Abstract
Objectives
Medical terminologies vary in the amount of concept information (the “density”) represented, even in the same sub-domains. This causes problems in terminology mapping, semantic harmonization and terminology integration. Moreover, complex clinical scenarios need to be encoded by a medical terminology with comprehensive content. SNOMED Clinical Terms (SNOMED CT), a leading clinical terminology, was reported to lack concepts and synonyms, problems that cannot be fully alleviated by using post-coordination. Therefore, a scalable solution is needed to enrich the conceptual content of SNOMED CT. We are developing a structure-based, algorithmic method to identify potential concepts for enriching the conceptual content of SNOMED CT and to support semantic harmonization of SNOMED CT with selected other Unified Medical Language System (UMLS) terminologies.
Methods
We first identified a subset of English terminologies in the UMLS that have ‘PAR’ relationship labeled with ‘IS_A’ and over 10% overlap with one or more of the 19 hierarchies of SNOMED CT. We call these “reference terminologies” and we note that our use of this name is different from the standard use. Next, we defined a set of topological patterns across pairs of terminologies, with SNOMED CT being one terminology in each pair and the other being one of the reference terminologies. We then explored how often these topological patterns appear between SNOMED CT and each reference terminology, and how to interpret them.
Results
Four viable reference terminologies were identified. Large density differences between terminologies were found. Expected interpretations of these differences were indeed observed, as follows. A random sample of 299 instances of special topological patterns (“2:3 and 3:2 trapezoids”) showed that 39.1% and 59.5% of analyzed concepts in SNOMED CT and in a reference terminology, respectively, were deemed to be alternative classifications of the same conceptual content. In 30.5% and 17.6% of the cases, it was found that intermediate concepts could be imported into SNOMED CT or into the reference terminology, respectively, to enhance their conceptual content, if approved by a human curator. Other cases included synonymy and errors in one of the terminologies.
Conclusion
These results show that structure-based algorithmic methods can be used to identify potential concepts to enrich SNOMED CT and the four reference terminologies. The comparative analysis has the future potential of supporting terminology authoring by suggesting new content to improve content coverage and semantic harmonization between terminologies.
Keywords: Biomedical Terminology, Semantic Interoperability, Semantic Harmonization, Structural Methodology, SNOMED CT, UMLS
1. Introduction
1.1. Motivation
Controlled terminologies and bio-ontologies provide structured domain knowledge as the foundation of various healthcare information systems as well as of biomedical research. They have been widely used for encoding clinical data for diagnoses, problem lists [1-3], billing [4], etc. The concepts linked by hierarchical and semantic relationships in controlled terminologies have also been facilitating major components of natural language processing systems, which lay a solid foundation for rule-based clinical decision support systems [5]. However, the field of biomedical informatics is increasingly suffering from the tension between numerous biomedical research information standards [6], available or under development, and their sparse adoption by researchers and vendors. Moreover, heterogeneous information models (e.g., Health Level Seven Reference Information Model [7], OpenEHR reference model [8]) that provide frameworks for structuring medical data are often required to use more than one terminology (e.g., ICD-9, LOINC) [9]. The same terminologies are also used with more than one information model [10], creating a barrier for semantic interoperability.
To address this issue, the informatics community has been putting efforts into semantic harmonization for both information models and terminologies [11], whose driving forces include improving semantic interoperability among heterogeneous healthcare systems. Cimino listed domain completeness (conceptual content) as the most desired property for a good controlled terminology [12], and pointed out that “any controlled terminology will necessarily lack the richness of detail available from the vocabulary of a natural language. [13]” Therefore, a harmonized core terminology is needed to serve as a common ground for heterogeneous medical systems [11]. However, due to varying structure and features, harmonization of different coding systems is difficult, labor-intensive and time-consuming, which poses a significant challenge for large-scale harmonization tasks [14]. Our interests lie in semi-automated methods for suggesting new concepts for existing terminologies to improve the domain coverage of each terminology.
SNOMED Clinical Terms (SNOMED CT), developed and managed by IHTSDO (International Health Terminology Standard Development Organization), is considered as the most comprehensive multilingual clinical healthcare terminology in the world [15, 16]. Internationally, SNOMED CT is being implemented as standard within IHTSDO member countries [17]. By 2015, SNOMED CT will be one United States standard for encoding diagnoses, procedures, and vital signs (e.g., height, weight, blood pressure, and smoking status) in electronic health records (EHRs) under Stage 2 of Meaningful Use, with the intention of promoting interoperability [18]. Specifically, SNOMED CT is to be used to “enable a user to electronically record, modify, and retrieve a patient’s problem list for longitudinal care (i.e., over multiple office visits)” [19, 20]. To accelerate the adoption and Meaningful Use of EHRs by providers, incentives and penalties were defined and later refined and adjusted [21, 22].
SNOMED CT arranges over 300,000 concepts in 19 top-level hierarchies (e.g., Substance, Procedure, Clinical finding) that are organized with IS_A relationships. Even though it provides rich conceptual content, researchers have advocated greater coverage of common problem statements with improved synonymy and conceptual content [23]. In a survey among its direct users, missing concepts and missing synonyms were encountered by 23% and 17% of the respondents, respectively [24]. To assess its effect in the clinical setting, we have previously simulated a primary care scenario and demonstrated some of the difficulties of choosing a proper SNOMED CT term when describing the symptoms of a patient [25].
Even though post-coordination could, to some extent, alleviate these issues, some clinical statements with complex and rare clinical scenarios could not be encoded using post-coordinated SNOMED CT terms [26]. We need to seek a scalable solution to improve the coverage of SNOMED CT. Toward this end, in this work, we focus on developing structural, algorithmic methods to enrich the conceptual content of SNOMED CT. We show that comparative analysis has the future potential to support terminology authoring by suggesting new concepts to improve content coverage. Increasing the number of concepts that are shared between two terminologies will make the task of semantic harmonization of these two terminologies easier.
1.2. Approach
The Unified Medical Language System (UMLS) [27], as the most comprehensive biomedical terminological system, has already inherently harmonized numerous well-established terminologies and ontologies within a coherent structure. The UMLS Metathesaurus [28, 29] integrates more than 8.9 million terms from 170 source vocabularies into 2.9 million concepts (in the 2013AB release). All terms with the same meaning have been mapped to the same UMLS concept with a distinct Concept Unique Identifier (CUI). The UMLS has been investigated for use in terminology alignment [30, 31] and integration [32]. It can serve as a source of pairs of terminologies with matched concepts. Importantly, SNOMED CT is also included in the UMLS. By using the UMLS, we can focus on the conceptual content regardless of the original concept model employed by its source terminologies.
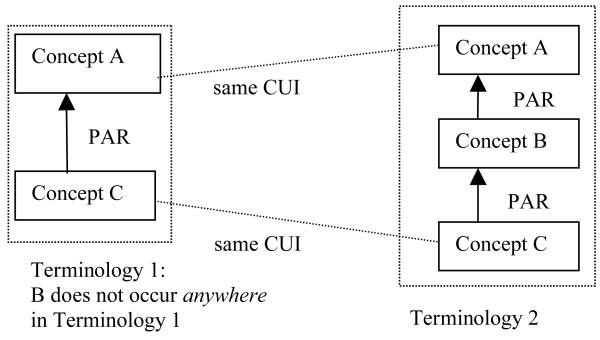
In our recently published paper [33], we introduced a semi-automated structural methodology that utilizes the common structure of the UMLS to find potential concepts for semantic harmonization. We defined “structurally congruent concept pairs” from pairs of terminologies in the UMLS [33]. In a structurally congruent topological pattern of concepts (illustrated in Figure 1), there are two different intermediate concepts in two UMLS terminologies that have identical parent concepts and identical child concepts in both terminologies (based on two IS_A paths). In previous work, we hypothesized that there are six ways how two congruent concepts can be related to each other. For example, the two concepts can be synonyms, alternative classifications of the same parent, or stand to each other in a parent child relationship, etc. Categorizing two structurally congruent concepts according to these six cases, semantic harmonization for each case can be conducted appropriately, e.g., by importing one concept as a parent of the other, importing it as a synonym, etc.
Figure 1.
An abstract layout of structurally congruent concepts
By analyzing a random sample of congruent concepts by a domain expert, we showed that it is feasible to use this semi-automated structural method to support semantic harmonization efforts. However, due to the simple layout of the structurally congruent topological pattern shown in Figure 1, the method does not scale well. Therefore, this method was not sufficient for a large-scale semantic harmonization task.
In this paper, we considerably extend the previously published structural method in terms of both complexity and depth: 1) We introduce a systematic method for identifying terminologies that are best suited for constructing topological patterns together with SNOMED CT concepts; 2) We identify and analyze considerably more complex topological patterns than in previous work; and 3) We use the occurrences and the complexity (i.e., number of intermediate concepts) of such topological patterns that were identified in 2) as a measure of density differences between SNOMED CT and the identified suitable terminologies. We hypothesized that there exist large density differences between the reference terminologies and SNOMED CT. Leveraging the common structure and native term mappings of source terminologies in the UMLS, this extended method identifies candidate concepts for semantic harmonization in a scalable fashion.
1.3. Related work
Previously, Bodenreider performed a study of redundant relations and similarity across families of terminologies and discussed the relationship between redundancy and semantic consistency [34]. Bodenreider observed that it is the policy in the UMLS that “PAR” represents an explicit parent-child relationship in a source, and “RB” indicates an implied one (as interpreted by the UMLS editorial team) [35].
To capture patient information precisely, it is required that the data is encoded with great detail. Previously, Arts et al. reviewed various methods for evaluating terminological systems and concept coverage was one of the most assessed metrics for a terminology [36]. Cornet suggested that semi-automatic terminology authoring could be based on information content, which can be used internally in one terminology, e.g., to balance the granularity level between hierarchies. However, the methods cannot be used to support harmonization across terminologies, which is the goal of our study [37].
The term density covers a number of distinct phenomena in biomedical informatics. Rector et al. distinguished between granularity and density in medical terminologies [38]. They start out by stating that “it is rarely made clear exactly what is meant by ’granularity,’ but stress that “a major challenge for bioinformatics is to bridge levels of granularity and scale…” Density is described by Rector as “The number of semantically ‘similar’ concepts in a particular conceptual region. How ‘bushy’ the subsumption graph is.” Rector et al.’s analysis provides logical formulations of important distinctions between density and related properties [38].
Kumar et al. use “granularity” as level of granularity in anatomy, e.g., single biological macromolecule versus the whole organism [39]. In this paper, we adopt “density” as our term, instead of “granularity.” The notion of density deals with the level of detail at which conceptual knowledge about the biomedical domain is represented in a medical terminology, which is not necessarily in terms of level of granularity in anatomy. Our approach is close to the comparative method of Sun and Zhang [40], however, they use the term “granularity” for this phenomenon. One case discovered in our analysis is that a density difference sometimes indicates the possibility of importing concepts from one terminology into the other terminology. MIREOT [41] defines a set of guidelines for importing classes from external ontologies. However, it only supports OBO foundry ontologies in Web Ontology Language (OWL) format. In this paper, all the terminologies are in UMLS Rich Release Format. Thus, the import guidelines introduced in MIREOT cannot be used here directly.
Omissions in terminologies are undesirable, and locating them is one of the goals of work in terminology auditing [42]. In past work, we have developed methods to recognize certain omissions in the UMLS and some of its source terminologies [43-45].
1.4. Objectives
Having established the need for a better understanding of inter-terminology relationships for semantic harmonization, the objectives of this work are best expressed by two connected research questions:
-
1)
What topological patterns occur between pairs of medical terminologies, and how often do these topological patterns occur?
-
2)
How can complex terminology patterns be interpreted?
The outcomes of this work are twofold: 1) We are identifying potential concepts for enhancing the conceptual content of SNOMED CT and of the selected reference terminologies. 2) We are identifying modeling errors, inconsistencies, overlooked synonyms and ambiguities in the UMLS and its source terminologies. With these outcomes, we intend to support semantic harmonization efforts between UMLS terminologies and help curators improve the quality of biomedical terminologies to better support their use in various applications.
The remainder of the paper is organized as follows. Section 2 describes the methods for identifying reference terminologies used for this work and the algorithms of finding topological patterns. In Section 3, we present the results of our experiments. Finally, we will discuss the results and implied future work in Section 4 and draw conclusions in Section 5.
2. Materials and methods
2.1. Identifying reference terminologies
In this work, we use the term “reference terminology” to refer to the selected UMLS terminologies that could potentially contribute concepts to SNOMED CT, and vice versa. We note that our use of this name is different from the standard use [46] (i.e., reference terminology versus interface terminology). Figure 2 illustrates the process of identifying reference terminologies for this work. We first identified English UMLS source terminologies with “PAR” relationships annotated with “IS_A” labels. The rationale for using these two criteria to identify an initial set of English candidate terminologies is (1) “PAR” relationships represent parent-child relationships between two concepts. (2) “IS_A” labels are used in well-defined terminologies to explicitly represent generality-specificity relationships, because a “PAR” relationship might encode, for example, meronymy (i.e., part_of).
Figure 2.

Process of identifying English reference terminologies for this work.
Next, we excluded repetitive terminologies, e.g., we used only one of two terminologies with common historical roots. We then analyzed the overlap of each of the remaining candidate terminologies with each of the 19 top-level hierarchies of SNOMED CT. If a candidate terminology has over 10% overlap with at least one hierarchy of SNOMED CT in terms of shared UMLS CUIs, it is included as a reference terminology in this work. We chose “10%” as the minimum overlap threshold, because only when a reference terminology has sufficiently many concepts in common with SNOMED CT (determined by the UMLS) can our algorithms yield a viable number of topological structures (as defined below) for semantic harmonization.
2.2. Analyzing 1:k and k:1 topological structures
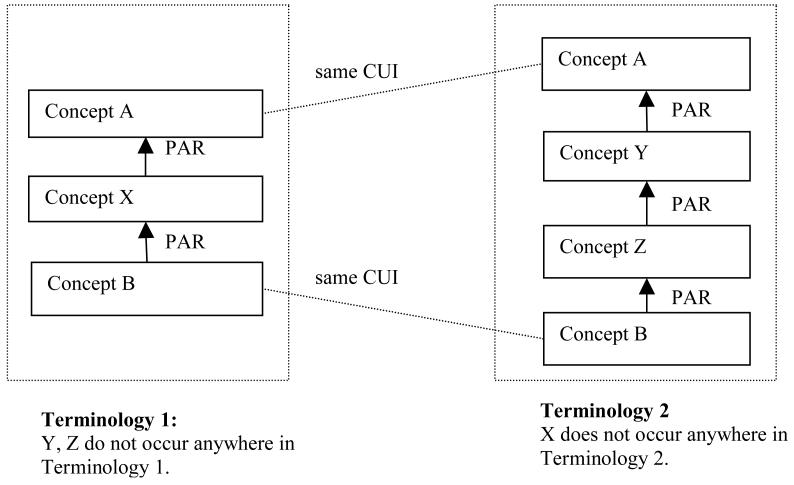
Figure 3 shows excerpts from two “hypothetical” terminologies. The instances of concept A have the same CUI in both terminologies, which means that UMLS curators regarded them as the same concept. The same is true for concept C. However, Terminology 2 has an additional concept B located on a path of PAR links from C to A. Thus, one can argue that in the limited scope of paths from C to A, Terminology 2 is of higher density than Terminology 1, because it represents more details, by including the additional concept B and its PAR links from C, and to A. Note that we ascertain that B does not appear anywhere in Terminology 1.
Figure 3.
The basic layout of a density difference.
The distinctions between Terminology 1 and Terminology 2 in Figure 3 appear in the vertical (PAR) structures of the two terminologies. Thus, this is referred to as a “vertical density difference.” The scope of this paper is limited to vertical density differences.
Due to the shape defined by the three PAR links and the two dotted lines (“same CUI”) indicating identity of the concepts from two source terminologies in the UMLS, this topological pattern is referred to as trapezoid in the balance of the paper. As the ratio of PAR links is 1:2, it is a 1:2 trapezoid. Similarly, 2:1 trapezoids are also defined. In this work, we exhaustively identify all the 1:k and k:1 trapezoids, i.e., with no intermediate concept in one terminology and multiple intermediate concepts in the other terminology (1:3, 1:4, 3:1, 4:1, etc.). We use trapezoids as a measure of density difference between the reference terminologies and SNOMED CT.
2.3. Analyzing m:n trapezoids
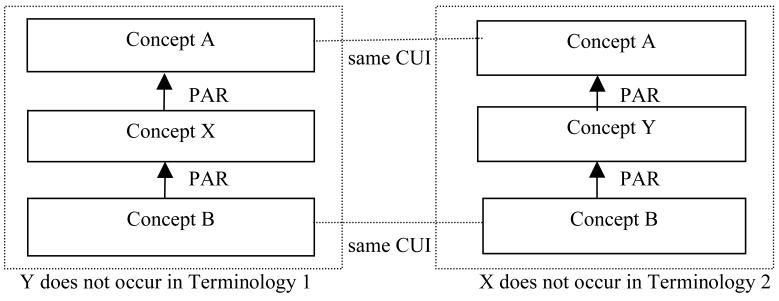
For m:n trapezoids where m >= 2 and n >= 2, the relationships of intermediate concepts from both terminologies need to be determined by domain experts. As the values of m and n grow, the possible relationships between intermediate concepts become more complex. In this study, we have conducted experiments to identify 2:n (n >= 3), 3:n (n >= 2), and 4:n (n > 1) trapezoids. Out of these kinds of trapezoids, 2:3 and 3:2 trapezoids were analyzed in detail.
For intermediate concepts X, Y and Z in a 2:3 trapezoid, as can be seen in Figure 4, it is hypothesized that there are six possible cases of how X, Y, and Z may relate to each other. Additionally, errors might be found in Terminology 1 and Terminology 2. We will not differentiate between different kinds of errors. Thus, in total, eight possibilities are defined.
Figure 4.
The layout of 2:3 topological patterns.
-
1)
The concepts X and Y are alternative classifications. That means that concept A may be validly assigned X and Y as its children. However, these two assignments are indicative of two different ways of clustering the grandchildren of A. Furthermore, concept B may be correctly classified as a child of X and as a child of Z. However, Terminology 1 omits the classification by Y and Terminology 2 omits the classification by X. (An example alternative classification would be by body location versus by population characteristics. A concrete example will be shown in Section 3.3.)
-
2)
It holds that B ➔ Z ➔ Y ➔ X ➔ A. The symbol “➔” is used to express the “IS_A” relationship. In other words, X may be inserted as a child of A and a parent of Y into Terminology 2, thereby adding more detailed information to Terminology 2. Similarly, Y may be inserted as a child of X into Terminology 1, and Z maybe inserted as child of Y into Terminology 1. Such insertions should only be done with approval of a domain expert.
-
3)
It holds that B ➔ Z ➔ X ➔ Y ➔ A, which is interpreted in a similar way as 2).
-
4)
It holds that B ➔ X ➔ Z ➔ Y ➔ A.
-
5)
Concept X is a real world synonym of concept Y, which was previously not recognized by the UMLS curators.
-
6)
Concept X is a real world synonym of concept Z, which was previously not recognized by the UMLS curators.
-
7)
There might be a structural error in Terminology 1, e.g., X is not really a child of A.
-
8)
There might be a structural error in Terminology 2, e.g., Y is an unrecognized synonym of Z.
For intermediate path concepts X, Y and Z in a 3:2 trapezoid, eight analogous hypotheses are defined. Samples of such topological patterns have been reviewed by a domain expert. The review consisted of identification of which of the eight types were present and how many of each. The results will be presented in Section 3.3. Note that 2:2 trapezoids (the notion of structurally congruent concepts in [33]) were analyzed previously and are therefore not included in this study.
2.4. Design and implementation of the trapezoid identification algorithm
We loaded the UMLS and SNOMED CT into our Oracle server. We separated SNOMED CT into 19 hierarchies for identifying reference terminologies that have sufficient overlap with at least one hierarchy of SNOMED CT to be included in our analysis. We then created a sub-table of the MRREL table (UMLS relationship table) containing all “PAR” relationships annotated with “IS_A” labels in the reference terminologies. This sub-table contained only the rows and columns absolutely necessary for the “IS_A” path construction.
We implemented the algorithms for finding all trapezoid topological patterns in pairs of terminologies in PL/SQL [47], the native procedural language of Oracle. One terminology was taken from the list of reference terminologies, the other one being SNOMED CT. The UMLS is well known to contain many cycles [35, 48]. Figure 5 illustrates an example hierarchical cycle with three concepts in the UMLS. According to our published method on auditing cycles in the UMLS [49], the thick arrow C ➔ B would be considered as erroneous and the cycle in this configuration should be broken by removing this IS_A relationship. In this study, we eliminated the hierarchical cycles during processing by detecting repeated concepts (CUIs) in the path of a terminology. In the example shown in Figure 5, the path B ➔ C ➔ B would be eliminated by our algorithms because the concept B appears twice in this path.
Figure 5.
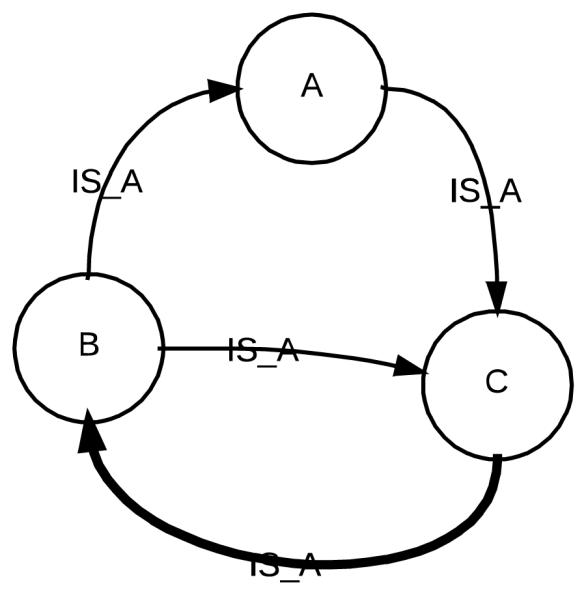
An example cycle of IS_A links with three concepts. Cycles are not allowed.
It should be noted that multiple parents may lead to overlapping trapezoids, which could in turn lead to counting the same intermediate concepts repeatedly. This problem was taken into account. When our algorithms identify various kinds of trapezoids, the same intermediate concept with multiple parents may be found in multiple trapezoids, each with a different parent. The combinations of CUIs (including the parent, the child, and the intermediate concepts) in the trapezoids are distinct. These intermediate concepts are collected, duplicates are eliminated, and counts are adjusted in our algorithms.
For each kind of trapezoid analyzed in this work, we wrote a separate PL/SQL procedure. The algorithm for identifying 1:2 trapezoids (Algorithm 1) is as follows. We matched the same concepts (concept A and concept B in this algorithm) by finding concepts in two source terminologies with the same CUIs. In the UMLS, an AUI (atom unique identifier) is the unique identifier of an atom, i.e., a term in the source terminology. Multiples AUIs in a source terminology can be mapped to the same UMLS concept with the same CUI. A relationship between two concepts in the source terminology is represented as a relationship between two AUIs in the UMLS. Therefore, we used AUIs to construct the hierarchical parent-child path within a terminology by matching the AUI of the target concept in a parent-child relationship to the AUI of the source concept in another parent-child relationship. As illustrated in Algorithm 1, the path A ➔ C ➔B was constructed by matching C’s AUI in two parent-child relationships “A IS_A C” and “C IS_A B.” In SNOMED CT, such a parent-child path resides in a single hierarchy, because all the ancestors and descendants of a given concept are located in the same hierarchy as the concept. The condition “C does not exist in Terminology 1” is true if and only if Terminology 1 does not contain the CUI of concept C. The algorithms for other trapezoid topologic patterns were written in a similar fashion.
For each reference terminology, we used the following algorithm (Algorithm 2) to identify all the m:n (where m>=1, n>=1) trapezoids. The outer loop controls the number of “PAR” relationships (“m” in this algorithm) in Terminology 1, whereas the inner loop controls the number of “PAR” relationships (“n” in this algorithm) in Terminology 2. The variable “n” is incremented whenever the algorithm finds any m:n or m:(n+1) trapezoids, while the variable “m” is incremented whenever the algorithm finds any m:1 or (m+1):1 trapezoids. For example, the algorithm will identify 1:2, 1:3, 1:4, …, 1:9 trapezoids first. If it cannot find any 1:10 and 1:11 trapezoids, it will look for 2:2, 2:3, …, 2:n trapezoids. Finally, k:1 trapezoids are identified after the algorithm finishes searching for k:n trapezoids.
3. Results
3.1. Identified reference terminologies
In the UMLS, we first identified eight candidate reference terminologies (Table 1), because they are in English and they use the “PAR” relationship annotated with the “IS_A” relationship label. The University of Washington Digital Anatomist (UWDA) was excluded, because it is a subset of FMA [50]. SCTUSX was excluded, because it has concepts in the US extension only. SNOMED CT was used as the focus terminology in this work.
Table 1.
Candidate reference terminologies.
| Terminology | Versioned source name in the UMLS |
Shortened name |
|---|---|---|
| MEDCIN | MEDCIN3_2012_07_16 | MEDCIN |
| National Cancer Institute Thesaurus | NCI2012_02D | NCI |
| Foundational Model of Anatomy Ontology | FMA3_1 | FMA |
| Gene ontology | GO2012_04_03 | GO |
| Medical Entity Dictionary | CPM2003 | CPM |
| Universal Medical Device Nomenclature System |
UMD2012 | UMD |
| University of Washington Digital Anatomist |
UWDA173 | UWDA |
| SNOMED CT US Extension | SCTUSX_2012_09_01 | SCTUSX |
According to our overlap analysis, among these six candidate terminologies, CPM and GO do not have >= 10% overlap with any of the 19 top-level hierarchies of SNOMED CT. In fact, they do not have >= 2% overlap with any of the 19 top-level hierarchies of SNOMED CT. Therefore, we removed them from the reduced candidate set of reference terminologies. The four remaining terminologies from the 2012AB release of the UMLS were used as reference terminologies for SNOMED CT. They are MEDCIN, NCI, UMD and FMA. Based on the results of our algorithmic overlap analysis, we verified that FMA may contribute concepts to the phenotypic structure and anatomical branch of SNOMED CT (Body structure hierarchy). NCI may contribute concepts to the carcinoma branch of SNOMED CT (Body structure, Pharmaceutical/biologic product, and Qualifier value hierarchies). MEDCIN, as a terminology for encoding data in EHRs, may contribute to the Clinical finding, Pharmaceutical/biologic product and Substance hierarchies of SNOMED CT. UMD may contribute concepts to the Physical object hierarchy of SNOMED CT.
3.2. Analysis of 1:k and k:1 trapezoids
Table 2 below shows the comparison of SNOMED CT with the four reference terminologies. When we calculated the numbers in columns 3 and 4, duplicate concepts were eliminated. One of the possible interpretations of a density difference is that a concept could be imported or exported into “the other” terminology. Table 2 is therefore ordered by the number of concepts that could be imported/exported (column 3).
Table 2.
Comparison of SNOMED CT with four reference terminologies.
| Reference terminology |
Size of reference terminology |
Additional concepts in reference terminology |
Number of k:1 trapezoids (reference terminology : SNOMED CT) |
Additional concepts in SNOMED CT |
Number of 1:k trapezoids (reference terminology : SNOMED CT) |
|---|---|---|---|---|---|
| NCI | 95523 | 504 | 608 | 2581 | 2125 |
| MEDCIN | 279529 | 324 | 511 | 2563 | 2304 |
| FMA | 82062 | 157 | 147 | 473 | 527 |
| UMD | 15956 | 24 | 16 | 35 | 42 |
The first column shows the name of the reference terminology and the second its size. The third column defines the number of concepts that the reference terminology could contribute to SNOMED CT. SNOMED CT could also contribute concepts to the reference terminologies. The numbers of those are in the fifth column. Columns 4 and 6 list the total numbers of 1:k trapezoids and k:1 trapezoids found by the algorithm. A path “on the right side” in a 1:3 trapezoid indicates that there are two concepts in SNOMED CT that could be contributed to the reference terminology. It can be seen from the table that NCI can contribute the largest number of concepts to SNOMED CT, while also receiving the largest number of concepts from SNOMED CT.
Table 3 shows the numbers of observed 1:k and k:1 trapezoids, ordered by increasing values of k. The table shows that 1:k trapezoids were found with k up to 9 (We did not find any 1:10 and 1:11 trapezoids, so the algorithm terminated after processing the case of 1:11). For the mirror image case, k:1 trapezoids, examples were found up to k = 6. Columns 3 and 6 show numbers of distinct additional concepts in each kind of trapezoid. The aggregated numbers of trapezoids together with the numbers of concepts can reveal to a terminology curator the association between the number of potential concepts to be imported and the complexity of the trapezoids. The number of trapezoids decreases with the increased height of the topological pattern. Importing concepts always requires approval of the responsible terminology curator. Since 2:1, 3:1, 4:1 trapezoids contribute over 90% of distinct additional concepts that could be imported into SNOMED CT, a curator should concentrate on these trapezoids to determine candidate concepts for import. In the online supplementary material (http://is.gd/pOFOJE), we have provided a spreadsheet including the CUIs of all the additional concepts and the kind of trapezoid for each identified additional concept. In the spreadsheet, we show distinct additional concepts for each pair of terminologies and each kind of trapezoid.
Table 3.
Observed trapezoids of various kinds between SNOMED CT and four reference terminologies.
| Path length ratio of reference terminology: SNOMED CT |
Number of trapezoids |
Additional concepts in SNOMED CT |
Path length ratio of reference terminology: SNOMED CT |
Number of trapezoids |
Additional concepts in reference terminology |
|---|---|---|---|---|---|
| 1:2 | 4998 | 2521 | 2:1 | 1282 | 734 |
| 1:3 | 1913 | 1754 | 3:1 | 208 | 257 |
| 1:4 | 707 | 922 | 4:1 | 27 | 67 |
| 1:5 | 439 | 628 | 5:1 | 7 | 23 |
| 1:6 | 223 | 444 | 6:1 | 1 | 5 |
| 1:7 | 94 | 174 | 7:1 | 0 | 0 |
| 1:8 | 37 | 59 | 8:1 | 0 | 0 |
| 1:9 | 4 | 10 | |||
| 1:10 | 0 | 0 | |||
| 1:11 | 0 | 0 |
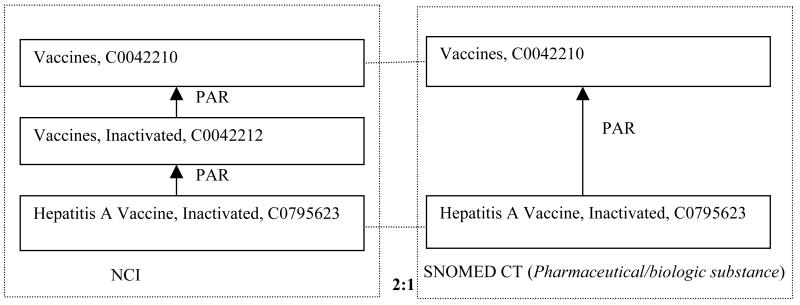
Figure 6 shows an example of a 2:1 trapezoid that was found in this research. The pair of concepts “Vaccines,” C0042210, and “Hepatitis A Vaccine, Inactivated,” C0795623, exists in the NCI and SNOMED CT. In SNOMED CT, “Hepatitis A Vaccine, Inactivated” is a child of “Vaccines.” In NCI the two concepts are separated by “Vaccines, Inactivated,” C0042212. Thus “Vaccines, Inactivated,” is a concept that could be imported into SNOMED CT (into the Pharmaceutical/biologic substance hierarchy), if a SNOMED CT curator agrees.
Figure 6.
An example of a 2:1 trapezoid that suggests a concept import into SNOMED CT.
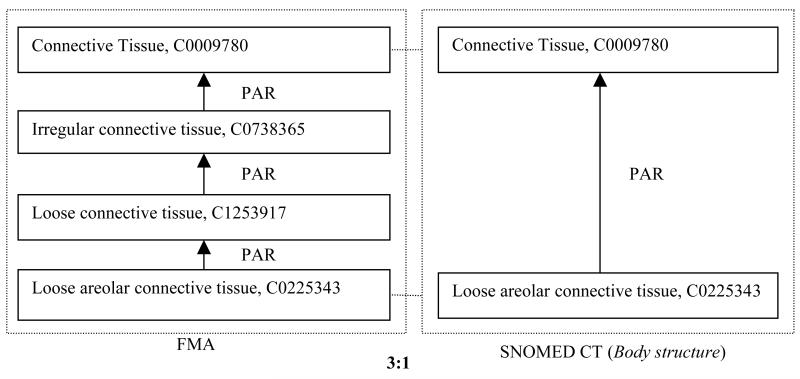
Figure 7 shows an example of a 3:1 trapezoid. Both the FMA and SNOMED CT contain the concepts “Connective Tissue” and “Loose areolar connective tissue.” There is a direct link between them in SNOMED CT, but there are two concepts “Irregular connective tissue” and “Loose connective tissue” between them in the FMA. Thus, it should be considered to import these two concepts into SNOMED CT (into the Body structure hierarchy). Table 4 shows three additional high density examples in a more space-conserving format, namely a 5:1 trapezoid with the FMA as the reference terminology, a 6:1 trapezoid with the NCI as the reference terminology and a 1:9 trapezoid with SNOMED CT having a much higher density than MEDCIN. Whether one wishes to import all those concepts depends on the domain and goals of SNOMED CT and of the reference terminologies and this decision needs to be made case by case, by the responsible ontology curators.
Figure 7.
An example of a 3:1 trapezoid that suggests two concept imports into SNOMED CT.
Table 4.
Three examples of high-ratio trapezoids
| Reference terminology | SNOMED CT |
|---|---|
| 5:1 | |
| FMA | SNOMED CT (Body structure) |
| Cell, C0007634 | Cell, C0007634 |
| Nucleated cell, C1180059 | |
| Diploid cell, C1257909 | |
| Connective Tissue Cells, C0009781 | |
| Epithelioid Cells, C0014603 | |
| Structure of interstitial cell of Leydig, C002362 | Structure of interstitial cell of Leydig, C002362 |
|
| |
| 6:1 | |
| NCI | SNOMED CT (Body structure) |
| Abnormal cell, C0333717 | Abnormal cell, C0333717 |
| Neoplastic cell, C0597032 | |
| Neoplastic Neuroepithelial Cell and Neoplastic | |
| Perineural Cell, C1514049 | |
| Neoplastic Neuroepithelial Cell, C1514048 | |
| Neoplastic Glial Cell, C1513978 | |
| Neoplastic Astrocyte, C1513925 | |
| Gemistocyte, C0333735 | Gemistocyte, C0333735 |
|
| |
| 1:9 | |
| MEDCIN | SNOMED CT (Procedure) |
| Biliary Tract Surgical Procedures, C0005427 | Biliary Tract Surgical Procedures, C0005427 |
| Bile duct operation, C0400634 | |
| Repair of bile duct, C0193566 | |
| Repair of hepatic duct, C1280034 | |
| Anastomosis of hepatic ducts, C0193540 | |
| Anastomosis of hepatic duct to gastrointestinal tract, C0193531 |
|
| Hepatojejunostomy, C0193425 | |
| Roux-en-Y hepaticojejunostomy, C0585537 | |
| Kasai procedure, C1536401 | |
| Portoenterostomy, Hepatic, C0032722 | Portoenterostomy, Hepatic, C0032722 |
3.3. Analysis of m:n trapezoids
Table 5 shows the numbers of 2:3 and 3:2 trapezoids found. In order to analyze the relationships of intermediate concepts in the trapezoids, for 2:3 trapezoids, random samples of 50 trapezoids were chosen from each of MEDCIN, NCI, and FMA, all of which have more than 50 2:3 trapezoids. For 3:2 trapezoids, random samples of 50 trapezoids were chosen from MEDCIN and NCI. If fewer than 50 trapezoids were found, i.e., for UMD and FMA, all the available trapezoids were reviewed by a domain expert.
Table 5.
2:3 and 3:2 trapezoids of SNOMED CT and reference terminologies.
| Reference terminologies | Size of reference terminology |
2:3 | Sample size |
3:2 | Sample size |
|---|---|---|---|---|---|
| MEDCIN | 279529 | 594 | 50 | 113 | 50 |
| NCI | 95523 | 335 | 50 | 219 | 50 |
| FMA | 82062 | 98 | 50 | 36 | 36 |
| UMD | 15956 | 1 | 1 | 12 | 12 |
| Total | 473,070 | 1028 | 151 | 380 | 148 |
YC, who graduated with a PhD on the topic of auditing medical terminologies and has training in Sports Medicine, was the expert who reviewed the sample. As explained in Section 2.3, we expected eight different possible cases for 2:3 trapezoids. The expert tried to identify additional cases but none were found. Table 6 shows the results, and all eight cases for intermediate path concepts in 2:3 trapezoids were observed. The results show that 39.1% are alternative classifications. Another 7.3% + 8.6% + 14.6% = 30.5 % fall into the three categories where the intermediate path concepts in the reference terminology could be imported into SNOMED CT, and vice versa.
Table 6.
Human review results of 2:3 trapezoids.
| Reference terminology |
Sample size |
Alter. classi- fication |
Z ➔ Y ➔ X | Z ➔ X ➔ Y | X ➔ Z ➔ Y | X is a synonym of Y |
X is a synonym of Z |
Error in terminology 1 |
Error in terminology 2 |
|---|---|---|---|---|---|---|---|---|---|
| MEDCIN | 50 | 20 | 5 | 4 | 10 | 2 | 6 | 3 | -- |
| NCI | 50 | 22 | 4 | 7 | 7 | 6 | 4 | -- | -- |
| UMD | 1 | -- | -- | -- | -- | 1 | -- | -- | -- |
| FMA | 50 | 17 | 2 | 2 | 5 | 4 | 19 | -- | 1 |
| Total | 151 | 59 | 11 | 13 | 22 | 13 | 29 | 3 | 1 |
| Percentage | 100% | 39.1% | 7.3% | 8.6% | 14.6% | 8.6% | 19.2% | 2.0% | 0.7% |
Table 7 shows the results for 3:2 trapezoids according to the eight hypotheses for intermediate path concepts. The results show that 59.5% are alternative classifications. Another 9.5% + 3.4% + 4.7% = 17.6% fall into the three categories where the intermediate path concepts in the reference terminology could be imported into SNOMED CT and vice versa.
Table 7.
Human review results of 3:2 trapezoids.
| Reference terminology |
Sample size |
Alter. classi- fication |
Y ➔ X ➔ Z | Y ➔ Z ➔ X | Z ➔ Y ➔ X | Z is a synonym of X |
Z is a synonym of Y |
Error in terminology 1 |
Error in terminology 2 |
|---|---|---|---|---|---|---|---|---|---|
| MEDCIN | 50 | 31 | 5 | 2 | 4 | 5 | 2 | 1 | -- |
| NCI | 50 | 30 | 3 | 3 | 2 | 8 | 4 | -- | -- |
| UMD | 12 | 2 | -- | -- | 1 | 9 | -- | -- | -- |
| FMA | 36 | 25 | 6 | -- | 0 | 2 | 3 | -- | -- |
| Total | 148 | 88 | 14 | 5 | 7 | 24 | 9 | 1 | 0 |
| Percentage | 100% | 59.5% | 9.5% | 3.4% | 4.7% | 16.2% | 6.1% | 0.7% | 0% |
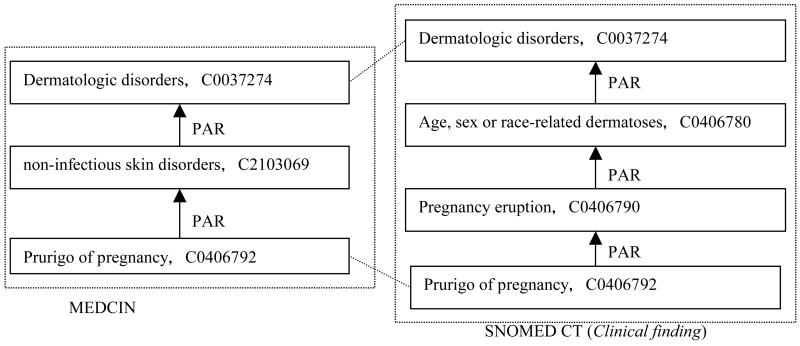
Figure 8 shows an example where intermediate concepts between MEDCIN and SNOMED CT (Clinical finding hierarchy) were deemed as alternative classifications. Thus, “non-infectious skin disorders” in MEDCIN is a classification by infectiousness, while in SNOMED CT, “Age, sex or race-related dermatoses” is a classification by patient characterization. Figure 9 shows another example topological configuration between FMA and SNOMED CT (Body structure hierarchy) that was also deemed an alternative classification.
Figure 8.
An example of alternative classifications between MEDCIN and SNOMED CT.
Figure 9.
An example of alternative classifications between FMA and SNOMED CT.
By making the implicit knowledge of different ways how to classify a concept explicit, terminology curators can contrast their view with other terminologies and possibly codify the alternative classifications in their terminology.
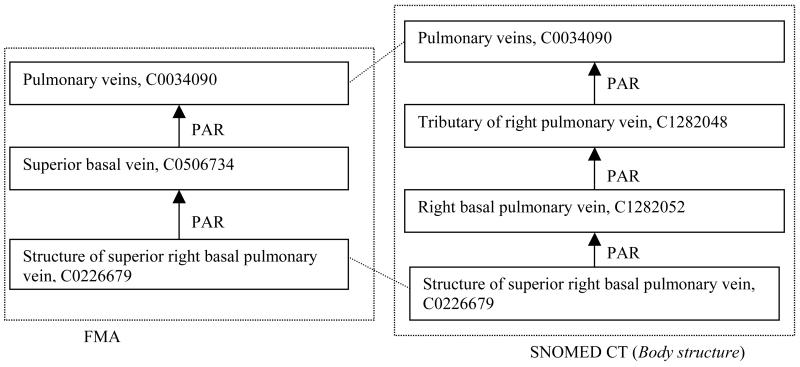
In SNOMED CT, pulmonary veins are apparently first divided by laterality, while FMA first distinguishes basal veins from other pulmonary veins. SNOMED CT then does a secondary classification of right pulmonary veins, with the result that both FMA and SNOMED CT contain (Structure of) right basal pulmonary vein.
The first example in Table 8 illustrates a case where the intermediate concept X was identified as a parent of the other concept Y by the domain expert. In this example, the intermediate concept “Systemic Congenital Disorder” can be a parent of “Congenital abnormality of lower limb AND/OR pelvic girdle,” thus the intermediate concept “Systemic Congenital Disorder” from NCI may be added as a parent of “Congenital abnormality of lower limb AND/OR pelvic girdle” in SNOMED CT (Clinical finding hierarchy), and vice versa, if this is needed according to the judgment of the curators of NCI and/or SNOMED CT.
Table 8.
Four examples for 2:3 trapezoids
| Reference terminology | SNOMED CT |
|---|---|
| NCI | SNOMED CT (Clinical finding) |
| Congenital Abnormality, C0000768 | Congenital Abnormality, C0000768 |
| X: Systemic Congenital Disorder, C3273258 |
Y: Congenital abnormality of lower limb AND/OR pelvic girdle, C0456309 |
| Z: Congenital anomaly of the pelvis, C0265708 | |
| Urogenital Abnormalities, C0042063 | Urogenital Abnormalities, C0042063 |
|
| |
| MEDCIN | SNOMED CT (Clinical finding) |
| Respiration Disorders, C0035204 | Respiration Disorders, C0035204 |
| X: pulmonary obstructive disorders, C2103594 | Y: Disorder of lower respiratory system, C1290325 |
| Z: Bronchial Diseases, C0006261 | |
| Stenosis of bronchus, C0151536 | Stenosis of bronchus, C0151536 |
|
| |
| FMA | SNOMED CT (Substance) |
| Body substance, C0504082 | Body substance, C0504082 |
| X: Ingested food, C1179481 | Y: Gastrointestinal Contents, C0017177 |
| Z: Intestinal Contents, C0226893 | |
| Small intestine contents, C0227258 | Small intestine contents, C0227258 |
|
| |
| MEDCIN | SNOMED CT (Clinical finding) |
| Gastrointestinal Diseases, C0017178 | Gastrointestinal Diseases, C0017178 |
| X: disorder of jejunum and ileum, C2103077 | Y: Disorder of upper gastrointestinal tract, C1290613 |
| Z: Duodenal Diseases, C0013289 | |
| Duodenal varices, C0580178 | Duodenal varices, C0580178 |
The second example in Table 8 shows a case where the intermediate concept X was identified as a parent of the concept Z and as a child of the concept Y by the domain expert. In this example, the intermediate concept “pulmonary obstructive disorders” can be a parent of “Bronchial Diseases,” and a child of “Disorder of lower respiratory system,” thus the intermediate concept “pulmonary obstructive disorders” from MEDCIN may be added as a parent of “Bronchial Diseases,” as well as a child of “Disorder of lower respiratory system” in SNOMED CT (Clinical finding hierarchy), and vice versa. (See our comments about this example in the Discussion section.)
The third example in Table 8 shows a case where Concept X was identified as a synonym of Y. In this example, the intermediate concepts “Ingested food” from the FMA and “Gastrointestinal Contents” from SNOMED CT (Substance hierarchy) were deemed by our domain expert to be synonyms that had not been recognized previously and thus should be merged.
The domain expert decided that there is an error in MEDCIN (the fourth example in Table 8). Duodenum, jejunum and ileum are the three different segments of the small intestine. Duodenal varices are located in the duodenum, not jejunum or ileum. Therefore, “Duodenal varices” are not a disorder of jejunum and ileum.
For 1:k and k:1 trapezoid topological patterns (k > 1) there is no intermediate concept in one terminology but there are multiple intermediate concepts in the other one. In these cases, semantic harmonization can be proposed to a domain expert by importing some or all of the intermediate concepts from one terminology into the other one. For 2:3 and 3:2 trapezoid topological patterns, domain experts are always needed to identify the relationships among intermediate concepts, chosen from the eight possibilities introduced in this paper.
We have also conducted experiments on cases of 2:n (n > 3), 3:n (n > 2), and 4:n (n > 1) trapezoids. Table 9 shows the number of trapezoids identified for each kind. With the values of m and n growing, the number of possible cases of relationships among intermediate path concepts grows very fast.
Table 9.
Numbers of 2:n. 3:n, and 4:n trapezoids identified.
| Trapezoid kind | # of trapezoids | Trapezoid kind | # of trapezoids | Trapezoid kind | # of trapezoids |
|---|---|---|---|---|---|
| 2:4 | 678 | 3:3 | 503 | 4:2 | 143 |
| 2:5 | 469 | 3:4 | 479 | 4:3 | 234 |
| 2:6 | 264 | 3:5 | 532 | 4:4 | 314 |
| 2:7 | 140 | 3:6 | 550 | 4:5 | 447 |
| 2:8 | 79 | 3:7 | 588 | 4:6 | 329 |
| 2:9 | 0 | 3:8 | 396 | 4:7 | 245 |
| 2:10 | 0 | 3:9 | 217 | 4:8 | 106 |
| 3:10 | 98 | 4:9 | 65 | ||
| 3:11 | 21 | 4:10 | 39 | ||
| 3:12 | 3 | 4:11 | 14 | ||
| 3:13 | 2 | 4:12 | 2 | ||
| 3:14 | 0 | 4:13 | 0 | ||
| 3:15 | 0 | 4:14 | 0 |
For example, in a 3:8 trapezoid, each of the two intermediate concepts on the left side might be a synonym of each of the seven intermediate concepts on the right side, allowing for fourteen possible connections. However, if both concepts on the left side have synonyms among the concepts on the right side, the number of correct possibilities is mutually constrained by the two concepts. In the same way, each of the two left concepts could potentially be inserted between any pair of concepts on the right side, but the possible insertions for one concept are not independent of the insertions of the other concept.
4. Discussion
In this work, we have considerably extended the previously introduced structural methodology for identifying potential concepts for a terminology. We found that this methodology is viable for identifying potentially useful concepts in one terminology for inclusion as parents, children, new synonyms, etc. in another terminology. Furthermore, we observed striking density differences between four reference terminologies and SNOMED CT, which suggests that there is ample room for applying this algorithmic methodology to terminology maintenance.
As a leading clinical terminology, SNOMED CT has recently attracted attention regarding harmonization with various other terminologies, e.g., ICD-11 [51] and LOINC [52]. The harmonization of ICD-11 and SNOMED CT includes a common concept model based on SNOMED CT. ICD-11 will be based on the common concept model using a linearization technique. ICD codes are mainly used for diagnoses and billing, whereas SNOMED CT is a rich reference terminology for, e.g., encoding problem lists. The ICD-11 project has the underlying philosophy that classifications and nomenclature are different from each other, with different purposes, echoing a well-known controversy [53].
The cooperative work of IHTSDO and the Regenstrief Institute to link SNOMED CT and LOINC was listed as one of the top 10 Informatics events in 2013 by the American Medical Informatics Association [54]. The goal of this initiative is to provide a common framework within which to use LONIC and SNOMED CT, so that the overlap between two terminologies should not be extended. In the LOINC project the underlying philosophy is that under a common concept model, concepts in LOINC should not have an identifier in SNOMED CT. Moreover, LOINC concepts can have links to post-coordinated SNOMED CT expressions, so that transformations between the representations are possible.
These studies on ICD-11 and LOINC have focused on harmonization of concept models, but not on enriching the content coverage of SNOMED CT. For concepts present in both, a link will be constructed. In this way, ICD-11 and LOINC can be used for what they are best at, with full interoperability but without creating redundant representations.1 Our study abstracts away from the purposes of terminologies and classifications as well as from the interoperability architectures. The methodology leverages harmonized terminologies in the UMLS to algorithmically suggest concepts, thereby supporting a concept enrichment mechanism as opposed to the aforementioned approaches [51, 52].
Previously, Weng et al. presented the BRIDG model as a community effort to harmonize existing information models and ontologies for clinical research [11]. It requires extensive in-person meetings, which are time-consuming and laborious. Tao et al. discussed the semantic harmonization of OWL-based ontologies, to be used for deriving temporal relations from clinical narratives [55]. The OWL conversion ensures the quality of the result, because it is based on a logical representation of concepts, which is an advantage of the approach. The UMLS approach in our study is dependent on the quality of the CUIs, and a curator must go through a large number of trapezoids to identify those that are relevant. However, Tao’s approach requires that the ontologies to be harmonized must be authored in the OWL format or converted to OWL format. The OWL conversion needs to be evaluated by domain experts. In contrast, the UMLS uses a uniform structure for all its source terminologies. Therefore, we can perform semantic harmonization of its source terminologies regardless of what their original formats are.
In the Result Section, we have demonstrated that by considerably extending the structural method introduced previously [33], large-scale semantic harmonization can be supported in a generalizable manner, but not completely automated. We also observed that one reference terminology can form trapezoids with one or several hierarchies of SNOMED CT. For example, NCI has over 10% overlap with the following hierarchies of SNOMED CT: Qualifier value, Substance, Pharmaceutical/biologic product, Linkage concept, Environment or geographic location, and Body structure. Because (1) our algorithm uses “IS_A” relationships and “AUIs” defined in the source terminology to construct a parent-child path in a terminology, and (2) SNOMED CT concepts are separated into 19 “IS_A” hierarchies, the parent-child path in SNOMED CT in a trapezoid resides completely in one hierarchy. It does not cross hierarchy boundaries of SNOMED CT. Therefore, the potential concept(s) found in a trapezoid would be suggested to be imported into one hierarchy of SNOMED CT.
Our method for enriching the conceptual content of a terminology also has the potential to improve semantic interoperability. In a hypothetical clinical scenario, if a doctor cannot identify a proper existing term or post-coordinated term to describe the clinical scenario of a patient, s/he would use free-text to record the findings, thereby reducing the accessibility of the record to computational processing. Natural Language Processing of clinical notes may help, however, a comprehensive terminology would be needed so that free-text notes can be transformed into a computer-recognizable format. Our methodology can help with enriching a terminology to achieve a higher level of comprehensiveness.
4.1. Limitations
A limitation of this research is that only vertical topological patterns of “PAR” links are used. The UMLS also supports “RB” (Relationship Broader) links that function in an analogous way to “PAR” links, but differ in the source of the relationships. Furthermore, we have only used “IS_A” annotations of “PAR” links. Many “PAR” relationships do not have any annotation (roughly half of them), but about 20,000 are annotated to indicate a part link, distinguishing those from relationships annotated in other ways, e.g., those expressing an “IS_A” link. A thorough analysis distinguishing between “PAR” relationships with different annotations and comparing the results with paths of “RB” relationships would provide deeper insights into the phenomenon of density differences.
An important limitation is caused by the UMLS itself. Over the past 25 years, our SABOC research group has devised numerous auditing techniques for the UMLS. Still, due to its size and complexity, while old modeling errors and inconsistencies were eliminated, new errors were introduced in new releases. Therefore, when using the UMLS as a backend for biomedical research, one needs to consider some intrinsic problems of the UMLS, which might affect the results of the experiments. Some findings in this work could also contribute to improving the UMLS, e.g., by making previously unrecognized synonyms explicit.
In this work, we only leveraged the native term mappings of the UMLS, i.e., terms with the same meaning were grouped together by the UMLS editors with the same CUI. It is possible that term mapping errors of the UMLS might have led to a few errors in our analysis. We did not use other techniques to identify concepts of similar meaning. Therefore, a suggested potential concept might have a semantically similar concept but with a different CUI in the target terminology, due to an oversight of the UMLS curators. For example, in the second example of Table 8, we suggested that the concept “pulmonary obstructive disorder” could be imported into SNOMED CT as a parent of “Bronchial Diseases” and a child of “Disorder of lower respiratory system.” However, there exists a concept “Respiratory obstruction” in SNOMED CT, which is similar to “pulmonary obstructive disorder,” but the two concepts have two different CUIs in the UMLS.
Another limitation of this work is that it uses SNOMED CT concepts and all reference terminology concepts in the format that they were provided in by the UMLS. Existing differences between the original concept representations of SNOMED CT (or the reference terminologies) and the representation of SNOMED CT that is accessible through the UMLS might cause subtle differences in the results.
4.3. Future work
We plan to report our results to the SNOMED CT curators, i.e., the content team of IHTSDO and the U.S. National Release Center at the U.S. National Library of Medicine. After getting their feedback and arguments for inclusion or exclusion of our suggested concepts, we will refine our methodology accordingly.
Additionally, in future work, experiments with other relationship annotations besides IS_A will be performed, although pilot experiments indicate that the expected yield is low. We plan to perform experiments between any two overlapping terminologies, i.e., Terminology 2 will not be limited to SNOMED CT anymore. In this work, we used only one release of the UMLS. As new versions of the source terminologies appear in new releases of the UMLS, we plan to perform a cross-release longitudinal study to assess how the density differences between two terminologies evolve over time, which may reveal more interesting patterns to support semantic harmonization between terminologies.
In this research, we analyzed only vertical density differences. In the future, we will investigate horizontal density differences according to numbers of corresponding sibling concepts between pairs of terminologies, which can potentially identify missing concepts for import.
Our methods identify concepts that may enrich the contents of SNOMED CT and four reference terminologies. One may question whether importing some of the intermediate concepts is really necessary. We plan to develop a data-driven method to evaluate the usefulness of potential concepts for a specific data set, e.g., free text clinical trial eligibility criteria of all the trial summaries on ClinicalTrials.gov. We will identify frequently used n-grams in the free-text eligibility criteria that are not SNOMED CT concepts and match them with the potential concepts identified in this study to find valuable concepts for import.
5. Conclusions
In this analysis of 1:k and k:1 trapezoid topological patterns, path length ratios of up to 1:9 and 6:1 were observed, i.e., a parent in MEDCIN was separated in SNOMED CT from the MEDCIN child by a path of nine “PAR” relationships. Six “PAR” relationships were found in NCI between two concepts that are connected by one “PAR” relationship in SNOMED CT. SNOMED CT curators could consider importing intermediate concepts found in these trapezoids to improve its coverage. SNOMED CT itself could function as a source for exporting concepts to the four reference terminologies.
In the analysis of 2:3 and 3:2 trapezoid topological patterns, 299 randomly selected cases were reviewed by a domain expert. It was shown that 39.1% and 59.5% of the intermediate concepts were deemed to be alternative classification in the 2:3 and 3:2 trapezoids, respectively. In 30.5% and 17.6% of the cases it was found that intermediate concepts could be imported into SNOMED CT and into reference terminologies, respectively, to enhance their conceptual content. This study contributes a structural method that has the potential to support semantic harmonization efforts.
Highlights.
We designed a structure-based algorithmic method to support semantic harmonization.
We defined a set of topological patterns across pairs of terminologies that are of different density.
We explored how these topological patterns appear for a subset of UMLS terminologies, focusing on SNOMED CT.
A human expert evaluated the method with a random sample generated by the algorithm.
Acknowledgments
We want to thank Drs. Yehoshua Perl, Michael Halper, Mei Liu, Gai Elhanan, and Chunhua Weng for providing their feedback and sharing their insights for this work. We also want to thank three anonymous referees for providing comprehensive comments that have significantly improved the quality of this manuscript. This work was partially supported by the U.S. National Cancer Institute of the National Institutes of Health under award number R01CA190779. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
We thank an anonymous reviewer for clarifying these points.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Agrawal A, He Z, Perl Y, Wei D, Halper M, Elhanan G, et al. The readiness of SNOMED problem list concepts for meaningful use of electronic health records. Artif Intell Med. 2013;58(2):73–80. doi: 10.1016/j.artmed.2013.03.008. [DOI] [PubMed] [Google Scholar]
- [2].Campbell JR, Xu J, Fung KW. Can SNOMED CT fulfill the vision of a compositional terminology? Analyzing the use case for problem list. AMIA Annu Symp Proc. 2011;2011:181–8. [PMC free article] [PubMed] [Google Scholar]
- [3].Matney SA, Warren JJ, Evans JL, Kim TY, Coenen A, Auld VA. Development of the nursing problem list subset of SNOMED CT(R). J Biomed Inform. 2012;45(4):683–8. doi: 10.1016/j.jbi.2011.12.003. [DOI] [PubMed] [Google Scholar]
- [4].Finnegan R. ICD-9-CM coding for physician billing. J Am Med Rec Assoc. 1989;60(2):22–3. [PubMed] [Google Scholar]
- [5].Demner-Fushman D, Chapman WW, McDonald CJ. What can natural language processing do for clinical decision support? J Biomed Inform. 2009;42(5):760–72. doi: 10.1016/j.jbi.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tenenbaum JD, Sansone SA, Haendel M. A sea of standards for omics data: sink or swim? J Am Med Inform Assoc. 2014;21(2):200–3. doi: 10.1136/amiajnl-2013-002066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. [Accessed: 30 December 2014];Health Level Seven International Homepage. Available from: http://www.hl7.org.
- [8]. [Accessed: 30 December 2014];OpenEHR Homepage. Available from: http://www.openehr.org.
- [9].Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010;43(3):451–67. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rector A, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Applied Ontology. 2009;4(1):51–69. [Google Scholar]
- [11].Weng C, Gennari JH, Fridsma DB. User-centered semantic harmonization: a case study. J Biomed Inform. 2007;40(3):353–64. doi: 10.1016/j.jbi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- [12].Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4-5):394–403. [PMC free article] [PubMed] [Google Scholar]
- [13].Cimino JJ. High-quality, standard, controlled healthcare terminologies come of age. Methods Inf Med. 2011;50(2):101–4. [PMC free article] [PubMed] [Google Scholar]
- [14].Richesson RL, Fung KW, Krischer JP. Heterogeneous but “standard” coding systems for adverse events: Issues in achieving interoperability between apples and oranges. Contemp Clin Trials. 2008;29(5):635–45. doi: 10.1016/j.cct.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Stearns MQ, Price C, Spackman KA, Wang AY. SNOMED clinical terms: overview of the development process and project status. Proc AMIA Symp. 2001:662–6. [PMC free article] [PubMed] [Google Scholar]
- [16].IHTSDO [Accessed: 30 December 2014];SNOMED CT Homepage. Available from: http://www.ihtsdo.org.
- [17].IHTSDO [Accessed: 30 December 2014];SNOMED CT Value Proposition. Available from: http://www.ihtsdo.org/snomed-ct/whysnomedct/snomedfeatures/
- [18].CMS [Accessed: 30 December 2014];Electronic Health Record Incentive Program—Stage 2. Available from: http://www.gpo.gov/fdsys/pkg/FR-2012-09-04/pdf/2012-21050.pdf.
- [19].CMS [Accessed: 30 December 2014];Eligible Professional Meaningful Use Core Measures. Available from: http://www.cms.gov/Regulations-and-Guidance/Legislation/EHRIncentivePrograms/Downloads/2013DefinitionEP_3_Maintain_Problem_List.pdf.
- [20]. [Accessed: 30 December 2014];Standards for Health IT: Meaningful Use and Beyond. Available from: http://www.hhs.gov/asl/testify/2012/11/t20121114a.html.
- [21].Medicare and Medicaid Programs. Electronic Health Record Incentive Program. CMS-0033-P. RIN 938-AP78 [Accessed: 1 January 2014]; Available from: http://healthit.hhs.gov/portal/server.pt/gateway/
- [22].Blumenthal D. Launching HITECH. N Engl J Med. 2010;362(5):382–5. doi: 10.1056/NEJMp0912825. [DOI] [PubMed] [Google Scholar]
- [23].Elkin PL, Brown SH, Husser CS, Bauer BA, Wahner-Roedler D, Rosenbloom ST, et al. Evaluation of the content coverage of SNOMED CT: ability of SNOMED clinical terms to represent clinical problem lists. Mayo Clin Proc. 2006;81(6):741–8. doi: 10.4065/81.6.741. [DOI] [PubMed] [Google Scholar]
- [24].Elhanan G, Perl Y, Geller J. A survey of SNOMED CT direct users, 2010: impressions and preferences regarding content and quality. J Am Med Inform Assoc. 2011;18(Suppl 1):i36–44. doi: 10.1136/amiajnl-2011-000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].He Z, Halper M, Perl Y, Elhanan G. Clinical clarity versus terminological order: the readiness of SNOMED CT concept descriptors for primary care; Proceedings of the 2nd international workshop on Managing interoperability and compleXity in health systems; Maui, Hawaii, USA. 2012; pp. 1–6. 2389674: ACM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Campbell WS, Campbell JR, West WW, McClay JC, Hinrichs SH. Semantic analysis of SNOMED CT for a post-coordinated database of histopathology findings. J Am Med Inform Assoc. 2014;21(5):885–92. doi: 10.1136/amiajnl-2013-002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tuttle M, Sherertz DD, M. E, Olson N, Nelson S. Implementing Meta-1: The First Version of the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care. 1989:483–7. [Google Scholar]
- [29].Schuyler PL, Hole WT, Tuttle MS, Sherertz DD. The UMLS Metathesaurus: representing different views of biomedical concepts. Bull Med Libr Assoc. 1993;81(2):217–22. [PMC free article] [PubMed] [Google Scholar]
- [30].Taboada M, Lalin R, Martinez D. An automated approach to mapping external terminologies to the UMLS. IEEE Trans Biomed Eng. 2009;56(6):1598–605. doi: 10.1109/TBME.2009.2015651. [DOI] [PubMed] [Google Scholar]
- [31].Marquet G, Mosser J, Burgun A. A method exploiting syntactic patterns and the UMLS semantics for aligning biomedical ontologies: the case of OBO disease ontologies. Int J Med Inform. 2007;76(Suppl 3):S353–61. doi: 10.1016/j.ijmedinf.2007.03.004. [DOI] [PubMed] [Google Scholar]
- [32].Huang KC, Geller J, Halper M, Perl Y, Xu J. Using WordNet synonym substitution to enhance UMLS source integration. Artif Intell Med. 2009;46(2):97–109. doi: 10.1016/j.artmed.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He Z, Geller J, Elhanan G. Categorizing the Relationships between Structurally Congruent Concepts from Pairs of Terminologies for Semantic Harmonization. AMIA Jt Summits Transl Sci Proc. 2014;2014:48–53. [PMC free article] [PubMed] [Google Scholar]
- [34].Bodenreider O. Strength in numbers: exploring redundancy in hierarchical relations across biomedical terminologies. AMIA Annu Symp Proc. 2003:101–5. [PMC free article] [PubMed] [Google Scholar]
- [35].Bodenreider O. Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and prevention. Proc AMIA Symp. 2001:57–61. [PMC free article] [PubMed] [Google Scholar]
- [36].Arts DG, Cornet R, de Jonge E, de Keizer NF. Methods for evaluation of medical terminological systems--a literature review and a case study. Methods Inf Med. 2005;44(5):616–25. [PubMed] [Google Scholar]
- [37].Cornet R. Information-content-based measures for the structure of terminological systems and for data recorded using these systems. Stud Health Technol Inform. 2010;160(Pt 2):1075–9. [PubMed] [Google Scholar]
- [38].Rector A, Rogers J, Bittner T. Granularity, scale and collectivity: when size does and does not matter. J Biomed Inform. 2006;39(3):333–49. doi: 10.1016/j.jbi.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [39].Kumar A, Smith B, Novotny DD. Biomedical informatics and granularity. Comp Funct Genomics. 2004;5(6-7):501–8. doi: 10.1002/cfg.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sun P, Zhang S. Identifying Granularity Differences between Large Biomedical Ontologies through Rules. AMIA Annu Symp Proc. 2010;2010:927–31. [PMC free article] [PubMed] [Google Scholar]
- [41].Courtot M, Gibson F, Lister AL, Malone J, Schober D, Brinkman RR, et al. In: Smith B, editor. MIREOT: The minimum information to reference an external ontology term; International Conference on Biomedical Ontology; Buffalo, New York, USA. 2009.pp. 87–90. [Google Scholar]
- [42].Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. J Biomed Inform. 2009;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. [DOI] [PubMed] [Google Scholar]
- [43].Wei D, Halper M, Elhanan G, Chen Y, Perl Y, Geller J, et al. Auditing SNOMED relationships using a converse abstraction network. AMIA Annu Symp Proc. 2009;2009:685–9. [PMC free article] [PubMed] [Google Scholar]
- [44].Gu H, Perl Y, Geller J, Halper M, Liu LM, Cimino JJ. Representing the UMLS as an object-oriented database: modeling issues and advantages. J Am Med Inform Assoc. 2000;7(1):66–80. doi: 10.1136/jamia.2000.0070066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen Y, Gu HH, Perl Y, Geller J. Structural group-based auditing of missing hierarchical relationships in UMLS. J Biomed Inform. 2009;42(3):452–67. doi: 10.1016/j.jbi.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. [Accessed: 30 December 2014];Definition of Reference Terminology. Available from: http://library.ahima.org/xpedio/groups/public/documents/ahima/bok1_022744.hcsp?dDocName=bok1_022744.
- [47]. [Accessed: 30 December 2014];Oracle. PL/SQL Homepage. Available from: http://www.oracle.com/technetwork/database/features/plsql/index.html.
- [48].Mougin F, Bodenreider O. Approaches to eliminating cycles in the UMLS Metathesaurus: naive vs. formal. AMIA Annu Symp Proc. 2005:550–4. [PMC free article] [PubMed] [Google Scholar]
- [49].Halper M, Morrey CP, Chen Y, Elhanan G, Hripcsak G, Perl Y. Auditing hierarchical cycles to locate other inconsistencies in the UMLS. AMIA Annu Symp Proc. 2011;2011:529–36. [PMC free article] [PubMed] [Google Scholar]
- [50].NLM University of Washington Digital Anatomist Source Information. 2013 May; Available from: http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/UWDA/
- [51].Rodrigues JM, Schulz S, Rector A, Spackman KA, Üstün B, Chute CG, et al. Sharing Ontology between ICD 11 and SNOMED CT will enable Seamless Re-use and Semantic Interoperability. Stud Health Technol Inform. 2013;192:343–6. [PubMed] [Google Scholar]
- [52].IHTSDO [Accessed: 1 March 2014];New Regenstrief and IHTSDO agreement to make EMRs more effective at improving health care. 2013 Available from: http://www.ihtsdo.org/fileadmin/user_upload/Docs_01/About_IHTSDO/Harmonization/IHTSDO_Regenstrief_2013_agreement_announcement_20130724.pdf.
- [53].Ingenerf J, Giere W. Concept-oriented standardization and statistics-oriented classification: continuing the classification versus nomenclature controversy. Methods Inf Med. 1998;37(4-5):527–39. [PubMed] [Google Scholar]
- [54].Informatics H. [Accessed: 30 December 2014];Live From AMIA: Top 10 Informatics Events of the Year 2013. Available from: http://www.healthcare-informatics.com/article/live-amia-top-10-informatics-events-year.
- [55].Tao C, Solbrig HR, Chute CG. CNTRO 2.0: A Harmonized Semantic Web Ontology for Temporal Relation Inferencing in Clinical Narratives. AMIA Summits Transl Sci Proc. 2011;2011:64–8. [PMC free article] [PubMed] [Google Scholar]