Abstract
The glycome constitutes the entire complement of free carbohydrates and glycoconjugates expressed on whole cells or tissues. ‘Systems Glycobiology’ is an emerging discipline that aims to quantitatively describe and analyse the glycome. Here, instead of developing a detailed understanding of single biochemical processes, a combination of computational and experimental tools are used to seek an integrated or ‘systems-level’ view. This can explain how multiple biochemical reactions and transport processes interact with each other to control glycome biosynthesis and function. Computational methods in this field commonly build in silico reaction network models to describe experimental data derived from structural studies that measure cell-surface glycan distribution. While considerable progress has been made, several challenges remain due to the complex and heterogeneous nature of this post-translational modification. First, for the in silico models to be standardized and shared among laboratories, it is necessary to integrate glycan structure information and glycosylation-related enzyme definitions into the mathematical models. Second, as glycoinformatics resources grow, it would be attractive to utilize ‘Big Data’ stored in these repositories for model construction and validation. Third, while the technology for profiling the glycome at the whole-cell level has been standardized, there is a need to integrate mass spectrometry derived site-specific glycosylation data into the models. The current review discusses progress that is being made to resolve the above bottlenecks. The focus is on how computational models can bridge the gap between ‘data’ generated in wet-laboratory studies with ‘knowledge’ that can enhance our understanding of the glycome.
INTRODUCTION
Glycosylation is one of the most ubiquitous and complex post-translational modifications in nature 1–3. A majority of secreted and cell-surface mammalian proteins bear at least one attached glycan (or carbohydrate structure). These macromolecules either absolutely control or finely-tune a very large number of diverse cellular processes in higher organisms 4. Studies of the glycome (collection of all glycans in a system) are also more complex compared to investigations of the genome and proteome since the number of natural monosaccharides and its variants across different species far exceeds the limited repertoire of naturally occurring nucleotides and amino acids. Due to this diversity and the branched nature of glycans, a bewilderingly diverse set of carbohydrate structures can be synthesized by a single cell. To add to this complexity, glycan expression on a single protein is heterogeneous both in terms of whether a particular peptide site is glycosylated (macroheterogeneity) and also in terms of the distribution of different glycans at a single site (microheterogeneity). This heterogeneity is in part due to diversity in the available metabolites, glycosyltransferases (glycoTs)footnote 1 and glycosidases which vary across cell-types and also cell growth conditions. Finally, the same glycan attached to different protein scaffolds may have different functions, and this highlights the need to consider context and site-specific glycosylation in functional assays. Due to these complexities, the application of computational tools in addition to high-throughput experiments may provide a more complete, quantitative understanding of the cellular glycosylation process and the glycome.
Systems Glycobiology is an emerging research theme that aims to examine glycosylation from a ‘systems perspective’ 1. Instead of traditional biochemical methods that focus on developing a detailed understanding of single biochemical processes, this field aims to develop experimental and computational tools that can provide insight into how different biochemical reaction and transport processes interact with each other to condition the ‘emergent properties’ of the system (Figure 1). While mathematical modeling is not a necessary criterion for the development of systems based approaches, computational frameworks and data analysis tools can aid the interpretation of complex experimental data. Such quantitative approaches allow the mathematical testing of different hypotheses, and quantification of the input-output response of the system in the face of perturbation. In general, systems glycobiology may study all aspects of the carbohydrate life-cycle including (Fig. 1): i. The biosynthesis reactions which occur in the cytoplasm and nuclear compartments to make various sugar-nucleotide donors (e.g. UDP-Galactose), ii. glycosylation reactions that occur in the endoplasmic reticulum and Golgi cisternae to guide the biosynthesis of a variety of glycoconjugates, iii. transport processes that result in the sub-cellular localization of glycoconjugates in their functional compartments, and iv. salvage pathways that aid the recycling of glycoconjugates back to their basic building blocks.
Figure 1. Carbohydrate life-cycle.
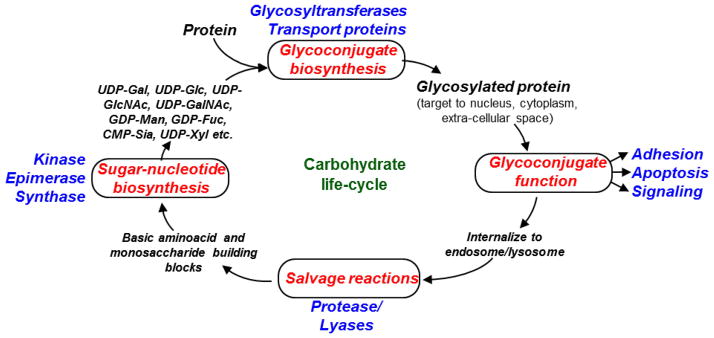
Carbohydrates are processed through various biosynthetic and degradative transformations in cells.
In terms of a ‘grand challenge’ for the field of Systems Glycobiology, one would ideally like to develop a computational framework where the glycoprotein and glycolipid profiles of an arbitrary cell could be quantitatively predicted based on limited experimental data at the gene, protein and/or functional level. Ideally, then, we would also like to predict a priori the outcome of specific cell/organism level perturbations without the need to perform corresponding wet-lab measurements. While this goal of collating data, information and knowledge is far from being achieved at the current time, efforts are being made to bridge the gap. This review article explains the basis for the biological complexity of the glycosylation machinery, summarizes areas were progress has been made, highlights current bottlenecks for the field, and describe directions that scientists are currently undertaking to address these shortcomings.
GLYCAN COMPLEXITY IN STRUCTURE AND FUNCTION
Glycans regulate a variety of fundamental biological processes. This includes structural features that control protein stability and conformation, and molecular recognition processes that control cellular binding. Such interactions are termed ‘intrinsic’ if they occur within or between cells of a single organism and ‘extrinsic’ if the molecular recognition involves external pathogenic microbes, agglutinins or toxins 5. For example, in the area of protein therapeutics, glycans attached to proteins have a dramatic effects on their half-life in circulation as exemplified by the case of erythropoietin (EPO) 6 and tissue plasminogen activator (tPA) 7. In these molecules, increasing the level of sialylation improves serum half-life by reducing clearance via the liver’s Ashwell-Morell receptor. In the case of IgG antibodies, the N-glycan structure at Asn-297 also regulates their efficacy, not in terms of altering antibody-antigen binding affinity but in terms of their binding to the Fcγ receptor expressed on immune cells and molecules of the complement pathway. Due to this, changes in glycan structures profoundly influence the efficacy of both antibody-dependent cellular toxicity (ADCC) and complement dependent cytotoxicity (CDC) 8. In the field of infectious diseases, glycosylation regulates host-virus interactions for a range of pathogens such as SARS-CoV, influenza, West Nile and Hendra 9. Here, the glycans regulate multiple steps during viral immune evasion and virulence including entry into host cell, proteolytic processing and protein trafficking. During immunity, glycans affect a number of steps including selectin dependent cell adhesion during immune cell trafficking, T-cell and B-cell receptor mediated signaling, lymphocyte development and a plethora of other signaling cascades 10. In the last example, cancer is accompanied by changes in cell-surface and secreted protein glycoconjugate structures. Due to such alterations, the unique cellular carbohydrate profiles of cancer cells serve not only as biomarkers or prognostic indicators of the disease, but it can also have functional impact on cell signaling, survival and metastasis 11, 12.
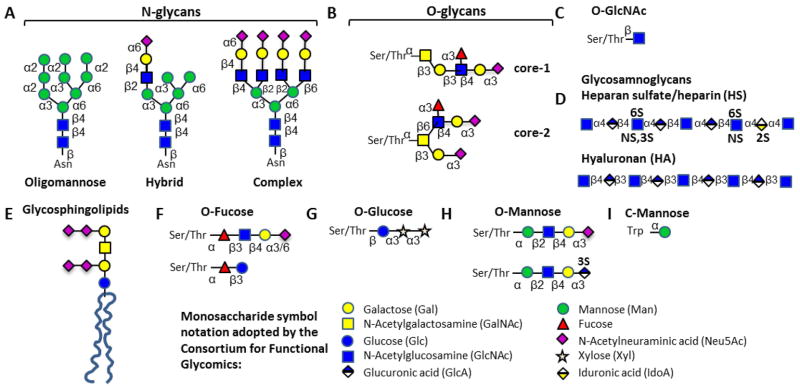
There is considerable structural diversity among the glycans expressed by mammals (Figure 2) 13. These macromolecules are principally divided into five categories: i. N-linked glycans, ii. O-linked or O-GalNAc (N-acetylgalactosamine) type glycans, iii. O-GlcNAc (N-acetylglucosamine) type glycans, iv. Proteoglycans, and v. glycosphingolipids (please see ref. 5, 14 for details). In this regard, most mammalian secreted and cell membrane proteins are commonly decorated by N- glycans that are attached to Asn residues in the sequon Asn-X-Ser/Thr (X ≠ Pro) and O-GalNAc linked glycans which appear at Ser and Thr. Here, the N-glycans are classified to be ‘oligomannose’ if all the antennae have terminal mannose (Fig. 2A). They are termed ‘complex’ if all antennae have GlcNAc extended chains, and ‘hybrid’ if they contain a mixture of terminal mannose and extended GlcNAc chains. The O-glycans appear in eight different core structures and these can be either extended/linear or branched. Fig 2B shows two examples of an extended core-1 glycan and a branched core-2 glycan containing the tetrasaccharide sialyl Lewis-X. Besides glycosylation on secreted proteins, the O-GlcNAc modification is one of the most abundant post-translational modifications in the nuclear and cytoplasmic compartments of metazoan cells, with more than 600 O-GlcNAcylated proteins identified to date 15 (Fig 2C). This is a terminal modification which is not further elaborated. In the case of proteoglycans, glycosaminoglycans (GAGs) containing repeating disaccharide units are attached to proteins via Xyl-O-Ser linkages (Fig. 2D). In the final category, glycosphingolipids or glycolipids are elaborated on lipid moieties called ceramides, a family of molecules containing long chain sphingosines in amide linkage with fatty acids. The first sugar linked to ceramide in higher animals is typically β-linked galactose in the case of the GalCer glycolipids and glucose for the GlcCers (Fig. 2E). In addition to the classical endoplasmic reticulum (ER)/Golgi derived O-glycans that are initiated by GalNAc, other important modifications have also been reported though these are less prevalent 3, 16. Notably, whereas fucose is typically observed at the terminal ends of glycans, more recently it has been observed to be O-linked to Ser/Thr on the peptide backbone (Fig. 2F). Such O-fucose modifications decorate and have functional impact on the EGF-like repeat domains of the Notch signaling receptor and thrombospondin (TSR) domains of various coagulation related proteins including members of the ADAMTS family. Coagulation factors like factor VII and factor IX, and also the EGF-like repeats of Notch are additionally decorated by O-glucose type chains (Fig. 2G). Additional unusual modifications include the O-mannose initiated chains on Ser/Thr that are common on the protein α-dystroglycan (Fig. 2H) and c-mannosylation on Trp residues of RNase 2 (Fig. 2I).
Figure 2. Glycan repertoire.
A variety of glycans with diverse structures are found in mammals. These include: A. N-linked glycans; B. O-GalNAc type O-linked glycans; C. O-GlcNAcylated glycans; D. Glycosaminoglycans; E. Glycosphingolipids; F–H. O-linked glycans initiated by Fucose (panel F), Glucose (G) or Mannose (H); and I. C-Mannosylated glycans attached to Trp. Note that only selected examples of each glycan class are shown.
Systems glycobiology aims to resolve the above complexity in structure and diversity in function with the goal of identifying key rate-limiting steps that regulate glycosylation pattern changes during the transition from the normal condition to disease states. Such critical steps may represent druggable targets for a range of pathologies.
CURRENT BOTTLENECKS FOR SYSTEMS GLYCOBIOLOGY
Whereas systems-based analysis have progressed relatively smoothly in studies involving signaling networks 17, metabolic processes 18 and physiological modeling 19, they are still gaining traction in the nascent field of Systems Glycobiology. This is potentially due to critical bottlenecks, both from the computational and analytical side. In particular:
There is no accepted standard for model building: A number of systems based models that simulate glycan biosynthesis have been developed over the last decade (reviewed in the next section). However, systematic model building has been lacking in this field since it is difficult to incorporate glycan structures and glycosylation-related enzyme specificity data into mathematical models. Further, few of the existing models are available in Systems Biology Markup Language (SBML) format 20. This limits the extent to which such computational models can be developed, shared and validated.
Glycoinformatics databases are under developed: A number of glycoscience-related databases have appeared in recent years. However, most of these repositories store glycan structure and taxonomy data, with only a limited amount of functional information. The development of systems-based models in the future will likely have greater reliance on these databases. Thus, the ability to relate carbohydrate structure with the specific enzymes that synthesize them, the rates of their synthesis and also their function will be key to future model building.
Insufficient quantitative data from glycoproteomics experiments: Two approaches are commonly used to measure the glycome. The first uses either enzymes or mild hydrolysis to separate the glycans from the peptide backbone. Following per-methylation of glycans, MS (mass spectrometry) analysis is performed to obtain semi-quantitative information regarding the composition and relative abundance of the carbohydrate structures 21. With the goal of gaining site-specific glycosylation information, which is lost in the above approach, the second method aims to analyze intact glycopeptides by adopting the conventional LC-MSn workflow that is common to the field of proteomics 22. The problem here is the lack of well-developed software and the complexity involved with the size of the search space since a single peptide may have many different sites of glycosylation and each site can bear a multitude of glycans. More sophisticated computational tools for glycoproteomics data analysis can accelerate systems-based model building and validation.
Below we provide a summary of ongoing developments to address each of the above challenges.
AN ENTITY-BASED MODELING FRAMEWORK
Several mathematical models of glycosylation have appeared in recent years (summarized in Table 1, detailed review in 23). Most of these focus on N-linked glycosylation pathways 24–31 with one of them also analyzing O-glycosylation 32. The various models of N-glycosylation were specifically developed to handle different aspects of the glycan biosynthesis process including the mechanism of N-glycosylation initiation 24, glycan branching via the action of different N-acetylglucosaminyl (GlcNAc)-transferases or GnT enzymes 25, 26, and chain extension by the action of galactosyltransferases, fucosyltransferases and sialyltransferases 27, 30, 31. While the primary focus of the N-glycosylation models is on protein production in the context of biotechnology, the model of O-glycosylation attempts to identify rate regulating steps controlling the kinetics of leukocyte adhesion to selectins in the context of human inflammation 32.
Table 1.
Network models of glycosylation
| Model description | Key outcome or conclusion | Reference |
|---|---|---|
| Model of N-glycosylation initiation | N-glycan macroheterogeneity is dependent on the relative rates of protein translation versus enzyme activity of oligosaccharyltransferase (OST) complex. | 24 |
| Model of N-glycan branching | Predicted distribution of complex-galactosylated glycoforms with varying numbers of antennae. Simulation results were consistent with recombinant protein biosynthesis data | 25 |
| Ultrasensitivity in the N-glycosylation branching pathway | Demonstrated ultrasensitivity in the N-glycan branching pathway due to: i) A sequential increase in KM among the N-glycan branching enzymes, and ii) Removal of intermediate products in this reaction pathway. | 26 |
| Modeling of N-glycan terminal fucosylation and sialylation | Compared model based heterogeneity predictions with experimental data for thrombopoietin expressed in CHO cells | 27 |
| Integration of the transcriptome and glycome for N-glycan modeling | Demonstrated that information from multiple datasets can be combined to better understand complex cellular processes | 28 |
| N-glycosylation models in Continuous Stirred Tank Reactors (CSTRs) and Plug Flow Reactors (PFRs) | Suggested that vesicular transport can be modeled as a series of CSTRs while Golgi maturation resembles a single plug-flow reactor (PFR) or a series of PFRs in series. | 29 |
| N-glycosylation Quality by design (QbD) simulations | Prediction of N-glycan microheterogeneity in a kinetic model that includes sugar-nucleotide biosynthesis/transport | 30, 31 |
| Model of O-linked glycosylation | Simulated glycan distribution on the protein PSGL-1 (P-selectin glycoprotein ligand-1) and matched it with experimental results | 32 |
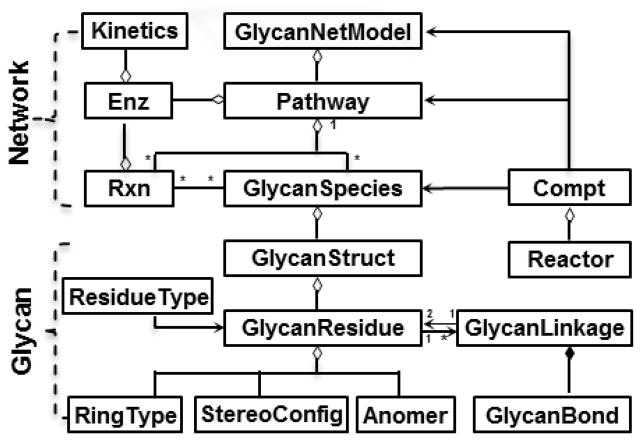
While the above approaches are valuable, unfortunately, most of these models are not written using SBML 20, the de facto standard for representing computational models in systems biology. Additionally, many of the previous manuscripts use rule-based synthesis of glycosylation networks that only partially account for detailed enzymatic specificity 27, 29. Thus, while useful for specific applications, the models cannot be readily shared among laboratories and developed further by the community. The reason for this is because SBML does not specifically handle glycan structure information, and it does not have facilities to define enzymes. Additionally, a streamlined strategy for the automated construction of glycosylation pathways has only recently been described with the development of the MATLAB based toolbox called the “Glycosylation Network Analysis Toolbox (GNAT)” 33, 34. Such model reconstruction is based on the object-oriented definitions of various ‘entities’ that participate in glycosylation reaction networks. These entities include glycans, enzymes, reactions, compartments and additional elements that are depicted in the UML diagram (Figure 3).
Figure 3. Entity-based modeling framework.
UML diagram of the classes used for the construction of glycosylation reaction networks in GNAT (http://sourceforge.net/projects/gnatmatlab/). (adapted from ref. 34 with permission)
The GlycanSpecies class
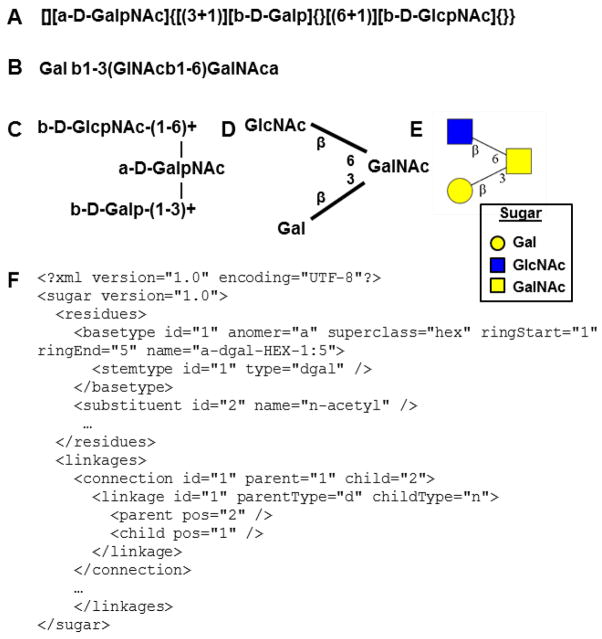
Linear, graphical and machine-readable formats have been utilized to describe glycans in literature (Figure 4). Among these, the LINUCS 35 (Fig. 4A), IUPAC (Fig. 4B) and Linear Code 36 nomenclatures are common methodologies for the linear representation of glycan structures. Two-dimensional graphical visualization allows the intuitive understanding of the branching pattern in carbohydrate structures. Thus, this approach has been used by the CarbBank, IUPAC and also the Consortium for Functional Glycomics (CFG) to render glycans (Fig. 4C–E). Finally, efforts have been undertaken to create machine readable descriptions for glycans using the Glyde-II 37 and GlycoCT 38 XML (eXtensible Markup Language) standards (Fig. 4F). In these last cases, the glycan structures contain descriptors for carbohydrate ring type, anomeric carbon position and stereoisomer configuration (“RingType”, “StereoConfig” and “Anomer”). Additional details are also provided regarding the “GlycanBond” and “GlycanLinkage” that join different monosaccharide residues (Fig. 3, bottom). These data are also included in the GlycanStruct class of GNAT, similar to the Glycan class in GlycanBuilder 39. This class utilizes a tree data structure to construct the branched sequence of a glycan. The final GlycanSpecies class in this modeling approach stores the entire GlycanStruct sequence, species short name, relative abundance and other data for the purpose of model building. To facilitate integration of glycosylation specific knowledge into SBML format files, GlycanStruct information is stored in the annotation field of the species-element definition (Box 1).
Figure 4. Representation of glycan structures.
Glycan representation in linear, graphical (2-dimensional) and data exchangeable formats: A. LINUCS; B. IUPAC; C. CarbBank 2-D representation; D. IUPAC graphics; E. CFG recommended graphics; and F. GlycoCT XML format.
The Enz, GTEnz and GHEnz classes
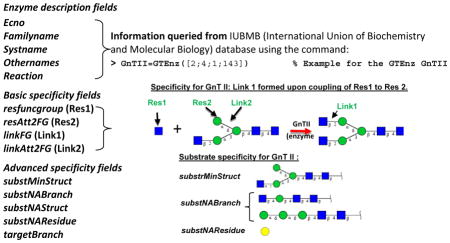
The “Enz” class describes various enzymes participating in glycosylation reactions within GNAT (Box 2). A number of facilities are available in this program to automatically populate the fields of the Enz class by directly querying the International Union of Biochemistry and Molecular Biology (IUBMB) enzyme database (Box 2). These fields include the Enzyme Commission (EC) number, enzyme names (including family, systematic and other names) and reaction description.
GTEnz and GHEnz are children classes of “Enz” that are used to describe the detailed specificity of the glycoTs and glycosidases. Besides the basic properties inherited from their parent Enz class, these structures contain additional features that fine tune the enzyme properties by either including or excluding specific types of acceptor substrates. Among these, resfuncgroup (or Residue 1, Box 2) and linkFG (or Link1) describe the monosaccharide and glycosidic bond that are either coupled by the GTEnz or that is released by the GHEnz. ResAtt2FG (or Residue 2) and linkAtt2FG (Link 2) define the substrate residue and glycosidic bond adjacent to the newly formed or removed linkage as depicted in the schematic in Box 2. This example presents the addition of GlcNAc (N-acetylglucosamine, Residue 1) to mannose (Residue 2) on N-linked glycan using the GTEnz β1,2-N-acetylglucosaminyltransferase or GnT II. Here, the ‘Residue’ and ‘Link’ described above represent the minimal fields necessary to describe the enzymatic activity. The additional properties of GnT II that constrain the feasibility of the reaction are also described in Box 2. In this regard, substNABranch describes specified sub-structures the presence of which prevents enzyme activity. All these properties can be viewed using an enzyme viewer (‘enzViewer’) provided in GNAT 33.
Automated Pathway object construction and simulation
The above systematic Enz definition rules enable the rapid construction of the Pathway object. Here, ‘Pathway’ depicts the entire glycosylation network, including the full complement of biochemical reactions (‘Rxn’ object), species (GlycanSpecies) and reactor compartments (‘Compt’ object, Fig. 3). During such Pathway construction, graph data structures are employed to represent the highly connected biochemical reactions, with each node representing a “GlycanSpecies” and edges corresponding to “Rxn” objects. Such data structures enable efficient calculation of the general properties of the network, e.g. the set of biosynthetic pathways connecting any two nodes or the identification of nodes with the highest degree of connections. Here, the kinetic rate law for the individual biochemical reactions can be mathematically expressed as elemental rate equations, reversible/irreversible Michaelis-Menten reactions, various type of Bi-Bi reactions (Ping-pong, random and sequential mechanisms) or transport equations. The compartments can be described as ideal continuous stirred tank reactors (CSTR), plug flow reactors (PFR) or batch reactors.
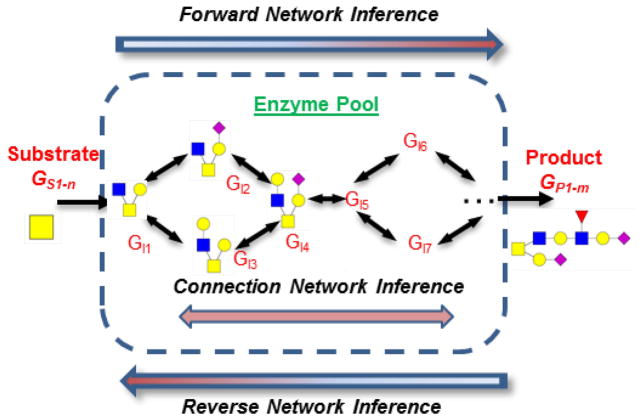
Various facilities are available in GNAT to automate the synthesis of Pathway objects including (Figure 5): i) ‘Forward network inference’: Here, starting with one or a small set of starting glycans, a product inference algorithm is applied in order to infer the potential reactions and products emanating from this starting material based on the enzymes available in the system. All newly generated glycans and reactions are then consolidated into a list by removing repeated elements. This process is then repeated until no additional new product(s) are formed in a given cycle. All unique structures at this point are incorporated into the Pathway object. ii) ‘Reverse network inference’: Here, the network is inferred given a set of final products and enzymes. This is similar to the ‘forward network inference’ algorithm, only it starts with the network products and applies backward substrate-inference to determine the starting material. iii) ‘Connection network inference’: This is applied to identify the intermediate glycans when a limited set of substrates and products are known. This algorithm considers pairs of glycans, one set to be the input/substrate and the second being the output/product based on the enzymes present in the system. For this substrate-product pair, starting with the substrate, a step-wise method is applied to identify all possible reaction pathways that link the substrate-product pair. This methodology is applied repeatedly until all substrate-product pairs are exhausted, with the final list of species and reactions being consolidated in the Pathway object.
Figure 5. Automated Pathway construction.
Three algorithms have been implemented in GNAT to automate network synthesis. These include the forward, reverse and connection network inference algorithms. (adapted from ref. 33 with permission)
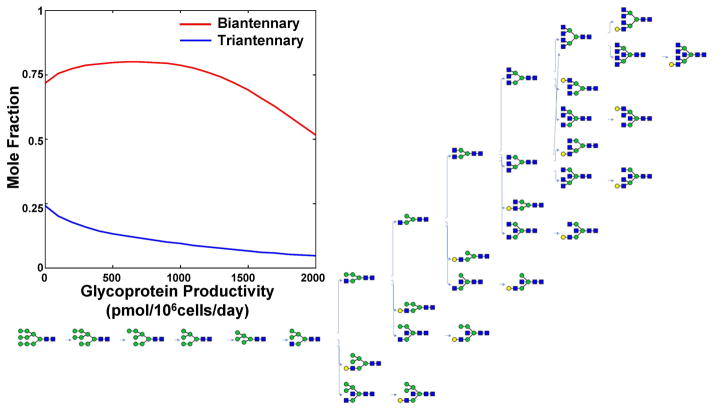
Once the Pathway object is constructed, computer simulations can be performed as appropriate for the system. The Pathway structure and simulation results are stored in the GlycanNetModel (Top of Fig. 3). Figure 6 depicts a sample glycosylation reaction network model visualized using the GNAT’s GlycanNetViewer (Fig. 6A) and corresponding simulation results (Fig. 6B).
Figure 6. Visualization and modeling of glycosylation reaction networks.
Synthesis of the N-linked glycosylation pathway described by Umana and Bailey 25. Glycosylation reaction network presented here was visualized using the GlycanNetViewer function of GNAT (main figure). The network can be exported into an SBML file that contains all glycan sequences. The computational simulation of this network using MATLAB predicts that the extent of N-linked glycosylation decreases upon increasing protein productivity (i.e. macroheterogeneity increases, see plot at top-left). This affects the relative concentration of bi- and tri-antennary N-glycans.
GLYCOINFORMATICS TO LINK GLYCAN STRUCTURE WITH FUNCTION
Over the years, a number of Glycoinformatics databases and related tools have appeared to serve as repositories for glycan structures (reviewed by 40–42). Many of these databases simply collate glycan structure, taxonomy and bibliography information. As there is considerable overlap among these resources, GlycomeDB (www.glycome-db.org) has undertaken the effort of collating at a single site, the data from multiple sources including CarbBank (Complex Carbohydrate Structure Database), GLYCOSCIENCES.de, KEGG (Kyoto Encyclopedia of Genes and Genomes) GLYCAN, CFG, JCGGDB (Japan Consortium for Glycobiology and Glycotechnology Database), BCSDB (Bacterial Carbohydrate Structure Database), GlycoBase, EuroCarbDB, and more recently also the Protein Data Bank 43. Many of the inconsistencies between the databases were resolved in the new databases. In all, >100,000 datasets from these different repositories were unified into 33,000 unique sequences and these are presented in the GlycoCT XML format 38.
With the goal of developing links between glycan structure and function, recent years have witnessed the growth of relational databases. While independent research laboratories have participated in this effort, there have also been notable contributions from international consortia formed in the USA (CFG, www.functionalglycomics.org) 44, Europe (EuroCarbDB, glycomics.ccrc.uga.edu/eurocarb/) 45 and Japan (JCGGDB, jcggdb.jp/index_en.html) (Table 3). Many of these repositories are at their formative stages currently, with data being collected only for a limited set of organisms. The array of experimental techniques employed in each of these efforts is also limited, and thus there remains considerable opportunity to expand these glycoinformatics resources. While some of the data stored here have to be downloaded manually, in many instances the query process can be automated. The following text summarizes these glycoinformatics activities with a focus on data that can feed into systems based model building:
Table 3.
Some computer programs for glycoproteomics data analysis
| Software Name |
mzXML/ mzML input |
High- throughput Processing |
Parallel Computing |
Decoy Database Generation |
False Discovery Calculation |
GlycoPeptide Database |
MS1 match |
N- and O- linked |
MS2 Scoring |
Fragmentation Mode |
Command Line/GUI/Web (Open-source) |
New Features | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i. Programs specializing in analysis of unfragmented glycopeptide (MS1 only) | |||||||||||||
| GlycoMod | No | No | No | No | No | Yes | Yes | N- and O- | No | - | Web application (Non-open-source) | part of ExPASy suite | 57 |
| GlycoSpectru mScan | No | No | No | No | No | Yes | Yes | N- and O- | No | - | Web application (Non-open-source) | Use glycan composition as inputs to generate glycopeptide database | 58 |
| GP(finder) GlycoX | No | No | No | No | No | No | Yes | N- and O- | No | - | Command Line (Available upon request) | Isotope filter/Diagnostic ion | 59 |
| GlycoPep DB | No | No | No | No | No | Limited | No | N | No | CID | Web application (Non-open-source) | Glycopeptide database | 60 |
| ii. MS/MS data analysis programs with limited ability to handle high-throughput data | |||||||||||||
| Sweet substitute | No | No | No | No | No | No | No | N- | No | - | GUI (Non-open-source) | In silico Fragmentation of glycopeptide | 61 |
| GlycoPep Grader | No (csv) | No | No | Yes | Yes | Yes | Yes | N-linked | Yes | CID | Web application (Non-open-source) | Glycan-type-dependent fragmentation rules | 62 |
| GlycoPep Evaluator | No | No | No | Yes | Yes | No | No | N-linked | Yes | ETD | Java-based GUI (Non-open-source) | FDR Prediction for small data sets | 63 |
| Peptoonist | No | No | No | No | No | Yes | Yes | N- | Yes | CID | GUI (Non-open-source) | Use cartoonist glycan database to create | 64 |
| iii. High-throughput algorithms/software | |||||||||||||
| a. Scoring without explicit glycopeptide database creation | |||||||||||||
| GlycoMiner | No | Yes | No | No | No | No | No | N- | Yes | CID | GUI (Non-open-source) | Spectra and glycan quality score | 65 |
| Medicel N- glycopeptide library | No | Yes | No | Yes | Yes | No | Yes | N-linked | Yes | CID | GUI (Propriety Software) | Branch and Bound algorithm for glycan identification | 66 |
| SweetSEQer | No (mgf file) | Yes | No | No | Yes | No | No | N-linked | No | HCD/CID | Command Line (Open-source) | De novo sequencing and annotation | 67 |
| Sweet-Heart | Yes | Yes | No | No | No | No | Yes | N-linked | Yes | CID | Command Line (Non-open-source) | Machine learning algorithm/MS3 Score | 68 |
| b. Scoring based on glycan/glycopeptide database creation | |||||||||||||
| GlycoPeptide Search | Yes | Yes | No | No | Yes | Yes | Yes | N-linked | No | CID | Command Line/GUI for inputs | Diagnostic Ion Filtering | 69 |
| GlycoMaster DB | No (mgf file) | Yes | No | No | No | No | Yes | N-linked | Yes | HCD/ETD | Web application (Non-open-source) | mixed HCD/ETD fragmentation | 70 |
| GlycoFrag- Work | Yes | Yes | No | Yes | Yes | Yes | Yes | N-linked | Yes | CID/ETD/HCD | Command Line (Non-open-source) | Mixed ETD-CID, mixed CID- HCD | 71 |
| Byonic | Yes | Yes | Yes | Yes | Yes | Yes | Yes | N- and O- | Yes | CID/ETD/HCD | GUI (Propriety) | Commercial software | 72 |
CFG
The CFG is largely focused on human and murine systems. This repository collates results from four complementary experimental methods: i. glycan array, ii. glycan profiling, iii. glyco-gene microarray and iv. mouse phenotyping. In the glycan array experiments, hundreds of glycans were printed on substrates and these were probed using an assortment of antibodies, serum samples, animal glycan-binding proteins, plant and microbial lectins and pathogens to measure binding specificity. In the glycan profiling studies, MALDI-TOF was applied to determine the N- and O-glycan profiles of a range of individual cells and also tissue samples from humans and murine knockouts. In some cases, MALDI-TOF-TOF-MS/MS and ESI-MS/MS were applied to distinguish between isomeric glycan structures. The gene expression data stored at the CFG database are based on custom Affymetrix glyco-gene microarray chips that focus on mouse and human glycosyltransferases, glycan-binding proteins, signaling and adhesion molecules, and other glycosylation related proteins that are relevant to cell communication. These studies measured gene expression data for both cell and tissue samples. Finally, the mouse phenotyping core stores histology, hematology, basic immunology and metabolism data that characterize a set of transgenic animals.
EUROCarbDB
The EUROCarbDB was designed to provide freely accessible informatics tools and databases to support glycomics research. With a focus on glycan sequence, this database was designed to collate experimental evidence for the existence and function of glycans based on high-performance liquid chromatography (HPLC), MS and nuclear magnetic resonance (NMR). This database contains 13,964 unique glycan sequences, 64 MS and 1261 NMR analysis. The GlycoBase database stores HPLC data 46.
Extending the informatics standards set out in EUROCarbDB, recently, UnicarbKB introduced a glycoproteomics knowledge base that includes data originally from GlycoSuiteDB 47 along with additional new entries 48. The detailed information about biological source, methods used and primary citations for these site-specific glycosylation data are also provided when available. The first release of this database includes 3740 glycan structure entries, 400 glycoproteins, and 598 protein glycosylation sites. These are largely annotated with experimental confirmation from over 890 literature references.
JCGGDB
This is a web portal for the storage of carbohydrate related data developed by different Japanese laboratories. Principally this database includes the following: i. ‘Lectin Frontier Database (LfDB)’ quantifies the binding affinity (Ka) of a few hundred lectins to a panel of pyridylaminated glycans using frontal affinity chromatography, and the ‘GlycoEpitope’ repository stores binding specificity information for ~613 glycosylation related antibodies to ~174 glycoepitopes; ii. ‘Glycogene Database’ includes glycosylation-related enzyme specificity and tissue-specific expression data for ~150 enzymes; iii. ‘Glycan Mass Spectral Database’ includes MSn spectra of O- and N-glycan standards in CID (collision induced dissociation) fragmentation mode; and iv. ‘Galaxy’ contains the elution profile of 500 pyridylamino-glycans separated by HPLC using three different columns.
Besides the above consortium driven efforts, there are several additional noteworthy databases that are relevant to systems glycobiology. In this regard, KEGG GLYCAN is an integrated knowledge base of pathway networks, genomic information, and chemical information 49. CAZY (Carbohydrate-Active enZYmes Database) 50 provides access to genomic, 3D-structural and biochemical data for glycosylation related enzymes including glycosyltransferases, glycosidases and polysaccharide lyases, carbohydrate esterase and auxiliary redox enzymes. The specific enzyme nomenclature defined by The IUBMB can be accessed through ExplorEnz database using the enzyme commission number 51. BRENDA is a twenty-five year old, rich enzyme database with 77,000 enzymes annotated from 135,000 references 52. This knowledgebase provides enzyme-disease relations, organism specific expression data, protein sequence, kinetic rate constants, catalytic reaction descriptions and genome annotations. Finally, the CBS prediction server (http://www.cbs.dtu.dk/services/) contains several resources for the prediction of N- and O-glycosylation sites, including the NetOGlc server which uses experimental data and neural networks to predict the location of mucin type GalNAc O-glycosylation sites in mammalian proteins 53.
While the above resources are all valuable, validation of the accuracy of data collated in these databases remains a challenge. In addition, data exchange between the resources is not straightforward due to the unique features of each database that do not allow a uniform machine-readable interface for easy extraction and cross-referencing of stored information. To address this, a new Resource Description Framework (RDF) called GlycoRDF is being implemented across several of the above databases 54. Once completed, this may speed-up the dynamic and automated construction of models for systems glycobiology using glycoinformatics databases.
GLYCOPROTEOMICS ANALYSIS ALGORITHMS AND SOFTWARE
The same glycan expressed on different protein scaffolds can have distinct functions. Thus, beyond glycomics profiling, which yields the overall distribution of carbohydrate structures on the cell surface, site-specific glycoproteomics data are necessary for the development of both glycoinformatics databases and systems glycobiology models. For such data collection, MS is the tool of choice due to recent technology breakthroughs and the high-throughput nature of such experiments (reviewed in 55). However, data interpretation remains a major challenge even for experienced users. Due to this necessity to collect quantitative glycoproteomics data for model building, we review many of glycoproteomics analysis algorithms available currently.
An ideal software for glycoproteomics analysis should have the following features. It should: i) Provide a complete set of functions for the analysis of every step in the experimental procedure, such as in silico protease digestion to create a list of all theoretically possible glycopeptides in the sample, facilities for MS1 precursor mass matching, and statistical scoring of MS/MS and MSn spectra to identify the most feasible structure; ii) Support MS data inputs in standard open formats such as mzXML 56, thus allowing usage with a variety of MS hardware; iii) Support multiple fragmentation modes, especially CID, HCD (High-energy collision dissociation) and ETD (electron transfer dissociation). In this regard, each of these collision modes fragment glycopeptides in a distinct manner. CID and HCD primarily fragment the glycan while leaving the peptide backbone largely intact, with HCD causing more intense fragmentation. ETD, on the other hand, prefers to fragment the peptide backbone while leaving the glycan intact. These different fragmentation modes thus provide complementary information regarding the underlying glycopeptide; iv) Provide facilities for both N- and O-linked glycopeptide analysis, ideally in a high-throughput manner, possibly with parallel computing facilities; v) Beyond being freely available, the ideal code should also be open-source so that it can be developed by the community for different applications. To date no software satisfies all the above criteria.
Programs for glycoproteomics analysis can be broken down into software that either: i. Focus on matching only the unfragmented glycopeptide mass; ii. Handle MS/MS fragmentation spectra but that only handle one or a limited number of spectra at a time; or iii. Process high throughput MS/MS data (Table 3).
Among the programs that are based on matching either the mass of the precursor ion or glycopeptide alone, GlycoMod 57 is popular since it was among the first to be developed, and also since it is hosted as part of the ExPASy Bioinformatics Resource Portal. This program is specifically designed to identify the glycan composition of glycopeptides by matching the experimentally measured mass to theoretical masses generated by varying the number and type of monosaccharide units attached to the peptide backbone. Prior knowledge of biochemistry regarding naturally occurring glycans is not necessary. Since the scope of the initial search in GlycoMod can be large, additional programs have appeared like GlycoSpectrumScan 58, GlycoX 59 and GlycoPep DB 60 that use a more focussed database.
While matching MS1 mass alone is useful, particularly if the data are from high-resolution MS instruments, MS/MS spectra analysis is necessary for assignment confirmation and also for glycan structure determination. Sweet Substitute 61 is the simplest example of the second class of programs and it is specifically designed to generate theoretical MS/MS spectra of tryptic digested peptides following the fragmentation of N-linked glycosidic bonds. GlycoPep Grader 62, Glycopeptide Evaluator 63 and Peptoonist 64 are additional programs that can perform CID or ETD fragmentation mode data analysis on either a single or a small set of MS/MS spectra.
The above programs that focus on limited MS/MS analysis perform relatively simple calculations and have few input and output fields. Due to their simplicity they are well suited for web-based interfaces. Increasing the complexity of the calculations and the handling of larger amounts of high-throughput data typically necessitates more computer resources and the development of stand-alone applications. To address this need, several programs focused on high-throughput data analysis have been developed either based on the application of principles akin to de novo sequencing, or algorithms that first generate a list of potential/candidate glycopeptides before scoring the tandem-MS data.
Glycominer 65 and Medicel 66 are two programs that utilize algorithms that do not create explicit glycopeptide databases. Among these, Glycominer is a GUI based software based on the premise that the CID mode fragmentation of glycopeptides results in the release of glycan oxonium ions 65. Thus, this program identifies spectra corresponding to glycopeptides by scoring the oxonium ions. Once a match is identified, the program discovers the underlying peptide sequence by attempting to identify the specific spectral peak that corresponds to the deglycosylated peptide. Once this is done, attempts are made to match the molecular weight of the putative peptide with databases, for example the FASTA database. Medicel utilizes a similar approach to identify glycopeptide spectra based on the appearance of carbohydrate oxonium ions and the glycan ladder pattern that is common to N-glycan. SweetSEQer facilitates the interpret of the ladder pattern seen in the MS/MS spectra by using a graph based approach 67. Sweet-Heart is another program that utilizes supervised machine learning to identify patterns in a training dataset prior to using this knowledge to identify N-glycopeptides 68.
Among the programs that match MS/MS spectra with respect to a database containing potential glycans and peptides, GlycoProteinSearch (GPS) identifies N-linked glycopeptides using CID MS/MS spectra data 69. Here, the peptide sequence is determined by using the MS1 precursor mass, glycan-loss ladder pattern in the MS/MS spectra and a library of potential peptides containing the Asn-X-Ser/Thr motif. Based on the overall glycan mass and fragments, the glycan is then conjectured by searching the GlycomeDB database. GlycoMasterDB is another program in this class that is specifically designed to handle high throughput HCD and HCD-ETD tandem MS data 70. The program uses the HCD spectra to identify spectra containing glycopeptides and potential glycans. Once this is done, peptide identification is done by either utilizing ETD data if this is available or alternatively simply matching the peptide molecular mass against a list of theoretically possible peptides. Neither of the above programs has a well-developed statistical scoring algorithm. Mayampurath et al. have developed GlycoFragWork 71 to handle CID, ETD and HCD fragmentation mode data. In the GlycoFragWork workflow, the oxonium ions in HCD mode are used to identify glycopeptide spectra, MS1 precursor ion matching is then applied to determine candidate glycopeptides that may explain the spectra, subsequently ETD mode data are used to confirm peptide identity and in the final step CID fragment spectra scoring is performed for a representative glycopeptide. Byonic is a software with a comprehensive scoring algorithm and a user-friendly GUI 72, but it is not freely available or open-source.
Conclusion
The human glycome is far more complex compared to the genome and proteome, both from the structural and regulatory perspective. While a study of such carbohydrates was complicated in the past, the development of new analytical technologies is beginning to provide large scale qualitative and quantitative ‘data’ for this field. This is then opening new possibilities since these data are now starting to be collated into glycoinformatics relational databases, which derive ‘information’ on the relation between glycan structure and function. More advanced analysis of these data and information is possible using systems based modeling since standardized tools for computational model synthesis and sharing have become available. Together, such analyses can lead to new experimentally testable hypotheses, for the iterative refinement of ‘knowledge’ in the field of systems glycobiology.
While the focus of the current review is on how quantitative protein and carbohydrate structure data can be used for such systems-based model building and analyses, data at the metabalome 26, 30, transcriptome 28 and genome 73 levels can also provide additional complementary information. For example, Lau et al. 26 studied the effect of feeding N-acetyl glucosamine (GlcNAc) to cells with a focus on understanding how this perturbation alters the pattern of N-glycan branching. These authors used mathematical models to fit MS based N-glycan profiling data following perturbation while simultaneously performing metabolome analysis to index the status of the cells. Bennun et al. 28 also fit experimental MS data of prostate cancer cells with computer models, while using gene microarray experiments to rationalize cellular enzymatic activities. Similarly, in the case of O-linked glycans, Liu et al. 32, 74 used enzymology based glycosyltransferase activity measurements to guide computer models that fit the distribution of O-glycans on specific human myeloid cell proteins. In the final example, recently, Agrawal et al. 75 compared lectin binding data for 55 cell lines taken from the NCI-60 (National Cancer Institute) panel with corresponding miRNA expression data using singular value decomposition. Their analysis reveals the critical miRNA controlling biosynthetic pathways that construct high mannose, fucose and βGalNAc bearing glycans. Thus, beyond simply fitting experimental data on glycan distribution, these efforts attempt to link glycan structure data to additional levels of biosynthetic control within cells. Such large-scale experimental strategies, along with a number of system-perturbation methods that use RNA interference 21, 76, miRNA perturbations 75, small molecule inhibitors 77 or genome editing 21, 53, may be applied in order to interrogate and further refine the computational models. Together, these approaches are likely to provide a more holistic view of the cellular glycosylation process. This may also lead to new exciting translational opportunities for this burgeoning field.
Table 2.
Glycoscience related databases *
| Data source | Gene and enzyme level data | Glycomics data | Glycan–protein binding data | Glycoproteomics data | Pathway data | Mouse phenotyping data |
|---|---|---|---|---|---|---|
| CFG | Custom Glycogene microarray data for human and murine cells/tissue | Profiling of N- and O-glycans from cells and tissue using MALDI-TOF | Glycan microarrays quantify binding to various glycan binding proteins | CFG Molecular Pages contains data on glycan structures, glycosyltransfera ses and glycan- binding proteins | Hematology, Immunology & histology phenotyping of transgenic mice | |
| EuroCarbDB & affiliated laboratories | GlycoBase, UniCarb-DB: LC, LC- MS/MS repository for N- & O- glycans. | UniCarb-KB: knowledgebase contains site-specific glycosylation data, built on the EUROCarbDB framework. | ||||
| JCGGDB | Tissue specific expression and enzyme specificity data for ~150 enzymes | MSn analysis of glycan standards | LfDB: frontal chromatography data for lectin binding; GlycoEpitope: knowledgebase with antibody specificity data |
|||
| Other data sources | ExplorEnz: gateway for IUBMB enzyme nomenclature; BRENDA: kinetic data for enzymes collated from literature |
CBS Predictor server: NetCGlyc, NetNGlyc, NetOGlyc & YinOYang for prediction of sites of c-mannosylation, N-glycosylation, O- glycosylation and O- GlcNAcylation, resp. | KEGG GLYCAN: Glycosylation pathway data |
All repositories do not contain all types of experimental data. Thus, there are several empty fields in the Table.
How can glycan structure data be incorporated into SBML files?
Glycan structures have been described using at least two XML standards, Glyde-II 37 and GlyoCT 38. Inter-conversion between these two standards and also additional formats described in Figure 4 is possible using GNAT 33, 34 and also other programs 39. These glycan XML structures can be incorporated into SBML format files within the species-annotation field as illustrated below. The addGlycanAnnotation command of GNAT can be used to automate the incorporation of multiple glycan structures rapidly into SBML files.
<sbml xmlns=“http://www.sbml.org/sbml/level2” level=“2” version=“1”>
<model id=“demo”>
....
<listOfSpecies>
<species id=“S1”.../>
<annotation>
<glycoct xmlns=“http://www.eurocarbdb.org/”>
<sugar version=“1.0”>
<residues>
....
</residues>
<linkages>
....
</linkages>
</sugar>
</glycoct>
</annotation>
</species>
....
</listOfSpecies>
....
</model>
</sbml>
How can glycosyltransferase specificity be described in silico?
It is desirable to have machine-readable definitions for the glycosyltransferases (glycoTs). GNAT captures this using the GTEnz class which contains various fields, some of which are shown below. Here, the enzyme description fields are automatically populated by querying the IUBMB database with the EC number. The ‘basic’ and ‘advanced’ specificity fields capture the absolute, group, linkage and stereochemical specificity of the glycoTs. Here, substrMinStruct presents the minimal substrate necessary for enzyme activity. substNABranch and substNAResidue describe structures and residues the presence of which prevent enzyme activity.
Acknowledgments
We acknowledge funding support from the National Institutes of Health (HL103411, HL63014 and Program of Excellence in Glycosciences award HL107146)
Footnotes
Glycosyltransferases (GlycoTs) are a family of ~250 enzymes that catalyze the biosynthesis of cellular glycoconjugates by transferring monosaccharides from nucleotide-sugar donors to carbohydrate/protein/lipid acceptors. Glycosidases are glycoside hydrolases that assist with the breakdown of glycosidic bonds. These enzymes typically remove monosaccharides from specific glycoconjugates.
The authors declare no conflicts of interest
References
- 1.Neelamegham S, Liu G. Systems Glycobiology: Biochemical Reaction Networks Regulating Glycan Structure and Function. Glycobiology. 2011;21:1541–1553. doi: 10.1093/glycob/cwr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343:1235681. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 3.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13:448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME. Essentials of Glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 6.Fukuda MN, Sasaki H, Lopez L, Fukuda M. Survival of recombinant erythropoietin in the circulation: the role of carbohydrates. Blood. 1989;73:84–89. [PubMed] [Google Scholar]
- 7.Weikert S, Papac D, Briggs J, Cowfer D, Tom S, Gawlitzek M, Lofgren J, Mehta S, Chisholm V, Modi N, et al. Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat Biotechnol. 1999;17:1116–1121. doi: 10.1038/15104. [DOI] [PubMed] [Google Scholar]
- 8.Beck A, Reichert JM. Marketing approval of mogamulizumab: a triumph for glyco-engineering. MAbs. 2012;4:419–425. doi: 10.4161/mabs.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vigerust DJ, Shepherd VL. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 2007;15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nat Rev Immunol. 2008;8:874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dall’Olio F, Malagolini N, Trinchera M, Chiricolo M. Mechanisms of cancer-associated glycosylation changes. Front Biosci (Landmark Ed) 2012;17:670–699. doi: 10.2741/3951. [DOI] [PubMed] [Google Scholar]
- 12.Patil SA, Bshara W, Morrison C, Chandrasekaran EV, Matta KL, Neelamegham S. Overexpression of alpha2,3sialyl T-antigen in breast cancer determined by miniaturized glycosyltransferase assays and confirmed using tissue microarray immunohistochemical analysis. Glycoconj J. 2014;31:509–521. doi: 10.1007/s10719-014-9548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 14.Taylor ME, Drickamer K. Introduction to Glycobiology. 3. New York: Oxford University Press; 2011. [Google Scholar]
- 15.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeze HH, Haltiwanger RS. Other Classes of ER/Golgi-derived Glycans. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2. Cold Spring Harbor; NY: 2009. [Google Scholar]
- 17.Papin JA, Hunter T, Palsson BO, Subramaniam S. Reconstruction of cellular signalling networks and analysis of their properties. Nat Rev Mol Cell Biol. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- 18.Hyduke DR, Lewis NE, Palsson BO. Analysis of omics data with genome-scale models of metabolism. Mol Biosyst. 2013;9:167–174. doi: 10.1039/c2mb25453k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mac Gabhann F, Qutub AA, Annex BH, Popel AS. Systems biology of pro-angiogenic therapies targeting the VEGF system. Wiley Interdiscip Rev Syst Biol Med. 2010;2:694–707. doi: 10.1002/wsbm.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 21.Mondal N, Buffone A, Jr, Stolfa G, Antonopoulos A, Lau JT, Haslam SM, Dell A, Neelamegham S. ST3Gal-4 is the primary sialyltransferase regulating the synthesis of E-, P-, and L-selectin ligands on human myeloid leukocytes. Blood. 2015;125:687–696. doi: 10.1182/blood-2014-07-588590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo CY, Antonopoulos A, Gupta R, Qu J, Dell A, Haslam SM, Neelamegham S. Competition between core-2 GlcNAc-transferase and ST6GalNAc-transferase regulates the synthesis of the leukocyte selectin ligand on human P-selectin glycoprotein ligand-1. J Biol Chem. 2013;288:13974–13987. doi: 10.1074/jbc.M113.463653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puri A, Neelamegham S. Understanding glycomechanics using mathematical modeling: a review of current approaches to simulate cellular glycosylation reaction networks. Ann Biomed Eng. 2012;40:816–827. doi: 10.1007/s10439-011-0464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shelikoff M, Sinskey AJ, Stephanopoulos G. A modeling framework for the study of protein glycosylation. Biotechnol Bioeng. 1996;50:73–90. doi: 10.1002/(SICI)1097-0290(19960405)50:1<73::AID-BIT9>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Umana P, Bailey JE. A mathematical model of N-linked glycoform biosynthesis. Biotechnol Bioeng. 1997;55:890–908. doi: 10.1002/(SICI)1097-0290(19970920)55:6<890::AID-BIT7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. S0092-8674(07)00315-7 [pii] [DOI] [PubMed] [Google Scholar]
- 27.Krambeck FJ, Betenbaugh MJ. A mathematical model of N-linked glycosylation. Biotechnol Bioeng. 2005;92:711–728. doi: 10.1002/bit.20645. [DOI] [PubMed] [Google Scholar]
- 28.Bennun SV, Yarema KJ, Betenbaugh MJ, Krambeck FJ. Integration of the transcriptome and glycome for identification of glycan cell signatures. PLoS Comput Biol. 2013;9:e1002813. doi: 10.1371/journal.pcbi.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hossler P, Mulukutla BC, Hu WS. Systems analysis of N-glycan processing in mammalian cells. PLoS One. 2007;2:e713. doi: 10.1371/journal.pone.0000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jedrzejewski PM, del Val IJ, Constantinou A, Dell A, Haslam SM, Polizzi KM, Kontoravdi C. Towards controlling the glycoform: a model framework linking extracellular metabolites to antibody glycosylation. Int J Mol Sci. 2014;15:4492–4522. doi: 10.3390/ijms15034492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.del Val IJ, Kyriakopoulos S, Polizzi KM, Kontoravdi C. An optimized method for extraction and quantification of nucleotides and nucleotide sugars from mammalian cells. Anal Biochem. 2013;443:172–180. doi: 10.1016/j.ab.2013.09.005. S0003-2697(13)00427-2 [pii] [DOI] [PubMed] [Google Scholar]
- 32.Liu G, Marathe DD, Matta KL, Neelamegham S. Systems-level modeling of cellular glycosylation reaction networks: O-linked glycan formation on natural selectin ligands. Bioinformatics. 2008;24:2740–2747. doi: 10.1093/bioinformatics/btn515. btn515 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu G, Neelamegham S. A computational framework for the automated construction of glycosylation reaction networks. PLoS One. 2014;9:e100939. doi: 10.1371/journal.pone.0100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu G, Puri A, Neelamegham S. Glycosylation Network Analysis Toolbox: a MATLAB-based environment for systems glycobiology. Bioinformatics. 2013;29:404–406. doi: 10.1093/bioinformatics/bts703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohne-Lang A, Lang E, Forster T, von der Lieth CW. LINUCS: linear notation for unique description of carbohydrate sequences. Carbohydr Res. 2001;336:1–11. doi: 10.1016/s0008-6215(01)00230-0. [DOI] [PubMed] [Google Scholar]
- 36.Banin E, Neuberger Y, Altshuler Y, Halevi A, Inbar O, Nir D, Dukler A. A novel Linear Code((R)) nomenclature for complex carbohydrates. Trends in Glycoscience and Glycotechnology. 2002;14:127–137. [Google Scholar]
- 37.Sahoo SS, Thomas C, Sheth A, Henson C, York WS. GLYDE-an expressive XML standard for the representation of glycan structure. Carbohydr Res. 2005;340:2802–2807. doi: 10.1016/j.carres.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Herget S, Ranzinger R, Maass K, Lieth CW. GlycoCT-a unifying sequence format for carbohydrates. Carbohydr Res. 2008;343:2162–2171. doi: 10.1016/j.carres.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol Chem. 2012;393:1357–1362. doi: 10.1515/hsz-2012-0135. [DOI] [PubMed] [Google Scholar]
- 40.Aoki-Kinoshita KF. Using databases and web resources for glycomics research. Mol Cell Proteomics. 2013;12:1036–1045. doi: 10.1074/mcp.R112.026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Artemenko NV, McDonald AG, Davey GP, Rudd PM. Databases and tools in glycobiology. Methods Mol Biol. 2012;899:325–350. doi: 10.1007/978-1-61779-921-1_21. [DOI] [PubMed] [Google Scholar]
- 42.Frank M, Schloissnig S. Bioinformatics and molecular modeling in glycobiology. Cell Mol Life Sci. 2010;67:2749–2772. doi: 10.1007/s00018-010-0352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ranzinger R, Herget S, von der Lieth CW, Frank M. GlycomeDB--a unified database for carbohydrate structures. Nucleic Acids Res. 2011;39:D373–376. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. Advancing glycomics: implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16:82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 45.von der Lieth CW, Freire AA, Blank D, Campbell MP, Ceroni A, Damerell DR, Dell A, Dwek RA, Ernst B, Fogh R, et al. EUROCarbDB: An open-access platform for glycoinformatics. Glycobiology. 2011;21:493–502. doi: 10.1093/glycob/cwq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24:1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 47.Cooper CA, Joshi HJ, Harrison MJ, Wilkins MR, Packer NH. GlycoSuiteDB: a curated relational database of glycoprotein glycan structures and their biological sources. 2003 update. Nucleic Acids Res. 2003;31:511–513. doi: 10.1093/nar/gkg099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. UniCarbKB: building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42:D215–221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. KEGG as a glycome informatics resource. Glycobiology. 2006;16:63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 50.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McDonald AG, Boyce S, Tipton KF. ExplorEnz: the primary source of the IUBMB enzyme list. Nucleic Acids Res. 2009;37:D593–597. doi: 10.1093/nar/gkn582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang A, Schomburg I, Placzek S, Jeske L, Ulbrich M, Xiao M, Sensen CW, Schomburg D. BRENDA in 2015: exciting developments in its 25th year of existence. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ranzinger R, Aoki-Kinoshita KF, Campbell MP, Kawano S, Lutteke T, Okuda S, Shinmachi D, Shikanai T, Sawaki H, Toukach P, et al. GlycoRDF: an ontology to standardize glycomics data in RDF. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alley WR, Jr, Mann BF, Novotny MV. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem Rev. 2013;113:2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedrioli PG, Eng JK, Hubley R, Vogelzang M, Deutsch EW, Raught B, Pratt B, Nilsson E, Angeletti RH, Apweiler R, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 57.Cooper CA, Gasteiger E, Packer NH. GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1:340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 58.Deshpande N, Jensen PH, Packer NH, Kolarich D. GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J Proteome Res. 2010;9:1063–1075. doi: 10.1021/pr900956x. [DOI] [PubMed] [Google Scholar]
- 59.An HJ, Tillinghast JS, Woodruff DL, Rocke DM, Lebrilla CB. A new computer program (GlycoX) to determine simultaneously the glycosylation sites and oligosaccharide heterogeneity of glycoproteins. J Proteome Res. 2006;5:2800–2808. doi: 10.1021/pr0602949. [DOI] [PubMed] [Google Scholar]
- 60.Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB: a tool for glycopeptide analysis using a “Smart Search”. Anal Chem. 2007;79:1708–1713. doi: 10.1021/ac061548c. [DOI] [PubMed] [Google Scholar]
- 61.Clerens S, Van den Ende W, Verhaert P, Geenen L, Arckens L. Sweet Substitute: a software tool for in silico fragmentation of peptide-linked N-glycans. Proteomics. 2004;4:629–632. doi: 10.1002/pmic.200300572. [DOI] [PubMed] [Google Scholar]
- 62.Woodin CL, Hua D, Maxon M, Rebecchi KR, Go EP, Desaire H. GlycoPep grader: a web-based utility for assigning the composition of N-linked glycopeptides. Anal Chem. 2012;84:4821–4829. doi: 10.1021/ac300393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Z, Su X, Go EP, Desaire H. New Glycoproteomics Software, GlycoPep Evaluator, Generates Decoy Glycopeptides de Novo and Enables Accurate False Discovery Rate Analysis for Small Data Sets. Anal Chem. 2014 doi: 10.1021/ac502176n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldberg D, Bern M, Parry S, Sutton-Smith M, Panico M, Morris HR, Dell A. Automated N-glycopeptide identification using a combination of single- and tandem-MS. J Proteome Res. 2007;6:3995–4005. doi: 10.1021/pr070239f. [DOI] [PubMed] [Google Scholar]
- 65.Ozohanics O, Krenyacz J, Ludanyi K, Pollreisz F, Vekey K, Drahos L. GlycoMiner: a new software tool to elucidate glycopeptide composition. Rapid Commun Mass Spectrom. 2008;22:3245–3254. doi: 10.1002/rcm.3731. [DOI] [PubMed] [Google Scholar]
- 66.Joenvaara S, Ritamo I, Peltoniemi H, Renkonen R. N-glycoproteomics - an automated workflow approach. Glycobiology. 2008;18:339–349. doi: 10.1093/glycob/cwn013. [DOI] [PubMed] [Google Scholar]
- 67.Serang O, Froehlich JW, Muntel J, McDowell G, Steen H, Lee RS, Steen JA. SweetSEQer, simple de novo filtering and annotation of glycoconjugate mass spectra. Mol Cell Proteomics. 2013;12:1735–1740. doi: 10.1074/mcp.O112.025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang SY, Wu SW, Pu TH, Chang FY, Khoo KH. An adaptive workflow coupled with Random Forest algorithm to identify intact N-glycopeptides detected from mass spectrometry. Bioinformatics. 2014;30:1908–1916. doi: 10.1093/bioinformatics/btu139. [DOI] [PubMed] [Google Scholar]
- 69.Chandler KB, Pompach P, Goldman R, Edwards N. Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search. J Proteome Res. 2013;12:3652–3666. doi: 10.1021/pr400196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: Software To Assist the Automated Identification of N-Linked Glycopeptides by Tandem Mass Spectrometry. J Proteome Res. 2014;13:3881–3895. doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- 71.Mayampurath A, Yu CY, Song E, Balan J, Mechref Y, Tang H. Computational framework for identification of intact glycopeptides in complex samples. Anal Chem. 2014;86:453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- 72.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics. 2012;Chapter 13(Unit13):20. doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spahn PN, Lewis NE. Systems glycobiology for glycoengineering. Curr Opin Biotechnol. 2014;30C:218–224. doi: 10.1016/j.copbio.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 74.Marathe DD, Chandrasekaran EV, Lau JT, Matta KL, Neelamegham S. Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes. FASEB J. 2008;22:4154–4167. doi: 10.1096/fj.07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu KL, Liang Y, et al. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci U S A. 2014;111:4338–4343. doi: 10.1073/pnas.1321524111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buffone A, Jr, Mondal N, Gupta R, McHugh KP, Lau JT, Neelamegham S. Silencing alpha1,3-fucosyltransferases in human leukocytes reveals a role for FUT9 enzyme during E-selectin-mediated cell adhesion. J Biol Chem. 2013;288:1620–1633. doi: 10.1074/jbc.M112.400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marathe DD, Buffone A, Jr, Chandrasekaran EV, Xue J, Locke RD, Nasirikenari M, Lau JT, Matta KL, Neelamegham S. Fluorinated per-acetylated GalNAc metabolically alters glycan structures on leukocyte PSGL-1 and reduces cell binding to selectins. Blood. 2010;115:1303–1312. doi: 10.1182/blood-2009-07-231480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading/Resources
- 1.Varki Ajit, Cummings Richard D, Esko Jeffrey D, Freeze Hudson H, Stanley Pamela, Bertozzi Carolyn R, Hart Gerald W, Etzler Marilynn E., editors. 2. Cold Spring Harbor (NY): NCBI Books; 2009. Essentials of Glycobiology. http://www.ncbi.nlm.nih.gov/books/NBK1908/] [PubMed] [Google Scholar]
- 2.Taylor Maureen E, Drickamer Kurt., editors. ‘Introduction to Glycobiology’. 3. Oxford University Press; 2011. [Google Scholar]