Abstract
The cysteine (Cys) proteome is a major component of the adaptive interface between the genome and the exposome. The thiol moiety of Cys undergoes a range of biologic modifications enabling biological switching of structure and reactivity. These biological modifications include sulfenylation and disulfide formation, formation of higher oxidation states, S-nitrosylation, persulfidation, metallation, and other modifications. Extensive knowledge about these systems and their compartmentalization now provides a foundation to develop advanced integrative models of Cys proteome regulation. In particular, detailed understanding of redox signaling pathways and sensing networks is becoming available to discriminate network structures. This research focuses attention on the need for atlases of Cys modifications to develop systems biology models. Such atlases will be especially useful for integrative studies linking the Cys proteome to imaging and other omics platforms, providing a basis for improved redox-based therapeutics. Thus, a framework is emerging to place the Cys proteome as a complement to the quantitative proteome in the omics continuum connecting the genome to the exposome.
Introduction
The sulfur atom of cysteine (Cys) provides a considerable range of chemical reactivity and structural flexibility in the proteome. The presence of conserved Cys in motifs of proteins found in essentially all life forms indicates that these chemical characteristics were harnessed in early evolution to support enzyme catalysis, transcriptional regulation, protein folding and 3-dimensional structure. A remarkable range of biologic functions is supported by Cys because sulfur is stable in multiple coordinate covalent bonds with the major atoms of living organisms (C, H, O, N, P), forms stable coordinate complexes with transition metal ions (Zn, Fe, Cu) and is stable at a range of oxidation states (-SH, -SS-, -SO2−1, -SO3−1). Additionally, Cys thiols differentially undergo reversible ionization to the negatively charged thiolate form over the physiologic range of pH to flexibly optimize the functions of specific peptidyl Cys.
The human genome encodes about 214,000 Cys. Cys is widely distributed among proteins, with most expressing at least one and many having multiple coordinated Cys and Cys-rich domains. The reactivity and diverse functions of Cys are mirrored by a spectrum of susceptibilities and dysfunctions of their respective proteins, resulting in central roles of the Cys proteome in development, signal transduction, biologic defenses, aging and disease. Considerable knowledge has accumulated for the symptoms and etiology of major human diseases, such as cardiovascular disease, Alzheimer’s disease, lung disease, metabolic syndrome, eye disease and diseases of other organ systems. These often share related oxidative phenotypes and mechanisms, and progress in redox systems biology is beginning to provide an understanding of the integration of the underlying redox systems.
Early photosynthetic organisms transformed the earth by fixing the carbon atom of CO2 into hydrocarbons and releasing bound atomic oxygen as gaseous molecular O2. This changed the atmosphere from <1% to about 21% O2 and had a profound effect on life forms by creating a diversity of external environments with different oxidative conditions. Contemporary redox proteomic systems have evolved as a result of the selective pressures of an oxygen environment and aerobic respiration, with extant genomes having selectively advantageous redox proteomic structures that protect the genome and ensure its replication despite environmental, organismal and developmental variation in oxidative and reductive challenges. Although oxidative pressure influences multiple critical loci of the redox proteome such as methionine (Met) and selenocysteine (Sec), the Cys proteome is central within this spectrum of biological plasticity and is the focus of this review.
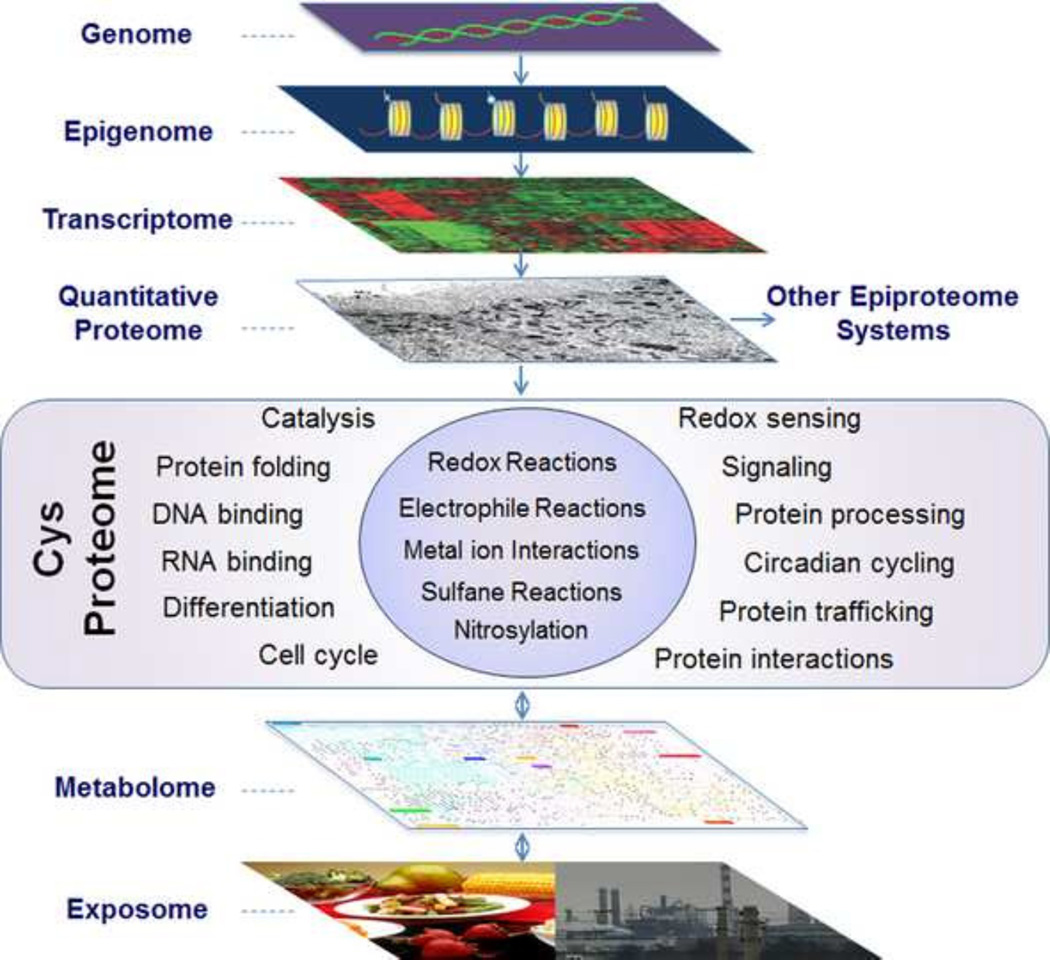
The Cys proteome (Fig. 1) serves as a central adaptive interface between the functional genome and the external environment of an organism [1]. The external environment is a critical determinant of individual survival; in human health, this is now discussed in terms of an individual’s “exposome” [2]. The exposome is defined as the cumulative measure of environmental influences and associated biological responses throughout the life span, including exposures from the environment, diet, behavior, and endogenous processes [3]. In addition to utility in contemporary environmental health research [3, 4], this provides a useful way to think about the evolution of complex multicellular organisms as the content of O2 increased in the atmosphere. Specifically, the redox sensing and signaling mechanisms supporting responses to challenge also provided sensing and signaling systems for temporal and spatial execution of differentiation and development in multicellular organisms.
Fig. 1.
The Cys proteome exists as an adaptive interface between the exposome and the functional genome. Research on integrated omics has emphasized the role of the metabolome and the proteome as intermediates between environmental exposures and the genome, epigenome and transcriptome. The Cys proteome has a more direct role to support sensing and signaling of nutritional and environmental conditions and also to provide an array of reactive nucleophiles to neutralize toxic electrophilic chemicals. Thus, the Cys proteome is an important functional component of the translated proteome, directly interacting with the metabolome to utilize environmental resources and defend against environmental threats.
In the present review of the Cys proteome, we start with a summary of lessons learned from study of the redox proteome, that portion of reversible and irreversible covalent modifications that link redox metabolism to biologic structure and function [1]. We follow this with a more specific consideration of the chemistry of sulfur and the spectrum of oxidative reactions that defines Cys proteomic functions. In a third section, we address the redox regulation systems, especially focusing on major reducing systems that balance the oxidative reactions and support a rapidly evolving understanding of the spatiotemporal nature of redox regulation. In this, we provide specific examples illustrating the organizational structure of stable, kinetically controlled functional redox networks and summarize other modifications such as S-nitrosylation and persulfidation. Finally, we discuss the Cys proteome within the context of integrated –omics approaches, underscoring the ongoing need for development of quantitative redox biology models.
Lessons from the redox proteome
Central dogma
Cys is incorporated into protein as the thiol (RSH) form, with apparently no exception. Thiols are oxidized to sulfenic acids (RSO−) intermediate to formation of disulfides (RSSR) and higher oxidation states (e.g., RSO2−). Thiols and disulfides also undergo exchange reactions in which the thiol reacts to form a new disulfide and liberates a different thiol (RSH + R1SSR1 ←→ RSSR1 + R1SH) [5]. Disulfide is a common post-translational modification for 3-dimensional structure, as a component of multi-step vectorial processing and transport, as a switching mechanism in regulation or signaling and as a consequence of oxidative stress. Research with redox western and mass spectrometry-based redox proteomics methods show that partial oxidation is common for Cys residues throughout the proteome of mammalian systems [6–10]. This contradicts earlier interpretations that Cys thiol oxidation only represents an artifact of extraction [11] and supports the hypothesis that significant speciation of different peptidyl Cys in basal oxidation, organization and function occurs within the Cys proteome.
Discussions of procedures and pitfalls in measurements of thiols and thiol/disulfide have been reviewed [12–14]. Measurements involving extraction and chemical modification reflect the efficiency of alkylation relative to oxidation in trapping the thiol form [See Hansen and Winther [13] for detailed discussion]. Importantly, processing must be rapid under conditions to minimize both oxidative and reductive artifacts. Artifacts are more likely in experimental studies with added oxidants, metal ions or electrophiles, especially if these are not removed prior to processing biologic materials for thiol analysis. Extraction at 0° slows reaction rates about 10-fold relative to 37°, and higher O2 in air-equilibrated solution increases rates about 5-fold that of tissues in vivo in mammals. Thus, in the absence of added oxidants and metals, oxidation of Cys during assay occurs at relatively slow rates, similar to the ongoing autoxidation rates in vivo. High concentrations of thiol-reactive chemicals can be readily achieved under most experimental conditions to minimize significant artifact [13]. An alternate reductive artifact can also occur with addition of thiol-trapping reagents under conditions where NADPH-dependent reduction continues.
Discussion of the redox activities of the Cys proteome requires care in consideration of assumptions upon which interpretations are based. For instance, redox potential refers to equilibrium conditions, which can be approximated experimentally in dilute solution with sufficient equilibration time. However, total protein concentration in biological systems is very high, many proteins are continuously undergoing reversible interaction with other proteins and many processes are kinetically limited and displaced from equilibrium. Local pH and metal ion concentrations may not be uniform, and dynamic processes of phosphorylation/dephosphorylation, acetylation/deacetylation, methylation/demethylation, reversible metal ion binding, etc., may impact in vivo properties. Most of the time, data and analytic methods are insufficient to fully evaluate these possibilities. Furthermore, site-directed mutagenesis studies to test function of a Cys can be misleading if experimental conditions do not replicate conditions under which the Cys impacts function. Such issues represent challenges for the future to improve integrated redox biology models.
Protein thiols exist in a dynamic steady state
Targeted redox analyses of thioredoxin-1 (Trx-1), thioredoxin reductase-1, peroxiredoxin-1, mitochondrial Trx-2, thioredoxin reductase-2, peroxiredoxin-3, NF-κB (p50), redox factor-1, protein disulfide isomerase and other proteins [for summary, see [15, 16]] show partial oxidation in cell culture under control conditions. Studies of glutathionylation also reveal modified residues under basal conditions, including Cys374 of actin in A431 cells [17] (more discussion of Cys modification below). However, these targeted studies mostly involve proteins that are regulatory or signaling in nature, leaving open the possibility that steady-state oxidation is an uncommon event. An important breakthrough during the past decade is derived from mass spectrometry-based proteomic methods, which provide much greater coverage of oxidation of the Cys proteome.
Sethuraman et al. [18] showed that Isotope-Coded Affinity Tag (ICAT) reagents [19] could be used to measure oxidation of individual Cys-containing peptides, specifically comparing differences between control and H2O2-treated samples. The design in this study did not permit evaluation of steady-state oxidation without oxidant. Related methods used by Le Moan et al [20], Leichert et al [21] and Go et al [22, 23] allow determination of the percentage reduction of Cys under control as well as experimentally treated conditions. For instance, proteins can be rapidly extracted, treated with a heavy stable isotopic ICAT reagent, treated with reductant and finally treated with a light stable isotopic ICAT reagent. With liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS), this allows measurement of the percent oxidation of specific residues in peptide sequences. Importantly, Le Moan et al [20] showed that in yeast, Cys in many proteins are partially oxidized under control conditions. Leichert et al [21] obtained similar results in studies of Escherichia coli, and Go et al found steady-state oxidation of proteins in mouse aortic endothelial cells [22], HT29 colon carcinoma cells [6], lung fibroblasts [8], mouse liver [24] and liver mitochondria [24].
More sensitive mass spectrometers and newer peptide tagging methods are becoming available to further enhance sensitivity and coverage of the Cys proteome [6] and efforts to identify oxidatively sensitive Cys, the collective “sulfenylome” of protein Cys that forms sulfenic acids (RSO−) in response to multiple stimuli, have yielded increasingly promising results [25, 26]. While there remains a need for caution until more independent analyses and confirmations are available, the mass spectrometry-based methods are enhancing ability to characterize both stable and transient, partially oxidized Cys residues within the mammalian Cys proteome.
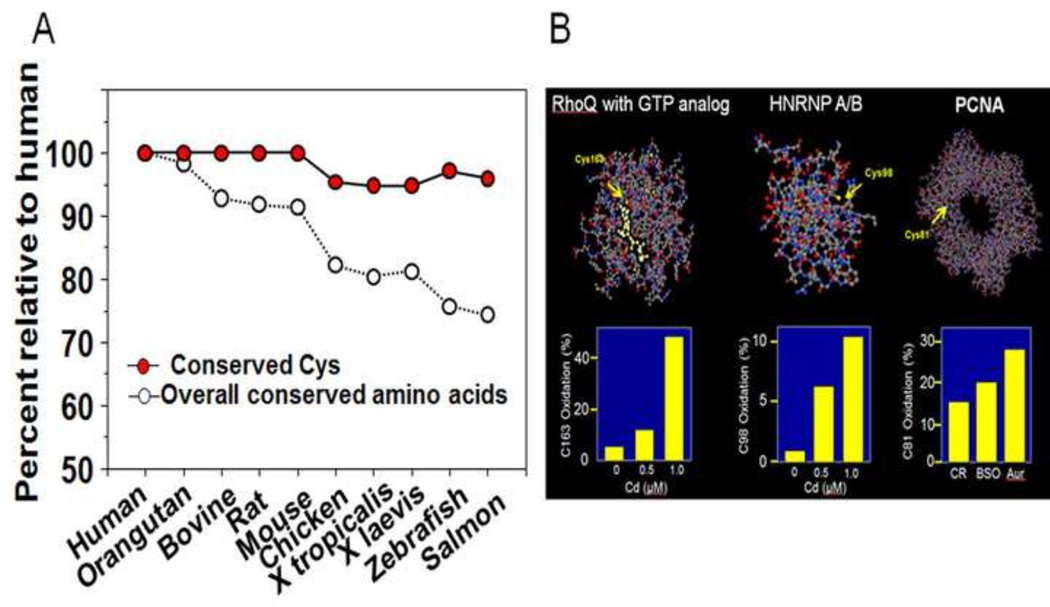
Knowledge of the functions of the redox-sensitive Cys residues is of critical importance. The peptides detected are from relatively abundant proteins and ionize well with available instrumentation. This leaves open the possibility that the Cys residues measured by LC-MS/MS are non-functional or function in a non-specific way to protect critical sites from thiol-reactive chemicals [27]. Three types of evidence suggest that many have more specific functions. Comparison among species of the measured protein Cys to other amino acid residues within a sampling of mitochondrial proteins showed that the Cys are more highly conserved (Fig. 2A; [6]). Examination of the positions of the measured protein Cys within the 3-dimensional crystal structures for homologous proteins shows that the Cys are often in positions expected to be functionally important (Fig. 2B). These include positions near catalytic sites, protein-protein interaction sites, and RNA-binding or DNA-interaction sites. Finally, the measured Cys peptides map to functional pathways according to percentage oxidation (Fig. 3) [6]. Some specific examples are discussed in more detail below. The results are sufficient to conclude that this is a general phenomenon, although not ubiquitous. Functional networks identified include the cytoskeleton, translation, nuclear import, RNA processing, energy metabolism and oxidative stress signaling [6, 8, 24].
Fig. 2.
Multiple lines of evidence support function of redox-sensitive Cys measured by mass spectrometry. A. Targeted searches show that Cys detected in redox proteomic analyses are highly conserved across metazoans compared to other amino acids in the same proteins. B. Examination of x-ray crystal structures shows that redox-sensitive Cys are commonly present in functional motifs of proteins. Examples shown include specific Cys adjacent to GTP-binding site in RhoQ, Cys in RNA binding site of heteronuclear ribonucleoprotein (HNRNP) A/B and DNA interaction site of PCNA. Lower panels show oxidation in response to challenge with cadmium or inhibition of thioredoxin with auranofin (Aur) or GSH depletion with buthionine sulfoximine (BSO).
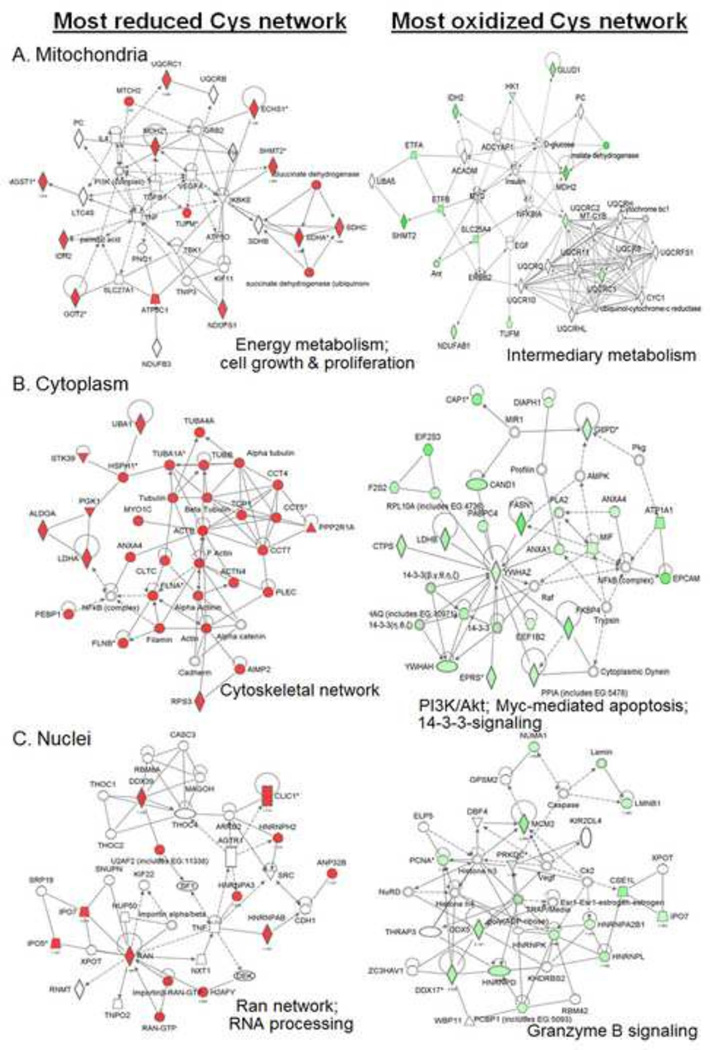
Fig. 3.
Protein Cys map to functional pathways based upon percent oxidation. Ingenuity Pathway Analysis of most reduced protein Cys in colon carcinoma HT29 cells under steady-state non-challenged conditions showed associations with mitochondrial pathways for energy metabolism, cell growth and proliferation; cytoplasmic cytoskeletal pathways and nuclear ran-import mechanisms and RNA processing. Most oxidized protein Cys were associated with mitochondrial intermediary metabolism, cytoplasmic cell survival, Myc-mediated apoptosis and 14-3-3 signaling, and nuclear granzyme B signaling. Data from [6, 8, 24, 49].
Selective pressure against Cys in protein
Cys codons represent 3.28% of all codons, yet the percentage of Cys in protein is typically only 2.2% or less [28], indicating that there is evolutionary selection against utilization of Cys. At the same time, the percentage of Cys in the proteome is substantially greater in complex eukaryotes (2.2%) than in archaebacteria and eubacteria (0.5%; [28]). The Cys residues are relatively conserved in metazoan evolution, leading to the interpretation that much of the Cys proteome is functional [16]. If such function were restricted to catalytic or other “active sites”, then one or two Cys within each of the 20,000 gene products would account for only 10–20% of the 214,000 Cys in the proteome. Instead, the data suggest that up to 80% could be functional, although most without essential “life/death” function. Alternative functions for Cys include stabilization of long-lived proteins by formation of Zn clusters [28], presentation of decoy thiols to protect critical thiols from oxidants and reactive electrophiles [27], and function as redox sensing elements for coordinate regulation of functional pathways [16]. These are linked together by the concept that the Cys proteome functions as an interface between the genome and the exposome (Fig. 1; [1]). Specifically, a Cys proteome network provides a defensive barrier and sensing system to enhance fitness to tolerate exogenous oxidants, reactive electrophiles, and toxic trace metal ions, while at the same time being used to optimize food utilization.
Early forms of life evolved in significantly more reducing conditions than in the current age of highly efficient photosynthesis. Prior to the evolution of an O2-rich atmosphere between 600 and 900 million years ago, living systems were already utilizing inorganic sulfur and Cys. Iron-sulfur clusters, for instance, are ancient prosthetic groups which may have functioned to abiotically catalyze formation of C-C bonds [29]. Along with the evolving Met proteome and associated redox reactions to regulate physiology [30, 31], Cys became increasingly utilized with organizational complexity [28]. Notably, selective pressure against the use of Cys due to vulnerability to oxidation is a result of the same chemical features that confer selective advantage when used as a redox sensor and switch [28]. The Cys proteome thus enabled evolution of systems to regulate O2 availability in tissues and subcellular compartments as well as support developmental systems to tolerate and adapt to complex oxidative environments.
Redox compartmentalization
Oxidation of protein Cys occurs within the secretory pathway as part of a vectorial processing system for proteins targeted to specific membrane regions and for protein export [32]. Measurements of steady-state oxidation of proteins show that during processing, protein disulfide isomerase (PDI) is partially oxidized [33, 34]. Recent studies show similar redox processing of proteins occurs with mitochondrial protein import [35, 36]. Dynamic redox processes are also involved in nuclear protein import and export [37–39]. These processes depend upon different control systems, discussed in more detail below, with many utilizing thioredoxins (Trx) and thioredoxin-like proteins.
Targeted studies of steady-state oxidation of Trx in subcellular compartments [40] show that oxidation of Trx differs among major compartments of mammalian cells, with oxidation in cytoplasm > nuclei > mitochondria [15, 41–44]. Dynamic control within organelles is illustrated by NF-κB reporter activity being decreased by generation of H2O2 within nuclei [9] and increased by targeting peroxiredoxin-1 to nuclei [45]. Increased abundance of nuclear Trx-1 in a transgenic mouse model showed increased NF-κB activity and a proinflammatory state potentiating cardiovascular disease [46] and increased sensitivity to influenza infection [46]. Thus, control of the Cys proteome within subcellular compartments impacts Cys reactivity and biologic function.
Functional networks within the Cys proteome
Differences in steady-state oxidation of Trx and GSH systems among compartments suggest an organizational structure in which the central thiol redox systems control subcellular functions. Studies using mass spectrometry-based redox proteomics revealed, however, that steady-state percent oxidation of Cys in proteins within cytosol, mitochondria and nuclei mapped to functional networks rather than to subcellular compartments [6]. These results support a role for the Cys proteome in metabolic and structural organization of cells, not just as a series of isolated elements in insulated pathways within subcellular compartments but rather as a redox network connected throughout the cell.
Emerging data indicate that redox networks coordinate large numbers of redox elements in function and resistance to environmental challenges. Barabasi and coworkers described metabolic organization in scale-free hierarchical structures that are inherently stable [47, 48]. A bilateral structure for the Cys proteome can accommodate different oxidant and reductant systems and has advantages over parallel branching pathways in which protein Cys residues are physically or kinetically isolated from each other [6, 7]. Furthermore, redox proteomics studies with inhibition of thioredoxin reductase (TrxR) show that many protein Cys are simultaneously oxidized [49]. Along with the above evidence for compartmentalization, and recent data showing that redox pathways can switch due to abundance of reactive targets [50], these observations indicate that interactions within the Cys proteome are best described as a network with spatial determinants [6, 7]. Redox signaling pathways are present within this network, characterized by being localized [51] and having rapid flux compared to the relatively slow and distributed election flow of the overall Cys proteome. We discuss this in more detail below under “Protein Cys redox regulation”, section on “Trx and GSH systems”.
Cys proteome chemistry and functions
Chemical properties of cysteinyl sulfur
The six valence electrons of sulfur in Cys are distributed among four sp3 orbitals, which consist of two lone pairs and two sigma bonds with C and H, respectively. Compared to the more abundant element oxygen, sulfur is less electronegative and has a valence shell radius nearly twice as large. In the absence of local effects in protein structure, the thiol of Cys is 5–6 orders of magnitude more acidic than the hydroxyl group of serine. At physiologic pH, local environments in proteins can result in predominantly the thiol (most common) or thiolate (most reactive) form. Sulfur also has greater nucleophilicity, with reactivity being substantially enhanced by ionization and local protein structure. These properties favor the use of cysteinyl sulfur in biological switching, as it has increased tendency both to donate electrons as well as to accept them [52].
Reactions of protein Cys thiol
Cys thiolate donates an electron pair to thermodynamically favorable targets resulting in the oxidation of the thiol group. Sulfenic acids (RSOH), present mostly as sulfenates (RSO−), are a common product of Cys oxidation and rapidly resolve by reaction with a thiol, eliminating water to form a disulfide (RSSR). The reaction of RSO− with RSH to form RSSR is >105 M−1 s−1 [52]. RSO− are central to the oxidative modification of large numbers of Cys targets [25]. Dimeric or internal disulfides often describe a biologically relevant half-cell with the reduced Cys species to form a redox couple maintained in non-equilibrium steady-state based on interacting redox couples, compartmentation, cell cycle progression and other parameters [40]. In addition to internal disulfides, protein Cys react with GSH or free Cys to form S-glutathionyl or S-cysteinyl derivatives, with protein Cys varying in selectivity for these modifications versus formation of internal disulfides [53].
The sulfur of Cys can be sequentially oxidized by strong oxidants, progressing from sulfenic to sulfinic and sulfonic acids (present as sulfinate, RSO2− and sulfonate, RSO3− respectively). These higher oxidation states of Cys are significantly more difficult to reduce and may be considered irreversible, although reduction of certain Cys-SO2− [e.g., peroxiredoxin (Prx)-2] is catalyzed by sulfiredoxin [54]. Higher oxidation states are detected in mass spectral analysis of proteins but the extent of natural occurrence and possible functions remain unclear.
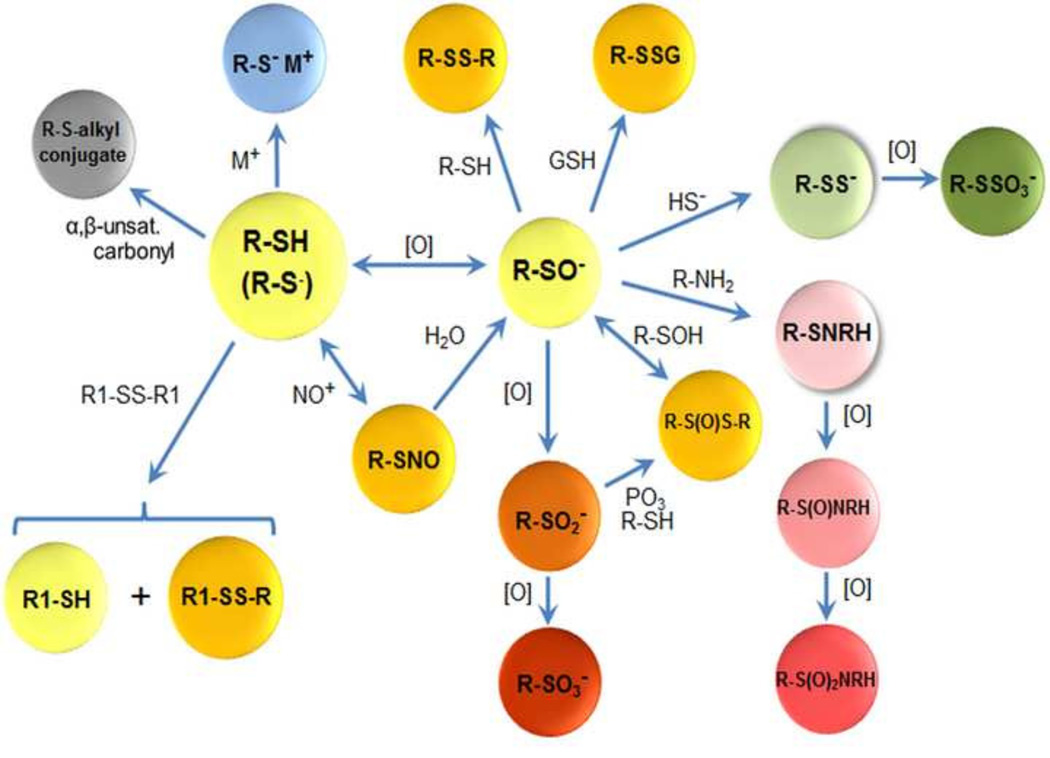
Cysteinyl sulfur undergoes a variety of other reversible and irreversible covalent modifications including nitrosylation, persulfidation, alkylation and arylation that can have unique effects on Cys redox regulation, function and signaling (Fig 4). At least 18 biologically occurring non-radical modifications of Cys thiol have been described, including the above reactions. These have widely varying stability and multiple reviews of their biological impacts are available [55–57]. A remarkable selectivity in reaction with different targets underscores the versatility of protein Cys in use. For instance, the redox sensor Keap1 has multiple Cys residues that react independently with alkenals, metals and nitric oxide to provide a selective detection system [58]. This system also discriminates acrylamide and H2O2 by reaction with different Cys residues [59, 60]. RS− can also be converted to thiyl radicals during ionizing radiation and other conditions. These radicals rapidly react with RS− and O2 in a series of reactions that convert more reactive radicals to superoxide [57, 61]. A Cys thiyl radical intermediate is used in the catalytic mechanism of all classes of ribonucleotide reductase, abstracting a hydrogen from the 3’ carbon of ribonucleotide ribose to permit deoxygenation at the 2’ position for deoxyribonucleotide synthesis [62, 63].
Fig. 4.
Functional modifications of the Cys proteome. Protein Cys (RSH) undergoes a range of enzymatic and non-enzymatic modifications, often mediated by pH-dependent ionization to Cys thiolate (RS−; large yellow sphere). Interaction of RS− with metal ions (blue) is common as a structural element, as a binding motif for nucleic acids and as a component of catalytic sites. RS− is subject to oxidation ([O]) to Cys sulfenate (RSO−, yellow) by H2O2 and other 2-electron oxidants. Thiol (RSH) also undergoes exchange with disulfide (R1S-SR1) to generate different thiol (R1SH) and disulfide (RS-SR1) in a process termed thiol-disulfide exchange. Non-equilibrium steady-state oxidation of specific RSH occurs due to the presence of low nanomolar concentrations of H2O2 in cells. RSO− reacts with RSH or GSH to produce the respective disulfides (RSSR, RSSG; dark yellow, top). Many RSO− can undergo hyperoxidation to sulfinate (RSO2−; orange, bottom) and sulfonate (RSO3−; red) in the presence of excess oxidant. RS− also reacts with hypothiocyanous acid (HOSCN) to produce sulfenyl thiocyanate (RSSCN; dark yellow, upper left), which rapidly hydrolyzes, and with nitric oxide through multiple means of nitronium ion transfer (NO+) to produce a corresponding nitrosothiol (RSNO; orange, bottom left). RSO− can also react with hydrogen sulfide (HS−) to form a cysteinyl persulfide (RSS−, green), which can be oxidized to a cysteinyl thiosulfate (RSSO3−). Alternatively, RSO− can react with primary amines of neighboring amino acids or separate biomolecules to form sulfenamide (RSNHR) which can be further oxidized to sulfinamide (RS(O)NHR) and sulfonamide (RS(O)2NHR), frequently resulting in cyclic intramolecular structures.
In addition to Cys, the genome includes special instructions for the translation of Sec, the so-called 21st amino acid [64]. Sec is identical to Cys except for the replacement of sulfur with selenium (Se). Cys and Sec undergo many comparable reactions, but Se is much more acidic and nucleophilic than sulfur [65]. Sec is incorporated into important thiol regulatory and antioxidant proteins, including TrxR, GPx and Met-R-sulfoxide reductase B1, formerly known as selenoprotein R [64, 66, 67]. These interactions warrant inclusion of the selenoproteome along with the Cys proteome and Met proteome in broader considerations of redox systems biology.
Factors affecting protein thiol reactivity
Common chemical features for thiol reactivity exist for reactions involving metal chelation, substrate reduction and/or conjugation or thiol-disulfide exchange. Specific Cys are, however, often limited to reaction with a fraction of potential thiol-reactive chemicals, e.g., individual thiols within the same monomeric protein are selective for thiol-disulfide exchange or S-nitrosylation [68]. Cys reactivity is mediated in part by the overall availability of the thiolate form because Cys thiol shares its donor electron pair in a sigma bond with hydrogen. As a consequence, pH and pKa interdependently regulate cysteinyl sulfur reactivity. For two hypothetical thiolates in solvent pH 7.4, sharing a reaction mechanism for H2O2 removal and having pKa 7.4 (50% RS−) and 8.4 (9.1% RS−) respectively, in absence of other influences, the more acidic thiol reacts approximately 5-fold more rapidly with the target species. Therefore, pKa exerts influence on the relationship of a thiol to solvent pH, which differs between subcellular compartments in eukaryotes. For instance, the mitochondrial matrix is alkaline at pH 7.8 compared to the intermembrane space, which is maintained approximately 1 unit lower [69]. This affects thiol redox potential (Eh) with specificity to each thiol. For a thiol-disulfide half-cell, the effect on the standard redox potential (Eo) is −5.9 mV/0.1 pH unit. In the case of vicinal dithiols, the effect is more limited (−2.95 mV/0.1 pH) [70]. In vicinal dithiols that collaborate in a single catalytic function, one is typically more acidic and the overall behavior depends upon the presence and characteristics of other reactants. Stable RSO− offer another means of differentiating pH regulation of redox couples, as they tend to be very strong acids [71]. However, the pKa- and pH-determined fraction of Cys available as thiolate varies over only a 102-fold range, and thus cannot solely explain the 106-fold range of reactivity of protein thiols with H2O2 and similar ranges for other oxidants [72, 73].
The nucleophilicity index (determined by highest occupied molecular orbital and lowest unoccupied molecular orbital) of a thiol also greatly contributes to its reactivity. Specifically, a strongly acidic thiol with poor nucleophilicity may be similar in reactivity to a weakly acidic thiol with high nucleophilicity [74]. The reactive thiolate species is a “soft” (polarizable) nucleophile and targets of Cys therefore tend to be “soft” (polarizable) electrophiles, e.g. 4-hydroxynonenal [74].
Protein Cys are also highly effective in coordinating metal ion complexes in enzyme active sites and are widely used to regulate protein structure, maintain metal ion homeostasis or sequester a metal toxicant [75, 76]. Cys cooperates with inorganic sulfur in >100 iron-sulfur (Fe-S) proteins, such as ferredoxins and Rieske proteins [77]. Proximal Cys interaction with heme iron is a conserved feature of cytochromes P450 (CYP) which enables monooxygenase activity [78]. Without the coordination of Cys234, CYP 2B4 is converted to a form with NADPH oxidase activity [79].
Cysteinyl sulfur (alone or in combination with histidine nitrogen) coordinates Zn2+ in 3% of human gene products [80]. Cu+ generally has higher affinity for Cys than other metals, and Cys-rich metal ion chaperones (e.g., metallothionein and ferritin) maintain metal ion homeostasis to prevent so-called “mis-metallation” of maturing enzymes [81]. Cys involved in metal ion coordination also participate in thiol-disulfide exchange during changes in the steady state potential of cellular antioxidants [82]. Subcellular compartmentalization of metal ions (particularly in the endomembrane system) is dependent on expression and oxidation of cysteinyl redox-active transporters such as sarcoplasmic reticulum Ca2+-ATPase (SERCA) [81, 83].
The role of Cys in catalytic sites of enzymes
The flexible reactivity of Cys allows it to confer a range of functions to enzyme catalytic mechanisms. A common role of Cys in enzyme catalysis is as a soft nucleophile, acting as the electron donor for a wide range of substrates. In addition, many proteins utilize Cys to facilitate protein-protein (e.g., intermolecular disulfide formation), protein-DNA (e.g., promoter binding) and protein-lipid interactions (e.g., S-palmitoylation) [84–86]. Furthermore, a number of proteins use Cys to permit the inclusion or function of enzyme prosthetic groups such as heme, Fe and Zn [78, 87, 88], discussed elsewhere in this review. Cys has direct catalytic involvement in at least five of the six Enzyme Commission classes of enzymes as briefly described here and summarized in Table 1.
Table 1.
Cys in catalytic sites of enzymes. The sulfur of Cys functions in the active site of at least five of the six classes of enzymes classified by Enzyme Commission. These include different types of oxidareductases linked to the NADP or NAD systems and also the radical-mediated ribonucleotide reduction. Cys functions in transfer of sulfane in some transferases. Cys is present in active sites of hydrolases, such as cysteine proteases and phosphatases. Protein disulfide isomerases (PDI) catalyze rearrangement of thiol and disulfide bonds without any net oxidation or reduction. Some ligases have active site Cys functioning in covalent bond formation.
| Function | Enzyme | Substrate(s) | Reference |
|---|---|---|---|
| Oxidoreductase | GR | GSSG | [251] |
| Grx | GSS-Cys, Trx-SS | [96, 252] | |
| GPx | Lipid, inorganic peroxides | [64] | |
| GST | Electophiles, lipid peroxides | [253] | |
| TrxR | Trx-SS | [64] | |
| Prx | Peroxides | [60] | |
| GAPDH | G3P | [59] | |
| ADH | Aldehydes | [93] | |
| RNR (radical-mediated reduction) | Ribonucleotides | [63, 254] | |
| Transferase | Thiosulfate sulfurtransferase, mercaptopyruvate sulfurtransferase | H2S, S2O3, mercaptopyruvate | [91] |
| Thymidylate synthase | dUMP | [255] | |
| Hydrolase | γ-Glu hydrolase | γ-Glu-peptides | [256] |
| Caspases, cathepsins, calpains | Polypeptides | [94] | |
| PTP, Cdc25 | pTyr | [176] | |
| Isomerase | PDI | RSSR-x-x-x-RSH | [89] |
| Ligase | Parkin E3 ubiquitin ligase | Cys-His-Glu | [257] |
Major subtypes of oxidoreductases involving Cys in enzyme catalysis include disulfide exchange, peroxidation, thiyl radical formation, thioesterification and persulfidation (Fig 4). Each of these reactions begins with reduced Cys thiol(ate) and involves the formation of covalent bonds between Cys thiol and substrates containing carbon, oxygen, sulfur or phosphorous but not nitrogen, which only bonds with catalytic Cys as a means of enzyme regulation. Many of these reactions transfer electrons to or from NAD(P)H via Cys. Note that while Cys thiol is oxidized in the initial step of all of these reactions, the substrates and catalytic cycle determine whether energy (often in the form of NADPH) is generated or consumed.
Protein Cys thiyl radicals are only known to occur in ribonucleotide reductases (RNR), which are essential for DNA synthesis in all kingdoms of life [63]. All RNR utilize a Cys thiyl radical to initiate the catalytic cycle but differ in how it is generated: class I RNR utilizes a stable tyrosyl radical, class II utilizes adenosylcobalamin and class III utilizes a glycyl radical generated by Fe-S clusters and S-adenosylmethionine [63]. The Cys thiyl radical initiates reaction by oxidizing C3 of ribonucleotide ribose. The carbon-centered radical shifts to C2, dehydrates and undergoes 1-electron reduction (again forming a radical) by a pair of additional Cys residues. Finally, the radical oxidizes the initiating Cys, regenerating the original thiyl radical and releasing deoxyribonucleotide [62].
Disulfide- and peroxide-reducing functions of catalytic Cys have been extensively reviewed. These reactions transfer electrons to substrate sulfur or oxygen in an oxidation state of 0 (RSOH sulfur) or −1 (RSSR and peroxides). The result is formation of disulfide or RSOH on the nucleophilic Cys and the liberation of RSH, H2O or ROH from the substrate. The initiating Cys, typically oxidized to RSSR, must then be reduced to regenerate the enzyme. These reactions make up the bulk of redox regulation by members of the TrxR (thioredoxin reductase) and glutathione reductase (GR) antioxidant nodes. These reactions are also found in important chaperone proteins, such as protein-disulfide isomerase, which are classified by the Enzyme Commission as isomerases because they rearrange protein disulfides rather than cause net oxidation or reduction [89].
Sulfurtransferases are proteins that catalyze the formation, interconversion and reactions of compounds containing sulfane sulfur atoms [90]. Although characterization of many is incomplete, Cys persulfide (Cys-SSH) is an important catalytic intermediate for some [91]. The catalytic Cys is persulfidated by thiosulfate, hydrogen sulfide or mercaptopyruvate, creating a highly reactive sulfur donor, which produces low molecular weight sulfur metabolites such as thiocyanate (SCN) and methanethiol [92]. H2S can be liberated from the Cys persulfide as well as the original persulfide donor [92]. As persulfidation is also a putative peptidyl Cys regulatory mechanism, it is covered elsewhere in this review.
Catalytic Cys-formed thioesters fall into two classes: thioacyl esters (S-CR bond) and thiophosphate esters (S-PO3− bond). Thioacyl esters are intermediates of aldehyde dehydrogenases (ALDH) and cysteine protease catalytic cycles. In the case of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ALDH, Cys thioacyl formation is preceded by a thiohemiacetal intermediate which donates a hydride to NADP+ [93]. Similarly, Cys proteases attack the amide carbon peptide backbones at select residues (e.g., aspartate), causing cleavage of the amide bond [94]. Cys proteases are unlike Asp proteases in that they do not participate in classical acid-base hydrolysis. Rather, the Cys thioacyl ester provides a good leaving group to facilitate hydrolysis and protease regeneration. Similarly, the Cys-SPO3− thiophosphate ester formed following nucleophilic attack on phosphotyrosine in the protein tyrosine phosphate (PTP) catalytic cycle is an excellent leaving group [95]. However, as in acid-base catalysis, these reactions are supported by negatively charged side chains such as Asp and Glu, which coordinate the protonation of water, substrates and Cys itself.
Protein Cys redox regulation
Trx and GSH systems
Extensive research in the last decades shows that redox responses of the Cys proteome occurring at the extracellular and cellular levels are critical in initiation and progression of disease. Two major cellular redox regulation systems, dependent upon thioredoxin (Trx) and glutathione (GSH), are central to regulation of the Cys proteome. These systems maintain a reduced redox environment in cells by catalyzing electron flux from NADPH through Trx reductase (TrxR) and glutathione disulfide reductase (GR) to Trx and GSH, respectively. In addition, mixed disulfides of GSH with protein Cys (S-glutathionylation) can have broad impact on protein function and disease [88]. S-Glutathionyl groups are removed by glutaredoxin (Grx) plus GSH. In some cases Grx may transfer electrons from the GR-GSH node into the TrxR-Trx node by reducing Trx disulfide [96]. Numerous studies of Cys modification and redox regulation provide evidence that redox-sensitive regulatory Cys residue(s) (see Fig 5 legend) play critical roles in cell signaling and control as part of a redox network structure. These systems are involved in multiple signaling pathways through direct or indirect interactions with a variety of target proteins controlled by the GSH and Trx redox systems (Fig. 5). Proteins and associated protein networks are often selectively regulated by Trx or GSH systems but also commonly regulated by both GSH and Trx systems, e.g., S-glutathionylation in C374 and S-nitrosylation in C285 of actin (Fig. 5).
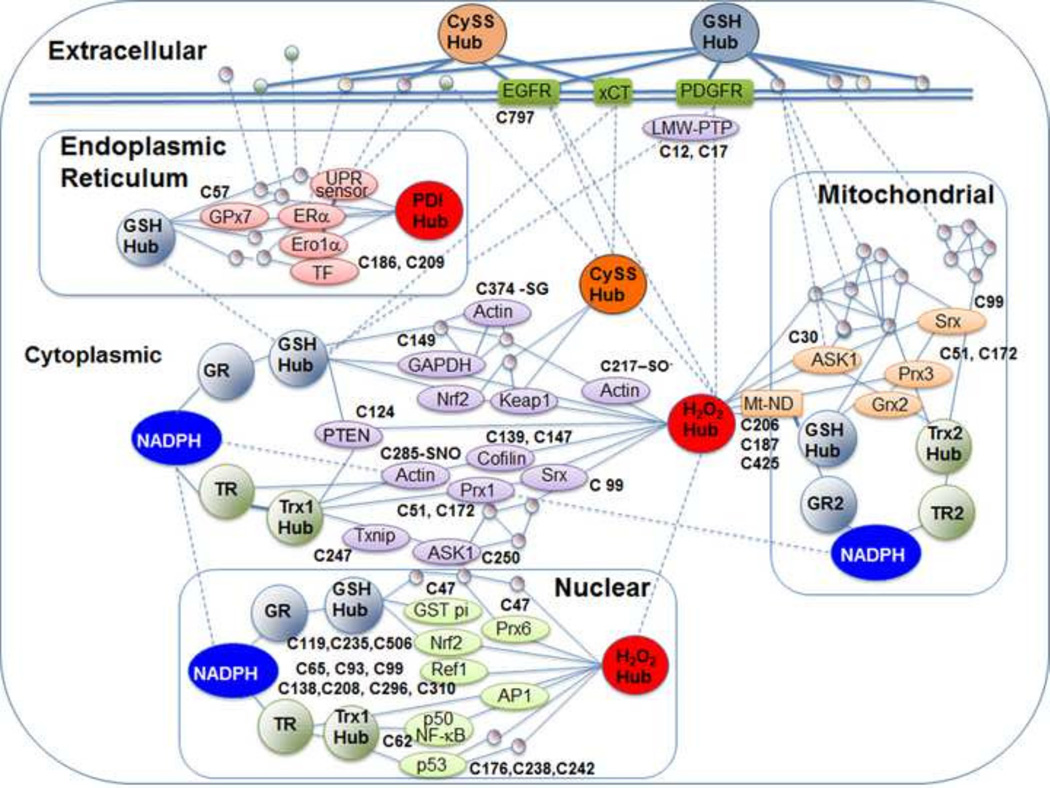
Fig. 5.
Cys proteome networks regulated by Trx and GSH systems. Accumulating evidence shows that redox networks exist in different subcellular compartments supported by NADPH as reductant functioning in opposition to oxidation by H2O2 and other oxidants. Trx is the most common intermediary reductant, with a smaller number of proteins supported by GSH. Cysteine and cystine constitute the most abundant extracellular low molecular weight thiol/disulfide couple, apparently functioning in regulation of protein Cys on the extracellular surface. References provide detailed information on interactions of Cys proteins and functional Cys residues illustrated in this figure. TrxR-actin [110]; Trx-dependent regulation of proteins [139]; Ref1-AP1 [258]; TrxR/AP1 [259]; PDGFR [260]; xCT [261]; PTEN-Trx [262, 263]; PTEN-GSH [264]; ASK1-Trx2 [264]; GSTp1-Prx6 [265]; GAPDH-GSH [266, 267]; ND-GSH [268]; p53-Trx [269]; Trx2-Prx3 [270]; Srx-Prx1 and Srx-Prx3 [271, 272]; Grx2-Prx3 [273]; Grx2-Prx3-Trx2 [274]; PDI-UPR sensor [275]; Era-PDI [276]; PDI-TF [277]; Ero-α-PDI and GSH [278]; GPx7-ER [279]; Nrf2 regulatory Cys residues [280]; Srx functional Cys residues [281].
As indicated above, mass spectrometry-based redox proteomics together with bioinformatics enables identification of redox-sensitive Cys residues, proteins and networks of Cys proteome in association with cellular conditions [6, 8, 49]. Studies in yeast showed that the redox proteome was preserved in cells deficient in the GSH system but not in cells with compromised Trx systems. These results show that the Trx system is most central in regulation of the yeast Cys proteome [97]. In mammalian cells, responses of the Cys proteome were tested by selectively disrupting the Trx- and GSH-dependent systems with inhibitors, auranofin for thioredoxin reductase and buthionine sulfoximine (BSO) for γ-glutamylcysteine synthetase, the first enzyme in the GSH synthesis pathway. Results showed that the Trx and GSH systems support distinct functions in cellular redox control by controlling different subsets of protein Cys. Consistent with the yeast findings, the Trx system regulated a much broader range of proteins than the GSH system [49]. However, GSH and Grx can act as a backup to the TrxR-Trx system to promote the reduction of Trx during aurothioglucose exposure [96]; the extent to which this alters redox proteomic effects of TrxR inhibition is unclear. Examples from studies with selective inhibition of TrxR and GSH systems are available [49, 98].
Regulation of actin cytoskeleton proteins
Cytoskeleton remodeling through actin dynamics regulates vital biological processes, e.g., cell morphogenesis, migration and growth, membrane trafficking, exocytosis and endocytosis [99–101]. Actin is susceptible to multiple post-translational modifications in Cys residues including reversible thiol oxidation (Cys217, 257, 272, 285, 374) [102–104], glutathionylation (Cys10, 217, 257, 374) [105–107] and nitrosylation (Cys217, 257, 285, 374) [108–110]. Oxidation disrupts actin dynamics, e.g. filamentous (assembled) and monomeric (disassembled) actin formation [111, 112]. Glutathionylation/deglutathionylation of actin, regulated by the concentration of GSH and Grx activity, is also associated with actin dynamics [113–115].
In addition to actin itself, a cohort of actin-associated cytoskeletal proteins control actin dynamics. Oxidation of Cys139 and Cys147 in actin-binding protein cofilin stimulates apoptotic signaling as a consequence of inhibiting interaction with actin as well as increasing mitochondrial translocation [116], and cofilin oxidation impairs cytoskeletal function in T cells [117]. Furthermore, destrin, a cofilin family member, is an actin depolymerizing factor regulated by Cys redox status. Destrin nuclear translocation was associated with redox disruption of actin cytoskeleton proteome [8]. Actin cytoskeleton proteins were also susceptible to oxidation during inflammatory signaling associated with mitochondrial oxidation [6, 22]. Redox-dependent control of actin dynamics was observed from interaction of actin with the Trx system, Trx1 and TrxR1 [110, 118, 119]. Release of Trx1 from actin resulted in degradation of Trx1, which then stimulated cell death [119]. Denitrosylation of actin by TrxR via focal adhesion kinase restored cellular function [110].
Extensive knowledge of the redox modulation of the cytoskeleton by GSH and Trx systems show the central importance of redox control in cell function. The data demonstrate that GSH and Trx do not indiscriminately “equilibrate” the peptidyl Cys environment to a consistent reduction potential but sensitively and selectively maintain discrete non-equilibrium dynamics for individual proteins and protein networks. The following examples further illustrate the use of kinetically controlled systems in nuclear protein trafficking and in maintenance of the integrity of the genome. Recent reviews are also available on the redox processing in the secretory pathway [33, 34, 120] and in mitochondrial import [35, 36]. Relatively less information is available on vectorial redox processing during endocytosis and proteolysis in lysosomes, but evidence is available suggesting that analogous systems are present [121, 122].
Cys proteome in nuclear protein trafficking
Redox-dependent transcription has been known for many years, with divergent mechanisms involved for AP-1, Nrf2, NF-κB, glucocorticoid receptor and other systems [38, 123–126]. Both Trx1 and its inhibitory binding partner Txnip are translocated into nuclei to regulate the Cys proteome. For instance, nuclear accumulation of Txnip was detected when cells were treated with the anti-cancer histone deacetylase inhibitor, suberoylanilide hydroxamic acid [127]. Rch1 (importin-α) is required for Txnip nuclear translocation and growth control, and this interaction is controlled by a Trx-dependent redox system [128]. Interestingly, Trx1 is also translocated into nuclei, and import required karyopherin-α (importin-α) and nitric oxide (NO). Site-directed mutagenesis of Lys81,82 of Trx1 showed that these residues are important for the translocation of Trx1 into nuclei [129], suggesting that the protein translocation mechanism is complex, requiring localized signal motifs in addition to redox-dependent regulation.
In addition to enhanced Trx1 translocation to nuclei, transport and translocation of a wide range of proteins are regulated in a redox-dependent manner. For instance, oxidative stress stimulates nuclear translocation of SBP 2 [SECIS (selenocysteine insertions sequence) binding protein 2] via oxidation of Cys within a redox-sensitive Cys-rich domain. Reversal by Trx1 or Grx suggests that redox state of SBP 2 is critical to compartmental translocation and selenoprotein synthesis [130]. In bronchopneumonia, protein S-glutathionylation significantly co-localized with Fas ligand, and Grx1 knockout mice had increased Fas S-glutathionylation and improved murine lung infection survival [131]. Mass spectrometry-based redox proteomics also showed functional redox networks of nuclear import proteins (Fig 3; [6]). Additionally, targeted expression of nuclear Trx1, stimulation of nuclear H2O2 production with nuclear-targeted D-amino acid oxidase, and enhanced nuclear peroxidase activity by expressing nuclear targeted Prx1 further show distinct redox control in nuclei [9, 45]. Thus, multiple approaches show the nuclear Cys proteome is controlled in a semiautonomous manner by transport and compartment-specific regulation.
Nuclear functions of the Cys proteome
The dynamic nature and functional importance of compartment-specific control of the Cys proteome are further illustrated by Trx-dependent protection against cellular stresses associated with growth inhibition and cell death induced by H2O2, TNFα and chemotherapeutic agents [132–134]. Trx-interacting protein [Txnip/Trx binding protein (TBP)/vitamin D3-upregulated protein] directly binds to Trx1 by forming a disulfide bond between Cys247 of Txnip and catalytically active Cys residues of Trx (Cys32 and Cys35), inhibiting redox activity of the Trx [135, 136]. The complex of Txnip-bound Trx contributes to pathogenesis of multiple diseases including diabetes, autoimmune disease and cancer [137–139]. Upregulation of Txnip under sustained high glucose exposure caused increased ROS generation and caspase-3 activation [140]. The pathological role of Txnip was revealed with release of Txnip from oxidized Trx1 to stimulate inflammation [141] and cell death [140]. Similarly, the reduced form of Trx1 binds to apoptosis signal-regulating kinase-1 (ASK-1) and inhibits its activity to prevent stress- and cytokine-induced cell death [142]. Activation of ASK-1 is stimulated under stress signaling after dissociation with Trx1 [142] and plays a key role in apoptosis with increased DNA damage [143, 144]. Direct interaction of Trx with Txnip (Cys247) and ASK-1 (Cys250) via reduced Cys32 (Cys30 of Trx2) supports a key role of Trx thiol-disulfide redox regulatory function against oxidative stress-mediated DNA damage and cell death signaling [141].
Trx1 also plays a key role in DNA repair mechanisms by regulating the base excision repair pathway via the p53 downstream gene, growth arrest, and DNA-damage-inducible protein 45–α and its interaction with apurinic/apyrimidinic endonuclease 1 [APE1, also called redox factor (Ref)-1] [145]. This mechanism is regulated by the interaction between Trx1, Ref-1 and redox sensitive p53 transcription factor [146, 147]. TrxR-Trx1 also interacts with Ref-1, and this interaction controls p53 activity supporting the Trx1 redox system in modulation of redox-sensitive transcription factors [148]. Moreover, Trx1 protection against cell death was shown to occur in growth regulation in B-cell chronic lymphocytic leukemia (CLL) by control of the interaction of protein disulfide isomerase with tumor necrosis factor receptor (TNFR) [149]. This finding additionally supports the beneficial role of Trx1 in survival, perhaps via activation of NF-κB transcription factor [147].
Oxidation of the Cys proteome by H2O2 and derivative species
Measurements of bulk protein Cys in mouse liver and HT29 cell culture show that 92% and 88%, respectively, are in the thiol form [6]. With procedures to minimize artifacts in mass spectrometry measurements, similar mean (84.3%) and median (87.7%) values were obtained for HT29 cells in culture. Specific protein Cys had a distribution of steady-state oxidation ranging from less than 8% to greater than 99% reduced. These observations raise key questions about the oxidants involved and the underlying enzymatic and non-enzymatic mechanisms contributing to steady-state oxidation.
The balance of oxidation and reduction of the Cys redox proteome is governed by the thermodynamics, selectivity and kinetics of different peptidyl Cys. As indicated above, details of the reduction systems are becoming clear. In contrast, the fine-tuned oxidative signaling has been ascribed extensively to H2O2, a species that is relatively unreactive with most Cys thiols. At least three factors impact specificity of oxidation, i.e., localized high activity generation of H2O2, catalytic mediators of oxidation, and generation of alternative low-molecular weight chemical oxidants. Enzymes with high reactivity with H2O2 [e.g., members of the Prx and GSH S-transferase(GST)] may act as special carriers of the oxidative signal to interacting proteins [150]. Alternatively, H2O2 is used to generate other oxidants with altered kinetics (e.g., hypohalous acids) [151], which in turn can form further derivative oxidizing species (e.g., haloamines). These latter processes support speciation of an H2O2 oxidative signal into various paths, which regulate not only the fate of H2O2 but also the effect of its oxidative signal.
H2O2
Although O2 is present at five orders of magnitude higher concentration than H2O2 in aerobic organisms, H2O2 is usually considered the most central oxidant in maintenance of steady-state oxidation of the Cys proteome. H2O2 is a product of at least 31 human enzymes (Table 2) and has been measured in mammalian tissues at concentrations in the range of 1 to 50 nM [152–155]. Measurements of H2O2 based upon steady-state kinetics of endogenous catalase in perfused rat liver developed by Sies [156] may be the most reliable tissue measurements available because there are no possible artifacts due to transfection or calibration of imaging reagents. Catalase concentration is too low to support this type of measurement in most cells and tissues, however, and more sensitive fluorescent probes have been widely used. Some, such as dichlorofluoroscein diacetate (DCFDA), are non-specific and require caution when evaluating cell and tissue H2O2 production. Other probes can also be non-specific, insensitive to rapid flux changes, or misleading due to redistribution of probes within cells [157]. In contrast to high reactivity of H2O2 with peroxiredoxins, GSH peroxidases and catalase, H2O2 reacts quite slowly with the majority of thiols. For low molecular weight thiolates, the rate constant of reaction with H2O2 is about 20 M−1 s−1 but the effective rate constant for total thiol (thiol+thiolate) is <5 M−1 s−1 [158]. Using 5 M−1s−1, total protein Cys at 20 mM and H2O2 at 10 nM, non-catalytic oxidation of protein Cys by H2O2 in cells occurs at 0.0003 % per minute. This rate is too slow to maintain the steady-state oxidation as indicated above.
Table 2.
Hydrogen peroxide-generating human enzymes. Enzymes producing superoxide anion (O2−) are included because O2− is commonly dismutated to produce H2O2.
| Name | Protein | Gene | Accession | Location | Product |
|---|---|---|---|---|---|
| Aldehyde oxidase | ADO | AOX1 | Q06278 | C | H2O2 |
| Amine oxidase (flavin-containing) A | AOFA | MAOA | P21397 | M | H2O2 |
| Amine oxidase (flavin-containing) B | AOFB | MAOB | P27338 | M | H2O2 |
| D-Amino acid oxidase | OXDA | DAO | P14920 | Px | H2O2 |
| L-Amino acid oxidase | OXLA | IL4I1 | Q96RQ9 | L | H2O2 |
| D-Aspartate oxidase | OXDD | DDO | Q99489 | Px | H2O2 |
| Amiloride-sensitive amino oxidase (copper containing | ABP1 | ABP1 | P19801 | S | H2O2 |
| FAD-linked sulfhydryl oxidase | ALR | GFER | P55789 | C, M, S | H2O2 |
| Hydroxyacid oxidase 1 | HAOX1 | HAO1 | Q9UJM8 | Px | H2O2 |
| Hydroxyacid oxidase 2 | HAOX2 | HAO2 | Q9NYQ3 | Px | H2O2 |
| Membrane primary amine oxidase | AOC3 | AOC3 | Q16853 | PM | H2O2 |
| Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | PAOX | PAOX | Q6QHF9 | Px, C | H2O2 |
| Peroxisomal acyl-coenzyme A oxidase 1 | ACOX1 | ACOX1 | Q15067 | Px | H2O2 |
| Peroxisomal acyl-coenzyme A oxidase 3 | ACOX3 | ACOX3 | Q15254 | Px | H2O2 |
| Peroxisomal sarcosine oxidase | SOX | PIPOX | Q9P0Z9 | Px | H2O2 |
| Prenylcysteine oxidase 1 | PCYOX | PCYOX1 | Q9UHG3 | L | H2O2 |
| Protein-lysine 6-oxidase | LYOX | LOX | P28300 | S | H2O2 |
| Pyridoxine-5-phosphate oxidase | PNPO | PNPO | Q9NVS9 | C | H2O2 |
| Retina-specific copper amine oxidase | AOC2 | AOC2 | O75106 | PM, C | H2O2 |
| Spermine oxidase | SMOX | SMOX | Q9NWM0 | C, N | H2O2 |
| Sulfhydryl oxidase 1 | QSOX1 | QSOX1 | O00391 | G | H2O2 |
| Sulfhydryl oxidase 2 | QSOX2 | QSOX2 | Q6ZRP7 | N, PM, S | H2O2 |
| Sulfite oxidase, mitochondrial | SUOX | SUOX | P51687 | M | H2O2 |
| Xanthine dehydrogenase/oxidase | XDH | XDH | P47989 | C, Px, S | H2O2 |
| NADPH oxidase 1 | NOX1 | NOX1 | Q9Y5S8 | PM | O2− |
| Cytochrome b-245 heavy chain (gp91-phox) | CY24B (NOX2) | CYBB | P04839 | PM | O2− |
| NADPH oxidase 3 | NOX3 | NOX3 | Q9HBY0 | PM | O2− |
| NADPH oxidase 4 | NOX4 | NOX4 | Q9NPH5 | E, PM, N | H2O2 |
| NADPH oxidase 5 | NOX5 | NOX5 | Q96PH1 | E | O2− |
| Dual oxidase 1 | DUOX1 | DUOX1 | Q9NRD9 | PM | H2O2 |
| Dual oxidase 2 | DUOX2 | DUOX2 | Q9NRD8 | PM | H2O2 |
Abbreviation used for subcellular localization: C, cytoplasm; E, endoplasmic reticulum; G, golgi; L, lysosome; M, mitochondria; N, nucleus; PM, plasma membrane; Px, peroxisome; S, secreted. Other proteins are likely to produce O2− or H2O2 but not included due to limited characterization. Enzymes generating lipid peroxides are not included.
Multiple alternatives exist which could account for higher steady-state oxidation, e.g., a fraction of protein Cys with higher rate constants, more reactive oxidants derived from H2O2, or catalytic oxidation of protein Cys by peroxiredoxins, glutathione S-transferases, or other enzymes in the presence of H2O2. Studies of different protein Cys show that rate constants for reaction with H2O2 vary over more than six orders of magnitude [57]. A network model of H2O2 clearance with 24 elements and 28 kinetic parameters, including pseudo-enzymatic oxidative turnover of protein thiols, the enzymatic actions of catalase, glutathione peroxidase, peroxiredoxin, and glutaredoxin, and the redox reactions of Trx and GSH, correctly reproduced experimentally measured oxidation profiles for the GSH and Trx redox couples [159]. The model suggested that approximately 10% of intracellular protein thiols react with H2O2 at substantial rates and predicted an unequal distribution of the intracellular anti-oxidative burden between Trx-dependent and GSH-dependent antioxidant pathways, with the former contributing the majority of the cellular antioxidant defense due to peroxiredoxins (Prx) and protein disulfides. Importantly, if enzymes with peroxidase activity, such as the Prx and GST families, use protein Cys as direct reductant, supported by Trx or GSH systems to reduce the protein Cys back to thiol form, then a complete enzymatic system exists to maintain the dynamic Cys proteome [7]. The overall organizational structure is one in which the Cys proteome is not collectively equilibrated with NADP+/NADPH, O2/H2O or at any single redox potential. Instead, a continuum of kinetically limited non-equilibrium steady states exist for protein Cys, varying in abundance and spatial and temporal distributions [40].
The system is dynamic and responds to changes in H2O2 during inflammation and signaling for proliferation, cell migration or host defense [151, 160–162]. Research shows substantial circadian variation [163], changes during development [164] and spatiotemporal temporal changes during wound healing [165]. The oxidative second messenger [166] with localized generation [51] is amplified by a “flood-gate” mechanism [167] involving excessive oxidation of Prx and Trx1 (see below) [168]. This mechanism could alter redox signaling via diverting H2O2 flux to oxidize other proteins or, perhaps more likely, support enzymatic oxidation or other redox-activated signaling mechanism [166].
Earlier analytic methods only detected protein disulfides; development of reagents for trapping RSO−, however, showed that RSO− is the predominant modification of the Cys proteome [25] and central to oxidative Cys regulation (Fig. 4). For instance, RSO− in multiple PTPs is associated with inhibition of CD8+ T cell activation [169], and formation of RSO− in IQGAP1 plays a direct role in endothelial cell migration and angiogenesis [170]. Recent bioinformatic analysis using a dimedone-tagged click chemistry probe has revealed multiple peptidyl Cys with basal RSO− modification, including Cys18 of SIRT6 and multiple Cys of pyruvate kinase M2 [26]. RSO− were detected in solvent-accessible peptidyl Cys from proteins associated with most subcellular compartments and included enzymes regulating phosphorylation, acetylation and ubiquitylation [26]. Peptidyl Cys with detectible RSO− modification were often associated with Glu and Lys residues and less often with other Cys residues [26].
Oxidation of RSO− to RSO2− (sulfinate) in peroxiredoxin has a special role in signaling because this oxidation inhibits peroxidase activity, redirects reducing equivalents from the Trx system away from Prx2 and changes protein Cys redox regulation [171]. This process is termed “hyperoxidation”, and detailed studies with Cys50 of Prx2 [172], including thermodynamics and contributions of proximal amino acids [60, 73, 173, 174] are available to describe the underlying mechanism. Ultimately, hyperoxidation of Prx2 diverts H2O2 to other antioxidant systems. Such diversion alters redox signaling by changing elements of the Cys proteome that react with oxidant [166]. Furthermore, hyperoxidation of Prx2 is biochemically regulated, as sulfiredoxin reduces Prx2 Cys sulfinate back to the sulfenate form, which may be reduced by Trx [54].
The Cys proteome includes sulfinic and sulfonic acids, as well as series of sulfenamides, sulfinamides and sulfonamides. With the exception of sulfinic acid and possible precursor RSONR2 [175] in peroxiredoxins, there is little evidence that these represent functional variations. Similarly, cyclic sulfonamide (isothiazolidinone) structures are formed from the catalytic Cys of certain phosphatases, e.g. Cys215 of PTP1B [175, 176], but the functional and physiologic relevance of these is unclear. Cysteinyl sulfur and amine interaction also provides means to cross-link molecules by strong oxidants, observed in model cysteinyl peptides and GSH and in vivo [177–179]. Thiosulfinates and thiosulfonates are documented in thiol chemistry [52], and thiosulfinate has been found as an intermediate of sulfiredoxin (Srx) Cys99 reduction of Prx2 sulfinic phosphoryl ester [180]. Thus, a diverse range of oxidized sulfur and sulfamine modifications of the Cys proteome can occur, but only a very small number have been characterized in terms of biologic functions.
Oxidant species derived from H2O2
The Cys proteome is subject to oxidation by peroxide-derived oxidants, some of which have similar thermodynamic potential but lesser kinetic limitations than H2O2. Hypohalous acids (hypochlorous acid, HOCl; hypobromous acid, HOBr) are strong oxidants generated by phagocytic myeloperoxidase (MPO) during inflammation and widely recognized as dual agents of host defense and host injury [151, 181]. HOCl and HOBr rapidly react with thiolates by transfer of Cl+ or Br+ [52]. The resulting sulfenyl halide is unstable and rapidly hydrolyzes to release Cl− or Br− and RSO− [182]. Similarly, eosinophil peroxidase is released in allergic events and generates HOBr, but little or no HOCl at physiologic pH [183, 184].
Low-level generation of HOCl and HOBr can also occur in tissues without inflammation via the activity of peroxidasins (also known as vascular peroxidases), which participate in collagen cross-linkage and are essential for tissue development [185–188]. However, the impact of constitutive HOCl and HOBr generation on Cys proteomic regulation has not been extensively characterized. In principle, these oxidants could oxidize selected subsets of the Cys proteome as suggested by the finding that HOCl independently activates Nrf2 and MAPK signaling in airway epithelia [189]. A potential limitation for specific Cys oxidation is that HOCl and HOBr have high and indiscriminate reactivity with most Cys [190] and also react with non-Cys residues, e.g., Met and primary amines [191, 192]. On the other hand, conversion of hypohalous acids into less reactive haloamines (e.g., RNHCl) may confer specificity. For instance, taurine chloramine oxidizes Cys282 of creatine kinase with 3-fold increased selectivity compared to HOCl [193].
Hypothiocyanous acid (HOSCN) is a related oxidant that is constantly produced from endogenous thiocyanate (SCN), lactoperoxidase (LPO) and H2O2 in human secretory fluids (e.g., saliva, airway lining fluid, tears and milk) [194–196]. HOSCN has been studied extensively in saliva and is maintained at concentrations >5-fold more abundant than the precursor H2O2 [197]. In addition to enzymatic production by LPO, MPO and other peroxidases (REF=Van Dalen et al Biochem J 1997)[186], HOSCN is produced by the direct reduction of HOCl or HOBr by SCN−, a reaction which competes effectively with physiologic concentrations of GSH [52, 198]. HOSCN reacts with protein Cys to form RSO− and SCN− [52, 198].
The most critical aspect of HOSCN is that it is significantly less reactive than H2O2, HOCl or HOBr [173, 199–201], which suggests greater specificity for Cys proteomic regulation. Support for this possibility is limited, but HOSCN, and not HOCl or HOBr, induces cell signaling by reaction with serum factor(s) [202, 203]. HOSCN selectively targets salivary GSTP, but not GSTA or GSTM [204]. Altered redox signaling of HOSCN may be reflected in altered cellular responses to this oxidant instead of others, e.g., removal of H2O2 and HOCl by SCN− protects against mammalian cell death and decreases inflammation [179, 205]. Furthermore, mammalian innate immunity exploits specificity of HOSCN reactivity to place selective oxidative pressure on microbes [206]. The biochemical and physiological effects of SCN− and HOSCN have been recently reviewed [207].
S-Nitrosylation
Nitric oxide (NO) is a precursor for S-nitrosylation of the Cys proteome, governed by multiple reaction pathways, e.g., metal catalysis of NO+ transfer, with the ultimate action being formation of protein S-nitrosocysteine (RSNO). S-Nitrosylation is regulated by GSH and Trx systems, which reduce RSNO [208, 209]. Trans-S-nitrosylation is important in NO signaling and as a negative feedback mechanism for NO synthases and effector proteins [209]. SERCA is nitrosylated at Cys344 and Cys349 following ONOO− exposure, which enhances subsequent protein glutathionylation [210]. The oxidation of critical SERCA Cys residues, e.g. S-nitrosylation and S-glutathionylation, may be a guanylate cyclase-independent means to alter vascular tone [211]. Skeletal muscle ryanodine receptor/Ca2+-release channel (RyR1) Cys3635 is regulated by S-nitrosylation while other Cys mediate redox sensitivity through SNO-independent means [209, 212, 213]. Several mitochondrial metabolic enzymes are targets of S-nitrosylation [214]. Thus, S-nitrosylation represents a series of modifications of the Cys proteome, which can be considered parallel to the oxidative systems in providing a network of regulatory elements impacting diverse aspects of cell function.
Persulfidation
Persulfidation, also termed S-sulfhydration [215, 216], is another mechanism for modification of the Cys proteome that can operate in parallel with the oxidative and S-nitrosylation systems. The protein persulfide (RSS−) is derived from H2S generated from Cys via intestinal microbes, mercaptopyruvate sulfotransferase, cystathionine β-synthase (CBS) and cystathionine γ-lyase (CGL) [217, 218]. At physiologic pH, H2S is principally ionized (HS−) and reacts with a disulfide to produce persulfide (RSS−) and release a thiol. For instance, GSH persulfide (GSS−) is formed from reaction of GSSG with H2S [219]. The sulfur added to the thiol in the persulfide moiety is present at the oxidation state of elemental sulfur (sulfane), which commonly exists as octasulfane (S8). Cysteinyl polysulfides have been documented in crystal structures of bacterial sulfide quinone oxidoreductases [215], while free polysulfides (H2Sn) in brain tissue can persulfidate multiple protein Cys targets [220]. Sulfane sulfur can be transferred between protein thiols and low molecular weight thiols (trans-persulfidation or trans-sulfanation). The extent of persulfide modification of proteins is currently unknown [221, 222].
Persulfide formation generally interferes with normal protein Cys reactivity in much the same way as S-nitrosylation or sulfenylation [223] but differs in that the highly nucleophilic auxiliary sulfane is more reactive toward electrophiles [224]. Inhibition of H2S generation leads to mitochondrial dysfunction and increased O2− flux, suggesting a role for H2S in regulation of mitochondrial function [225]. Persulfidation of 39 proteins, including GAPDH Cys150 was detected by mass spectrometry [226]. Persulfidation of GAPDH Cys150 by treatment with NaSH increased enzyme activity, with an EC50 of 15 µM; GAPDH is also a S-nitrosylation target, highlighting a potential for H2S/NO cross-talk [226]. After TNF-α stimulation of CGL, increased persulfidation of p65 Cys38 regulated its DNA binding and anti-apoptotic function [227]. The C59S mutant of P66Shc is resistant to anti-phosphorylation effects of H2S or CBS overexpression, implicating regulation by persulfidation [228]. H2S also regulates eNOS activity by persulfidation of Cys443, which promotes activity of the NOS in opposition to NO binding [216].
Alkylation
Many modifications of the Cys proteome are irreversible under normal circumstances in vivo. Some of these, such as modification by endogenously derived reactive lipids, function to provide a response to stress [229]. Reactive aldehydes make up a highly significant and well-recognized fraction of permanent Cys adduction [230]. Many α/β-unsaturated aldehydes such as 4-hydroxynonenal (4-HNE) are “soft” electrophiles that react readily with soft nucleophiles and are chiefly regulated in vivo by Cys thiolate [74, 174]. 4-HNE reacts with Cys residues of many important proteins. The thioredoxin system, including TrxR (Cys496 along with Sec497 [65]) and Trx (Cys32, Cys35), is very sensitive to inhibition by 4-HNE [231]. Similarly, the catalytic and modifying subunits of Glu-Cys ligase (GCLC/M) are targets of 4-HNE (GCLC Cys553, GCLM Cys35) [232]. 4-HNE also induces antioxidant genes (e.g., TrxR1; heme oxygenase-1, HO-1) through induction of the Nrf2 promoter by modifying Cys288 of Keap1 [58, 233]. Conversely, 4-HNE inhibits the release of NF-κB by modifying Cys179 of IKK-β [234]. Together, these examples emphasize that irreversible modification of Cys in signaling proteins is functionally important to respond to electrophile exposures [229].
Selectivity of protein Cys to electrophilic modification has been demonstrated by comparisons of proteins modified by iodoacetyl- and maleimido- reagents [235]. For instance, of 1255 distinct Cys-containing peptides from isolated mitochondria, the electrophiles only overlapped 35% in Cys adduction and had distinctly different effects on cytotoxicity and apoptosis. Similarly in nuclear-cytosolic fractions, there was only 14% peptidyl Cys overlap between the electrophiles [236]. The targeted residues represented proteins involved in a variety of central cellular pathways including Trx-2, heat shock proteins, permeability transition pore complex, nuclear/cytosolic cytoskeleton, DNA replication/repair, protein synthesis and RNA processing [236].
As described above, the Cys proteome is part of an adaptive interface of an organism with its environment, and covalent modifications of the Cys proteome are useful to characterize environmental exposures to pollutants, dietary metabolites, drug metabolites and endogenous toxicants [237]. In human serum albumin, 66 adducts of Cys34 have been detected [238]. Similarly in drug metabolism, quinone structures from metabolites of acetaminophen (APAP), diclofenac and related drugs have distinct 1,4-Michael addition reaction with thiols to form a covalently modified Cys34 of albumin. The APAP metabolite N-acetyl-p-benzoquinone imine (NAPQI) targets peptidyl Cys of essential metabolic and regulatory proteins, including TrxR1 which is covalently modified by NAPQI at residues Cys59 (GR-homologous active residue), Cys497 and Sec498 (vicinal thiol-selenol pair), associates these modifications with loss of TrxR activity in APAP overdose mouse liver [239]. Other modifications also occur, such as, GAPDH Cys149 modification by NAPQI [240].
Integrated function of the Cys proteome
The proteome is the embodiment of the genome, serving to translate the genetic and epigenetic systems into function and interaction with the environment. The series of post-translational modifications of the translated proteome, collectively termed the epiproteome [241], dramatically expands the structural variation and range of reactivity beyond that found in the translated proteome. Perhaps most importantly, bi- and multiphasic switches of the Cys proteome provide organizational hubs to broadly control and integrate complex biologic functions.
While not fully elucidated, recent studies have begun to define multiphasic switching by the Cys proteome through detection of proteins with S-nitrosylation, S-glutathionylation, persulfidation and sequential oxygenation [242]. Some Cys residues can undergo multiple modifications, potentially providing pathway switches to change biologic function. For instance, Cys374 of actin can be nitrosylated as well as glutathionylated [107, 109]. However, comparison of sulfenylated Cys residues in 193 proteins with previously identified targets of glutathionylation, disulfide formation and S-nitrosylation shows minimal overlap [25]. These results suggest that different Cys modifications are selectively targeted toward different subsets of the Cys proteome rather than serving as multidirectional switches. Expansion of this pioneering research is needed to provide an understanding of the integrated function of the Cys proteome.
At the same time, multiple omics platforms are being used to translate basic science into medical applications [243]. Comparison to genomics and transcriptomics illustrates current limitations at the level of the Cys proteome. Unlike the genome and epigenome, where systematic methods provide nearly comprehensive coverage, only a few percent of the Cys in the proteome are captured in the best mass-spectrometry proteomics analyses. In studies of gene-expression-redox proteomic interaction [244] common pathway responses were observed, but the data were too sparse to allow direct correlation. On the other hand, more targeted analysis of redox proteomic associations with metabolomics in isolated mitochondria provided direct correlations [24]. The data indicate that with improved coverage of the proteome, integrated omics research will enable mechanistic understanding at the level of systems biology, i.e., directly linking changes in the Cys proteome with altered biologic function.
Perspective and future directions
A conceptual framework is emerging for elucidation of the Cys proteome as a central omics layer within the continuum of interactive systems linking the genome and exposome (Fig. 1). Major gaps exist, however, in the hierarchy of interactions of Cys proteome sub-networks, i.e., how do the different oxidation, nitrosylation, persulfidation, metallation and alkylation sub-networks synergize or antagonize each other? Sufficient detailed understanding of individual Cys proteomics systems is available to initiate construction of more complex systems models using a “bottom-up” approach. However, a limitation of a bottom-up approach is that an atlas of Cys modifications is required to place individual signaling elements within the context of others. This highlights a need for systematic development of resources for systems biology of the Cys proteome. With advances in mass spectrometry-based methods, increased coverage of large segments of the Cys proteome is possible. But consortium efforts will be needed to approach the coverage currently available for the genome and transcriptome. Such resources could allow development of quantitative models to test function of the different Cys proteome sub-networks with existing physiologic, pharmacologic, toxicologic and genetic tools.
Such quantitative descriptions of Cys proteome pathways and networks are essential to understand sites of vulnerability in disease mechanisms and provide a foundation for a new generation of network-based redox therapeutics. Many manipulations of the Cys proteome have been used in human research and some are effective in clinical practice. For instance, global regulation of Trx-1 and GSH systems occurs through activation of the Nrf2 system, a system targeted to prevent neurodegeneration [245], in cancer prevention [246], cardiovascular disease [247] and other disease processes [248].
While optimism is appropriate concerning such applications, there is also need for recognition of the failures of free radical scavenging antioxidant trials [249, 250], as manipulations of the Cys proteome are pursued for health outcomes. The research discussed in the present review shows that the Cys proteome is a complex network, apparently with an evolved function to support adaption to diet and environment. Therapeutic manipulations are, in effect, environmental perturbations. Hence, outcomes from treatments to modify the Cys proteome may suffer the same fate as the antioxidant trials if network structures are not considered in study design.
Fortunately, the range of modifications for biological switching of structure and reactivity is now well established. Methods are available to quantify abundance and reactivity, to incorporate reactions with electrophilic chemicals and binding to metal ions, to elucidate the details of the switching systems and to define their compartmentalized functions. With these capabilities, advanced integrative models, with links to imaging and other omics platforms, will provide capabilities to recognize the therapeutic potential available through targeting the cysteine proteome.
Highlights.
The cysteine proteome provides an interface for an organism to sense and adapt to its environment, including resources such as food and O2, and threats, such as infection and toxicants.
Functional switching of protein structure and reactivity occurs through purposeful sulfenylation, disulfide formation, oxidation to higher oxidation states, S-nitrosylation, persulfidation, metallation, and other modifications
Systematic mapping of cysteine proteome reactivities and modifications will support integrated systems biology models connecting cell signaling and other complex functions; this mapping will enable new therapeutic approaches targeting redox networks.
Acknowledgments
The authors acknowledge funding from NIH R01 ES023485 (Y.G. and D.J.), ES009047 (D.J.) and AG0387 (D.J.), and NIH P20 HL113451 (D.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Go YM, Jones DP. Redox biology: interface of the exposome with the proteome, epigenome and genome. Redox Biol. 2014;2:358–360. doi: 10.1016/j.redox.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild CP. Complementing the genome with an "exposome": the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 3.Miller GW, Jones DP. The Nature of Nurture: Refining the Definition of the Exposome. Toxicol Sci. 2013 doi: 10.1093/toxsci/kft251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 6.Go YM, Duong DM, Peng J, Jones DP. Protein Cysteines Map to Functional Networks According to Steady-state Level of Oxidation. J Proteomics Bioinform. 2011;4:196–209. doi: 10.4172/jpb.1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Go YM, Jones DP. Thiol/disulfide redox states in signaling and sensing. Crit Rev Biochem Mol Biol. 2013;48:173–181. doi: 10.3109/10409238.2013.764840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Go YM, Orr M, Jones DP. Actin cytoskeleton redox proteome oxidation by cadmium. Am J Physiol Lung Cell Mol Physiol. 2013;305:L831–L843. doi: 10.1152/ajplung.00203.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halvey PJ, Hansen JM, Johnson JM, Go YM, Samali A, Jones DP. Selective oxidative stress in cell nuclei by nuclear-targeted D-amino acid oxidase. Antioxid Redox Signal. 2007;9:807–816. doi: 10.1089/ars.2007.1526. [DOI] [PubMed] [Google Scholar]
- 10.Halvey PJ, Watson WH, Hansen JM, Go YM, Samali A, Jones DP. Compartmental oxidation of thiol-disulphide redox couples during epidermal growth factor signalling. Biochem J. 2005;386:215–219. doi: 10.1042/BJ20041829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu Rev Biochem. 1985;54:305–329. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 13.Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 14.Jones DP, Liang Y. Measuring the poise of thiol/disulfide couples in vivo. Free Radic Biol Med. 2009;47:1329–1338. doi: 10.1016/j.freeradbiomed.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–C868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DP. Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Intern Med. 2010;268:432–448. doi: 10.1111/j.1365-2796.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, Mieyal JJ, Chock PB. Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem. 2001;276:47763–47766. doi: 10.1074/jbc.C100415200. [DOI] [PubMed] [Google Scholar]
- 18.Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic Biol Med. 2007;42:823–829. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 20.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 21.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go YM, Park H, Koval M, Orr M, Reed M, Liang Y, Smith D, Pohl J, Jones DP. A key role for mitochondria in endothelial signaling by plasma cysteine/cystine redox potential. Free Radic Biol Med. 2010;48:275–283. doi: 10.1016/j.freeradbiomed.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Go YM, Pohl J, Jones DP. Quantification of redox conditions in the nucleus. Methods Mol Biol. 2009;464:303–317. doi: 10.1007/978-1-60327-461-6_17. [DOI] [PubMed] [Google Scholar]
- 24.Go YM, Roede JR, Orr M, Liang Y, Jones DP. Integrated redox proteomics and metabolomics of mitochondria to identify mechanisms of cd toxicity. Toxicol Sci. 2014;139:59–73. doi: 10.1093/toxsci/kfu018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem Biol. 2009;4:783–799. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy MP. Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxid Redox Signal. 2012;16:476–495. doi: 10.1089/ars.2011.4289. [DOI] [PubMed] [Google Scholar]
- 28.Miseta A, Csutora P. Relationship between the occurrence of cysteine in proteins and the complexity of organisms. Mol Biol Evol. 2000;17:1232–1239. doi: 10.1093/oxfordjournals.molbev.a026406. [DOI] [PubMed] [Google Scholar]
- 29.Huber C, Wachtershauser G. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science. 1997;276:245–247. doi: 10.1126/science.276.5310.245. [DOI] [PubMed] [Google Scholar]
- 30.Drazic A, Winter J. The physiological role of reversible methionine oxidation. Biochim Biophys Acta. 2014;1844:1367–1382. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 31.Kim G, Weiss SJ, Levine RL. Methionine oxidation and reduction in proteins. Biochim Biophys Acta. 2014;1840:901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cenci S, Sitia R. Managing and exploiting stress in the antibody factory. FEBS Lett. 2007;581:3652–3657. doi: 10.1016/j.febslet.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Yu J, Huo L, Wang L, Feng W, Wang CC. Human protein-disulfide isomerase is a redox-regulated chaperone activated by oxidation of domain a'. J Biol Chem. 2012;287:1139–1149. doi: 10.1074/jbc.M111.303149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chambers JE, Tavender TJ, Oka OB, Warwood S, Knight D, Bulleid NJ. The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1alpha. J Biol Chem. 2010;285:29200–29207. doi: 10.1074/jbc.M110.156596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Ali M, Fischer M, Riemer J. Human copper chaperone for superoxide dismutase 1 mediates its own oxidation-dependent import into mitochondria. Nat Commun. 2013;4:2430. doi: 10.1038/ncomms3430. [DOI] [PubMed] [Google Scholar]
- 36.Wrobel L, Trojanowska A, Sztolsztener ME, Chacinska A. Mitochondrial protein import: Mia40 facilitates Tim22 translocation into the inner membrane of mitochondria. Mol Biol Cell. 2013;24:543–554. doi: 10.1091/mbc.E12-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huh YJ, Kim JM, Kim H, Song H, So H, Lee SY, Kwon SB, Kim HJ, Kim HH, Lee SH, Choi Y, Chung SC, Jeong DW, Min BM. Regulation of osteoclast differentiation by the redox-dependent modulation of nuclear import of transcription factors. Cell Death Differ. 2006;13:1138–1146. doi: 10.1038/sj.cdd.4401793. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto K, Tanaka H, Ogawa H, Makino Y, Eguchi H, Hayashi S, Yoshikawa N, Poellinger L, Umesono K, Makino I. Redox-dependent regulation of nuclear import of the glucocorticoid receptor. J Biol Chem. 1999;274:10363–10371. doi: 10.1074/jbc.274.15.10363. [DOI] [PubMed] [Google Scholar]
- 39.Qu J, Liu GH, Huang B, Chen C. Nitric oxide controls nuclear export of APE1/Ref-1 through S-nitrosation of cysteines 93 and 310. Nucleic Acids Res. 2007;35:2522–2532. doi: 10.1093/nar/gkl1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp M, Go YM, Jones DP. Nonequilibrium thermodynamics of thiol/disulfide redox systems: a perspective on redox systems biology. Free Radic Biol Med. 2008;44:921–937. doi: 10.1016/j.freeradbiomed.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson CL, Iyer SS, Ziegler TR, Jones DP. Control of extracellular cysteine/cystine redox state by HT-29 cells is independent of cellular glutathione. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1069–R1075. doi: 10.1152/ajpregu.00195.2007. [DOI] [PubMed] [Google Scholar]
- 42.Go YM, Ziegler TR, Johnson JM, Gu L, Hansen JM, Jones DP. Selective protection of nuclear thioredoxin-1 and glutathione redox systems against oxidation during glucose and glutamine deficiency in human colonic epithelial cells. Free Radic Biol Med. 2007;42:363–370. doi: 10.1016/j.freeradbiomed.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DP, Go YM, Anderson CL, Ziegler TR, Kinkade JM, Jr, Kirlin WG. Cysteine/cystine couple is a newly recognized node in the circuitry for biologic redox signaling and control. FASEB J. 2004;18:1246–1248. doi: 10.1096/fj.03-0971fje. [DOI] [PubMed] [Google Scholar]
- 44.Kirlin WG, Cai J, Thompson SA, Diaz D, Kavanagh TJ, Jones DP. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic Biol Med. 1999;27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 45.Hansen JM, Moriarty-Craige S, Jones DP. Nuclear and cytoplasmic peroxiredoxin-1 differentially regulate NF-kappaB activities. Free Radic Biol Med. 2007;43:282–288. doi: 10.1016/j.freeradbiomed.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Go YM, Kang SM, Roede JR, Orr M, Jones DP. Increased inflammatory signaling and lethality of influenza H1N1 by nuclear thioredoxin-1. PLoS One. 2011;6:e18918. doi: 10.1371/journal.pone.0018918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barabasi AL. Scale-free networks: a decade and beyond. Science. 2009;325:412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 48.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 49.Go YM, Roede JR, Walker DI, Duong DM, Seyfried NT, Orr M, Liang Y, Pennell KD, Jones DP. Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol Cell Proteomics. 2013;12:3285–3296. doi: 10.1074/mcp.M113.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pader I, Sengupta R, Cebula M, Xu J, Lundberg JO, Holmgren A, Johansson K, Arner ES. Thioredoxin-related protein of 14 kDa is an efficient L-cystine reductase and Sdenitrosylase. Proc Natl Acad Sci U S A. 2014;111:6964–6969. doi: 10.1073/pnas.1317320111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.D'Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 52.Nagy P, Ashby MT. Reactive sulfur species: kinetics and mechanisms of the oxidation of cysteine by hypohalous acid to give cysteine sulfenic acid. J Am Chem Soc. 2007;129:14082–14091. doi: 10.1021/ja0737218. [DOI] [PubMed] [Google Scholar]
- 53.Lim A, Wally J, Walsh MT, Skinner M, Costello CE. Identification and location of a cysteinyl posttranslational modification in an amyloidogenic kappa1 light chain protein by electrospray ionization and matrix-assisted laser desorption/ionization mass spectrometry. Analytical biochemistry. 2001;295:45–56. doi: 10.1006/abio.2001.5187. [DOI] [PubMed] [Google Scholar]
- 54.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 55.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roede JR, Jones DP. Reactive species and mitochondrial dysfunction: mechanistic significance of 4-hydroxynonenal. Environ Mol Mutagen. 2010;51:380–390. doi: 10.1002/em.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagy P, Winterbourn CC, editors. Redox chemistry of biological thiols. Elsevier; 2010. [Google Scholar]
- 58.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martyniuk CJ, Fang B, Koomen JM, Gavin T, Zhang L, Barber DS, Lopachin RM. Molecular mechanism of glyceraldehyde-3-phosphate dehydrogenase inactivation by alpha,beta-unsaturated carbonyl derivatives. Chem Res Toxicol. 2011;24:2302–2311. doi: 10.1021/tx200437y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagy P, Karton A, Betz A, Peskin AV, Pace P, O'Reilly RJ, Hampton MB, Radom L, Winterbourn CC. Model for the exceptional reactivity of peroxiredoxins 2 and 3 with hydrogen peroxide: a kinetic and computational study. J Biol Chem. 2011;286:18048–18055. doi: 10.1074/jbc.M111.232355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wardman P, von Sonntag C. Kinetic factors that control the fate of thiyl radicals in cells. Methods Enzymol. 1995;251:31–45. doi: 10.1016/0076-6879(95)51108-3. [DOI] [PubMed] [Google Scholar]
- 62.Jordan A, Reichard P. Ribonucleotide reductases. Annual review of biochemistry. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 63.Nordlund P, Reichard P. Ribonucleotide reductases. Annual review of biochemistry. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 64.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arner ES. Selenoproteins-What unique properties can arise with selenocysteine in place of cysteine? Exp Cell Res. 2010;316:1296–1303. doi: 10.1016/j.yexcr.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 66.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci U S A. 2002;99:4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 68.Wu C, Parrott AM, Liu T, Jain MR, Yang Y, Sadoshima J, Li H. Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. J Proteomics. 2011;74:2498–2509. doi: 10.1016/j.jprot.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Porcelli AM, Ghelli A, Zanna C, Pinton P, Rizzuto R, Rugolo M. pH difference across the outer mitochondrial membrane measured with a green fluorescent protein mutant. Biochem Biophys Res Commun. 2005;326:799–804. doi: 10.1016/j.bbrc.2004.11.105. [DOI] [PubMed] [Google Scholar]
- 70.Moutevelis E, Warwicker J. Prediction of pKa and redox properties in the thioredoxin superfamily. Protein Sci. 2004;13:2744–2752. doi: 10.1110/ps.04804504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferrer-Sueta G, Manta B, Botti H, Radi R, Trujillo M, Denicola A. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 73.Winterbourn CC. The biological chemistry of hydrogen peroxide. Methods Enzymol. 2013;528:3–25. doi: 10.1016/B978-0-12-405881-1.00001-X. [DOI] [PubMed] [Google Scholar]
- 74.Lopachin RM, Gavin T, Decaprio A, Barber DS. Application of the Hard and Soft, Acids and Bases (HSAB) theory to toxicant--target interactions. Chem Res Toxicol. 2012;25:239–251. doi: 10.1021/tx2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Babula P, Masarik M, Adam V, Eckschlager T, Stiborova M, Trnkova L, Skutkova H, Provaznik I, Hubalek J, Kizek R. Mammalian metallothioneins: properties and functions. Metallomics. 2012;4:739–750. doi: 10.1039/c2mt20081c. [DOI] [PubMed] [Google Scholar]
- 76.Rubino JT, Chenkin MP, Keller M, Riggs-Gelasco P, Franz KJ. A comparison of methionine, histidine and cysteine in copper(I)-binding peptides reveals differences relevant to copper uptake by organisms in diverse environments. Metallomics. 2011;3:61–73. [PubMed] [Google Scholar]
- 77.Johnson MK. Iron-sulfur proteins: new roles for old clusters. Curr Opin Chem Biol. 1998;2:173–181. doi: 10.1016/s1367-5931(98)80058-6. [DOI] [PubMed] [Google Scholar]
- 78.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 79.Vatsis KP, Peng HM, Coon MJ. Replacement of active-site cysteine-436 by serine converts cytochrome P450 2B4 into an NADPH oxidase with negligible monooxygenase activity. J Inorg Biochem. 2002;91:542–553. doi: 10.1016/s0162-0134(02)00438-5. [DOI] [PubMed] [Google Scholar]
- 80.Maret W. Zinc and sulfur: a critical biological partnership. Biochemistry. 2004;43:3301–3309. doi: 10.1021/bi036340p. [DOI] [PubMed] [Google Scholar]
- 81.Blaby-Haas CE, Merchant SS. Lysosome-related Organelles as Mediators of Metal Homeostasis. J Biol Chem. 2014;289:28129–28136. doi: 10.1074/jbc.R114.592618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakagawa I, Suzuki M, Imura N, Naganuma A. Involvement of oxidative stress in paraquat-induced metallothionein synthesis under glutathione depletion. Free Radic Biol Med. 1998;24:1390–1395. doi: 10.1016/s0891-5849(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 83.Tong X, Evangelista A, Cohen RA. Targeting the redox regulation of SERCA in vascular physiology and disease. Curr Opin Pharmacol. 2010;10:133–138. doi: 10.1016/j.coph.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dietrich LE, Ungermann C. On the mechanism of protein palmitoylation. EMBO Rep. 2004;5:1053–1057. doi: 10.1038/sj.embor.7400277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Furtmuller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. Active site structure and catalytic mechanisms of human peroxidases. Arch Biochem Biophys. 2006;445:199–213. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 86.Perkins ND. Cysteine 38 holds the key to NF-kappaB activation. Mol Cell. 2012;45:1–3. doi: 10.1016/j.molcel.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 87.Langeland BT, Morris DL, McKinley-McKee JS. Metal binding properties of thiols; complexes with horse liver alcohol dehydrogenase. Comp Biochem Physiol B Biochem Mol Biol. 1999;123:155–162. doi: 10.1016/s0305-0491(99)00048-6. [DOI] [PubMed] [Google Scholar]
- 88.Mailloux RJ, Willmore WG. S-glutathionylation reactions in mitochondrial function and disease. Front Cell Dev Biol. 2014;2:68. doi: 10.3389/fcell.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ali Khan H, Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2014;2:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Westley J, Adler H, Westley L, Nishida C. The sulfurtransferases. Fundam Appl Toxicol. 1983;3:377–382. doi: 10.1016/s0272-0590(83)80008-6. [DOI] [PubMed] [Google Scholar]
- 91.Nagahara N. Catalytic site cysteines of thiol enzyme: sulfurtransferases. J Amino Acids. 2011;2011:709404. doi: 10.4061/2011/709404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li L, Rose P, Moore PK. Hydrogen sulfide and cell signaling. Annu Rev Pharmacol Toxicol. 2011;51:169–187. doi: 10.1146/annurev-pharmtox-010510-100505. [DOI] [PubMed] [Google Scholar]
- 93.Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64:520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stennicke HR, Salvesen GS. Catalytic properties of the caspases. Cell Death Differ. 1999;6:1054–1059. doi: 10.1038/sj.cdd.4400599. [DOI] [PubMed] [Google Scholar]
- 95.Kolmodin K, Aqvist J. The catalytic mechanism of protein tyrosine phosphatases revisited. FEBS Lett. 2001;498:208–213. doi: 10.1016/s0014-5793(01)02479-6. [DOI] [PubMed] [Google Scholar]
- 96.Du Y, Zhang H, Lu J, Holmgren A. Glutathione and glutaredoxin act as a backup of human thioredoxin reductase 1 to reduce thioredoxin 1 preventing cell death by aurothioglucose. J Biol Chem. 2012;287:38210–38219. doi: 10.1074/jbc.M112.392225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toledano MB, Delaunay-Moisan A, Outten CE, Igbaria A. Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid Redox Signal. 2013;18:1699–1711. doi: 10.1089/ars.2012.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 99.Godin LM, Vergen J, Prakash YS, Pagano RE, Hubmayr RD. Spatiotemporal dynamics of actin remodeling and endomembrane trafficking in alveolar epithelial type I cell wound healing. Am J Physiol Lung Cell Mol Physiol. 2011;300:L615–L623. doi: 10.1152/ajplung.00265.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lacayo CI, Pincus Z, VanDuijn MM, Wilson CA, Fletcher DA, Gertler FB, Mogilner A, Theriot JA. Emergence of large-scale cell morphology and movement from local actin filament growth dynamics. PLoS Biol. 2007;5:e233. doi: 10.1371/journal.pbio.0050233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zuo X, Zhang J, Zhang Y, Hsu SC, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]
- 102.Fedorova M, Kuleva N, Hoffmann R. Identification of cysteine, methionine and tryptophan residues of actin oxidized in vivo during oxidative stress. J Proteome Res. 2010;9:1598–1609. doi: 10.1021/pr901099e. [DOI] [PubMed] [Google Scholar]
- 103.Ishiwata S. Freezing of actin. Reversible oxidation of a sulfhydryl group and structural change. J Biochem. 1976;80:595–609. doi: 10.1093/oxfordjournals.jbchem.a131315. [DOI] [PubMed] [Google Scholar]
- 104.Lassing I, Schmitzberger F, Bjornstedt M, Holmgren A, Nordlund P, Schutt CE, Lindberg U. Molecular and structural basis for redox regulation of beta-actin. J Mol Biol. 2007;370:331–348. doi: 10.1016/j.jmb.2007.04.056. [DOI] [PubMed] [Google Scholar]
- 105.Berndt C, Poschmann G, Stuhler K, Holmgren A, Brautigam L. Zebrafish heart development is regulated via glutaredoxin 2 dependent migration and survival of neural crest cells. Redox Biol. 2014;2:673–678. doi: 10.1016/j.redox.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamnell-Pamment Y, Lind C, Palmberg C, Bergman T, Cotgreave IA. Determination of site-specificity of S-glutathionylated cellular proteins. Biochem Biophys Res Commun. 2005;332:362–369. doi: 10.1016/j.bbrc.2005.04.130. [DOI] [PubMed] [Google Scholar]
- 107.Stournaras C, Drewes G, Blackholm H, Merkler I, Faulstich H. Glutathionyl(cysteine-374) actin forms filaments of low mechanical stability. Biochim Biophys Acta. 1990;1037:86–91. doi: 10.1016/0167-4838(90)90105-o. [DOI] [PubMed] [Google Scholar]
- 108.Chen SC, Huang B, Liu YC, Shyu KG, Lin PY, Wang DL. Acute hypoxia enhances proteins' S-nitrosylation in endothelial cells. Biochem Biophys Res Commun. 2008;377:1274–1278. doi: 10.1016/j.bbrc.2008.10.144. [DOI] [PubMed] [Google Scholar]
- 109.Dalle-Donne I, Milzani A, Giustarini D, Di Simplicio P, Colombo R, Rossi R. S-NO-actin: S-nitrosylation kinetics and the effect on isolated vascular smooth muscle. J Muscle Res Cell Motil. 2000;21:171–181. doi: 10.1023/a:1005671319604. [DOI] [PubMed] [Google Scholar]
- 110.Thom SR, Bhopale VM, Milovanova TN, Yang M, Bogush M. Thioredoxin reductase linked to cytoskeleton by focal adhesion kinase reverses actin S-nitrosylation and restores neutrophil beta(2) integrin function. J Biol Chem. 2012;287:30346–30357. doi: 10.1074/jbc.M112.355875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dalle-Donne I, Rossi R, Giustarini D, Gagliano N, Di Simplicio P, Colombo R, Milzani A. Methionine oxidation as a major cause of the functional impairment of oxidized actin. Free Radic Biol Med. 2002;32:927–937. doi: 10.1016/s0891-5849(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 112.Farah ME, Sirotkin V, Haarer B, Kakhniashvili D, Amberg DC. Diverse protective roles of the actin cytoskeleton during oxidative stress. Cytoskeleton (Hoboken) 2011;68:340–354. doi: 10.1002/cm.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. Actin S-glutathionylation: evidence against a thiol-disulphide exchange mechanism. Free Radic Biol Med. 2003;35:1185–1193. doi: 10.1016/s0891-5849(03)00504-5. [DOI] [PubMed] [Google Scholar]
- 114.Johansson M, Lundberg M. Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem. 2007;8:26. doi: 10.1186/1471-2091-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sakai J, Li J, Subramanian KK, Mondal S, Bajrami B, Hattori H, Jia Y, Dickinson BC, Zhong J, Ye K, Chang CJ, Ho YS, Zhou J, Luo HR. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 2012;37:1037–1049. doi: 10.1016/j.immuni.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klamt F, Zdanov S, Levine RL, Pariser A, Zhang Y, Zhang B, Yu LR, Veenstra TD, Shacter E. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat Cell Biol. 2009;11:1241–1246. doi: 10.1038/ncb1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Klemke M, Wabnitz GH, Funke F, Funk B, Kirchgessner H, Samstag Y. Oxidation of cofilin mediates T cell hyporesponsiveness under oxidative stress conditions. Immunity. 2008;29:404–413. doi: 10.1016/j.immuni.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 118.Wang X, Ling S, Zhao D, Sun Q, Li Q, Wu F, Nie J, Qu L, Wang B, Shen X, Bai Y, Li Y. Redox regulation of actin by thioredoxin-1 is mediated by the interaction of the proteins via cysteine 62. Antioxid Redox Signal. 2010;13:565–573. doi: 10.1089/ars.2009.2833. [DOI] [PubMed] [Google Scholar]
- 119.Zschauer TC, Kunze K, Jakob S, Haendeler J, Altschmied J. Oxidative stressinduced degradation of thioredoxin-1 and apoptosis is inhibited by thioredoxin-1-actin interaction in endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:650–656. doi: 10.1161/ATVBAHA.110.218982. [DOI] [PubMed] [Google Scholar]
- 120.McCarthy DA, Clark RR, Bartling TR, Trebak M, Melendez JA. Redox control of the senescence regulator interleukin-1alpha and the secretory phenotype. J Biol Chem. 2013;288:32149–32159. doi: 10.1074/jbc.M113.493841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dembla M, Wahl S, Katiyar R, Schmitz F. ArfGAP3 is a component of the photoreceptor synaptic ribbon complex and forms an NAD(H)-regulated, redox-sensitive complex with RIBEYE that is important for endocytosis. J Neurosci. 2014;34:5245–5260. doi: 10.1523/JNEUROSCI.3837-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arunachalam B, Phan UT, Geuze HJ, Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT) Proc Natl Acad Sci U S A. 2000;97:745–750. doi: 10.1073/pnas.97.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bloom D, Dhakshinamoorthy S, Jaiswal AK. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene. 2002;21:2191–2200. doi: 10.1038/sj.onc.1205288. [DOI] [PubMed] [Google Scholar]
- 124.Christman JW, Blackwell TS, Juurlink BH. Redox regulation of nuclear factor kappa B: therapeutic potential for attenuating inflammatory responses. Brain Pathol. 2000;10:153–162. doi: 10.1111/j.1750-3639.2000.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. AP-1 transcriptional activity is regulated by a direct association between thioredoxin and Ref-1. Proc Natl Acad Sci U S A. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xanthoudakis S, Curran T. Redox regulation of AP-1: a link between transcription factor signaling and DNA repair. Adv Exp Med Biol. 1996;387:69–75. [PubMed] [Google Scholar]
- 127.Butler LM, Zhou X, Xu WS, Scher HI, Rifkind RA, Marks PA, Richon VM. The histone deacetylase inhibitor SAHA arrests cancer cell growth, up-regulates thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc Natl Acad Sci U S A. 2002;99:11700–11705. doi: 10.1073/pnas.182372299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nishinaka Y, Masutani H, Oka S, Matsuo Y, Yamaguchi Y, Nishio K, Ishii Y, Yodoi J. Importin alpha1 (Rch1) mediates nuclear translocation of thioredoxin-binding protein-2/vitamin D(3)-up-regulated protein 1. J Biol Chem. 2004;279:37559–37565. doi: 10.1074/jbc.M405473200. [DOI] [PubMed] [Google Scholar]
- 129.Schroeder P, Popp R, Wiegand B, Altschmied J, Haendeler J. Nuclear redoxsignaling is essential for apoptosis inhibition in endothelial cells--important role for nuclear thioredoxin-1. Arterioscler Thromb Vasc Biol. 2007;27:2325–2331. doi: 10.1161/ATVBAHA.107.149419. [DOI] [PubMed] [Google Scholar]
- 130.Papp LV, Lu J, Striebel F, Kennedy D, Holmgren A, Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol Cell Biol. 2006;26:4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Anathy V, Aesif SW, Hoffman SM, Bement JL, Guala AS, Lahue KG, Leclair LW, Suratt BT, Cool CD, Wargo MJ, Janssen-Heininger YM. Glutaredoxin-1 attenuates S-glutathionylation of the death receptor fas and decreases resolution of Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2014;189:463–474. doi: 10.1164/rccm.201310-1905OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eriksson SE, Prast-Nielsen S, Flaberg E, Szekely L, Arner ES. High levels of thioredoxin reductase 1 modulate drug-specific cytotoxic efficacy. Free Radic Biol Med. 2009;47:1661–1671. doi: 10.1016/j.freeradbiomed.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 133.Myers JM, Myers CR. The effects of hexavalent chromium on thioredoxin reductase and peroxiredoxins in human bronchial epithelial cells. Free Radic Biol Med. 2009;47:1477–1485. doi: 10.1016/j.freeradbiomed.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhao F, Yan J, Deng S, Lan L, He F, Kuang B, Zeng H. A thioredoxin reductase inhibitor induces growth inhibition and apoptosis in five cultured human carcinoma cell lines. Cancer Lett. 2006;236:46–53. doi: 10.1016/j.canlet.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 135.Yoshihara E, Masaki S, Matsuo Y, Chen Z, Tian H, Yodoi J. Thioredoxin/Txnip: redoxisome, as a redox switch for the pathogenesis of diseases. Front Immunol. 2014;4:514. doi: 10.3389/fimmu.2013.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Abdelsaid MA, Matragoon S, El-Remessy AB. Thioredoxin-interacting protein expression is required for VEGF-mediated angiogenic signal in endothelial cells. Antioxid Redox Signal. 2013;19:2199–2212. doi: 10.1089/ars.2012.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Masutani H, Yoshihara E, Masaki S, Chen Z, Yodoi J. Thioredoxin binding protein (TBP)-2/Txnip and alpha-arrestin proteins in cancer and diabetes mellitus. J Clin Biochem Nutr. 2012;50:23–34. doi: 10.3164/jcbn.11-36SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Watanabe R, Nakamura H, Masutani H, Yodoi J. Anti-oxidative, anti-cancer and anti-inflammatory actions by thioredoxin 1 and thioredoxin-binding protein-2. Pharmacol Ther. 2010;127:261–270. doi: 10.1016/j.pharmthera.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 139.Yoshihara E, Chen Z, Matsuo Y, Masutani H, Yodoi J. Thiol redox transitions by thioredoxin and thioredoxin-binding protein-2 in cell signaling. Methods Enzymol. 2010;474:67–82. doi: 10.1016/S0076-6879(10)74005-2. [DOI] [PubMed] [Google Scholar]
- 140.Devi TS, Hosoya K, Terasaki T, Singh LP. Critical role of TXNIP in oxidative stress, DNA damage and retinal pericyte apoptosis under high glucose: implications for diabetic retinopathy. Exp Cell Res. 2013;319:1001–1012. doi: 10.1016/j.yexcr.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thompson JK, Westbom CM, MacPherson MB, Mossman BT, Heintz NH, Spiess P, Shukla A. Asbestos modulates thioredoxin-thioredoxin interacting protein interaction to regulate inflammasome activation. Part Fibre Toxicol. 2014;11:24. doi: 10.1186/1743-8977-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lu J, Holmgren A. Thioredoxin system in cell death progression. Antioxid Redox Signal. 2012;17:1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 143.Lee YJ, Kim JH, Chen J, Song JJ. Enhancement of metabolic oxidative stress-induced cytotoxicity by the thioredoxin inhibitor 1-methylpropyl 2-imidazolyl disulfide is mediated through the ASK1-SEK1-JNK1 pathway. Mol Pharmacol. 2002;62:1409–1417. doi: 10.1124/mol.62.6.1409. [DOI] [PubMed] [Google Scholar]
- 144.Yang W, Tiffany-Castiglioni E, Koh HC, Son IH. Paraquat activates the IRE1/ASK1/JNK cascade associated with apoptosis in human neuroblastoma SH-SY5Y cells. Toxicol Lett. 2009;191:203–210. doi: 10.1016/j.toxlet.2009.08.024. [DOI] [PubMed] [Google Scholar]
- 145.Kim HL, Koedrith P, Lee SM, Kim YJ, Seo YR. Base excision DNA repair defect in thioredoxin-1 (Trx1)-deficient cells. Mutat Res. 2013;751–752:1–7. doi: 10.1016/j.mrfmmm.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 146.Malik G, Gorbounov N, Das S, Gurusamy N, Otani H, Maulik N, Goswami S, Das DK. Ischemic preconditioning triggers nuclear translocation of thioredoxin and its interaction with Ref-1 potentiating a survival signal through the PI-3-kinase-Akt pathway. Antioxid Redox Signal. 2006;8:2101–2109. doi: 10.1089/ars.2006.8.2101. [DOI] [PubMed] [Google Scholar]
- 147.Go YM, Jones DP. Redox control systems in the nucleus: mechanisms and functions. Antioxid Redox Signal. 2010;13:489–509. doi: 10.1089/ars.2009.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seemann S, Hainaut P. Roles of thioredoxin reductase 1 and APE/Ref-1 in the control of basal p53 stability and activity. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- 149.Soderberg A, Hossain A, Rosen A. A protein disulfide isomerase/thioredoxin-1 complex is physically attached to exofacial membrane tumor necrosis factor receptors: overexpression in chronic lymphocytic leukemia cells. Antioxid Redox Signal. 2013;18:363–375. doi: 10.1089/ars.2012.4789. [DOI] [PubMed] [Google Scholar]
- 150.Adnan H, Antenos M, Kirby GM. The effect of menadione on glutathione S-transferase A1 (GSTA1): c-Jun N-terminal kinase (JNK) complex dissociation in human colonic adenocarcinoma Caco-2 cells. Toxicol Lett. 2012;214:53–62. doi: 10.1016/j.toxlet.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 151.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 152.Jones DP, Eklow L, Thor H, Orrenius S. Metabolism of hydrogen peroxide in isolated hepatocytes: relative contributions of catalase and glutathione peroxidase in decomposition of endogenously generated H2O2. Arch Biochem Biophys. 1981;210:505–516. doi: 10.1016/0003-9861(81)90215-0. [DOI] [PubMed] [Google Scholar]
- 153.Jones DP. Intracellular catalase function: analysis of the catalatic activity by product formation in isolated liver cells. Arch Biochem Biophys. 1982;214:806–814. doi: 10.1016/0003-9861(82)90087-x. [DOI] [PubMed] [Google Scholar]
- 154.Sies H, Bucher T, Oshino N, Chance B. Heme occupancy of catalase in hemoglobin-free perfused rat liver and of isolated rat liver catalase. Arch Biochem Biophys. 1973;154:106–116. doi: 10.1016/0003-9861(73)90039-8. [DOI] [PubMed] [Google Scholar]
- 155.Oshino N, Chance B, Sies H, Bucher T. The role of H 2 O 2 generation in perfused rat liver and the reaction of catalase compound I and hydrogen donors. Arch Biochem Biophys. 1973;154:117–131. doi: 10.1016/0003-9861(73)90040-4. [DOI] [PubMed] [Google Scholar]
- 156.Sies H, Chance B. The steady state level of catalase compound I in isolated hemoglobin-free perfused rat liver. FEBS Lett. 1970;11:172–176. doi: 10.1016/0014-5793(70)80521-x. [DOI] [PubMed] [Google Scholar]
- 157.Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ, 2nd, Ischiropoulos H. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med. 2012;52:1–6. doi: 10.1016/j.freeradbiomed.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Winterbourn CC, Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med. 1999;27:322–328. doi: 10.1016/s0891-5849(99)00051-9. [DOI] [PubMed] [Google Scholar]
- 159.Adimora NJ, Jones DP, Kemp ML. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid Redox Signal. 2010;13:731–743. doi: 10.1089/ars.2009.2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choi MH, Lee IK, Kim GW, Kim BU, Han YH, Yu DY, Park HS, Kim KY, Lee JS, Choi C, Bae YS, Lee BI, Rhee SG, Kang SW. Regulation of PDGF signalling and vascular remodelling by peroxiredoxin II. Nature. 2005;435:347–353. doi: 10.1038/nature03587. [DOI] [PubMed] [Google Scholar]
- 161.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 162.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 163.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O'Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich AK, Jakob U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell. 2012;47:767–776. doi: 10.1016/j.molcel.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol. 2013;15:222–228. doi: 10.1038/ncb2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 168.Watson WH, Pohl J, Montfort WR, Stuchlik O, Reed MS, Powis G, Jones DP. Redox potential of human thioredoxin 1 and identification of a second dithiol/disulfide motif. J Biol Chem. 2003;278:33408–33415. doi: 10.1074/jbc.M211107200. [DOI] [PubMed] [Google Scholar]
- 169.Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–6467. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 170.Kaplan N, Urao N, Furuta E, Kim SJ, Razvi M, Nakamura Y, McKinney RD, Poole LB, Fukai T, Ushio-Fukai M. Localized cysteine sulfenic acid formation by vascular endothelial growth factor: role in endothelial cell migration and angiogenesis. Free Radic Res. 2011;45:1124–1135. doi: 10.3109/10715762.2011.602073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA. Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell. 2012;45:398–408. doi: 10.1016/j.molcel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 172.Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The high reactivity of peroxiredoxin 2 with H(2)O(2) is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem. 2007;282:11885–11892. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 173.Arnhold J, Monzani E, Furtmüller PG, Zederbauer M, Casella L, Obinger C. Kinetics and Thermodynamics of Halide and Nitrite Oxidation by Mammalian Heme Peroxidases. European Journal of Inorganic Chemistry. 2006;2006:3801–3811. [Google Scholar]
- 174.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid Redox Signal. 2005;7:560–577. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 176.Sarma BK, Mugesh G. Redox regulation of protein tyrosine phosphatase 1B (PTP1B): a biomimetic study on the unexpected formation of a sulfenyl amide intermediate. J Am Chem Soc. 2007;129:8872–8881. doi: 10.1021/ja070410o. [DOI] [PubMed] [Google Scholar]
- 177.Fu X, Mueller DM, Heinecke JW. Generation of intramolecular and intermolecular sulfenamides, sulfinamides, and sulfonamides by hypochlorous acid: a potential pathway for oxidative cross-linking of low-density lipoprotein by myeloperoxidase. Biochemistry. 2002;41:1293–1301. doi: 10.1021/bi015777z. [DOI] [PubMed] [Google Scholar]
- 178.Kettle AJ, Turner R, Gangell CL, Harwood DT, Khalilova IS, Chapman AL, Winterbourn CC, Sly PD. Oxidation contributes to low glutathione in the airways of children with cystic fibrosis. Eur Respir J. 2014;44:122–129. doi: 10.1183/09031936.00170213. [DOI] [PubMed] [Google Scholar]
- 179.Chandler JD, Min E, Huang J, McElroy CS, Dickerhof N, Mocatta T, Fletcher AA, Evans CM, Liang L, Patel M, Kettle AJ, Nichols DP, Day BJ. Anti-Inflammatory and Anti-Microbial Effects of Thiocyanate in a Cystic Fibrosis Mouse Model. Am J Respir Cell Mol Biol. 2014 doi: 10.1165/rcmb.2014-0208OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jonsson TJ, Tsang AW, Lowther WT, Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mocatta TJ, Pilbrow AP, Cameron VA, Senthilmohan R, Frampton CM, Richards AM, Winterbourn CC. Plasma concentrations of myeloperoxidase predict mortality after myocardial infarction. J Am Coll Cardiol. 2007;49:1993–2000. doi: 10.1016/j.jacc.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 182.Nagy P, Beal JL, Ashby MT. Thiocyanate is an efficient endogenous scavenger of the phagocytic killing agent hypobromous acid. Chem Res Toxicol. 2006;19:587–593. doi: 10.1021/tx050338c. [DOI] [PubMed] [Google Scholar]
- 183.van Dalen CJ, Kettle AJ. Substrates and products of eosinophil peroxidase. Biochem J. 2001;358:233–239. doi: 10.1042/0264-6021:3580233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wu W, Chen Y, Hazen SL. Eosinophil peroxidase nitrates protein tyrosyl residues. Implications for oxidative damage by nitrating intermediates in eosinophilic inflammatory disorders. J Biol Chem. 1999;274:25933–25944. doi: 10.1074/jbc.274.36.25933. [DOI] [PubMed] [Google Scholar]
- 185.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG. Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. Nat Chem Biol. 2012;8:784–790. doi: 10.1038/nchembio.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Li H, Cao Z, Zhang G, Thannickal VJ, Cheng G. Vascular peroxidase 1 catalyzes the formation of hypohalous acids: characterization of its substrate specificity and enzymatic properties. Free Radic Biol Med. 2012;53:1954–1959. doi: 10.1016/j.freeradbiomed.2012.08.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ronsein GE, Winterbourn CC, Di Mascio P, Kettle AJ. Cross-linking methionine and amine residues with reactive halogen species. Free Radic Biol Med. 2014;70:278–287. doi: 10.1016/j.freeradbiomed.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 188.Peterfi Z, Geiszt M. Peroxidasins: novel players in tissue genesis. Trends Biochem Sci. 2014;39:305–307. doi: 10.1016/j.tibs.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 189.Zhu L, Pi J, Wachi S, Andersen ME, Wu R, Chen Y. Identification of Nrf2-dependent airway epithelial adaptive response to proinflammatory oxidant-hypochlorous acid challenge by transcription profiling. Am J Physiol Lung Cell Mol Physiol. 2008;294:L469–L477. doi: 10.1152/ajplung.00310.2007. [DOI] [PubMed] [Google Scholar]
- 190.Peskin AV, Winterbourn CC. Kinetics of the reactions of hypochlorous acid and amino acid chloramines with thiols, methionine, and ascorbate. Free Radic Biol Med. 2001;30:572–579. doi: 10.1016/s0891-5849(00)00506-2. [DOI] [PubMed] [Google Scholar]
- 191.Pattison DI, Davies MJ. Absolute rate constants for the reaction of hypochlorous acid with protein side chains and peptide bonds. Chem Res Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- 192.Pattison DI, Davies MJ. Kinetic analysis of the reactions of hypobromous acid with protein components: implications for cellular damage and use of 3-bromotyrosine as a marker of oxidative stress. Biochemistry. 2004;43:4799–4809. doi: 10.1021/bi035946a. [DOI] [PubMed] [Google Scholar]
- 193.Peskin AV, Winterbourn CC. Taurine chloramine is more selective than hypochlorous acid at targeting critical cysteines and inactivating creatine kinase and glyceraldehyde-3-phosphate dehydrogenase. Free Radic Biol Med. 2006;40:45–53. doi: 10.1016/j.freeradbiomed.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 194.Bjorck L, Rosen C, Marshall V, Reiter B. Antibacterial activity of the lactoperoxidase system in milk against pseudomonads and other gram-negative bacteria. Appl Microbiol. 1975;30:199–204. doi: 10.1128/am.30.2.199-204.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.van Haeringen NJ, Ensink FT, Glasius E. The peroxidase-thiocyanatehydrogenperoxide system in tear fluid and saliva of different species. Exp Eye Res. 1979;28:343–347. doi: 10.1016/0014-4835(79)90095-2. [DOI] [PubMed] [Google Scholar]
- 196.Wijkstrom-Frei C, El-Chemaly S, Ali-Rachedi R, Gerson C, Cobas MA, Forteza R, Salathe M, Conner GE. Lactoperoxidase and human airway host defense. Am J Respir Cell Mol Biol. 2003;29:206–212. doi: 10.1165/rcmb.2002-0152OC. [DOI] [PubMed] [Google Scholar]
- 197.Pruitt KM, Tenovuo J, Mansson-Rahemtulla B, Harrington P, Baldone DC. Is thiocyanate peroxidation at equilibrium in vivo? Biochim Biophys Acta. 1986;870:385–391. doi: 10.1016/0167-4838(86)90245-1. [DOI] [PubMed] [Google Scholar]
- 198.Ashby MT, Aneetha H. Reactive sulfur species: aqueous chemistry of sulfenyl thiocyanates. J Am Chem Soc. 2004;126:10216–10217. doi: 10.1021/ja048585a. [DOI] [PubMed] [Google Scholar]
- 199.Nagy P, Jameson GN, Winterbourn CC. Kinetics and mechanisms of the reaction of hypothiocyanous acid with 5-thio-2-nitrobenzoic acid and reduced glutathione. Chem Res Toxicol. 2009;22:1833–1840. doi: 10.1021/tx900249d. [DOI] [PubMed] [Google Scholar]
- 200.Skaff O, Pattison DI, Davies MJ. Hypothiocyanous acid reactivity with low-molecular-mass and protein thiols: absolute rate constants and assessment of biological relevance. Biochem J. 2009;422:111–117. doi: 10.1042/BJ20090276. [DOI] [PubMed] [Google Scholar]
- 201.Skaff O, Pattison DI, Morgan PE, Bachana R, Jain VK, Priyadarsini KI, Davies MJ. Selenium-containing amino acids are targets for myeloperoxidase-derived hypothiocyanous acid: determination of absolute rate constants and implications for biological damage. Biochem J. 2012;441:305–316. doi: 10.1042/BJ20101762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang JG, Mahmud SA, Nguyen J, Slungaard A. Thiocyanate-dependent induction of endothelial cell adhesion molecule expression by phagocyte peroxidases: a novel HOSCN-specific oxidant mechanism to amplify inflammation. J Immunol. 2006;177:8714–8722. doi: 10.4049/jimmunol.177.12.8714. [DOI] [PubMed] [Google Scholar]
- 203.Wang JG, Mahmud SA, Thompson JA, Geng JG, Key NS, Slungaard A. The principal eosinophil peroxidase product, HOSCN, is a uniquely potent phagocyte oxidant inducer of endothelial cell tissue factor activity: a potential mechanism for thrombosis in eosinophilic inflammatory states. Blood. 2006;107:558–565. doi: 10.1182/blood-2005-05-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fabrini R, Bocedi A, Camerini S, Fusetti M, Ottaviani F, Passali FM, Topazio D, Iavarone F, Francia I, Castagnola M, Ricci G. Inactivation of human salivary glutathione transferase P1-1 by hypothiocyanite: a post-translational control system in search of a role. PLoS One. 2014;9:e112797. doi: 10.1371/journal.pone.0112797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Xu Y, Szep S, Lu Z. The antioxidant role of thiocyanate in the pathogenesis of cystic fibrosis and other inflammation-related diseases. Proc Natl Acad Sci U S A. 2009;106:20515–20519. doi: 10.1073/pnas.0911412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chandler JD, Nichols DP, Nick JA, Hondal RJ, Day BJ. Selective metabolism of hypothiocyanous acid by mammalian thioredoxin reductase promotes lung innate immunity and antioxidant defense. J Biol Chem. 2013;288:18421–18428. doi: 10.1074/jbc.M113.468090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chandler JD, Day BJ. Biochemical mechanisms and therapeutic potential of pseudohalide thiocyanate in human health. Free Radic Res. 2015:1–16. doi: 10.3109/10715762.2014.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Anand P, Stamler JS. Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J Mol Med (Berl) 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 210.Viner RI, Williams TD, Schoneich C. Peroxynitrite modification of protein thiols: oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 1999;38:12408–12415. doi: 10.1021/bi9909445. [DOI] [PubMed] [Google Scholar]
- 211.Cohen RA, Adachi T. Nitric-oxide-induced vasodilatation: regulation by physiologic s-glutathiolation and pathologic oxidation of the sarcoplasmic endoplasmic reticulum calcium ATPase. Trends Cardiovasc Med. 2006;16:109–114. doi: 10.1016/j.tcm.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 212.Eu JP, Sun J, Xu L, Stamler JS, Meissner G. The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell. 2000;102:499–509. doi: 10.1016/s0092-8674(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 213.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chang AH, Sancheti H, Garcia J, Kaplowitz N, Cadenas E, Han D. Respiratory substrates regulate S-nitrosylation of mitochondrial proteins through a thiol-dependent pathway. Chem Res Toxicol. 2014;27:794–804. doi: 10.1021/tx400462r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kabil O, Banerjee R. Enzymology of H2S biogenesis, decay and signaling. Antioxid Redox Signal. 2014;20:770–782. doi: 10.1089/ars.2013.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Altaany Z, Ju Y, Yang G, Wang R. The coordination of S-sulfhydration, S-nitrosylation, and phosphorylation of endothelial nitric oxide synthase by hydrogen sulfide. Sci Signal. 2014;7:ra87. doi: 10.1126/scisignal.2005478. [DOI] [PubMed] [Google Scholar]
- 217.Gadalla MM, Snyder SH. Hydrogen sulfide as a gasotransmitter. J Neurochem. 2010;113:14–26. doi: 10.1111/j.1471-4159.2010.06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kolluru GK, Shen X, Kevil CG. A tale of two gases: NO and H2S, foes or friends for life? Redox Biol. 2013;1:313–318. doi: 10.1016/j.redox.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rohwerder T, Sand W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology. 2003;149:1699–1710. doi: 10.1099/mic.0.26212-0. [DOI] [PubMed] [Google Scholar]
- 220.Kimura H. Hydrogen sulfide and polysulfides as biological mediators. Molecules. 2014;19:16146–16157. doi: 10.3390/molecules191016146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hanschmann EM, Godoy JR, Berndt C, Hudemann C, Lillig CH. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: from cofactors to antioxidants to redox signaling. Antioxid Redox Signal. 2013;19:1539–1605. doi: 10.1089/ars.2012.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Lu C, Kavalier A, Lukyanov E, Gross SS. S-sulfhydration/desulfhydration and S-nitrosylation/denitrosylation: a common paradigm for gasotransmitter signaling by H2S and NO. Methods. 2013;62:177–181. doi: 10.1016/j.ymeth.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Francoleon NE, Carrington SJ, Fukuto JM. The reaction of H(2)S with oxidized thiols: generation of persulfides and implications to H(2)S biology. Arch Biochem Biophys. 2011;516:146–153. doi: 10.1016/j.abb.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 224.Ono K, Akaike T, Sawa T, Kumagai Y, Wink DA, Tantillo DJ, Hobbs AJ, Nagy P, Xian M, Lin J, Fukuto JM. Redox chemistry and chemical biology of HS, hydropersulfides, and derived species: Implications of their possible biological activity and utility. Free Radic Biol Med. 2014 doi: 10.1016/j.freeradbiomed.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wang CN, Liu YJ, Duan GL, Zhao W, Li XH, Zhu XY, Ni X. CBS and CSE Are Critical for Maintenance of Mitochondrial Function and Glucocorticoid Production in Adrenal Cortex. Antioxid Redox Signal. 2014;21:2192–2207. doi: 10.1089/ars.2013.5682. [DOI] [PubMed] [Google Scholar]
- 226.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Xie ZZ, Shi MM, Xie L, Wu ZY, Li G, Hua F, Bian JS. Sulfhydration of p66Shc at Cysteine59 Mediates the Antioxidant Effect of Hydrogen Sulfide. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Higdon A, Diers AR, Oh JY, Landar A, Darley-Usmar VM. Cell signalling by reactive lipid species: new concepts and molecular mechanisms. Biochem J. 2012;442:453–464. doi: 10.1042/BJ20111752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fritz KS, Petersen DR. An overview of the chemistry and biology of reactive aldehydes. Free Radic Biol Med. 2013;59:85–91. doi: 10.1016/j.freeradbiomed.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fang J, Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J Am Chem Soc. 2006;128:1879–1885. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 232.Backos DS, Fritz KS, Roede JR, Petersen DR, Franklin CC. Posttranslational modification and regulation of glutamate-cysteine ligase by the alpha,beta-unsaturated aldehyde 4-hydroxy-2-nonenal. Free Radic Biol Med. 2011;50:14–26. doi: 10.1016/j.freeradbiomed.2010.10.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chen ZH, Saito Y, Yoshida Y, Sekine A, Noguchi N, Niki E. 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J Biol Chem. 2005;280:41921–41927. doi: 10.1074/jbc.M508556200. [DOI] [PubMed] [Google Scholar]
- 234.Ji C, Kozak KR, Marnett LJ. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J Biol Chem. 2001;276:18223–18228. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 235.Wong HL, Liebler DC. Mitochondrial protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2008;21:796–804. doi: 10.1021/tx700433m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dennehy MK, Richards KA, Wernke GR, Shyr Y, Liebler DC. Cytosolic and nuclear protein targets of thiol-reactive electrophiles. Chem Res Toxicol. 2006;19:20–29. doi: 10.1021/tx050312l. [DOI] [PubMed] [Google Scholar]
- 237.Rappaport SM, Li H, Grigoryan H, Funk WE, Williams ER. Adductomics: characterizing exposures to reactive electrophiles. Toxicol Lett. 2012;213:83–90. doi: 10.1016/j.toxlet.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Li H, Grigoryan H, Funk WE, Lu SS, Rose S, Williams ER, Rappaport SM. Profiling Cys34 adducts of human serum albumin by fixed-step selected reaction monitoring. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004606. M110 004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jan YH, Heck DE, Dragomir AC, Gardner CR, Laskin DL, Laskin JD. Acetaminophen reactive intermediates target hepatic thioredoxin reductase. Chem Res Toxicol. 2014;27:882–894. doi: 10.1021/tx5000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dietze EC, Schafer A, Omichinski JG, Nelson SD. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by a reactive metabolite of acetaminophen and mass spectral characterization of an arylated active site peptide. Chem Res Toxicol. 1997;10:1097–1103. doi: 10.1021/tx970090u. [DOI] [PubMed] [Google Scholar]
- 241.Go YM, Jones DP. The redox proteome. J Biol Chem. 2013;288:26512–26520. doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Doulias PT, Tenopoulou M, Raju K, Spruce LA, Seeholzer SH, Ischiropoulos H. Site specific identification of endogenous S-nitrosocysteine proteomes. J Proteomics. 2013;92:195–203. doi: 10.1016/j.jprot.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Topol EJ. Individualized medicine from prewomb to tomb. Cell. 2014;157:241–253. doi: 10.1016/j.cell.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Go YM, Craige SE, Orr M, Gernert KM, Jones DP. Gene and protein responses of human monocytes to extracellular cysteine redox potential. Toxicol Sci. 2009;112:354–362. doi: 10.1093/toxsci/kfp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Joshi G, Johnson JA. The Nrf2-ARE pathway: a valuable therapeutic target for the treatment of neurodegenerative diseases. Recent Pat CNS Drug Discov. 2012;7:218–229. doi: 10.2174/157488912803252023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Balogun E, Hoque M, Gong P, Killeen E, Green CJ, Foresti R, Alam J, Motterlini R. Curcumin activates the haem oxygenase-1 gene via regulation of Nrf2 and the antioxidant-responsive element. Biochem J. 2003;371:887–895. doi: 10.1042/BJ20021619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li J, Ichikawa T, Villacorta L, Janicki JS, Brower GL, Yamamoto M, Cui T. Nrf2 protects against maladaptive cardiac responses to hemodynamic stress. Arterioscler Thromb Vasc Biol. 2009;29:1843–1850. doi: 10.1161/ATVBAHA.109.189480. [DOI] [PubMed] [Google Scholar]
- 248.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 250.Goodman M, Bostick RM, Kucuk O, Jones DP. Clinical trials of antioxidants as cancer prevention agents: past, present, and future. Free Radic Biol Med. 2011;51:1068–1084. doi: 10.1016/j.freeradbiomed.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 251.Hirt RP, Muller S, Embley TM, Coombs GH. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 2002;18:302–308. doi: 10.1016/s1471-4922(02)02293-6. [DOI] [PubMed] [Google Scholar]
- 252.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid Redox Signal. 2000;2:811–820. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 253.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 254.Nagahara N, Okazaki T, Nishino T. Cytosolic mercaptopyruvate sulfurtransferase is evolutionarily related to mitochondrial rhodanese. Striking similarity in active site amino acid sequence and the increase in the mercaptopyruvate sulfurtransferase activity of rhodanese by site-directed mutagenesis. J Biol Chem. 1995;270:16230–16235. doi: 10.1074/jbc.270.27.16230. [DOI] [PubMed] [Google Scholar]
- 255.Phan J, Mahdavian E, Nivens MC, Minor W, Berger S, Spencer HT, Dunlap RB, Lebioda L. Catalytic cysteine of thymidylate synthase is activated upon substrate binding. Biochemistry. 2000;39:6969–6978. doi: 10.1021/bi000367g. [DOI] [PubMed] [Google Scholar]
- 256.Schneider E, Ryan TJ. Gamma-glutamyl hydrolase and drug resistance. Clin Chim Acta. 2006;374:25–32. doi: 10.1016/j.cca.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 257.Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Nguyen C, Teo JL, Matsuda A, Eguchi M, Chi EY, Henderson WR, Jr, Kahn M. Chemogenomic identification of Ref-1/AP-1 as a therapeutic target for asthma. Proc Natl Acad Sci U S A. 2003;100:1169–1173. doi: 10.1073/pnas.0437889100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Karimpour S, Lou J, Lin LL, Rene LM, Lagunas L, Ma X, Karra S, Bradbury CM, Markovina S, Goswami PC, Spitz DR, Hirota K, Kalvakolanu DV, Yodoi J, Gius D. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene. 2002;21:6317–6327. doi: 10.1038/sj.onc.1205749. [DOI] [PubMed] [Google Scholar]
- 260.Chiarugi P, Fiaschi T, Taddei ML, Talini D, Giannoni E, Raugei G, Ramponi G. Two vicinal cysteines confer a peculiar redox regulation to low molecular weight protein tyrosine phosphatase in response to platelet-derived growth factor receptor stimulation. J Biol Chem. 2001;276:33478–33487. doi: 10.1074/jbc.M102302200. [DOI] [PubMed] [Google Scholar]
- 261.Ishii T, Mann GE. Redox status in mammalian cells and stem cells during culture in vitro: critical roles of Nrf2 and cystine transporter activity in the maintenance of redox balance. Redox Biol. 2014;2:786–794. doi: 10.1016/j.redox.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lee SR, Yang KS, Kwon J, Lee C, Jeong W, Rhee SG. Reversible inactivation of the tumor suppressor PTEN by H2O2. J Biol Chem. 2002;277:20336–20342. doi: 10.1074/jbc.M111899200. [DOI] [PubMed] [Google Scholar]
- 263.Schwertassek U, Haque A, Krishnan N, Greiner R, Weingarten L, Dick TP, Tonks NK. Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. Febs J. 2014;281:3545–3558. doi: 10.1111/febs.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Zhang R, Al-Lamki R, Bai L, Streb JW, Miano JM, Bradley J, Min W. Thioredoxin-2 inhibits mitochondria-located ASK1-mediated apoptosis in a JNK-independent manner. Circ Res. 2004;94:1483–1491. doi: 10.1161/01.RES.0000130525.37646.a7. [DOI] [PubMed] [Google Scholar]
- 265.Manevich Y, Hutchens S, Tew KD, Townsend DM. Allelic variants of glutathione S-transferase P1-1 differentially mediate the peroxidase function of peroxiredoxin VI and alter membrane lipid peroxidation. Free Radic Biol Med. 2013;54:62–70. doi: 10.1016/j.freeradbiomed.2012.10.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 267.Zaffagnini M, Michelet L, Marchand C, Sparla F, Decottignies P, Le Marechal P, Miginiac-Maslow M, Noctor G, Trost P, Lemaire SD. The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. Febs J. 2007;274:212–226. doi: 10.1111/j.1742-4658.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 268.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, Chen YR. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46:5754–5765. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Stoner CS, Pearson GD, Koc A, Merwin JR, Lopez NI, Merrill GF. Effect of thioredoxin deletion and p53 cysteine replacement on human p53 activity in wild-type and thioredoxin reductase null yeast. Biochemistry. 2009;48:9156–9169. doi: 10.1021/bi900757q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Newick K, Cunniff B, Preston K, Held P, Arbiser J, Pass H, Mossman B, Shukla A, Heintz N. Peroxiredoxin 3 is a redox-dependent target of thiostrepton in malignant mesothelioma cells. PLoS One. 2012;7:e39404. doi: 10.1371/journal.pone.0039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Rhee SG, Jeong W, Chang TS, Woo HA. Sulfiredoxin, the cysteine sulfinic acid reductase specific to 2-Cys peroxiredoxin: its discovery, mechanism of action, and biological significance. Kidney Int Suppl. 2007:S3–S8. doi: 10.1038/sj.ki.5002380. [DOI] [PubMed] [Google Scholar]
- 272.Noh YH, Baek JY, Jeong W, Rhee SG, Chang TS. Sulfiredoxin Translocation into Mitochondria Plays a Crucial Role in Reducing Hyperoxidized Peroxiredoxin III. J Biol Chem. 2009;284:8470–8477. doi: 10.1074/jbc.M808981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Pedrajas JR, Padilla CA, McDonagh B, Barcena JA. Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-CYS peroxiredoxin in Saccharomyces cerevisiae. Antioxid Redox Signal. 2010;13:249–258. doi: 10.1089/ars.2009.2950. [DOI] [PubMed] [Google Scholar]
- 274.Hanschmann EM, Lonn ME, Schutte LD, Funke M, Godoy JR, Eitner S, Hudemann C, Lillig CH. Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J Biol Chem. 2010;285:40699–40705. doi: 10.1074/jbc.M110.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Eletto D, Dersh D, Gidalevitz T, Argon Y. Protein disulfide isomerase A6 controls the decay of IRE1alpha signaling via disulfide-dependent association. Mol Cell. 2014;53:562–576. doi: 10.1016/j.molcel.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Xiong Y, Manevich Y, Tew KD, Townsend DM. S-Glutathionylation of Protein Disulfide Isomerase Regulates Estrogen Receptor alpha Stability and Function. Int J Cell Biol. 2012;2012:273549. doi: 10.1155/2012/273549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Manukyan D, von Bruehl ML, Massberg S, Engelmann B. Protein disulfide isomerase as a trigger for tissue factor-dependent fibrin generation. Thromb Res. 2008;122(Suppl 1):S19–S22. doi: 10.1016/S0049-3848(08)70013-6. [DOI] [PubMed] [Google Scholar]
- 278.Araki K, Iemura S, Kamiya Y, Ron D, Kato K, Natsume T, Nagata K. Ero1-alpha and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J Cell Biol. 2013;202:861–874. doi: 10.1083/jcb.201303027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang L, Zhang L, Niu Y, Sitia R, Wang CC. Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1alpha to promote oxidative protein folding. Antioxid Redox Signal. 2014;20:545–556. doi: 10.1089/ars.2013.5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.He X, Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol Pharmacol. 2009;76:1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Jonsson TJ, Lowther WT. The peroxiredoxin repair proteins. Subcell Biochem. 2007;44:115–141. doi: 10.1007/978-1-4020-6051-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]