Abstract
Background
Expression of the neuronal membrane glycoprotein M6a (GPM6A), the proteolipid protein (PLP/DM20) family member, is downregulated in the hippocampus of chronically stressed animals. Its neuroplastic function involves a role in neurite formation, filopodium outgrowth and synaptogenesis through an unknown mechanism. Disruptions in neuroplasticity mechanisms have been shown to play a significant part in the etiology of depression. Thus, the current investigation examined whether GPM6A expression is also altered in human depressed brain.
Methods
Expression levels and coexpression patterns of GPM6A, GPM6B, and PLP1 (two other members of PLP/DM20 family) as well as of the neuroplasticity-related genes identified to associate with GPM6A were determined using quantitative polymerase chain reaction (qPCR) in postmortem samples from the hippocampus (n =18) and the prefrontal cortex (PFC) (n= 25) of depressed suicide victims and compared with control subjects (hippocampus n= 18; PFC n =25). Neuroplasticity-related proteins that form complexes with GPM6A were identified by coimmunoprecipitation technique followed by mass spectrometry.
Results
Results indicated transcriptional downregulation of GPM6A and GPM6B in the hippocampus of depressed suicides. The expression level of calcium/calmodulin-dependent protein kinase II alpha (CAMK2A) and coronin1A (CORO1A) was also significantly decreased. Subsequent analysis of coexpression patterns demonstrated coordinated gene expression in the hippocampus and in the PFC indicating that the function of these genes might be coregulated in the human brain. However, in the brain of depressed suicides this coordinated response was disrupted.
Conclusions
Disruption of coordinated gene expression as well as abnormalities in GPM6A and GPM6B expression and expression of the components of GPM6A complexes were detected in the brain of depressed suicides.
Keywords: postmortem, mRNA expression, coexpression analysis, glycoprotein M6a, qPCR
INTRODUCTION
Neuroplasticity is the mechanism by which information is stored and maintained within individual synapses, neurons, and neuronal circuits to guide organism behavior. Several lines of evidence demonstrate impairment of neuroplasticity in depression (Pittenger and Duman, 2008; Christoffel et al., 2011). For example, in patients with different types of depression, the hippocampus and the prefrontal cortex (PFC) are both reduced in size and activity, and alterations in synaptic and morphological plasticity have been reported (Rajkowska et al., 1999; Stockmeier et al., 2004; Drevets et al., 2008; Pittenger and Duman, 2008; Kang et al., 2012). The histopathological correlates include reductions in synapses or synaptic proteins, reductions in neuronal size, and in neuropil (Drevets et al., 2008). Similar alterations have been observed in animal models of chronic stress: reductions in dendritic arborizations and a loss of highly specialized dendritic spines and synapses in regions that appear homologous to the areas where reductions are evident in depressed humans (i.e., PFC, hippocampus) (McKittrick et al., 2000; Radley et al., 2004; Drevets et al., 2008; Pittenger and Duman, 2008). The intracellular mechanisms underlying these alterations and their relevance to human depression are poorly understood.
The neuronal membrane glycoprotein M6a (GPM6A), a member of the myelin proteolipid protein (PLP/DM20) family, has been shown to play a role in stress response in different animal models (Alfonso et al., 2004a,b; Cooper et al., 2009; Monteleone et al., 2014). For example, chronic social and physical stress decreases Gpm6a mRNA levels in the hippocampus, and this downregulation is prevented by administration of antidepressants (Alfonso et al., 2004b, 2006). An association of the GPM6A gene with a depression subgroup of schizophrenia patients (Boks et al., 2008) as well as a critical role of GPM6A expression levels for cognitive function have been reported recently (Gregor et al., 2014). Gpm6a knockout mouse model is viable and shows no gross malformations or behavioral abnormalities (El-Kordi et al., 2013). However, after mild social stress by single housing, these mice displayed a claustrophobia-like phenotype. Interestingly, in humans a 3′UTR variant of GPM6A has been linked to claustrophobia in two pedigrees (El-Kordi et al., 2013).
Apart from Gpm6a, other members of the family, the closely related Gpm6b and Dm20 (Plp1 transcript variant), but not Plp1 itself, have been shown to be downregulated by chronic stress (Fernandez et al., 2010). Remarkably, the myelin proteolipid protein (PLP/DM20) family members, such as GPM6A, GPM6B, and PLP1 transcript variant DM20, but not PLP1 (Fernandez et al., 2010), have been shown to be involved in the processes of neurite outgrowth and filopodium formation (Lagenaur et al., 1992; Mukobata et al., 2002; Alfonso et al., 2005; Michibata et al., 2008; Zhao et al., 2008; Fuchsova et al., 2009; Brocco et al., 2010; Scorticati et al., 2011). GPM6A, in particular, is also required for filopodium motility and synaptogenesis (Fuchsova et al., 2009; Brocco et al., 2010), and it has been implicated in neuronal differentiation of human stem cells (Michibata et al., 2009) and PC12 cells (Mukobata et al., 2002). When siRNA methodology is used, GPM6A low-expressing neurons display decreased filopodia numbers and a lower density of synaptophysin clusters (Alfonso et al., 2005).
Neurite growth and remodeling, as well as filopodium and spine formation, represent fundamental processes during neuroplasticity. Thus, we hypothesized that alterations in the expression of the stress responsive neuroplasticity-related genes such as the members of the PLP family could suggest that the cellular pathways that involve these genes are sensitive to disease condition. This would result in dysregulation of neuroplasticity mechanisms involved in the etiology of this disease. Therefore, we examined in the present study the expression of PLP family members GPM6A, GPM6B, and PLP1 in the hippocampus (n=18) and the PFC (n=25) of depressed suicides. The mechanisms and signaling pathways that mediate GPM6A neuroplastic effects are still unknown. Thus, we also identified the neuroplasticity-related proteins that form complexes with GPM6A and examined their expression as well.
EXPERIMENTAL PROCEDURES
Subjects
The study was performed in PFC (Brodmann area 9) samples from suicide (n=25) and matched nonpsychiatric control subjects (n=25). Brain tissues from the same cohort have been used previously in various studies published by our group (Dwivedi et al., 2008; Dwivedi et al., 2009, 2010; Pandey et al., 2014). Dissected regions of interest included predominantly cortical gray matter. The white matter was removed as much as possible, but there was still some white matter left. Hippocampi were available for 18 suicide subjects and 18 nonpsychiatric controls. Postmortem brain samples were obtained from the Maryland Brain Collection at the Maryland Psychiatric Research Center, Baltimore. Tissues were collected only after a family member gave informed consent. Brain samples were free of neuropathologic abnormalities or human immunodeficiency virus antibodies. Toxicological data were obtained by an analysis of urine and blood samples. Psychiatric diagnoses in suicide and control subjects were evaluated with the Diagnostic Evaluation After Death (Salzman et al., 1983) and the Structured Clinical Interview for DSM-IV (Spitzer et al., 1995). Family members gave permission for clinical records to be obtained from mental health treatment providers in all cases of suicide. Control subjects were verified to be free from mental illnesses using a consensus diagnostic procedure. All procedures were approved by the Institutional Review Board of the University of Illinois at Chicago. Detailed demography of subjects is provided in Table 1.
Table 1.
Demographic characteristics of depressed suicide victims and normal control subjects
| Age (years) | Race | Gender | PMI (hours) | Brain pH | Cause of death | Psychotropic drugs (at the time of death) | Psychiatric diagnosis | ||
|---|---|---|---|---|---|---|---|---|---|
| Normal control subjectsa | |||||||||
| 1 | CONTROL | 22 | Black | Male | 19 | 6.9 | GSW | None | Normal |
| 2 | CONTROL | 42 | White | Female | 23 | 7.2 | Pneumonia | None | Normal |
| 3 | CONTROL | 37 | Black | Male | 5 | 7.1 | ASCVD | None | Normal |
| 4 | CONTROL | 31 | Black | Male | 8 | 7.2 | GSW | None | Normal |
| 5 | CONTROL | 46 | Black | Male | 9 | 7.1 | Multiple injuries | None | Normal |
| 6 | CONTROL | 33 | White | Male | 15 | 7.0 | GSW | None | Normal |
| 7 | CONTROL | 48 | White | Male | 26 | 6.9 | ASCVD | None | Normal |
| 8 | CONTROL | 40 | White | Female | 7 | 7.0 | ASCVD | None | Normal |
| 9 | CONTROL | 23 | Black | Male | 15 | 6.8 | GSW | None | Normal |
| 10 | CONTROL | 38 | Black | Male | 16 | 6.9 | Lung Sarcoidosis | None | Normal |
| 11 | CONTROL | 65 | Black | Female | 23 | 6.9 | ASCVD | None | Normal |
| 12 | CONTROL | 35 | White | Male | 24 | 6.9 | Crush injury to abdomen and chest | None | Normal |
| 13 | CONTROL | 52 | White | Male | 30 | 7.3 | ASCVD | None | Normal |
| 14 | CONTROL | 63 | White | Female | 30 | 7.1 | Ovarian cancer | None | Normal |
| 15 | CONTROL | 37 | White | Male | 24 | 7.0 | ASCVD | None | Normal |
| 16 | CONTROL | 45 | White | Male | 22 | 7.3 | ASCVD | None | Normal |
| 17 | CONTROL | 26 | White | Male | 12 | 6.9 | Arrhythmia | None | Normal |
| 18 | CONTROL | 72 | White | Female | 23 | 6.9 | MVA | None | Normal |
| 19 | CONTROL | 42 | White | Female | 23 | 6.9 | Mitral valve prolapse | None | Normal |
| 20 | CONTROL | 47 | White | Male | 10 | 7.0 | ASCVD | None | Normal |
| 21 | CONTROL | 31 | White | Male | 16 | 7.2 | MVA | None | Normal |
| 22 | CONTROL | 60 | White | Male | 15 | 7.1 | Accidental drowning | None | Normal |
| 23 | CONTROL | 28 | White | Male | 13 | 6.8 | Electrocution | None | Normal |
| 24 | CONTROL | 45 | White | Female | 16 | 6.9 | Cardiac arrhythmia | None | Normal |
| 25 | CONTROL | 62 | White | Male | 19 | 7.0 | Cardiac arrest | None | Normal |
| Depressed suicide victimsb | |||||||||
| 1 | SUICIDE | 27 | White | Male | 24 | 7.0 | GSW | None | MDD, Ethanol abuse |
| 2 | SUICIDE | 44 | White | Female | 11 | 7.2 | Drug OD | Nortriptyline | MDD, Ethanol abuse |
| 3 | SUICIDE | 36 | White | Female | 10 | 7.1 | GSW | None | MDD |
| 4 | SUICIDE | 24 | White | Male | 7 | 7.1 | GSW | Ethanol | MDD |
| 5 | SUICIDE | 43 | White | Male | 12 | 7.0 | Drug OD | None | MDD, Polysubstance Abuse |
| 6 | SUICIDE | 53 | White | Male | 23 | 6.9 | Jump from height | None | MDD |
| 7 | SUICIDE | 41 | White | Female | 27 | 7.1 | Drug OD | Amitriptyline, Desipramine, Nortriptyline, Ethanol | MDD, Ethanol abuse |
| 8 | SUICIDE | 22 | Black | Female | 16 | 7.3 | Drug OD | None | MDD |
| 9 | SUICIDE | 46 | White | Female | 21 | 6.9 | Drug OD | Amitriptyline, Desipramine, Ethanol | MDD |
| 10 | SUICIDE | 36 | White | Female | 18 | 7.2 | GSW | None | MDD |
| 11 | SUICIDE | 38 | White | Male | 24 | 7.0 | Drug OD, Ethanol intoxication | Ethanol | MDD, Ethanol abuse |
| 12 | SUICIDE | 46 | White | Female | 16 | 6.8 | Drug OD | Nortriptyline | MDD, Panic disorder with agoraphobia |
| 13 | SUICIDE | 23 | White | Male | 12 | 7.0 | Hanging | Paroxetine | MDD |
| 14 | SUICIDE | 30 | White | Male | 17 | 7.1 | Hanging | Venlafaxine | MDD |
| 15 | SUICIDE | 44 | White | Female | 30 | 7.2 | Drug OD, Ethanol intoxication | Fluoxetine, Ethanol | MDD, Ethanol abuse, Opioid abuse |
| 16 | SUICIDE | 74 | White | Female | 27 | 7.0 | Venlafaxine OD | Venlafaxine, Ethanol | MDD, Ethanol abuse |
| 17 | SUICIDE | 25 | White | Male | 14 | 6.8 | Hanging | Ethanol | MDD |
| 18 | SUICIDE | 23 | Black | Male | 23 | 6.9 | Hanging | None | MDD |
| 19 | SUICIDE | 63 | White | Male | 19 | 6.9 | Drug OD, Ethanol intoxication | Ethanol | MDD |
| 20 | SUICIDE | 67 | White | Male | 22 | 7.0 | GSW | Fluoxetine, Venlafaxine | MDD |
| 21 | SUICIDE | 40 | White | Female | 20 | 7.0 | Drug OD | Xanax | MDD |
| 22 | SUICIDE | 53 | White | Male | 26 | 7.1 | Stabbing | Sertraline | MDD |
| 23 | SUICIDE | 68 | White | Female | 26 | 6.8 | GSW | Amitriptyline | Bipolar Disorder |
| 24 | SUICIDE | 31 | White | Female | 31 | 6.8 | Hanging | Trazadone, Ethanol | Bipolar Disorder, Bulimia, OCD |
| 25 | SUICIDE | 51 | White | Female | 28 | 6.9 | Amitriptyline OD | Amitriptyline, Ethanol | Bipolar Disorder, Ethanol abuse |
Abbreviations: ASCVD, atherosclerotic cardiovascular disease; CO, carbon monoxide; DKA, diabetic ketoacidosis; GSW, gunshot wound; MDD, major depressive disorder; MVA, motor vehicle accident; OCD, obsessive–compulsive disorder; OD, overdose; PMI, postmortem interval.
Mean±SD age is 42.8±13.58 years; PMI, 17.72±7.00 h; and brain pH 7.01±0.15.
Mean±SD age is 41.92±15.07 years; PMI, 20.16±6.68 h; and brain pH 7.00±0.14.
RNA isolation and reverse transcription
Total RNA was extracted from 100 mg of tissue using the Trizol® (Invitrogen, Carlsbad, California) according to the manufacturer’s directions. RNA concentration and purity were determined by measuring the OD A260/A280 and A260/A230 using NanoDrop®ND-1000 (NanoDrop Technologies, Montchanin, Delaware). All samples were free of contaminants with absorbance ratios close to two. RNA quality was assessed using Agilent Bioanalyzer 2100. All samples had 28S/18S ratios >1.2 and RNA integrity number (RIN) above 6.6. First-strand cDNA was synthesized from 1 μg of total RNA using MMLV-reverse transcriptase (Invitrogen) in the presence of random hexamers (2.5 μM) (Invitrogen) according to manufacturer’s instructions.
Oligonucleotide primers
Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA) was used to design primers for the amplification of human GPM6A, GPM6B, PLP1, CORO1A, GIT1, CAMK2A, BDNF and five candidate internal reference genes (GAPDH, YWHAZ, CYC1, PPIA, and EIF4A2). Primer sequence, full gene name, accession number, function, and chromosomal localization are listed in Table 2 (reference genes) and Table 3 (target genes).
Table 2.
Primer sequences for reference genes selected for evaluation of expression stability
| Symbol | Entrez Gene ID | Name | Function | Forward primer | Reverse primer | Location |
|---|---|---|---|---|---|---|
| GAPDH | 2597 | glyceraldehyde-3-phosphate dehydrogenase | Catalysis of oxidative phosphorylation of glyceraldehyde-3-phosphate in the presence of NAD | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 12p13 |
| PPIA | 5478 | peptidylprolylisomerase A (cyclophilin A) | Peptidyl-prolyl cis–trans isomerase | GGCAAATGCTGGACCCAACACA | TGCTGGTCTTGCCATTCCTGGA | 7p13 |
| CYC1 | 1537 | cytochrome c-1 | Electron transporter | CCAGATAGCCAAGGATGTGTGC | GACTGACCACTTGTGCCGCTTT | 8q24.3 |
| EIF4A2 | 1974 | eukaryotic translation initiation factor 4A2 | Translation initiation factor | CTCTCCTTCGTGGCATCTATGC | TGGCTGTCTTGCCAGTACCTGA | 3q28 |
| YWHAZ | 7534 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Signal transduction by binding to phosphoserine-containing proteins | ACCGTTACTTGGCTGAGGTTGC | CCCAGTCTGATAGGATGTGTTGG | 8q23.1 |
Table 3.
Primer sequences for target genes
| Symbol | Entrez Gene ID | Name | Forward primer | Reverse primer | Location |
|---|---|---|---|---|---|
| BDNF | 627 | Brain-derived neurotrophic factor | CATCCGAGGACAAGGTGGCTTG | GCCGAACTTTCTGGTCCTCATC | 11p13 |
| GPM6A | 2823 | Glycoprotein M6A | ACCTTGACGCTCTTCTTTTCATTATT | CAATGGTCAAATCAGTGCACAA | 4q34 |
| GPM6B | 2824 | glycoprotein M6B | GAATGTTCGTTTTCCTCACCTATGT | GGCACCGCTGAGAAACCA | Xp22.2 |
| PLP1 | 5354 | proteolipid protein 1 | GGCCAACATCAAGCTCATTCTT | AGGTGATGCCCACAAACTTGT | Xq22 |
| CAMK2A | 815 | Calcium/calmodulin-dependent protein kinase II alpha | AGCCAAGGATCTGATCAATAAGATG | GGCTTCGGCAGCTGTGA | 5q32 |
| GIT1 | 28,964 | G protein-coupled receptor kinase interacting ArfGAP 1 | CCGAGAGTTTGCCACCTTGATC | ACGCTGTCGTAGTCGTGTTGGT | 17p11.2 |
| CORO1A | 11,151 | Coronin, actin binding protein, 1A | CCAACATCGTCTACCTCTGTGG | CTCACACTTGTTCACCTCCAGG | 16p11.2 |
Quantitative polymerase chain reaction (qPCR)
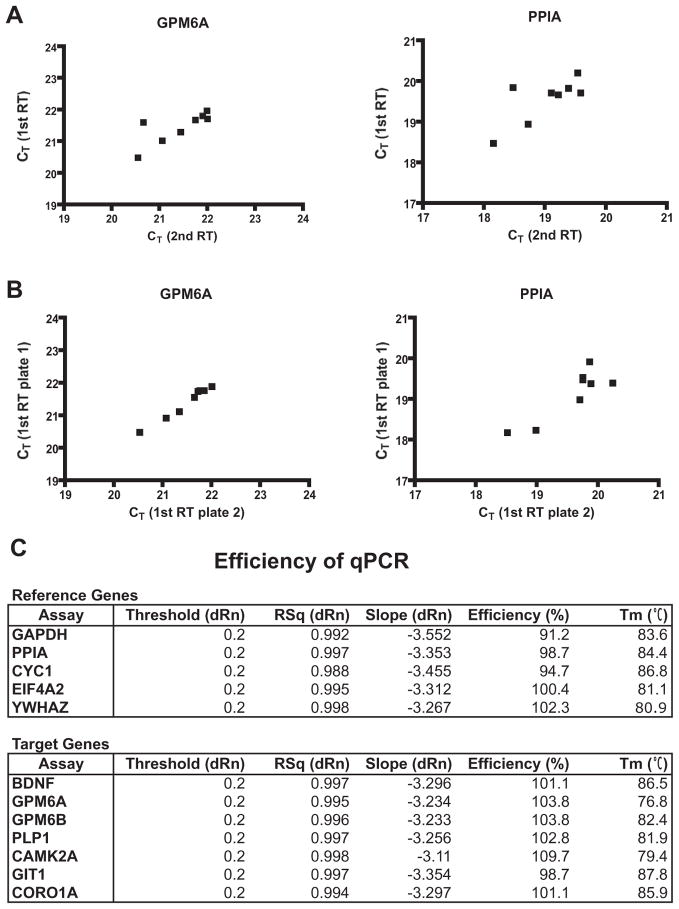
To determine mRNA levels, qPCR reactions were performed using SYBR®Select Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Measurements were done on Stratagene Mx3005P equipped with MxPro software (Stratagene, La Jolla, CA, USA). All reactions were performed in duplicate. No-RT control, no-template control and three different inter-run calibrators were always included for each individual plate and gene assay. After the final cycle of PCR, reactions were subjected to a heat dissociation protocol to verify primer specificity. A single peak corresponding to the melting temperature (Tm) of expected qPCR assay product was observed for all pairs of primers assayed (Fig. 1C). Reproducibility of the reverse transcription (RT) reaction and inter-run reproducibility of qPCR was evaluated by measuring twice the expression of GPM6A and PPIA in eight randomly selected samples as shown in the Fig. 1A (reproducibility of two independent RT reactions) and 1B (reproducibility of two independent qPCR runs) (Fig. 1A, B). Relative expression levels, reported as fold change (FC), were determined by the relative standard curve method as described in the Applied Biosystems Guide to Performing Relative Quantitation of Gene Expression using Real-Time Quantitative PCR. Standard curves were constructed using 10-fold serial dilutions of cDNA derived from all subjects and the linear range and PCR reaction efficiency of each gene assay were determined. All gene assays were found to have r2 ≥ 0.988 and efficiencies between 90 and 110 percent (Fig. 1C). Raw expression data (CT values) were analyzed by qBasePLUS software (Biogazelle, Zwijnaarde, Belgium). Data are normalized to the normalization factor calculated as a geometric mean of the expression of three reference genes and normalized values are used for further statistical analysis (Vandesompele et al., 2002). Outliers were excluded before analysis if the normalized values were greater than two standard deviations from the group mean.
Fig. 1.
(A) Correlation studies to evaluate reproducibility of reverse transcription (RT) reaction. Equal amounts of RNA extracted from eight samples were reverse transcribed in two independent RT reactions (1st RT and 2nd RT) and used as a template for qPCR to measure twice the expression of GPM6A and PPIA. Both genes show a statistically significant correlation (GPM6A Pearson coefficient=0.7702, *p value (two-tailed)=0.0254; PPIA Pearson coefficient=0.7452, *p value (two-tailed)=0.0339). (B) Correlation studies to evaluate inter-run reproducibility qPCR reaction. Two identical replica plates containing eight cDNA samples obtained from 1st RT reaction were prepared and used to measure the expression of GPM6A and PPIA in two independent runs. Both genes show a statistically significant correlation (GPM6A Pearson coefficient=0.9882, ***p value (two-tailed)< 0.0001; PPIA Pearson coefficient=0.8709, **p value (two-tailed)=0.0049). (C) Efficiency data for candidate reference genes and target genes. CT values obtained from 10-fold serial dilutions of cDNA were plotted against dilution factors and the reaction efficiency was calculated using qBasePLUS software. RT, reverse transcription; CT, treshold cycle; RSq, r-squared; Rn, normalized reporter signal; dRn, delta Rn (Rn minus the baseline); Tm, melting temperature.
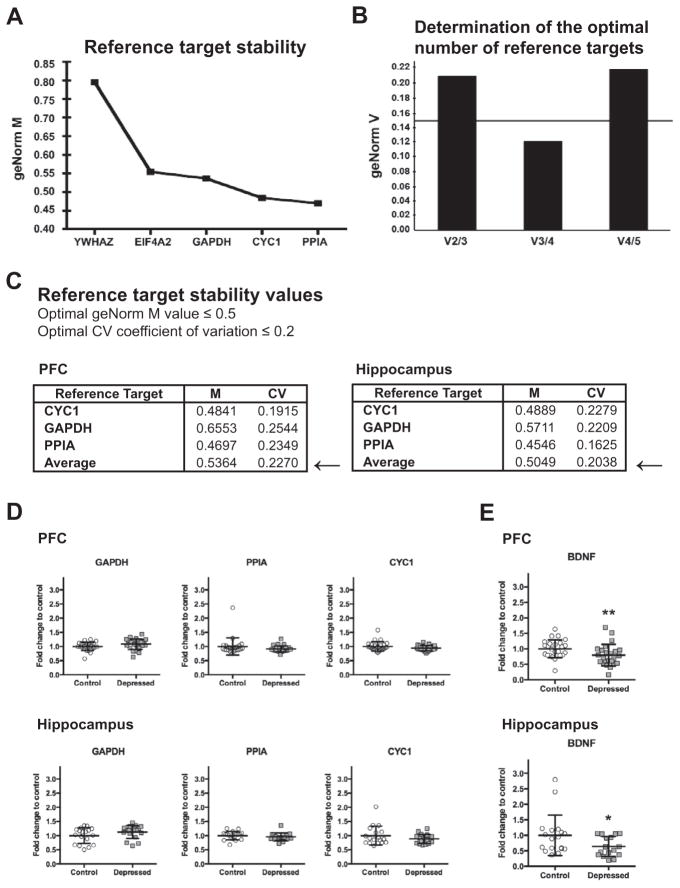
Determination of reference targets for qPCR data normalization
The algorithm geNorm of qBasePLUS software was used to evaluate expression stability of reference genes used for qPCR data normalization as described (Vandesompele et al., 2002). Gene stability of five candidates (EIF4A2, YWHAZ, GAPDH, CYC1, and PPIA) was evaluated. Special attention was paid to selecting genes that belong to different functional classes, which significantly reduces the chance that genes might be coregulated (see the Table 2 for full gene name, accession number, function, chromosomal localization, and primer sequences).
The reference targets PPIA, CYC1, and GAPDH were determined as the most stably expressed genes and the optimum number of three reference candidates was found adequate for the optimal normalization of data in our experimental setting (Fig. 2A–C). Fig. 2D shows that control and suicide samples did not differ with respect to GAPDH, CYC1 and PPIA mRNA expression levels. The geometric mean of GAPDH, CYC1, and PPIA was further used for normalization in all relative quantitation assays.
Fig. 2.
Expression stability of five candidate reference genes in human brain samples calculated by geNorm algorithm using qBasePLUS software (Biogazelle). GeNorm analysis was performed on the gene expression data from PFC and hippocampal postmortem samples obtained from all depressed suicide subjects and matched nonpsychiatric controls. (A) Average expression stability values (M) of five candidate genes from the least stable to the most stable in PFC. The gene stability measure M is defined as the average pairwise variation of a particular gene with all other control genes (Vandesompele et al., 2002). Genes with lowest values have the most stable expression (optimal geNorm M ≤ 0.5). In the PFC samples, the geNorm gene stability measurement values M were ranked in the following order (from the least to the most stable): YWHAZ, EIF4A2, GAPDH, CYC1, and PPIA. (B) Determination of the optimum number of reference genes required for adequate data normalization based on pairwise variation (Vn/Vn+1) between candidate genes in the PFC (optimal geNorm V<0.15). Normalization to a single control gene can lead to erroneous normalization. For this reason, geNorm also calculates the pairwise variation value (Vn/n+1) that shows the effect of adding further reference genes on the normalization factor. When analyzing Vn/n+1, we observed that the stepwise inclusion of individual reference genes showed a decrease until a minimal value (V3/4). The next addition of other reference gene determined an increase of V4/5, suggesting that there was a decrease of expression stability due to the inclusion of a relatively unstable fifth gene. Thus, the adequate number of reference targets in this experimental situation is three when comparing a normalization factor based on three or four most stable targets. (C) Reference target stability values (M and CV) determined for the combination of the reference targets GAPDH, CYC1, and PPIA in human brain samples from the PFC and hippocampus. To assess that the genes with the lowest M values have indeed the most stable expression, the gene-specific variation of each control gene is determined as the variation coefficient (CV) of the expression levels after normalization. Mean CV values lower than 0.2 are typically observed for stably expressed reference genes in relatively homogenous sample panels. As such, in human postmortem PFC and hippocampal samples, the optimal normalization factor can be calculated as the geometric mean of reference targets PPIA, CYC1, and GAPDH. (D) Expression levels of reference genes GAPDH, PPIA andCYC1 in the PFC and in the hippocampus of non-psychiatric controls (Control: PFC n=25 for PPIA and CYC1, n=24 for GAPDH; Hippocampus n=18 for GAPDH, PPIA and CYC1) and depressed (Depressed: PFC n=25 for GAPDH, PPIA and CYC1; Hippocampus n=18 for GAPDH, PPIA and CYC1) suicide subjects. Results are expressed as fold change in mRNA levels. Values are fold change±SD. No significant differences between the depressed suicide group and the control group were determined for any of the genes. (E) Expression levels of BDNF in the PFC and in the hippocampus of non-psychiatric controls (Control: PFC n=25, Hippocampus n=18) and depressed (Depressed: PFC n=24, Hippocampus n=16) suicide subjects. Results are expressed as fold change in mRNA levels. Values are fold change±SD. BDNF mRNA levels were significantly decreased in the depressed subjects compared with controls in both the PCF and the hippocampus (F=16.39, p=0.0001, and F=12.65, p=0.0006, respectively). Significant effects are marked by asterisks (Bonferroni corrected significance threshold, after correction: asterisk, P<0.05; two asterisks, P<0.01).
Immunoprecipitation (IP) and mass spectrometry
Hippocampal tissue from a two-month-old female Wistar rat was dissected and collected in lysis buffer (150 mM NaCl, 50 mM Tris/HCl, 1% deoxycholate sodium salt, 0.1% SDS, pH 8) supplemented with protease inhibitor cocktail (Sigma–Aldrich, St Louis, MO, USA) to prepare the protein extract. The precleared protein extract was then incubated with the polyclonal antibody against the C-terminus of rat GPM6A developed in our laboratory (Fernandez and Frasch, unpublished results) covalently bound to Protein A–Sepharose via dimethyl pimelimidate (Sigma–Aldrich) as described (Harlow and Lane, 1988). As a control, non-immune rabbit serum was used. Alkylation of the samples to block cysteine residues was performed by treatment with iodoacetamide (Sigma–Aldrich) at the final concentration of 20 mM. The immunoprecipitates were analyzed by SDS–PAGE followed either by Western blotting or by mass spectrometry. In the second case, polyacrylamide gels were silver stained by the mass spectrometry compatible staining method. Only specifically bound proteins not detectable in control immunoprecipitates were excised from the gel, subjected to tryptic digest and analyzed by mass spectrometry.
Peptide sequencing
In order to obtain internal peptide sequences, in-gel tryptic digestion was performed as described (Hellman et al., 1995), peptides were separated using reversed-phase HPLC in a 150-mm × 1-mm Kromasil C18 column, and sequenced in an Applied Biosystems, model 494 A Procise Automatic Sequencer (Applied Biosystems), run according to the manufacturer’s instructions.
Matrix-assisted laser desorption-ionization time-of-flight-MS (MALDI-TOF-MS)
MALDI-TOF-MS was performed on a Bruker Autoflex, operated for analysis of intact proteins in linear mode and externally calibrated with trypsinogen (23.98 kDa), protein A (44.61 kDa) and BSA (66.43 kDa). Prior toanalysis, the sample was desalted by micro-reversed-phase chromatography. The crystals were prepared by the double-layer technique using sinapinic acid as matrix. For peptide mass fingerprinting, tryptic digests were analyzed using a dried droplet preparation with alfa-cyano 4-hydroxy cinnamic acid as matrix. Calibration was done internally, with autolytic peptides from the porcine trypsin used for digestion. The manufacturer’s instructions were followed, and the resulting peptide mass lists were used to scan the latest NCBInr sequence database for protein identity using the search engine ProFound.
Plasmid transfection, SDS–PAGE, and Western blot
COS7 cells cultured in Dulbecco’s modified Eagle’s medium with 10% (v/v) fetal bovine serum, penicillin, and streptomycin were transfected with plasmids encoding a green fluorescent protein (GFP) or a fusion protein GPM6A-GFP using Lipofectamine®2000 (Invitrogen) following the manufacturer’s instructions. Twenty-four hours later, cells were lysed and the protein samples for SDS–PAGE were prepared as described (Fuchsova et al., 2009). The protein samples were separated on 10% SDS–PAGE gels and transferred to a nitro-cellulose membrane by electroblotting. The membranes were blocked in Tris-buffered saline containing 5% nonfat dried milk and incubated with the rabbit anti-GFP polyclonal serum (1/750) (Invitrogen) or with the polyclonal anti-GPM6A antibody (1/400) developed in our laboratory. Antigen–antibody complexes were detected using an anti-rabbit horseradish peroxidase-linked secondary antibody (1/10,000) (Dako, Glostrup, Denmark) according to standard Enhanced Chemiluminescence protocol using SuperSignal West Pico/Femto chemiluminescent substrate (Pierce, Rockford, IL, USA).
Statistical analysis
Data were analyzed by analysis of covariance (ANCOVA) independently for each brain region and gene of interest to compare control subjects and depressed suicide victims adjusting the effects of age, postmortem interval (PMI), and brain pH. For multiple comparisons, Bonferroni correction to maintain alpha at 0.05 was used to adjust the type I error rates. Results are reported as individual values and group mean±SD. For correlation analysis of mRNA expression levels, pairwise Pearson correlation coefficients along with its 95% confidence intervals, and p value (two-tailed) were calculated on normalized mRNA expression levels. Pearson correlation values p<0.05 were considered significant. Chi-square analyses were conducted to determine whether the frequency of significant correlations differed between groups. Statistical analysis and graphs were done with GraphPad Prism software.
RESULTS
Demographic characteristics
Detailed demographic characteristics of suicide and control subjects are provided in Table 1. The age range of suicide subjects and controls was between 22 and 74 years, and the PMI was between 5 and 31 h. There were no significant differences between suicide subjects and their matched controls in age (p=0.8292, unpaired t-test, two-tailed) or PMI (p=0.2132, unpaired t-test, two-tailed). The mean brain pH±SD in controls was 7.01±0.15 and in suicides was 7.00±0.14, which was not significantly different (p=0.8436, unpaired t-test, two-tailed). The mean RIN±SD in the PFC was 7.21±0.56 in the control group and 7.23±0.61 in the depressed group. In the hippocampus, the mean RIN±SD was 7.16±0.72 in the control group and 7.45±0.83 in the depressed group. There was no difference in RIN values between controls and depressed suicide subjects in both the PCF and the hippocampus (p=0.7 and p=0.37; respectively, unpaired t-test, two-tailed).
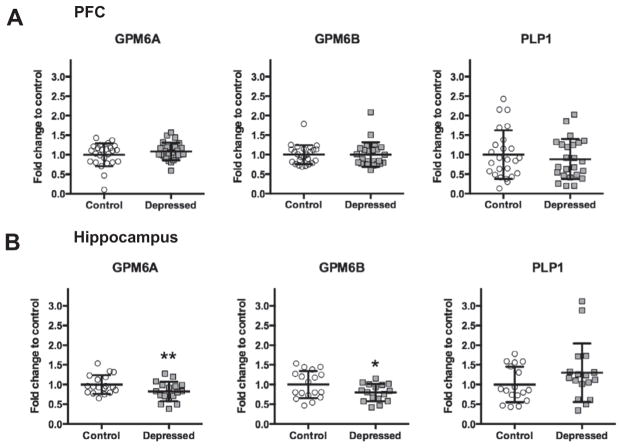
GPM6A expression is downregulated in the hippocampus of depressed suicides
To determine whether the altered expression of GPM6A is associated with the pathophysiology of human depression, its mRNA level was measured in the entire cohort by qPCR. Stress responsiveness and cellular function is conserved among the members of the PLP/DM20 family (Fernandez et al., 2010). Thus, we also examined GPM6B and PLP1 expression.
Fig. 3A, B show mRNA levels of GPM6A, GPM6B, and PLP1 in the PFC (Fig. 3A) and in the hippocampus (Fig. 3B) of depressed suicides and matched non-psychiatric controls. In the PFC, no significant differences were observed between the control and depressed groups for any of the members. On the other hand, F-test of ANCOVA showed that in the hippocampus, the mean GPM6A mRNA level was significantly decreased in the depressed group (F=14.55, p=0.0002, FC=0.82). Closely related GPM6B also demonstrated a significant decrease (F=12.98, p=0.0005, FC =0.80) and no changes were observed for PLP1.
Fig. 3.
mRNA levels of GPM6A, GPM6B, and PLP1 in PFC (A) and in the hippocampus (B) of non-psychiatric controls (Control: PFC n=25 for GPM6A, GPM6B, and PLP1; Hippocampus n=18 for GPM6A and GPM6B, n=17 for PLP1) and depressed (Depressed: PFC n=25 for GPM6A and GPM6B, n=24 for PLP1; Hippocampus n=17 for GPM6A and PLP1, n=16 for GPM6B) suicide subjects normalized to the geometric mean of the three reference targets (GAPDH, CYC1, PPIA). Results are expressed as fold change in mRNA levels. Values are fold change±SD. Significant differences between the depressed suicides and the control group were determined for GPM6A (F=14.55, p=0.0002) and GPM6B (F=12.98, p=0.0005) in the hippocampal tissue. Significant effects are marked by asterisks (Bonferroni corrected significance threshold, after correction: asterisk, P<0.05; two asterisks, P<0.01).
As a control, we evaluated the expression of BDNF because the decreased BDNF mRNA level in postmortem PFC and hippocampus of depressed suicides has been reported frequently in the past (Dwivedi et al., 2003; Duman and Monteggia, 2006). As expected, BDNF mRNA levels were significantly decreased in the depressed subjects compared with controls in both the PCF and the hippocampus (F=16.39, p=0.0001, FC=0.79, and F=12.65, p=0.0006, FC=0.64; respectively) (Fig. 2E). The age, PMI and brain pH had no significant effects on mRNA expressions of GPM6A, GPM6B, and PLP1.
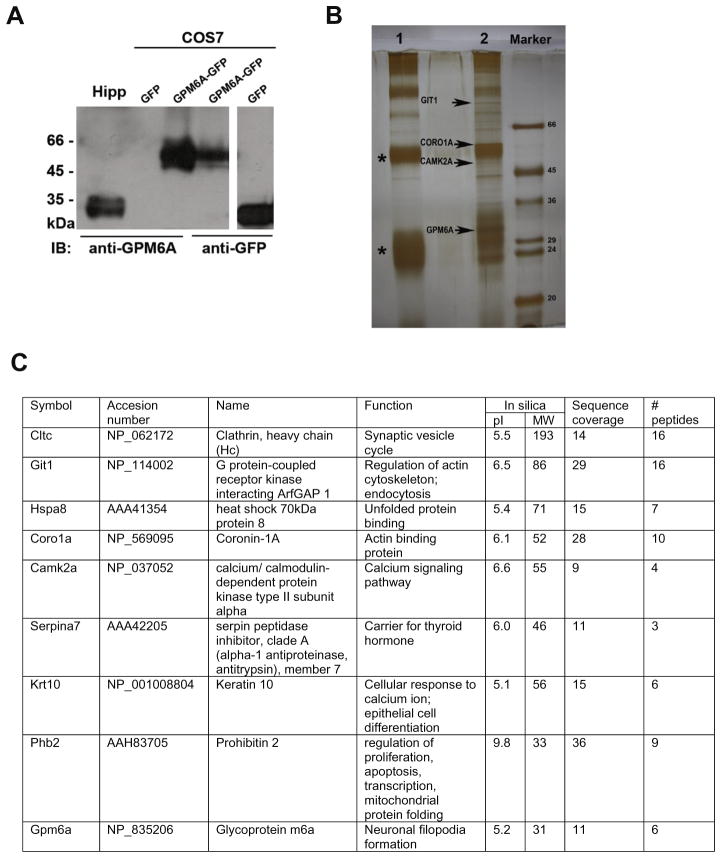
Coronin1A (CORO1A), calcium/calmodulin-dependent protein kinase II alpha (CAMK2A), and GIT1 co-immunoprecipitate with the stress-responsive glycoprotein GPM6A
To identify the underlying molecular mechanism through which GPM6A affects neuronal remodeling and plasticity, we searched for proteins that associate with GPM6A. For this purpose, first, the polyclonal antibody against the C-terminus of GPM6A was developed and purified. Western blot in the Fig. 4A shows that the anti-GPM6A antibody recognizes the endogenous GPM6A (~35 kDa) in the lysate from rat hippocampus (Hipp; 1st lane) and the exogenous GPM6A overexpressed as a GFP-fusion protein (~62 kDa) in non-neuronal cell line COS7 (3rd lane). No signal was detected when COS7 were transfected with GFP alone (2nd lane). The polyclonal anti-GFP antibody recognizes both GPM6A-GFP and GFP when overexpressed in COS7 cells (4th–5th lane; respectively).
Fig. 4.
(A) Polyclonal antibody against C-terminus of GPM6A specifically recognizes endogenous as well as exogenous GPM6A protein. Western blot analysis of lysates from adult rat hippocampus (Hipp) and from non-neuronal cell line COS7 overexpressing exogenous GPM6A as a GFP fusion protein (COS7) shows a protein migrating at ~35 kDa (1st lane) corresponding to endogenous GPM6A and a protein migrating at ~62 kDa (3rd lane) corresponding to exogenous GPM6A overexpressed as a GFP fusion protein (COS7). No additional nonspecific bands were detected. No signal was detected when COS7 cells were transfected with GFP alone (2nd lane). The polyclonal anti-GFP antibody recognizes both GPM6A-GFP as well as GFP when overexpressed in COS7 cells (4th and 5th lane respectively). (B) SDS-PAGE analysis of proteins co-immunoprecipitating with GPM6A. The endogenous GPM6A was immunoprecipitated from adult rat hippocampus using the polyclonal antibody against C-terminal end of GPM6A coupled to protein A-Sepharose (lane 2). As a control nonimmune rabbit serum covalently coupled to protein A–Sepharose was used (lane 1). Coimmunoprecipitating proteins were visualized by silver staining technique compatible with mass spectrometry. The proteins of interest were excised from the gel, subjected to tryptic digest and analyzed by mass spectrometry. G protein-coupled receptor kinase-interacting protein 1 (GIT1), coronin-1A (CORO1A), and calcium/calmodulin-dependent protein kinase 2 aplha (CAMK2A) were identified among proteins that coimmunoprecipitate with GPM6A (arrows). (C) Complete list of proteins co-immunoprecipitating with GPM6A as identified by mass spectrometry.
Subsequently, immunoprecipitation of the endogenous GPM6A from the rat hippocampal lysate was performed using the anti-GPM6A antibody covalently coupled to protein A-Sepharose (Fig. 4B, lane 2). As a control, non-immune rabbit serum covalently coupled to protein A-Sepharose was used (Fig. 4B, lane 1). Using mass spectrometry, GPM6A protein was correctly identified in the immunoprecipitate. In addition, among other proteins, cytoskeleton- and neuroplasticity-related proteins, such as GIT1, CORO1A, and CAMK2A, were identified to coimmunoprecipitate with GPM6A (Fig. 4B, arrows; see the table in the Fig. 4C for the complete list of identified proteins).
Altered expression of CORO1A, GIT1, and CAMK2A in the postmortem brain of depressed suicides
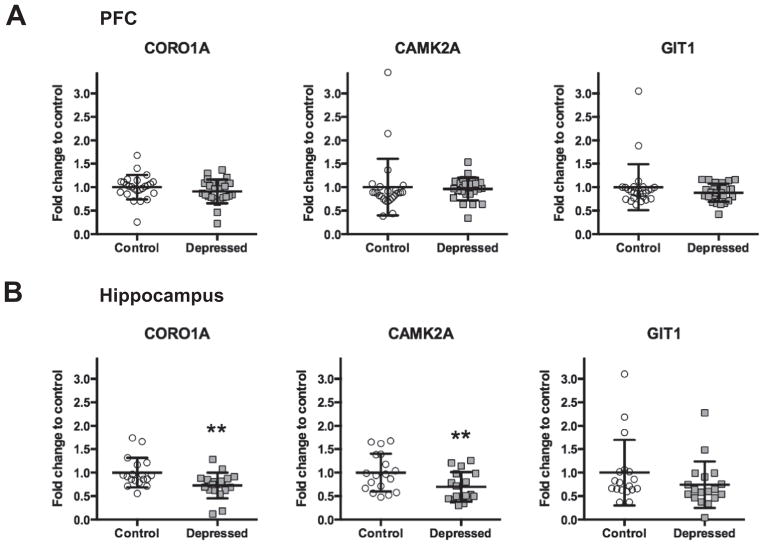
Next, we evaluated whether the expression of the genes encoding the proteins identified to form complexes with GPM6A (namely GIT1, CORO1A, and CAMK2A) is altered in the postmortem brain of depressed suicide subjects. Fig. 5 shows mRNA levels of CORO1A, GIT1, and CAMK2A in the PFC (Fig. 5A) and in the hippocampus (Fig. 5B) of depressed suicides and matched non-psychiatric controls.
Fig. 5.
mRNA levels of CORO1A, GIT1, and CAMK2A in the PFC (A) and in the hippocampus (B) of non-psychiatric controls (Control: PFC n=25 for CORO1A, GIT1, and CAMK2A; Hippocampus n=18 for CORO1A, GIT1, and CAMK2A) and depressed (Depressed: PFC n=25 for CORO1A, GIT1, and CAMK2A; Hippocampus n=18 for CORO1A and GIT1, n=17 for CAMK2A) suicide subjects normalized to the geometric mean of the three reference targets (GAPDH, CYC1, PPIA). Results are expressed as fold change in mRNA levels. Values are fold change±SD. Significant differences between the depressed suicide group and the control group were determined for CORO1A and for CAMK2A in the hippocampal tissue (F=25.45, p<0.0001, and F =18.47, p<0.0001; respectively). Significant effects are marked by asterisks (Bonferroni corrected significance threshold, after correction: asterisk, P <0.05; two asterisks, P<0.01).
For CORO1A and CAMK2A, our ANCOVA analysis showed significant differences between the depressed and the control group in the hippocampus (F=25.45, p<0.0001, FC=0.73, and F=18.47, p<0.0001, FC=0.70; respectively). We did not observe any significant changes for CORO1A and CAMK2A in the PFC. Expression of GIT1 mRNA did not differ in either of the tissues. The age, PMI and brain pH had no significant effects on mRNA expressions of CORO1A, CAMK2A, and GIT1.
Coordinated expression
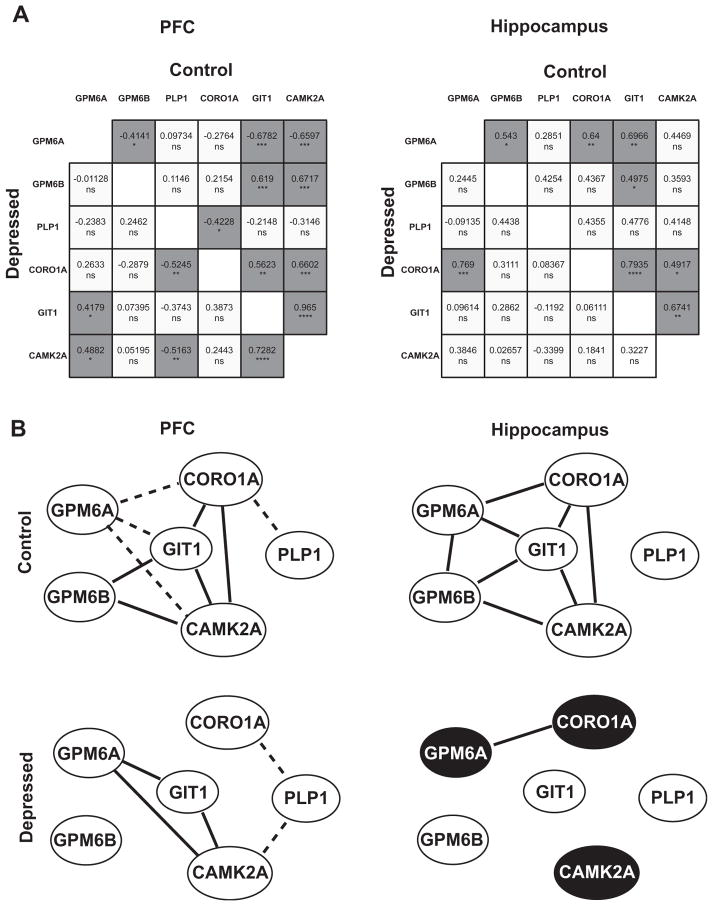
To determine whether interrelationships existed among the GPM6A levels and GIT1, CORO1A, and CAMK2A expression, and among the expression of the two other members of PLP/DM20 family—GPM6B and PLP1—cross-correlation analyses were conducted independently for each brain region among the depressed suicides and in the control group. This type of analysis allows us to detect coordinated changes in gene expression if the pathway is affected by disease. Pearson correlation coefficients and p values were calculated for all genes on normalized mRNA expression levels for the control group and the depression group separately in the PFC and in the hippocampus.
Fig. 6A shows correlation matrices where statistically significant Pearson correlation p values are indicated by asterisks. In both the hippocampus and the PFC of control individuals, significant interrelationships were frequent. Specifically, in the control PFC, 9 of 15 possible correlations were statistically significant. Within the hippocampus of control individuals, 7 of 15 correlations were statistically significant. The correlations evident in controls differed from the correlations that were significant in the suicide brain. The reduction of correlations was observed in the PFC, where only five correlations were significant, as well as in the hippocampus, where the number of significant correlations declined to one. In the hippocampus, the difference in the frequency of significant correlations between the depressed suicide group and the control group was statistically significant (chi-square=6.136, p=0.0132).
Fig. 6.
Coordinated expression is decreased in the brain of depressed suicides. (A) Correlation analysis of mRNA expression levels in the PFC and in the hippocampus. In the matrix all pairwise Pearson correlation coefficients calculated on normalized mRNA expression levels are shown. Statistically significant Pearson correlation p values (two-tailed) are indicated by asterisks (*<0.05, **<0.01, ***<0.001, ns=no significant; dark gray indicates a statistically significant correlation, light gray indicates a non significant correlation). In suicide brain (lower portion of the correlation matrix) significant correlations appear less frequently than in controls (upper portion of the correlation matrix). Decrease in the frequency of significant correlations between controls and depressed suicides is statistically significant in the hippocampus (chi-square=6.136, *p=0.013; in PFC chi-square=2.143, p=0.1432). (B) Network of the gene coexpression pattern in the PFC and in the hippocampus in control and depressed groups. Gene pairs that show a significant correlation are indicated: a positive significant correlation (full line); a negative significant correlation (dashed line). Dark circles indicate differentially expressed genes. Differences between control and depressed groups are observed in both tissues.
Fig. 6B depicts the network of the gene coexpression patterns in the control and depressed groups to illustrate our findings. In both tissues of the control group, correlated genes formed an extensive inter-connected network, suggesting they function in common signaling cascades. In the depressed group, this coordinated gene expression is considerably altered in the PFC and notably, it completely disappears in the hippocampus, where the only correlation detected was between GPM6A and CORO1A.
Effect of psychotropic drugs and gender
Since several subjects in the depressed suicide group were on antidepressant medication at the time of death and/or suffered from comorbid substance abuse, we examined if the presence of psychotropic drugs had any influence on the mRNA expression of studied genes. In the depressed suicide group, there were 14 subjects who were on different types of antidepressants and seven subjects who were free of any psychotropic medication. When we compared the mRNA expression of studied genes, we did not find any significant differences between these two groups, suggesting that the presence of antidepressants did not have a significant influence on the expression of GPM6A, GPM6B, CORO1A, PLP1, GIT1, and CAMK2A. Thus, the differences in the GPM6A, GPM6B, CORO1A, and CAMK2A expression in the hippocampus between the depressed suicide group and normal controls do not appear to be related to the presence of psychotropic drugs.
To assess the effect of gender on gene expression we analyzed the data by a two-way ANOVA independently for each brain region with diagnosis and gender as categorical independent variables adjusting the effects of age, PMI, and brain pH. The only gene expression where a significant gender by diagnosis interaction was revealed was GPM6B expression in the PFC (F=8.31, p=0.0062). No effect on GPM6B expression was detected in the hippocampus. To further investigate this gender by diagnosis effect on GPM6B expression in PFC, we conducted ANCOVA analysis on males and females separately. A significant decrease in the GPM6B expression was observed in depressed male subjects (F=10.63, p=0.0016) but not in female subjects.
DISCUSSION
In our previous studies, we showed that chronic stress, a factor with a clear role in the etiology of depression, reduces the hippocampal mRNA levels of PLP/DM20 family members in animal models of chronic psychosocial or physical stress (Alfonso et al., 2004b, 2006). In the present work, we demonstrate that, GPM6A mRNA levels are significantly reduced in the hippocampus of depressed suicides. Moreover, the closely related GPM6B, but not PLP1, is downregulated.
Previous studies by in situ hybridization have indicated that PLP1, the major component of myelin sheets, is expressed in glial cells but not neurons, GPM6B is expressed in glial cells and neurons, whereas GPM6A is restricted to the latter (Yan et al., 1996). Extensive evidence supports the misfunction of oligodendrocytes in major depression. Among other alterations, reduced oligodendrocyte numbers in amygdala and PFC have been observed and downregulation of oligodendrocyte-related gene transcripts has been reported in the brain of depressed subjects in regions including the amygdala and PFC (Edgar and Sibille, 2012). In addition, PLP1 itself was shown to be downregulated in the temporal cortex of depressed subjects (Aston et al., 2005). Taking into account the overlap of the functions of PLP1 and GPM6B in developmental myelination (Jahn et al., 2009; Werner et al., 2013), we could hypothesize that reduced GPM6B expression might participate in oligodendrocyte misfunction associated with depression even though the PLP1 expression was not altered in our study.
Chronic stress induces profound behavioral changes in humans and rodents, manifested as depressive-like symptoms, a hyperanxious state, and learning/memory deficits, paralleled by structural damage and impaired synaptic plasticity, mainly in the hippocampus and PFC (Pittenger and Duman, 2008). Stress-induced functional deficits have been shown to propagate from a hippocampus-dependent task to a PFC-dependent task (Cerqueira et al., 2007). We could speculate that region-specific downregulation of PLP family member genes in the hippocampus may reflect a region’s vulnerability to the diseases and involvement of their gene products in underlying disruptions of hippocampus-dependent tasks (Pittenger and Duman, 2008). For example, GPM6A potential involvement in the hippocampus-dependent memory would be in agreement with the impaired long-term memory performance of M6 (GPM6A ortholog in fly)-deficient and overexpressing flies (Gregor et al., 2014) and identification of de novo duplication of GPM6A in a patient with learning disability. In fly, M6 shows particularly strong expression in the fly mushroom body (Zappia et al., 2012), which is important for learning and memory processes in Drosophila (McGuire et al., 2001). Another potential mechanism involved in observed region-specific effects could be neurogenesis known to play a critical role in the pathophysiology of depression. Findings by Michibata et al. (2008) suggest that expression level of GPM6A is directly or indirectly associated with the differentiation of neurons derived from undifferentiated embryonic stem cells. However, the involvement of PLP1 family members in adult neurogenesis has not been explored at the moment.
GPM6A functions in processes of neuronal remodeling and plasticity, such as neurite outgrowth, filopodium formation, and synaptogenesis (Alfonso et al., 2005; Fuchsova et al., 2009; Brocco et al., 2010; Scorticati et al., 2011). Its overexpression induces neurite formation and increases filopodia density in hippocampal neurons. Similarly, GPM6B and DM20, but not PLP1, induce filopodium formation in primary hippocampal neurons (Fernandez et al., 2010).
Dendrites, dendritic filopodia (precursors to spine synapses), and spines are crucial components for synaptic function and plasticity. Changes in their density or morphological features can result in significant alterations in the connectivity of neural systems (Segal, 2005; Chen et al., 2007) and would be expected to result in changes in the neurobehavioral functions subserved by those systems. We suppose that the reduction in GPM6A expression in the depressed suicide brain might be related to the morphological alterations found in the brain of depressed humans.
Synapses between neurons can change functionally and structurally in response to activity in a process known as synaptic plasticity. Apart from alterations in structural plasticity, disruptions in functional synaptic plasticity have also been reported in depression. Among the molecules implicated in synaptic plasticity, CAMK2A has been established as an enzyme of central importance (Lisman et al., 2002; Zhang et al., 2008; Lucchesi et al., 2011). Previous studies have suggested the involvement of CAMK2 in the pathophysiology of psychiatric conditions, including bipolar disorder (Xing et al., 2002), Alzheimer disease (Amada et al., 2005), schizophrenia, and depression (Novak et al., 2006) as well as in the mechanism of action of antidepressants (Lisman et al., 2002; Celano et al., 2003; Tiraboschi et al., 2004; Barbiero et al., 2007; Lucchesi et al., 2011). CAMK2A is expressed postnatally during brain maturation and stabilizes dendritic arbor structure (Wu and Cline, 1998). It is the most abundant protein in the postsynaptic density (PSD) (Cheng et al., 2006), where the activated CAMK2A phosphorylates AMPA receptors and other PSD proteins, which likely contributes to enhancement of synaptic strength (Lisman et al., 2002). In our study, CAMK2A has been identified as one of the proteins that form complexes with GPM6A. Moreover, we observed significantly lower CAMK2A expression in the hippocampus of depressed suicides. We hypothesize that the observed downregulation of CAMK2A could decrease the potential for synaptic plasticity in agreement with reductions in neuroplasticity in depression. Other findings show that synaptic CAMK2A targeting depends on an intact F-actin cytoskeleton (Jalan-Sakrikar et al., 2012). Indeed, actin is essential for functional and structural plasticity and specific mechanisms of cytoskeleton regulation are integral to the processes of neuronal remodeling (Hotulainen and Hoogenraad, 2010; Svitkina et al., 2010). These pathways mainly converge on the Rho family of small GTPases such as RhoA, Rac1, or Cdc42. In line with these observations, the other two proteins that we identified as components of GPM6A complexes are CORO1A and GIT1. CORO1A belongs to a family of WD40 domain-containing proteins that control diverse aspects of F-actin polymerization and branching cycle. It binds filamentous actin, Arp2/3 complex (Chan et al., 2011) and regulates the plasma membrane localization and activation of Rac1 (Castro-Castro et al., 2011). On the other hand, GIT1 functions as an integrator of signaling pathways controlling vesicle trafficking, adhesion, and cytoskeletal organization. It also regulates spine morphogenesis and synapse formation by targeting actin regulators and locally modulating Rac activity at synapses (Zhang et al., 2003, 2005; Webb et al., 2007). Here, we observed that CORO1A, but not GIT1 expression is downregulated in the hippocampus in the depressed group. Our findings are consistent with the previously described role of structural plasticity in depression (Christoffel et al., 2011). The pathway analyses of gene ontology groups commonly identify structural plasticity- and cytoskeleton-related signaling pathways as being highly modulated by stress and depression-like behavioral paradigms (Piubelli et al., 2011; Andrus et al., 2012). Furthermore, genetic association studies among individuals with lifetime depression have shown that a pathway consisting of genes that regulate cytoskeletal dynamics significantly associates with anhedonic depression (van Veen et al., 2012).
Gene products that function together in common signaling cascades or protein complexes are expected to show greater similarities in their expression patterns than random sets of gene products (Vidal et al., 2011). Indeed, in the present study we observe significant correlations among the expression levels of GPM6A, GPM6B, CORO1A, GIT1, and CAMK2A, but not PLP1 in the hippocampus of control subjects. Positive significant correlations are also observed among GPM6B, CORO1A, GIT1, and CAMK2A in the PFC. Here, in addition, negative correlations among GPM6A, CORO1A, GIT1, CAMK2A, and PLP1 are detected. These observations support the fact that these gene products might be co-regulated in the hippocampus and PFC of humans. And although further studies are needed to define detailed mechanisms of interactions of GPM6A with these proteins and will be described elsewhere (B Fuchsova and AC Frasch, unpublished results), we suggest that the neuroplastic function of GPM6A in filopodia and neurite outgrowth could be mediated by signaling pathways that involve GIT1, CORO1A, and/or CAMK2A.
Notably, we observed that the coordinated gene expression is disrupted or reorganized in depressed suicides. In the hippocampus, the only significant correlation detected is between GPM6A and CORO1A and, in the PFC considerable alterations compared to the control group are observed. Various studies have applied gene coexpression network analysis to associate coexpression modules with psychiatric diseases, providing important insights into the pathways disbalanced in the disease (Merali et al., 2004; Poulter et al., 2010; Zhurov et al., 2012; Gaiteri et al., 2014). Our findings are in agreement with observations that pathological mechanisms leading to depression may affect the coordination of gene expression in the brain (Merali et al., 2004; Gaiteri et al., 2010, 2014; Poulter et al., 2010; Zhurov et al., 2012). This may reflect dysregulation of gene expression and loss of coordinated transcriptional response, although observed changes may i result from other regulatory processes such as altered mRNA stability or microRNA actions.
Depression and stress may lead to the downregulation of our targets genes and their functional consequences through different mechanisms. Epigenetic mechanisms could be a possible explanation since GPM6A has been identified as a novel target for epigenetic regulation during prenatal stress through changes in methylation status and in posttranscriptional regulation by microRNAs. GPM6A mRNA expression has been shown to be modulated by miR-133b (Monteleone et al., 2014) and by miR-124 (El-Kordi et al., 2013). Interestingly, miR-133b expression is modulated by prenatal stress exposure in rats both in the hippocampus and PFC (Monteleone et al., 2014). Whether miR-133b and/or miR-124 regulate GPM6A expression in major depression remains to be established. Since one miRNA can target several mRNAs, dysregulation of one or just a few miRNAs could cause differential expression of a wide network of genes, as observed in complex pathologies such as depression (Dwivedi, 2014). Another possible mechanism could involve posttranscriptional regulation similar to what is observed for PLP1 transcript in schizophrenia patients (Aberg et al., 2006). It has been shown that QKI, an RNA-binding protein previously identified as a new candidate gene for schizophrenia, modulates mRNA levels of human oligodendrocyte-related genes, including PLP1, in schizophrenia patients through disturbance in its splicing (Aberg et al., 2006). It would be interesting to evaluate association of QKI with depression as well as its involvement in modulation of mRNA stability of other members of the PLP family.
Major depressive disorder and other neuropsychiatric disorders are complex diseases implicating a large number of genes. Each of them confers a small and incremental risk that potentially converges in dysregulated biological pathways, cellular functions, local circuit changes, and, eventually scales up to brain region pathophysiology (Belmaker and Agam, 2008). Even if differential expression is low or absent, the genetics of complex disease can led to alterations in the molecular interactions of cellular pathways. Thus, changes in gene–gene correlation may occur and their collective effect may become clear through the coexpression networks (Gaiteri et al., 2014). In this context, changes in coexpression network structure can help to identify candidate disease genes. For example, Rhinn et al. identified a novel mechanism for the alpha synuclein regulation in the context of Parkinson’s disease pathology by using differential coexpression to prioritize disease-related molecular targets. The alpha synuclein variant aSynL, containing a long 3′UTR, was identified as the most differentially coexpressed gene in several Parkinson’s disease datasets; however, aSynL was not among the most differentially expressed transcripts between patients and controls (Rhinn et al., 2012) and thus would have likely been over-looked by traditional analysis. From this point of view, we consider the loss of coordinated expression in our study of high interest. It suggests that the cellular pathways that involve studied genes are sensitive to disease condition. As for the mechanistic insight, it is tempting to speculate that as a consequence of their altered coexpression, GPM6A morphogenetic function in neurite outgrowth, filopodium formation and synaptogenesis as well as the synaptic plasticity function of CAMK2A and CORO1A would be affected with consequences for neuronal remodeling and connectivity. This would fit with the observations that dendritic complexity and synapse density are reduced in depression (Drevets et al., 2008).
The conclusions that can be drawn on the basis of the present investigation are subject to several limitations. Among other things, our cohort is confounded by the fact that the samples come from those who committed suicide. Although all individuals that had died by suicide had a history of depression (depressed phase in case of bipolar disorder), it is not necessarily the case that the present findings are generalizable to depression where suicidal intent was not present, nor is it the case that suicide itself was without effect (van Heeringen, 2001). Recent studies by Turecki and others have shown that there may be gene expression patterns that are associated with suicidality (Sequeira et al., 2007, 2009) albeit with some overlap in those associated with major depression. Thus, our findings may be relevant to suicidality and/or depression. Moreover, follow-up studies are necessary to determine the impact on the protein levels of studied genes. Despite these limitations, we consider that the observed alterations in mRNA levels and disturbance in coordinated gene expression of the neuroplasticity-related genes represent an important contribution in relation to the efforts of identifying potential candidates associated with mental illness. The observed changes in the balance between mRNA levels of studied genes might result in significant alterations in the neuronal connectivity between brain regions resulting in pathological behaviors.
In the future, global assessment of changes in gene expression would be of great value to help us to identify key molecular networks associated with depression. For example, high-throughput RNAseq detects both coding and noncoding RNAs, is superior for gene network construction, detects alternative spliced transcripts, detects allele-specific expression and can be used to extract genotype information, e.g. nonsynonymous coding single nucleotide polymorphisms. In this sense, it could allow us to look at the system as a whole thus deepening our understanding of such complex diseases such as depression.
CONCLUSION
Disruption of coordinated gene expression and abnormalities in mRNA expression of GPM6A, GPM6B and components of GPM6A protein complexes were detected in the brain of depressed suicides.
Highlights.
GPM6A and GPM6B are transcriptionally downregulated in the hippocampus of depressed suicides.
Neuroplasticity-related proteins that form complexes with GPM6A were identified.
mRNA of the components of GPM6A complexes is decreased in the depressed group.
Coexpression pattern analysis demonstrates coordinated gene expression in the brain.
In the brain of depressed suicides this coordinated response is disrupted.
Acknowledgments
This study was supported by grants to GNP (RO1 MH 48153 and RO1 MH 98554) from the National Institute of Mental Health, Rockville, MD. This work was also supported by grants to ACF and to BF from the ANPCyT and by grants to BF from the UNSAM, CONICET, and Fulbright Commission. AAJ is recipient of a doctoral fellowship from CONICET. BF and ACF are researchers from CONICET. The funding sources had no role in study design, acquisition, and interpretation of data or writing of the report. We thank Ulf Hellman from the Ludwig Institute for Cancer Research, Uppsala, Sweden for help with mass spectrometry analysis.
Abbreviations
- ANCOVA
analysis of covariance
- CAMK2A
calcium/calmodulin-dependent protein kinase II alpha
- CORO1A
coronin1A
- FC
fold change
- GFP
green fluorescent protein
- GPM6A
neuronal membrane glycoprotein M6a
- PFC
prefrontal cortex
- PLP
proteolipid protein
- PMI
postmortem interval
- qPCR
quantitative polymerase chain reaction
- RT
reverse transcription
Footnotes
CONFLICT OF INTEREST
The authors report no biomedical financial interests or potential conflicts of interest. Dr. Pandey reports grants from USA: National Institutes of Health, during the conduct of the study.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: BF. Performed the experiments: BF AAJ HR. Analyzed the data: BF AAJ. Interpretation of data: BF ACF GNP. Wrote the paper: BF.
References
- Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso J, Aguero F, Sanchez DO, Flugge G, Fuchs E, Frasch AC, Pollevick GD. Gene expression analysis in the hippocampal formation of tree shrews chronically treated with cortisol. J Neurosci Res. 2004a;78:702–710. doi: 10.1002/jnr.20328. [DOI] [PubMed] [Google Scholar]
- Alfonso J, Fernandez ME, Cooper B, Flugge G, Frasch AC. The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc Natl Acad Sci U S A. 2005;102:17196–17201. doi: 10.1073/pnas.0504262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso J, Frick LR, Silberman DM, Palumbo ML, Genaro AM, Frasch AC. Regulation of hippocampal gene expression is conserved in two species subjected to different stressors and antidepressant treatments. Biol Psychiatry. 2006;59:244–251. doi: 10.1016/j.biopsych.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Alfonso J, Pollevick GD, Van Der Hart MG, Flugge G, Fuchs E, Frasch AC. Identification of genes regulated by chronic psychosocial stress and antidepressant treatment in the hippocampus. Eur J Neurosci. 2004b;19:659–666. doi: 10.1111/j.1460-9568.2004.03178.x. [DOI] [PubMed] [Google Scholar]
- Amada N, Aihara K, Ravid R, Horie M. Reduction of NR1 and phosphorylated Ca2+/calmodulin-dependent protein kinase II levels in Alzheimer’s disease. Neuroreport. 2005;16:1809–1813. doi: 10.1097/01.wnr.0000185015.44563.5d. [DOI] [PubMed] [Google Scholar]
- Andrus BM, Blizinsky K, Vedell PT, Dennis K, Shukla PK, Schaffer DJ, Radulovic J, Churchill GA, Redei EE. Gene expression patterns in the hippocampus and amygdala of endogenous depression and chronic stress models. Mol Psychiatry. 2012;17:49–61. doi: 10.1038/mp.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. 2005;10:309–322. doi: 10.1038/sj.mp.4001565. [DOI] [PubMed] [Google Scholar]
- Barbiero VS, Giambelli R, Musazzi L, Tiraboschi E, Tardito D, Perez J, Drago F, Racagni G, Popoli M. Chronic antidepressants induce redistribution and differential activation of alphaCaM kinase II between presynaptic compartments. Neuropsychopharmacology. 2007;32:2511–2519. doi: 10.1038/sj.npp.1301378. [DOI] [PubMed] [Google Scholar]
- Belmaker RH, Agam G. Major depressive disorder. N Engl J Med. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- Boks MP, Hoogendoorn M, Jungerius BJ, Bakker SC, Sommer IE, Sinke RJ, Ophoff RA, Kahn RS. Do mood symptoms subdivide the schizophrenia phenotype? Association of the GMP6A gene with a depression subgroup. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:707–711. doi: 10.1002/ajmg.b.30667. [DOI] [PubMed] [Google Scholar]
- Brocco MA, Fernandez ME, Frasch AC. Filopodial protrusions induced by glycoprotein M6a exhibit high motility and aids synapse formation. Eur J Neurosci. 2010;31:195–202. doi: 10.1111/j.1460-9568.2009.07064.x. [DOI] [PubMed] [Google Scholar]
- Castro-Castro A, Ojeda V, Barreira M, Sauzeau V, Navarro-Lerida I, Muriel O, Couceiro JR, Pimentel-Muinos FX, Del Pozo MA, Bustelo XR. Coronin 1A promotes a cytoskeletal-based feedback loop that facilitates Rac1 translocation and activation. EMBO J. 2011;30:3913–3927. doi: 10.1038/emboj.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celano E, Tiraboschi E, Consogno E, D’Urso G, Mbakop MP, Gennarelli M, de Bartolomeis A, Racagni G, Popoli M. Selective regulation of presynaptic calcium/calmodulin-dependent protein kinase II by psychotropic drugs. Biol Psychiatry. 2003;53:442–449. doi: 10.1016/s0006-3223(02)01491-9. [DOI] [PubMed] [Google Scholar]
- Cerqueira JJ, Mailliet F, Almeida OF, Jay TM, Sousa N. The prefrontal cortex as a key target of the maladaptive response to stress. J Neurosci. 2007;27:2781–2787. doi: 10.1523/JNEUROSCI.4372-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KT, Creed SJ, Bear JE. Unraveling the enigma: progress towards understanding the coronin family of actin regulators. Trends Cell Biol. 2011;21:481–488. doi: 10.1016/j.tcb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–5372. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Russo SJ. Structural and synaptic plasticity in stress-related disorders. Rev Neurosci. 2011;22:535–549. doi: 10.1515/RNS.2011.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper B, Fuchs E, Flugge G. Expression of the axonal membrane glycoprotein M6a is regulated by chronic stress. PLoS One. 2009;4:e3659. doi: 10.1371/journal.pone.0003659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci. 2014;16:43–61. doi: 10.31887/DCNS.2014.16.1/ydwivedi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Conley RR, Roberts RC, Tamminga CA, Pandey GN. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch Gen Psychiatry. 2003;60:804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Teppen T, Zhang H, Mondal A, Roberts RC, Conley RR, Pandey GN. Lower phosphoinositide 3-kinase (PI 3-kinase) activity and differential expression levels of selective catalytic and regulatory PI 3-kinase subunit isoforms in prefrontal cortex and hippocampus of suicide subjects. Neuropsychopharmacology. 2008;33:2324–2340. doi: 10.1038/sj.npp.1301641. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Aberrant extracellular signal-regulated kinase (ERK)1/2 signalling in suicide brain: role of ERK kinase 1 (MEK1) Int J Neuropsychopharmacol. 2009;12:1337–1354. doi: 10.1017/S1461145709990575. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Modulation in activation and expression of phosphatase and tensin homolog on chromosome ten, Akt1, and 3-phosphoinositide-dependent kinase 1: further evidence demonstrating altered phosphoinositide 3-kinase signaling in postmortem brain of suicide subjects. Biol Psychiatry. 2010;67:1017–1025. doi: 10.1016/j.biopsych.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar N, Sibille E. A putative functional role for oligodendrocytes in mood regulation. Transl Psychiatry. 2012;2:e109. doi: 10.1038/tp.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kordi A, Kastner A, Grube S, Klugmann M, Begemann M, Sperling S, Hammerschmidt K, Hammer C, Stepniak B, Patzig J, de Monasterio-Schrader P, Strenzke N, Flugge G, Werner HB, Pawlak R, Nave KA, Ehrenreich H. A single gene defect causing claustrophobia. Transl Psychiatry. 2013;3:e254. doi: 10.1038/tp.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez ME, Alfonso J, Brocco MA, Frasch AC. Conserved cellular function and stress-mediated regulation among members of the proteolipid protein family. J Neurosci Res. 2010;88:1298–1308. doi: 10.1002/jnr.22298. [DOI] [PubMed] [Google Scholar]
- Fuchsova B, Fernandez ME, Alfonso J, Frasch AC. Cysteine residues in the large extracellular loop (EC2) are essential for the function of the stress-regulated glycoprotein M6a. J Biol Chem. 2009;284:32075–32088. doi: 10.1074/jbc.M109.012377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Guilloux JP, Lewis DA, Sibille E. Altered gene synchrony suggests a combined hormone-mediated dysregulated state in major depression. PLoS One. 2010;5:e9970. doi: 10.1371/journal.pone.0009970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor A, Kramer JM, van der Voet M, Schanze I, Uebe S, Donders R, Reis A, Schenck A, Zweier C. Altered GPM6A/M6 dosage impairs cognition and causes phenotypes responsive to cholesterol in human and drosophila. Hum Mutat. 2014;35:1495–1505. doi: 10.1002/humu.22697. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. Coupling antibodies to protein A beads; pp. 521–523. [Google Scholar]
- Hellman U, Wernstedt C, Gonez J, Heldin CH. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol. 2010;189:619–629. doi: 10.1083/jcb.201003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol. 2009;40:55–72. doi: 10.1007/s12035-009-8071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalan-Sakrikar N, Bartlett RK, Baucum AJ, 2nd, Colbran RJ. Substrate-selective and Calcium-independent Activation of CaMKII by alpha-Actinin. J Biol Chem. 2012;287:15275–15283. doi: 10.1074/jbc.M112.351817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, Lepack A, Majik MS, Jeong LS, Banasr M, Son H, Duman RS. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagenaur C, Kunemund V, Fischer G, Fushiki S, Schachner M. Monoclonal M6 antibody interferes with neurite extension of cultured neurons. J Neurobiol. 1992;23:71–88. doi: 10.1002/neu.480230108. [DOI] [PubMed] [Google Scholar]
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci. 2002;3:175–190. doi: 10.1038/nrn753. [DOI] [PubMed] [Google Scholar]
- Lucchesi W, Mizuno K, Giese KP. Novel insights into CaMKII function and regulation during memory formation. Brain Res Bull. 2011;85:2–8. doi: 10.1016/j.brainresbull.2010.10.009. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- McKittrick CR, Magarinos AM, Blanchard DC, Blanchard RJ, McEwen BS, Sakai RR. Chronic social stress reduces dendritic arbors in CA3 of hippocampus and decreases binding to serotonin transporter sites. Synapse. 2000;36:85–94. doi: 10.1002/(SICI)1098-2396(200005)36:2<85::AID-SYN1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Merali Z, Du L, Hrdina P, Palkovits M, Faludi G, Poulter MO, Anisman H. Dysregulation in the suicide brain: mRNA expression of corticotropin-releasing hormone receptors and GABA(A) receptor subunits in frontal cortical brain region. J Neurosci. 2004;24:1478–1485. doi: 10.1523/JNEUROSCI.4734-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michibata H, Okuno T, Konishi N, Kyono K, Wakimoto K, Aoki K, Kondo Y, Takata K, Kitamura Y, Taniguchi T. Human GPM6A is associated with differentiation and neuronal migration of neurons derived from human embryonic stem cells. Stem Cells Dev. 2009;18:629–639. doi: 10.1089/scd.2008.0215. [DOI] [PubMed] [Google Scholar]
- Michibata H, Okuno T, Konishi N, Wakimoto K, Kyono K, Aoki K, Kondo Y, Takata K, Kitamura Y, Taniguchi T. Inhibition of mouse GPM6A expression leads to decreased differentiation of neurons derived from mouse embryonic stem cells. Stem Cells Dev. 2008;17:641–651. doi: 10.1089/scd.2008.0088. [DOI] [PubMed] [Google Scholar]
- Monteleone MC, Adrover E, Pallares ME, Antonelli MC, Frasch AC, Brocco MA. Prenatal stress changes the glycoprotein GPM6A gene expression and induces epigenetic changes in rat offspring brain. Epigenetics. 2014;9:152–160. doi: 10.4161/epi.25925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukobata S, Hibino T, Sugiyama A, Urano Y, Inatomi A, Kanai Y, Endo H, Tashiro F. M6a acts as a nerve growth factor-gated Ca(2+) channel in neuronal differentiation. Biochem Biophys Res Commun. 2002;297:722–728. doi: 10.1016/s0006-291x(02)02284-2. [DOI] [PubMed] [Google Scholar]
- Novak G, Seeman P, Tallerico T. Increased expression of calcium/calmodulin-dependent protein kinase IIbeta in frontal cortex in schizophrenia and depression. Synapse. 2006;59:61–68. doi: 10.1002/syn.20211. [DOI] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Ren X, Bhaumik R, Dwivedi Y. Toll-like receptors in the depressed and suicide brain. J Psychiatr Res. 2014;53:62–68. doi: 10.1016/j.jpsychires.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C, Duman RS. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- Piubelli C, Carboni L, Becchi S, Mathe AA, Domenici E. Regulation of cytoskeleton machinery, neurogenesis and energy metabolism pathways in a rat gene-environment model of depression revealed by proteomic analysis. Neuroscience. 2011;176:349–380. doi: 10.1016/j.neuroscience.2010.12.043. [DOI] [PubMed] [Google Scholar]
- Poulter MO, Du L, Zhurov V, Palkovits M, Faludi G, Merali Z, Anisman H. Altered Organization of GABA(A) Receptor mRNA Expression in the Depressed Suicide Brain. Front Mol Neurosci. 2010;3:3. doi: 10.3389/neuro.02.003.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Sisti HM, Hao J, Rocher AB, McCall T, Hof PR, McEwen BS, Morrison JH. Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex. Neuroscience. 2004;125:1–6. doi: 10.1016/j.neuroscience.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY, Overholser JC, Roth BL, Stockmeier CA. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45:1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- Rhinn H, Qiang L, Yamashita T, Rhee D, Zolin A, Vanti W, Abeliovich A. Alternative alpha-synuclein transcript usage as a convergent mechanism in Parkinson’s disease pathology. Nat Commun. 2012;3:1084. doi: 10.1038/ncomms2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman S, Endicott J, Clayton P, Winokur G. Diagnostic evaluation after death (DEAD) Rockville, MD: National Institute of Mental Health, Neuroscience Research Branch; 1983. [Google Scholar]
- Scorticati C, Formoso K, Frasch AC. Neuronal glycoprotein M6a induces filopodia formation via association with cholesterol-rich lipid rafts. J Neurochem. 2011 doi: 10.1111/j.1471-4159.2011.07252.x. [DOI] [PubMed] [Google Scholar]
- Segal M. Dendritic spines and long-term plasticity. Nat Rev Neurosci. 2005;6:277–284. doi: 10.1038/nrn1649. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer R, Williams J, Gibbon M, First M. Structural clinical interview for DSM-IV (SCID) New York, NY: New York State Psychiatric Institute Biometrics Research; 1995. [Google Scholar]
- Stockmeier CA, Mahajan GJ, Konick LC, Overholser JC, Jurjus GJ, Meltzer HY, Uylings HB, Friedman L, Rajkowska G. Cellular changes in the postmortem hippocampus in major depression. Biol Psychiatry. 2004;56:640–650. doi: 10.1016/j.biopsych.2004.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T, Lin WH, Webb DJ, Yasuda R, Wayman GA, Van Aelst L, Soderling SH. Regulation of the postsynaptic cytoskeleton: roles in development, plasticity, and disorders. J Neurosci. 2010;30:14937–14942. doi: 10.1523/JNEUROSCI.4276-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraboschi E, Giambelli R, D’Urso G, Galietta A, Barbon A, de Bartolomeis A, Gennarelli M, Barlati S, Racagni G, Popoli M. Antidepressants activate CaMKII in neuron cell body by Thr286 phosphorylation. Neuroreport. 2004;15:2393–2396. doi: 10.1097/00001756-200410250-00018. [DOI] [PubMed] [Google Scholar]
- van Heeringen C. Suicide, serotonin, and the brain. Crisis. 2001;22:66–70. doi: 10.1027//0227-5910.22.2.66. [DOI] [PubMed] [Google Scholar]
- van Veen T, Goeman JJ, Monajemi R, Wardenaar KJ, Hartman CA, Snieder H, Nolte IM, Penninx BW, Zitman FG. Different gene sets contribute to different symptom dimensions of depression and anxiety. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:519–528. doi: 10.1002/ajmg.b.32058. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb DJ, Zhang H, Majumdar D, Horwitz AF. Alpha5 integrin signaling regulates the formation of spines and synapses in hippocampal neurons. J Biol Chem. 2007;282:6929–6935. doi: 10.1074/jbc.M610981200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner HB, Kramer-Albers EM, Strenzke N, Saher G, Tenzer S, Ohno-Iwashita Y, De Monasterio-Schrader P, Mobius W, Moser T, Griffiths IR, Nave KA. A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia. 2013;61:567–586. doi: 10.1002/glia.22456. [DOI] [PubMed] [Google Scholar]
- Wu GY, Cline HT. Stabilization of dendritic arbor structure in vivo by CaMKII. Science. 1998;279:222–226. doi: 10.1126/science.279.5348.222. [DOI] [PubMed] [Google Scholar]
- Xing G, Russell S, Hough C, O’Grady J, Zhang L, Yang S, Zhang LX, Post R. Decreased prefrontal CaMKII alpha mRNA in bipolar illness. Neuroreport. 2002;13:501–505. doi: 10.1097/00001756-200203250-00029. [DOI] [PubMed] [Google Scholar]
- Yan Y, Narayanan V, Lagenaur C. Expression of members of the proteolipid protein gene family in the developing murine central nervous system. J Comp Neurol. 1996;370:465–478. doi: 10.1002/(SICI)1096-9861(19960708)370:4<465::AID-CNE4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Zappia MP, Bernabo G, Billi SC, Frasch AC, Ceriani MF, Brocco MA. A role for the membrane protein M6 in the Drosophila visual system. BMC Neurosci. 2012;13:78. doi: 10.1186/1471-2202-13-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YP, Holbro N, Oertner TG. Optical induction of plasticity at single synapses reveals input-specific accumulation of alphaCaMKII. Proc Natl Acad Sci U S A. 2008;105:12039–12044. doi: 10.1073/pnas.0802940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Iida A, Ouchi Y, Satoh S, Watanabe S. M6a is expressed in the murine neural retina and regulates neurite extension. Mol Vis. 2008;14:1623–1630. [PMC free article] [PubMed] [Google Scholar]
- Zhurov V, Stead JD, Merali Z, Palkovits M, Faludi G, Schild-Poulter C, Anisman H, Poulter MO. Molecular pathway reconstruction and analysis of disturbed gene expression in depressed individuals who died by suicide. PLoS One. 2012;7:e47581. doi: 10.1371/journal.pone.0047581. [DOI] [PMC free article] [PubMed] [Google Scholar]