Abstract
Objective
Microbial invasion of the amniotic cavity is associated with spontaneous preterm labor and adverse pregnancy outcome, and Mycoplasma hominis often is present. However, the pathogenic process by which M. hominis invades the amniotic cavity and gestational tissues, often resulting in chorioamnionitis and preterm birth, remains unknown. We hypothesized that strains of M. hominis vary genetically with regards to their potential to invade and colonize the amniotic cavity and placenta.
Study Design
We sequenced the entire genomes of 2 amniotic fluid isolates and a placental isolate of M. hominis from pregnancies that resulted in preterm births and compared them with the previously sequenced genome of the Type strain PG21. We identified genes that were specific to the amniotic fluid/placental isolates. We then determined the microbial burden and the presence of these genes in another set of subjects from whom samples of amniotic fluid had been collected and were positive for M. hominis.
Results
We identified 2 genes that encode surface-located membrane proteins (Lmp1 and Lmp-like) in the sequenced amniotic fluid/placental isolates that were severely truncated in PG21. We also identified, for the first time, a microbial gene of unknown function that is referred to in this study as gene of interest C that was significantly associated with bacterial burden in amniotic fluid and the risk of preterm delivery in patients with preterm labor.
Conclusion
A gene in M. hominis was identified that is associated significantly with colonization and/or infection of the upper reproductive tract during pregnancy and with preterm birth.
Keywords: chorioamnionitis, microbial invasion of the amniotic cavity, genital mycoplasmas, pathogenicity
Introduction
M. hominis is a common vaginal inhabitant that is associated with bacterial vaginosis (BV)1–3. The bacterium is considered harmless for the most part in non-pregnant women, but can cause intra-amniotic infections, which are associated with inflammation, preterm premature rupture of membranes (PPROM) and preterm birth4–15. A cohort study involving more than 10,000 pregnant women, found that women with BV had an increased risk for preterm birth, and among women with BV, those who were colonized with both bacteroides and Mycoplasma hominis had the greatest increase in risk16. More recent studies that employ 16S surveys to assess vaginal and intrauterine bacteria support the association between preterm birth and BV and/or M. hominis17, 18.
One of every three preterm births is associated with microbial invasion of the amniotic cavity (MIAC)19–33. Intra-amniotic infections can lead to inflammation, which triggers spontaneous preterm labor34–36. Therapeutic intervention for intra-amniotic infection is being studied, but treatment is currently largely unsuccessful37–41. Intra-amniotic infection is particularly common in births that occur prior to 32 completed weeks of gestation. This observation is significant because morbidity and mortality increase with decreasing gestational age at delivery. M. hominis is frequently isolated from infected fetal membranes and amniotic fluid, and genital mycoplasmas (including Ureaplasma spp.) are isolated from umbilical cord blood in approximately 20% of very preterm (<32 weeks gestation) newborns6, 42. However, the etiology and pathogenesis of infectious preterm birth remain poorly understood. Bacteria likely invade the amniotic cavity by ascending from the vagina through the cervix, or via hematogenous spread from more remote sources such as the oral cavity43–50. While microbial invasion of the amniotic cavity by M. hominis is clearly associated with preterm birth, the reported relative risk associated with vaginal colonization varies widely in the literature and the association is not strong enough or sufficiently consistent to render vaginal colonization as a predictor for poor pregnancy outcome9, 51–56. This and the lack of a strong association between other vaginal colonizers and preterm birth, has hindered early prediction of risk and effective medical intervention. However, a very recent study found that treatment of genital mycoplasmas late in pregnancy improves pregnancy outcome57.
Closely related bacterial taxa can vary widely with respect to their gene content, even within a single species. With the advent of next-generation sequencing and the increasing speed and ease with which whole bacterial genomes can be sequenced, has come the ability to observe the gain, loss, and modification of genes and evidence of the evolution of pathogens from non-pathogenic predecessors58. For example, uropathogenic Escherichia coli differ from commensal gastrointestinal E. coli strains in that they harbor adhesins that facilitate adherence to urinary epithelial cells, secreted toxins, and iron-acquisition systems that promote survival in the iron-limited environment of the urinary tract59. Recently, Whidbey et al. found that a nonsense mutation in a negative regulator of the hemolytic ornithine rhamnolipid pigment of Group B Streptococcus led to increased hemolysis, cytotoxicity, and penetration of fetal membranes60. In the same study, they also found that the majority of GBS isolates from amniotic fluid and chorioamnion from women in preterm labor were hyper-hemolytic and contained mutations in this regulator suggesting a genetic basis for the ability to cause ascending infection and preterm birth. These findings support the existence of genotypically distinct strains of bacterial species that possess virulence factor genes that increase their potential to cause infection or to infect a particular niche. We hypothesized that, similarly, distinct strains of M. hominis that have increased genetic potential to invade the amniotic cavity exist, and that identification of the genetic determinants involved in this heightened virulence could advance preterm risk assessment associated with vaginal colonization and lead to a better understanding of the pathogenesis of ascending M. hominis infections. To test this hypothesis, we sequenced the genomes of M. hominis amniotic fluid/placental isolates from three pregnant women who had episodes of spontaneous preterm labor that resulted in preterm delivery, and compared their genomes to the only previously sequenced and publicly available complete genome of strain PG21 (ATCC 23114), a rectal isolate from a healthy individual2.
Materials and Methods
Amniotic fluid and placenta isolates
Three M. hominis amniotic fluid/placental isolates from pregnancies that resulted in preterm birth were collected as part of a previous study from 1991–199616, 61. Two of the isolates were from amniotic fluid (AF1, AF3) and one was from placenta (PL5). Gestational ages at delivery are listed in Table 1. The isolates were cultured in modified arginine broth or on arginine agar (mycoplasma broth or agar containing 0.5% arginine, 20% horse serum, 2.5% yeast extract (Oxoid; Hampshire, UK), and 150 µg ampicillin/mL) stationary at 37°C in air supplemented with 5% CO262.
Table 1.
M. hominis strains analyzed in this study
| Features | PG21a | AF1b | AF3b | PL5b |
|---|---|---|---|---|
| Reference | Pereyre 2 | |||
| Body site | Rectum | Amniotic fluid | Amniotic fluid | Placenta |
| Gestational age at delivery | N/A | 31 weeks | 28 weeks | 33 weeks |
| Genome size | 665,445 bp | 704,093 bp | 680,135 bp | 721,886 bp |
| GC contentc | 27.1% | 27.2% | 27.2% | 27.2% |
| Protein encoding genes | 537 | 581 | 576 | 627 |
| tRNA genes | 33 | 33 | 33 | 33 |
N/A, not applicable
Strain PG21, also known as ATCC 23114, is a control. It was isolated from the rectum of a healthy individual, not from a case of intra-amniotic infection.
Strains AF1, AF3, and PL5 were isolated from intra-amniotic infections in pregnancies in which episodes of spontaneous preterm labor resulted in preterm births16, 61.
Percent of the nucleotides in the whole genome that are either guanines or cytosines.
Preparation of sequencing libraries
Bacteria were grown in 50 mL arginine broth, collected by centrifugation, and DNA was isolated using the Genomic-tip 500/G (Qiagen; Valencia, CA) according to manufacturer’s instructions. Genome sequencing was performed using Roche 454 pyrosequencing technology (Roche 454; 454 Life Sciences, Branford, CT), with a combined strategy of whole genome shotgun and 8-kilobase pair (kbp) paired-end reads as previously described63.
Genome assembly
Roche’s Newbler assembly software was used to perform de novo genome assemblies, using 454-FLX sequence data. The contigs were ordered by alignment to the reference genome of strain PG21 (Accession: PRJNA41875) using Mauve (Genome Evolution Laboratory, University of Wisconsin-Madison, WI)64. The genome of AF1 was closed by polymerase chain reaction (PCR) and Sanger sequencing.
Gene calling and analysis
Rapid Annotation using Subsystems Technology (RAST) was used to annotate the AF1, AF3, PL5, and PG21 genomes and to compare the genes and identify genes specific to one or more strains65. OrthoMCL (Eukaryotic Pathogen Database Resources) was also used to identify unique genes66. Absence of these genes in PG21 was confirmed by the tBLASTn database at NCBI (National Center for Biotechnology Information, Bethesda, MD). The circular chromosomes were visualized and compared using the BLAST Ring Image Generator (BRIG) 67. Amino acid sequences of surface proteins were compared with the use of Geneious software (version 7.1.5; Biomatters Inc., San Francisco, CA).
Vaginal microbiome samples
Participants were recruited from outpatient clinics at the Virginia Commonwealth University Medical Center and the Virginia Department of Health following written, informed consent from 2009–2013. Inclusion criteria included women age 18–44 years who were able to provide informed consent and who were willing or already scheduled to undergo a vaginal examination using a speculum. The Institutional Review Boards for Human Subjects Research at Virginia Commonwealth University and the Virginia Department of Health reviewed and approved this study. Participants filled out a detailed questionnaire that included questions about ethnicity, education, employment, health habits, dietary habits, and sexual history. Clinicians used CultureSwab™EZ polyurethane foam swabs (BD) to obtain specimens from the mid-vaginal wall during a speculum examination. DNA was extracted from the swabs within 4 hours of collection using the Powersoil® kit (MO BIO Laboratories Inc, Carlsbad, CA). The swabs were swirled directly in the Powerbead tubes (MO BIO Laboratories Inc) supplied with the kit and processing was according to manufacturer’s instructions. The 16S primers contain the A or B Titanium sequencing adapter (shown in italics), followed immediately by a unique variable (6–9 base) barcode sequence and finally the 5’ end of primer. The forward primer was a mixture (4:1) of the primers Fwd-P1 (5’ - CCATCTCATCCCTGCGTGTCTCCGACTCAG BBBBBB AGAGTTYGATYMTGGCTYAG) and Fwd-P2 (5’ - CCATCTCATCCCTGCGTGTCTCCGACTCAG BBBBBB AGARTTTGATCYTGGTTCAG). The reverse primer was Rev1B (5’ – CCTATCCCCTGTGTGCCTTGGCAGTCTCAG ATTACCGCGGCTGCTGG). PCR products were sequenced using the Roche 454 GS FLX Titanium platform (454 Life Sciences, Branford, CT). These data were generated as part of the Vaginal Human Microbiome Project68. Raw sequence data from the project is available from the Short Read Archive at NCBI (projectID phs000256)68. We used a deep sequencing approach with a median 24,030 reads/sample. All processed samples were represented by > 5,000 reads. Sequences were classified using a local installation of the RDP (NCBI) classifier (0.8 cutoff) and the STIRRUPS analysis platform69. All samples for which gestational age at delivery was known that contained M. hominis reads, were used in this study69, 70.
Detection of genes specific to sequenced amniotic fluid/placental isolates
A portion of an overnight culture of AF1 was serially diluted and plated for enumeration. Another portion of the same culture was reserved for DNA extraction using the Powersoil kit. This DNA standard was used to quantify the amount of DNA amplified in realtime PCR assays. Primers for the 16S gene (5’-ATGAGGGTGCGGAACATTAG-3’, 5’-TAATTCCGGATAACGCTTGC-3’), arl (5’-CTGGCGGAAATTCACTAAGC-3’, 5’-ATCGCATCAAACATCGTGTC-3’), goiB (5’-CGCCAAAACTATGCACGCATTTAT-3’, 5’-GGTTAGCCTTTGGCCTCATAGTA-3’), and goiC (5’-CCTTACGGATATATGGTTGTTTCG-3’, 5’-CTAACTTAAATCATCAAGAGTACGG-3’), were validated for efficiencies between 97–100%. Quantitative realtime PCR was performed using iTaq™ Universal® SYBR Green Supermix and an iQ thermal cycler (Biorad).
Statistical Analyses
Read counts were converted to proportions for all samples. Alpha diversity was measured using the inverse Simpson’s index. Differences in diversity between groups of samples were tested using a two-sided t-test. Effect sizes of bacterial species that correlate with ethnicity were created using LEfSe (Linear discriminant analysis Effect Size)71. LEfSe uses the Kruskal-Wallis rank sum test to detect taxa that distinguish groups of subjects, and uses linear discriminant analysis (LDA) to calculate an LDA score for the effect size as described71
Results
Genetic differences in M. hominis strains associated with intra-amniotic infection
We sequenced the genomes of 3 M. hominis amniotic fluid/placental isolates from pregnancies with episodes of spontaneous preterm labor that resulted in preterm births. Isolates AF1 and AF3 were from amniotic fluid, and PL5 was from placenta. The AF1 genome was finished and circularized (accession number: CP009677) and AF3 (accession number: JRWZ00000000) and PL5 (accession number: JRXA00000000) were assembled into 12 and 10 contigs, respectively. Features of the genomes are shown in Table 1. The genomes of all three amniotic fluid/placental isolates were slightly larger than the PG21 genome. The largest, from PL5, contained ~722 kb and 627 open reading frames. The average nucleotide identify between the three amniotic fluid/placental isolates and PG21 ranged from 98.1–98.6%. The percentages of guanine and cytosine nucleotides (%GC), a variable used in bacterial systematics to classify taxa, was similar for the genomes of all four strains.
Mobile genetic elements
Virulence and antibiotic resistance determinants are often transferred between bacterial strains within mobile elements such as pathogenicity islands, transposons and phages. The PG21 genome does not appear to contain any transposons or prophages. Strain AF3 apparently lacks phage genes as well. However, strains AF1 and PL5 each harbor a distinct mobile element. A putative prophage in AF1 is 15.2 kb and exhibits similarity to the M. fermentans phiMFV1 prophage 72. A putative 26.6 kb mobile element in PL5 shares 96% identity with a transposon from Streptococcus agalactiae and other streptococci and enterococci. It contains 18 genes that include several predicted to encode proteins involved in the assembly of a conjugative pilus and conjugative transfer, a TetM tetracycline resistance protein, and 6 genes that are predicted to function in drug efflux or transport. We tested the minimum inhibitory concentrations (MIC) of tetracycline for all four strains and found that the MIC for PL5 was 5 µg/ml, whereas the MIC for AF1 was 0.08 µg/ml, and AF3 and PG21 were 0.16 µg/ml, suggesting that the TetM protein is active in PL5.
Variations in adhesins and virulence factors
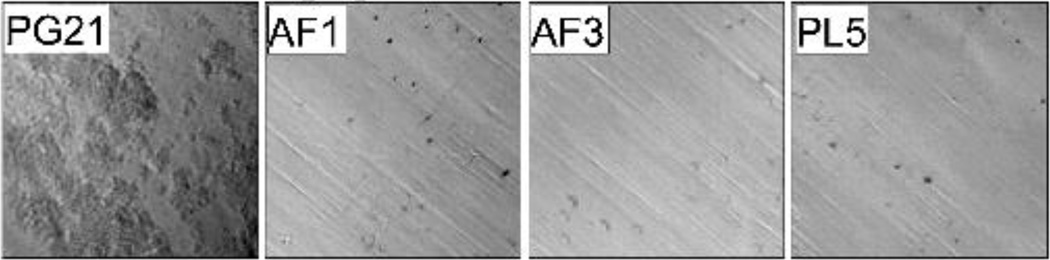
PG21 aggregated and adhered to the culture vessel wall in liquid culture whereas the cultures of the amniotic fluid/placental isolates were turbid and homogeneous. Adherence to the culture tubes was visualized by low-power microscopy and is shown in Figure 1. The variable surface protein Lmp1 appears to play a role in suppressing autoaggregation in M. hominis73. We therefore examined the major surface lipoproteins including Vaa, the variable membrane protein (Vmp), the Lmp proteins, P120, and P75 to determine whether genetic differences could explain the differential phenotype74–77.
Figure 1. Aggregative properties of isolates from intra-amniotic infection in liquid culture.
The M. hominis strains were cultured overnight in polystyrene culture tubes and the wall of the tube was imaged using a light microscope with a 10X objective. The rectal isolate PG21 aggregated and adhered to the walls of the polystyrene culture tubes (visible in the first panel as clusters of bacteria), whereas all three amniotic fluid/placental isolates failed to aggregate or adhere to the culture vessel (bare polystyrene), suggesting differences in the expression of surface proteins.
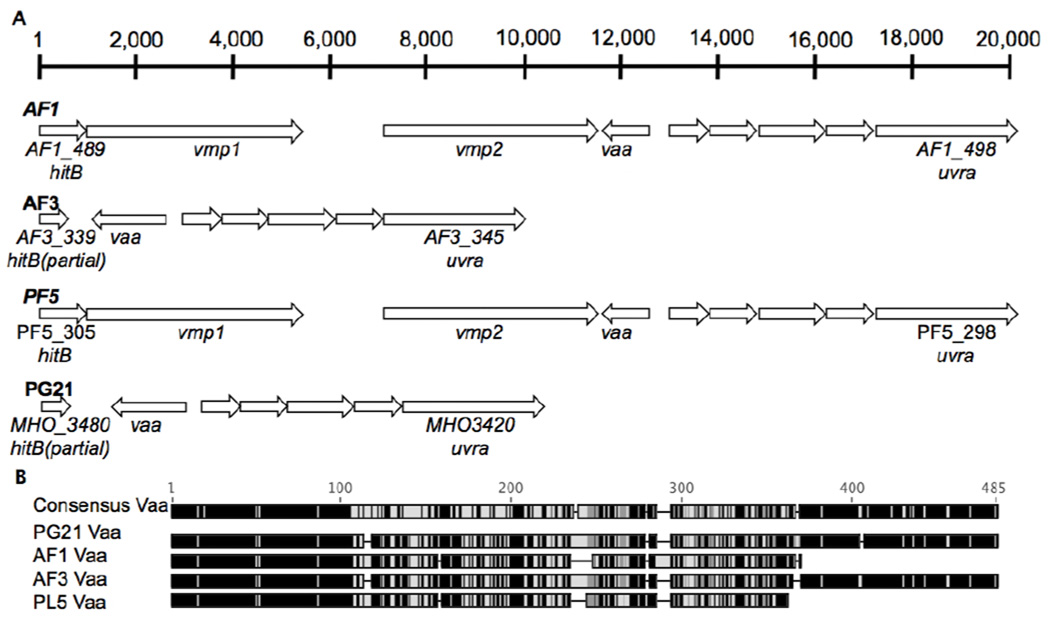
The Vmp loci of the four strains are illustrated in Figure 2A. As indicated in Figure 2B, the Vaa protein encoded by AF3 and PG21 shared high identities and similarities (93% and 95%, respectively) whereas Vaa in PL5 was shorter and exhibited lower identities and similarities with PG21 (57% and 73%). The length of Vaa in AF1 was similar to PL5 and the identities and similarities with the protein from this strain were 79% and 87% and it exhibited low identities and similarities with PG21 (57% and 71%), AF1 and PL5 each encoded two copies of vmp, whereas AF3 and PG21 lacked vmp (Figure 2). In sum, the Vaa loci of the amniotic fluid isolates were not more similar to one another than they were to PG21.
Figure 2. Variable membrane protein loci.
A. The variable membrane protein loci from the 3 amniotic fluid/placental strains and PG21 are depicted. The scale bar at the top indicates the number of nucleotides within the longest loci (AF1 and PF5). The loci within the genomes of AF1 and PF5 each contained two vmp genes whereas the AF3 and PG21 genomes lacked vmp. B. The Vaa amino acid sequences were compared using Geneious. The heatmaps indicate amnio acid similarity and range from 100% similar (black) to <60% similar (light grey).
All 3 amniotic fluid/placental isolates encoded Lmp1 proteins (AF1_245, AF3_439, and PL5_1) that were severely truncated (537 amino acid versus 1,522 amnio acids) with respect to Lmp1 in PG21 (MHO_0530). All three amniotic fluid/placental strains encoded a 668 amino acid Lmp-like protein (AF1_409, AF3_259, and PL5_148) that was truncated at 482 amino acids in PG21 (MHO_4280). The Lmp3 proteins encoded by the 3 amniotic fluid/placental isolates and PG21 varied in size. The P120 proteins encoded within PG21 (MHO_3660) and PL5 (PL5_410) shared 99% identities, whereas AF3_321 shared only 84% identities with PL5_410 and MHO_3660. The P75 proteins of AF3 (AF3_315) and PL5 (PL5_416) shared higher identities with PG21 (MHO_3720) (91% and 96%, respectively) than with each other (89%). In sum, the amniotic fluid/placental isolates encoded similar, truncated Lmp1 proteins and similar Lmp-like proteins, but the remaining surface-associated proteins were not more similar amongst the amniotic fluid/placental isolates versus PG21.
Genes exclusive to M. hominis strains associated with intra-amniotic infection
Figure 3 depicts the AF1 chromosome as the benchmark (central red circle) to which the AF3, PL5 and PG21 were compared using the analysis tool BRIG67 to facilitate identification of genes that were present in the amniotic fluid/placental isolates but absent in PG21. Genes present in the amniotic fluid/placental isolates but absent in PG21 are visualized as gaps restricted to the PG21 (green) chromosome. Overall, there are relatively few gaps in the chromosome suggesting that all four strains generally bear a similar complement of highly related genes. All 3 amniotic fluid/placental isolates contain 3 genes that are absent in PG21 so we investigated these 3 genes further.
Figure 3. Whole genome comparison.
The chromosome of the amniotic fluid isolate AF1 (red) was used as the benchmark, and the chromosomes from the amniotic fluid/placental isolates AF3 (blue), PF5 (purple), and the rectal isolate PG21 (green) were compared to it. Genes that are absent within a given chromosome appear as gaps. The genes present in all three MIAC strains, but absent in PG21 (alr, goiB, goiC) appear as gaps exclusive to the green circle and are denoted with black arrows. A prophage that was exclusive to AF1 and absent in the other 3 strains in noted with a grey arrow. The membrane protein Lmp1 is present in PG21 but appears as a gap because it is in a different location on the chromosome, and it is noted with a grey arrow.
One of the genes that was in the amniotic fluid/placental isolates and absent in PG21 appears to encode alanine racemase (designated for the purposes of this study as alr). Alanine racemase converts L-alanine to D-alanine, a component of peptidoglycan. Mycoplasmas lack peptidoglycan but some species do encode alanine racemase. The function of this enzyme in mycoplasmas is unknown.
The second gene encodes a 379 amino acid protein (AF1_212, AF3_405, PL5_34) of unknown function (designated for the purposes of this study as gene of interest B, goiB). The protein encoded by goiB aligns over 97% of its length to a hypothetical protein from U. urealyticum (41% identity and 63% similarity). This protein appears to have a signal peptide and is predicted by the PSORTb bacterial subcellular localization prediction tool (Brinkman Laboratory, Simon Fraser University, British Columbia, Canada) to be secreted78. It was analyzed using Phyre2 (Structural Bioinformatics Group, Imperial College, London, UK), which modeled 46% of the amino acid residues with >90% confidence and the closest structural match was a putative c39-like peptidase (96.2% confidence)79. Therefore, the gene could encode a secreted protease. There are no predicted secreted proteases annotated in the PG21 genome.
The third gene (AF1_518, AF3_365, PL5_294) encodes a protein that is not similar to any proteins of known function (designated for the purposes of this study as gene of interest C, goiC). The region surrounding this gene is similar in strains AF1 and AF3 but portions of the region are absent in PL5 and PG21 (Figure 4). The amniotic fluid/placental isolates also contain multiple direct repeats in this region. All three amniotic fluid/placental strains exhibit 547 nt and 1,884 nt repeats and AF1 and AF3 also contain a 246 nt repeat and three additional DNA methyltransferase genes. The variable region is flanked by a conserved HAD hydrolase (MHO_3310 in PG21) and an ABC transporter (MHO_3250).
Figure 4. Genetic locus of M hominis goiC.
The variable locus surrounding the goiC gene is shown for all 4 strains. Strains AF1 and AF3 contained highly similar loci, with 3 sets of direct repeats (shown in the consensus map at the top as colored rectangles). PL5 lacked one set of repeats and 10 of the genes present in AF1 and AF3. The PG21 genome lacked all 3 sets of repeats and goiC.
All of the genes that were unique to one or more amniotic fluid/placental isolates are listed in Table 2. We also analyzed and compared the lengths of all genes within each strain to facilitate the detection of truncations that might result in loss of protein function. Aside from the membrane protein-encoding genes, we did not detect any genes that were truncated in all three amniotic fluid/placental strains. However, there were 4 genes that encoded longer proteins in the amniotic fluid/placental strains than in PG21, suggesting possible loss of function of these proteins in PG21. Two of these were annotated by RAST as hypothetical proteins and two are likely involved in transport (Table 2).
Table 2.
Putative genes found exclusively in one or more strains of M. hominis isolated from cases of intra-amniotic infection
| Gene identifiera | Putative function | Additional information |
|---|---|---|
| AF3_360 | Type II restriction enzyme | |
| PL5_435 | Vmp | |
| AF1_110, AF3_18 | Type II restriction enzyme | |
| AF1_210, AF3_363 | Type II restriction enzyme | |
| AF1_231, PL5_326 | Vmp | |
| AF1_511-AF1_517, AF3_358-AF3_364 | Hypothetical proteins | Not present in PL5 |
| AF1_23, AF3_18, PL5_476 | Alanine racemase | amniotic fluid/placental isolate gene A (alr)* |
| AF1_212, AF3_405, PL5_34 | Hypothetical protein from U. urealyticum (low similarity). | amniotic fluid/placental isolate gene B (goiB)* |
| AF1_518, AF3_365, PL5_284 | Hypothetical protein | amniotic fluid/placental isolate gene C (goiC)* |
| AF1_135, AF3_121, PL5_112, MHO_1580 | Hypothetical protein | Present in PG21 but truncated due to nonsense mutation |
| AF1_330, AF3_184, PL5_343 | Cation-transporting ATPase, E1-E2 family | Present in PG21 but truncated due to nonsense mutation (not annotated) |
| AF1_401, AF3_251, PL5_156, MHO_4360 | ABC transporter ATP- binding protein | Present in PG21 but truncated due to nonsense mutation |
| AF1_248 AF3_442 PL5_555 | Hypothetical protein | Present in PG21 but truncated due to nonsense mutation (not annotated) |
The strains harboring the gene are listed. Amino acid and nucleotide sequences corresponding to the gene identifiers can be found through NCBI (Accession numbers CP009677, JRWZ00000000, and JRXA00000000).
The presence of these genes in vaginal swab samples and amniotic fluid samples was determined in this study.
Association of specific genes with preterm labor and delivery
We performed 16S gene surveys on DNA extracted from vaginal swab samples from 58 pregnant subjects, 10 of whom gave birth preterm and 48 of whom gave birth after >=37 weeks gestation. Of the 58 subjects, 37 (64%) were self-reported African American, 8 were self-reported Caucasian (14%), 11 were self-reported Hispanic (19%), and 2 reported more than one ethnicity (3%). M. hominis was detected in vaginal swabs of 17 of the subjects, 15 of whom were African American (88%) and 2 were Hispanic (12%). Of the 10 preterm births, 4 were positive for M. hominis, and of the 48 of the full-term births, 13 were positive for M. hominis. There was no significant difference in the frequency of detection of M. hominis and preterm birth (Fisher’s exact test P=0.46).
We analyzed DNA extracted from the 17 samples that contained M. hominis by quantitative PCR using 16S ribosomal RNA gene-specific primers to determine the approximate numbers of M. hominis present per mL vaginal fluid. We also analyzed the samples for the presence of alr, goiB, and goiC. The results are shown in Table 3. Two of the 4 preterm samples and 5 of the 13 full-term vaginal samples contained alr, but presence of this gene was not associated with preterm birth (Fisher’s exact test P=1). PCR revealed that 3 out of the 4 M. hominis–positive samples from preterm deliveries contained goiB whereas 6 out of 13 of the full-term pregnancies contained the gene and this association did not reach statistical significance (Fisher’s exact test P>0.05). Interestingly, PCR revealed that 4 out of the 4 samples from preterm pregnancies with M. hominis contained goiC whereas only 4 out of 13 of the samples from full-term pregnancies contained the gene. Accordingly, there was a statistically significant association between presence of the gene in the vaginal samples and preterm birth (Fisher’s exact test P<0.05).
Table 3.
The presence of M. hominis genes specific to amniotic fluid/placental isolates in M. hominis present in vaginal samples
| Vaginal sample # |
GA at deliverya |
Ethnicityb | Agec | Ruptured | CFU/ml (SD)e |
alrf | goiBf | goiCf |
|---|---|---|---|---|---|---|---|---|
| 1 | 34 | AA | 36 | No | 10,608 (722) | − | + | + |
| 2 | 34 | AA | 23 | No | 308,247 (49,687) | + | + | + |
| 3 | 34.2 | AA | 24 | Yes | 439,288 (17,654) | + | − | + |
| 4 | 35.4 | AA | 23 | ND | 2,637,620 (309,422) | − | + | + |
| 5 | 38 | AA | 26 | No | 85,841 (16662) | − | + | − |
| 6 | 38.2 | AA | 36 | No | 32,114 (720) | + | + | + |
| 10 | 38.5 | AA | 22 | No | 2,153 (128) | − | − | − |
| 7 | 39.0 | His | 26 | No | 151,221 (29,232) | − | − | − |
| 8 | 39.1 | His | 26 | No | 1,867 (261) | − | − | − |
| 9 | 39.2 | AA | 28 | No | 1,232 (198) | + | − | − |
| 11 | 39.3 | AA | 41 | No | 527 (26) | + | − | − |
| 12 | 39.3 | AA | 21 | No | 1726 (129) | − | − | − |
| 13 | 39.6 | AA | 19 | No | 310 (54) | + | + | + |
| 14 | 39.6 | AA | 23 | No | 981 (142) | − | + | − |
| 15 | 40.5 | AA | 21 | No | 3,950 (341) | + | + | + |
| 16 | 41 | AA | 26 | Yes | 64,711 (6,053) | − | − | + |
| 17 | 41 | AA | 26 | Yes | 32,595 (1,924) | − | + | − |
Gestational age at delivery (weeks.days).
Self-reported maternal ethnicity.
Self-reported maternal age.
Ruptured membranes noted at time of sampling.
Estimated number of M. hominis per mL vaginal fluid as determined by 16S ribosomal RNA gene qPCR (standard deviation).
Presence (+) or absence (−) of the amniotic fluid/placental isolate genes A, B, or C (alr, goiB, goiC) as determined by realtime PCR.
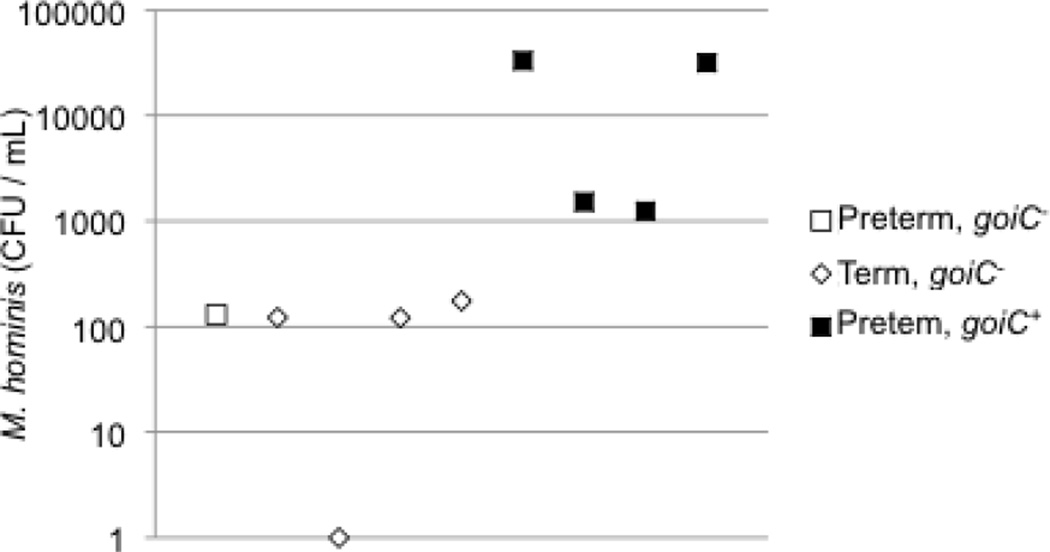
The numbers of M. hominis per mL and presence of the 3 genes found only in the amniotic fluid/placental isolates were also determined in 14 amniotic fluid samples (Table 4). The samples were selected from a larger study based on presence of M. hominis by culture and gestational age at delivery27. Of the samples, 9 contained M. hominis by culture. Five of the pregnancies with M. hominis-positive amniotic fluid resulted in term deliveries and these samples were selected as controls for association between the 3 genes found in the amniotic fluid/placental isolates and preterm birth. None of the amniotic fluid samples tested positive by PCR for goiB. The alanine racemase gene (alr) was detected in 4 of the 9 samples that were culture-positive for M. hominis but did not correlate significantly with preterm birth (two tailed P=1 Fisher’s exact test) or bacterial burden. However, goiC was significantly associated (two tailed P<0.05 Fisher’s exact test) with preterm birth. The gene was also significantly associated with bacterial burden, and all of the samples estimated to contain at least 1,000 bacteria per mL were positive for goiC (P<.01) (Figure 5). There was no association between bacterial load and presence of goiC in vaginal samples, suggesting that the gene may be more likely to contribute to colonization and/or growth on placenta or in amniotic fluid rather than the vagina.
Table 4.
Presence of M. hominis genes found in amniotic fluid/placental isolates in a different set of amniotic fluid samples
| AMNIOTIC FLUIDa | GA Amniob |
GA deliveryc |
16S CFU/ml (SD)d |
arle | goiBe | goiCe |
|---|---|---|---|---|---|---|
| Positive M. hominis culture | ||||||
| Preterm | 30.0 | 30.2 | 130(41) | + | − | − |
| 28.2 | 28.2 | 33,056 (2462) | + | − | + | |
| 24.3 | 24.5 | 1,516 (140) | − | − | + | |
| 33.6 | 33.6 | 1,242 (263) | − | − | + | |
| 30.0 | 30.0 | 31,584 (1981) | − | − | + | |
| Term | 34.6 | 38.6 | 122(37) | + | − | − |
| 28.3 | 38.4 | − | + | − | − | |
| 34.4 | 39.0 | 121(22) | − | − | − | |
| 33.0 | 37.0 | 175(50) | − | − | − | |
| Negative M. hominis culture | ||||||
| Term | 31.3 | 38.5 | − | − | − | − |
| 34.2 | 38.5 | − | − | − | − | |
| 31.2 | 38.5 | − | − | − | − | |
| 25.4 | 30.5 | − | + | − | − | |
| 33.6 | 34.2 | − | − | − | − | |
Amniotic fluid samples were culture positive or negative for M. hominis and the women delivered at term or preterm.
Gestational age at the time of amniocentesis (weeks.days).
Gestational age at the time of delivery (weeks.days).
Estimated number of M. hominis per mL vaginal fluid as determined by 16S rRNA gene qPCR (standard deviation).
Presence (+) or absence (−) of the amniotic fluid/placental isolate genes arl, goiB, and goiC as determined by realtime PCR.
Figure 5. M hominis goiC is associated with preterm delivery and bacterial burden.
The y-axis (logarithmic scale) represents the microbial burden in amniotic fluid in colony forming units per mL amniotic fluid. The open square represents amniotic fluid from a case of spontaneous preterm labor and preterm birth in which the M. hominis goiC gene was absent. The open diamonds represent amniotic fluid from cases of spontaneous preterm labor and term birth in which goiC was absent. The black squares represent amniotic fluid from cases of spontaneous preterm labor and preterm birth in which goiC gene was present.
Because infections of fetal membranes and amniotic fluid are often polymicrobial, we reasoned that goiC could potentially serve a competitive fitness function in this niche. To test this, we analyzed the vaginal 16S rRNA profiles for significant positive and negative associations between goiC and other bacterial species. There was no association between goiC and overall alpha diversity (goiC present=2.73±0.489, goiC absent=1.77±0.259, P=0.107). However, LefSe analysis suggested a positive association between the presence of goiC and the genera Microaerophilus and Dialister and an association between the absence of goiC and the genus Ureaplasma.
Comment
Principal findings of this study
In an effort to find M. hominis virulence determinants involved in amniotic invasion, we sequenced three amniotic fluid/placental isolates from pregnancies with episodes of spontaneous preterm labor resulting in preterm births. We compared the genomic sequences to that of the only previously sequenced and publicly available reference strain, PG21, a rectal isolate from a healthy individual, in an attempt to find genes unique to the amniotic fluid/placental strains. We found several genotypic differences between the amniotic fluid/placental isolates and PG21. We identified a gene that appears to be significantly associated with preterm birth. The gene was not absolutely required for invasion of the amniotic cavity as strains without the gene were detected in amniotic fluid. However, presence of the gene was associated with bacterial burden in the amniotic fluid suggesting that it could play a role in survival or fitness in this niche. This observation is clinically relevant because the bacterial load of mycoplasmas in amniotic fluid correlates with histologic chorioamnionitis80, 81.
Genetic differences detected in amniotic fluid/placental isolates
Very little is currently known about the pathogenesis of MIAC, but in order for a bacterium in the vagina to invade the amniotic cavity, it would presumably need to colonize the vagina, ascend through the cervix to the uterine cavity, colonize the uterine cavity, and evade immune defenses and potentially other, antagonistic bacterial species. In order to cause amnionitis, it would also need to traverse the fetal membranes or placenta, and survive and grow in nutrient-poor, iron-limited amniotic fluid. Presumably, to induce preterm labor, it would then need to elicit a maternal and/or fetal inflammatory response. Randis et al recently investigated the role for the GBS β-hemolysin/cytolysin in the pathogenesis of ascending GBS infection using a mouse model82. They found that the toxin played a role in vaginal colonization and in inducing inflammation, host tissue damage, and preterm birth and/or fetal demise, but was not required for ascending infection. In this study we found a gene that was associated with bacterial burden in amniotic fluid and with preterm birth but was not required for amnionitis. It was not significantly associated with presence of bacterial load of M. hominis in the vagina, suggesting that it does not play a role in survival or fitness in the vagina. Therefore, it may be more likely that the product of this gene is involved in bacterial survival and/or growth on placenta and in the amniotic cavity rather than being required for ascension or invasion. Although the sample size was very small, absence of goiC correlated with the presence of Ureaplasma in the vagina. This is particularly interesting because Ureaplasma and Mycoplasma are frequently detected together in amnionitis. Therefore, goiC could confer a survival or growth advantage upon M. hominis in the face of competition from Ureaplasma. Of note, we also found that the two isolates from amniotic fluid are more similar to each other than they are to the placental isolate, suggesting that these two strains may have a greater genetic potential for traversing fetal membranes or surviving in amniotic fluid. We also detected differences in genes encoding surface-associated lipoproteins that have been previously implicated in adherence to host cells and colonization.
We noted that upon liquid culture, the amniotic fluid/placental strains appeared dispersed, whereas PG21 appeared flocculated and adhered to the plastic culture vessel, suggesting a variation on the bacterial surface. Variation in the sequence and structure of surface lipoproteins likely contributes to the avidity of the bacteria for host cells and tissues, and therefore, may be associated with disease outcome83. As expected, we found considerable variation in these genes. The genes encoding an Lmp-like protein and Lmp1 are more similar among the amniotic fluid/placental strains than they are to homologs in PG21. These lipoproteins could contribute to adherence to host cells in some way that promotes ascension from the lower to the upper genitourinary tract or colonization of the uterine cavity.
Clinical implications
We hypothesized that a particular gene or set of genes are involved in invasion of the amniotic cavity and that these gene/s may be absent or genotypically nonfunctional in strains that colonize the vagina but fail to invade. Such a finding would be clinically relevant because genetic identification of bacteria capable of invading the amniotic cavity and eliciting preterm labor could result in earlier detection and improvements in therapeutic intervention. Furthermore, identification of gene/s that are involved in invasion of the amniotic cavity could contribute to a better understanding of the role of this bacterial species in preterm birth.
Limitations of the study
Many strain-to-strain differences cannot be easily detected by comparative genomic sequence analysis. Complex interactions between proteins and regulatory pathways can be difficult to predict, meaning that the loss of one gene can affect the expression of other genes or the function of other proteins. Furthermore, gene regulation at the transcriptional and post-transcriptional levels can influence phenotype. Finally, changes that are more subtle than gene deletions, such as polymorphisms or missense mutations can lead to unpredicted changes in protein function. Therefore, other differences could exist between PG21 and the amniotic fluid/placental strains that were not detected in this study that could be responsible for microbial burden and preterm birth. The small sample size of the sequenced strains (n=3), and the relatively small size of the amniotic fluid samples (n=14) also limit the study.
Future directions
The role of goiC in uterine invasion, MIAC, and spontaneous preterm birth should be confirmed using a larger sample size. From this study it appears that it might be involved in survival and growth on placenta and in the amniotic cavity rather than invasion of this site, however, elucidation of its function and role will require further study.
Very little is known about the role of bacteriophage in M. hominis biology and pathogenesis84, 85. Thus, the role of bacteriophage in pathogenic strains of M. hominis deserves further examination.
Differences in outer surface proteins were also noted in this study. These differences could affect adherence to epithelial cells or other components of the chorioamnion or they could affect resistance to host immune defenses. Further study to confirm that all amniotic fluid/placental strains share similar outer membrane proteins and analysis of their role in adherence and immune evasion will be required to address these knowledge gaps.
Conclusion
Sequence analysis of M. hominis isolates from amniotic fluid and placenta identified a gene that is significantly associated with M. hominis density in amniotic fluid and with preterm birth. This gene may contribute to the pathogenic potential of M. hominis.
Acknowledgments
Financial support: This research was supported by the Perinatology Research Branch, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services (NICHD/NIH) and National Institutes of Health grants 4UH3AI083263, “The Vaginal Microbiome: Disease, Genetics and the Environment”, P60 MD002256, the “VCU NIMHD Comprehensive Center of Excellence”, U54 DE023786-01 “A Multi-’omic Analysis of the Vaginal Microbiome during Pregnancy”, and AI 31871.
Footnotes
Disclosure statement: The authors report no conflict of interest.
References
- 1.Thorsen P, Jensen IP, Jeune B, et al. Few microorganisms associated with bacterial vaginosis may constitute the pathologic core: a population-based microbiologic study among 3596 pregnant women. Am J Obstet Gynecol. 1998;178:580–587. doi: 10.1016/s0002-9378(98)70442-9. [DOI] [PubMed] [Google Scholar]
- 2.Pereyre S, Sirand-Pugnet P, Beven L, et al. Life on arginine for Mycoplasma hominis: clues from its minimal genome and comparison with other human urogenital mycoplasmas. PLoS Genet. 2009;5:e1000677. doi: 10.1371/journal.pgen.1000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kharsany AB, Hoosen AA, Moodley J. Bacterial vaginosis and lower genital tract infections in women attending out-patient clinics at a tertiary institution serving a developing community. J Obstet Gynaecol. 1997;17:171–175. doi: 10.1080/01443619750113807. [DOI] [PubMed] [Google Scholar]
- 4.Aaltone R, Jalava J, Laurikainen E, Karkkainen U, Alanen A. Cervical Ureaplasma urealyticum colonization: comparison of PCR and culture for its detection and association with preterm birth. Scand J Infect Dis. 2002;34:35–40. doi: 10.1080/00365540110077074. [DOI] [PubMed] [Google Scholar]
- 5.Breugelmans M, Vancutsem E, Naessens A, Laubach M, Foulon W. Association of abnormal vaginal flora and Ureaplasma species as risk factors for preterm birth: a cohort study. Acta Obstet Gynecol Scand. 2010;89:256–260. doi: 10.3109/00016340903418769. [DOI] [PubMed] [Google Scholar]
- 6.Goldenberg RL, Andrews WW, Goepfert AR, et al. The Alabama Preterm Birth Study: umbilical cord blood Ureaplasma urealyticum and Mycoplasma hominis cultures in very preterm newborn infants. Am J Obstet Gynecol. 2008;198:43. doi: 10.1016/j.ajog.2007.07.033. e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kataoka S, Yamada T, Chou K, et al. Association between preterm birth and vaginal colonization by mycoplasmas in early pregnancy. J Clin Microbiol. 2006;44:51–55. doi: 10.1128/JCM.44.1.51-55.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsen B, Hwang J. Mycoplasma, Ureaplasma, and adverse pregnancy outcomes: a fresh look. Infect Dis Obstet Gynecol. 2010;2010 doi: 10.1155/2010/521921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwak DW, Hwang HS, Kwon JY, Park YW, Kim YH. Co-infection with vaginal Ureaplasma urealyticum and Mycoplasma hominis increases adverse pregnancy outcomes in patients with preterm labor or preterm premature rupture of membranes. J Matern Fetal Neonatal Med. 2014;27:333–337. doi: 10.3109/14767058.2013.818124. [DOI] [PubMed] [Google Scholar]
- 10.Oh KJ, Lee KA, Sohn YK, et al. Intraamniotic infection with genital mycoplasmas exhibits a more intense inflammatory response than intraamniotic infection with other microorganisms in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 2010;203:211. doi: 10.1016/j.ajog.2010.03.035. e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen A, Srinivasan U, Goldberg D, et al. Selected vaginal bacteria and risk of preterm birth: an ecological perspective. J Infect Dis. 2014;209:1087–1094. doi: 10.1093/infdis/jit632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat G, Peltier MR, Syed TA, Drobek CO, Saade G, Menon R. Fetal membrane biomarker network diversity and disease functions induced by intra-amniotic pathogens. Am J Reprod Immunol. 2013;69:124–133. doi: 10.1111/aji.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marconi C, de Andrade Ramos BR, Peracoli JC, Donders GG, da Silva MG. Amniotic fluid interleukin-1 beta and interleukin-6, but not interleukin-8 correlate with microbial invasion of the amniotic cavity in preterm labor. Am J Reprod Immunol. 2011;65:549–556. doi: 10.1111/j.1600-0897.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 14.Taylor-Robinson D, Lamont RF. Mycoplasmas in pregnancy. BJOG. 2011;118:164–174. doi: 10.1111/j.1471-0528.2010.02766.x. [DOI] [PubMed] [Google Scholar]
- 15.Massaro G, Scaravilli G, Simeone S, et al. Interleukin-6 and Mycoplasma hominis as markers of preterm birth and related brain damage: our experience. J Matern Fetal Neonatal Med. 2009;22:1063–1067. doi: 10.3109/14767050903026473. [DOI] [PubMed] [Google Scholar]
- 16.Hillier SL, Nugent RP, Eschenbach DA, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 17.Doyle RM, Alber DG, Jones HE, et al. Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta. 2014;35:1099–1101. doi: 10.1016/j.placenta.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Fichorova RN, Beatty N, Sassi RR, Yamamoto HS, Allred EN, Leviton A. Systemic Inflammation in the Extremely Low Gestational Age Newborn Following Maternal Genitourinary Infections. Am J Reprod Immunol. 2014 doi: 10.1111/aji.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 20.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DiGiulio DB, Romero R, Kusanovic JP, et al. Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes. Am J Reprod Immunol. 2010;64:38–57. doi: 10.1111/j.1600-0897.2010.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobitt JR, Ledger WJ. Amniotic fluid analysis. Its role in maternal neonatal infection. Obstet Gynecol. 1978;51:56–62. [PubMed] [Google Scholar]
- 23.Miller JM, Jr, Pupkin MJ, Hill GB. Bacterial colonization of amniotic fluid from intact fetal membranes. Am J Obstet Gynecol. 1980;136:796–804. doi: 10.1016/0002-9378(80)90458-5. [DOI] [PubMed] [Google Scholar]
- 24.Wallace RL, Herrick CN. Amniocentesis in the evaluation of premature labor. Obstet Gynecol. 1981;57:483–486. [PubMed] [Google Scholar]
- 25.Bobitt JR, Hayslip CC, Damato JD. Amniotic fluid infection as determined by transabdominal amniocentesis in patients with intact membranes in premature labor. Am J Obstet Gynecol. 1981;140:947–952. doi: 10.1016/0002-9378(81)90090-9. [DOI] [PubMed] [Google Scholar]
- 26.Wahbeh CJ, Hill GB, Eden RD, Gall SA. Intra-amniotic bacterial colonization in premature labor. Am J Obstet Gynecol. 1984;148:739–743. doi: 10.1016/0002-9378(84)90558-1. [DOI] [PubMed] [Google Scholar]
- 27.Romero R, Sirtori M, Oyarzun E, et al. Infection and labor. V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol. 1989;161:817–824. doi: 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- 28.Hameed C, Tejani N, Verma UL, Archbald F. Silent chorioamnionitis as a cause of preterm labor refractory to tocolytic therapy. Am J Obstet Gynecol. 1984;149:726–730. doi: 10.1016/0002-9378(84)90111-x. [DOI] [PubMed] [Google Scholar]
- 29.Gravett MG, Hummel D, Eschenbach DA, Holmes KK. Preterm labor associated with subclinical amniotic fluid infection and with bacterial vaginosis. Obstet Gynecol. 1986;67:229–237. doi: 10.1097/00006250-198602000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Skoll MA, Moretti ML, Sibai BM. The incidence of positive amniotic fluid cultures in patients preterm labor with intact membranes. Am J Obstet Gynecol. 1989;161:813–816. doi: 10.1016/0002-9378(89)90407-9. [DOI] [PubMed] [Google Scholar]
- 31.Watts DH, Krohn MA, Hillier SL, Eschenbach DA. The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet Gynecol. 1992;79:351–357. doi: 10.1097/00006250-199203000-00005. [DOI] [PubMed] [Google Scholar]
- 32.Coultrip LL, Grossman JH. Evaluation of rapid diagnostic tests in the detection of microbial invasion of the amniotic cavity. Am J Obstet Gynecol. 1992;167:1231–1242. doi: 10.1016/s0002-9378(11)91694-9. [DOI] [PubMed] [Google Scholar]
- 33.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science (New York, N Y ) 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Combs CA, Gravett M, Garite TJ, et al. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol. 2014;210:125. doi: 10.1016/j.ajog.2013.11.032. e1–125 e15. [DOI] [PubMed] [Google Scholar]
- 35.Hofer N, Kothari R, Morris N, Muller W, Resch B. The fetal inflammatory response syndrome is a risk factor for morbidity in preterm neonates. Am J Obstet Gynecol. 2013;209:542. doi: 10.1016/j.ajog.2013.08.030. e1–542 e11. [DOI] [PubMed] [Google Scholar]
- 36.Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G. Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth. Am J Obstet Gynecol. 2004;190:1509–1519. doi: 10.1016/j.ajog.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Kemp MW, Miura Y, Payne MS, et al. Repeated maternal intramuscular or intraamniotic erythromycin incompletely resolves intrauterine Ureaplasma parvum infection in a sheep model of pregnancy. Am J Obstet Gynecol. 2014;211:134. doi: 10.1016/j.ajog.2014.02.025. e1–9. [DOI] [PubMed] [Google Scholar]
- 38.Grigsby PL, Novy MJ, Sadowsky DW, et al. Maternal azithromycin therapy for Ureaplasma intraamniotic infection delays preterm delivery and reduces fetal lung injury in a primate model. Am J Obstet Gynecol. 2012;207:475. doi: 10.1016/j.ajog.2012.10.871. e1–475 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont RF, Nhan-Chang CL, Sobel JD, Workowski K, Conde-Agudelo A, Romero R. Treatment of abnormal vaginal flora in early pregnancy with clindamycin for the prevention of spontaneous preterm birth: a systematic review and metaanalysis. Am J Obstet Gynecol. 2011;205:177–190. doi: 10.1016/j.ajog.2011.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mercer BM, Crouse DT, Goldenberg RL, et al. The antibiotic treatment of PPROM study: systemic maternal and fetal markers and perinatal outcomes. Am J Obstet Gynecol. 2012;206:145. doi: 10.1016/j.ajog.2011.08.028. e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basraon SK, Menon R, Makhlouf M, et al. Can statins reduce the inflammatory response associated with preterm birth in an animal model? Am J Obstet Gynecol. 2012;207:224. doi: 10.1016/j.ajog.2012.06.020. e1–7. [DOI] [PubMed] [Google Scholar]
- 42.Romero R, Garite TJ. Twenty percent of very preterm neonates (23–32 weeks of gestation) are born with bacteremia caused by genital Mycoplasmas. Am J Obstet Gynecol. 2008;198:1–3. doi: 10.1016/j.ajog.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 43.Donders GG, Van Calsteren K, Bellen G, et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG. 2009;116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- 44.Prince AL, Antony KM, Chu DM, Aagaard KM. The microbiome, parturition, and timing of birth: more questions than answers. J Reprod Immunol. 2014 doi: 10.1016/j.jri.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boggess KA, Moss K, Madianos P, Murtha AP, Beck J, Offenbacher S. Fetal immune response to oral pathogens and risk of preterm birth. Am J Obstet Gynecol. 2005;193:1121–1126. doi: 10.1016/j.ajog.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 46.Boggess KA. Treatment of localized periodontal disease in pregnancy does not reduce the occurrence of preterm birth: results from the Periodontal Infections and Prematurity Study (PIPS) Am J Obstet Gynecol. 2010;202:101–102. doi: 10.1016/j.ajog.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 47.Macones GA, Parry S, Nelson DB, et al. Treatment of localized periodontal disease in pregnancy does not reduce the occurrence of preterm birth: results from the Periodontal Infections and Prematurity Study (PIPS) Am J Obstet Gynecol. 2010;202:147. doi: 10.1016/j.ajog.2009.10.892. e1–8. [DOI] [PubMed] [Google Scholar]
- 48.Polyzos NP, Polyzos IP, Mauri D, et al. Effect of periodontal disease treatment during pregnancy on preterm birth incidence: a metaanalysis of randomized trials. Am J Obstet Gynecol. 2009;200:225–232. doi: 10.1016/j.ajog.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 49.Jeffcoat M, Parry S, Gerlach RW, Doyle MJ. Use of alcohol-free antimicrobial mouth rinse is associated with decreased incidence of preterm birth in a high-risk population. Am J Obstet Gynecol. 2011;205:382. doi: 10.1016/j.ajog.2011.07.016. e1–6. [DOI] [PubMed] [Google Scholar]
- 50.Xiong X, Buekens P, Goldenberg RL, Offenbacher S, Qian X. Optimal timing of periodontal disease treatment for prevention of adverse pregnancy outcomes: before or during pregnancy? Am J Obstet Gynecol. 2011;205:111. doi: 10.1016/j.ajog.2011.03.017. e1–6. [DOI] [PubMed] [Google Scholar]
- 51.Choi SJ, Park SD, Jang IH, Uh Y, Lee A. The prevalence of vaginal microorganisms in pregnant women with preterm labor and preterm birth. Ann Lab Med. 2012;32:194–200. doi: 10.3343/alm.2012.32.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Capoccia R, Greub G, Baud D. Ureaplasma urealyticum, Mycoplasma hominis and adverse pregnancy outcomes. Curr Opin Infect Dis. 2013;26:231–240. doi: 10.1097/QCO.0b013e328360db58. [DOI] [PubMed] [Google Scholar]
- 53.Foxman B, Wen A, Srinivasan U, et al. Mycoplasma, bacterial vaginosis-associated bacteria BVAB3, race, and risk of preterm birth in a high-risk cohort. Am J Obstet Gynecol. 2014;210:226. doi: 10.1016/j.ajog.2013.10.003. e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perni SC, Vardhana S, Korneeva I, et al. Mycoplasma hominis and Ureaplasma urealyticum in midtrimester amniotic fluid: association with amniotic fluid cytokine levels and pregnancy outcome. Am J Obstet Gynecol. 2004;191:1382–1386. doi: 10.1016/j.ajog.2004.05.070. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen DP, Gerber S, Hohlfeld P, Sandrine G, Witkin SS. Mycoplasma hominis in mid-trimester amniotic fluid: relation to pregnancy outcome. J Perinat Med. 2004;32:323–326. doi: 10.1515/JPM.2004.060. [DOI] [PubMed] [Google Scholar]
- 56.Carroll SG, Papaioannou S, Ntumazah IL, Philpott-Howard J, Nicolaides KH. Lower genital tract swabs in the prediction of intrauterine infection in preterm prelabour rupture of the membranes. Br J Obstet Gynaecol. 1996;103:54–59. doi: 10.1111/j.1471-0528.1996.tb09515.x. [DOI] [PubMed] [Google Scholar]
- 57.Vouga M, Greub G, Prod’hom G, et al. Treatment of genital mycoplasma in colonized pregnant women in late pregnancy is associated with a lower rate of premature labour and neonatal complications. Clin Microbiol Infect. 2014;20:1074–1079. doi: 10.1111/1469-0691.12686. [DOI] [PubMed] [Google Scholar]
- 58.Reuter S, Connor TR, Barquist L, et al. Parallel independent evolution of pathogenicity within the genus Yersinia . Proc Natl Acad Sci U S A. 2014;111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agarwal J, Srivastava S, Singh M. Pathogenomics of uropathogenic Escherichia coli . Indian J Med Microbiol. 2012;30:141–149. doi: 10.4103/0255-0857.96657. [DOI] [PubMed] [Google Scholar]
- 60.Whidbey C, Harrell MI, Burnside K, et al. A hemolytic pigment of Group B Streptococcus allows bacterial penetration of human placenta. J Exp Med. 2013;210:1265–1281. doi: 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hitti J, Krohn MA, Patton DL, et al. Amniotic fluid tumor necrosis factor-alpha and the risk of respiratory distress syndrome among preterm infants. Am J Obstet Gynecol. 1997;177:50–56. doi: 10.1016/s0002-9378(97)70437-x. [DOI] [PubMed] [Google Scholar]
- 62.Henrich B, Feldmann RC, Hadding U. Cytoadhesins of Mycoplasma hominis . Infect Immun. 1993;61:2945–2951. doi: 10.1128/iai.61.7.2945-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harwich MD, Jr, Serrano MG, Fettweis JM, et al. Genomic sequence analysis and characterization of Sneathia amnii sp. nov. BMC Genomics. 2012;13(Suppl 8):S4. doi: 10.1186/1471-2164-13-S8-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overbeek R, Olson R, Pusch GD, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fettweis J, Alves J, Borzelleca J, et al. The Vaginal Microbiome: Disease, Genetics and the Environment. [Accessed 02/26/15];Nature Preceedings. 2011 Available at: http://dx.doi.org/10.1038/npre.2011.5150.2. [Google Scholar]
- 69.Fettweis JM, Serrano MG, Sheth NU, et al. Species-level classification of the vaginal microbiome. BMC Genomics. 2012;13(Suppl 8):S17. doi: 10.1186/1471-2164-13-S8-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roske K, Calcutt MJ, Wise KS. The Mycoplasma fermentans prophage phiMFV1: genome organization, mobility and variable expression of an encoded surface protein. Mol Microbiol. 2004;52:1703–1720. doi: 10.1111/j.1365-2958.2004.04087.x. [DOI] [PubMed] [Google Scholar]
- 73.Ladefoged SA, Jensen LT, Brock B, Birkelund S, Christiansen G. Analysis of 0.5-kilobase-pair repeats in the Mycoplasma hominis lmp gene system and identification of gene products. J Bacteriol. 1996;178:2775–2784. doi: 10.1128/jb.178.10.2775-2784.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boesen T, Emmersen J, Baczynska A, Birkelund S, Christiansen G. The vaa locus of Mycoplasma hominis contains a divergent genetic islet encoding a putative membrane protein. BMC Microbiol. 2004;4:37. doi: 10.1186/1471-2180-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Q, Wise KS. Molecular basis of size and antigenic variation of a Mycoplasma hominis adhesin encoded by divergent vaa genes. Infect Immun. 1996;64:2737–2744. doi: 10.1128/iai.64.7.2737-2744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mygind T, Birkelund S, Christiansen G. Characterization of the variability of a 75-kDa membrane protein in Mycoplasma hominis . FEMS Microbiol Lett. 2000;190:167–176. doi: 10.1111/j.1574-6968.2000.tb09281.x. [DOI] [PubMed] [Google Scholar]
- 77.Nyvold C, Birkelund S, Christiansen G. The Mycoplasma hominis P120 membrane protein contains a 216 amino acid hypervariable domain that is recognized by the human humoral immune response. Microbiology. 1997;143(Pt 2):675–688. doi: 10.1099/00221287-143-2-675. [DOI] [PubMed] [Google Scholar]
- 78.Yu NY, Wagner JR, Laird MR, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 80.Kacerovsky M, Pliskova L, Bolehovska R, et al. The microbial load with genital mycoplasmas correlates with the degree of histologic chorioamnionitis in preterm PROM. Am J Obstet Gynecol. 2011;205:213. doi: 10.1016/j.ajog.2011.04.028. e1–7. [DOI] [PubMed] [Google Scholar]
- 81.Kacerovsky M, Pliskova L, Menon R, et al. Microbial load of umbilical cord blood Ureaplasma species and Mycoplasma hominis in preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med. 2014 doi: 10.3109/14767058.2014.887068. [DOI] [PubMed] [Google Scholar]
- 82.Randis TM, Gelber SE, Hooven TA, et al. Group B Streptococcus beta-hemolysin/Cytolysin Breaches Maternal-Fetal Barriers to Cause Preterm Birth and Intrauterine Fetal Demise in Vivo. J Infect Dis. 2014;210:265–273. doi: 10.1093/infdis/jiu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jansson E, Backman A, Hakkarainen K, von Bonsdorff CH, Seniusova B, Miettinen A. Isolation and preliminary characterisation of mycoplasmavirus 20-P. Med Biol. 1982;60:116–120. [PubMed] [Google Scholar]
- 85.Robertson J, Gomersall M, Gill P. Virus-like particles in Mycoplasma hominis. Can J Microbiol. 1972;18:1971–1972. doi: 10.1139/m72-307. [DOI] [PubMed] [Google Scholar]