Abstract
High-throughput experimental techniques for generating genomes, transcriptomes, proteomes, metabolomes, and interactomes have provided unprecedented opportunities to interrogate biological systems and human diseases on a global level. Systems biology integrates the mass of heterogeneous high-throughput data and predictive computational modeling to understand biological functions as system-level properties. Most human diseases are biological states caused by multiple components of perturbed pathways and regulatory networks rather than individual failing components. Systems biology not only facilitates basic biological research, but also provides new avenues through which to understand human diseases, identify diagnostic biomarkers, and develop disease treatments. At the same time, systems biology seeks to assist in drug discovery, drug optimization, drug combinations, and drug repositioning by investigating the molecular mechanisms of action of drugs at a system’s level. Indeed, systems biology is evolving to systems medicine as a new discipline that aims to offer new approaches for addressing the diagnosis and treatment of major human diseases uniquely, effectively, and with personalized precision.
INTRODUCTION
Reductionism is a ‘divide and conquer’ approach and assumes that complex problems in cellular systems are solvable by reducing biological processes into more basic units 1. This research strategy has dominated biomedical research for many years and made great progress in identifying many critical components accounting for specific cellular phenotypes and human diseases. Owing to the complexity of biological systems, however, many important questions cannot be answered using reductionist approaches alone that typically focus on individual molecular components 2. In the past two decades, a variety of high-throughput technologies have been developed, such as cDNA microarrays, next-generation sequencing, precision mass spectrometry, and yeast two-hybrid assays 3–6. These technologies are capable of measuring the abundance of numerous components of biological systems simultaneously and have generated massive amounts of ‘omics data. With the accumulation of such experimental data, systems biology has emerged as a new approach to biology that bridges quantitative sciences and experimental biology in order to derive biological functions as system-level properties. Researchers increasingly realize that the functions of biological systems are not fully accounted for by independent individual components, but, rather, by the complex interactions between molecular components and their environment (the exposome) 7, 8. Reductionist approaches are, therefore, insufficient for fully addressing biological phenomena in this way. High-throughput technologies drive systems biology to become a valuable approach for investigating multi-dimensional molecular biology in complex human diseases 9.
One of the major challenges in the era of ‘omics is how to mine biological knowledge and generate novel mechanistic insights from the sea of high-throughput data. Obviously, handling such biological data requires multidisciplinary expertise from different fields, often including mathematics, physics, engineering, and computer science. Systems biology coupled with such multidisciplinary collaboration has developed many tools and approaches that have the potential to make a significant impact in biomedical science. These approaches apply a wide spectrum of mathematical formalisms across different scales, from data-driven methods to model-based methods, from static qualitative models to dynamic quantitative models, and from statistical analysis to network modeling 9. The choice among different approaches depends on the question to be addressed by the modeling, the availability of experimental data, and the intricacy of the system under consideration. Such computational modeling approaches play a vital role in systems biology and enable efficient in silico predictions that have the potential to enhance the design of mechanistic experiments.
Human diseases result from the complex interplay between perturbed molecular pathways and environmental factors rather than individual failing components 10, 11. Systems-based approaches are particularly valuable in complex diseases that have multifaceted causative factors, such as cancers, diabetes mellitus, and cardiovascular diseases. The rapid accumulation of high-throughput data and sophisticated computational modeling methodology in systems biology offer new opportunities to understand human diseases, identify diagnostic biomarkers, and develop disease treatments. For example, traditional disease biomarkers involve individual proteins or metabolites, without emphasizing the importance of changes to the system induced by interactions between gene or gene products that may occur in different states. Differential network analysis between diseased and normal conditions allows for the identification of network biomarkers and disease modules that account for the sensors or drivers of a disease 11, 12. In addition, pharmacology has also begun to apply systems biology principles to consider the effect(s) of a drug as the result of network interaction perturbations rather than one specific drug-protein interaction (e.g., termed the “silver bullet theory” of conventional pharmacology) 13–15. By investigating the molecular mechanisms of action of drugs at a system’s level, systems biology seeks to assist conventional pharmacology in a variety of drug development processes, including drug discovery, drug combination, and drug repurposing.
One of the valuable resources that has been overlooked by systems biologists in investigating human diseases is physiological and clinical data (the phenome) 16. Systems medicine is not simply the application of systems biology in medicine; rather, it is the logical next step and necessary extension of systems biology with more emphasis on clinically relevant applications 17. Building on the success of systems biology, systems medicine is defined as an emerging discipline that integrates comprehensively computational modeling, ‘omics data, clinical data, and environmental factors to model and predict disease expression (the pathophenome) 17, 18. In this review, we will introduce high-throughput technologies that drive the emergence and development of systems biology and computational modeling methods that are being developed for systems biology. We will also consider how complex human diseases and pharmacology benefit from systems-based approaches. Finally, we will discuss the evolution of systems biology and the early phases of systems medicine in the context of aiding physicians in addressing human disease complexity and, ultimately, improving clinical practice for patients.
HIGH-THROUGHPUT TECHNOLOGIES DRIVING SYSTEMS BIOLOGY
A complex system is composed of a large number of interconnected components whose interplay accounts for a variety of system functions. A key factor that promoted the emergence of systems biology is the development of various high-throughput technologies. These biotechnologies not only allow quantification of individual components (e.g., genes, proteins, microRNAs, and metabolites) of a biological system, but also afford the generation of massive interactomes describing the complex interactions of these components, and even decipher the function of the system.
DNA sequencing techniques determine the complete DNA sequence of an organism’s genome and the entire set of genes at a single time. Moreover, next-generation sequencing (NGS) now can generate DNA sequences of many organisms at a very low cost 4. Gene-expression microarray allows global quantification of mRNA transcripts of thousands of genes 3. Furthermore, NGS-based RNA sequencing (RNA-seq) can not only be used to measure gene expression levels at a higher resolution and sample throughput, but also can reveal alternative gene spliced transcripts 6. For example, Gene Expression Omnibus (GEO) and other database repositories store massive microarray- and sequence-based gene expression datasets that can be reused as a basis for new biological studies 19. Similarly, mass spectrometry (MS) and isobaric tags for relative and absolute quantification (iTRAQ) can be used to determine the concentration of thousands of proteins in a single experiment 20, 21. The Human Protein Atlas (www.proteinatlas.org) contains immunohistochemistry-based maps of protein expression and localization profiles for a large majority of all human protein-coding genes based on both RNA and protein data in normal tissue, cancer, subcellular organelles, and cell lines 22, 23. Such datasets offer the possibility to explore tissue-specific proteomes and analyze tissue profiles for specific protein classes. Global metabolomic profiling by nuclear magnetic resonance (NMR) and liquid chromatography (LC) or gas chromatography (GC) coupled with MS is used to measure the composition and concentration of both targeted and untargeted metabolites 24. In particular, mass cytometry facilitates high-dimensional quantitative analysis of the effects of molecules at single-cell resolution 25. Such single-cell genomic analyses greatly enhance diagnostic and experimental analyses. Experimental data generated by these biotechnologies represent the levels or abundance of individual biological elements and have been deposited in major databases, as listed in Table 1.
Table 1.
A list of high-throughput technologies and the data they generated, with representative databases
| Biotechnologies | Experimental data | Representative databases |
|---|---|---|
| DNA-seq, NGS | DNA sequences, exome sequences, genomes, genes | GenBank109, DDBJ110, Ensembl111 |
| Microarray, RNA-seq | Gene expression levels, microRNA levels, transcripts | GEO112, Expression Atlas113 |
| MS, iTRAQ | Protein concentration, phosphorylations | GPMdb, PRIDE, Human Protein Atlas22 |
| C-MS, GC-MS, NMR | Metabolite levels | HMDB, GMD |
| ChIP-chip, ChIP-seq | Protein-DNA interactions, transcript factor binding sites | GEO112, TRANSFAC, JASPAR, ENCODE, modENCODE |
| CLIP-seq, PAR-CLIP, iCLIP | MicroRNA-mRNA regulations | StarBase114, miRTarBase |
| Y2H, AP/MS, MaMTH, maPPIT | Protein-protein interactions | HPRD115, BioGRID116, DIP, IntAct, and MINT, CCSB interactome database |
| Protein microarray | Kinase–substrate interactions | RegPhos, PhosphoPOINT |
| SGA, E-MAP, RNAi | Genetic interactions | HPRD115, BioGRID116 |
| SNP genotyping array | GWAS loci, eQTL, aberrant SNPs | GWAS Catalog, GWASdb, GTEx, dbGAP, dbSNP HGMD |
| LUMIER, data integration | Signaling pathways, metabolic pathways, molecular signatures | KEGG, ConsensusPathDB, BioCart, Pathway Commons, MSigDB, Reactome, BiGG |
There are also biotechnologies that can detect the interactions between biological elements in a high-throughput manner. For example, chromatin immunoprecipitation assay (ChIP) is a technology that utilizes DNA microarray technology for investigating interactions between proteins and DNA 26, while ChIP-seq technology profiles genome-wide protein-DNA interactions by utilizing next-generation parallel DNA sequencing 27. ENCODE (Encyclopedia of DNA Elements, www.encodeproject.org/) is an international collaborative project whose goal is to identify all functional elements in the human genome sequence. All the data generated by ENCODE, including ChIP-seq, RNA-seq, and Dnase-seq datasets in differential tissues, can be freely downloadable for research purposes. Experimental strategies for discovering transcriptome-wide microRNA-mRNA regulatory interactions include Crosslinking and Immunoprecipitation followed by high-throughput sequencing (CLIP-seq), Photoactivatable-Ribonucleoside-Enhanced CLIP (PAR-CLIP), and individual-CLIP (iCLIP) 28. Protein-protein interactions (PPIs) can be mapped by improving variations of yeast two-hybrid screening (Y2H) for direct binary interactions or by affinity- or immuno-purification to isolate protein complexes, followed by mass spectrometry (AP/MS) to identify indirect associations between proteins 5, 29. Rolland and colleague recently performed a systematic map of 13,944 high-quality human binary protein-protein interactions among 4,303 distinct proteins, providing unprecedented opportunities for understanding human diseases through the interactome 5, 30. In addition, mammalian-membrane two-hybrid assay (MaMTH) is a technique developed recently for detection of integral membrane PPIs 31. Some functional protein microarrays and mass spectroscopy-based assays can also be used to identify the phosphorylation targets of individual protein kinases (i.e., kinase–substrate interactions) 32. Recently, Saliba and colleagues developed a liposome microarray–based assay (LiMA) that measures protein recruitment to membranes in a quantitative and high-throughput manner, generating a large number of protein-lipid interactions 33. An automated high-throughput technology, LUMIER (luminescence-based mammalian interactome mapping), was also developed for mapping dynamic signaling networks in mammalian cells 34. The interactome data generated by these technologies have been stored in some databases, as well (Table 1).
The ultimate goal of molecular biology is to interpret how genotypes account for different phenotypes and diseases. High-throughput genotyping and phenotyping approaches have made great steps towards determining genotype determinants and their interactions in model organisms. For example, yeast genetic interaction studies are well established by Synthetic Genetic Array analysis (SGA) and Epistatic Miniarray Profiling (E-MAP) 35. Systematic analysis of genetic interactions in human cells is still in early stages of developmental application; however, Laufer and colleagues provided a detailed protocol for large-scale mapping of genetic interactions in human cells by combining RNA interference (RNAi) and automated imaging 36. In addition, advances in DNA sequencing technologies allow us to monitor the effects of common genetic variations in sequences at population levels. For example, single nucleotide polymorphism (SNP) genotyping arrays can measure genetic variations of SNPs among a population. Genome-wide association studies (GWAS) focus on examining statistical associations between common SNPs and complex phenotypic traits in a population, and have identified a large number of genetic loci that may be causally associated with major human diseases 37. SNP genotyping arrays also produce massive amounts of expression Quantitative Trait Loci (eQTL), which expose potential mechanisms by which to explain observed associations. Such high-throughput genetic data further enable the investigation of complex relationships between genotypes and phenotypes (Table 1). In doing so, the accuracy of information is very important as a system or model works only if the input data are sound. There are some available software tools that assist in using the correct phenotype information. For example, Genome-Phenome Analyzer, launched by SimulConsult (www.simulconsult.com/), links curated phenome databases, clinical findings, and associated variants generated by whole-exome sequencing to compute a differential diagnosis for patients. PhenoDB (http://phenodb.net) is a Web-based portal for integration and analysis of phenotypic features, whole exome/genome sequence data, knowledge of pedigree structure, and previous clinical testing. It can also be used to format phenotypic data for submission to dbGaP.
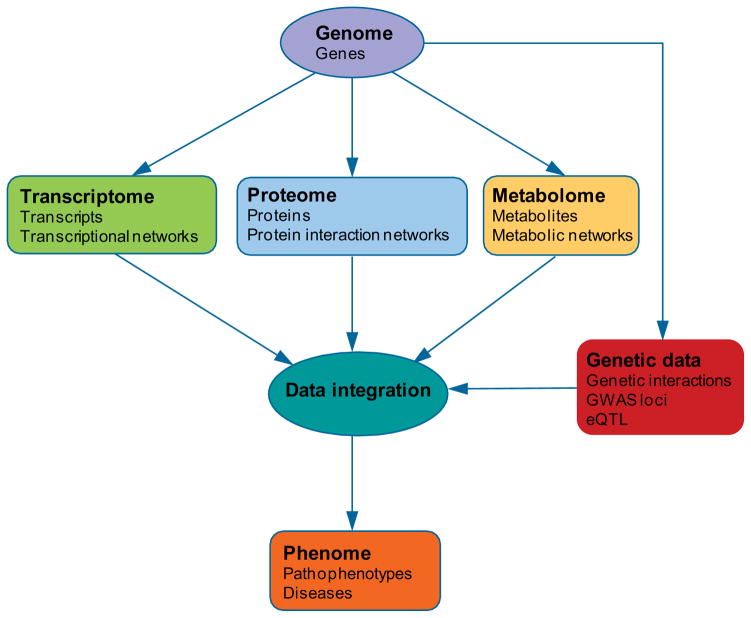
Biological systems are highly dynamic and hierarchical. Each technology can only generate the data at one dimension of complex biological systems. However, any single type of high-throughput data cannot fully interpret a variety of system functions. Therefore, how to integrate heterogeneous and large ‘omics data and mine useful knowledge to interpret phenotypes is critical for the success of systems biology (Figure 1). Thus, at the very least, systems biology must also borrow quantitative modeling approaches from multidisciplinary fields, which we will discuss in next section.
Figure 1. High-throughput data and their hierarchical relationships in describing cellular phenotypes or human diseases.
Each type of biological data represents a certain dimension of complex biological systems. Interpretation of cellular phenotypes or human diseases using systems biology approaches requires integration of all heterogeneous high-throughput data types.
COMPUTATIONAL METHODS IN SYSTEMS BIOLOGY
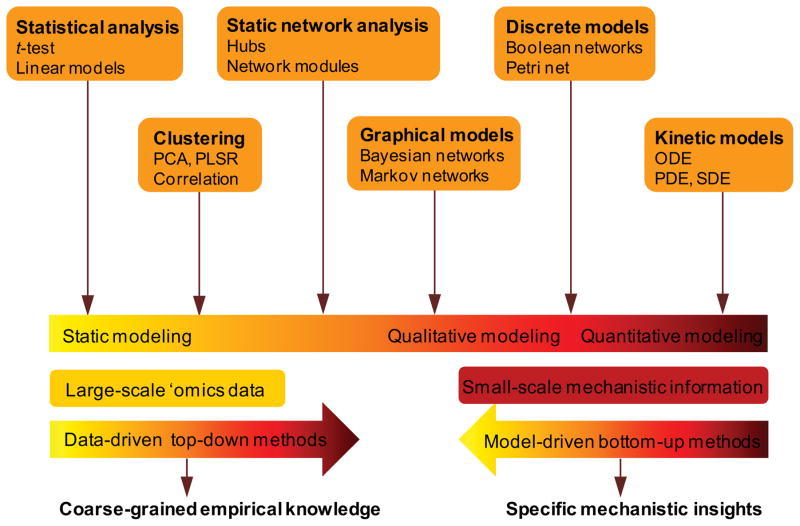
High-throughput technologies highlight the challenge of how to mine biological knowledge and generate testable hypotheses from the massive amount of available data. Tackling this challenge requires sophisticated quantitative modeling methods and multidisciplinary expertise from different fields, such as mathematics, physics, and computer science. One of the hallmarks of systems biology is the use of computational approaches from quantitative science to develop a wide spectrum of models and tools for analyzing large-scale data (Figure 2).
Figure 2. An overview of computational modeling methods used in systems biology.
Computational approaches in systems biology apply a wide spectrum of mathematical formalisms across different scales, ranging from data-driven top-down methods to model-driven bottom-up methods, and from static qualitative models to dynamic quantitative models.
Computational methods that have been used in systems biology can be classified into data-driven top-down methods and model-driven bottom-up methods 9. In general, high-throughput multi-parametric ‘omics data characterize the abundance of biological elements across different system states. Data-driven top-down approaches integrate and analyze experimental data to reveal biomarkers and biologically meaningful patterns. These approaches can be applied to the analysis of unbiased genome-scale data with thousands of components to obtain coarse-grained knowledge about biological systems. For example, various statistical analyses have been used for identifying differentially expressed genes, proteins, or metabolites 38. One can further examine whether the resulting component lists are enriched for known gene signatures or signaling pathways 39. Statistical methods, such as Principle Component Analysis (PCA), Partial Linear-square Regression (PLSR), and Canonical Ccorrelation Analysis (CCA), are then utilized to identify functional relationships by checking the expression correlations between components or clustering the expression profiles of individual elements 40, 41. For example, Dewey and colleagues assembled all myocardial transcript data from the Gene Expression Omnibus (GEO) database and used gene coexpression network analysis to derive functional modules and regulatory mediators in developing and failing myocardium that were not present in normal adult tissue 42.
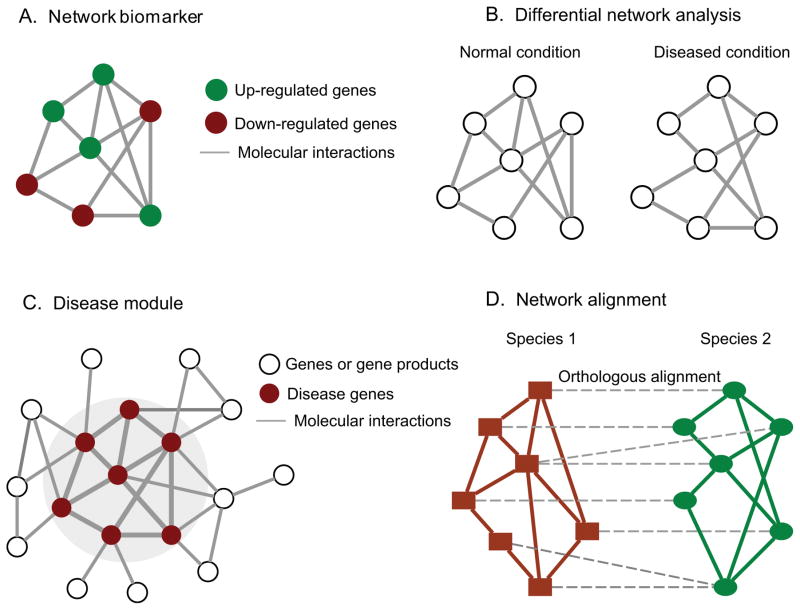
Biological elements do not function in isolation; rather molecules and their dynamic interactions determine the function of a complex biological system. These interactions form different types of cellular (and subcellular) networks with characteristic topology, such as gene regulatory networks, microRNA-mRNA target networks, protein-protein interaction networks, metabolic networks, and signal transduction networks 7. Network representation of the interactome data simplifies complex systems and focuses on the elements and their interactions, enabling use of various tools from network science and graph theory to analyze the data 7. Investigators increasingly realize that the topological structure of biological networks is closely related to their functions. Therefore, local and global structural features can reveal key properties of biological systems. For example, it has been shown that the number of interactors of a protein is highly correlated with its lethality associated with any variation in its expression (e.g., adverse consequences of protein over- or under-expression) and essentiality (e.g., protein functionality), with hubs (nodes with many edges) tending to play important biological roles 43. Groups of densely connected proteins in the protein interactome (called functional modules) often correspond to protein complexes 44. Similarly, disease modules are groups of densely connected biological elements in the human interactome whose perturbation or dysfunction can be linked to a particular disease phenotype 8, 45. As an example, starting from a small set of seed genes relevant to asthma, Sharma and colleague used a network-based approach based on the comprehensive human interactome to determine the local neighborhood of the interactome whose perturbation is associated with asthma, i.e., the asthma disease module 46. Network topology can be also augmented with functional regulatory rules to predict the essentiality of biological components more accurately 47, 48. Differential network analysis, which compares the topological changes of biological networks over different conditions, may help to identify key players or disease markers 12. Network alignment across different species can identify conserved orthologous functional regions beyond individual genes or interactions 49. Figure 3 illustrates some concepts of network analysis. Excellent discussions of the application of network modeling in biology and medicine are reviewed in references 7, 8, 11, 14, 30.
Figure 3. Illustration of some concepts of network analysis.
(A) Network biomarker. Different from traditional individual biomarkers, a network biomarker is a subnetwork consisting of two or more differentially expressed components in control samples vs. disease samples. (B) Differential network analysis examines the same network over two different conditions, highlighting the topological changes induced by diseases. (C) A disease module represents a group of nodes whose perturbation can be linked to a particular disease phenotype. (D) Network alignment compares two networks from different species and aligns orthologous components and their interactions.
While the above-mentioned top-down methods are used for analysis of unbiased high-throughput data, the published biological literature is also a valuable source that must be considered for the construction of networks since the literature covers numerous small-scale experiments central to specific biological processes. Probabilistic graphical models, such as Bayesian networks and Markov networks, can incorporate prior knowledge from the literature and be used to construct (imputed) causal networks from observational biological data 50. They can also serve as gene regulatory network models to learn network structures from gene expression data 51. Chu and colleagues proposed a partial correlation network method based on a Gaussian graphical model to analyze the association between chronic obstructive pulmonary disease (COPD) and other factors, including case-control status, disease severity, and genetic variants (see below for detailed discussions) 52. Probabilistic graphical models have many successful applications in systems genetics, as well 53.
In contrast to data-driven approaches, model-driven bottom-up approaches to characterizing complex biological systems are used to simulate the dynamics of the system and, in turn, model various perturbations to the system by using relevant mathematical models. Biological networks that drive various biological processes are condition-specific and highly dynamic. Bottom-up methods model how interacting elements achieve the temporal patterns of cellular systems. This class of methods usually originates with the availability of data pertaining to biological mechanisms coupled with observational data generated from individual small-scale experiments and complementary information from high-throughput data. Continuous dynamic modeling approaches, such as those involving deterministic ordinary differential equations (ODE) and partial differential equations (PDE) or stochastic differential equations (SDE), have been widely used as bottom-up methods. These modeling approaches can be used to explain quantitative behaviors of a system; however, the construction of these models is typically hampered by a lack of temporally resolved experimental data and/or sufficient mechanistic details, including kinetic parameters, such as synthesis/degradation rates, and absolute intracellular concentrations of macromolecular or metabolic species, which, collectively, make these methods practical only in small or simple systems. By contrast, knowledge about biological networks from the experimental literature and high-throughput technologies is often of a qualitative nature, which has promoted the widespread use of discrete qualitative modeling approaches, such as Boolean network models, multi-valued logical models, and Petri nets 54, 55. Based on reasonable simplification of biological reality, discrete dynamic modeling can make qualitative dynamic predictions of system behaviors. As they do not require quantitative kinetic parameters, these approaches can be employed for relatively large and complex systems. For example, Ryall and colleagues developed a computational model of the cardiac myocyte hypertrophy signaling network with 106 species and 193 reactions, integrating 14 established pathways regulating cardiac myocyte growth 56. They used the model to determine how the individual components and their interactions lead to differential regulation of transcription factors, gene expression, and myocyte size, and validated a majority of model predictions using published experimental data. Dynamic modeling can simulate a variety of perturbations of a biological system; specifically, knocking down or over-expressing certain genetic nodes and interactions, which may attract the system to a new phenotypic state or diseased condition. In this way, dynamic modeling informs in silico predictions to generate testable hypotheses, guiding targeted experimental validation follow-up studies.
In addition to the aforementioned methods of mathematical formalism, high-throughput data sets are often heterogeneous, and, thus, integration techniques from computer science and statistical learning are required to fuse them. To address the issue of shared and integrated mass data, we also need a variety of computational platforms, such as biological ontology databases and semantic webs. Among different computational approaches in systems biology, whether static modeling, qualitative modeling, or quantitative modeling should be chosen hinges on the question to be addressed by the modeling, the availability of experimental data, and the complexity of the systems under consideration. For example, longitudinal or time-series biological data contain more dynamic information than snapshot data. The time aspect reflects the temporal activity of biological components and can be used to construct continuous dynamic models. When time-dependent quantitative experimental data are not sufficient, only qualitative models can be constructed. In addition to making qualitative predictions of system behaviors, these models can also serve as a basis for developing a corresponding quantitative continuous model once more time-series data are available 57.
APPLIED SYSTEMS BIOLOGY: INFLUENCES IN CLINICAL MEDICINE
In the context of systems biology, diseases are viewed as the results of the complex interplay between perturbed molecular pathways and environmental factors rather than individual failing components 8, 10, 11. Systems-based approaches are particularly valuable in complex diseases that have multifaceted causative factors and clinical presentations, such as cancer, diabetes mellitus, respiratory diseases, and cardiovascular diseases 2, 58. Systems biology can provide new avenues for understandimg human diseases; for example, identification of diagnostic disease biomarkers, development of disease treatments by revealing disease subtypes, and identification of novel therapeutic targets for diseases.
The penetration of systems biology to the medical science literature is escalating; for example, the number of PubMed-indexed citations relevant to this field has increased by ten-fold over the previous decade 59. While the preponderance of these contributions aims to characterize the translational relevance of novel subcellular physical interactions (i.e., protein-protein interactions, miRNA-mRNA interactions, and others as described in greater detail earlier) to patients clinically, this is not uniformly the case. A number of recent reports describe the application of systems biology strategies to the characterization of the relationships between complex diseases according to symptomatology, prevalence, and associated co-morbidities in the absence of consideration to pathobiological mechanism per se or as a method by which to validate elements of the interactome potentially relevant to the disease phenotype 16.
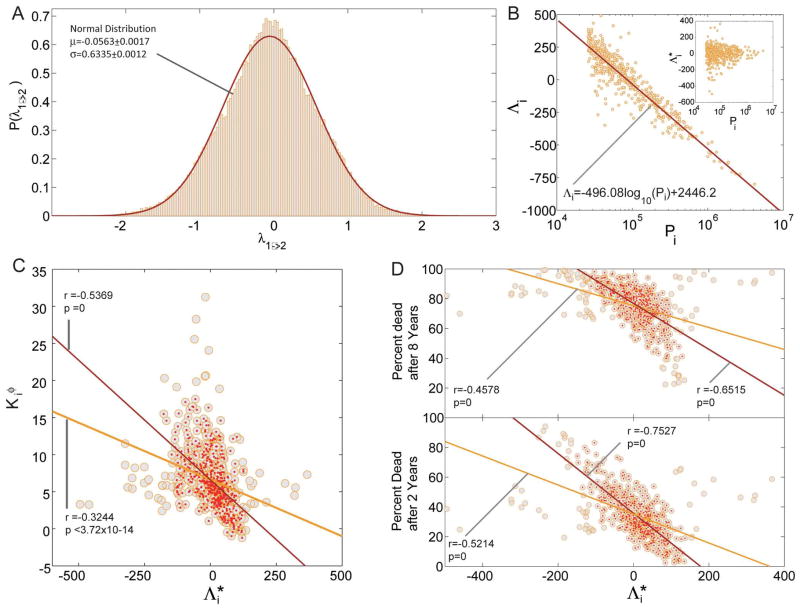
Zhou and colleagues synthesized a symptom-based network of human disease based on large-scale medical bibliographic records derived from Medical Subject Heading (MeSH) metadata and PubMed databases to demonstrate that symptom variability for a particular disease correlates with the density of protein-protein interactions linked to the pathobiology of that disease 16. For example, they observed that overlap for 78 symptoms between the inflammatory bowel disorders ulcerative colitis and Crohn’s disease was also common to a number of infectious diseases linked to the development of intestinal inflammation and colonic mucosal effacement that define these diseases pathologically. Specifically, symptom linkage paralleled a pattern of gene network connectivity between ulcerative colitis and Crohn’s disease and various intestinal viral, bacterial, and parasitic infections whose incidence is, in turn, implicated in the clinical expression of bowel inflammation in patients 60, 61. These findings are conceptually similar to evidence by others indicating that the probability among patients diagnosed with a single disease developing another specific condition is not random. Analyses of the Phenotypic Disease Network, which links disease groups within the human disease interactome according to common phenotype modules, suggest connected co-morbidities follow along the lines of proteomic connectivity and may be relevant to inform disease prognosis (Figure 4) 62. Furthermore, network analyses have exposed disease similarities based on genetic connectivity not identified by classical population genomics alone. In a proof-of-concept analysis of 1.5 million data records, Rzhetsky and colleagues reported polymorphism overlap between bipolar disorder and schizophrenia, and between bipolar disorder and autism by up to 60% and 75%, respectively 59.
Figure 4. Directionality of disease progression.
(figure from Hidalgo et al. 62)
A. Distribution of λ1→2 B. Disease precedence Λi as a function of disease prevalence Pi. The inset shows the same plot after removing the trend from disease precedence (Λi* = Λi+496.08log10(Pi)-2446.2) C. Disease connectivity calculated from the φ-PDN as a function of Λi*. The yellow line shows the best fit for the 518 diseases with a prevalence larger than 1/500 (yellow circles) while the red line shows the best fit for the 463 diseases at the center of the cloud (red points). The correlation coefficient is represented by r and its associated p-value by p. D. Percentage of patients who died 2 and 8 years after being diagnosed with a disease with a given detrended precedence Λi*. The yellow lines show the best fit for all the 518 diseases (yellow circles) while the red lines show the fit for the 434 (top panel) and 465 (bottom panel) diseases at the bulk of the cloud.
Despite the incompleteness of the human interactome, Menche and colleagues studied disease-disease relationships using network topological analysis and provided evidence that interactome network-based location of each disease module reflects its pathobiological relationship to other diseases 63. This approach has illuminated unexpected gene overlap between diseases that are, by convention, regarded as unrelated clinical entities. For example, SMARCA4 is a protein associated with myocardial infarction, which in their interactome is linked with the proteins ALK, MYC, and NFKB2 that are implicated in the pathogenesis of lymphoma. Associations such as these may account for heretofore incompletely explained epidemiological associations between diseases that are seemingly unrelated biologically, including in this instance large cell lymphoma and myocardial infarction, which share a comorbidity rate that is higher than anticipated based on current understanding of their respective pathobiologies. Furthermore, a number of diseases occupying overlapping modules within the interactome, but for which known pathobiological relationships are lacking, were also reported, including glomerulonephritis and biliary cirrhosis, glioma and myocardial infarction, hepatic cirrhosis and spondylitis, albuminuria and respiratory disease, among other pairs. Forthcoming empiric efforts are required to crystalize the mechanisms by which to account for disease interrelatedness among these phenotypes.
The extent to which these early observations may redefine the epidemiology of complex syndromes remains to be determined. Nevertheless, these contributions illuminate overlap in the biological substrate underlying convergent pathophenotypes and by so doing provide a novel framework for predicting disease incidence and potentially refining the natural history of certain syndromes. This section of the review will discuss systems biology observations that have already set such a course for selected lung diseases, cardiovascular diseases, cancer, and inflammatory disorders of the digestive tract.
Systems biology and cardiovascular medicine
Thrombosis, inflammation, cellular proliferation, and fibrosis are among the fundamental pathobiological mechanisms implicated in the genesis of vascular diseases that are also the subject of recent systems biology investigations. One general approach to investigating these mechanisms involves emphasis first on lynchpin signaling intermediaries that are known to i) regulate a particular pathobiological process, and ii) promote a rare complex human disease. For example, hereditary hemorrhagic telangiectasia (HHT) is a condition characterized by arteriovenous malformations, dysregulated fibrinolysis, and various vascular complications including arteriovenous shunts and thrombosis that is driven, in part, by dysfunctional endothelial nitric oxide synthase 64. The transforming growth factor-β (TGF-β) superfamily ligands are critically involved in vascular development by regulating endothelial cell signaling, including the co-receptors endoglin and ACVRL1. High-throughput interactome mapping recently identified 181 novel interactors between ACVRL1, the TGF-β receptor-2, and endoglin, including protein phosphatase subunit beta (PPP2RB). In turn, PPP2RB was shown to disrupt endothelial nitric oxide synthase signaling in endoglin-deficient cells in vitro, identifying a potential role for PPP2RB in the pathobiology of HHT 65.
Others have reported that secondary analyses of genome-wide association studies using a systems approach is useful for identifying key characteristics defining common, but complex, cardiovascular disease pathophenotypes. By establishing a network comprising SNPs linked to various measures of dyslipidemia (i.e., abnormal serum total cholesterol [TC], low-density lipipoprotein cholesterol [LDL-C], high-density lipoprotein cholesterol, and/or triglyceride levels) derived from the Global Lipids Genetics Consortium (P< 5×10−8), Sharma and colleagues identified rs234706 as a novel cystathionine beta synthase SNP involved in expression of the total cholesterol and LDL-C trait (i.e., measurably elevated levels of each) 66. These findings were validated through a linkage study analyzing data from an unrelated registry, the Malmö Diet and Cancer Cardiovascular Cohort; liver tissue from CBS-deficient mice in vivo; and healthy human livers biopsied at the time of surgery (in which the minor allele of rs234706 was detectable). Although CBS deficiency was established previously to play a role in lipid metabolism, the biological significance of the specific SNP was not known prior to the original GWAS and its systems analysis.
An alternative methodology by which to target human disease using network medicine methodology involves the initial construction of a large-scale interactome, which may be derived from analysis of the curated literature, biosample data, or a combination thereof according to methods described earlier. A substantial effort is underway to assemble interactomes relevant to vascular inflammation and thrombosis in order to characterize further the pathogenesis of relevant cardiovascular diseases, particularly myocardial infarction (MI). The National Institutes of Health-sponsored consortium MAPGen (www.mapgenprogram.org), for example, consists of five university centers with access to large human sample repositories and clinical data from international, multi-centered cardiovascular trials that are anticipated to generate broad and unbiased inflammasome and thrombosome networks. These large-scale individual networks and sub-networks created by overlap between them are currently being analyzed to define unrecognized protein-protein interactions pertinent to stroke, MI, and venous thromboemoblic disease. The selection of specific protein(s) or protein product(s) from this data set or other networks of similar scale for validation experimentally is likely to hinge on the strength of association, location of targets within the network, their proximity to other important protein/products, and/or data linking naturally-occurring loss- or gain-of-function mutations of the putative target to relevant clinical disorders, among other factors.
While systematic analysis of data from the MAPGen project is forthcoming, other reports from smaller cardiovascular disease datasets have emerged. For example, proteomic analysis of circulating microvesicles harvested from patients with acute ST-segment elevation myocardial infarction or stable coronary artery disease was performed by mass spectrometry 67. Using this approach, investigators were able to identify 117 proteins that varied by at least 2-fold between groups, such as α2-macroglobulin isoforms and fibrinogen. Protein discovery was then subjected to Ingenuity® pathway analysis to generate a protein-protein interaction network. Findings from this work suggest that a majority of microvesicle-derived proteins are located within inflammatory and thrombosis networks, affirming the contemporary view that myocardial infarction is a consequence of these interrelated processes.
Parenchymal lung disease
Owing to the complex interplay between numerous cell types comprising the lung-pulmonary vascular axis, a number of important pathophenotypes affecting these systems have evolved as attractive fields for systems biology investigations 68. Along these lines, chronic obstructive pulmonary disease (COPD), which comprises a heterogeneous range of parenchymal lung disorders, has been increasingly studied using network analyses to parse out differences and similarities among patients with respect to gene expression profiles and subpathophenotypes. Using the novel diVIsive Shuffling Approach (VIStA) designed to optimize identification of patient subgroups through gene expression differences, it was demonstrated that characterizing COPD subtypes according to many common clinical characteristics was inefficacious at grouping patients according to overlap in gene expression differences 69. Important exceptions to this observation were airflow obstruction and emphysema severity, which proved to be drivers of COPD patients’ gene expression clustering. Among the most noteworthy of the secondary characteristics (i.e., functional to inform the genetic signature of COPD) was walk distance, raising the possibility that unrecognized biomarkers could be identified through a research approach that predicts functional capacity in this or other similar diseases.
Along these lines, Davidsen and colleagues reported their recent findings using a systems biology approach to identify novel factors that promote skeletal muscle dysfunction in COPD 70. They analyzed differences in gene expression profiles in whole lung and hind limb skeletal muscle tissue harvested from swine exposed to chronic hypoxia, chronic cigarette smoke, or a combination of these factors to develop a genome-wide transcriptome (151,072 transcript sequences) relevant to COPD. These data were subjected to enriched KEGG pathway analyses, which, in turn, predicted that the systemic cytokines CXCL10 and CXCL9 may be important markers of dysregulated metabolic function in skeletal muscle in COPD. The investigators confirmed their hypothesis by demonstrating that circulating CXCL10/9 levels are significantly different in plasma from COPD patients as compared to healthy controls, providing evidence in support of these factors as potential biomarkers indicative of extrapulmonary end-organ damage in chronic lung disease.
The mechanistic underpinnings of lung injury responses have also been addressed using a combination of network biology strategies in order to understand better the pathways resulting in airway remodeling. In a unique experimental design, airway biopsy samples from lungs of patients exposed remotely to sulfur mustard or unexposed controls were subjected to microarray gene expression analysis and combined with genes corresponding to biological factors linked to airway remodeling identified from curated literature searches 71. From these datasets, protein-protein interaction and gene regulatory networks could be synthesized, which were ultimately merged to create functional modules. In that study, matrix remodeling proteins, particularly matrix metalloproteinase (MMP)-9, were centrally located and well connected within the network. The observation identifying CYP11B1 (11-β hydroxylase), STARD10 (steroidogenic acute regulator protein-related lipid transfer protein), and pro-fibrotic proteins (i.e., TGF-β) as putative airway remodeling proteins in that analysis was consistent with findings from whole lung transcriptome analysis involving patients with pulmonary arterial hypertension 72. Moreover, these findings are in agreement with data from our laboratory 73 demonstrating activation of steroidogenesis signaling pathways, including aldosterone biosynthesis, in pulmonary vascular cells subjected to chronic hypoxia in vitro and its link to upregulation of TGF-β- and MMP-dependent signaling pathways that promote pulmonary vascular remodeling.
Cancer biology
An overarching goal of contemporary cancer therapeutics is the design of personalized drugs to match treatment targets with patients’ particular disease pathobiology. Angiogenesis and cellular plasticity are principal processes implicated in tumor growth, and, therefore, have evolved as the subject of key investigations aiming to identify targets within the framework of this personalized medicine approach. Intrinsic disorder proteins (IDPs) contain unique amino acid sequences that inhibit energetically favorable three-dimensional structures. Relative to normal proteins, IDPs demonstrate increased plasticity and tend to participate in the dysregulation of many cellular processes that define cancer biology, including cellular proliferation and dedifferentiation. Building on this concept, Malaney and colleagues studied the protein suppressor and IDP, PTEN 74. They used PONDR-FIT software to develop a series of interactomes comprising the IDP network that included PTEN and associated interactors, including PTEN phosphorylating kinases. Other levels in the analysis accounted for mutated amino acid combinations favoring abnormal protein function by introducing hydrophobicity, aromaticity, and redox-sensitive properties properties. Forty PTEN-associated proteins emerged from the analysis, of which 25 appear to interact with the intrinsically disordered region of PTEN at the carboxy-tail. The interactome was also in agreement with a number of previous publications in the cancer literature: 13 cancer-related proteins were also identified as strong IDP candidates and, in turn, formed a small, but potentially important, “PTEN-Cancer interactome.”
One evolving area of converging research streams is that of factors influencing treatment resistance to some cancers. As one example of this property of many malignancies, genetic data were collected from 71 patients registered in the Long-HER study, which characterized clinical responsiveness to the monoclonal antibody, trastuzumab, for the treatment of metastistic breast cancer. From this dataset, a number of expression profile differences involving PTEN and PTEN-associated genes were observed between treatment responders and non-responders, including intermediates involved in activation of the proliferative and anti-apoptotic kinase mammalian target of rapamycin (mTOR) 75. A number of reports have aimed to use similar methodologies to identify generic markers that distinguish tumor benignity from malignancy. For example, elevated concordance rates were observed in one study between tissue and plasma proteins differentially expressed in benign vs. malignant serous ovarian tumors and measured by liquid chromatography-mass spectrometry. Subsequent hierarchical pathway analysis focusing on 20 proteins suggested that 14-3-3 zeta/delta, 14-3-3 beta/alpha, alpha-actinin 4, HSP60, and PCBP1 are candidate markers of tumor malignancy 76.
Unopposed angiogenesis is yet another fundamental pathological mechanism responsible for growth and propagation of various solid tumors that has also been the subject of network analyses. Indeed, characterizing the protein-protein response pattern to vascular endothelial growth factor (VEGF) treatment in vascular endothelial cells in vitro has contributed to early iterations of the “angiome” 77. Others have developed networks focusing on identifying interactors of alternative, but critical, proangiogenic proteins 78, including MMPs 79, epidermal growth factor 80, vonWillebrand factor 81, and hypoxia-inducible factor (HIF)-1 82 to expand the number of potential treatment targets for various cancer subtypes, including prostate, pancreatic, and breast adenocarcinoma.
Diseases of the gastrointestinal tract
Ulcerative colitis and Crohn’s disease are two overlapping clinical pathophenotypes characterized by inflammatory changes to the colon in the former, and the colon, small intestine, and/or other (extra)intestinal sites in the latter that together affect 1:250 individuals 83. In the setting of particular clinical clues or epidemiological factors, the diagnosis of one of these disease entities is often suspected. However, demonstrating specific pathological findings on mucosal biopsy is often required to reach a definitive diagnosis. Despite some gains in the therapeutic approach to these diseases, including monoclonal antibody therapy in the case of Crohn’s disease, the pathobiological substrate of either is poorly understood and in the absence of effective risk stratification methods or non-invasive disease trajectory modifying interventions, surgical bowel resection remains the definitive treatment in many patients.
Owing, in part, to observations indicating differences in levels of sulfur-reducing bacteria in ulcerative colitis patients, one contemporary pathophysiology paradigm for these diseases points to differences in the gut microbiome profile 84. In support of this hypothesis is a recent deep sequencing analysis of fecal flora from a large cohort of controls and treatment-naïve Crohn’s disease patients prior to the initiation of antibiotic therapy illustrating key contributors of the mucosal microbome in new-onset disease. Specifically, dysbiosis involving bacteria linked to oxidative resistance, gastrointestinal ulcer formation, and inflammatory invasion of intestinal epithelial cells to include Escherichia, Fusobacterium, Haemophilus and Veillonella among others comprised the microbial signature of untreated Crohn’s patients. Interestingly, concordance in the dysbiotic signature of rectal and illeal samples demonstrated through network methodologies in that study raises the possibility that options other than colonoscopy (i.e., invasive)-requiring biopsy exist for disease diagnosis 85.
Tuller and colleagues demonstrated significant overlap in the protein-protein interaction network derived from circulating peripheral lymphocytes harvested from patients with Crohn’s disease and ulcerative colitis 86. This observation matches genome studies identifying 163 loci common to various forms of inflammatory bowel disease 61 and clinical practice experience in which distinguishing these entities is not possible in up to 15% of cases despite multi-modality assessment. By contast, early efforts in the complex process of leveraging ‘omics-based methods for the purposes of diagnostics in these diseases appear promising. In one large-scale proteomic project that aimed to validate the clinical diagnosis of Crohn’s disease and ulcerative colitis by spectral analysis of mucosal tissue from 312 spectral peaks distinguishing these diseases using conventional statistical analyses, a (non-probabilistical) Support Vector Machine (SVM) algorithm weighted signal relevance for 25 peaks. Using this methodology, spectral accuracy was 60.4% and 93.3% for diagnosing Crohn’s disease and ulcerative colitis, respectively 87. Additional efforts are required to refine and validate these and other similar techniques 88, identify the spectra-linked proteins, and assess their diagnostic applicability to real world practice.
SYSTEMS PHARMACOLOGY
Systems-based approaches that integrate data from multiple levels can not only facilitate human disease studies, but also are beneficial to drug design, drug combination, and drug repurposing. Traditional drug discovery involves cell-based or target-focused screening of chemical compounds in a very expensive and lengthy process. By contrast, many drugs exert their effects by modulating biological pathways rather than individual targets. Large-scale genomes, transcriptomes, proteome, interactome data, and their integration with metabolomic data and computational modeling have now enabled a systems-level view of drug discovery and development 14. In Table 2, we provide a list of public resources that can support drug discovery by using systems-based approaches. Systems pharmacology aims to understand the actions and adverse effects of drugs by considering targets in the context of their biological pathways and regulatory networks 13, 15.
Table 2.
Publicly available data resources that could be used for drug discovery, drug combination, and drug repositioning.
| Resources | Descriptions | URL |
|---|---|---|
| DrugBank | A database that combines detailed drug data with comprehensive target information over 7740 drug entries. | http://www.drugbank.ca/ |
| PharmGKB | A comprehensive resource that curates knowledge linking genetic variations, drug response, and pathways. | https://www.pharmgkb.org/ |
| FDA Orange Book | A list of approved drugs and drug combinations with their therapeutic equivalence evaluations. | http://www.accessdata.fda.gov/scripts/cder/ob/ |
| European Medicines Agency (EMA) | An agency of the European Union that report 950 human medicines and their therapeutic areas. | http://www.ema.europa.eu/ema/ |
| SIDER | A side effect database that contains information on 996 marketed medicines and their recorded adverse drug reactions. | http://sideeffects.embl.de/ |
| DrugMatrix | A toxicology reference database that store the results of thousands of highly controlled and standardized toxicological experiments. | https://ntp.niehs.nih.gov/drugmatrix |
| CMap | A collection of over 7,000 genome-wide transcriptional expression profiles from cultured human cells treated with bioactive small molecules. | https://www.broadinstitute.org/cmap/ |
| STITCH | A database of protein–chemical interactions that integrates many sources of experimental and manually curated evidence. | http://stitch.embl.de/ |
| Therapeutic Target Database (TTD) | A database to provide information about therapeutic protein and nucleic acid targets, targeted diseases, pathway information and the drugs directed at each of these targets. | http://bidd.nus.edu.sg/group/cjttd/ |
| PROMISCUOUS | A resource of protein-protein and drug- protein interactions that aims to provide a uniform data set for drug repositioning and further analysis. | http://bioinformatics.charite.de/promiscuous/ |
Drug combination therapy is a therapeutic intervention in which more than one drug therapy is administered to the patient. Mathematical modeling and clinical data show that some drug combination treatments have higher efficacy, fewer side effects, and less toxicity compared to single-drug treatment (rational polypharmacy) 89, 90. However, experimental screening of drug combinations is very costly and often only identifies a small number of synergistic combinations due to the large search space. Complex dependencies of drug-induced transcription profiles explored by mathematical models provide rich information for drug synergy identification. The DREAM consortium launched an open challenge to develop computational methods for ranking 91 compound pairs based on gene-expression profiles of human B cells treated with individual compounds at multiple time points and concentrations 91. Among the 32 methods the consortium assessed, four performed significantly better than random guessing, indicating that computational prediction of drug combination is possible. Zhao and colleagues reported a simple correlation-based strategy to reveal the synergistic effects of drug combinations by exploring the same data set 92. Jin and colleagues developed an enhanced Petri net model to recognize the synergistic effects of drug combinations from drug-treated microarray data 93. Rosiglitazone is an anti-diabetic drug that has been reported to increase the risk of cardiovascular complications, including myocardial infarction (MI). Zhao and colleagues searched for usage of a second drug in the FDA’s Adverse Event Reporting System (FAERS) that could mitigate the risk of rosiglitazone–associated MI and found that the combination of rosiglitazone with exenatide significantly reduces rosiglitazone-associated MI. Using cell biological networks and the data from a mouse model, they identified the regulatory mechanism underlying the mitigating effect of exenatide on rosiglitazone-associated MI 94.
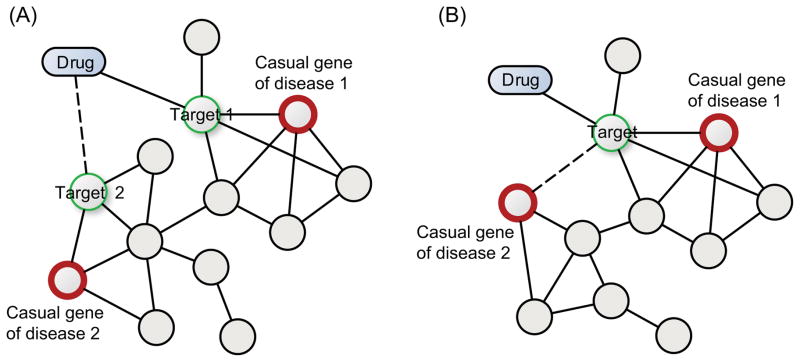
Owing to the high cost and lengthy time necessary for developing a new drug, drug repurposing, which aims to identify new indications of existing drugs, offers a promising alternative to de novo drug discovery. Many network-based methods have been developed for predicting drug repurposing. Two core concepts that support drug repurposing are drug-target interactions and target-disease associations. As shown in Figure 5A, a single drug may have multiple targets, and identification of new drug-target interactions that connects with causal genes for another disease may, therefore, be helpful for drug repositioning. In addition, by revealing new relationships of an existing target with another disease, a drug may be repositioned. Some methods utilize drug-induced transcriptional profiles for drug repurposing. For example, to pursue a systematic approach to the discovery of functional connections among diseases, genetic perturbation, and drug action, Lamb and colleagues have created a reference collection of gene-expression profiles from cultured human cells treated with bioactive small molecules 95. By using pattern matching methods to mine the data, this Connectivity Map (also known as CMap) resource can be used to find connections among small molecules sharing a mechanism of action, or structural or physiological processes. One of the successful applications of CMap for drug repositioning was conducted by Iorio and colleagues 96. In this study, an automatic approach that exploits similarity in gene expression profiles following drug treatment was developed to predict similarities in drug effect and mode of action. A drug network displaying similarities between pair of drugs was next constructed and partitioned into groups of densely interconnected nodes. Based on this network, Iorio and colleagues correctly predicted the mode of action for nine anticancer compounds and discovered an unreported effect for a well-known drug, fasudil (a Rho-kinase inhibitor). Using CMap data, a large set of drug-induced transcriptional modules was identified in another study 97. By utilizing conserved and cell-type-specific drug-induced modules, the investigators further predicted gene functions of some regulators and revealed new mechanisms-of-action for existing drugs, providing a starting point for drug repositioning.
Figure 5. An example of network-based drug repositioning.
(A) Drug repositioning by identifying new drug-target interactions (the dotted line). (B) Drug repositioning by identifying new target-disease associations (the dotted line).
Examples mentioned above demonstrate that drug-induced high-throughput gene expression profiles combined with proper computational methods are very useful for drug combination and drug repositioning. In addition to transcriptional profiles, drug-target networks and protein-protein interaction networks have been widely utilized for drug target identification 98. Such methods often use node similarity or structural features of biological networks. For example, Keiser and colleagues constructed drug-target networks and used a statistics-based chemoinformatics approach that explores the chemical similarities between drugs and ligand sets to predict thousands of drug-target unanticipated associations 99. Hwang and colleagues developed a novel network metric called bridging centrality to identify bridging nodes critically involved in connecting modular subregions of a protein interaction network. They showed that bridging nodes are promising drug targets from the standpoints of efficacy and side effects 100. Metabolite profiles and metabolic networks have been used in drug discovery studies, as well 101. In addition, some methods have been developed for predicting the adverse side effects of drugs using network models 102, 103.
PERSONALIZED MEDICINE
Personalized medicine, a medical model of customized healthcare in which an individual patient is provided with treatments tailored to his/her genomic makeup, has been discussed for many years. Advances in next generation sequencing and DNA variation arrays facilitate the generation of personalized patient data, such as individual human genomes and individual SNP profiles, which offers unprecedented opportunities for personalized medicine. By integrating various ‘omics data sets and GWAS loci/eQTLs, personalized medicine is making promising progress. For example, the Cancer Genome Atlas (TCGA) research team has used the latest sequencing technologies and sophisticated bioinformatic analytical methods to identify somatic variants in the genomes of thousands of tumor samples from at least 20 tumor types 104. Chen and colleagues presented an integrative personal ‘omics profile that combines genomic, transcriptomic, proteomic, metabolomic, and autoantibody profiles from a single individual over a 14-month period 105. This longitudinal analysis revealed various medical risks and individual disease states, including type II diabetes, rhinovirus, and respiratory syncytial virus infections, demonstrating the possibility of predictive and preventive medicine enabled by systems biology and whole genome sequencing.
EVOLUTION OF SYSTEMS BIOLOGY INTO SYSTEMS MEDICINE
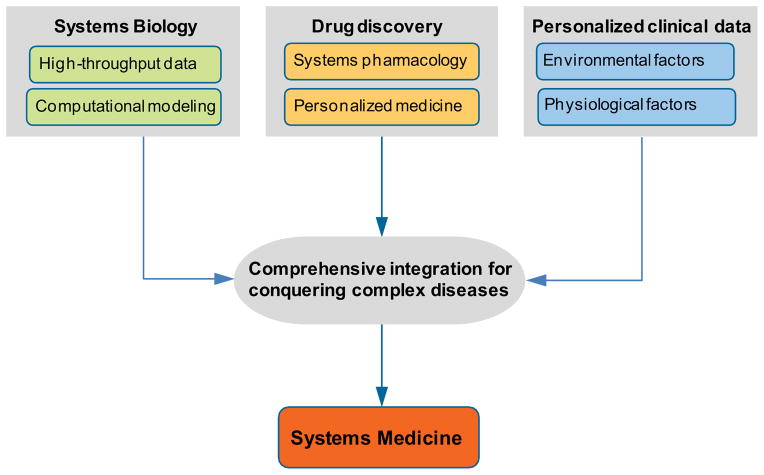
Building on the successes of systems biology, systems medicine is defined as an emerging discipline that more comprehensively integrates computational modeling, ‘omics data, physiological data, clinical data, and environmental factors to model disease expression predictively 17, 18. Systems medicine integrates basic research and clinical practice, and emphasizes translational and clinical research. As shown in Figure 6, the basic elements of systems medicine include systems-based approaches to human diseases and pharmacology and exploration of personalized patient clinical data space, including physiological data and environmental data. In addition to systems biology and network-based drug discovery, new high-dimensional patient data are another factor driving the emergence of systems medicine. Parallel to an explosion in the number of high-throughput molecular and cellular data sets, the digital revolution has induced a massive accumulation of electronic heath data that capture thousands of clinical measurements collected in medical practice 106. This valuable resource of longitudinal patient records has been overlooked by systems biologists in investigating human diseases due to tight privacy-based regulation of medical data. However, if made publically available, this data resource will provide rich information about individual disease states after integration with molecular data. In addition, each patient is different and needs personalization of medical treatment. To this end, environmental factors, such as diet, gender, age, and family histories, and physiological factors, such as tissues, organs, or whole body must be considered in a clinical practice context for systems medicine. This means that physicians must deal with large-scale non-linear, multi-dimensional data. Integration of such heterogeneous clinical data requires more sophisticated computational modeling strategies, but will make personalized medicine and personalized healthcare highly possible.
Figure 6. Basic elements of systems medicine.
System medicine is far more than systems biology of human diseases. It comprehensively integrates computational modeling, ‘omics data, physiological data, clinical data, and environmental factors to address major human diseases uniquely, efficiently, and with personalized precision.
Systems medicine is highly comprehensive and integrative, and utilizes all types of nonlinear information. Different from the collaborations in systems biology that focus on data integration and experimental validation, systems medicine is expected to be led by a team covering much broader range of expertise and requires large-scale interdisciplinary collaborative efforts from clinicians, patients, biomedical researchers, computational scientists, government, pharmaceutical industry and police makers. For example, a physician cannot make diagnostic decisions even if he/she is faced with thousands of data points of ‘omics data and clinical data. Multidisciplinary collaboration should utilize expertise from quantitative sciences to make the data readily accessible and easy to understand by physicians, and develop friendly computational tools for physicians to use the data. Also, sharing personal medical records will have great personal and societal benefits, but requires necessary ethical regulations, the cooperation of patients, and the participation of policy-makers. As a practical example, an innovative integrated health system has been proposed to combat major non-communicable diseases (NCDs) (cardiovascular diseases, cancer, chronic respiratory diseases, diabetes, rheumatologic diseases and mental health) by using systems medicine approaches and strategic partnerships 107. It includes several key components, such as understanding environmental, genetic, and molecular determinants of the diseases; practice-based interprofessional collaboration; carefully phenotyped patients; development of unbiased and accurate biomarkers for comorbidities; etc. The strategy takes a holistic systems medicine approach to tackle NCDs as a common group of diseases, and is designed to allow the results to be used globally and also adapted to local needs and specificities. In short, the ultimate goal of systems medicine is to transform reactive medicine and healthcare to a P4 medicine that is predictive, preventive, personalized, and participatory 108, and provide a powerful approach to developing novel therapeutic interventions and addressing major human diseases uniquely, efficiently, and with personalized precision.
Conclusion
Many diseases involve the complex interaction between genetic and environmental factors that are difficult to dissect using reductionist approaches. High-throughput technologies and predictive computational modeling drive the emergence and development of systems biology. Ever since its inception, the application of systems biology has penetrated biomedical disciplines rapidly, from basic research to human diseases and pharmacology. With the accumulation of individualized clinical measures, genetic variants and environmental data, systems biology is evolving from bench to bedside by integrating more types of heterogeneous data and recruiting diverse expertise from broader fields, which have promoted the emergence of systems medicine. Systems medicine aims to offer a powerful set of methodologies to improve our understanding of disease pathogenesis and to design personalized therapies to address the complexity of human diseases. Although systems medicine is in its early stages and faces many challenges, it will no doubt revolutionize the practice of medicine and healthcare.
Acknowledgments
The authors wish to thank Stephanie Tribuna for expert assistance. This work was supported in part by NIH grants 1K08HL111207-01A1 (to BAM), and HL061795, HL108630 (MAPGen Consortium), HG007690 (to JL), and the Pulmonary Hypertension Association (to BAM).
Footnotes
Conflicts of interest: The authors have declared no conflicts of interest for this article.
References
- 1.Ahn AC, Tewari M, Poon CS, Phillips RS. The limits of reductionism in medicine: could systems biology offer an alternative? PLoS Med. 2006;3:e208. doi: 10.1371/journal.pmed.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacLellan WR, Wang Y, Lusis AJ. Systems-based approaches to cardiovascular disease. Nat Rev Cardiol. 2012;9:172–184. doi: 10.1038/nrcardio.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 4.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 5.Rolland T, Ta An M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 8.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobie EA, Lee YS, Jenkins SL, Iyengar R. Systems biology--biomedical modeling. Sci Signal. 2011;4:tr2. doi: 10.1126/scisignal.2001989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 12.Ideker T, Krogan NJ. Differential network biology. Mol Syst Biol. 2012;8:565. doi: 10.1038/msb.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wist AD, Berger SI, Iyengar R. Systems pharmacology and genome medicine: a future perspective. Genome Med. 2009;1:11. doi: 10.1186/gm11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 15.Antman E, Weiss S, Loscalzo J. Systems pharmacology, pharmacogenetics, and clinical trial design in network medicine. Wiley Interdiscip Rev Syst Biol Med. 2012;4:367–383. doi: 10.1002/wsbm.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, Menche J, Barabasi AL, Sharma A. Human symptoms-disease network. Nat Commun. 2014;5:4212. doi: 10.1038/ncomms5212. [DOI] [PubMed] [Google Scholar]
- 17.Wolkenhauer O, Auffray C, Jaster R, Steinhoff G, Dammann O. The road from systems biology to systems medicine. Pediatr Res. 2013;73:502–507. doi: 10.1038/pr.2013.4. [DOI] [PubMed] [Google Scholar]
- 18.Auffray C, Chen Z, Hood L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 2009;1:2. doi: 10.1186/gm2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 20.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 21.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7:340–350. doi: 10.1002/pmic.200600422. [DOI] [PubMed] [Google Scholar]
- 22.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 23.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 24.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 27.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 29.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petschnigg J, Groisman B, Kotlyar M, Taipale M, Zheng Y, Kurat CF, Sayad A, Sierra JR, Mattiazzi Usaj M, Snider J, et al. The mammalian-membrane two-hybrid assay (MaMTH) for probing membrane-protein interactions in human cells. Nat Methods. 2014;11:585–592. doi: 10.1038/nmeth.2895. [DOI] [PubMed] [Google Scholar]
- 32.Smith MG, Ptacek J, Snyder M. Kinase substrate interactions. Methods Mol Biol. 2011;723:201–212. doi: 10.1007/978-1-61779-043-0_13. [DOI] [PubMed] [Google Scholar]
- 33.Saliba AE, Vonkova I, Ceschia S, Findlay GM, Maeda K, Tischer C, Deghou S, van Noort V, Bork P, Pawson T, et al. A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat Methods. 2014;11:47–50. doi: 10.1038/nmeth.2734. [DOI] [PubMed] [Google Scholar]
- 34.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 35.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 36.Laufer C, Fischer B, Huber W, Boutros M. Measuring genetic interactions in human cells by RNAi and imaging. Nat Protoc. 2014;9:2341–2353. doi: 10.1038/nprot.2014.160. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 38.Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S, Chen X, Jin L, Xiong M. Canonical correlation analysis for RNA-seq co-expression networks. Nucleic Acids Res. 2013;41:e95. doi: 10.1093/nar/gkt145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewey FE, Perez MV, Wheeler MT, Watt C, Spin J, Langfelder P, Horvath S, Hannenhalli S, Cappola TP, Ashley EA. Gene coexpression network topology of cardiac development, hypertrophy, and failure. Circ Cardiovasc Genet. 2011;4:26–35. doi: 10.1161/CIRCGENETICS.110.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 44.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan SY, Loscalzo J. The emerging paradigm of network medicine in the study of human disease. Circ Res. 2012;111:359–374. doi: 10.1161/CIRCRESAHA.111.258541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma A, Menche J, Huang C, Ort T, Zhou X, Kitsak M, Sahni N, Thibault D, Voung L, Guo F, et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang RS, Albert R. Elementary signaling modes predict the essentiality of signal transduction network components. BMC Syst Biol. 2011;5:44. doi: 10.1186/1752-0509-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang RS, Sun Z, Albert R. Minimal functional routes in directed graphs with dependent edges. International Transactions in Operational Research. 2013;20:391–409. [Google Scholar]
- 49.Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, Uetz P, Sittler T, Karp RM, Ideker T. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGeachie MJ, Chang HH, Weiss ST. CGBayesNets: conditional Gaussian Bayesian network learning and inference with mixed discrete and continuous data. PLoS Comput Biol. 2014;10:e1003676. doi: 10.1371/journal.pcbi.1003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman N, Linial M, Nachman I, Pe’er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 52.Chu JH, Hersh CP, Castaldi PJ, Cho MH, Raby BA, Laird N, Bowler R, Rennard S, Loscalzo J, Quackenbush J, et al. Analyzing networks of phenotypes in complex diseases: methodology and applications in COPD. BMC Syst Biol. 2014;8:78. doi: 10.1186/1752-0509-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mourad R, Sinoquet C, Leray P. Probabilistic graphical models for genetic association studies. Brief Bioinform. 2012;13:20–33. doi: 10.1093/bib/bbr015. [DOI] [PubMed] [Google Scholar]
- 54.Wang RS, Saadatpour A, Albert R. Boolean modeling in systems biology: an overview of methodology and applications. Phys Biol. 2012;9:055001. doi: 10.1088/1478-3975/9/5/055001. [DOI] [PubMed] [Google Scholar]
- 55.Samaga R, Klamt S. Modeling approaches for qualitative and semi-quantitative analysis of cellular signaling networks. Cell Commun Signal. 2013;11:43. doi: 10.1186/1478-811X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryall KA, Holland DO, Delaney KA, Kraeutler MJ, Parker AJ, Saucerman JJ. Network reconstruction and systems analysis of cardiac myocyte hypertrophy signaling. J Biol Chem. 2012;287:42259–42268. doi: 10.1074/jbc.M112.382937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wittmann DM, Krumsiek J, Saez-Rodriguez J, Lauffenburger DA, Klamt S, Theis FJ. Transforming Boolean models to continuous models: methodology and application to T-cell receptor signaling. BMC Syst Biol. 2009;3:98. doi: 10.1186/1752-0509-3-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang RS, Oldham WM, Loscalzo J. Network-based association of hypoxia-responsive genes with cardiovascular diseases. New J Phys. 2014;16:105014. doi: 10.1088/1367-2630/16/10/105014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc Natl Acad Sci U S A. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hidalgo CA, Blumm N, Barabasi AL, Christakis NA. A dynamic network approach for the study of human phenotypes. PLoS Comput Biol. 2009;5:e1000353. doi: 10.1371/journal.pcbi.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zucco L, Zhang Q, Kuliszewski MA, Kandic I, Faughnan ME, Stewart DJ, Kutryk MJ. Circulating angiogenic cell dysfunction in patients with hereditary hemorrhagic telangiectasia. PLoS One. 2014;9:e89927. doi: 10.1371/journal.pone.0089927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu G, Barrios-Rodiles M, Jerkic M, Turinsky AL, Nadon R, Vera S, Voulgaraki D, Wrana JL, Toporsian M, Letarte M. Novel protein interactions with endoglin and activin receptor-like kinase 1: potential role in vascular networks. Mol Cell Proteomics. 2014;13:489–502. doi: 10.1074/mcp.M113.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma A, Gulbahce N, Pevzner SJ, Menche J, Ladenvall C, Folkersen L, Eriksson P, Orho-Melander M, Barabasi AL. Network-based analysis of genome wide association data provides novel candidate genes for lipid and lipoprotein traits. Mol Cell Proteomics. 2013;12:3398–3408. doi: 10.1074/mcp.M112.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Velez P, Parguina AF, Ocaranza-Sanchez R, Grigorian-Shamagian L, Rosa I, Alonso-Orgaz S, de la Cuesta F, Guitian E, Moreu J, Barderas MG, et al. Identification of a circulating microvesicle protein network involved in ST-elevation myocardial infarction. Thromb Haemost. 2014;112:716–726. doi: 10.1160/TH14-04-0337. [DOI] [PubMed] [Google Scholar]
- 68.Faner R, Cruz T, Lopez-Giraldo A, Agusti A. Network medicine, multimorbidity and the lung in the elderly. Eur Respir J. 2014;44:775–788. doi: 10.1183/09031936.00078714. [DOI] [PubMed] [Google Scholar]
- 69.Menche J, Sharma A, Cho MH, Mayer RJ, Rennard SI, Celli B, Miller BE, Locantore N, Tal-Singer R, Ghosh S, et al. A diVIsive Shuffling Approach (VIStA) for gene expression analysis to identify subtypes in Chronic Obstructive Pulmonary Disease. BMC Syst Biol. 2014;8 (Suppl 2):S8. doi: 10.1186/1752-0509-8-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davidsen PK, Herbert JM, Antczak P, Clarke K, Ferrer E, Peinado VI, Gonzalez C, Roca J, Egginton S, Barbera JA, et al. A systems biology approach reveals a link between systemic cytokines and skeletal muscle energy metabolism in a rodent smoking model and human COPD. Genome Med. 2014;6:59. doi: 10.1186/s13073-014-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Najafi A, Masoudi-Nejad A, Ghanei M, Nourani MR, Moeini A. Pathway reconstruction of airway remodeling in chronic lung diseases: a systems biology approach. PLoS One. 2014;9:e100094. doi: 10.1371/journal.pone.0100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao Y, Peng J, Lu C, Hsin M, Mura M, Wu L, Chu L, Zamel R, Machuca T, Waddell T, et al. Metabolomic heterogeneity of pulmonary arterial hypertension. PLoS One. 2014;9:e88727. doi: 10.1371/journal.pone.0088727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maron BA, Oldham WM, Chan SY, Vargas SO, Arons E, Zhang YY, Loscalzo J, Leopold JA. Upregulation of steroidogenic acute regulatory protein by hypoxia stimulates aldosterone synthesis in pulmonary artery endothelial cells to promote pulmonary vascular fibrosis. Circulation. 2014;130:168–179. doi: 10.1161/CIRCULATIONAHA.113.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malaney P, Pathak RR, Xue B, Uversky VN, Dave V. Intrinsic disorder in PTEN and its interactome confers structural plasticity and functional versatility. Sci Rep. 2013;3:2035. doi: 10.1038/srep02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gamez-Pozo A, Perez Carrion RM, Manso L, Crespo C, Mendiola C, Lopez-Vacas R, Berges-Soria J, Lopez IA, Margeli M, Calero JL, et al. The Long-HER Study: Clinical and Molecular Analysis of Patients with HER2+ Advanced Breast Cancer Who Become Long-Term Survivors with Trastuzumab-Based Therapy. PLoS One. 2014;9:e109611. doi: 10.1371/journal.pone.0109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wegdam W, Argmann CA, Kramer G, Vissers JP, Buist MR, Kenter GG, Aerts JM, Meijer D, Moerland PD. Label-free LC-MSe in tissue and serum reveals protein networks underlying differences between benign and malignant serous ovarian tumors. PLoS One. 2014;9:e108046. doi: 10.1371/journal.pone.0108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu LH, Lee E, Bader JS, Popel AS. Angiogenesis interactome and time course microarray data reveal the distinct activation patterns in endothelial cells. PLoS One. 2014;9:e110871. doi: 10.1371/journal.pone.0110871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Finley SD, Chu LH, Popel AS. Computational systems biology approaches to anti-angiogenic cancer therapeutics. Drug Discov Today. 2014 doi: 10.1016/j.drudis.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vempati P, Mac Gabhann F, Popel AS. Quantifying the proteolytic release of extracellular matrix-sequestered VEGF with a computational model. PLoS One. 2010;5:e11860. doi: 10.1371/journal.pone.0011860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li P, Liu Y, Wang H, He Y, Wang X, Lv F, Chen H, Pang X, Liu M, Shi T, et al. PubAngioGen: a database and knowledge for angiogenesis and related diseases. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karagiannis GS, Saraon P, Jarvi KA, Diamandis EP. Proteomic signatures of angiogenesis in androgen-independent prostate cancer. Prostate. 2014;74:260–272. doi: 10.1002/pros.22747. [DOI] [PubMed] [Google Scholar]
- 82.Yoon DY, Buchler P, Saarikoski ST, Hines OJ, Reber HA, Hankinson O. Identification of genes differentially induced by hypoxia in pancreatic cancer cells. Biochem Biophys Res Commun. 2001;288:882–886. doi: 10.1006/bbrc.2001.5867. [DOI] [PubMed] [Google Scholar]
- 83.Huang H, Vangay P, McKinlay CE, Knights D. Multi-omics analysis of inflammatory bowel disease. Immunol Lett. 2014 doi: 10.1016/j.imlet.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 84.Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, Shah M, Halfvarson J, Tysk C, Henrissat B, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS One. 2012;7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tuller T, Atar S, Ruppin E, Gurevich M, Achiron A. Common and specific signatures of gene expression and protein-protein interactions in autoimmune diseases. Genes Immun. 2013;14:67–82. doi: 10.1038/gene.2012.55. [DOI] [PubMed] [Google Scholar]
- 87.Seeley EH, Washington MK, Caprioli RM, M’Koma AE. Proteomic patterns of colonic mucosal tissues delineate Crohn’s colitis and ulcerative colitis. Proteomics Clin Appl. 2013;7:541–549. doi: 10.1002/prca.201200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.M’Koma AE, Seeley EH, Washington MK, Schwartz DA, Muldoon RL, Herline AJ, Wise PE, Caprioli RM. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflamm Bowel Dis. 2011;17:875–883. doi: 10.1002/ibd.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol. 2006;2:458–466. doi: 10.1038/nchembio817. [DOI] [PubMed] [Google Scholar]
- 90.Sun X, Vilar S, Tatonetti NP. High-throughput methods for combinatorial drug discovery. Sci Transl Med. 2013;5:205rv201. doi: 10.1126/scitranslmed.3006667. [DOI] [PubMed] [Google Scholar]
- 91.Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, Xiao G, Li Y, Allen J, Zhong R, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014 doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao J, Zhang XS, Zhang S. Predicting cooperative drug effects through the quantitative cellular profiling of response to individual drugs. CPT Pharmacometrics Syst Pharmacol. 2014;3:e102. doi: 10.1038/psp.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jin G, Zhao H, Zhou X, Wong ST. An enhanced Petri-net model to predict synergistic effects of pairwise drug combinations from gene microarray data. Bioinformatics. 2011;27:i310–316. doi: 10.1093/bioinformatics/btr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao S, Nishimura T, Chen Y, Azeloglu EU, Gottesman O, Giannarelli C, Zafar MU, Benard L, Badimon JJ, Hajjar RJ, et al. Systems pharmacology of adverse event mitigation by drug combinations. Sci Transl Med. 2013;5:206ra140. doi: 10.1126/scitranslmed.3006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 96.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iskar M, Zeller G, Blattmann P, Campillos M, Kuhn M, Kaminska KH, Runz H, Gavin AC, Pepperkok R, van Noort V, et al. Characterization of drug-induced transcriptional modules: towards drug repositioning and functional understanding. Mol Syst Biol. 2013;9:662. doi: 10.1038/msb.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farkas IJ, Korcsmaros T, Kovacs IA, Mihalik A, Palotai R, Simko GI, Szalay KZ, Szalay-Beko M, Vellai T, Wang S, et al. Network-based tools for the identification of novel drug targets. Sci Signal. 2011;4(pt3) doi: 10.1126/scisignal.2001950. [DOI] [PubMed] [Google Scholar]
- 99.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hwang WC, Zhang A, Ramanathan M. Identification of information flow-modulating drug targets: a novel bridging paradigm for drug discovery. Clin Pharmacol Ther. 2008;84:563–572. doi: 10.1038/clpt.2008.129. [DOI] [PubMed] [Google Scholar]
- 101.Zhu M, Zhang H, Humphreys WG. Drug metabolite profiling and identification by high-resolution mass spectrometry. J Biol Chem. 2011;286:25419–25425. doi: 10.1074/jbc.R110.200055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cami A, Arnold A, Manzi S, Reis B. Predicting adverse drug events using pharmacological network models. Sci Transl Med. 2011;3:114ra127. doi: 10.1126/scitranslmed.3002774. [DOI] [PubMed] [Google Scholar]
- 103.Tatonetti NP, Ye PP, Daneshjou R, Altman RB. Data-driven prediction of drug effects and interactions. Sci Transl Med. 2012;4:125ra131. doi: 10.1126/scitranslmed.3003377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HY, Miriami E, Karczewski KJ, Hariharan M, Dewey FE, Cheng Y, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Roden DM, Xu H, Denny JC, Wilke RA. Electronic medical records as a tool in clinical pharmacology: opportunities and challenges. Clin Pharmacol Ther. 2012;91:1083–1086. doi: 10.1038/clpt.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bousquet J, Jorgensen C, Dauzat M, Cesario A, Camuzat T, Bourret R, Best N, Anto JM, Abecassis F, Aubas P, et al. Systems medicine approaches for the definition of complex phenotypes in chronic diseases and ageing. From concept to implementation and policies. Curr Pharm Des. 2014;20:5928–5944. doi: 10.2174/1381612820666140314115505. [DOI] [PubMed] [Google Scholar]
- 108.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 109.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014;42:D32–37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kodama Y, Mashima J, Kosuge T, Katayama T, Fujisawa T, Kaminuma E, Ogasawara O, Okubo K, Takagi T, Nakamura Y. The DDBJ Japanese Genotype-phenotype Archive for genetic and phenotypic human data. Nucleic Acids Res. 2015;43:D18–22. doi: 10.1093/nar/gku1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, et al. Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–932. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading/Resources
- Emmert-Streib F, Dehmer M. Enhancing systems medicine beyond genotype data by dynamic patient signatures: having information and using it too. Front Genet. 2013;4:241. doi: 10.3389/fgene.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M, Nestor CE, Zhang H, Barabási AL, Baranzini S, Brunak S, Chung KF, Federoff HJ, Gavin AC, Meehan RR, et al. Modules, networks and systems medicine for understanding disease and aiding diagnosis. Genome Med. 2014;6(10):82. doi: 10.1186/s13073-014-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn AC, Tewari M, Poon C-S, Phillips RS. The Clinical Applications of a Systems Approach. PLoS Med. 2006;3(7):e209. doi: 10.1371/journal.pmed.0030209. [DOI] [PMC free article] [PubMed] [Google Scholar]