Abstract
Objective
We conducted a genome-wide linkage analysis to identify pelvic organ prolapse (POP) predisposition genes using a resource of high-risk POP pedigrees.
Study Design
Cases are defined as women who reported bothersome symptoms of POP based on standardized symptom questions (Pelvic Floor Distress Inventory (PFDI), moderately or quite bothered), and/or received treatment for POP documented in medical records. Our complete pedigree resource contains 299 familial POP cases in 83 high-risk pedigrees. Genotype data were obtained from Illumina HumanHap550, 610Q, the Human1M-Duo, Human Omni1-Quad, or the Human Omni 2.5 platforms. A set of single nucleotide polymorphism (SNP) markers common to all platforms was identified and markers in high linkage disequilibrium were removed. We performed a genome-wide linkage analysis under general dominant and recessive models using a Markov Chain, Monte Carlo linkage analysis method implemented in MCLINK software. Because 70 individuals in 32 pedigrees were used in a previously published linkage analysis for a phenotype of POP requiring treatment/surgery, we also performed linkage only including the 225 newly recruited and genotyped cases in 61 pedigrees.
Results
Linkage analysis using our complete pedigree resource for the loosened criteria of bothersome POP showed evidence for significant genome-wide linkage on chromosome 10q24-26 (recessive model, max HLOD 3.4); suggestive evidence was identified on chromosomes 6, 17, and an additional region on chromosome 10. In the subset of only the newly recruited familial POP cases, significant evidence for genome-wide linkage was observed on chromosome 17q25 (recessive model, max HLOD 3.3), and suggestive evidence for linkage was observed on chromosomes 10 and 11. Neither analysis duplicated the previously published linkage evidence for the POP-requiring-treatment/surgery phenotype observed on chromosome 9.
Conclusion
While the etiology of this common condition is unknown, this study provides evidence that loci on chromosomes 10q and 17q may contribute to POP etiology.
Keywords: chromosome 10q24-26, chromosome 17q25, genetic linkage analysis, high-risk familial cases, pelvic organ prolapse
Introduction
Pelvic organ prolapse (POP) is a common condition in adult women over age 50 years; the estimated lifetime risk of surgery for POP is 12.6% by the age of 80 years.1 Over 200,000 surgeries are performed annually in the United States for POP, exceeding 1 billion dollars in direct costs;2, 3 many more women manage their symptoms without surgery.4
Established environmental risk factors for POP include age, race, BMI, family history of POP, chronic intra-abdominal pressure and childbirth factors such as the number of full term pregnancies, infant birth weight, and forceps delivery.5, 6 However, the underlying molecular mechanism remains unknown. It is well recognized that POP clusters in families, giving credence to a hereditary contribution to the phenotype.7, 8 Genetic contributions to POP are under investigation.9, 10, 11
To better understand the genetics of POP, we have identified and recruited women with a family history of POP to be part of a genetic study of high-risk pedigrees. We previously performed an initial linkage analysis of this resource including primarily sibling pairs, both of whom had been surgically treated for POP. Significant linkage evidence for POP was identified on chromosome 9q21.11 This same resource was analyzed for a genome-wide association study of POP; six SNPs significantly associated with POP were identified.10 We have continued to expand this Utah genetics resource and recruit women with symptoms and a diagnosis of POP in high-risk POP families, regardless of their surgical treatment status.
The purpose of this study was to assess evidence of genetic linkage for a POP phenotype that included bothersome symptoms of POP in Utah high-risk POP pedigrees. In contrast to our prior linkage publication where subjects were required to have had surgery to treat their POP, the POP case definition used for this analysis included women with bothersome symptoms of POP and/or women who had been treated for POP. This expansion of the POP case definition allows inclusion of younger women in pedigrees who are symptomatic but have not yet been surgically treated for POP, which may increase power to detect additional evidence for linkage. Because we previously published linkage evidence on a subset of the complete resource analyzed here, we repeated the linkage analysis using only cases and pedigrees not included in the original publication.
Materials and Methods
Subjects
We initially recruited women who underwent surgical repair of POP on the urogynecology service at the University of Utah from 1996–2008 and who had at least one sister who also received surgical treatment for POP and was willing to participate in our study.11 Documentation of surgical management was required for women who had surgery outside the University of Utah system. Our original published linkage analysis included 70 women surgically treated with POP, most of whom were part of sibships.11 Since that publication we have expanded our resource in a number of different ways. First, using the Utah Population Database (UPDB), a population-based computerized genealogical resource that has been linked to electronic medical records at the University of Utah,12 extended relationships between some previously recruited sibpairs were recognized and these sibships were combined into larger, extended pedigrees which have increased power to detect regions of chromosomal sharing. As siblings tend to share on average half their genomes, it can be difficult to differentiate chance sharing from excess sharing due to a shared disease phenotype. Larger pedigrees have the potential to identify chromosomal sharing between distant relatives. We have also identified additional related POP cases in some of the original pedigrees by using POP ICD-9 diagnosis codes and CPT procedure codes in the electronic medical records; all diagnoses have been verified through examination of the medical record, and eligible subjects have been invited to participate in the study. Finally, we have identified new high-risk pedigrees with a significant excess number of POP cases (p<0.05) among descendants of founders in the UPDB. For the new high-risk pedigrees, the number of observed POP cases among descendants within a pedigree was compared to expected numbers of POP cases calculated within UPDB cohorts based on a 5-year birth year, sex, and birth-place (in or out of Utah). Recruitment efforts have been targeted to expand the high-risk pedigree resource with a primary focus of recruiting women with symptoms, diagnosis or treatment of POP who also have a family history of POP.
Study subjects have completed a number of questionnaires including the standardized Pelvic Floor Distress Inventory (PFDI). 13 We defined POP affection status for this study as a self-report of bothersome symptoms of POP on the PFDI (moderately or quite bothered) and/or treatment for POP documented in medical records. Eligible families for the linkage analysis had to have at least two female relatives meeting the definition for POP affection. This study was approved by the University of Utah IRB, and informed consent was obtained from all study participants prior to participation in the study.
Genotype data
DNA was extracted from all eligible subjects and genome-wide genotyping was performed using the Illumina HumanHap550, 610Q, the Human1M-Duo, Human Omni1-Quad, or the Human Omni 2.5 platforms. We identified a set of single nucleotide polymorphism (SNP) markers common to all platforms and used this as the genome-wide marker set. To avoid inflation of linkage statistics due to linkage disequilibrium and false positive results, the marker set was pruned by eliminating SNPs in high linkage disequilibrium. The 25,436 markers selected had a minimum spacing of 0.1 cM, a minimum heterozygosity of 0.3, and a maximum r2 of 0.16 over a sliding 500,000 basepair window in the publically available HapMap CEPH/Utah (CEU) data, and exceeded an individual call rate of 98% for genotyped subjects.
Linkage Analysis
Linkage analysis was performed using a multipoint Markov-chain Monte Carlo (MCMC) linkage method implemented in MCLINK.14 MCLINK is capable of analyzing extended pedigrees with multilocus markers and was used previously for linkage analysis of POP.11 MCLINK calculates multipoint LOD scores (i.e., TLOD and HLOD 15) which are robust to model misspecification. We performed an affecteds-only analysis using general dominant and recessive models. Phenotypes of all “non-affected” individuals were considered “unknown” for the analysis; some of the women were too young to be affected and males, although they cannot express the phenotype, may be carriers of a predisposition gene(s). We assumed a disease allele frequency of 0.001 for the dominant model and 0.01 for the recessive model. The penetrance estimates for carriers and non-carriers were 0.5 and 0.0005, respectively. We have used general dominant and recessive models for the linkage analysis; general models will be less powerful than the true underlying genetic model, but they will not have an increase in Type I error rates.16, 17 We report heterogeneity LOD (HLOD) scores for all pedigrees combined as they allow multiple heterogeneous loci to contribute to a complex disease trait, like POP. Linkage significance was defined using the Lander and Kruglyak genome-wide criteria, which assumes a dense genotype map.18 Suggestive linkage evidence was defined by HLOD scores ≥ 1.86 and significant genome-wide linkage evidence was defined by HLOD scores ≥ 3.30. For regions where significant genome-wide linkage evidence was observed, we report the number of pedigrees that obtained nominal point-wise significance (i.e., pedigree-specific TLOD>0.5) on a by-pedigree basis at that specific locus.
Results
The complete Utah high-risk pedigree resource contains 299 familial cases eligible for linkage analysis; this includes 225 newly recruited subjects who fit pedigree criteria (i.e., at least two related women who meet POP case definition), 70 subjects who are mostly sibpairs used in the previous linkage analysis, and 4 additional, newly recruited subjects who are part of previously recruited sibship pedigrees but do not constitute a pedigree on their own. The complete POP pedigree resource contains 83 pedigrees that range in size from 2 to 22 genotyped and affected cases; 19 of the pedigrees contain 5 or more cases (Table 1). The majority of all Utah POP pedigree subjects (78.6%) have been treated for POP (see Table 2).
Table 1.
Number of Genotyped, Affected Subjects by Family
| Families | Linkage analysis using newly recruited subjects | Linkage analysis using complete resource1 |
|---|---|---|
| 2 affected females | 26 | 37 |
| 3 affected females | 16 | 21 |
| 4 affected females | 3 | 6 |
| 5 affected females | 7 | 10 |
| >5 affected females | 9 | 9 |
| Total | 61 | 83 |
Linkage analysis using the complete resource includes the 225 newly recruited subjects, 70 individuals analyzed previously for linkage evidence and 4 additional, newly recruited individuals who are part of existing sibship pedigrees, but do not constitute a pedigree on their own.
Table 2.
Pelvic floor disorder severity among affected POP cases with at least bothersome symptoms of POP
| Pelvic Floor Disorder | Linkage Analysis using newly recruited subjects N=225 (% of POP cases) | Linkage Analysis using complete dataset N=299 (% of POP cases) |
|---|---|---|
| Pelvic Organ Prolapse | ||
| Bothersome symptoms | 62 (27.6%) | 64 (21.4%) |
| Primary treatment | 116 (51.6%) | 167 (55.9%) |
| Recurrent treatment | 47 (20.9%) | 68 (22.7%) |
| TOTAL | 225 (100%) | 299 (100%) |
| Stress Urinary Incontinence | ||
| Bothersome symptoms | 43 (19.1%) | 53 (17.7%) |
| Primary treatment | 85 (37.8%) | 121 (40.5%) |
| Recurrent treatment | 14 (6.2%) | 23 (7.7%) |
| TOTAL | 142 (63.1%) | 197 (65.9%) |
| Urge Urinary Incontinence | ||
| Bothersome symptoms | 49 (21.8%) | 68 (22.7%) |
| Treatment with anticholinergic agents | 43 (19.1%) | 65 (21.7%) |
| Treatment with Botox or neuromodulation | 3 (1.3%) | 3 (1.0%) |
| TOTAL | 95 (42.2%) | 136 (45.5%) |
| Hernia | ||
| Bothersome symptoms | 10 (4.4%) | 10 (3.3%) |
| Surgical treatment | 19 (8.4%) | 23 (7.7%) |
| Recurrent treatment | 3 (1.3%) | 3 (1.0%) |
| TOTAL | 32 (14.2%) | 36 (12.0%) |
The subset of newly recruited POP cases not used in the previously published linkage analysis includes 225 affected subjects in 61 pedigrees. The pedigrees range in size from 2 to 19 genotyped and affected individuals; 16 of the pedigrees had 5 or more genotyped cases (Table 1). The majority of newly recruited subjects (72.4%) had received treatment for POP (Table 2).
Table 2 summarizes the presence and severity of other pelvic floor disorders among affected POP cases including stress urinary incontinence (SUI), overactive bladder (OAB), and hernia. The majority of POP subjects had more than one pelvic floor disorder; 63.1% of newly recruited subjects and 65.9% of subjects in the complete pedigree resource also had bothersome symptoms of SUI or had been treated for SUI. Overactive bladder was slightly less common among familial POP cases; 42.2% of newly recruited subjects and 45.5% of subjects in the complete resource had bothersome symptoms or had received treatment for OAB. Approximately 10% of newly recruited cases and cases had hernias.
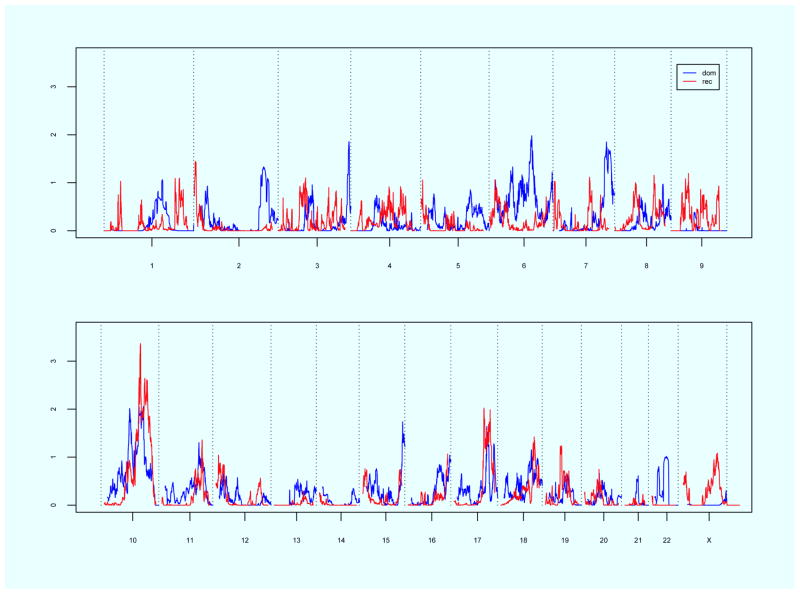
Linkage results for the complete Utah POP pedigree resource are displayed in Figure 1. Significant genome-wide linkage evidence was observed on chromosome10q24-26 under a recessive model (maximum HLOD = 3.4). Thirty of the 83 total pedigrees (36.1%) attained nominal linkage evidence (TLOD > 0.5) in this region. Two of the pedigrees had TLOD scores that individually exceeded 2.0 in this region. Suggestive evidence was observed on chromosome 6q21-22 (max HLOD 2.0, dominant model), 10q21 (max HLOD 2.0, dominant model), 10q23-25 (max HLOD 2.1, dominant model), 17q24 (max HLOD 2.0, recessive model), and 17q25 (max HLOD 2.0, recessive model).
Figure 1.
Genome-wide linkage results for recessive and dominant models using the complete Pelvic Floor Disorder Resource. Chromosome number shown on the horizontal axis; HLOD score shown on the vertical axis. Blue line = dominant model, Red line = recessive model.
Using the complete Utah POP pedigree resource there were also two pedigrees that approached significance by themselves. One pedigree with nine affected and genotyped cases had a TLOD score of 2.9 on chromosome 2q32-36, a region which includes the gene fibronectin 1 (FN1), involved in cell adhesion, wound healing, and host defense. As one of the extracellular matrix molecules, fibronectin has been shown to be involved in collagen I regulation19 and regulation of smooth muscle cell growth.20 Mutations in the FN1 gene are associated with glomerulopathy with fibronectin deposits,21 and in the lens of zebrafish mutations have resulted in cataracts and defects in fiber adhesion, organization, elongation and packing.22 The other pedigree with five affected and genotyped cases had a TLOD score of 2.9 on chromosome 7q34-36.1.
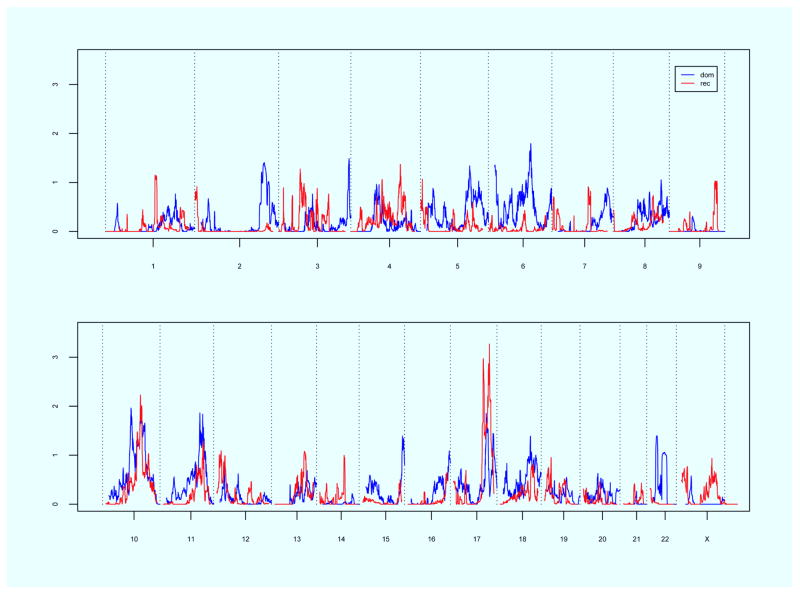
Linkage results for the subset of newly recruited POP cases are displayed in Figure 2. Significant linkage evidence was observed on chromosome 17q25 under a recessive model (maximum HLOD = 3.3). There were 13 pedigrees (21.3%) that had nominal linkage evidence in this region, including one pedigree with a TLOD score of 2.0 by itself. Suggestive linkage evidence was observed on chromosomes 10q21 (max HLOD 2.0), dominant model), 10q23 (max HLOD 1.9, dominant model), 10q24-25 (max HLOD 2.2, recessive model), 11q22 (max HLOD 1.9, dominant model) and 17q24 (max HLOD 3.0, recessive model).
Figure 2.
Genome-wide linkage results for recessive and dominant models using newly recruited subjects. Chromosome number shown on the horizontal axis; HLOD score shown on the vertical axis. Blue line = dominant model, Red line = recessive model.
Discussion
Using the Utah high-risk POP pedigree resource we have identified evidence for linkage on chromosomes 10q24-26 and 17q25. These regions may harbor POP predisposition genes. The strength of this study is the large number of extended pedigrees with a statistical excess of POP cases.
This analysis represents the third genome-wide linkage analysis reported to date for familial POP. Nikolova et al. performed the first genome-wide linkage scan for POP on a three-generation Filipino family with early onset POP.23 They identified ten regions with LOD scores near 1.5, the maximum possible for this single family and marker density. None of the chromosomal regions identified by us in this current analysis are among the ten regions observed by Nikolova’s study. As POP is a complex disease, there are likely to be multiple genes that contribute to the phenotype. Differences in familial POP genetics between our current study and the study by Nikolova study may arise because of racial and ethnic variation in allele frequencies, or perhaps because some cases of POP may be attributable to rare mutations present in only a limited number of families.
The second linkage analysis for POP reported to date was performed on an earlier set of Utah POP sibships.11 Using 32 mostly sibpairs (70 total subjects), both of whom had been surgically treated for POP, we reported significant evidence for linkage on chromosome 9q21 (max HLOD 3.41). In this current study, we did not see significant or suggestive linkage evidence in this chromosome 9q21 region, despite the fact that all of the same individuals in the original linkage analysis were used in the analysis of the complete resource. We previously observed that 17 of 32 pedigrees had at least nominal linkage evidence in the 9q21 region, two of which exceeded a TLOD score of 1.0. In our current analysis of the complete pedigree set, there were 14 pedigrees with at least nominal linkage evidence in the 9q21 region, four of which exceeded a TLOD score of 1.0. However, because new and more extended pedigrees in the current analysis provide evidence against linkage in the chromosome 9q21 region, this region is no longer significant overall. Because the original linkage analysis required very strictly defined POP concordant sibpairs, both of whom had received surgical treatment for their disease, it is possible that a POP predisposition gene in this chromosomal region is rare and highly penetrant. Larger high-risk pedigrees with multiple POP cases may harbor more common risk variants with lower individual effect. As POP is a common and complex disease, it is likely that there are multiple genes that contribute to this disease, and different study designs are likely to identify the combination of rare and common variants, with multiple effect sizes that underlie predisposition to POP.24 Sequencing of cases within linked pedigrees defined by study design may further understanding of POP genetic architecture.
There are a number of candidate genes in the 10q24-26 region that may predispose to POP. The most notable candidate gene is lysyl oxidase-like 4 (LOXL4), a member of the lysyl oxidase (LOX) family of genes which play a substantial role in the biogenesis of connective tissue matrix by catalyzing lysine derived from cross-links in collagen and elastin.25 While LOXL4 gene expression levels in severe POP patients have not been observed to be significantly different from levels in asymptomatic postmenopausal control patients,26 it is possible that regulatory elements of this gene 27 may be involved in POP etiology. Furthermore, a knock-out mouse model for another lysyl oxidase gene family member (i.e., LOXL1) has frequently been found to have pelvic floor dysfunction after pregnancy and delivery.28 Other candidate genes also in the 10q24-25 region that may predispose to POP include ankyrin repeat domain 2 (ANKRD2), heparanase 2 (HPSE2), collagen type 17A1 (COL17A1), and carbohydrate sulfotransferase 15 (CHST15). ANKRD2 is a gene involved in slow type 1 muscle fibers, and results of a knock-out mouse model showed an increased resting sarcomere length and greater muscle injury following eccentric contraction exercise.29 The HPSE2 gene functions to degrade heparin sulfate proteoglycans in the extracellular matrix. Mutations in the HPSE2 gene have been identified in patients with Ochoa (Urofacial) syndrome, which includes a reported case with both Ochoa syndrome and rectal prolapse.30 The COL17A1 gene encodes the alpha chain of the transmembrane type XVII collagen protein and is a major component of hemidesmosomes, which mediate the adhesion of keratinocytes and other epithelial cells to the underlying basement membrane.31 Mutations in the COL17A1 gene are associated with junctional epidermolysis bullosa in which congenital malformations of the urogenital tract have been observed to occur.32 CHST15 is an important structural component of the extracellular matrix and assists in forming proteogylcans. Mutations in the CHST15 gene have been observed in Kashin-Beck disease, a degenerative joint disease.33
The most notable candidate gene for POP in the chromosome 17q25 region is TIMP metallopeptidase inhibitor 2 (TIMP2), a member of the TIMP gene family and an endogenous inhibitor of matrix metalloproteinases, as well as a major factor in extracellular matrix remodeling.34 TIMP2 has been found associated with connective tissue disorders, vascular disorders, and cancer.35–37
There are limitations of this study. While we used bothersome symptoms of POP to define the phenotype, it is possible that this phenotype definition may include more heterogeneous cases; symptomatic POP based on bothersome symptoms is a subjective measure. However, the majority of the POP cases studied have received treatment for POP warranting a diagnosis that is both bothersome and clinically significant. The subjects included in this study are affected with multiple pelvic floor disorders including SUI, UUI, and hernias. We cannot definitively say that these results apply only to POP. Furthermore, our results do not contradict a multi-factorial etiology for POP that includes both genetic and environmental risk factors. These results suggest that there are some high-risk familial POP cases with likely stronger genetic underpinnings. Confirmation of these results can aid in future POP predisposition gene finding efforts and lead to enhanced understanding of POP etiology and lead efforts to prevent and treat this condition.
In conclusion, we have identified chromosomes 10q24-26 and 17q25 as chromosomal regions with significant linkage evidence for POP and which contain biologically plausible genes. These results can be used to enhance future gene finding efforts, and they provide further evidence that genetic factors contribute to POP etiology. Sequencing of appropriate related cases in high-risk pedigrees with evidence for linkage is underway.
Acknowledgments
Financial Support: Supported by a grant from the Eunice Kennedy Shriver National Institute of Child Health and Human Development RO1 HD061821
Footnotes
Presented in a preliminary form at the Society of Gynecologic Surgeons 40th Annual Scientific Meeting, Scottsdale, Arizona, March 23-26, 2014
COI: The authors report no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kristina ALLEN-BRADY, Email: kristina.allen@utah.edu.
Lisa A. CANNON-ALBRIGHT, Email: lisa.albright@utah.edu.
James M. FARNHAM, Email: jim.farnham@utah.edu.
Peggy A. NORTON, Email: peggy.norton@hsc.utah.edu.
References
- 1.Wu JM, Matthews CA, Conover MM, Pate V, Jonsson Funk M. Lifetime risk of stress urinary incontinence or pelvic organ prolapse surgery. Obstet Gynecol. 2014;123(6):1201–6. doi: 10.1097/AOG.0000000000000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith FJ, Holman CD, Moorin RE, Tsokos N. Lifetime risk of undergoing surgery for pelvic organ prolapse. Obstet Gynecol. 2010;116(5):1096–100. doi: 10.1097/AOG.0b013e3181f73729. [DOI] [PubMed] [Google Scholar]
- 3.Subak LL, Waetjen LE, van den Eeden S, Thom DH, Vittinghoff E, Brown JS. Cost of pelvic organ prolapse surgery in the United States. Obstet Gynecol. 2001;98(4):646–51. doi: 10.1016/s0029-7844(01)01472-7. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence JM, Lukacz ES, Nager CW, Hsu JW, Luber KM. Prevalence and co-occurrence of pelvic floor disorders in community-dwelling women. Obstet Gynecol. 2008;111(3):678–85. doi: 10.1097/AOG.0b013e3181660c1b. [DOI] [PubMed] [Google Scholar]
- 5.Bump RC, Norton PA. Epidemiology and natural history of pelvic floor dysfunction. Obstet Gynecol Clin North Am. 1998;25(4):723–46. doi: 10.1016/s0889-8545(05)70039-5. [DOI] [PubMed] [Google Scholar]
- 6.Hendrix SL, Clark A, Nygaard I, Aragaki A, Barnabei V, McTiernan A. Pelvic organ prolapse in the Women’s Health Initiative: gravity and gravidity. Am J Obstet Gynecol. 2002;186(6):1160–6. doi: 10.1067/mob.2002.123819. [DOI] [PubMed] [Google Scholar]
- 7.Lince SL, van Kempen LC, Vierhout ME, Kluivers KB. A systematic review of clinical studies on hereditary factors in pelvic organ prolapse. Int Urogynecol J. 2012;23(10):1327–36. doi: 10.1007/s00192-012-1704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norton PA, Allen-Brady K, Cannon-Albright LA. The familiality of pelvic organ prolapse in the Utah Population Database. Int Urogynecol J. 2013;24(3):413–8. doi: 10.1007/s00192-012-1866-0. [DOI] [PubMed] [Google Scholar]
- 9.Campeau L, Gorbachinsky I, Badlani GH, Andersson KE. Pelvic floor disorders: linking genetic risk factors to biochemical changes. BJU Int. 2011;108(8):1240–7. doi: 10.1111/j.1464-410X.2011.10385.x. [DOI] [PubMed] [Google Scholar]
- 10.Allen-Brady K, Cannon-Albright L, Farnham JM, et al. Identification of six loci associated with pelvic organ prolapse using genome-wide association analysis. Obstet Gynecol. 2011;118(6):1345–53. doi: 10.1097/AOG.0b013e318236f4b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen-Brady K, Norton PA, Farnham JM, Teerlink C, Cannon-Albright LA. Significant linkage evidence for a predisposition gene for pelvic floor disorders on chromosome 9q21. Am J Hum Genet. 2009;84(5):678–82. doi: 10.1016/j.ajhg.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedigree and Population Resource: Utah Population Database. Available from: http://www.huntsmancancer.org/research/shared-resources/utah-population-database/overview.
- 13.Barber MD, Neubauer NL, Klein-Olarte V. Can we screen for pelvic organ prolapse without a physical examination in epidemiologic studies? Am J Obstet Gynecol. 2006;195(4):942–8. doi: 10.1016/j.ajog.2006.02.050. [DOI] [PubMed] [Google Scholar]
- 14.Thomas A, Gutin A, Abkevich V, Bansal A. Multipoint linkage analysis by blocked Gibbs sampling. Stat Comput. 2000;10:259–69. [Google Scholar]
- 15.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors I: complex-valued recombination fractions and complex phenotypes. Am J Hum Genet. 2000;66(3):1095–106. doi: 10.1086/302797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clerget-Darpoux F, Bonaiti-Pellie C, Hochez J. Effects of misspecifying genetic parameters in lod score analysis. Biometrics. 1986;42(2):393–9. [PubMed] [Google Scholar]
- 17.Pal DK, Durner M, Greenberg DA. Effect of misspecification of gene frequency on the two-point LOD score. Eur J Hum Genet. 2001;9(11):855–9. doi: 10.1038/sj.ejhg.5200724. [DOI] [PubMed] [Google Scholar]
- 18.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11(3):241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 19.Shi F, Harman J, Fujiwara K, Sottile J. Collagen I matrix turnover is regulated by fibronectin polymerization. Am J Physiol Cell Physiol. 2010;298(5):C1265–75. doi: 10.1152/ajpcell.00341.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi F, Long X, Hendershot A, Miano JM, Sottile J. Fibronectin matrix polymerization regulates smooth muscle cell phenotype through a Rac1 dependent mechanism. PloS One. 2014;9(4):e94988. doi: 10.1371/journal.pone.0094988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ertoy Baydar D, Kutlugun AA, Bresin E, Piras R. A case of familial glomerulopathy with fibronectin deposits caused by the Y973C mutation in fibronectin. Am J Kidney Dis. 2013;61(3):514–8. doi: 10.1053/j.ajkd.2012.08.050. [DOI] [PubMed] [Google Scholar]
- 22.Hayes JM, Hartsock A, Clark BS, Napier HR, Link BA, Gross JM. Integrin alpha5/fibronectin1 and focal adhesion kinase are required for lens fiber morphogenesis in zebrafish. Mol Biol Cell. 2012;23(24):4725–38. doi: 10.1091/mbc.E12-09-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikolova G, Lee H, Berkovitz S, et al. Sequence variant in the laminin gamma1 (LAMC1) gene associated with familial pelvic organ prolapse. Hum Genet. 2007;120(6):847–56. doi: 10.1007/s00439-006-0267-1. [DOI] [PubMed] [Google Scholar]
- 24.Agarwala V, Flannick J, Sunyaev S, Go TDC, Altshuler D. Evaluating empirical bounds on complex disease genetic architecture. Nat Genet. 2013;45(12):1418–27. doi: 10.1038/ng.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith-Mungo LI, Kagan HM. Lysyl oxidase: properties, regulation and multiple functions in biology. Matrix Biol. 1998;16(7):387–98. doi: 10.1016/s0945-053x(98)90012-9. [DOI] [PubMed] [Google Scholar]
- 26.Shynlova O, Bortolini MA, Alarab M. Genes responsible for vaginal extracellular matrix metabolism are modulated by women’s reproductive cycle and menopause. Int Braz J Urol. 2013;39(2):257–67. doi: 10.1590/S1677-5538.IBJU.2013.02.15. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Wang C, Huang DY, et al. TGF-beta1 induces the different expressions of lysyl oxidases and matrix metalloproteinases in anterior cruciate ligament and medial collateral ligament fibroblasts after mechanical injury. J Biomech. 2013;46(5):890–8. doi: 10.1016/j.jbiomech.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Gustilo-Ashby AM, Lee U, Vurbic D, et al. The impact of cesarean delivery on pelvic floor dysfunction in lysyl oxidase like-1 knockout mice. Female Pelvic Med Reconstr Surg. 2010;16(1):21–30. doi: 10.1097/SPV.0b013e3181d00035. [DOI] [PubMed] [Google Scholar]
- 29.Barash IA, Bang ML, Mathew L, Greaser ML, Chen J, Lieber RL. Structural and regulatory roles of muscle ankyrin repeat protein family in skeletal muscle. Am J Physiol Cell Physiol. 2007;293(1):C218–27. doi: 10.1152/ajpcell.00055.2007. [DOI] [PubMed] [Google Scholar]
- 30.Al Badr W, Al Bader S, Otto E, et al. Exome capture and massively parallel sequencing identifies a novel HPSE2 mutation in a Saudi Arabian child with Ochoa (urofacial) syndrome. J Pediatr Urol. 2011;7(5):569–73. doi: 10.1016/j.jpurol.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franzke CW, Tasanen K, Schacke H, et al. Transmembrane collagen XVII, an epithelial adhesion protein, is shed from the cell surface by ADAMs. EMBO J. 2002;21(19):5026–35. doi: 10.1093/emboj/cdf532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kajbafzadeh AM, Elmi A, Mazaheri P, Talab SS, Jan D. Genitourinary involvement in epidermolysis bullosa: clinical presentations and therapeutic challenges. BJU Int. 2010;106(11):1763–6. doi: 10.1111/j.1464-410X.2010.09399.x. [DOI] [PubMed] [Google Scholar]
- 33.Luo M, Chen J, Li S, et al. Changes in the metabolism of chondroitin sulfate glycosaminoglycans in articular cartilage from patients with Kashin-Beck disease. Osteoarthritis Cartilage. 2014;22(7):986–95. doi: 10.1016/j.joca.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 34.Brew K, Dinakarpandian D, Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim Biophys Acta. 2000;1477(1–2):267–83. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 35.Braicu EI, Fotopoulou C, Chekerov R, et al. Role of serum concentration of VEGFR1 and TIMP2 on clinical outcome in primary cervical cancer: results of a companion protocol of the randomized, NOGGO-AGO phase III adjuvant trial of simultaneous cisplatin-based radiochemotherapy vs. carboplatin and paclitaxel containing sequential radiotherapy. Cytokine. 2013;61(3):755–8. doi: 10.1016/j.cyto.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 36.Givvimani S, Kundu S, Narayanan N, et al. TIMP-2 mutant decreases MMP-2 activity and augments pressure overload induced LV dysfunction and heart failure. Arch Physiol Biochem. 2013;119(2):65–74. doi: 10.3109/13813455.2012.755548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stetler-Stevenson WG, Seo DW. TIMP-2: an endogenous inhibitor of angiogenesis. Trends Mol Med. 2005;11(3):97–103. doi: 10.1016/j.molmed.2005.01.007. [DOI] [PubMed] [Google Scholar]