Abstract
Background
Cutaneous T-Cell Lymphomas (Mycosis Fungoides and its leukemic variant, Sézary Syndrome) are rare malignancies. Reports of occurrence of Mycosis Fungoides in married couples and families raise the possibility of an environmental trigger for this cancer. While it was suggested that CTLC arises from inappropriate T cell stimulation, currently no preventable trigger has been identified.
Methods
We analyzed by region, zip code, age, sex and ethnicity the demographic data of 1047 patients from Texas, who were seen in a CTCL clinic at the MD Anderson Cancer Center during 2000-2012 (the MDACC database) and 1990 patients that were recorded in the population-based Texas Cancer Registry (TCR) between 1996-2010. Subsequently data from both databases was cross analyzed and compared.
Results
Our findings, based on the MDACC database, document geographic clustering of patients in three communities within the Houston metropolitan area, where CTCL incidence rates were 5-20 times higher than the expected population rate. Analysis of the TCR database defined the CTCL population rate for the state to be 5.8 [95% CI 5.5, 6.0] cases per million individuals per year, confirmed the observations from the MDACC database and further highlighted additional areas of geographic clustering and regions spared by CTCL in Texas.
Conclusions
Our study documents geographic clustering of CTCL cases in Texas and argues for the existence of yet unknown external causes/triggers for this rare malignancy.
Keywords: Cutaneous T Cell Lymphoma (CTCL), Mycosis Fungoides, Sézary Syndrome, patient clustering, geographic clustering and disease hotspots
INTRODUCTION
Cutaneous T-Cell Lymphomas (CTCL) are a rare group of non-Hodgkin’s lymphomas with the documented incidence of ~4-8 cases per million1-4. A number of studies documented a ~3 fold increase in the incidence of CTCL in the last 25-30 years1, 3. Reports indicate that recently the incidence of this cancer in the United States has stabilized at ~10 cases per million per year4. However, in different parts of the country the incidence rate can vary from ~4 to 13 cases per million individuals per year4.
Mycosis Fungoides (MF) and its leukemic variant, Sézary Syndrome (SS), are the two common subtypes of CTCL5. In Caucasians MF/SS primarily affects individuals over 55 years of age, while in African-American and Hispanic individuals this disease presents at a significantly younger age2, 6. Furthermore, CTCL was reported to have a higher predilection for males and African-Americans, where disease typically presents with higher clinical stage and follows a more aggressive clinical course2, 6.
The pathogenesis of CTCL remains only partially understood. Recent reports elucidated the nature of cancer initiating cells for MF and SS7. Multiple studies attempted to clarify the genetic multistep carcinogenesis of CTCL8-10. Also, notably, certain HLA class II alleles were associated with CTCL, therefore suggesting that one of the molecular pathogenesis mechanisms may involve inapropriate T-cell activation via antigen presentation followed by accumulation of neoplastic memory T cells11, 12.
The majority of skin cancers are caused by external and sometimes preventable agents including Human Papilloma Virus (HPV), Merkel cell polyomavirus or exposure to sun, arsenic and radiation4, 13, 14. Previous reports suggested that CTCL may occur in married couples15 and clusters in families16. These and similar findings triggered an extensive search for a viral, chemical or an occupational disease trigger, but failed to yield any conclusive etiologic agent17-19. Some patients with smoldering HTLV-1 associated Adult T-Cell Lymphoma present with MF skin lesions20, 21, but based on other studies, viruses have not been identified in the vast majority of MF cases22, 23.
In the current work, we analyze the demographic data on CTCL in Houston and Texas using two distinct databases, The MD Anderson Cancer Center (MDACC) CTCL Clinic Patient Database and the statewide population-based Texas Cancer Registry (TCR), to demonstrate the existence of disease clustering in a number of communities in Texas.
MATERIALS AND METHODS
Patient demographics and chart review
This study was approved by the MDACC IRB (IRB protocols: PA12-0497, PA12-0267 and Lab97-256). All patients signed an IRB-approved consent24. Based on the periodic IRB review of this study, the participation rate was >90%. Patient information on sex, race, date of diagnosis as well as age, clinical stage and residential address at the time of diagnosis were analyzed for patients seen in the clinic during 2000-2012 (i.e., MDACC database). The residential addresses provided by patients were compared to the addresses at the time of referral recorded by the MDACC electronic medical record (EMR) system.
Detailed individual chart review for Katy and Spring, as well as in the Houston Memorial area (zip code 77024) patients was performed and available pathological slides were retrieved and reviewed by at least two pathologists in order to confirm the diagnosis and identify important pathological features as recently reviewed in 25.
Texas Cancer Registry (TCR) is a population-based registry and it collects data on all cancers, including CTCL, for the entire state of Texas. Hence, to confirm our results we obtained de-identified data from this public database. ICD-O codes 9700/3, 9709/3, 9701/3 and ICD-10 codes C84.0, C84.1, C84.8 were used to identify cases of CTCL diagnosed statewide during 1996-2010 (dates of data availability). Data was provided for the entire state and for each individual zip code. TCR does not provide data by city since city limits frequently change. TCR was not able to provide the data by clinical stage at the time of diagnosis. Comparative analysis between the two databases was conducted using the overlapping data sets for 2002-2010 years.
Mapping Analysis
Maps indicating the residence of all patients recorded by the MDACC database and zip codes identified by the TCR were created using GIS software (ArcMap 10.1 from Environmental Systems Research Institute-ESRI, Redlands, CA). For the map of Texas, a standard ESRI template was generated and zip code information was added. To build a Houston metropolitan area map, a standard ESRI template was used and individual addresses were entered. In mapping of the TCR results, only zip codes with populations >10,000 were selected in order to reduce erroneous false-positive hits, where a single case of CTCL in a zip code with <500 residents might have artificially inflated the incidence rate.
Statistical Analyses
Unless otherwise specified, analysis of the complete data on all patients seen at the MDACC CTCL clinic during 2000-2012 is presented throughout the paper. Incidence rates and 95% confidence intervals (CI) were calculated and reported overall, by year of diagnosis and specific regions that were identified by the mapping analysis. Unless otherwise specified, 2000 and 2010 US Census data was used for all population analyses, where ≤2005 results were compared to the year 2000 US Census, while ≥2006 results were compared to 2010 US census. Confidence intervals were based on Poisson distributions. Incidence rates were plotted using linear regression model to assess trends over time. Standardized Mortality Ratio (SMR) with standardization for age and gender for 2000-2010 was calculated as previously described26. For all analyses standard model selection procedures were used to select the final models 27.
RESULTS
Comparison of the MDACC CTCL clinic database with the TCR public database
During the study period (2000-2012), 1047 CTCL patients were seen in our MDACC CTCL clinic with on average ~80 new cases being evaluated and treated each year from Texas24. Also, to corroborate our findings from the MDACC database, we obtained de-identified data from the TCR for 1996-2010 years (dates of data availability). During this time 1990 cases of CTCL were recorded in the registry with ~132 cases on average being documented each year. Linear regression analysis documented a steady increase in the annual incidence rate over the past 15 years (Figure 1A). The overall annual incidence rate for the state of Texas for 1996-2010 years was 5.8; 95% CI [5.5, 6.0] cases per million individuals per year. The rate of increase for CTCL incidence was calculated to be 0.16 cases per million individuals per year (Figure 1A).
Figure 1. Incidence of CTCL in Texas over time.
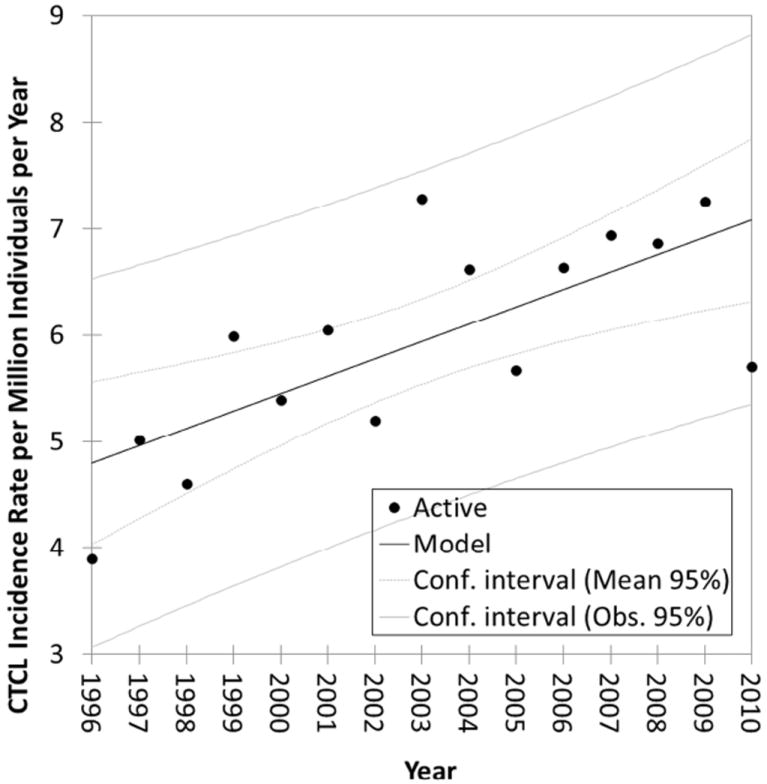
Linear regression analysis of CTCL population incidence rate over time (R2=0.526, p=0.02). The slope of the line is 0.16 cases per million individuals per year (TCR database results).
We also conducted a comparative analysis between the two databases using the overlapping data sets for 2002-2010 years. During these years, MDACC database documented 717 cases, while the TCR documented 1366 cases (i.e., MDACC had a catch rate of ~52% for the entire state of Texas). Within the state of Texas the MDACC and TCR databases had 400 zip codes in common. For these zip codes the TCR database documented 816 patients, while the MDACC database documented 710 patients, which corresponds to 87% correlation rate between the two databases.
Demographic characteristics of patients in both databases revealed that they came from various racial groups that were reflective of the demographic representation of the state (Table 1A and B). Most patients were diagnosed in their late 50s. Slight predominance of this disease was noted in males and the majority of patients presented with stage I disease (Table 1A and B).
Table 1.
Clinical characteristics of CTCL patients in the study based on (A) MDACC and (B) TCR databases
| A. | MDACC Database | State of Texas | Spring, TX | Katy, TX | Memorial Area | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| n | % | n | % | p* | n | % | p* | n | % | p* | ||||
|
|
||||||||||||||
| Number of Patients | 1047 | 40 | N/A | 25 | N/A | 16 | N/A | |||||||
|
|
|
|
||||||||||||
| Age at Diagnosis | 56.3±16.6 | 49.7±16.9 | 52.4±13.4 | 61.8±18.9 | ||||||||||
|
|
||||||||||||||
| Sex | Female | 491 | 46.9 | 24 | 60 | 0.10 | 12 | 48 | 0.91 | 5 | 31.3 | 0.21 | ||
|
|
|
|
||||||||||||
| Male | 556 | 53.1 | 16 | 40 | 13 | 52 | 11 | 68.7 | ||||||
|
|
||||||||||||||
| Race | Caucasian | 718 | 68.6 | 25 | 62.5 | 0.73 | 21 | 84 | 0.25 | 13 | 81.3 | 0.41 | ||
|
|
|
|
||||||||||||
| African-American | 144 | 13.8 | 8 | 20 | 2 | 8 | 1 | 6.2 | ||||||
|
|
|
|
||||||||||||
| Hispanic | 156 | 14.9 | 6 | 15 | 1 | 4 | 1 | 6.3 | ||||||
|
|
|
|
||||||||||||
| Other | 29 | 2.8 | 1 | 2.5 | 1 | 4 | 1 | 6.2 | ||||||
|
|
||||||||||||||
| CTCL Clinical Stage | I | 794 | 75.8 | 35 | 87.5 | 0.16 | 22 | 88 | 0.70 | 13 | 81.2 | 0.85 | ||
|
|
|
|
||||||||||||
| II | 112 | 10.7 | 2 | 5 | 1 | 4 | 2 | 12.5 | ||||||
|
|
|
|
||||||||||||
| III | 32 | 3.1 | 2 | 5 | 0 | 0 | 0 | 0 | ||||||
|
|
|
|
||||||||||||
| IV | 109 | 10.4 | 1 | 2.5 | 2 | 8 | 1 | 6.3 | ||||||
| B. | TCR Database | State of Texas | |||
|---|---|---|---|---|---|
|
| |||||
| n | % | p* | |||
|
|
|||||
| Number of Patients | 1990 | N/A | |||
|
|
|||||
| Age at Diagnosis | 58.7±17.2 | ||||
|
|
|||||
| Sex | Female | 892 | 44.8 | 0.28 | |
|
|
|||||
| Male | 1098 | 55.2 | |||
|
|
|||||
| Race | Caucasian | 1308 | 65.7 | 0.35 | |
|
|
|||||
| African-American | 318 | 16.0 | |||
|
|
|||||
| Hispanic | 303 | 15.2 | |||
|
|
|||||
| Asian Pacific | 24 | 1.2 | |||
|
|
|||||
| Other | 37 | 1.9 | |||
We also analysed CTCL incidence rates in all major cities in the state. The rate for Houston was documented to be 6.4 [5.8, 7.1] cases per million per year. The rates for Austin, Dallas, Fort Worth and San Antonio were 6.3 [5.3, 7.6]; 6.6 [5.8, 7.5]; 5.1[4.2, 6.2] and 4.7 [3.9, 5.6] per million individuals per year, respectively.
Mapping analysis of CTCL cases in Houston metropolitan area
MDACC patients were mapped based on their residential zip codes. This analysis revealed that patients were clustering in several communities (Figure 2). Specifically, analysis of the Houston metropolitan area demonstrated that communities in the north of the city (Spring, population~54,500), west of the city (Katy, population~14,000) and Houston Memorial area (zip code 77024, population~35,000) contained a higher number of cases than would be expected from population data (Figure 2, inset 3 and Figure 3). Mapping of patient residential addresses from these communities revealed a striking clustering of cases, where in certain instances patients resided along the same highway/street and/or near streams (Figure 3). Calculated incidence rates in Katy, Spring and Houston Memorial area were 136, 52 and 34 cases per million individuals per year, respectively. Furthermore, standardized Mortality Ratio (SMR) analysis adjusted by age and sex for the 2000-2010 demonstrated that the incidence rates of this cancer in Katy and Spring were ~9-35 fold higher than documented Houston metropolitan rate (Table 2).
Figure 2. Geographic distribution of CTCL in Texas (MDACC database results).
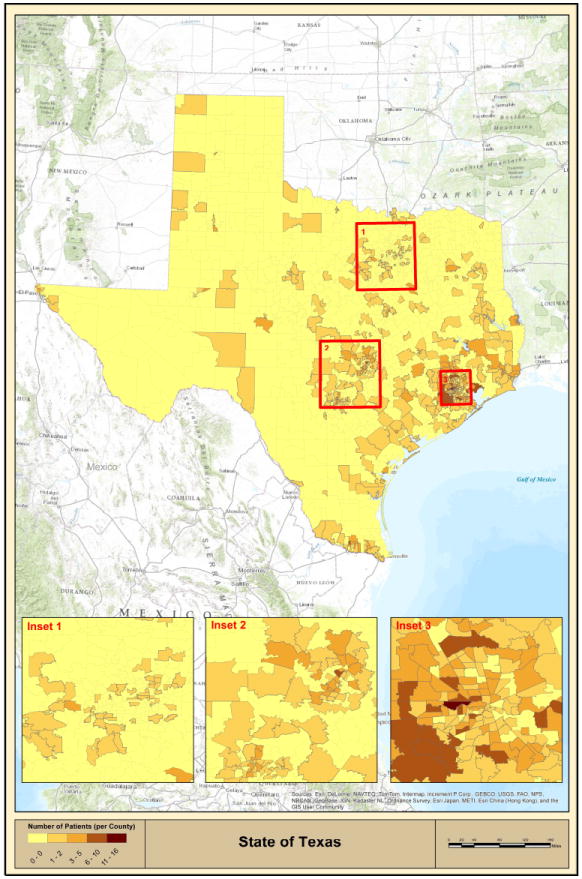
Geographic distribution of CTCL patients in the state of Texas based on zip code mapping analysis. Insets 1, 2 and 3 show incidence of disease in Dallas-Fort Worth, Austin-San Antonio and Houston metropolitan areas, respectively.
Figure 3. Detailed map of high CTCL incidence areas in Houston (MDACC database results).
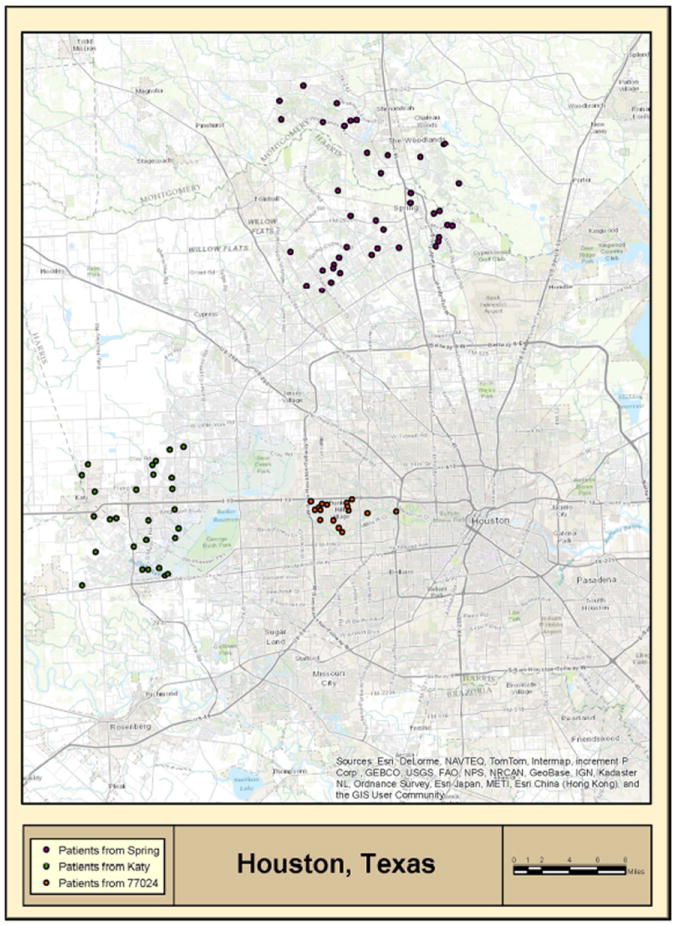
Geographic mapping reveals CTCL high incidence areas, where patients with this rare cancer were residing along the same street/highway and/or streams. Patients in Spring are marked in violet, patients in Katy are marked in green, while patients residing in Houston Memorial area (zip code 77024) are marked in orange.
Table 2.
Standardized Mortality Ratio (SMR) analysis by age and sex was used to compare disease incidence in the areas of CTCL geographic clustering. MDACC database results were used for this analysis. Data presented as folds higher than the Houston metropolitan incidence rate (6.4 cases per million per year).
| Standardized Mortality Ratio Analysis | |||||
|---|---|---|---|---|---|
| Age standardization | Rate ratio: Observed/Expected=SMR | 95% CI of SMR | |||
| Houston Memorial Area (77024) | 16/2.9 = 5.5 | 3.2 | 9.0 | ||
| KATY | 24/0.8 = 31.2 | 20.0 | 46.4 | ||
| SPRING | 40/2.2 = 18.0 | 12.9 | 24.5 | ||
| Age-Sex standardization | Sex | Rate ratio: Observed/Expected=SMR | |||
| Houston Memorial Area (77024) | Male | 11/1.1 = 10.0 | 5.0 | 17.8 | |
| Female | 5/1.26 = 4.0 | 1.3 | 9.3 | ||
| KATY | Male | 12/0.34 = 34.9 | 18.0 | 60.9 | |
| Female | 12/0.41 = 29.6 | 15.3 | 51.7 | ||
| SPRING | Male | 16/1.03 = 15.5 | 8.9 | 25.2 | |
| Female | 24/1.29 = 18.6 | 11.9 | 27.7 | ||
Detailed chart review was performed for patients from these communities. Based on past address history in the MDACC database, we have confirmed that these patients were living in these areas prior to their diagnosis, precluding the possibility of clustering caused by an intentional migration to an area close to our cancer center. Also, we confirmed that they were referred by a diverse group of practicing physicians (General Practitioners, Dermatologists and Oncologists) from the Houston metropolitan area. From the review of pathological slides, we observed that the spectrum of disease observed in patients from these communities was representative of the overall spectrum of CTCL seen in our center. In particular, the majority of cases (i.e., > 80% of cases) exhibited typical changes of patch/plaque CD4+ MF. Tumor lesions, folliculotropic MF, large cell transformation or younger patients with CD8+ disease did not appear to be either over- or under-represented in this population.
Mapping analysis of cases for the state of Texas
Similar mapping and statistical analyses by zip code were conducted using the TCR database results. Based on the conducted statistical analyses, 93 zip codes were identified to have a significantly higher incidence rates than recorded for the entire state. We subsequently mapped only selected zip codes with populations >10,000 in order to reduce erroneous false-positive hits. Mapping analysis of these 48 zip codes confirmed 2 out of 3 disease clusters (i.e., Houston Memorial area and Spring, TX) within the Houston metropolitan area (Figure 4, black arrows in Inset 3). Population incidence rates for Spring, Katy and Houston Memorial areas are presented in table 3A.
Figure 4. Geographic clustering of multiple high incidence zip codes across Texas (TCR Database Results).
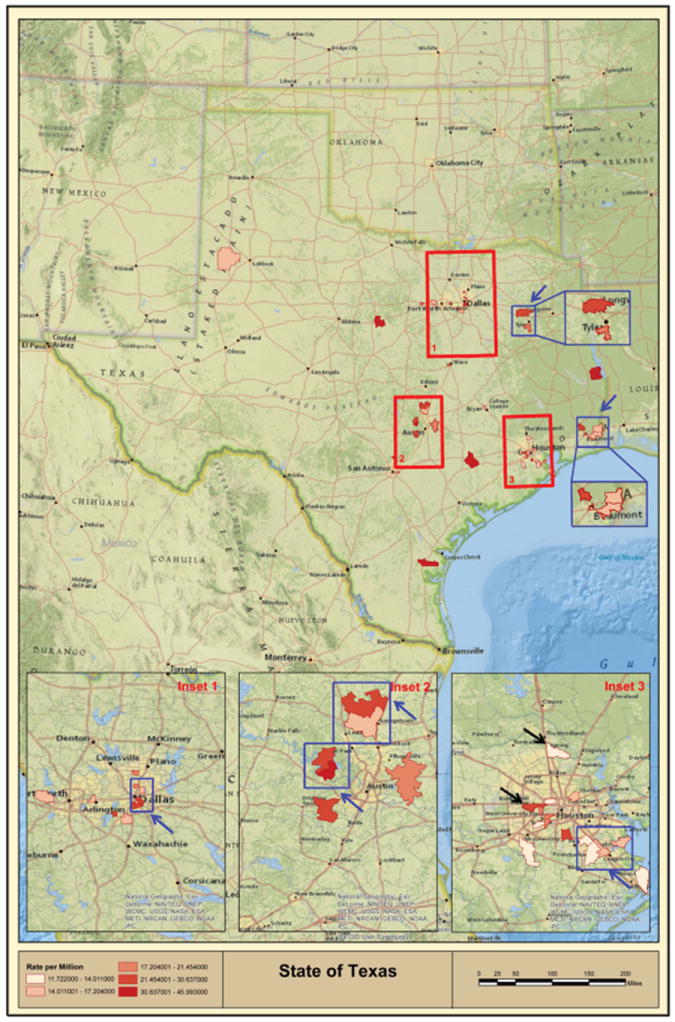
Geographic distribution of CTCL high incidence zip code clusters (i.e., multiple high incidence zip codes located together) as indicated by blue squares and blue arrows for zip codes with total population >10,000 individuals. Inset 3 shows Houston Metropolitan area, where TCR analysis confirmed two out of three high incidence areas (Spring and Memorial area zip code 77024) as identified using the MDACC database and indicated with black arrows.
Table 3.
Incidence of CTCL across Texas. A. Calculated incidence rates for the selected communities using TCR database zip code results. B. List of clusters of high incidence zip codes and corresponding population incidence rates by geographic area (TCR database results). C. List of identified low incidence zip codes and corresponding population incidence rates (TCR database results).
| A. | TCR Database | Incidence Rate (1996-2010) [95% CI] |
|---|---|---|
|
|
||
| Spring, TX (all zip codes) | 10.6 [7.2, 15.8]* | |
|
|
||
| Memorial area (77024) | 26.3 [16.1, 43.0]* | |
|
|
||
| Katy, TX all zip codes | 9.2 [5.5, 15.2] | |
| B. | TCR Database | Zip codes | Incidence rate (1996-2010) [95% CI] |
|---|---|---|---|
|
|
|||
| Beaumont area | 77630 | 14.4 [6.5, 32.1] | |
|
|
|||
| 77632 | 15.0 [6.2, 36.0] | ||
|
|
|||
| 77651 | 20.0 [7.5, 53.4] | ||
|
|
|||
| 77657 | 22.8 [10.3, 50.8] | ||
|
|
|||
| 77701 | 17.1 [6.4, 45.7] | ||
|
|
|||
| 77659 | 28.1 [7.0, 112.4] | ||
|
|
|||
| Tyler/Lindale area | 75703 | 17.8 [9.2. 34.1] | |
|
|
|||
| 75771 | 28.7 [13.7, 60.2] | ||
|
|
|||
| Dallas | 75115 | 15.4 [8.3, 28.6] | |
|
|
|||
| 75214 | 15.6 [7.8, 31.1] | ||
|
|
|||
| 75215 | 28.0 [13.3, 58.7] | ||
|
|
|||
| 75216 | 22.9 [14.2, 36.8] | ||
|
|
|||
| 75225 | 19.4 [8.7, 43.2] | ||
|
|
|||
| 75248 | 18.1 [9.4, 34.7] | ||
|
|
|||
| 75249 | 23.6 [8.9, 64.0] | ||
|
|
|||
| North Austin | 78628 | 16.1 [7.3, 35.9] | |
|
|
|||
| 78633 | 24.1 [11.5, 50.6] | ||
|
|
|||
| West Austin | 78645 | 30.6 [11.5, 81.6] | |
|
|
|||
| 78734 | 34.9 [17.4, 69.8] | ||
|
|
|||
| Central Houston | 77008 | 13.5 [6.1, 30.1] | |
|
|
|||
| 77024 | 23.7 [13.5, 41.7] | ||
|
|
|||
| 77056 | 20.4 [8.5, 49.0] | ||
|
|
|||
| 77025 | 21.5 [10.7, 42.9] | ||
|
|
|||
| 77096 | 14.0 [6.7, 29.4] | ||
|
|
|||
| 77005 | 19.1 [9.1, 40.1] | ||
|
|
|||
| Southeast Houston | 77048 | 22.6 [9.4, 54.2] | |
|
|
|||
| 77089 | 12.5 [6.3, 25.1] | ||
|
|
|||
| 77546 | 12.3 [6.2, 24.6] | ||
|
|
|||
| 77598 | 15.6 [6.5, 37.5] | ||
|
|
|||
| 77586 | 16.8 [7.0, 40.3] | ||
|
|
|||
| All Texas | 5.8 [5.5; 6.0] | ||
| C. | TCR Database | Zip codes | Population | Incidence rate (1996-2010) [95% CI] |
|---|---|---|---|---|
|
|
||||
| Areas of low incidence of CTCL | 79936 | 101,500 | 0 [0, 2.5] | |
|
|
||||
| 79928 | 49,500 | 0 [0, 5.2] | ||
|
|
||||
| 78596 | 57,500 | 0 [0, 4.5] | ||
|
|
||||
| 78240 | 47,500 | 0 [0, 5.4] | ||
|
|
||||
| 78046 | 54,000 | 0 [0, 4.7] | ||
|
|
||||
| 77573 | 56,500 | 0 [0, 4.5] | ||
denotes statistical significance.
Most strikingly, this analysis showed that a number of identified zip codes clustered together and highlighted additional areas, where multiple high incidence zip codes were adjacent or contiguous (Figure 4, blue squares and blue arrows). The complete list of these zip code clusters is provided in table 3B. The most remarkable agglomeration of these high incidence zip codes outside of major cities was found in Beaumont, TX (population ~118,000), where 6 high incidence zip codes covered a major part of the city (Figure 4 and Table 3B).
We also conducted rate analysis in order to identify areas, where CTCL incidence was less than expected. This analysis revealed only 6 zip codes (Table 3C). Notably, 2 out of 6 zip codes (79936 and 79928) were adjacent geographically and were located in the hot desert climate near El Paso, TX. These zip codes have sizable populations with 101,500 people residing in 79936 zip code and 49,500 people residing in 79928 zip code. No cases of CTCL were detected in either zip code during 1996-2010, which is highly significant. According to the city data, the population in these zip codes was well educated (>70% of individuals with High-School degrees) and median household incomes (>$45,000) were comparable to the state average. These zip codes are served by numerous hospitals including, The University Medical Center tertiary hospital. According to the American Academy of Dermatology, there are 13 dermatologists practicing in El Paso, TX, which would make local diagnosis and treatment of CTCL easily accessible. Hispanics represent over 80% of the population in these areas and these zip codes are located adjacent to the borders of New Mexico and Mexico.
DISCUSSION
CTCL is a rare malignancy and previous epidemiologic studies based on the Surveillance, Epidemiology and End Results (SEER) databases established that until recently this disease was on the rise in the United States4. A literature search revealed a two reports, where Git et al. in 1977 demonstrated clustering of patients in the Västernorrland county of Sweden28 and more recently Dr. Geskin’s group demonstrated non-random distribution of CTCL patients in the Greater Pittsburgh Area29.
A retrospective analysis of our MDACC database documented that the incidence of CTCL is elevated in the three areas of Houston and is significantly higher than the incidence in all of Houston. In order to corroborate this interesting observation we compared our data to the TCR population-based database, which confirmed our findings and identified additional areas of clustering. Also, our analysis of the TCR database demonstrated two adjacent low CTCL incidence zip codes near El Paso, Texas, a hot desert area adjacent to the borders of New Mexico and Mexico. One can potentially speculate that ~150,000 residents in this area were not exposed to a potential disease trigger.
The MDACC database is directly connected to each patient and, hence, enables us to access and analyze a wealth of clinical information, while the TCR database only includes very limited de-identified data by zip code. This is very important since a small city of Katy (population of ~14,000) is served by 4 zip codes (77449, 77450, 77493 and 77494). The total combined population of these zip codes is ~182,000 people. Hence, when the zip code analysis is conducted the number of cases identified in the cities is significantly diluted into a larger population encompassed by these zip codes and does not accurately capture the relative geographic proximity of the affected patients. In light of this, it is not surprising that the TCR database could not validate Katy, TX as a CTCL cluster. On the on the other hand, it is striking that the TCR was able to validate Spring, TX as a disease cluster despite this dilution effect. Since Houston Memorial area was defined as a single zip code, 77024, this problem did not arise and the TCR database easily validated this high incidence area.
One of the important limitations of the current study at this time is the absence of a distinctive etiologic agent as a possible trigger for CTCL in these communities. Identifying such a trigger will require extensive evidence from around the world before one can reliably assign causality. Possible factors may include contaminated water supply, air pollution or industrial exposure in these communities. Furthermore, exposure to an inciting infectious, radioactive or chemical agent cannot be excluded. Spring and Memorial areas are highly wooded with multiple small streams. Memorial area and Katy share the Buffalo Bayou and the I-10 interstate highway. Houston, Beaumont and Tyler areas house multiple oil refineries. A number of potential pollution sources were documented across Texas. Geographic maps combining the identified high incidence zip codes and radioactive/oil refinery pollution are presented in supplementary figures 1 and 2.
In conclusion, our study combined with previous reports 28, 29 strongly argues for the existence of environmental and potentially preventable trigger for this cancer.
Supplementary Material
Supplementary Figure 1. Geographic map of radiation pollution sources in Texas.
Supplementary Figure 2. Geographic map of oil refinery locations in Texas.
Acknowledgments
We thank the Texas Cancer Registry for providing de-identified data on patients in Texas diagnosed with CTCL. We thank Mr. Brendan Cordeiro and Mr. Gregory Cormack for their technical assistance.
Details of all funding sources: This work was supported by the NIH Research Grants (R21CA074117 and K24CA08681) to Dr. Duvic. Fonds de la recherche en santé du Québec (FRSQ) research grant #22648 to Dr. Sasseville and the Canadian Dermatology Foundation Research grant to Dr. Litvinov, Dr. Pehr and Dr. Sasseville. The development of CTCL database was supported by Sherry L. Anderson and Stanton CTCL Patient Research Funds.
Abbreviations
- CTCL
Cutaneous T Cell Lymphoma
- MF
Mycosis Fungoides
- SS
Sézary Syndrome
- TCR
Texas Cancer Registry
- MDACC
MD Anderson Cancer Center
- SMR
Standardized Mortality Ratio
- CI
Confidence Intervals
Footnotes
Conflicts of interest/Financial disclosures: Authors declare no competing interests.
References
- 1.Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood. 2009;113:5064–5073. doi: 10.1182/blood-2008-10-184168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson LD, Hinds GA, Yu JB. Age, race, sex, stage, and incidence of cutaneous lymphoma. Clin Lymphoma Myeloma Leuk. 2012;12:291–296. doi: 10.1016/j.clml.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch Dermatol. 2007;143:854–859. doi: 10.1001/archderm.143.7.854. [DOI] [PubMed] [Google Scholar]
- 4.Buzzell RA. Carcinogenesis of cutaneous malignancies. Dermatol Surg. 1996;22:209–215. doi: 10.1111/j.1524-4725.1996.tb00311.x. [DOI] [PubMed] [Google Scholar]
- 5.Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105:3768–3785. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 6.Sun G, Berthelot C, Li Y, et al. Poor prognosis in non-Caucasian patients with early-onset mycosis fungoides. J Am Acad Dermatol. 2009;60:231–235. doi: 10.1016/j.jaad.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 7.Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood. 2010;116:767–771. doi: 10.1182/blood-2009-11-251926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litvinov IV, Jones DA, Sasseville D, Kupper TS. Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma. Clin Cancer Res. 2010;16:2106–2114. doi: 10.1158/1078-0432.CCR-09-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J, Monti S, Aires DJ, et al. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110:3015–3027. doi: 10.1182/blood-2006-12-061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Kester MS, Borg MK, Zoutman WH, et al. A meta-analysis of gene expression data identifies a molecular signature characteristic for tumor-stage mycosis fungoides. J Invest Dermatol. 2012;132:2050–2059. doi: 10.1038/jid.2012.117. [DOI] [PubMed] [Google Scholar]
- 11.Jackow CM, McHam JB, Friss A, Alvear J, Reveille JR, Duvic M. HLA-DR5 and DQB1*03 class II alleles are associated with cutaneous T-cell lymphoma. Journal of Investigative Dermatology. 1996;107:373–376. doi: 10.1111/1523-1747.ep12363352. [DOI] [PubMed] [Google Scholar]
- 12.Hodak E, Lapidoth M, Kohn K, et al. Mycosis fungoides: HLA class II associations among Ashkenazi and non-Ashkenazi Jewish patients. Br J Dermatol. 2001;145:974–980. doi: 10.1046/j.1365-2133.2001.04496.x. [DOI] [PubMed] [Google Scholar]
- 13.Czapiewski P, Biernat W. Merkel cell carcinoma - Recent advances in the biology, diagnostics and treatment. Int J Biochem Cell Biol. 2014 doi: 10.1016/j.biocel.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Fabbrocini G, Triassi M, Mauriello MC, et al. Epidemiology of skin cancer: role of some environmental factors. Cancers (Basel) 2010;2:1980–1989. doi: 10.3390/cancers2041980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazen PG, Michel B. Hodgkin’s disease and mycosis fungoides in a married couple. Dermatologica. 1977;154:257–260. doi: 10.1159/000251078. [DOI] [PubMed] [Google Scholar]
- 16.Hodak E, Klein T, Gabay B, et al. Familial mycosis fungoides: report of 6 kindreds and a study of the HLA system. J Am Acad Dermatol. 2005;52:393–402. doi: 10.1016/j.jaad.2003.12.052. [DOI] [PubMed] [Google Scholar]
- 17.Morales-Suarez-Varela MM, Olsen J, Johansen P, et al. Occupational risk factors for mycosis fungoides: a European multicenter case-control study. J Occup Environ Med. 2004;46:205–211. doi: 10.1097/01.jom.0000116819.01813.8c. [DOI] [PubMed] [Google Scholar]
- 18.Wohl Y, Tur E. Environmental risk factors for mycosis fungoides. Curr Probl Dermatol. 2007;35:52–64. doi: 10.1159/000106410. [DOI] [PubMed] [Google Scholar]
- 19.Mirvish ED, Pomerantz RG, Geskin LJ. Infectious agents in cutaneous T-cell lymphoma. J Am Acad Dermatol. 2011;64:423–431. doi: 10.1016/j.jaad.2009.11.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reitz MS, Jr, Poiesz BJ, Ruscetti FW, Gallo RC. Characterization and distribution of nucleic acid sequences of a novel type C retrovirus isolated from neoplastic human T lymphocytes. Proc Natl Acad Sci U S A. 1981;78:1887–1891. doi: 10.1073/pnas.78.3.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zucker-Franklin D, Pancake BA. The role of human T-cell lymphotropic viruses (HTLV-I and II) in cutaneous T-cell lymphomas. Semin Dermatol. 1994;13:160–165. [PubMed] [Google Scholar]
- 22.Detmar M, Pauli G, Anagnostopoulos I, et al. A case of classical mycosis fungoides associated with human T-cell lymphotropic virus type I. Br J Dermatol. 1991;124:198–202. doi: 10.1111/j.1365-2133.1991.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 23.Pawlaczyk M, Filas V, Sobieska M, Gozdzicka-Jozefiak A, Wiktorowicz K, Breborowicz J. No evidence of HTLV-I infection in patients with mycosis fungoides and Sezary syndrome. Neoplasma. 2005;52:52–55. [PubMed] [Google Scholar]
- 24.Talpur R, Singh L, Daulat S, et al. Long-term outcomes of 1,263 patients with mycosis fungoides and Sezary syndrome from 1982 to 2009. Clinical Cancer Research. 2012;18:5051–5060. doi: 10.1158/1078-0432.CCR-12-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part II. Prognosis, management, and future directions. J Am Acad Dermatol. 2014;70:223 e221–217. doi: 10.1016/j.jaad.2013.08.033. quiz 240-222. [DOI] [PubMed] [Google Scholar]
- 26.Stevenson EK, Rubenstein AR, Radin GT, Wiener RS, Walkey AJ. Two decades of mortality trends among patients with severe sepsis: a comparative meta-analysis*. Crit Care Med. 2014;42:625–631. doi: 10.1097/CCM.0000000000000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosmer DW, L . Applied Logistic Regression. 2. New York, NY: John Wiley & Sons Inc.; 2000. [Google Scholar]
- 28.Gip L, Nilsson E. Clustering of mycosis fungoides in the County of Vasternorrland. Lakartidningen. 1977;74:1174–1176. [PubMed] [Google Scholar]
- 29.Moreau JF, Buchanich JM, Geskin JZ, Akilov OE, Geskin LJ. Non-random geographic distribution of patients with cutaneous T-cell lymphoma in the Greater Pittsburgh Area. Dermatol Online J. 2014;20 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Geographic map of radiation pollution sources in Texas.
Supplementary Figure 2. Geographic map of oil refinery locations in Texas.


