Abstract
It would be very beneficial if the status of cancers could be determined from a blood specimen. However, peripheral blood leukocytes are very heterogeneous between individuals and thus high resolution technologies are likely required. We used cytometry by time-of-flight (CyTOF) and next generation sequencing to ask whether a plasma cell cancer (multiple myeloma) and related pre-cancerous states had any consistent effect on the peripheral blood mononuclear cell phenotypes of patients. Analysis of peripheral blood samples from 13 cancer patients, 9 pre-cancer patients, and 9 healthy individuals revealed significant differences in the frequencies of the T, B, and natural killer cell compartments. Most strikingly, we identified a novel B-cell population that normally accounts for 4.0±0.7% (mean±SD) of total B cells and is up to 13-fold expanded in multiple myeloma patients with active disease. This population expressed markers previously associated with both memory (CD27+) and naïve (CD24loCD38+) phenotypes. Single-cell immunoglobulin gene sequencing showed polyclonality, indicating that these cells are not precursors to the myeloma, and somatic mutations, a characteristic of memory cells. SYK, ERK, and p38 phosphorylation responses, and the fact that most of these cells expressed isotypes other than IgM or IgD, confirmed the memory character of this population, defining it as a novel type of memory B cells.
Keywords: Multiple Myeloma, Tumor Immunology, B-Lymphocyte Biology, Immune Phenotyping, Peripheral Blood
Introduction
The human immune system has a unique role in cancer pathogenesis, modulating therapeutic responses and disease outcome. Especially with emerging immune therapeutics and treatment regimens, the characterization of cancer-associated immunological signatures has been of considerable interest. However, most studies of tumor “immunomes” rely on gene expression data and focus on the tumor microenvironment (1–3). To some extent this may be due to the limited resolving power of standard cell analysis methods such as flow cytometry and the heterogeneity of peripheral blood cells. However, recent advances in cell phenotyping technology, particularly cytometry by time-of-flight (CyTOF), which supports 40-label analysis (4–6), and next generation sequencing (NGS) of immunologlobulin (Ig) genes (7,8) have greatly increased the possible depth of analysis at the single-cell level and expanded the resolving power of immune phenotyping tremendously. This suggests that it may be possible to detect disease-related immune signatures in human peripheral blood using these methods. As peripheral blood is readily and safely obtainable from almost any patient, finding useful signatures of disease in this type of specimen could be very valuable both clinically and in understanding the immune response to particular cancers at a systems-biology level.
For this reason we analyzed the peripheral blood of patients with hematopoietic cancer (multiple myeloma) and its corresponding pre-cancer (asymptomatic myeloma (AM) and monoclonal gammopathy of undetermined significance (MGUS)). Multiple myeloma (MM), a malignancy of the B lineage, is characterized by the accumulation of clonal plasma cells in the bone marrow and the production of monoclonal Ig. The corresponding pre-cancer diseases, MGUS and AM are characterized by the production of monoclonal Ig and the lack of symptoms in often undiagnosed patients. They are not considered malignant because they do not always progress into MM, however, patients have an approximate risk of 1% per year to develop MM (9), and many, if not all, MM cases develop from a preceding MGUS or AM (10,11).
MM represents a prototypical disease model for tumor – microenvironment interactions (12,13) and recent studies of the MM microenvironment have underlined its role in promoting tumor growth and survival (14), as well as protection from T-cell responses (15) and chemotherapeutics (16,17). The myeloma cells “reprogram” their environment, influencing many chemokine and cytokine levels including interleukin (IL)6, IL1α, IL1β, MIP-1α, TNFα, M-CSF, and VEGF (13). With this profound effect on the bone marrow – the organ in which hematopoiesis takes place in adults – we reasoned that MM and perhaps its precursor diseases might have a significant impact on immune cells in the peripheral blood that could be detectable with these new technologies.
Here we report that we have found major effects of MM, but not MGUS or AM, that are detectable in peripheral blood cells of patients with active disease, in particular significant changes in the T, B and NK cell compartments, and most notably, the expansion of a novel, polyclonal B-cell subset.
Materials and Methods
Subjects
13 MM, 9 MGUS/AM patients (either untreated or without cancer-specific treatment during the last 6 months, Table 1 and Suppl. Table 1) and 5 treatment-naïve colorectal cancer patients (Suppl. Fig. 10) were recruited in accordance with federal and local human subjects regulations (IRB protocol ID 25310). 9 Age- and sex-matched healthy individuals were recruited through the Stanford Blood Center. The median ages of the MM, pre-cancer, and healthy control groups were 67, 69, and 68 years, respectively. Male subjects accounted for 54% in the cancer cohort, 56% in the pre-cancer cohort, and 56% in the healthy control group.
Table 1. Clinical characteristics of MM, AM, and MGUS patients.
None of the patients with indicated last treatment have been treated during the last six months before the study blood draw. MM: multiple myeloma, AM: asymptomatic myeloma, MGUS: monoclonal gammopathy of undetermined significance, BM: bone marrow, ISS: International Staging System, n.a.: not applicable, n.d.: not determined, RVD: revlimide velcade dexamethasone, TD: thalidomide dexamethasone, MP: melphalan prednisone
| Subject | Disease | Age (years) | Sex | M-component type | Stage (ISS) | BM infiltration (%) | Cytogenetics | Last treatment |
|---|---|---|---|---|---|---|---|---|
| 1 | AM | 56 | F | IgA λ | n/a | 15–20 | n.d. | - |
| 2 | MGUS or AM | 77 | M | IgG λ | n/a | n.d. | n.d. | - |
| 5 | MGUS or AM | 70 | M | IgG κ | n/a | n.d. | n.d | - |
| 9 | MGUS | 70 | M | IgA κ | n/a | 9 | normal karyotype | - |
| 23 | AM | 69 | M | IgG λ | n/a | 20 | extra copy of 1q21, partial deletion of chromosome 13, +1, dic(1;8), t(8;22), del13 and t(14;20) | - |
| 24 | MGUS or AM | 69 | F | IgG κ | n/a | n.d. | n.d. | - |
| 26 | MGUS or AM | 62 | M | IgG κ | n/a | 5 | n.d. | - |
| 27 | MGUS or AM | 47 | F | IgG λ | n/a | n.d. | n.d. | - |
| 28 | AM | 51 | F | IgG λ | n/a | 17 | n.d. | - |
| 3 | MM | 69 | F | IgG κ | n.d. | 40 | n.d. | melphalan/BCNU, autologous transplant |
| 4 | MM | 65 | F | IgA κ | n.d. | 30 | hyperdiploidy | RVD |
| 6 | MM | 46 | M | IgA λ | 3 | 60 | normal karyotype | - |
| 7 | MM | 63 | F | n.k. | n.k. | n.k. | n.k. | TD, high dose melphalan |
| 8 | MM | 57 | M | IgA κ | 1 | 30 | normal karyotype | - |
| 10 | MM | 56 | F | κ light chain | 3 | 50 | Del13q, t(11;14) | - |
| 11 | MM | 72 | M | IgG λ | 2 | 48 | del Y | - |
| 18 | MM | 81 | M | IgG κ | 1 | 30 | normal karyotype | - |
| 19 | MM | 61 | F | κ light chain | 3 | 50 | normal karyotype | - |
| 20 | MM | 77 | M | IgA κ | 3 | 15 | t(1;16), gain of IgH | - |
| 21 | MM | 67 | M | IgG κ | 3 | 25 | normal karyotype | - |
| 25 | MM | 85 | M | IgG κ | 3 | 10–90 | n/a | MP |
| 32 | MM | 77 | F | IgG κ | 3 | 40 | n/a | - |
PBMC preparation
Peripheral blood mononuclear cells (PBMC) were isolated from up to 40ml of freshly drawn heparin anticoagulated blood using Ficoll-Paque™ PLUS (GE Healthcare Bio-Sciences AB) centrifugation and resuspended in cell culture medium (Suppl. Table 2). Freshly isolated PBMCs were immediately used for in vitro stimulation and CyTOF staining; remaining cells were frozen after the addition of an equal volume of FBS containing 20% DMSO (both Sigma Aldrich).
Cell stimulation
For CyTOF, PBMCs were stimulated in 1ml cell culture medium containing 20ng/ml PMA, 1uM ionomycin (Sigma Aldrich), 5μg/ml R848, or 3μg/ml CpG ODN2216 (both InvivoGen) or left unstimulated for 6h. 1μl GolgiPlug and 0.7μl GolgiStop (both BD Biosciences) were added at the beginning of the stimulation for PMA/ionomycin or to unstimulated samples, or added after 2h for R848 or after 3h for CpG ODN2216/DOTAP. DOTAP liposomal transfection reagent (Roche) was added at 1μl/ml for CpG ODN2216 stimulation. Stimulation was done at 37°C and 5% CO2.
For phosphorylation analysis, stimulation was done in reverse time order in 250μl pre-warmed cell culture medium containing 50uM CpG ODN2006 (InvivoGen), or 10μg/ml goat anti-human IgM (life technologies) and 10μg/ml goat F(ab)2 anti-human IgG (AbD Serotec). For B-cell receptor (BCR) stimulation H2O2 (MP Biomedicals) was added within 10s after addition of the stimulating antibodies to a final concentration of 3.3mM.
CyTOF antibody labeling and staining
Purified antibodies were labeled using MaxPar® DN3 kits (Fluidigm) and stored at 4°C at 0.2mg/ml in W buffer (Fluidigm) containing antibody stabilizer (Candor).
For staining 1–10×106 cells were washed in CyFACS buffer (Suppl. Table 2) and stained in 50ul CyFACS buffer containing a surface antibody cocktail (Suppl. Table 3) for 30min. The γδ T cell antibody stain was done separately and the metal-labeled anti-PE antibody added to the surface antibody cocktail. Cells were washed in CyPBS, PBS (Ambion), and stained with maleimide-DOTA loaded with 115 In for 20min at RT. After washing in CyFACS and CyPBS cells were fixed in 150μl of 2% paraformaldehyde (PFA) (Electron Microscopy Sciences) over night. Cells were washed twice in permeabilization buffer (eBioscience) and stained in 50μl intracellular antibody cocktail (Suppl. Table 3) for 30min on ice. After another wash in permeabilization buffer and CyPBS cells were stained with iridium DNA intercalator (Fluidigm) for 20min at RT, washed 2x in CyFACS, 2x in CyPBS, 2x in H2O and resuspended in H2O for analysis on a CyTOF® instrument (Fluidigm).
Cell signaling analysis
Cells were thawed, washed 2x in pre-warmed cell culture medium and rested for 2h at 37°C, 5% CO2. Cells were washed in pure PBS and stained with zombie aqua (BioLegend), washed 1x in pure PBS and stained with CD24 and CD38 antibodies (Suppl. Table 4) for 15min at 4°C. After washing in pre-warmed cell culture medium cells were re-suspended in 250μl warm cell culture medium and immediately stimulated as described in the cell stimulation section. Stimulation was stopped by adding 150μl of 4% PFA and incubated for 15min at RT. Cells were washed with pure PBS and permeabilized in methanol at −80°C overnight. After 2x washing in pure PBS, cells were stained with an intracellular staining cocktail of antibodies specific for phosphorylated signaling molecules and additional phenotyping markers (Suppl. Table 4), washed, and finally stained with AF488-labeled goat anti-rabbit antibody (Suppl. Table 4) before analysis on an LSRII flow cytometer (BD Biosciences).
Single-cell immunoglobulin sequencing
PBMCs were thawed and stained with fluorochromes according to Supplementary Table 4. Single B cells were identified by their scatter (FSC/SSC) characteristics, CD19, and CD20 expression and sorted into RT-PCR buffer in 96-well plates according to the gating strategy in Figure 4A and Supplementary Figure 8. Ig genes were amplified and sequenced as previously described (7,8).
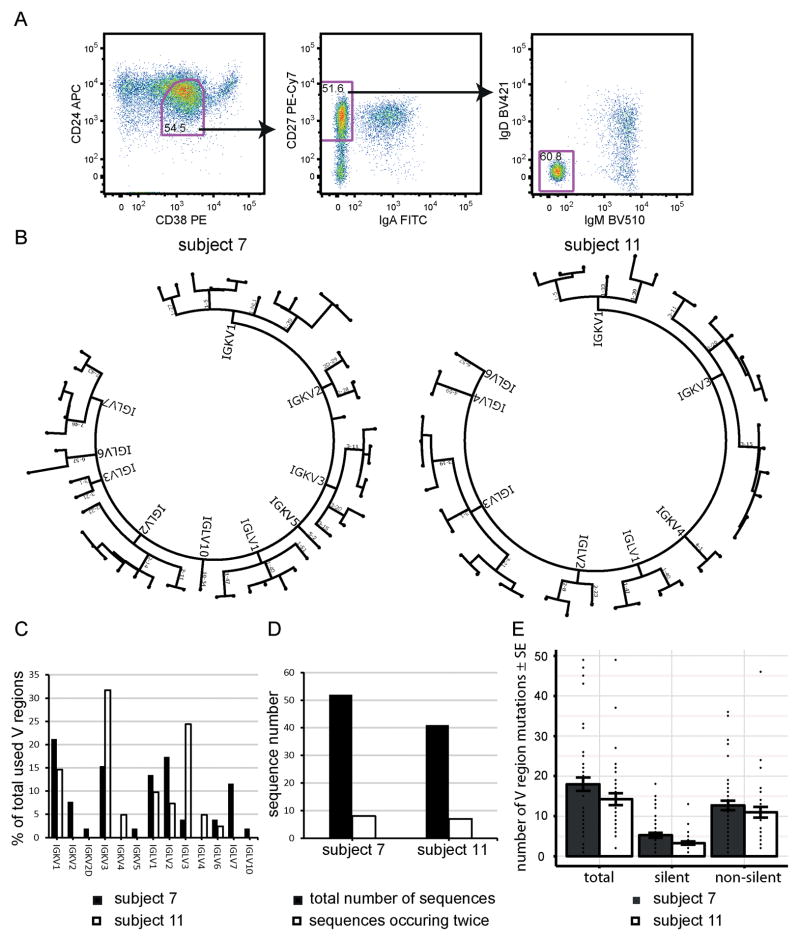
Figure 4. Clonal restriction and somatic mutations of CD24loCD38+CD27+ B cells.
(A) Gating strategy for single-cell FACS sorts. The plots are pre-gated on CD19+CD20+CD14− single live cells and CD24loCD38+CD27+IgA−IgD−IgM− single cells were sorted. (B) Light chain dendrograms of the V region usage of single sorted B cells. Each dot in the end of a branch of the phylogenetic trees represents one single cell. (C) Overlap of V region usage between subjects 7 and 11. (D) Clonality within the V regions of the two patients. (E) Number of V region mutations per analyzed cell. Each dot represents a single cell. Bars indicate means±SE. Subject numbers correspond to the numbers in the clinical characteristics Table 1.
Fluorescence in situ hybridization (FISH)
Cells were stained as described for single-cell Ig sequencing and bulk sorted according to the gating scheme in Supplementary Figure 8. Cells were stained with Vysis LSI CCND1 and Vysis LSI IGH probes (both Abbott Molecular) according to standard protocols, and 200 cells per population (105 for CD19+CD20+CD24hiCD38−CD27−IgD−IgM− B cells) were analyzed.
Cytometry data analysis and statistics
All two-dimensional gating analysis was done using FlowJo (Treestar).
For heatmaps (Fig. 1A, Suppl. Fig. 2, Suppl. Fig. 9) data in FlowJo-exported tables were trimmed to the 95th and 5th percentiles per gated population and visualized using heatmap.2 (gplots version 2.11.0).
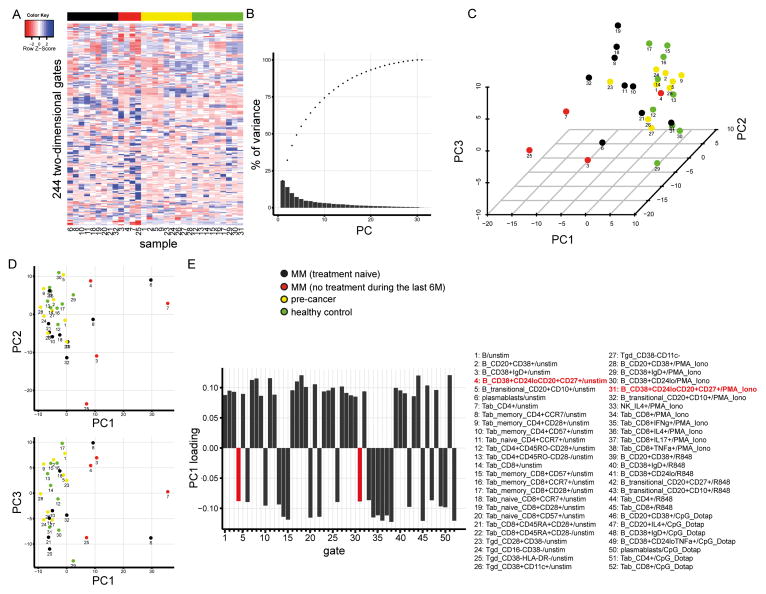
Figure 1. Phenotypic diversity of MM, AM, and MGUS patients.
Displayed is the CyTOF data from 13 MM patients, 11 pre-cancer patients, and 9 healthy individuals. The sample numbers in figure parts A, C, and D correspond to the subject numbers in Table 1. (A) Normalized cell frequencies (%) in 244 two dimensional gates per sample are visualized as a heatmap. Gates are arranged in rows, samples in columns. No clustering was applied to the columns. (B–E) Show principal components analysis (PCA) on the cell frequencies displayed in the gates in A. (B) Cumulative (dots) variance explained by the individual (bars) principal components. (C–D) 3D and 2D visualizations of the first three PCs. (E) Loadings of PC1 > 0.085. X axis labels and bar colors correspond to the numbered and colored gate names in the lower right panel of the figure.
Principal component analysis (PCA) was done using prcomp (stats version 2.15.3) and visualized with ggplot2 (version 0.9.3.1) and scatterplot3d (version 0.3–34).
For statistics in scatterplots (Fig. 2B and Suppl. Fig. 3) we used ANOVA. P values were adjusted using the Benjamini Hochberg correction and all differences that reached q values below 0.05 were followed up with TukeyHSD, the results of which are indicated as p values in the respective plots. All statistics were done in R version 2.15.3.(18)
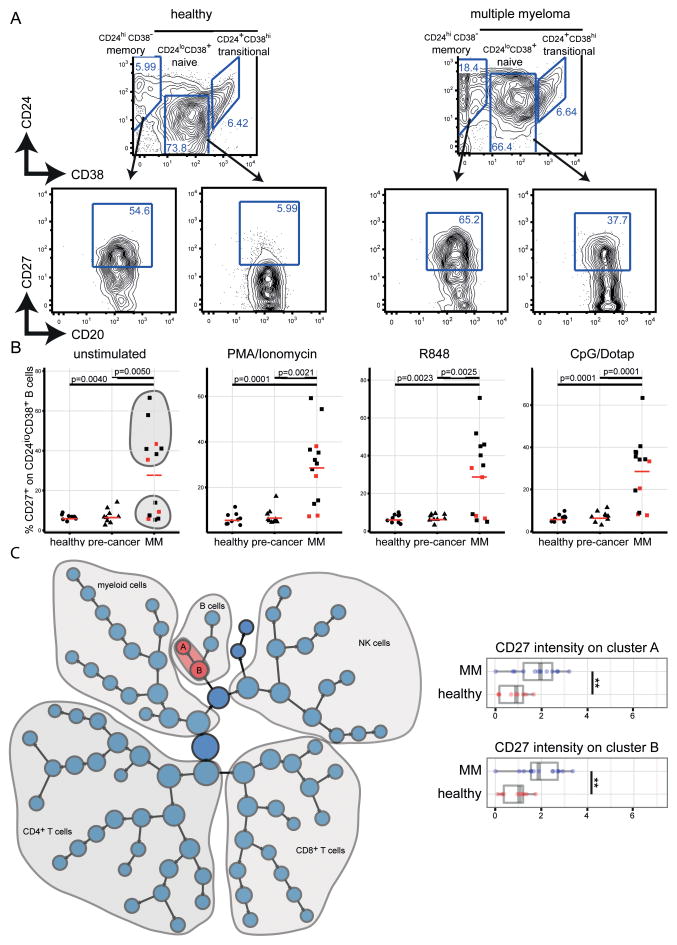
Figure 2. Expansion of CD24loCD38+CD27+ B cells in the peripheral blood of MM patients.
(A) The two-dimensional B-cell phenotype characteristics in one MM and one healthy control sample analyzed with CyTOF. The plots are pre-gated on CD19+ cells. CD27 gates were defined based on the CD27 expression in total CD19+ B cells. (B) Summarizes frequencies of CD19+CD20+CD24loCD38+CD27+ B cells in all samples included in the study analyzed with CyTOF. Red squares indicate samples from patients that have received MM-specific treatment in the past but not during the last six months. Red bars represent frequency means. Stimuli conditions are indicated at the top of the plot. (C) Citrus analysis comparing the expression of CD27 between 13 MM and 9 healthy control samples of our CyTOF dataset. Highlighted in red are the nodes with significantly different CD27 expression (FDR < 0.01) which is normalized and plotted for each of the two nodes (A and B) in the box plots in the right panel of the figure. ** q < 0.01
Citrus version 0.05 (https://github.com/nolanlab/citrus) (19) was used on all healthy control and all MM samples. Each file was pre-gated on live CD45+ cells in FlowJo. The file sample size was set to 10000 events and CD33, CD4, CD20, CD8, CD16, CD19, CD11c, CD24, CD10, CD1c, CD14, CCR7, CD7, CD28, CD5, CD38, CD45RA, CD123, CD45RO, CD56, HLA-DR, CD25, TCR-αβ, and CD127 were used as clustering markers. Median differences (median columns) were calculated for IgM, CD57, IgD, CD27, CCR7, CD28, TNF-α, IL17A, and HLA-DR. The minimum cluster size was set to 0.02% and SAM was used as model type.
Results
Phenotypic diversity of PBMC subsets in multiple myeloma
We used CyTOF with a 39-marker panel (Suppl. Table 3) to analyze the human immune system in freshly isolated PBMCs from MM patients, from pre-cancer MGUS and AM patients (Table 1 and Suppl. Table 1), and from healthy individuals. Frequencies of all major PBMC subsets and their cytokine expression patterns were determined by two-dimensional gating as percentages of the corresponding parent populations in four different stimulus conditions (see Suppl. Fig. 1 and Suppl. Table 5 for typical gating and a list of all applied gates).
We visualized cell frequencies in each of the 244 two-dimensional gates in a heatmap representing the immune phenotypic diversity in the peripheral blood of patients with MM, pre-cancer patients, and healthy controls (Fig. 1A). Samples were arranged by disease group and no clustering was applied to the columns. Emphasizing the phenotypic heterogeneity of the dataset, sample clustering by Euclidian distances of relative cell frequencies did not lead to the formation of clear clusters for cancer patients, pre-cancer patients, and healthy individuals (Suppl. Fig. 2).
The direct comparison of individual population frequencies between disease groups revealed significant differences in the T (CD57+CD8+CD45RA+ αβ and regulatory) and B lymphocyte (total, CD27+, and transitional) compartments in different stimulus conditions as summarized in Supplementary Figure 3.
To analyze whether individual markers or combinations of those could distinguish cancer samples from pre-cancer or healthy samples, we applied principal component analysis (PCA) to cell frequencies in the same 244 two-dimensional gates. The first four principal components (PC) described close to 50% of the variance in our dataset (Fig. 1B) and plotting PCs 1–3 confirmed phenotypic heterogeneity in samples from the cancer group whereas the pre-cancer and healthy control samples tended to cluster (Fig. 1C–D). Nevertheless, PC1 separated a subset of cancer samples from pre-cancer and healthy donor samples. The loadings for PC1 were dominated by αβ T-cell gates including co-stimulatory molecules (CD28) and markers that have been associated with T-cell exhaustion (CD57), or homing (CCR7), both on CD4+ and CD8+ T cells (Fig. 1E). Surprisingly, CD19+CD20+ B cells with a distinct marker combination (CD24loCD38+) and expression of CD27 contributed to PC1 across different stimuli conditions (unstimulated and after PMA/ionomycin stimulation, Fig. 1E in red). This population was particularly interesting because it combined two markers (CD24loCD38+) that have been associated with the naïve stage of B-cell differentiation (20), and CD27, which is associated with memory B cells (21). This is a novel phenotype that has not been reported previously.
CD24loCD38+CD27+ B cells are uniquely expanded in the peripheral blood of MM patients and associated with active disease
On human peripheral blood B cells, CD24 and CD38 expression levels can be measured to identify memory (CD24+CD38−), naïve (CD24loCD38+), and transitional (CD24+CD38hi) subsets (20).
As expected, the majority of CD24+CD38− B cells expressed CD27 in the peripheral blood of all three groups (MM, pre-cancer, and healthy individuals) and we detected only minor amounts of CD24loCD38+ B cells expressing CD27 in pre-cancer patients and healthy individuals (Fig. 2A/B). Strikingly, in the MM samples we identified an up to 13-fold expanded CD27+ population among the CD24loCD38+ B cells when compared to that in healthy controls (Fig. 2A/B). Notably, the expression of CD27 on CD24loCD38+ B cells in MM samples showed a very broad range (5.3–66.6%) with samples clustering at the high and low ends of this distribution, suggesting the existence of two patient populations (Fig. 2B, shaded in grey). This dichotomy was already noted in the PCA where PC1 separated only a subset of MM samples from pre-cancer and healthy control samples (Fig. 1C). Nevertheless, the frequency of CD24loCD38+CD27+ B cells was significantly higher in MM samples than in pre-cancer samples (p=0.005) and never exceeded 10% in healthy controls (p=0.004) (Fig. 2B, p values for unstimulated samples). In vitro stimulation showed only minor influences on CD27 expression on CD24loCD38+ B cells, suggesting that the observed phenotype is not due to transient activation. CD24loCD38+CD27+ B cells in MM samples showed mixed characteristics of other B-cell development-associated markers: 15.3±9.5% expressed CD10; 27.5±18.3% were IgD+, and 63.1±27.3 expressed TNF-α after PMA/ionomycin stimulation (all mean±SD) (Suppl. Fig. 4). Notably, none of the patients were diagnosed with plasma cell leukemia or showed phenotypic characteristics of malignant or de-differentiated plasma cells in the CD24loCD38+ gate (Fig. 2A and Suppl. Fig. 5).
Given the 39 dimensional space of our CyTOF dataset, two-dimensional gating fails to capture the richness that comes from including all possible marker combinations. We therefore used a new analysis method, Citrus (19) for the identification of significantly different marker expressions on cell populations from high-dimensional data from MM patients and healthy individuals (Fig. 2C and Suppl. Fig. 6). Among the different expression levels of CD57 on T-cell and NK-cell subsets (Suppl. Fig. 6B/C), Citrus detected significantly higher CD27 levels in two B-cell subsets (Fig. 2C). Cluster A (Fig. 2C) showed lower CD24 expression and therefore most likely represents the CD38+CD24lo B-cell subset identified with two-dimensional gating and PCA.
Taken together, CyTOF data analyzed using two different methods enabled us to identify a uniquely expanded B-cell population with aberrant expression levels of CD24, CD38, and CD27 in the peripheral blood of many MM patients. (Fig. 2B). This population was not evident in MGUS or AM patients and healthy individuals.
We then verified these findings with flow cytometry, analyzing PBMCs from all MM samples, which confirmed the expansion of the CD24loCD38+CD27+ B cells (Suppl. Fig. 7). In addition, we also analyzed IgA, IgM and IgD expression levels on B-lineage cells, and serum levels of IgG, IgA, IgM, kappa, and lambda immunoglobulin light chains (Fig. 3 and Table 2).
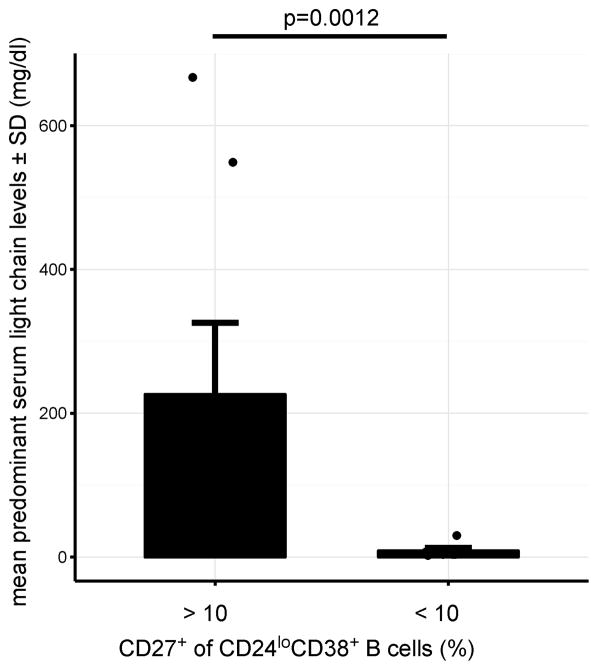
Figure 3. Association of CD24loCD38+CD27+ B-cell expansion with MM activity.
Elevated predominant serum light chain levels are associated with expansion (>10 %) of CD27+ population within CD24loCD38+ B cells. The unpaired two-sided Wilcoxon Rank-Sum test was used to calculate the p-value. Raw data for this figure are included in Table 2. The frequencies of CD24loCD38+CD27+ B cells and surface immunoglobulin expression were determined with FACS and plots for each of the 13 MM samples are shown in Supplementary Figure 7.
Table 2. Association of CD24loCD38+CD27+ B cell expansion with MM activity.
Displayed are light chain and immunoglobulin serum concentrations and the percentages of CD24loCD38+CD27+ B cells that expressed the indicated immunoglobulin. FACS data for each individual sample is shown in Supplementary Figure 7. Serum K and λ levels of the predominant light chain are printed in bold. Clinical data for each subject can be found in Table 1.
| Subject | % pos on CD24loCD38+ PB B cells | Serum levels (mg/dl) | % pos on CD24lo CD38+CD27+ PB B cells | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| CD27 | K | Λ | K/Λ | IgG | IgA | IgM | IgA | IgM | IgD | |
| 3 | 5.2 | 3.9 | 1.2 | 1.9 | 1410 | 58 | 23 | 3.0 | 41.9 | 41.4 |
|
|
||||||||||
| 4 | 2.2 | 3.7 | 1.0 | 3.7 | 688 | 61 | 37 | 8.2 | 56.1 | 56.5 |
|
|
||||||||||
| 6 | 48.5 | 1.9 | 667 | 0.0 | 1160 | 4800 | 96 | 17.1 | 13.6 | 12.4 |
|
|
||||||||||
| 7 | 5.4 | 1.6 | 2.2 | 0.9 | 1250 | 526 | 121 | 20.2 | 32.4 | 36.1 |
|
|
||||||||||
| 8 | 6.1 | 7.3 | 0.6 | 12.1 | 342 | 3690 | 15 | 26.7 | 34.7 | 33.3 |
|
|
||||||||||
| 10 | 32.7 | 549.0 | 0.2 | 2745.0 | 528 | 24 | <6 | 39.9 | 9.7 | 7.0 |
|
|
||||||||||
| 11 | 86.0 | 0.2 | 42.1 | 0.0 | 1350 | 24 | <6 | 34.7 | 28.5 | 14.9 |
|
|
||||||||||
| 18 | 56.3 | 85.7 | 1.0 | 85.7 | 3580 | 20 | 25 | 21.7 | 20.6 | 19.3 |
|
|
||||||||||
| 19 | 68.0 | 98.1 | 0.2 | 490.5 | 397 | 16 | 10 | 18.2 | 30.3 | 25.2 |
|
|
||||||||||
| 20 | 4.7 | 30.3 | 2.5 | 12.1 | 992 | 1060 | 22 | 23.8 | 44.9 | 45.5 |
|
|
||||||||||
| 21 | 5.6 | 3.6 | 1.1 | 3.3 | 3060 | 82 | 39 | 10.9 | 57.1 | 56.5 |
|
|
||||||||||
| 25 | 78.2 | 41.6 | 1.9 | 21.9 | 1410 | 28 | 13 | 26.5 | 39.8 | 26.2 |
|
|
||||||||||
| 32 | 33.2 | 99.0 | 0.6 | 165 | 709 | 15 | 19 | 19.3 | 8.32 | 7.8 |
|
|
||||||||||
We found that only a minority of this novel B-cell population expressed IgD (29.4±17.2% IgD+) or IgM (32.1±16.0% IgM+, both mean±SD), indicating that they largely represent a differentiation stage after class-switch recombination (Table 2). With 20.8±10.1% of this population expressing IgA, the majority can be assumed to be IgG+. Strikingly, the expansion of CD27+ B cells among CD24loCD38+ cells was associated with high serum levels of the patient’s predominant Ig light chain and therefore might be useful as a measure of disease activity. Disease activity was also reflected in an either very high or very low K/Λ ratio depending on the M protein type of the disease (Table 2).
Immunoglobulin genes of single CD24loCD38+CD27+ B cells from MM patients are polyclonal and show somatic mutations
To analyze clonality and whether the CD24loCD38+CD27+ population is a memory or naïve B-cell population we sequenced the Ig genes from single CD24loCD38+CD27+IgD−IgM−IgA− B cells from two MM patients (Sort gates in Fig. 4A).
In total, light chain sequences from 93 individual cells were sequenced and further analyzed. Both patients used a variety of different V region genes that partly overlapped between patients (Fig. 4B and C). The direct comparison of individual cells’ Ig sequences within the same sample revealed eight cells in subject 7 and seven cells in subject 11 whose sequences occurred twice (Fig. 4D). Nevertheless, based on different Ig sequences and the variety of V regions used, the CD24loCD38+CD27+ B-cell compartment has to be considered polyclonal although an underlying antigen-driven process responsible for the expansion of this B-cell subset is possible. Notably, there were no identical sequences across both patients.
We detected an average of 18 (subject 7) or 14 (subject 11) somatic mutations per V region, 5 (subject 7) and 3 (subject 11) of which were silent and 13 (subject 7) or 11 (subject 11) were non-silent mutations. None of the V region sequences were germline in either patient (Fig. 4E), a further indication that this B-cell subset is a memory population.
To further strengthen the case that the investigated B-cell populations are not directly related to the malignant plasma-cell clone, we FACS-sorted bulk CD24loCD38+CD27+, CD24loCD38+CD27−IgM+, CD24hiCD38−CD27+, and CD24hiCD38−CD27−IgM−IgD− B cells (Suppl. Fig. 8) from one patient whose malignant plasma-cell population contained the t(11;14). Sorted cells were analyzed with FISH and the t(11;14) could not be detected in any of these populations (Suppl. Fig. 8).
CD24loCD38+CD27+ B cells show phosphorylation patterns similar to total CD27+ memory B cells after B-cell receptor and TLR9 stimulation
Aberrant intracellular signaling has already been demonstrated in cancer, especially in lymphoma B cells (22). We measured the phosphorylation of one BCR proximal (SYK) and two downstream (ERK and p38) signaling molecules in response to BCR stimulation or BCR-bypassing TLR9 stimulation. The BCR was stimulated with polyclonal anti-human IgG and IgM in the presence of H2O2. The addition of H2O2 can amplify early signaling events by inhibiting protein tyrosine phosphatase (PTP) activity (23), thereby increasing signal strength. Phosphorylation levels in either unstimulated cells or after the addition of H2O2 will be referred to as baseline phosphorylation.
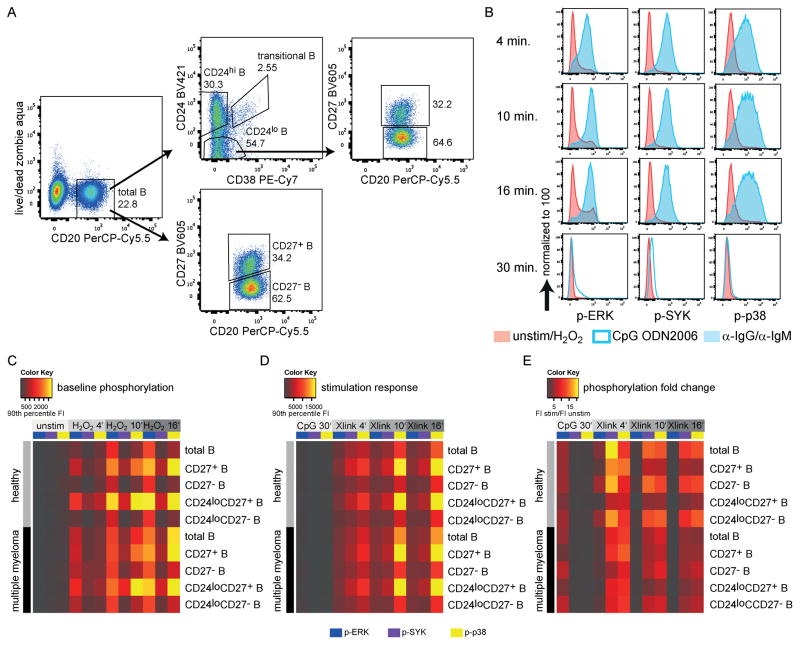
The quantification of signaling molecule phosphorylation with flow cytometry requires intracellular staining after PFA fixation and methanol permeabilization which particularly affects CD38 PE-Cy7 staining. Due to this decrease in the CD38 signal, CD24loCD38+ and CD24hiCD38− B cells will be referred to as CD24lo and CD24hi B cells (Fig. 5A). CD24hi and CD24lo populations corresponded to CD24+CD38− and CD24loCD38+ populations respectively after staining without fixation/permeabilization, as shown by the predominant expression of CD27 on CD24hi (memory) B cells in healthy individuals after methanol permeabilization (Suppl. Fig. 9A). Phosphorylation responses to anti-human IgG/IgM or TLR9 ligand CpG ODN2006 stimulation were clearly distinguishable from baseline phosphorylation, as shown in Figure 5B.
Figure 5. Intracellular signaling in B-cell subsets of MM patients and healthy individuals.
(A) FACS gating on different B-cell populations after stimulation, fixation, permeabilization and intracellular staining. (B) Typical phosphorylation responses in total CD20+ B cells of one healthy individual measured with FACS. (C–E) Mean phosphorylation responses of MM patients (n=3) and sex- and age-matched healthy controls (n=3). Unstim: cells cultured without any additional additives for 30 minutes; CpG: CpG ODN2006; Xlink: Addition of anti-human IgG and anti-human IgM; FI: fluorescence intensity; For the calculation of fold-changes 90th percentile FI of stimulated samples were divided by 90th percentile FI values of unstimulated or H2O2 samples.
No significant phosphorylation was detectable at baseline in any subpopulations of unstimulated B cells. The addition of H2O2 resulted in slightly increased phosphorylation over time that was most pronounced in ERK and p38 (Fig. 5C). Interestingly, total CD27+ and CD24loCD27+ B cells showed very similar phosphorylation levels in both healthy individuals and PBMCs from MM patients. Stimulation with CpG ODN2006 or anti-human IgG/IgM elicited up to 21-fold higher phosphorylation when compared to that of H2O2 controls (Fig. 5D/E). The unique phosphorylation patterns of CD27+, CD27−, and total B-cell populations were consistent at baseline and after stimulation, suggesting essentially different intracellular signaling responses in these subsets; however, significant differences between healthy and MM B-cell phosphorylation levels could be detected neither at baseline nor after stimulation. Overall, CpG ODN2006 stimulation caused mostly ERK phosphorylation, whereas BCR stimulation elicited predominantly SYK and p38 phosphorylation in the analyzed populations (Fig. 5E). The unique phosphorylation patterns of CD27+, CD27−, and total B-cell subsets at baseline and after BCR stimulation were confirmed in two additional sample pairs (Suppl. Fig. 9).
Discussion
In this study we present high-dimensional cytometry data on the human immunological landscape of peripheral blood cells across most of the known developmental stages of MM (MM, AM, MGUS, and healthy individuals). We manually determined the frequencies of all major PBMC subsets (αβ T, γδ T, B, NK, myeloid dendritic cells, plasmacytoid dendritic cells, and monocytes) and the expression of additional phenotypic and functional markers in four different stimulus conditions. With 434 two-dimensional initial gates per sample, to the best of our knowledge, this is the most detailed phenotypic and functional analysis of PBMC subsets in this disease. After removing mutually exclusive gates (e.g. from CD38+CD4+ and CD38−CD4+ T-cell gates only the CD38+CD4+ was kept), 244 of these gates (listed in Suppl. Table 5) were used for statistical analyses and data visualization. Cell frequencies in each of these gates were compared between disease groups, one at a time, and using principal component analysis. The manual comparisons allowed a very detailed view of particular cell subsets. Elevated numbers of CD57+CD45RA+CD8+ T cells (Suppl. Fig. 3A) could be expected given the previous work of Sze and colleagues (24); cells sharing this phenotype are considered to be clonally-restricted and associated with a favorable disease outcome. In addition, we detected lower numbers of regulatory T cells (Treg) in the peripheral blood of MM patients (Suppl. Fig. 3A). In MM the Treg/Th17 balance correlates with disease outcome (25,26); however, differences in frequencies of IL17-producing cells between patients with MM, pre-cancer patients, and healthy individuals did not reach statistical significance and Treg frequencies in the peripheral blood of MM patients are a matter of debate (26,27).
We detected significantly lower B-cell frequencies in total leukocytes of MM patients when compared to that in healthy individuals (Suppl. Fig. 3A). In a study involving hundreds of patients, low CD19+ cell frequencies have been associated with an advanced disease stage and higher B-cell numbers with a better prognosis of MM (28). The low percentages in our study may therefore reflect an advanced stage of disease in our patients, most of whom were in International Staging System (ISS) stadium 3 (Table 1), and/or the limited sample size. Memory (CD20+CD27+) B-cell frequencies were also elevated (Suppl. Fig. 3A) as shown along with all other direct subset comparisons in Supplementary Figure 3B.
The most striking finding was the selective expansion of CD24loCD38+CD27+ B cells in the peripheral blood of multiple myeloma patients with active disease as measured by serum immunoglobulin light chain levels. This B-cell phenotype has not been described before and attracted our attention for two reasons. First, because of a curiously mixed phenotype, in that while CD24loCD38+ expression is a characteristic of naïve B cells; CD27 is currently one of the most well established memory B cell markers and has been associated with the presence of somatic mutations in Ig genes (21,29,30). Second, this phenotype was stable even after 6 hours of in vitro stimulation. CD24 expression on CD24loCD38+CD27+ B cells in MM patients appeared to be higher than in their healthy counterparts but was still lower than in CD24+CD38− memory B cells (Fig. 2A). Even though this population shows variable CD24 expression (Suppl. Fig. 7), CD38 expression was clearly present while (CD27+) memory B cells have been shown to be CD38− in various studies (20,31).
Citrus is a new analysis tool that compares marker expression in individual multi-dimensional cell clusters between sample groups in a statistically rigorous fashion (19). Besides differences in CD27 expression on B-cell subsets (Fig. 2C), Citrus confirmed differences in CD57 expression on T-cell and NK-cell subsets (Suppl. Fig. 6B–C). Automated gating/clustering strategies confirmed our findings from data analyst-influenced manual gating with high statistical significance (FDR<0.01) but did not add any categorically new findings. Thus, the two-dimensional analysis approach even in this 39-dimensional dataset was still able to reveal most of the significant differences between sample groups, although it could not address all marker combinations on every single cell. The advantages of Citrus are represented in the “unsupervised” nature of the approach and the enormous reduction of analysis time.
MM plasma cells are usually not highly proliferative and the dividing MM precursor pool is assumed to comprise post germinal center B cells (31–34), which could match the phenotype of CD24loCD38+CD27+ B cells (Suppl. Fig. 4). Yet, the existence and phenotype of clonotypic B cells in the peripheral blood of MM patients is still a matter of debate (33, 35–37). Taken into account the phenotype of CD24loCD38+CD27+ B cells (Fig. 2A and Suppl. Fig. 5), the polyclonality of immunoglobulin genes, and the absence of MM clone-specific translocations (Suppl. Fig. 8), this B-cell population is unlikely to be directly derived from or preceding the MM plasma-cell population. The presence of somatic mutations in all single cells analyzed characterized this population as a memory-cell population, questioning the exclusive use of CD24 and CD38 for the identification of naïve and memory B cells. To determine whether CD24loCD38+CD27+ B cells can also be expanded in malignancies other than multiple myeloma we analyzed peripheral blood samples from five colorectal cancer patients (Suppl. Fig. 10) and could not detect any significant expansion of this population. Therefore, we suggest that the polyclonal expansion of CD24loCD38+CD27+ B cells in MM is most likely a phenomenon driven by active disease that results in some cues to which this B-cell type is responsive.
Earlier studies have shown aberrant intracellular signaling in different types of lymphoma B cells and that BCR signaling can be phenotype-dependent (22,38). Our intracellular signaling data indicated that the MM B-cell compartments are not obviously different from their counterparts in healthy individuals on a functional level. In this context CD24loCD38+CD27+ B cells behaved very similarly to “conventional” CD27+ B cells, further indicating that these cells are not in some unusual state. The trigger for their expansion in MM can be assumed in some cues exposed during active disease.
Supplementary Material
Acknowledgments
Financial support: Single cell sorting was performed in the Stanford Shared FACS Facility using NIH S10 Shared Instrument Grant (S10RR025518-01). Leo Hansmann was supported by a research fellowship by the German Research Foundation (DFG). This research was funded by grants to MMD from the NIH (U19 AI00019) and the Howard Hughes Medical Institute.
We thank Jonathan M. Irish at Vanderbilt University, William O’Gorman now at Genentech, and Michael Leipold at the Human Immune Monitoring Center at Stanford for very helpful phosphoflow and CyTOF advice, Dana Bangs at Stanford Cytogenetics for help with FISH analyses, Cindy Kin for help with consenting patients, as well as Alessandra Aquilanti and Murad Mamedov for critically reading the manuscript. CyTOF analysis was performed in the Human Immune Monitoring Center at Stanford and single-cell sorting was performed in the Stanford Shared FACS Facility using NIH S10 Shared Instrument Grant (S10RR025518-01). Leo Hansmann is supported by a research fellowship by the German Research Foundation.
This research was funded by grants to MMD from the NIH (U19 AI00019) and the Howard Hughes Medical Institute.
Footnotes
Conflict of Interest Disclosures
L.H., L.B., C.J., M.L., M.M.D. have no relevant conflicts of interest to disclose.
W.H.R. is a consultant to, a member of the Board of Directors of, and owns equity in Atreca, Inc.
Authorship Contributions
L.H. conceived the project and experiments, performed experiments, analyzed data, generated figures and wrote the manuscript. L.B., C.J. and W.H.R. performed experiments, analyzed data, and generated figures. M.L. identified and accrued patients, provided clinical data and clinical background. M.M.D. contributed to the project design, and analysis, and helped to write the manuscript.
References
- 1.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Olechnowicz SW, Edwards CM. Contributions of the host microenvironment to cancer-induced bone disease. Cancer Res. 2014;74:1625–31. doi: 10.1158/0008-5472.CAN-13-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Restifo NP. A “big data” view of the tumor “immunome”. Immunity. 2013;39:631–2. doi: 10.1016/j.immuni.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–52. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–9. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu DR, Tan YC, Kongpachith S, Cai X, Stein EA, Lindstrom TM, et al. Identifying functional anti-Staphylococcus aureus antibodies by sequencing antibody repertoires of patient plasmablasts. Clin Immunol. 2014;152:77–89. doi: 10.1016/j.clim.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan YC, Blum LK, Kongpachith S, Ju CH, Cai X, Lindstrom TM, et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin Immunol. 2014;151:55–65. doi: 10.1016/j.clim.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zingone A, Kuehl WM. Pathogenesis of monoclonal gammopathy of undetermined significance and progression to multiple myeloma. Semin Hematol. 2011;48:4–12. doi: 10.1053/j.seminhematol.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, et al. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113:5412–7. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss BM, Abadie J, Verma P, Howard RS, Kuehl WM. A monoclonal gammopathy precedes multiple myeloma in most patients. Blood. 2009;113:5418–22. doi: 10.1182/blood-2008-12-195008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitsiades CS, Mitsiades N, Munshi NC, Anderson KC. Focus on multiple myeloma. Cancer Cell. 2004;6:439–44. doi: 10.1016/j.ccr.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Mitsiades CS, Mitsiades NS, Richardson PG, Munshi NC, Anderson KC. Multiple myeloma: a prototypic disease model for the characterization and therapeutic targeting of interactions between tumor cells and their local microenvironment. J Cell Biochem. 2007;101:950–68. doi: 10.1002/jcb.21213. [DOI] [PubMed] [Google Scholar]
- 14.Hideshima T, Nakamura N, Chauhan D, Anderson KC. Biologic sequelae of interleukin-6 induced PI3-K/Akt signaling in multiple myeloma. Oncogene. 2001;20:5991–6000. doi: 10.1038/sj.onc.1204833. [DOI] [PubMed] [Google Scholar]
- 15.de Haart SJ, van de Donk NW, Minnema MC, Huang JH, Aarts-Riemens T, Bovenschen N, et al. Accessory cells of the microenvironment protect multiple myeloma from T-cell cytotoxicity through cell adhesion-mediated immune resistance. Clin Cancer Res. 2013;19:5591–601. doi: 10.1158/1078-0432.CCR-12-3676. [DOI] [PubMed] [Google Scholar]
- 16.Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104:607–18. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- 17.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–30. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 18.Team RC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 19.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ, Nolan GP. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111:E2770–7. doi: 10.1073/pnas.1408792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–8. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–89. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irish JM, Czerwinski DK, Nolan GP, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor-infiltrating nonmalignant B cells. Blood. 2006;108:3135–42. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nat Immunol. 2002;3:1129–34. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 24.Sze DM, Giesajtis G, Brown RD, Raitakari M, Gibson J, Ho J, et al. Clonal cytotoxic T cells are expanded in myeloma and reside in the CD8(+)CD57(+)CD28(−) compartment. Blood. 2001;98:2817–27. doi: 10.1182/blood.v98.9.2817. [DOI] [PubMed] [Google Scholar]
- 25.Bryant C, Suen H, Brown R, Yang S, Favaloro J, Aklilu E, et al. Long-term survival in multiple myeloma is associated with a distinct immunological profile, which includes proliferative cytotoxic T-cell clones and a favourable Treg/Th17 balance. Blood Cancer J. 2013;3:e148. doi: 10.1038/bcj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favaloro J, Brown R, Aklilu E, Yang S, Suen H, Hart D, et al. Myeloma skews regulatory T and pro-inflammatory T helper 17 cell balance in favor of a suppressive state. Leuk Lymphoma. 2014;55:1090–8. doi: 10.3109/10428194.2013.825905. [DOI] [PubMed] [Google Scholar]
- 27.Feng P, Yan R, Dai X, Xie X, Wen H, Yang S. The Alteration and Clinical Significance of Th1/Th2/Th17/Treg Cells in Patients with Multiple Myeloma. Inflammation. 2014 Jul 18; doi: 10.1007/s10753-014-9980-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Kay NE, Leong T, Kyle RA, Greipp P, Billadeau D, Van Ness B, et al. Circulating blood B cells in multiple myeloma: analysis and relationship to circulating clonal cells and clinical parameters in a cohort of patients entered on the Eastern Cooperative Oncology Group phase III E9486 clinical trial. Blood. 1997;90:340–5. [PubMed] [Google Scholar]
- 29.Seifert M, Kuppers R. Molecular footprints of a germinal center derivation of human IgM+(IgD+)CD27+ B cells and the dynamics of memory B cell generation. J Exp Med. 2009;206:2659–69. doi: 10.1084/jem.20091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agematsu K, Nagumo H, Yang FC, Nakazawa T, Fukushima K, Ito S, et al. B cell subpopulations separated by CD27 and crucial collaboration of CD27+ B cells and helper T cells in immunoglobulin production. Eur J Immunol. 1997;27:2073–9. doi: 10.1002/eji.1830270835. [DOI] [PubMed] [Google Scholar]
- 31.Caraux A, Klein B, Paiva B, Bret C, Schmitz A, Fuhler GM, et al. Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138− and CD138+ plasma cells. Haematologica. 2010;95:1016–20. doi: 10.3324/haematol.2009.018689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukreja A, Hutchinson A, Dhodapkar K, Mazumder A, Vesole D, Angitapalli R, et al. Enhancement of clonogenicity of human multiple myeloma by dendritic cells. J Exp Med. 2006;203:1859–65. doi: 10.1084/jem.20052136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahindra A, Hideshima T, Anderson KC. Multiple myeloma: biology of the disease. Blood Rev. 2010;24 (Suppl 1):S5–11. doi: 10.1016/S0268-960X(10)70003-5. [DOI] [PubMed] [Google Scholar]
- 34.Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–7. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonsalves WI, Rajkumar SV, Gupta V, Morice WG, Timm MM, Singh PP, et al. Quantification of clonal circulating plasma cells in newly diagnosed multiple myeloma: implications for redefining high-risk myeloma. Leukemia. 2014;28:2060–5. doi: 10.1038/leu.2014.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsui W, Huff CA, Wang Q, Malehorn MT, Barber J, Tanhehco Y, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–6. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiago LS, Perez-Andres M, Balanzategui A, Sarasquete ME, Paiva B, Jara-Acevedo M, et al. Circulating clonotypic B-cells in multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica. 2014;99:155–62. doi: 10.3324/haematol.2013.092817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irish JM, Czerwinski DK, Nolan GP, Levy R. Kinetics of B cell receptor signaling in human B cell subsets mapped by phosphospecific flow cytometry. J Immunol. 2006;177:1581–9. doi: 10.4049/jimmunol.177.3.1581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.