Abstract
The rise of antibiotic resistance is a growing public health crisis. Novel antimicrobials are sought, preferably developing nontraditional chemical scaffolds that do not inhibit standard targets such as cell wall synthesis or the ribosome. Iron scavenging has been proposed as a viable target, because bacterial and fungal pathogens must overcome the nutritional immunity of the host to be virulent. This review highlights the recent work toward exploiting the biosynthetic enzymes of siderophore production for the design of next generation antimicrobials.
Keywords: siderophore, nonribosomal peptide synthetase, polyketide synthase, NRPS-independent synthetase
1. Introduction
Antibiotic resistance is of major concern, and novel therapies are in urgent need to combat this growing health crisis. In response to the United States Presidential Executive Order to combat antibiotic resistant bacteria [1], the Centers for Disease Control and Prevention have announced the Antibiotic Resistance Solutions Initiative [2]. The World Health Organization is drafting a Global Action Plan on antimicrobial resistance [3]. These national and international proposals include plans for promoting awareness and education of the problem, directives for improved antibiotic stewardship, goals for prevention and response to outbreaks, and investment in the design of new antibiotics and diagnostics.
One viable mechanism for the development of novel antimicrobial agents is targeting bacterial pathways responsible for acquisition of essential nutrients [4]. Indeed, our defense systems against bacterial pathogens include “nutritional immunity” – withholding of nutrients [5]. Iron serves as an important cofactor for a variety of enzymes that perform crucial reactions, including roles in electron transfer, resistance to reactive oxygen intermediates, and RNA synthesis. Fe(III) is very insoluble and biologically inaccessible such that the concentration of free ferric iron available to pathogens in the human host ranges in estimation from 10−15 to 10−24 M [6, 7], whereas a typical pathogenic bacterium requires ~1 μM iron for optimal growth [7]. In response, bacterial pathogens have developed a variety of systems to scavenge iron from the host, including methods to import hemoproteins and other iron binding proteins such as transferrin, and the use of hemophores, heme-scavenging molecules [5]. The focus of this review is the biosynthesis of low molecular weight iron chelators called siderophores. Bacteria synthesize, secrete, and then selectively take up the iron-loaded siderophore to colonize human tissues [8–10]. Siderophore biosynthetic enzymes frequently have no human homologues, making them attractive antimicrobial targets [11].
2. Siderophore biosynthesis
Compounds from primary metabolism serve as a source of building blocks to generate siderophore natural products with differing levels of complexity both chemically and biologically. Diagrams of the siderophores are included as they appear in the discussion, detailing their chemical simplicity (ex. pyochelin) or complexity (ex. pyoverdin). Siderophores sometimes have roles not only in iron chelation, but can also have roles in other biological processes such as quorum sensing. To start our discussion, an overview of the biosynthetic process is provided. Bacterial pathogens synthesize siderophores using nonribosomal peptide synthetase (NRPS) enzymes, polyketide synthase (PKS) enzymes, and/or by NRPS independent siderophore (NIS) synthetase enzymes.
2.1. Nonribosomal peptide synthetase (NRPS) biosynthesis
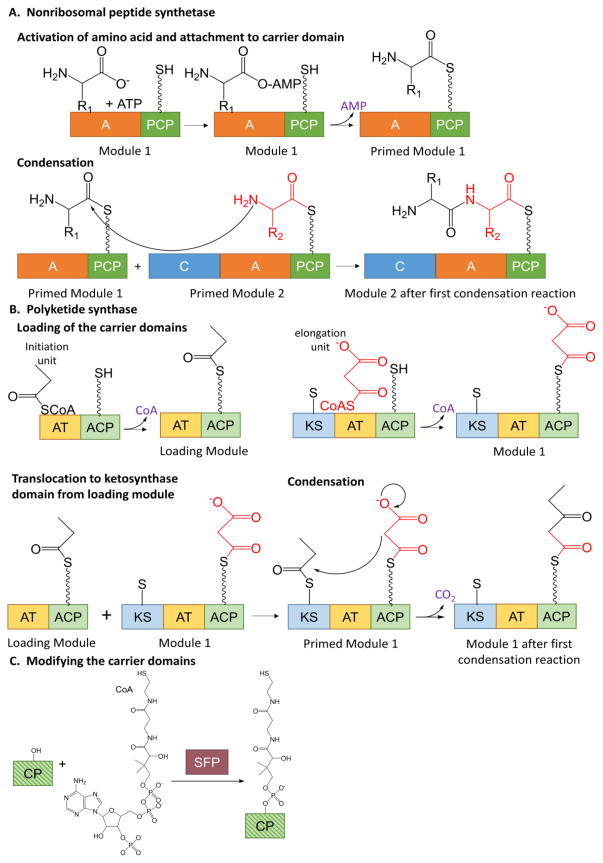
Siderophores generated by NRPS enzymes are primarily composed of amino acids, including nonproteinogenic amino acids, linked by peptide bonds. Significant work in the field has detailed the mechanisms for peptide bond formation in NRPS enzymes, which are multidomain, multifunction assembly line enzymes (for excellent reviews, see [12, 13]). In short, a module consists of a condensation domain, an adenylation domain and a carrier domain (except for the first, which lacks a condensation domain). The amino acid is activated by conversion to an amino acyl-AMP by the adenylation domain and then covalently attached (with the loss of the AMP) to the carrier domain, priming the module (Figure 1A, top). The carrier domain can accept the amino acid because it was previously post-translationally modified by the addition of a phosphopantetheinyl moiety on a conserved serine (Figure 1C). When two adjacent modules are primed, the condensation domain forms a peptide bond between the two amino acids, leaving the dipeptide on the second carrier domain (Figure 1A, bottom). When the third carrier domain is primed, a second peptide bond is formed, generating a tripeptide on the third carrier domain. This assembly line mechanism continues until the chain is complete. A thioesterase domain, frequently in the final module, removes the complete chain from the final carrier domain. The secondary metabolites generated by NRPS systems are not merely short chain peptides. To generate the bioactive properties that make them useful, the peptides must be tailored. There are tailoring domains, both incorporated into the NRPS modules and stand-alone, that decorate or modify the amino acids of the chain. These modifications can be epimerization, methylation, oxidation, reduction, hydroxylation, lipidation and glycosylation, among others.
Figure 1. Siderophore biosynthesis.
A. Nonribosomal peptide synthetase chain elongation. The adenylation domain activates an amino acid and attaches it to the carrier domain, thereby priming the module (top line). The condensation domain forms a peptide bond between two primed modules (bottom line). The process continues with more modules in an assembly line fashion. A = adenylation, C = condensation, PCP = peptidyl carrier protein. The wavy line in the PCP domain denotes the phosphopantethienyl post-translational modification. B. Polyketide synthase bond formation. The acyl transferase domains load acyl groups onto the carrier domains of the loading module and module 1 (top). The acyl group from the loading module is transferred to the ketosynthase domain of module 1 thereby priming the module (bottom left). The ketosynthase domain performs the condensation reaction (bottom right). The growing chain is transferred to the ketosynthase domain of the next module for the assembly line to continue. AT = acyl transferase, ACP = acyl carrier protein, KS = ketosynthase. The wavy line in the ACP domain denotes the phosphopantethienyl post-translational modification. The short, straight line in the KS domain denotes an active site cysteine. C. Post translational modification of carrier domains to generate phosphopantetheinyl swinging arm. CP = carrier protein is striped in shades of green to represent that this is common to both NRPS (dark green) and PKS (light green) carrier domains. SFP is a promiscuous PPTase from Bacillus subtilis commonly used to perform this reaction in vitro.
2.2. Polyketide synthase (PKS) biosynthesis
Some siderophores are constructed from assembly line systems that include polyketide synthase (PKS) modules to include other functionalities into the backbone. A PKS module consists of a ketosynthase domain, an acyltransferase domain and a carrier domain (the loading module lacks a ketosynthase domain). An initiation unit (in Figure 1B, top, the example is propionyl-CoA) is covalently attached to the carrier domain by the acyltransferase domain with the loss of the CoA. An elongation unit (malonyl-CoA shown here) is similarly attached in module 1. In both cases, the modules are prepared to receive the chain because of the same phosphopantetheinyl post-translational modification noted in the NRSP system (Figure 1C). The acyl chain is transferred from the carrier domain of the loading module to a cysteine in the ketosynthase domain, priming module 1 (Figure 1B, bottom). The ketosynthase domain catalyzes the condensation reaction and the growing chain is now attached to the carrier domain of the first module, ready to translocate to the ketosynthase domain of the next module. The assembly line process continues, with differing tailoring domains incorporated into each module to allow for the addition of differing functionalities. Tailoring chemistries include but are not limited to ketoreductases, dehydratases, methyltransferases, and oxidases. A thioesterase domain removes the complete chain from the final carrier domain, by reduction, hydrolysis or sometimes cyclization. For excellent reviews see references [14, 15]. The carrier domains in both PKS and NRPS proteins allow for an easy transition between incorporation of ketides and amino acids into the backbones of siderophore chains.
2.3. NRPS-independent siderophore (NIS) synthetase biosynthesis
Some siderophores are not built in the assembly line fashion using NRSP or PKS modules. Instead, precursors are prepared and linked using NRPS-independent siderophore (NIS) synthetases [16]. The synthetases of these biosynthetic clusters generate siderophores that incorporate citric acid, α-ketoglutarate, and succinic acid. The NIS synthetase serves as an acyl adenylation domain (for example, generating a citryladenylate [citrate-AMP]) to provide an energy rich bond for a condensation reaction with an amino acid or polyamine. There is at least one known example of a siderophore generated by a combination of NIS and NRPS proteins [17].
3. Inhibitors under iron-limiting conditions
Inhibitors of siderophore biosynthesis may be identified by screening for compounds that prevent bacterial growth in iron-limiting media. This is especially true for pathogens that are dependent on a single siderophore for iron scavenging, or on a limited number of iron acquisition routes. One may hypothesize that pathogens that have developed and/or acquired several iron acquisition pathways may be less susceptible to drugs developed against a single system. However, the interplay between siderophore biosynthesis and other virulence mechanisms is complex, including the interplay of the regulation for production of different siderophores in the same organism [18]. As seen for several inhibitors in the coming sections, inhibition of a single pathway can cause growth inhibition even for pathogens with seemingly redundant iron scavenging systems.
3.1. A generic screen for siderophore biosynthetic inhibitors
A screen has been described that was designed to identify inhibitors of siderophore biosynthesis by finding compounds that inhibit pathogen growth depending on the iron content of the media. Aspergillus fumigatus, a fungus that is the causative agent of invasive aspergillosis, was the model organism used. The screen requires two steps [19]. In the first step, A. fumigatus is grown in iron-poor or iron-replete media. Compounds that slow growth in iron-poor media but allow growth in iron-replete media carry forward to the second step. The fungi are grown again in iron-poor media with the compounds identified in the first step, and those that show no production of siderophores are to be considered hits. After this second growth, the cultures are filtered, and siderophore production is monitored as a color change upon the addition of iron since the Fe(III)-siderophore complexes are red. The method is described, but the results of a screen are not reported.
3.2. Siderophore mimics as inhibitors
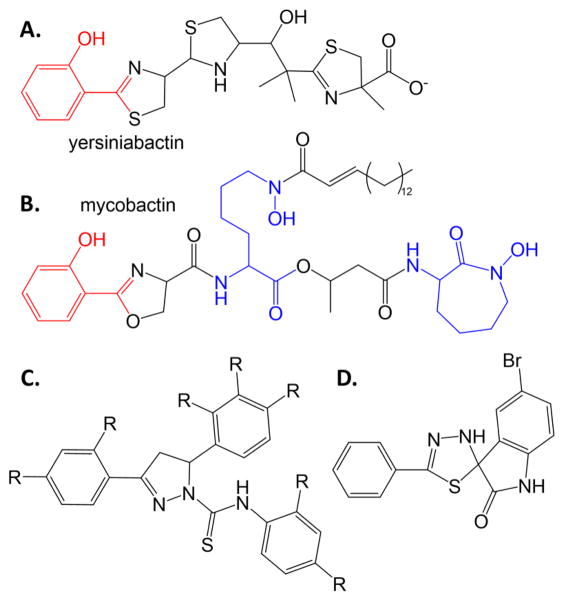
Another screen aimed at growth inhibition under iron-limiting environments was based on the premise that compounds that resemble the structure of the siderophore may make good inhibitors of siderophore production, and focused on the pathogens Mycobacterium tuberculosis (causative agent of tuberculosis; siderophore: mycobactin) and Yersinia pestis (causative agent of plague; siderophore: yersiniabactin) [20]. As can be seen in Figures 2A and 2B, these siderophores share a common hydroxyphenyl-oxazoline/thiazoline scaffold which was the basis of the mimicking potential inhibitor library (scaffold shown in Figure 2C). In this case, growth inhibitory concentrations were determined against M. tuberculosis and Y. pestis, with much better results for the latter organism showing high nanomolar EC50 values1 and low micromolar MIC90 values2. Compounds were labelled as bacteriostatic (impairing the siderophore system and effective only in iron-limiting conditions), bactericidal (inhibiting other functions that are essential in iron-limiting conditions), and antimicrobial (inhibiting regardless of iron concentration), with all three categories identified by the chemically similar molecules from this screen.
Figure 2. Siderophore mimics.
Salicylate-capped siderophores yersiniabactin (A) and mycobactin (B). The salicylate caps are shown as red. Hydroxylysine residues are blue. C. Scaffold for siderophore mimics with antimicrobial activity against Y. pestis and M. tuberculosis [20]. D. Spiro-indoline-thiadiazole inhibitor that converts to a merocyanine metal chelator and has antimicrobial activity against E. coli [21].
A separate high throughput screen was built on the idea of seeking inhibitors in iron limited media, but not necessarily seeking inhibitors of siderophore production. The assay monitored growth inhibition of E. coli in the presence of the iron chelator ethylenediaminetetraacetic acid (EDTA) [21]. Compounds that allowed growth in the absence of EDTA were further considered, and compounds that took advantage of the membrane-compromising properties of EDTA were eliminated. Finally, the successful compounds were again screened in minimal media treated with chelex-resin to removed iron. A screen of 30,000 compounds identified three compounds that met all criteria. The best compound (low micromolar MIC90 value) was a spiro-indoline-thiadiazole (Figure 2D), which showed intracellular antibacterial activity. Intriguingly, the compound appears to be a siderophore mimic: in the presence of iron, the compound converts to a merocyanine metal chelator and was hypothesized to interrupt iron homeostasis.
3.3. Repurposing known drugs to target siderophore biosynthesis
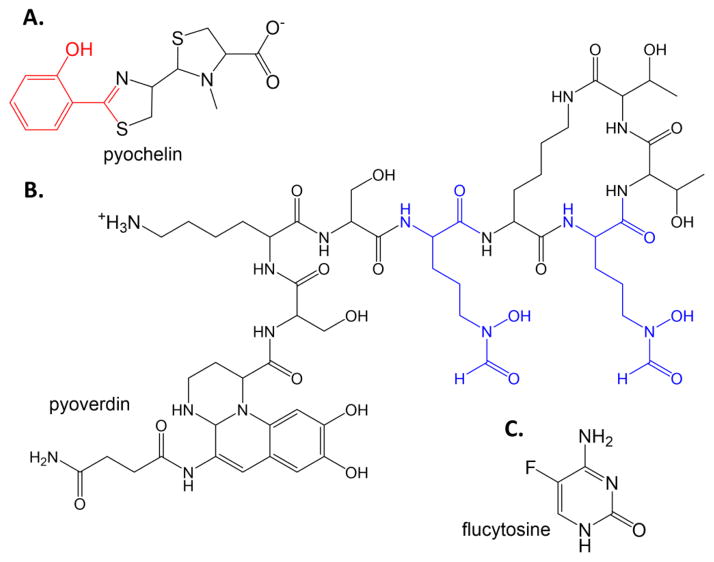
Pseudomonas aeruginosa causes nosocomial infections in immune deficient patients, including chemotherapy patients and burn patients, and causes chronic lung infections in cystic fibrosis patients. P. aeruginosa produces two siderophores under iron-limiting conditions: pyochelin and pyoverdin (Figure 3). Using a commercial library of bioactive compounds, a screen was developed to find inhibitors of pyoverdin production by finding compounds that prevent the expression of the iron-starvation alternate signaling factor PvdS [22]. PvdS promotes the expression of the exoprotease and exotoxin A virulence factors, and the expression of the pyoverdin biosynthesis genes when iron is limiting. The compound flucytosine (Figure 3C), an antimycotic fluorinated pyrimidine, was found to be effective in preventing pvdS gene expression, thereby causing a down-regulation of virulence genes including the pyoverdin biosynthetic genes, and was found to be effective in reducing P. aeruginosa pathogenicity in a mouse model.
Figure 3. Repurposing drugs.
The P. aeruginosa siderophores pyochelin (A) and pyoverdin (B). In pyochelin, the salicylate cap is again red. In pyoverdin, the formyl-hydroxyornithine residues are blue. C. Flucytosine.
4. Inhibitors of NRPS biosynthesis
Inhibitors of the catalytic activities of NRPS domains have focused on two activities: the production of the thiol tether on the carrier domain and the activation of the amino acids by adenylation. The logic of the researchers for choosing these proteins is likely two-fold: a desire for broadly applicable drugs and the ease or cost of the experiment. All of the assembly lines require the phosphopantetheinyl post-translational modification, and the enzymes that catalyze the reaction have been shown to be promiscuous across assembly lines and species [23]. Indeed, Sfp was originally cloned from Bacillus subtilis (a model organism) [24], and is commonly used in the laboratory to add the phosphopantetheinyl group to carrier domains from a wide variety of assembly lines. Therefore, compounds developed against Sfp and its homologues may be applicable to a wide array of pathogens that utilize NRPS and PKS dependent virulence factors. In contrast, adenylation domains are not promiscuous; indeed, the amino acid sequence of the peptide backbone made by NRPS enzymes has been hypothesized to be largely the result of adenylation domain substrate specificity [25]. Nevertheless, inhibitors designed to block the activation of amino and hydroxy acids by the adenylation domains are potentially also broadly applicable, because many chains contain common building blocks (for example, the salicylate and dihydroxybenzoate caps noted later in this section). Finally, most NRPS protein targets are multidomain, multifunctional polypeptides that contain at least one module and are quite large, ~120 kDa for an adenylation, carrier and condensation domain. In contrast, Sfp is a two domain protein of ~26 kDa and the adenylation domains exploited in drug design are stand-alone domains of ~60 kDa, making assay development easier and more economical.
4.1. Phosphopantetheinyl transferase inhibitors
The carrier domains must be post-translationally modified to be ready to accept the building blocks of NRPS and PKS siderophore biosynthesis. The phosphopantetheine moiety of CoA is attached to a serine residue producing a “swinging arm” [26] (Figure 1C). The amino acids (NRPS biosynthesis) or ketides (PKS) are covalently attached through the thiol, providing an anchor for chain extension and tailoring reactions. This linkage is denoted as a wavy line in Figure 1. The Sfp-type phosphopantetheinyl transferases (PPTases) are responsible for modifying the carrier domains of NRPS and PKS enzymes (as opposed to the AcpS-PPTases for fatty acid biosynthesis), and have been targeted for inhibition. A traditional assay for determining activity in PPTase enzymes is to measure radiolabel incorporation from the addition of tritiated-CoA to a carrier domain. This assay has been miniaturized for high throughput screening with the development of a scintillation proximity assay for M. tuberculosis PPTase [27]. The radiolabelled carrier domain, when in proximity of the scintillation beads, causes the emission of light. Inhibitors of the PPTase prevent the transfer of the radiolabel to the carrier domain, thereby eliminating the scintillation signal.
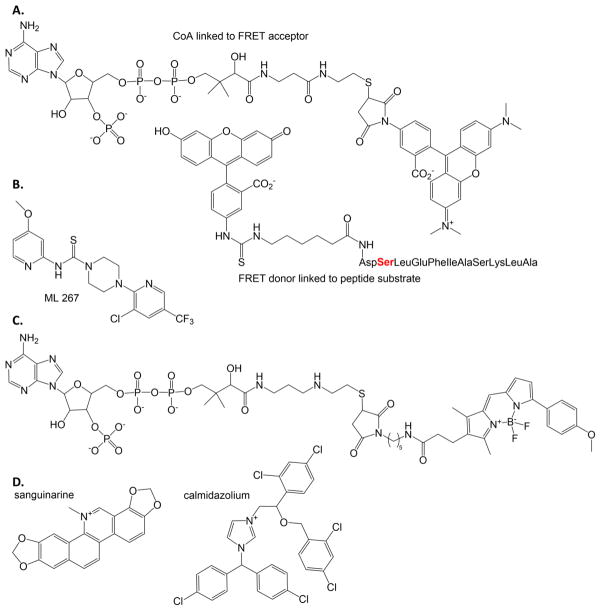
A high throughput assay dependent on fluorescence resonance energy transfer (FRET) for the Sfp-PPTase from B. subtilis has been developed (Figure 4A) [28, 29]. The FRET acceptor (a TAMRA-labeled CoA) is covalently attached by the PPTase enzyme to the serine residue (red) in the FITC-labeled peptide [30], a FRET donor. When the two are linked by the PPTase enzyme, FRET occurs. Inhibitors of the PPTase enzyme will prevent the attachment of the TAMRA-CoA to the peptide, preventing the FRET signal [28, 29]. A subsequent high throughput screen of over 300,000 compounds yielded the identification of ML267 (Figure 4B) as an inhibitor of Sfp-PPTase, with approximately 30-fold selectivity over the bacterial fatty acid synthesis PPTase and greater than 500-fold relative to the human orthologue [31, 32]. ML267 is an allosteric, reversible inhibitor with a high nanomolar IC50 value 3 that is bactericidal against Gram positive pathogens including B. subtilis and methicillin-sensitive and -resistant strains of Staphylococcus aureus (MRSA).
Figure 4. Phosphopantetheinyl transferase probes and inhibitor.
A. The FRET probes developed to determine inhibitors of PPTase enzymes. [28] The serine where the PPTase attaches the FRET acceptor to the FRET donor-labeled peptide is shown in red. B. ML267 inhibitor of Sfp-PPTase identified by high throughput screening using the FRET assay using probes in part A. [31, 32] C. BODIPY-TMR fluorescence polarization probe developed to assay PPTase inhibitors. [33] D. The two most inhibitory compounds of the PPTase from M. tuberculosis are shown, determined when using the fluorescence polarization assay where the probe is the FRET acceptor in part A. [34, 35]
Two fluorescence polarization assays have also been described for the detection of inhibitors of PPTase enzymes [33, 34]. In the first example, a BODIPY-TMR-labeled CoA serves as a fluorescence polarization probe (Figure 4C). The fluorescent CoA produces a polarized signal when bound to the Sfp enzyme (tumbling in solution is slow because the probe is bound to the PPTase). Inhibitors displace the probe from the PPTase active site leading to a decrease in fluorescence polarization (the tumbling rate is increased because probe is no longer bound to protein) [33]. In a second fluorescence polarization assay, the probe used is the TAMRA-labeled FRET acceptor in Figure 4A. The TAMRA-CoA is attached to a carrier domain by a PPTase, causing an increase in fluorescence polarization (the tumbling rate is slowed, because probe becomes bound to protein) [34]. A limited screen was performed using the PPTase from M. tuberculosis to label a carrier domain of VibB for vibriobactin biosynthesis (Vibrio cholera, causative agent of cholera) or a carrier domain of mycocerosic acid synthase from M. tuberculosis [35]. The most effective inhibitors were calmidazolium and sanguinarine with low micromolar IC50 values (Figure 4D). Sanguinarine will appear again as an inhibitor of the ornithine hydroxylase, identified using a remarkably similar fluorescent probe, despite a lack of similarity in the target enzyme structure or function (Section 7.3).
4.2. Adenylation domain inhibitors
4.2.1 Salicylate adenylation inhibitors
The first step in the NRPS assembly line is the priming of carrier domains with the initiator or elongation units. This is accomplished by the adenylation domains (Figure 1), which can be embedded in the NRPS, or can be stand-alone domains. For ease in inhibitor design, the stand-alone domains have been predominantly used, because the multidomain NRPS modules are large and can be difficult to express and purify, especially in sufficient quality and quantity. The adenylation domains that activate the hydroxy acid salicylate for initiation of salicylate-capped siderophores have been the focus of much work. These include siderophores from Y. pestis and Y. enterocolitica (causative agent of yersinosis, a diarrheal disease; siderophore: yersiniabactin, Figure 2A), M. tuberculosis (mycobactin, Figure 2B), and P. aeruginosa (pyochelin, Figure 3A).
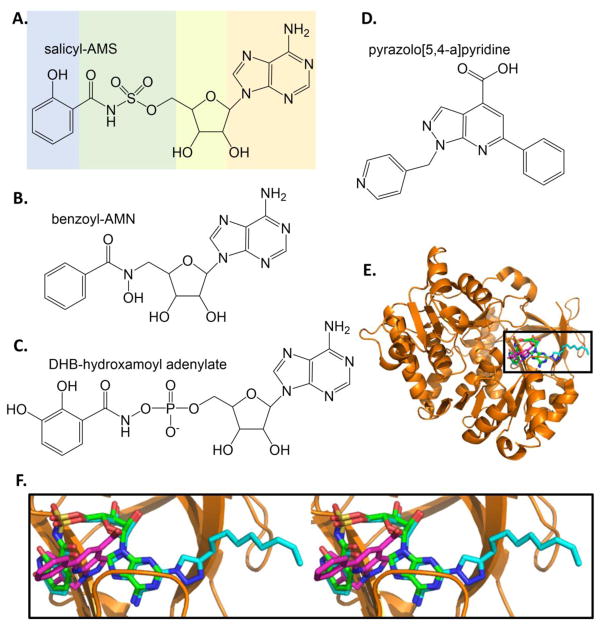
The most advanced study is related to the inhibitor called salicyl-AMS for 5′-O-(N-salicylsulfamoyl)adenosine (Figure 5A). This nonhydrolyzable reaction intermediate was shown to have low nanomolar IC50 values for the salicylate adenylation enzymes MbtA (M. tuberculosis), YbtE (Y. pestis), and PchD (P. aeruginosa), and to have low micromolar EC50 values for growth inhibition of M. tuberculosis and Y. pestis [36]. Salicyl-AMS is sometimes also called a bisubstrate inhibitor (as opposed to a reaction intermediate), because it links the salicylate ring to the sugar nucleotide. Using primarily MbtA as the model enzyme and M. tuberculosis as the organism, there have been many variations on the salicyl-AMS theme: i) exchanging the acylsulfamate linkage with a β-ketosulfonamide linkage (green in Figure 5A) [37]; ii) differing glycosyl modifications (yellow) [38]; iii) modification at the salicyl ring position (blue) [39]; iv) decorations to the salicyl ring in combination with salicyl-sugar linkages (blue and green) [40]; v) modification of the nucleobase (orange) [41]; and vi) triazole derivatives of the nucleobase (orange) [39]. These extensive structure-activity studies yielded compounds of comparable or worse values for enzyme inhibition and bacterial growth inhibition when compared to the parent compound. Salicyl-AMS has subsequently been shown to be effective in significantly inhibiting M. tuberculosis growth in a mouse model; however, the compound was toxic to the mice at the higher dose (~17 mg/kg), which was proposed to be the result of off-target effects [42].
Figure 5. Inhibitors of salicylate and dihydroxybenzoate adenylation enzymes.
A. Salicyl-AMS, a rationally designed reaction intermediate analogue. [36]. B. Benzoyl-AMN [43]. C. DHB-hydroxamoyl adenylate [44]. D. Inhibitor from high throughput screen. [46]. E. Three structures of BasE are overlayed: BasE with DHB-AMS bound is the orange cartoon with green stick inhibitor (PDB ID: 3O82), with a triazole derivative of DHB-AMS shown in cyan sticks (PDB ID: 3O83), and with inhibitor from part D shown in magenta sticks (PDB ID: 3O84). F. The area in the box in part E is shown in stereo (all stereo images are wall-eye or divergent stereo). Note the unexpected binding mode of the magenta HTS inhibitor relative the green and cyan substrate analogue inhibitors. Structure figures made in PyMOL [121].
4.2.2 Dihydroxybenzolate adenylation inhibitors
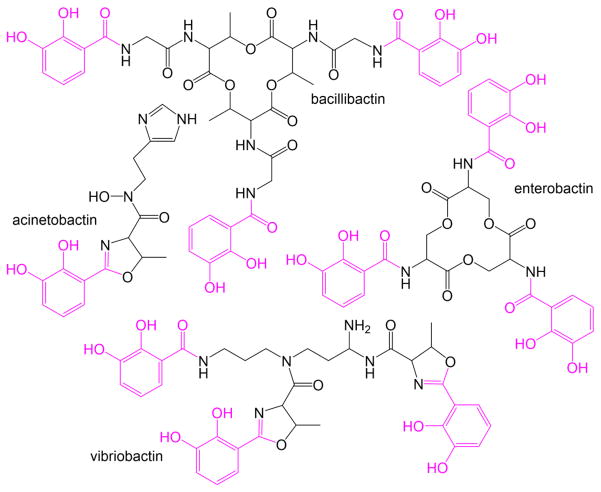
Inhibitors have also been designed to the dihydroxybenzoate-adenylating enzymes. Dihydroxybenzoate (DHB) capped siderophores are generated by E. coli (enterobactin), V. cholera (vibriobactin), and B. subtilis (bacillibactin) (Figure 6). Early work included the design of a variety of AMS derivatives with differing decorations on the benzoyl ring, and also to hydroxamoyl adenosine (AMN) derivatives (Figure 5B), yielding nanomolar inhibition constants when tested against both the dihydroxybenzoate-adenylating enzyme from B. subtilis (DhbE) and the salicylate-adenylating enzyme from Y. pestis (YbtE) [43]. Nanomolar inhibition was reported for 2,3-dihydroxybenzohydroxamoyl adenylate when tested against the dihydroxybenzoate-adenylating enzyme from E. coli (EntE) (Figure 5C) [44]. Inhibition of EntE by salicyl-AMS and dihydroxybenzoate-AMS were reported to be of a one-step, slow-onset, tight-binding mechanism, again with low nanomolar inhibition [45].
Figure 6. Dihydroxbenzoate-capped siderophores.
Bacillobactin from B. subtilis, acinetobactin from A. baumannii, enterobactin from E. coli, and vibrobactin from V. cholera. DHB caps are shown in pink.
The dihydroxybenzoate-adenylating enzyme from Acinetobacter baumannii (a nosocomial, opportunistic pathogen) provides the DHB cap for the siderophore acinetobactin (Figure 6). A fluorescence polarization based high throughput screen was developed for BasE, in which a probe was generated that consisted of a fluorescein attached to the 2′-O of the ribosyl moiety of salicyl-AMS thru a linker [46]. Displacement of the probe from the enzyme active site by an inhibitor leads to increased fluorescence depolarization. A screen of ~84,000 compounds produced several inhibitor chemotypes that were effective not only against BasE, but also against the DHB adenylation domains EntE (E. coli), VibE (V. cholera), DhbE (B. subtilis), and the salicylate adenylation enzymes MbtA (M. tuberculosis) and YbtE (Y. pestis). The most promising compound contains a pyrazolo[5,4-a]pyridine scaffold (KD of 78 nM) is shown in Figure 5D. Since that time, the structure of BasE has been determined in three states: a) with the compound from the screen bound (magenta in Figures 5E and 5F), b) with DHB-AMS bound (green), and c) with a triazole derivative of DHB-AMS bound (cyan)[47]. The bisubstrate inhibitors bound as predicted by a previous MbtA homology model derived from the DhbE structure, driven by product (adenylated-DHB) binding modes found in DhbE [41]. Binding in the active site of the pyrazolo[5,4-a]pyridine was expected because of the documented competitive inhibition; however, the new chemotype did not bind as predicted by docking algorithms. The phenyl ring of pyrazolopyridine-inhibitor was anticipated to align with the DHB ring of the AMS-inhibitor structures, whereas the pyridine ring (pyrazolopyridine-inhibitor) was predicted to align with the adenine (AMS-inhibitor) [46]. Instead, the crystal structure shows a unique pose: note the change in the orientation of the pyraxolopyridine ring system in magenta in Figure 5F [47].
4.2.3 Macrocyclic inhibitors of cysteine adenylation
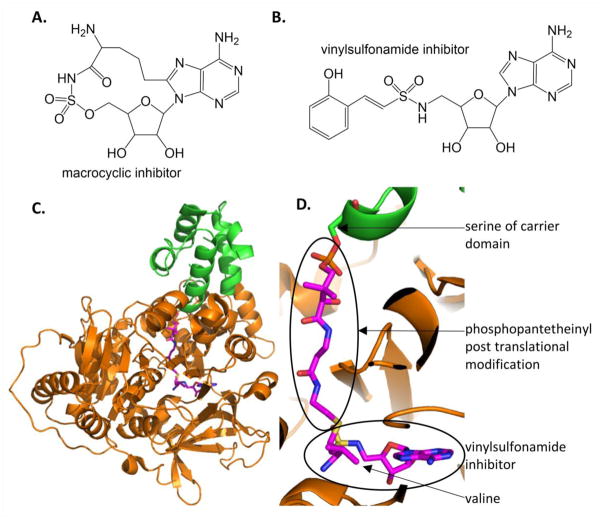
The AMS inhibitors have not been expanded to amino acid substrates because aminoacyl-tRNA synthetases perform very similar chemistry. Therefore, aminoacyl-AMP analogues, such as aminoacyl-AMS, will inhibit both ribosomal protein formation [48] and NRPS synthesis of natural products. Indeed, [(phenlylalanine)sulfamoyl] adenosine inhibits the phenylalanine adenylation domain (PheA) in gramicidin biosynthesis (Bacillus brevis) and [(leucine)sulfamoyl] adenosine inhibits the leucine adenylation domain (LeuA) in surfactin biosynthesis (B. subtilis) with nanomolar Ki values4, and both also inhibit tRNA synthetases [49]. Tan and colleagues [50] very cleverly generated cysteine- and alanine-AMS, and also macrocyclic alanine-AMS where the alanine sidechain is linked back to the nucleobase by 2 or 3 carbon units (Figure 7A). They first showed that the cysteine- and alanine-AMS inhibited both the adenylation domain of HMWP2 (High Molecular Weight Protein 2 – a two module NRPS in the biosynthesis of yersiniabactin) and inhibited in vitro translation. Next, they showed that the macrocyclic alanine-AMS inhibited HMWP2 adenylation activity but not in vitro translation.
Figure 7. Inhibitors of adenylation domains in NRPS modules.
A. Macrocyclic inhibitor of cysteine adenylation domain in HWMP2 for yersiniabactin biosynthesis. [50] B. Mechanism based inhibitor used to trap adenylation and carrier domains in stable conformation. The one shown here was designed for the DHB-specific EntE [53]. C. The structure of the carrier domain (green cartoon) and adenylation domain (orange) with the vinylsulfonamide inhibitor (magenta sticks) of PA1221 from P. aeruginosa, a two domain NRPS specific for the incorporation of valine (PDB ID: 4DG9). D. Close up of the active site showing the covalent linkage of the adenylation domain inhibitor (similar to that in part B) to the phosphopantetheinyl post translation modification of the carrier domain.
4.2.4 Vinylsulfonamide inhibitors for trapping adenylation-carrier domain interaction
It should be noted that adenylation domains of NRPS domains are remarkably mobile, having distinct conformations for the activation of the amino or hydroxy acid followed by a large conformational change to attach the amino/hydroxy acid to the carrier domain on the pantetheine group [51–54]. This flexibility can make adenylation domains problematic in crystallographic studies. Vinylsulfonamide inhibitors are mechanism based inhibitors that covalently attach the carrier domain panthetheine group to a dead end analogue of the AMS compounds (Figure 7B). The vinylsylfonamide inhibitors have been used to lock an adenylation domain in a closed conformation in a valine specific adenylation-carrier protein didomain from P. aeruginosa called PA1221 [55] (Figure 7C and 7D) and similarly for an EntE (adenylation)- EntB (carrier domain) artificial construct [53]. EntE, a stand alone adenylation domain, was cloned so that the resulting peptide was linked by a four amino acid linker to the carrier domain of EntB. While the vinylsufonamide compounds may be unlikely drug leads, they are excellent chemical biology probes for understanding protein-protein or domain-domain interfaces in these complex multi-domain, multi-protein molecular machines.
4.3. An NRPS enzyme as inhibitor
Chillar and colleagues [56] tested whether microbial competition could be exploited to find new antifungal proteins. Cell free extracts containing secretory proteins or cytoplasmic proteins were tested for antifungal properties against A. fumigatus, A. flavus (causes aspergillosis in immunocompromised patients) and A. niger (a food contaminant, causes black mold on fruits and vegetables). A protein from E. coli was identified that inhibited fungal growth. While the tests were not conducted under conditions of iron limitation, the protein was identified as an NRPS in a siderophore biosynthetic cluster, and the protein was subsequently shown to inhibit siderophore production by A. fumigatus.
5. Inhibitors of PKS biosynthesis
Inhibition of PKS biosynthesis is a difficult target for drug design because of the similar chemistry, logic and structure shared with the enzymes of fatty acid biosynthesis. Indeed, PKS and fatty acid synthase (FAS) enzymes have been hypothesized to have evolved from a common ancestor [14]. As described above for PKS biosynthesis, FAS tether initiation and elongation units (malonyl or methylmalonyl CoA) to carrier domains for condensation by ketosynthase domains [14, 57]. To generate the fully unsaturated hydrocarbon of a fatty acid, each module contains ketoreductase, dehydratase, and enoylreductase domains. As mentioned previously, PKS modules may have some, all, or none of these tailoring domains to generate more varied natural products. Therefore, inhibitors of pathogenic PKS enzymes will likely also be inhibitors of host FAS enzymes, making design of selective inhibitors for the complex and quite large PKS enzymes problematic. Instead, PKS drug design has focused on identifying and developing novel polyketides with therapeutic properties [58, 59].
6. Inhibitors of NIS biosynthesis
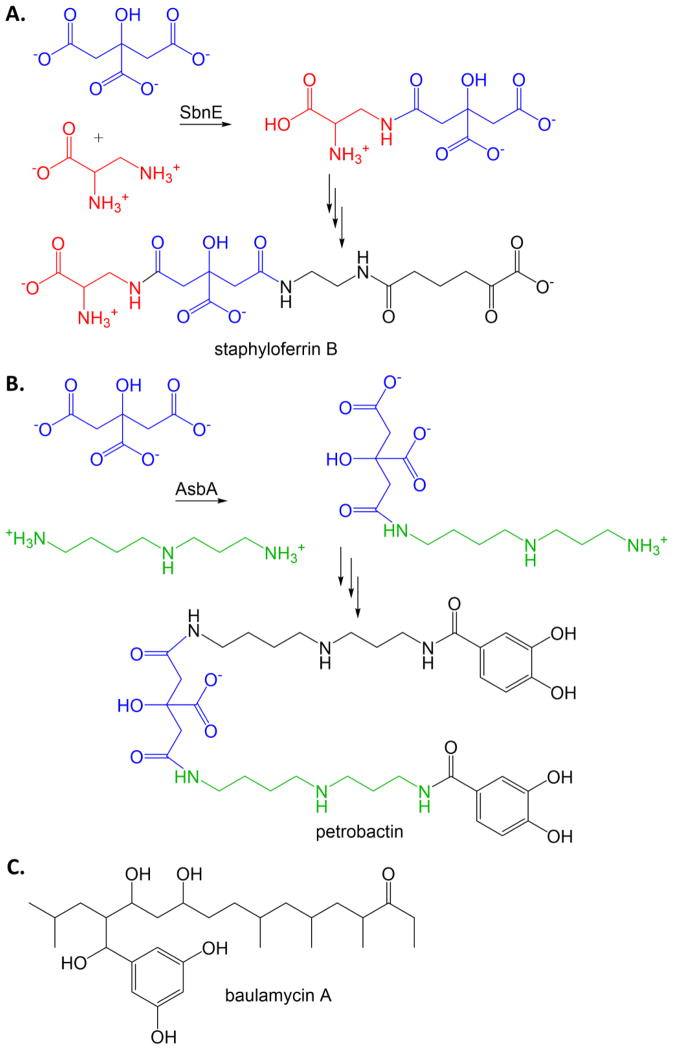
S. aureus, including MRSA, and Bacillus anthracis (causative agent of anthrax) make siderophores using a nonribosomal peptide synthetase independent pathway, with the production of staphyloferrin B and petrobactin (respective siderophores) being strongly linked to survival in iron limiting conditions. The NIS synthetases, SbnE for S. aureus and AsbA for B. anthracis, link citric acid to an amine alcohol (former) or a polyamine (latter) (Figures 8A and 8B). This process is ATP dependent, adenylating the citrate with elimination of pyrophosphate before linking to the citrate to the amine, with loss of AMP, reminiscent of the chemistry performed by NRPS adenylation domains but without the thiotemplate assembly line. An activity assay was developed for high throughput screening (originally intended for NRPS enzymes) that converts the pyrophosphate to inorganic phosphate which is detected by malachite green [60]. A screen was designed to only identify compounds from a marine microbial-derived natural product library that inhibited both SbnE and AsbA [61]. The result was the identification of the baulamycins (Figure 8C) as competitive, reversible inhibitors with IC50 values in the micromolar range. The baulamycins, however, inhibit growth of the pathogens in both iron-replete and iron-limiting media, indicating the compounds are nonspecific.
Figure 8. NRPS-independent siderophore (NIS) synthetase inhibitor.
A. Condensation of citric acid (blue) with diaminopropionic acid (red) by NIS synthetase SbnE from S. aureus for the production of staphyloferrin. B. Condensation of citric acid (blue) with spermidine (green) by NIS synthetase AsbA from B. anthrasis for the production of petrobactin. C. Micromolar antibiotic inhibitor of SbnE and AsbA [61].
7. Generation of secondary metabolites for incorporation into siderophores
Frequently, siderophores include building blocks such as modified amino acids or hydroxy acids that must be produced for incorporation into the backbone. Some building blocks are modified by tailoring domains that are incorporated into the NRPS or PKS modules or alternatively found as stand-alone domains that interact with an NRPS module. In other cases, there are enzymes in the biosynthetic cluster that generate the building blocks in advance of their use by the synth(et)ases. Several of these accessory enzymes have been the targets of rational and high throughput drug design. In particular, these include enzymes that generate isochorismate, salicylate, and hydroxyornithine.
7.1. Isochorismate synthase and salicylate synthase
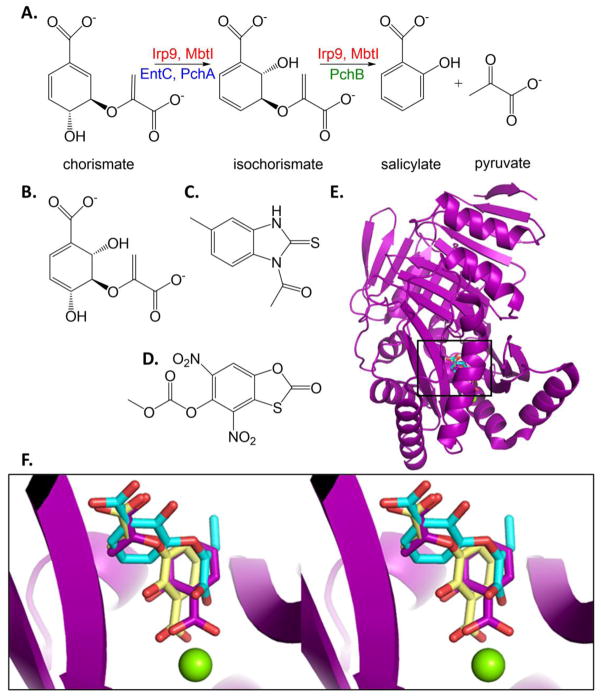
The enzymes that produce isochorismate and salicylate for the production of salicylate- and dihydroxybenzoate-capped siderophores are all structural homologues of the Menaquinoe, Siderophore, Tryptophan (MST) class. Chorismate is the substrate for these enzymes, thereby also earning the name “chorismate-utilizing” enzymes, and is at the branchpoint of the shikimate pathway for the production of aromatic amino acids by bacteria, fungi and plants, but this pathway is not found in animals. The MST enzymes are magnesium dependent and all perform the isomerization of the chorismate ring to form isochorismate by general acid-general base chemistry [62–64]. Some of the enzymes are capable of only this chemistry, and are thus called isochorismate synthases. In contrast, the salicylate synthases then cleave the pyruvylenol tail from the ring to generate salicylate and pyruvate (Figure 9A). Both reactions occur in the same active site. The cleavage reaction has been hypothesized to be a pericyclic reaction [64, 65]; however, there is a recent resurgence for the idea that the elimination reaction is also general acid-general base catalyzed [66].
Figure 9. Isochorismate synthase, salicylate synthase, isochorismate-pyruvate lyase.
A. Salicylate production for the generation of salicylate-capped siderophores. B. Potential transition state analogue inhibitor for isochorismate synthase activity. [68] C. Salicylate synthase inhibitor from HTS. [82] D. Isochorismate-pyruvate lyase inhibitor from HTS that also is effective against salicylate synthase and chorismate mutase. [86]. E. The structure of the salicylate synthase from Y. enterocolitica (Irp9, purple cartoon, PDB ID: 2FN1) with the catalytic Mg ion (green sphere) and products salicylate and pyruvate (purples sticks) was used to align two inhibitor-bound structures of the salicylate synthase from M. tuberculosis, MbtI. An MbtI structure with the required Mg has never been determined. The structure of MbtI with the aromatized isochorismate analogue known as AMT is shown in yellow sticks (PDB ID: 3ST6). MbtI with methyl-AMT is shown in cyan sticks (PDB ID: 3VEH). The methyl group addition is at the –ene of the pyruvylenol tail. Cartoons for 3ST6 and 3VEH are not included for the sake clarity. F. Close up of the active site is shown in stereo illustrating the binding modes of the inhibitors relative to products. Note that AMT (yellow) binds similarly to the substrate/products, with the salicyl ring analogue coordinated to the Mg2+. By contrast, additions to the pyruvylenol tail cause a flipping of the binding mode (methyl-AMT as one example, cyan) with the salicyl ring analogue now binding in the location where the pyruvate product is normally found.
The isochorismate synthase from E. coli (EntC for the production of enterobactin), and salicylate synthases from Y. enterocolita (Irp9, yersiniabactin) and M. tuberculosis (MbtI, mycobactin) have been the subject of many rational inhibitor design initiatives. Frequently, the investigators tested their compounds on several enzymes simultaneously, and sometimes in conjunction with structurally and functionally related chorismate-utilizing enzymes that generate anthranilate for tryptophan biosynthesis and 4-amino-4-deoxychorismate for folate biosynthesis. The earliest investigations were based on a mechanism described for EntC in which the hydroxyl groups of the isomerization are both chelated by magnesium ion at the transition state [67], yielding the rationally designed inhibitors such as the one shown in Figure 9B [68, 69]. The racemic compounds were strong competitive inhibitors with a high nanomolar Ki value.
The first structures for the MST enzymes (anthranilate synthase [70–72] and 4-amino-4-deoxychorismate synthase [73]) were solved showing that the magnesium does not bind the chorismate substrate through the hydroxyls, but rather through the ring carboxylate, leading to the hypothesis of acid-base chemistry. With this information in hand, a larger array of chorismate and isochorismate analogues were made and tested, including: i) variations at the hydroxyl sites [74]; ii) variations at the pyruvylenol tail [74–77], and iii) use of benzoate, pyridine and pyridone rings [75, 76, 78]. These compounds were tested against Irp9, MbtI, and/or EntC with the best compounds, with modifications to the terminal alkene of the pyruvylenol tail, exhibiting low micromolar inhibition (Ki values). The benzoate analogues were predicted by docking experiments to bind in a pose similar to chorismate which the ring carboxylate bound to the magnesium for the salicylate adenylation enzymes [75, 78], but in a flipped pose where the ring carboxylate bound in the site usually occupied by the pyruvyl-carboxylate for isochorismate synthase EntC (the change in pose is based on the enzyme) [78]. When the crystal structures of the salicylate synthase MbtI were determined, the benzoate compound bound in the chorismate-like pose, but inhibitors with additions to the pyruvylenol tail bound flipped (the change in pose is based on inhibitor) [79]. Figures 9E and 9F show the product-bound structure of Irp9 (salicylate and pyruvate in purple) [80] overlayed with the benzoate inhibitor structure (yellow, same pose as products) [79], and the benzoate inhibitor with a methylated pyruvylenol tail (cyan, flipped pose) in MbtI.
In another rational design effort, an elaborate construct was generated in which a chorismate mimic (“payload”) was connected by a “spacer” to a “combi” section intended to extend out of the active site and interact with the surface of the protein. These constructs yielded compounds that are millimolar to high micromolar EntC inhibitors [81]. Finally, inhibitors of MbtI were sought through a high throughput screen that exploits the fluorescent nature of the product salicylate [82]. Sixteen compounds from the screen were confirmed with an orthogonal assay and two of these compounds, a diarylsulfone and a benzimidazole-2-thione, were considered good leads and structure activity relationships were determined. The diarylsulfone is a pan assay interference compound [83] and an unlikely candidate; however, a benzimidazole-2-thione exhibits low micromolar inhibition and is shown in Figure 9C.
7.2 Isochorismate pyruvate lyase
P. aeruginosa generates a salicylate-capped siderophore, but the MST enzyme in the siderophore biosynthetic pathway for pyochelin (Figure 3A) is an isochorismate synthase (PchA, Figure 9A, blue) that is incapable of the lyase reaction [84]. A second enzyme in the pathway, PchB, is an isochorismate-pyruvate lyase performing the pericyclic elimination reaction (Figure 9A, green) [85]. A high throughput screen was designed that exploits salicylate fluorescence, and the hits were tested for inhibition in PchB, the Y. enterocolitica salicylate synthase Irp9 (Section 7.1) and also E. coli chorismate mutase (EcCM) [86]. The rationale was that Irp9 performs the same reaction, and EcCM is a structural homologue performing a similar reaction with a similar transition state. Mid nanomolar PchB inhibitors (IC50 values) were identified. Two demonstrated growth inhibition of P. aeruginosa in the millimolar range (EC50 values) in iron-poor media, but were ineffective in iron-replete media, as would be expected for an inhibitor specific for siderophore biosynthesis. One of the compounds was also a nanomolar inhibitor of Irp9 and a micromolar inhibitor of EcCM (IC50 values) that demonstrated growth inhibition for Y. enterocolitica and E. coli in the millimolar range (EC50 values). This most successful compound is shown in Figure 9D. The similarity of this scaffold to that in Figure 9C, which was found in a high throughput screen for MbtI (Section 7.1), should be noted.
6.3. Ornithine hydroxylase
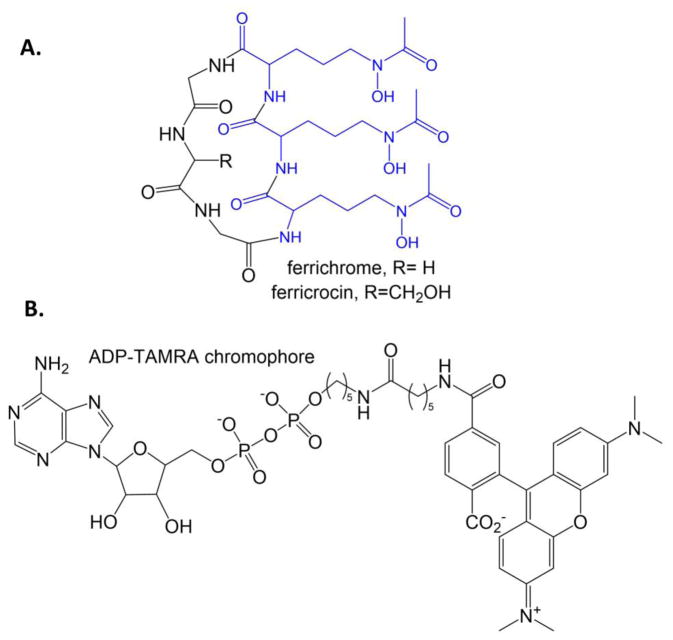
Hydroxamate siderophores are constructed by NRPS enzymes using modified lysine, ornithine, or polyamines such as spermine. The first step is the hydroxylation of the sidechain amine followed by formylation of the same nitrogen to make the hydroxamate moiety that chelates the iron in the fully formed siderophore. Examples include the formyl-hydroxyornithine residues of pyoverdin (blue in Figure 3B) and the hydroxylysine residues, one acylated and one cyclized, in mycobactin (blue in Figure 2B). The hydroxylation reaction is carried out by an N-hydroxylating flavin monooxygenase, and has been the target for the development of a fluorescence polarization screen [87]. The goal was to make a probe that would bind in the NADPH binding site linked to a fluorophore (rhodamine or TAMRA) (Figure 10B). The result was a compound that served as a competitive inhibitor to NADPH (as designed) and also ornithine, suggesting that the fluorophore bound across the substrate binding site. The assays were conducted against SidA, the ornithine hydroxylase of A. fumigatus required for the production of the siderophores ferricrocin and ferrichrome (Figure 10A), and also the lysine hydroxylase from the non-pathogenic Mycobacterium smegmatis (MbsG) for the production of the mycobactin siderophores. A very limited initial screen (160 compounds) identified sanguinarine (Figure 4D) has a weak inhibitor of SidA (high micromolar IC50 value). The fluorescence polarization probe for these experiments (Figure 10B) is very similar to the probes designed for the Sfp-PPTase experiments (Figures 4A and 4C), differing only in the linker between the sugar-nucleotide and the fluorophore. Screens for Sfp and SidA identified the same compound, sanguinarine, as an inhibitor, despite the assayed enzymes being structurally and functionally unrelated. It may be important to determine if the sanguinarine is primarily interrupting an interaction of the fluorophore with the enzymes in each case, instead of inhibiting binding of the portion of the probe designed to mimic substrate. Additionally, sanguinarine has been shown to be thiol-reactive [88] and has been reported as an inhibitor of many diverse targets (some examples [89–98]).
Figure 10. Ornithine hydroxylase.
A. Ferrichrome and ferricrocin siderophores from A. fumigatus. The formyl-hydroxyornithine residues are blue. B. ADP-TAMRA chromophore designed for a high throughput assay for the flavin-dependent N-hydroxylating monooxygenases, such as ornithine and lysine hydroxylases.
7.4. Ornithine decarboxylase
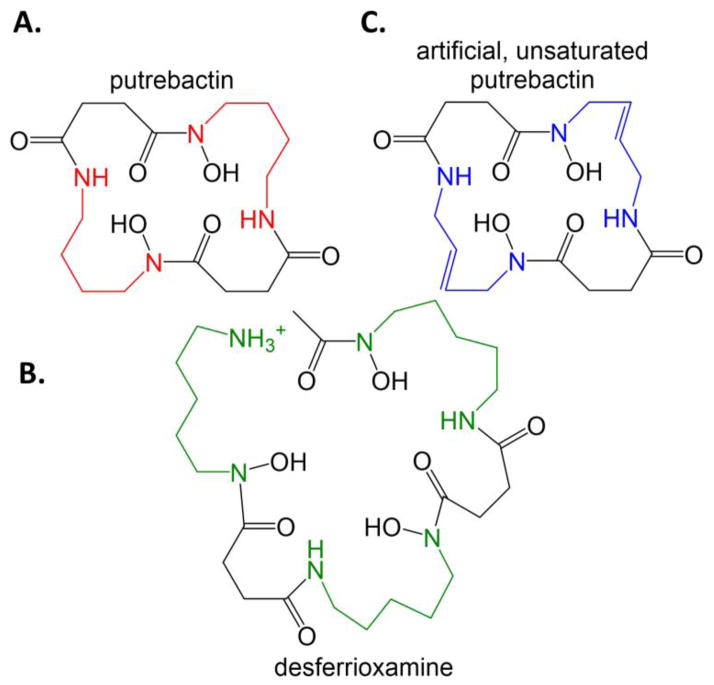
Putrebactin, a macrocyclic dihydroxamate siderophore, is generated by Shewanella putrefaciens (a marine bacteria that rarely causes bacteremia) using NIS synthetases from precursors putrescine and succinate (Figure 11A). Putrescine is generated from ornithine by the enzyme ornithine decarboxylase. Unnatural siderophores can be generated by S. putrefaciens when an ornithine decarboxylase inhibitor is added to the culture. In particular, desferrioxamine B, a cadaverine-substituted form of putrebactin, was generated with the addition of the inhibitor 1,4-diamino-2-butanone (Figure 11B) [99]. Further, an unsaturated macrocycle was made in the presence of the inhibitor co-administered with the putrescine analogue 1,4-diamino-2-butene (Figure 11C) [100].
Figure 11. Inhibiting ornithine decarboxylase to generate novel siderophores.
A. Putrebactin, the natural siderophore of S. putrefaciens with putrescine (diaminobutane, red) incorporated. B. When ornithine decarboxylase is inhibited, preventing the production of putrescine, then S. putrefaciens makes desferrioxamine, using cadaverine (diaminopentane, green) instead. C. When ornithine decarboxylase is inhibited and an external source of diaminobutene (blue) is added to the culture, then an unsaturated form of putrebactin is formed.
8. Siderophore maturation
Some siderophores require maturation after the synthesis of the backbone is complete. Pyoverdin from P. aeruginosa (Figure 3B), is one such siderophore: the peptide backbone is synthesized by NRPS enzymes in the cytoplasm, and maturation of the fluorescent chromophore occurs in the periplasm before export [101]. PvdQ is one of the maturation enzymes, acting as an NTN hydrolase [102] to cleave off the fatty acid myristate incorporated by the first module of the first NRPS (PvdL) [103]. PvdQ also has a role in quorum-quenching by degrading long-chain N-acylhomoserine lactones, which are major communication molecules of Gram-negative bacteria, and their destruction causes a decrease in the expression of virulence factors and a decreased ability to form biofilms [104]. In this latter capacity, an inhalable preparation of freeze-dried PvdQ powder has been prepared as a potential treatment for cystic fibrosis patients where P. aeruginosa causes chronic infections [105].
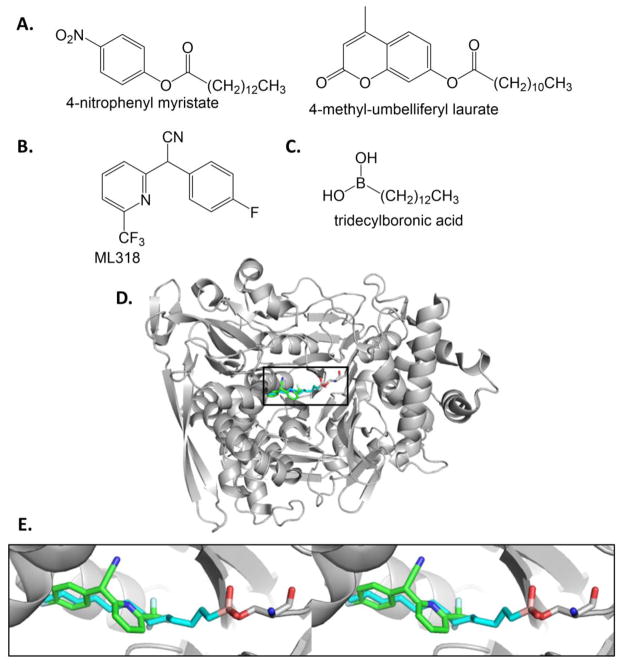
A high throughput screen was developed against PvdQ hydrolase activity for the maturation of pyoverdin [103, 106]. Two reporter probes were identified: the colorimetric 4-nitrophenyl myristate and the fluorescent 4-methyl-umbelliferyl laurate (Figure 12A), which served as substrates in the screen. An initial screen of 1280 compounds produced two compounds with micromolar IC50 values [103]. The inhibitors were shown by crystallography to have overlapping binding sites with myristate. A subsequent screen of more than 300,000 compounds using the 4-methyl-umbelliferyl laurate substrate yielded a 40 nM inhibitor (IC50 value) which was subsequently refined by structure activity relationship analysis to produce a 20 nM inhibitor called ML318 (Figure 12B) [106, 107]. ML318 inhibited the production of pyoverdin and inhibited P. aeruginosa growth under iron-limiting conditions with an EC50 value less than 50 μM [106]. Furthermore, ML318 showed no apparent toxicity in HeLa cells at the same concentration. The crystal structure shows ML318 binding in the acyl-binding site (Figures 12D and 12E) [107]. A combination treatment of ML318 with flucytosine (Figure 3C) yielded a complementary effect, with an EC50 value of ~2 μM. A rationally designed boronic acid inhibitor demonstrated a picomolar Ki value (Figure 12C) [108]. This transition state analogue binds covalently to the active site serine nucleophile (shown crystallographically, Figure 12D and 12E), and inhibits P. aeruginosa growth under iron-limiting conditions when co-administered with an efflux pump inhibitor. A series of n-alkylboronic acid compounds were tested kinetically and structures determined, with the C12- and C13-B(OH)2 compounds remaining the most potent [109].
Figure 12. Reporter substrates and inhibitors for PvdQ.
A. Substrate analogue probes. 4-nitrophenyl myristate provides an absorbance readout, whereas 4-methyl-umbelliferyl laurate is fluorescent when cleaved. [103] B. Inhibitor of PvdQ identified by high throughput screening and SAR. [106, 107] C. Rationally designed transition state analogue inhibitor [108]. D. The PvdQ structures with ML318 (PDB ID: 4K2G, inhibitor in green sticks) and tridecylboronic acid (PDB ID: 4M1J, inhibitor cyan) are overlayed. Only the latter cartoon is shown for clarity. E. Stereoview of active site with inhibitors bound. Note that the boronic acid inhibitor (boron is pink) forms a covalent transition state analogue with the serine nucleophile (grey sticks).
8. Conclusions
The focus of research for inhibiting siderophore biosynthesis has been directed at a variety of accessory enzymes that generate secondary metabolites for incorporation into siderophores, production of the phosphopantethienyl posttranslational modification of carrier domains, incorporation of hydroxy acids by NRPS enzymes, and a siderophore maturation fatty acid hydrolase enzyme. Targeting the NRPS synthetase, PKS synthase and NIS synthetase enzymes has the confounding problem of selectivity with the need to avoid inhibiting either aminoacyl-tRNA synthetases or fatty acid synthases. The macrocyclic inhibitor effective against the cysteine adenylation domains but selective so as not to inhibit the aminoacyl-tRNA synthetases demonstrates that even this difficult problem is not out of reach [50]. An avenue not yet exploited is inhibition of tailoring activities, those domains that modify peptide or ketide units while still attached to the assembly line enzymes.
Finding potent and selective inhibitors of siderophore biosynthetic enzymes is just one requirement for developing a successful antibiotic treatment. Many bacteria have redundant iron-scavenging systems, including the use of more than one siderophore and/or heme uptake. Some express multi-drug efflux pumps, contain porin variants, or exist in biofilms that might impact the ability of inhibitors to reach their targets. The most successful projects have produced pico- and nanomolar inhibitors that show iron-dependent growth inhibition of the targeted pathogen when the pathogen is grown in culture. Testing in animal models, like that highlighted herein for the salicyl-AMS inhibitor [42], is the next necessary step for most of these translational projects [110].
Biosynthesis is but one target when considering how to develop antimicrobials directed at or exploiting siderophore-dependent iron nutritional immunity. Considerable effort has been expended in tricking siderophore uptake systems to import antibiotics, the Trojan horse strategy [111, 112]. Interestingly, nature (in the form of Streptomyces) was the first designer of Trojan Horse antibiotics, linking the siderophore ferrichrome to a peptidylnucleoside antibiotic to generate albomycins [113]. Conversely, blocking siderophore export yields a toxic cytoplasmic buildup of siderophores [114]. Development of natural products with novel bioactive properties by engineering PKS and NRPS assembly lines has long been a goal of metabolic engineers with significant recent advances (some examples are found in references [115–120]). Regardless of how the systems for siderophore iron-scavenging are exploited, iron scavenging pathways provide a key access point for fighting the looming health crisis of antibiotic resistance in pathogenic microbes. The collected work of this review shows the promise and recent successes for targeting inhibitors at the biosynthesis of siderophores in the design of new antimicrobial therapies.
Acknowledgments
Funding Sources.
This work was supported by K02 AI093675 from the National Institute for Allergy and Infectious Disease of the National Institutes of Health and CHE 1403293 from the National Science Foundation.
Footnotes
An EC50 value reports the half maximal effective concentration for inhibiting bacterial growth.
An MIC90 value reports the minimum inhibitory concentration for inhibiting 90% of bacterial growth.
An IC50 value reports the half maximal enzyme inhibitory concentration.
A Ki value reports the equilibrium dissociation constant of the inhibitor.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The White House Executive Order -- Combating Antibiotic-Resistant Bacteria. https://www.whitehouse.gov/the-press-office/2014/09/18/executive-order-combating-antibiotic-resistant-bacteria.
- 2.Centers for Disease Control and Prevention Antibiotic Resistance Solutions Initiative. http://www.cdc.gov/drugresistance/solutions-initiative/index.html.
- 3.World Health Organization. Antimicrobial Resistance: Global Report on Surveillance. 2014. [Google Scholar]
- 4.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3(9):541–8. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 5.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS pathogens. 2010;6(8):e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(7):3584–8. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasil ML, Ochsner UA. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Molecular microbiology. 1999;34(3):399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis P, Matthijs S. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ Microbiol. 2002;4(12):787–98. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]
- 9.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66(2):223–49. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97(7):2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 11.Quadri LE. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infectious disorders drug targets. 2007;7(3):230–7. doi: 10.2174/187152607782110040. [DOI] [PubMed] [Google Scholar]
- 12.Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Molecular pharmaceutics. 2008;5(2):191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koglin A, Walsh CT. Structural insights into nonribosomal peptide enzymatic assembly lines. Natural product reports. 2009;26(8):987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keatinge-Clay AT. The structures of type I polyketide synthases. Natural product reports. 2012;29(10):1050–73. doi: 10.1039/c2np20019h. [DOI] [PubMed] [Google Scholar]
- 15.Khosla C, Herschlag D, Cane DE, Walsh CT. Assembly line polyketide synthases: mechanistic insights and unsolved problems. Biochemistry. 2014;53(18):2875–83. doi: 10.1021/bi500290t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oves-Costales D, Kadi N, Challis GL. The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chemical communications. 2009;(43):6530–41. doi: 10.1039/b913092f. [DOI] [PubMed] [Google Scholar]
- 17.Cendrowski S, MacArthur W, Hanna P. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Molecular microbiology. 2004;51(2):407–17. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 18.Dumas Z, Ross-Gillespie A, Kummerli R. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proceedings Biological sciences/The Royal Society. 2013;280(1764):20131055. doi: 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinto LJ, Moore MM. Screening method to identify inhibitors of siderophore biosynthesis in the opportunistic fungal pathogen, Aspergillus fumigatus. Letters in applied microbiology. 2009;49(1):8–13. doi: 10.1111/j.1472-765X.2009.02582.x. [DOI] [PubMed] [Google Scholar]
- 20.Stirrett KL, Ferreras JA, Jayaprakash V, Sinha BN, Ren T, Quadri LE. Small molecules with structural similarities to siderophores as novel antimicrobials against Mycobacterium tuberculosis and Yersinia pestis. Bioorganic & medicinal chemistry letters. 2008;18(8):2662–8. doi: 10.1016/j.bmcl.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falconer SB, Wang W, Gehrke SS, Cuneo JD, Britten JF, Wright GD, Brown ED. Metal-induced isomerization yields an intracellular chelator that disrupts bacterial iron homeostasis. Chemistry & biology. 2014;21(1):136–45. doi: 10.1016/j.chembiol.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Imperi F, Massai F, Facchini M, Frangipani E, Visaggio D, Leoni L, Bragonzi A, Visca P. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(18):7458–63. doi: 10.1073/pnas.1222706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootz HD, Finking R, Marahiel MA. 4′-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. The Journal of biological chemistry. 2001;276(40):37289–98. doi: 10.1074/jbc.M103556200. [DOI] [PubMed] [Google Scholar]
- 24.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chemistry & biology. 1996;3(11):923–36. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 25.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chemistry & biology. 1999;6(8):493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 26.Walsh CT, Gehring AM, Weinreb PH, Quadri LE, Flugel RS. Post-translational modification of polyketide and nonribosomal peptide synthases. Current opinion in chemical biology. 1997;1(3):309–15. doi: 10.1016/s1367-5931(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 27.Leblanc C, Prudhomme T, Tabouret G, Ray A, Burbaud S, Cabantous S, Mourey L, Guilhot C, Chalut C. 4′-Phosphopantetheinyl transferase PptT, a new drug target required for Mycobacterium tuberculosis growth and persistence in vivo. PLoS pathogens. 2012;8(12):e1003097. doi: 10.1371/journal.ppat.1003097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foley TL, Burkart MD. A homogeneous resonance energy transfer assay for phosphopantetheinyl transferase. Analytical biochemistry. 2009;394(1):39–47. doi: 10.1016/j.ab.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yasgar A, Foley TL, Jadhav A, Inglese J, Burkart MD, Simeonov A. A strategy to discover inhibitors of Bacillus subtilis surfactin-type phosphopantetheinyl transferase. Molecular bio Systems. 2010;6(2):365–75. doi: 10.1039/b913291k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(44):15815–20. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Foley TL, Rai G, Yasgar A, Attene-Ramos MS, Burkart MD, Simeonov A, Jadhav A, Maloney DJ. Discovery of ML 267 as a Novel Inhibitor of Pathogenic Sfp phosphopantetheinyl transferase (PPTase). Probe Reports from the NIH Molecular Libraries Program; Bethesda (MD). 2010. [PubMed] [Google Scholar]
- 32.Foley TL, Rai G, Yasgar A, Daniel T, Baker HL, Attene-Ramos M, Kosa NM, Leister W, Burkart MD, Jadhav A, Simeonov A, Maloney DJ. 4-(3-Chloro-5-(trifluoromethyl)pyridin-2-yl)-N-(4-methoxypyridin-2-yl)piperazine- 1-carbothioamide (ML267), a potent inhibitor of bacterial phosphopantetheinyl transferase that attenuates secondary metabolism and thwarts bacterial growth. Journal of medicinal chemistry. 2014;57(3):1063–78. doi: 10.1021/jm401752p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duckworth BP, Aldrich CC. Development of a high-throughput fluorescence polarization assay for the discovery of phosphopantetheinyl transferase inhibitors. Analytical biochemistry. 2010;403(1–2):13–9. doi: 10.1016/j.ab.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Kosa NM, Foley TL, Burkart MD. Fluorescent techniques for discovery and characterization of phosphopantetheinyl transferase inhibitors. The Journal of antibiotics. 2014;67(1):113–20. doi: 10.1038/ja.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vickery CR, Kosa NM, Casavant EP, Duan S, Noel JP, Burkart MD. Structure, biochemistry, and inhibition of essential 4′-phosphopantetheinyl transferases from two species of Mycobacteria. ACS chemical biology. 2014;9(9):1939–44. doi: 10.1021/cb500263p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreras JA, Ryu JS, Di Lello F, Tan DS, Quadri LE. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nature chemical biology. 2005;1(1):29–32. doi: 10.1038/nchembio706. [DOI] [PubMed] [Google Scholar]
- 37.Vannada J, Bennett EM, Wilson DJ, Boshoff HI, Barry CE, 3rd, Aldrich CC. Design, synthesis, and biological evaluation of beta-ketosulfonamide adenylation inhibitors as potential antitubercular agents. Organic letters. 2006;8(21):4707–10. doi: 10.1021/ol0617289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somu RV, Wilson DJ, Bennett EM, Boshoff HI, Celia L, Beck BJ, Barry CE, 3rd, Aldrich CC. Antitubercular nucleosides that inhibit siderophore biosynthesis: SAR of the glycosyl domain. Journal of medicinal chemistry. 2006;49(26):7623–35. doi: 10.1021/jm061068d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupte A, Boshoff HI, Wilson DJ, Neres J, Labello NP, Somu RV, Xing C, Barry CE, Aldrich CC. Inhibition of siderophore biosynthesis by 2-triazole substituted analogues of 5′-O-[N-(salicyl)sulfamoyl]adenosine: antibacterial nucleosides effective against Mycobacterium tuberculosis. Journal of medicinal chemistry. 2008;51(23):7495–507. doi: 10.1021/jm8008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somu RV, Boshoff H, Qiao C, Bennett EM, Barry CE, 3rd, Aldrich CC. Rationally designed nucleoside antibiotics that inhibit siderophore biosynthesis of Mycobacterium tuberculosis. Journal of medicinal chemistry. 2006;49(1):31–4. doi: 10.1021/jm051060o. [DOI] [PubMed] [Google Scholar]
- 41.Neres J, Labello NP, Somu RV, Boshoff HI, Wilson DJ, Vannada J, Chen L, Barry CE, 3rd, Bennett EM, Aldrich CC. Inhibition of siderophore biosynthesis in Mycobacterium tuberculosis with nucleoside bisubstrate analogues: structure-activity relationships of the nucleobase domain of 5′-O-[N-(salicyl)sulfamoyl]adenosine. Journal of medicinal chemistry. 2008;51(17):5349–70. doi: 10.1021/jm800567v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lun S, Guo H, Adamson J, Cisar JS, Davis TD, Chavadi SS, Warren JD, Quadri LE, Tan DS, Bishai WR. Pharmacokinetic and in vivo efficacy studies of the mycobactin biosynthesis inhibitor salicyl-AMS in mice. Antimicrobial agents and chemotherapy. 2013;57(10):5138–40. doi: 10.1128/AAC.00918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miethke M, Bisseret P, Beckering CL, Vignard D, Eustache J, Marahiel MA. Inhibition of aryl acid adenylation domains involved in bacterial siderophore synthesis. The FEBS journal. 2006;273(2):409–19. doi: 10.1111/j.1742-4658.2005.05077.x. [DOI] [PubMed] [Google Scholar]
- 44.Callahan BP, Lomino JV, Wolfenden R. Nanomolar inhibition of the enterobactin biosynthesis enzyme, EntE: synthesis, substituent effects, and additivity. Bioorganic & medicinal chemistry letters. 2006;16(14):3802–5. doi: 10.1016/j.bmcl.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Sikora AL, Wilson DJ, Aldrich CC, Blanchard JS. Kinetic and inhibition studies of dihydroxybenzoate-AMP ligase from Escherichia coli. Biochemistry. 2010;49(17):3648–57. doi: 10.1021/bi100350c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neres J, Wilson DJ, Celia L, Beck BJ, Aldrich CC. Aryl acid adenylating enzymes involved in siderophore biosynthesis: fluorescence polarization assay, ligand specificity, and discovery of non-nucleoside inhibitors via high-throughput screening. Biochemistry. 2008;47(45):11735–49. doi: 10.1021/bi801625b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drake EJ, Duckworth BP, Neres J, Aldrich CC, Gulick AM. Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis. Biochemistry. 2010;49(43):9292–305. doi: 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueda H, Shoku Y, Hayashi N, Mitsunaga J, In Y, Doi M, Inoue M, Ishida T. X-ray crystallographic conformational study of 5′-O-[N-(L-alanyl)-sulfamoyl]adenosine, a substrate analogue for alanyl-tRNA synthetase. Biochimica et biophysica acta. 1991;1080(2):126–34. doi: 10.1016/0167-4838(91)90138-p. [DOI] [PubMed] [Google Scholar]
- 49.Finking R, Neumuller A, Solsbacher J, Konz D, Kretzschmar G, Schweitzer M, Krumm T, Marahiel MA. Aminoacyl adenylate substrate analogues for the inhibition of adenylation domains of nonribosomal peptide synthetases. Chembiochem: a European journal of chemical biology. 2003;4(9):903–6. doi: 10.1002/cbic.200300666. [DOI] [PubMed] [Google Scholar]
- 50.Cisar JS, Ferreras JA, Soni RK, Quadri LE, Tan DS. Exploiting ligand conformation in selective inhibition of non-ribosomal peptide synthetase amino acid adenylation with designed macrocyclic small molecules. Journal of the American Chemical Society. 2007;129(25):7752–3. doi: 10.1021/ja0721521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gulick AM. Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS chemical biology. 2009;4(10):811–27. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM. Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism. Biochemistry. 2012;51(33):6493–5. doi: 10.1021/bi300934s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chemistry & biology. 2012;19(2):188–98. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu R, Reger AS, Cao J, Gulick AM, Dunaway-Mariano D. Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range. Biochemistry. 2007;46(50):14487–99. doi: 10.1021/bi701609w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Structure of PA1221, a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry. 2012;51(15):3252–63. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balhara M, Ruhil S, Kumar M, Dhankhar S, Chhillar AK. An anti-Aspergillus protein from Escherichia coli DH5alpha: putative inhibitor of siderophore biosynthesis in Aspergillus fumigatus. Mycoses. 2014;57(3):153–62. doi: 10.1111/myc.12119. [DOI] [PubMed] [Google Scholar]
- 57.Meier JL, Burkart MD. The chemical biology of modular biosynthetic enzymes. Chemical Society reviews. 2009;38(7):2012–45. doi: 10.1039/b805115c. [DOI] [PubMed] [Google Scholar]
- 58.Cummings M, Breitling R, Takano E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS microbiology letters. 2014;351(2):116–25. doi: 10.1111/1574-6968.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams GJ. Engineering polyketide synthases and nonribosomal peptide synthetases. Current opinion in structural biology. 2013;23(4):603–12. doi: 10.1016/j.sbi.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McQuade TJ, Shallop AD, Sheoran A, Delproposto JE, Tsodikov OV, Garneau-Tsodikova S. A nonradioactive high-throughput assay for screening and characterization of adenylation domains for nonribosomal peptide combinatorial biosynthesis. Analytical biochemistry. 2009;386(2):244–50. doi: 10.1016/j.ab.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 61.Tripathi A, Schofield MM, Chlipala GE, Schultz PJ, Yim I, Newmister SA, Nusca TD, Scaglione JB, Hanna PC, Tamayo-Castillo G, Sherman DH. Baulamycins A and B, broad-spectrum antibiotics identified as inhibitors of siderophore biosynthesis in Staphylococcus aureus and Bacillus anthracis. Journal of the American Chemical Society. 2014;136(4):1579–86. doi: 10.1021/ja4115924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J Am Chem Soc. 2004;126(8):2378–85. doi: 10.1021/ja0389927. [DOI] [PubMed] [Google Scholar]
- 63.Meneely KM, Luo Q, Dhar P, Lamb AL. Lysine221 is the general base residue of the isochorismate synthase from Pseudomonas aeruginosa (PchA) in a reaction that is diffusion limited. Archives of biochemistry and biophysics. 2013;538(1):49–56. doi: 10.1016/j.abb.2013.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwahlen J, Kolappan S, Zhou R, Kisker C, Tonge PJ. Structure and mechanism of MbtI, the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2007;46(4):954–64. doi: 10.1021/bi060852x. [DOI] [PubMed] [Google Scholar]
- 65.DeClue MS, Baldridge KK, Kunzler DE, Kast P, Hilvert D. Isochorismate Pyruvate Lyase: A Pericyclic Reaction Mechanism? J Am Chem Soc. 2005;127(43):15002–15003. doi: 10.1021/ja055871t. [DOI] [PubMed] [Google Scholar]
- 66.Culbertson JE, Chung DH, Ziebart KT, Espiritu E, Toney MD. Conversion of Aminodeoxychorismate Synthase into Anthranilate Synthase with Janus Mutations: mechanism of pyruvate elimination catalyzed by chorismate enzymes. Biochemistry. 2015 doi: 10.1021/acs.biochem.5b00013. [DOI] [PubMed] [Google Scholar]
- 67.Walsh CT, Liu J, Rusnak F, Sakaitani M. Molecular studies on enzymes in chorismate metabolism and the enterobactin biosynthetic pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 68.Kozlowski MC, Bartlett PA. Synthesis of a potential transition-state analog inhibitor of isochorismate synthase. Journal of the American Chemical Society. 1991;113:5987–5898. [Google Scholar]
- 69.Kozlowski MC, Tom NJ, Seto CT, Sefler AM, Bartlett PA. Chorismate-utilizing enzymes isochorismate synthase, anthranilate synthase, and p-aminobenzoate synthase: mechanistic insight through inhibitor design. Journal of the American Chemical Society. 1995;117:2128–2140. [Google Scholar]
- 70.Knochel T, Ivens A, Hester G, Gonzalez A, Bauerle R, Wilmanns M, Kirschner K, Jansonius JN. The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(17):9479–84. doi: 10.1073/pnas.96.17.9479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morollo AA, Eck MJ. Structure of the cooperative allosteric anthranilate synthase from Salmonella typhimurium. Nature structural biology. 2001;8(3):243–7. doi: 10.1038/84988. [DOI] [PubMed] [Google Scholar]
- 72.Spraggon G, Kim C, Nguyen-Huu X, Yee MC, Yanofsky C, Mills SE. The structures of anthranilate synthase of Serratia marcescens crystallized in the presence of (i) its substrates, chorismate and glutamine, and a product, glutamate, and (ii) its end-product inhibitor, L-tryptophan. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(11):6021–6. doi: 10.1073/pnas.111150298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parsons JF, Jensen PY, Pachikara AS, Howard AJ, Eisenstein E, Ladner JE. Structure of Escherichia coli aminodeoxychorismate synthase: architectural conservation and diversity in chorismate-utilizing enzymes. Biochemistry. 2002;41(7):2198–208. doi: 10.1021/bi015791b. [DOI] [PubMed] [Google Scholar]
- 74.Payne RJ, Kerbarh O, Miguel RN, Abell AD, Abell C. Inhibition studies on salicylate synthase. Organic & biomolecular chemistry. 2005;3(10):1825–7. doi: 10.1039/b503800f. [DOI] [PubMed] [Google Scholar]
- 75.Manos-Turvey A, Bulloch EM, Rutledge PJ, Baker EN, Lott JS, Payne RJ. Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI) Chem Med Chem. 2010;5(7):1067–79. doi: 10.1002/cmdc.201000137. [DOI] [PubMed] [Google Scholar]
- 76.Manos-Turvey A, Cergol KM, Salam NK, Bulloch EM, Chi G, Pang A, Britton WJ, West NP, Baker EN, Lott JS, Payne RJ. Synthesis and evaluation of M. tuberculosis salicylate synthase (MbtI) inhibitors designed to probe plasticity in the active site. Organic & biomolecular chemistry. 2012;10(46):9223–36. doi: 10.1039/c2ob26736e. [DOI] [PubMed] [Google Scholar]
- 77.Payne RJ, Bulloch EM, Toscano MM, Jones MA, Kerbarh O, Abell C. Synthesis and evaluation of 2,5-dihydrochorismate analogues as inhibitors of the chorismate-utilising enzymes. Organic & biomolecular chemistry. 2009;7(11):2421–9. doi: 10.1039/b901694e. [DOI] [PubMed] [Google Scholar]
- 78.Payne RJ, Bulloch EM, Kerbarh O, Abell C. Inhibition of chorismate-utilising enzymes by 2-amino-4-carboxypyridine and 4-carboxypyridone and 5-carboxypyridone analogues. Organic & biomolecular chemistry. 2010;8(15):3534–42. doi: 10.1039/c004062b. [DOI] [PubMed] [Google Scholar]
- 79.Chi G, Manos-Turvey A, O’Connor PD, Johnston JM, Evans GL, Baker EN, Payne RJ, Lott JS, Bulloch EM. Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis. Biochemistry. 2012;51(24):4868–79. doi: 10.1021/bi3002067. [DOI] [PubMed] [Google Scholar]
- 80.Kerbarh O, Chirgadze DY, Blundell TL, Abell C. Crystal structures of Yersinia enterocolitica salicylate synthase and its complex with the reaction products salicylate and pyruvate. Journal of molecular biology. 2006;357(2):524–34. doi: 10.1016/j.jmb.2005.12.078. [DOI] [PubMed] [Google Scholar]
- 81.Ziebart KT, Dixon SM, Avila B, El-Badri MH, Guggenheim KG, Kurth MJ, Toney MD. Targeting multiple chorismate-utilizing enzymes with a single inhibitor: validation of a three-stage design. Journal of medicinal chemistry. 2010;53(9):3718–29. doi: 10.1021/jm100158v. [DOI] [PubMed] [Google Scholar]
- 82.Vasan M, Neres J, Williams J, Wilson DJ, Teitelbaum AM, Remmel RP, Aldrich CC. Inhibitors of the salicylate synthase (MbtI) from Mycobacterium tuberculosis discovered by high-throughput screening. Chem Med Chem. 2010;5(12):2079–87. doi: 10.1002/cmdc.201000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baell JB, Holloway GA. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. Journal of medicinal chemistry. 2010;53(7):2719–40. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 84.Gaille C, Reimmann C, Haas D. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J Biol Chem. 2003;278(19):16893–8. doi: 10.1074/jbc.M212324200. [DOI] [PubMed] [Google Scholar]
- 85.Gaille C, Kast P, Haas D. Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J Biol Chem. 2002;277(24):21768–75. doi: 10.1074/jbc.M202410200. [DOI] [PubMed] [Google Scholar]
- 86.Meneely KM, Luo Q, Riley AP, Taylor B, Roy A, Stein RL, Prisinzano TE, Lamb AL. Expanding the results of a high throughput screen against an isochorismate-pyruvate lyase to enzymes of a similar scaffold or mechanism. Bioorganic & medicinal chemistry. 2014;22(21):5961–9. doi: 10.1016/j.bmc.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qi J, Kizjakina K, Robinson R, Tolani K, Sobrado P. A fluorescence polarization binding assay to identify inhibitors of flavin-dependent monooxygenases. Analytical biochemistry. 2012;425(1):80–7. doi: 10.1016/j.ab.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belyaeva T, Leontieva E, Shpakov A, Mozhenok T, Faddejeva M. Sensitivity of lysosomal enzymes to the plant alkaloid sanguinarine: comparison with other SH-specific agents. Cell biology international. 2003;27(11):887–95. doi: 10.1016/s1065-6995(03)00161-6. [DOI] [PubMed] [Google Scholar]
- 89.Basini G, Bussolati S, Santini SE, Grasselli F. Sanguinarine inhibits VEGF-induced angiogenesis in a fibrin gel matrix. Bio Factors. 2007;29(1):11–8. doi: 10.1002/biof.5520290102. [DOI] [PubMed] [Google Scholar]
- 90.Basini G, Santini SE, Bussolati S, Grasselli F. Sanguinarine inhibits VEGF-induced Akt phosphorylation. Annals of the New York Academy of Sciences. 2007;1095:371–6. doi: 10.1196/annals.1397.040. [DOI] [PubMed] [Google Scholar]
- 91.Chen Y, Wang H, Zhang R, Wang H, Peng Z, Sun R, Tan Q. Microinjection of sanguinarine into the ventrolateral orbital cortex inhibits Mkp-1 and exerts an antidepressant-like effect in rats. Neuroscience letters. 2012;506(2):327–31. doi: 10.1016/j.neulet.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 92.Dong XZ, Zhang M, Wang K, Liu P, Guo DH, Zheng XL, Ge XY. Sanguinarine inhibits vascular endothelial growth factor release by generation of reactive oxygen species in MCF-7 human mammary adenocarcinoma cells. Bio Med research international. 2013;2013:517698. doi: 10.1155/2013/517698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H, Zhai Z, Liu G, Tang T, Lin Z, Zheng M, Qin A, Dai K. Sanguinarine inhibits osteoclast formation and bone resorption via suppressing RANKL-induced activation of NF-kappaB and ERK signaling pathways. Biochemical and biophysical research communications. 2013;430(3):951–6. doi: 10.1016/j.bbrc.2012.12.051. [DOI] [PubMed] [Google Scholar]
- 94.Park SY, Jin ML, Kim YH, Lee SJ, Park G. Sanguinarine inhibits invasiveness and the MMP-9 and COX-2 expression in TPA-induced breast cancer cells by inducing HO-1 expression. Oncology reports. 2014;31(1):497–504. doi: 10.3892/or.2013.2843. [DOI] [PubMed] [Google Scholar]
- 95.Seifen E, Adams RJ, Riemer RK. Sanguinarine: a positive inotropic alkaloid which inhibits cardiac Na+, K+-ATPase. European journal of pharmacology. 1979;60(4):373–7. doi: 10.1016/0014-2999(79)90245-0. [DOI] [PubMed] [Google Scholar]
- 96.Sun M, Lou W, Chun JY, Cho DS, Nadiminty N, Evans CP, Chen J, Yue J, Zhou Q, Gao AC. Sanguinarine suppresses prostate tumor growth and inhibits survivin expression. Genes & cancer. 2010;1(3):283–92. doi: 10.1177/1947601910368849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu JY, Meng QH, Chong Y, Jiao Y, Zhao L, Rosen EM, Fan S. Sanguinarine inhibits growth of human cervical cancer cells through the induction of apoptosis. Oncology reports. 2012;28(6):2264–70. doi: 10.3892/or.2012.2024. [DOI] [PubMed] [Google Scholar]
- 98.Ying L, Li G, Wei SS, Wang H, An P, Wang X, Guo K, Luo XJ, Gao JM, Zhou Q, Li W, Yu Y, Li YG, Duan JL, Wang YP. Sanguinarine inhibits Rac1b-rendered cell survival enhancement by promoting apoptosis and blocking proliferation. Acta pharmacologica Sinica. 2015;36(2):229–40. doi: 10.1038/aps.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soe CZ, Pakchung AA, Codd R. Directing the biosynthesis of putrebactin or desferrioxamine B in Shewanella putrefaciens through the upstream inhibition of ornithine decarboxylase. Chemistry & biodiversity. 2012;9(9):1880–90. doi: 10.1002/cbdv.201200014. [DOI] [PubMed] [Google Scholar]
- 100.Soe CZ, Codd R. Unsaturated macrocyclic dihydroxamic acid siderophores produced by Shewanella putrefaciens using precursor-directed biosynthesis. ACS chemical biology. 2014;9(4):945–56. doi: 10.1021/cb400901j. [DOI] [PubMed] [Google Scholar]
- 101.Visca P, Imperi F, Lamont IL. Pyoverdine siderophores: from biogenesis to biosignificance. Trends in microbiology. 2007;15(1):22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Bokhove M, Nadal Jimenez P, Quax WJ, Dijkstra BW. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):686–91. doi: 10.1073/pnas.0911839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Drake EJ, Gulick AM. Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis. ACS chemical biology. 2011;6(11):1277–86. doi: 10.1021/cb2002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nadal Jimenez P, Koch G, Papaioannou E, Wahjudi M, Krzeslak J, Coenye T, Cool RH, Quax WJ. Role of PvdQ in Pseudomonas aeruginosa virulence under iron-limiting conditions. Microbiology. 2010;156(Pt 1):49–59. doi: 10.1099/mic.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 105.Wahjudi M, Murugappan S, van Merkerk R, Eissens AC, Visser MR, Hinrichs WL, Quax WJ. Development of a dry, stable and inhalable acyl-homoserine-lactone-acylase powder formulation for the treatment of pulmonary Pseudomonas aeruginosa infections. European journal of pharmaceutical sciences: official journal of the European Federation for Pharmaceutical Sciences. 2013;48(4–5):637–43. doi: 10.1016/j.ejps.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 106.Theriault JR, Wurst J, Jewett I, Verplank L, Perez JR, Gulick AM, Drake EJ, Palmer M, Moskowitz S, Dasgupta N, Brannon MK, Dandapani S, Munoz B, Schreiber S. Identification of a small molecule inhibitor of Pseudomonas aeruginosa PvdQ acylase, an enzyme involved in siderophore pyoverdine synthesis. Probe Reports from the NIH Molecular Libraries Program; Bethesda (MD). 2010. [PubMed] [Google Scholar]
- 107.Wurst JM, Drake EJ, Theriault JR, Jewett IT, VerPlank L, Perez JR, Dandapani S, Palmer M, Moskowitz SM, Schreiber SL, Munoz B, Gulick AM. Identification of inhibitors of PvdQ, an enzyme involved in the synthesis of the siderophore pyoverdine. ACS chemical biology. 2014;9(7):1536–44. doi: 10.1021/cb5001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clevenger KD, Wu R, Er JA, Liu D, Fast W. Rational design of a transition state analogue with picomolar affinity for Pseudomonas aeruginosa PvdQ, a siderophore biosynthetic enzyme. ACS chemical biology. 2013;8(10):2192–200. doi: 10.1021/cb400345h. [DOI] [PubMed] [Google Scholar]
- 109.Clevenger KD, Wu R, Liu D, Fast W. n-Alkylboronic acid inhibitors reveal determinants of ligand specificity in the quorum-quenching and siderophore biosynthetic enzyme PvdQ. Biochemistry. 2014;53(42):6679–86. doi: 10.1021/bi501086s. [DOI] [PubMed] [Google Scholar]
- 110.Foley TL, Simeonov A. Targeting iron assimilation to develop new antibacterials. Expert opinion on drug discovery. 2012;7(9):831–47. doi: 10.1517/17460441.2012.708335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mislin GLA, Schalk IJ. Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics. 2014;6(3):408–420. doi: 10.1039/c3mt00359k. [DOI] [PubMed] [Google Scholar]
- 112.Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. Microbial siderophores and their potential applications: a review. Environmental science and pollution research international. 2015 doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- 113.Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S, Chen S. Biosynthesis of albomycin delta(2) provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS chemical biology. 2012;7(9):1565–75. doi: 10.1021/cb300173x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones CM, Wells RM, Madduri AV, Renfrow MB, Ratledge C, Moody DB, Niederweis M. Self-poisoning of Mycobacterium tuberculosis by interrupting siderophore recycling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(5):1945–50. doi: 10.1073/pnas.1311402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Calcott MJ, Ackerley DF. Genetic manipulation of non-ribosomal peptide synthetases to generate novel bioactive peptide products. Biotechnology letters. 2014;36(12):2407–16. doi: 10.1007/s10529-014-1642-y. [DOI] [PubMed] [Google Scholar]
- 116.Calcott MJ, Owen JG, Lamont IL, Ackerley DF. Biosynthesis of novel Pyoverdines by domain substitution in a nonribosomal peptide synthetase of Pseudomonas aeruginosa. Applied and environmental microbiology. 2014;80(18):5723–31. doi: 10.1128/AEM.01453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Das A, Szu PH, Fitzgerald JT, Khosla C. Mechanism and engineering of polyketide chain initiation in fredericamycin biosynthesis. Journal of the American Chemical Society. 2010;132(26):8831–3. doi: 10.1021/ja102517q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(11):4110–5. doi: 10.1073/pnas.1118734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Muchiri R, Walker KD. Taxol biosynthesis: tyrocidine synthetase A catalyzes the production of phenylisoserinyl CoA and other amino phenylpropanoyl thioesters. Chemistry & biology. 2012;19(6):679–85. doi: 10.1016/j.chembiol.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 120.Zheng J, Piasecki SK, Keatinge-Clay AT. Structural studies of an A2-type modular polyketide synthase ketoreductase reveal features controlling alpha-substituent stereochemistry. ACS chemical biology. 2013;8(9):1964–71. doi: 10.1021/cb400161g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]