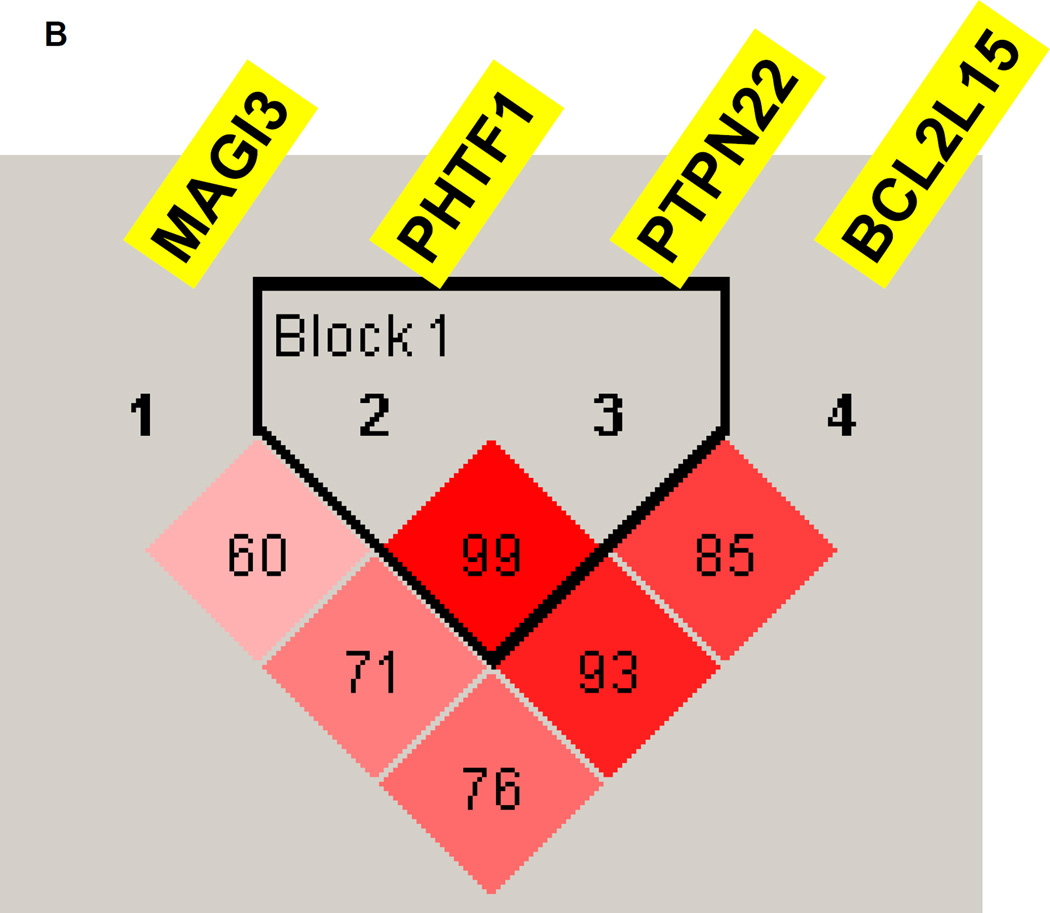
3.3 Linkage disequilibrium (LD) analysis
LD analysis for the MHC region showed that the MHC gene-locus where the 16 replicated genes were located consisted of 4 LD blocks, suggesting that while some genes may show association with APS3v because of LD others may have independent contributions (Figure 3A). LD analysis for the 1p13 locus showed tight LD between PHTF1 and PTPN22 and weaker LD between these two genes and BCL2L15, while MAGI3 did not show significant LD with the other 3 genes (Figure 3B), suggesting that more than one gene in this locus may have independent effects.
Figure 3.
(A) Linkage disequilibrium (LD) analysis of SNPs within 16 MHC genes that were replicated. LD analysis showed that the MHC region, where the replicated genes are located, consisted of 4 LD blocks. These results suggested that while some genes that are located within the same LD block (e.g. HCP5 and MICA) may show association with APS3v because of LD, others that are outside these blocks may have independent contributions. (B) LD analysis of the 1p13 locus. The PHTF1 and PTPN22 genes show tight LD with a D’ of 0.99 while the BCL2L15 gene was in weaker LD with PTPN22 (D’=0.85) and PHTF1 (D’=0.93). In contrast, MAGI3 was not in LD with the other 3 genes at the locus showing D’<0.80 with all 3 genes.
3.4 Logistic regression analysis (Conditional haplotype-based testing)
MHC locus (6p)
The overall variation of the haplotypes formed by the 23 SNPs located within associated MHC genes influenced the presence of the APS3v phenotype (omnibus association LR test p-value < 1×10−8). Intriguingly, even though the MHC region had 4 LD blocks only one gene, HSPA1L showed independent effect on disease risk after adjusting for the effect of the entire haplotype (Supplemental Table C1). SNP rs2227956, located within HSPA1L, showed the strongest independent effect (conditional LR test p-value=0.00227). Moreover, for rs2227956 (HSPA1L) we found that the other SNPs jointly contributed to the association with APS3v beyond the effect of rs2227956 alone (marginal omnibus association LR test p-value = 1.72×10−42, Supplemental Table C1).
Chromosome 1p13 locus
Logistic regression analysis was performed on the 4 most associated SNPs within 4 genes in this locus, MAGI3, PHTF1, PTPN22, BCL2L15, in order to determine whether any of these genes had independent effects. The analysis showed that only PTPN22 had independent effect on the susceptibility to APS3v (conditional LR test p-value=0.008), suggesting that it is the causative gene and not the other 3 genes in this locus (Supplemental Table C2).
Chromosome 4q27 locus
3 SNPs in this locus showed significant association with APS3v by the logistic regression analysis. Analyses for allelic variation, as well as genotypic variation under dominant and recessive models all showed significant associations (Supplemental Table C3).
3.5 Pathway analysis
Pathway analysis was performed to provide insight into mechanisms underlying the etiology of APS3v. Several pathways were found to be significantly associated with APS3v (Supplemental Table F). The three most strongly associated pathways were cell cycle, B-cell development and CD40 signaling.
4. DISCUSSION
Organ-specific autoimmune diseases, such as T1D and AITD, share common underlying pathogenetic mechanisms. Indeed, genetic studies have demonstrated the existence of common autoimmunity genes predisposing to several autoimmune diseases [1;22]. One of the strongest phenotypic associations between two different organ-specific autoimmune diseases is the association between T1D and AITD [1], suggesting a significant shared genetic susceptibility. In the current study we dissected the joint genetic susceptibility for T1D and AITD using a genome-wide approach, analyzing patients who developed both T1D and AITD (i.e. APS3v). Our data demonstrated that the majority of SNPs showing genome-wide significant associations were located within or near the MHC locus. These findings confirm previous studies by us [13] and others [23;24] demonstrating the key role the MHC region plays in the etiology of APS3v.
The mechanism by which HLA class II proteins predispose to APS3v was shown by us to be through alteration of antigen presentation [13]. Antigen presentation is determined by the HLA class II pocket structure, which is genetically determined by its inherited sequence that has high population variability. Certain autoantigenic peptides that fit within the HLA class II pocket are more likely to be presented to T-cells that escaped tolerance thereby triggering disease [25].
We also identified SNPs in 16 additional non-HLA genes within the MHC region that were strongly associated with APS3v. In order to determine if they are associated with APS3v by virtue of LD with HLA class II or whether they confer independent effects we performed a conditional logistic regression analysis. Our logistic regression analysis suggested that at least one of these 16 non-HLA genes in the MHC region, HSPA1L, had an independent effect on the risk for disease (Supplemental Table C1). Moreover, LD analysis (Figure 3A) showed 4 LD blocks in the MHC region suggesting that other MHC genes may have independent effects that could not be detected in our dataset. Heat shock 70kDa protein 1-like (HSPA1L) is a member of the heat shock proteins (HSPs) that are upregulated in cells in response to stressful stimuli, and have been previously shown to play a role in AITD and T1D [26]. Recently, the HSPA1L protein was identified in islets of enterovirus-associated fulminant type 1 diabetes suggesting a role in T1D [27].
In addition to the 16 MHC genes demonstrating significant associations with APS3v, we identified SNPs in 5 non-MHC genes that were associated with APS3v: BCL2L15 (B-cell lymphoma 2-like protein 5), MAGI3 (membrane associated guanylate kinase WW and PDZ domain containing 3), PHTF1 (putative homeodomain transcription factor 1), PTPN22 (protein tyrosine phosphatase non-receptor type 22), and GPR103 (G protein-coupled receptor 103). Interestingly, 4 of the 5 non-MHC genes found to be associated with APS3v in our GWAS (MAGI3, PHTF1, PTPN22, BCL2L15) are located within a 350 Kb region on chromosome 1p13 suggesting that they are all in LD. LD analysis showed that only PHTF1 and PTPN22 are in tight LD (Figure 3B). This suggested that more than one gene in this locus may have an independent effect on the etiology of APS3. However, logistic regression analysis demonstrated that only the PTPN22 gene had an independent effect on disease risk confirming earlier studies showing that PTPN22 is a major autoimmunity gene [1;28]. These results confirmed our previous studies, using linkage analysis that showed strong linkage of PTPN22 with APS3v [11;12]. The PTPN22 gene encodes the lymphoid tyrosine phosphatase (LYP) protein, a potent inhibitor of T-cell activation [29]. A non-synonymous SNP causing a tryptophan to arginine change at position 620 has been associated with several autoimmune diseases [1;28], but the substitution results in gain-of-function and increases suppression of T-cell receptor signaling. It is hypothesized that this substitution may allow escape of self-reactive T-cells from deletion by central tolerance [30].
The GPR103 gene is especially interesting since it is a novel gene-locus identified to be unique for the APS3v phenotype. The GPR103 gene (also designated as QRFPR gene) is a G-protein-coupled receptor that is expressed mostly in brain, but also in human pancreatic islets and it may have an effect on insulin secretion [31]. Moreover, the ligand for GPR103 (P518) is expressed in thyroid cells [32]. Taken together these expression data suggest a role for GPR103 and/or its signaling pathway in APS3v. The GPR103 gene is located on chromosome 4q27 and it has not been previously reported to be associated with autoimmunity. However, a locus on chromosome 4q27 that is approximately 625 Kb downstream the GPR103 gene was previously reported to be associated with T1D [33], as well as with other autoimmune diseases including rheumatoid arthritis [34], Celiac disease [35], and ulcerative colitis [36]. Currently it is unclear whether this is the same locus as the one associated with APS3v, and further studies are needed to explore this possibility.
No previous studies tested the GPR103 locus on 4q27 for association with AITD, and therefore we tested the two GPR103 SNPs showing association with APS3v, for association with AITD in our AITD dataset (251 GD patients, 114 HT patients and 365 matched controls). The analysis showed that SNPs rs1513695 and rs7679475 were both associated with HT (p=0.04 and p=0.02, respectively) but not with GD. These data are consistent with the fact that most APS3v patients have HT and only a minority has GD [13].
Previous studies by us [12] and others [37] showed strong linkage and association of CTLA-4 with APS3v, while in the current study CTLA-4 markers did not reach genome wide significance. However, our pathway analysis did show significant association of the CTLA-4 signaling pathway with APS3v confirming earlier results. These data show the power of pathway analysis to identify important mechanisms underlying common diseases. Other pathways that were significantly associated with APS3v included cell cycle, B-cell development, CD40 signaling, death receptor signaling, IL12 and IL-2 signaling pathways. The association with CD40 signaling pathway is consistent with previous data showing that CD40 plays a role in the etiology of both AITD [38] and T1D [39]. The association with the IL-2 and IL-12 signaling pathways suggests that abnormalities in T-cell activation are also important to the etiology of APS3v.
The main limitation of our study is the small cohort size that may have limited our ability to identify common variants with weak effects. Even though our study had a relatively small sample size its main strength was the phenotype selection. All the patients in the discovery set developed both type 1 diabetes and TPO antibodies before age 20. The development of two autoimmune diseases in childhood supports a strong genetic contribution to the T1D+AITD phenotype of our patients (since children have less time for environmental exposures), and therefore enabled us to identify the strongest genetic effects in our cohort.
Several GWAS studies have been performed in T1D (reviewed in [40]) identifying over 30 genes-loci. By far the strongest association is with the MHC locus. Similarly, in our study the HLA class II genes showed the strongest association (Supplemental Table D1), and 16 out of the 21 genes we identified were located within the MHC region. Therefore, for both phenotypes, T1D alone and T1D+AITD (APS3v), the MHC region is the most important susceptibility locus. Among non-MHC genes the PTPN22 overlapped between T1D and APS3v, suggesting that some T1D susceptibility genes have a major contribution to the APS3v phenotype, as well. Several genes are specific to the APS3v phenotype. These include the CTLA-4 gene that has been previously shown to be unique for APS3v [11;37], and the GPR103 gene identified in the current study. These data support the notion that APS3v is a unique phenotype with specific genetic susceptibility.
In summary, this is the first GWAS study performed in patients that developed both T1D+AITD in childhood. Our data support a major role for MHC genes with contributions from non-MHC genes. Identifying the mechanisms by which these genes predispose to disease will hopefully facilitate the development of novel mechanism-based therapeutic strategies.
Supplementary Material
Highlights.
Autoimmune thyroiditis (AITD) and Type 1 diabetes (T1D) are genetically associated.
We mapped multiple loci predisposing to T1D+AITD using a genome wide approach.
Sixteen genes were non-HLA genes located in the MHC locus.
We identified GPR103 as a novel susceptibility gene for T1D+AITD.
Major pathways predisposing to T1D+AITD included CD40 and CTLA-4 signaling.
Acknowledgements
We thank the Human Biological Data Interchange (Philadelphia, PA) for assisting with the recruitment of the replication set. This work was supported in part by grants DK61659, DK067555 & DK073681 from NIH-NIDDK (to YT). This study was also supported in part by the Charles Bronfman Institute for Personalized Medicine at the Icahn School of Medicine at Mount Sinai with funding provided by the Andrea and Charles Bronfman Philanthropies.
The SEARCH for Diabetes in Youth Study is indebted to the many youth and their families, and their health care providers, whose participation made this study possible.
Grant Support: SEARCH for Diabetes in Youth is funded by the Centers for Disease Control and Prevention (PA numbers 00097, DP-05-069, and DP-10-001) and supported by the National Institute of Diabetes and Digestive and Kidney Diseases.
Site Contract Numbers: Kaiser Permanente Southern California (U48/CCU919219, U01 DP000246, and U18DP002714), University of Colorado Denver (U48/CCU819241-3, U01 DP000247, and U18DP000247-06A1), Kuakini Medical Center (U58CCU919256 and U01 DP000245), Children’s Hospital Medical Center (Cincinnati) (U48/CCU519239, U01 DP000248, and 1U18DP002709), University of North Carolina at Chapel Hill (U48/CCU419249, U01 DP000254, and U18DP002708), University of Washington School of Medicine (U58/CCU019235-4, U01 DP000244, and U18DP002710-01), Wake Forest University School of Medicine (U48/CCU919219, U01 DP000250, and 200-2010-35171).
The authors wish to acknowledge the involvement of General Clinical Research Centers (GCRC) at the South Carolina Clinical & Translational Research (SCTR) Institute, at the Medical University of South Carolina (NIH/NCRR Grant number UL1RR029882); Seattle Children’s Hospital (NIH CTSA Grant UL1 TR00423 of the University of Washington); University of Colorado Pediatric Clinical and Translational Research Center (CTRC) (Grant Number UL1 TR000154) and the Barbara Davis Center at the University of Colorado at Denver (DERC NIH P30 DK57516); and the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 8 UL1 TR000077; and the Children with Medical Handicaps program managed by the Ohio Department of Health.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention and the National Institute of Diabetes and Digestive and Kidney Diseases. The authors acknowledge the contributions of Desmond E. Williams, MD, PhD, Henry S. Kahn, MD, Bernice Moore, MBA, Edward W. Gregg, PhD from the Centers for Disease Control and Prevention.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Huber A, Menconi F, Corathers S, Jacobson EM, Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev. 2008;29:697–725. doi: 10.1210/er.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKenna MJ, Herskowitz R, Wolfsdorf JI. Screening for thyroid disease in children with IDDM. Diabetes Care. 1990;13:801–803. doi: 10.2337/diacare.13.7.801. [DOI] [PubMed] [Google Scholar]
- 3.Kordonouri O, Klinghammer A, Lang EB, Gruters-Kieslich A, Grabert M, Holl RW. Thyroid autoimmunity in children and adolescents with type 1 diabetes: a multicenter survey. Diabetes Care. 2002;25:1346–1350. doi: 10.2337/diacare.25.8.1346. [DOI] [PubMed] [Google Scholar]
- 4.Bright GM, Blizzard RM, Kaiser DL, Clarke WL. Organ-specific autoantibodies in children with common endocrine diseases. J Pediatr. 1982;100:8–14. doi: 10.1016/s0022-3476(82)80227-8. [DOI] [PubMed] [Google Scholar]
- 5.Eisenbarth GS, Gottlieb PA. Autoimmune polyendocrine syndromes. N Engl J Med. 2004;350:2068–2079. doi: 10.1056/NEJMra030158. [DOI] [PubMed] [Google Scholar]
- 6.McCanlies E, O'Leary LA, Foley TP, Kramer MK, Burke JP, Libman A, Swan JS, Steenkiste AR, McCarthy BJ, Trucco M, Dorman JS. Hashimoto's thyroiditis and insulin-dependent diabetes mellitus: differences among individuals with and without abnormal thyroid function. J Clin Endocrinol Metab. 1998;83:1548–1551. doi: 10.1210/jcem.83.5.4769. [DOI] [PubMed] [Google Scholar]
- 7.Dorman J, Kramer MK, O'Lear LA, Burke JP, McCanlies E, McCarthy BJ, Trucco M, Swan JS, Steenkiste A, Koehler AN, Foley TP. Molecular epidemiology of autoimmune thyroid disease. Gac Med Mex. 1997;133(Supp 1):97–103. [PubMed] [Google Scholar]
- 8.Tait KF, Marshall T, Berman J, Carr-Smith J, Rowe B, Todd JA, Bain SC, Barnett AH, Gough SC. Clustering of autoimmune disease in parents of siblings from the Type 1 diabetes Warren repository. Diabet Med. 2004;21:358–362. doi: 10.1111/j.1464-5491.2004.01162.x. [DOI] [PubMed] [Google Scholar]
- 9.Anaya JM, Castiblanco J, Tobon GJ, Garcia J, Abad V, Cuervo H, Velasquez A, Angel ID, Vega P, Arango A. Familial clustering of autoimmune diseases in patients with type 1 diabetes mellitus. J Autoimmun. 2006;26:208–214. doi: 10.1016/j.jaut.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Levin L, Ban Y, Concepcion E, Davies TF, Greenberg DA, Tomer Y. Analysis of HLA genes in families with autoimmune diabetes and thyroiditis. Hum Immunol. 2004;65:640–647. doi: 10.1016/j.humimm.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 11.Golden B, Levin L, Ban Y, Concepcion E, Greenberg DA, Tomer Y. Genetic analysis of families with autoimmune diabetes and thyroiditis: evidence for common and unique genes. J Clin Endocrinol Metab. 2005;90:4904–4911. doi: 10.1210/jc.2004-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villano MJ, Huber AK, Greenberg DA, Golden BK, Concepcion E, Tomer Y. Autoimmune thyroiditis and diabetes: dissecting the joint genetic susceptibility in a large cohort of multiplex families. J Clin Endocrinol Metab. 2009;94:1458–1466. doi: 10.1210/jc.2008-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menconi F, Osman R, Monti MC, Greenberg DA, Concepcion ES, Tomer Y. Shared molecular amino acid signature in the HLA-DR peptide binding pocket predisposes to both autoimmune diabetes and thyroiditis. Proc Natl Acad Sci U S A. 2010;107:16899–16903. doi: 10.1073/pnas.1009511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmar M, Kahaly GJ. Genetics of the autoimmune polyglandular syndrome type 3 variant. Thyroid. 2010;20:737–743. doi: 10.1089/thy.2010.1639. [DOI] [PubMed] [Google Scholar]
- 15.Hamman RF, Bell RA, Dabelea D, D'Agostino RB, Dolan L, Imperatore G, Lawrence JM, Linder B, Marcovina S, Mayer-Davis EJ, Pihoker C, Rodriguez BL, Saydah S SEARCH for Diabetes in Yough Study Group. The SEARCH for diabetes in the youth study: Rationale, findings, and future directions. Diabetes Care. 2014;37:3336–3344. doi: 10.2337/dc14-0574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 17.Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, Ballman KV, Berger M, Buckner JC, Chang S, Giannini C, Halder C, Kollmeyer TM, Kosel ML, Lachance DH, McCoy L, O'Neill BP, Patoka J, Pico AR, Prados M, Quesenberry C, Rice T, Rynearson AL, Smirnov I, Tihan T, Wiemels J, Yang P, Wiencke JK. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mells GF, Floyd JA, Morley KI, Cordell HJ, Franklin CS, Shin SY, Heneghan MA, Neuberger JM, Donaldson PT, Day DB, Ducker SJ, Muriithi AW, Wheater EF, Hammond CJ, Dawwas MF, Jones DE, Peltonen L, Alexander GJ, Sandford RN, Anderson CA. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Pettitt DJ, Talton J, Dabelea D, Divers J, Imperatore G, Lawrence JM, Liese AD, Linder B, Mayer-Davis EJ, Pihoker C, Saydah SH, Standiford DA, Hamman RF. Prevalence of diabetes in U.S youth in 2009: the SEARCH for diabetes in youth study. Diabetes Care. 2014;37:402–408. doi: 10.2337/dc13-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becker KG, Simon RM, Bailey-Wilson JE, Freidlin B, Biddison WE, McFarland HF, Trent JM. Clustering of non-major histocompatibility complex susceptibility candidate loci in human autoimmune diseases. Proc Natl Acad Sci U S A. 1998;95:9979–9984. doi: 10.1073/pnas.95.17.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santamaria P, Barbosa JJ, Lindstrom AL, Lemke TA, Goetz FC, Rich SS. HLA-DQB1-associated susceptibility that distinguishes Hashimoto's thyroiditis from Graves' disease in type I diabetic patients. J Clin Endocrinol Metab. 1994;78:878–883. doi: 10.1210/jcem.78.4.8157715. [DOI] [PubMed] [Google Scholar]
- 24.Wallaschofski H, Meyer A, Tuschy U, Lohmann T. HLA-DQA1*0301-associated susceptibility for autoimmune polyglandular syndrome type II and III. Horm Metab Res. 2003;35:120–124. doi: 10.1055/s-2003-39059. [DOI] [PubMed] [Google Scholar]
- 25.Faas S, Trucco M. The genes influencing the susceptibility to IDDM in humans. J Endocrinol Invest. 1994;17:477–495. doi: 10.1007/BF03347743. [DOI] [PubMed] [Google Scholar]
- 26.Birk OS, Douek DC, Elias D, Takacs K, Dewchand H, Gur SL, Walker MD, van der Zee R, Cohen IR, Altmann DM. A role of Hsp60 in autoimmune diabetes: analysis in a transgenic model. Proc Natl Acad Sci U S A. 1996;93:1032–1037. doi: 10.1073/pnas.93.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishida Y, Aida K, Kihara M, Kobayashi T. Antibody-validated proteins in inflamed islets of fulminant type 1 diabetes profiled by laser-capture microdissection followed by mass spectrometry. PLoS One. 2014;9:e107664. doi: 10.1371/journal.pone.0107664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, Kern M, Moser KL, Begovich AB, Carlton VE, Li W, Lee AT, Ortmann W, Behrens TW, Gregersen PK. Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet. 2005;76:561–571. doi: 10.1086/429096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T. Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol. 1999;29:3845–3854. doi: 10.1002/(SICI)1521-4141(199912)29:12<3845::AID-IMMU3845>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 30.Vang T, Miletic AV, Bottini N, Mustelin T. Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity. 2007;40:453–461. doi: 10.1080/08916930701464897. [DOI] [PubMed] [Google Scholar]
- 31.Granata R, Settanni F, Trovato L, Gallo D, Gesmundo I, Nano R, Gallo MP, Bergandi L, Volante M, Alloatti G, Piemonti L, Leprince J, Papotti M, Vaudry H, Ong H, Ghigo E. RFamide peptides 43RFa and 26RFa both promote survival of pancreatic beta-cells and human pancreatic islets but exert opposite effects on insulin secretion. Diabetes. 2014;63:2380–2393. doi: 10.2337/db13-1522. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Luo L, Gustafson EL, Yadav D, Laverty M, Murgolo N, Vassileva G, Zeng M, Laz TM, Behan J, Qiu P, Wang L, Wang S, Bayne M, Greene J, Monsma F, Jr, Zhang FL. Identification and characterization of a novel RF-amide peptide ligand for orphan G-protein-coupled receptor SP9155. J Biol Chem. 2003;278:27652–27657. doi: 10.1074/jbc.M302945200. [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, Plagnol V, Pociot F, Schuilenburg H, Smyth DJ, Stevens H, Todd JA, Walker NM, Rich SS. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-Van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP, Wijmenga C, Karlson EW, Toes RE, de VN, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK, Feighery C, Jewell D, Kelleher D, Kumar P, Travis S, Walters JR, Sanders DS, Howdle P, Swift J, Playford RJ, McLaren WM, Mearin ML, Mulder CJ, McManus R, McGinnis R, Cardon LR, Deloukas P, Wijmenga C. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827–829. doi: 10.1038/ng2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, De VM, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howson JM, Dunger DB, Nutland S, Stevens H, Wicker LS, Todd JA. A type 1 diabetes subgroup with a female bias is characterised by failure in tolerance to thyroid peroxidase at an early age and a strong association with the cytotoxic T-lymphocyte-associated antigen-4 gene. Diabetologia. 2007;50:741–746. doi: 10.1007/s00125-007-0603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber AK, Finkelman FD, Li CW, Concepcion E, Smith E, Jacobson E, Latif R, Keddache M, Zhang W, Tomer Y. Genetically Driven Target Tissue Overexpression of CD40: A Novel Mechanism in Autoimmune Disease. J Immunol. 2012;189:3043–3053. doi: 10.4049/jimmunol.1200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Homann D, Jahreis A, Wolfe T, Hughes A, Coon B, van Stipdonk MJ, Prilliman KR, Schoenberger SP, von Herrath MG. CD40L blockade prevents autoimmune diabetes by induction of bitypic NK/DC regulatory cells. Immunity. 2002;16:403–415. doi: 10.1016/s1074-7613(02)00290-x. [DOI] [PubMed] [Google Scholar]
- 40.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.