Abstract
Ablation of glycosylated lysosomal membrane protein (GLMP, formerly known as NCU-G1) has been shown to cause chronic liver injury which progresses into liver fibrosis in mice. Both lysosomal dysfunction and chronic liver injury can cause metabolic dysregulation. Glmpgt/gt mice (formerly known as Ncu-g1gt/gtmice) were studied between 3 weeks and 9 months of age. Body weight gain and feed efficiency of Glmpgt/gt mice were comparable to wild type siblings, only at the age of 9 months the Glmpgt/gt siblings had significantly reduced body weight. Reduced size of epididymal fat pads was accompanied by hepatosplenomegaly in Glmpgt/gt mice. Blood analysis revealed reduced levels of blood glucose, circulating triacylglycerol and non-esterified fatty acids in Glmpgt/gt mice. Increased flux of glucose, increased de novo lipogenesis and lipid accumulation were detected in Glmpgt/gt primary hepatocytes, as well as elevated triacylglycerol levels in Glmpgt/gt liver homogenates, compared to hepatocytes and liver from wild type mice. Gene expression analysis showed an increased expression of genes involved in fatty acid uptake and lipogenesis in Glmpgt/gt liver compared to wild type. Our findings are in agreement with the metabolic alterations observed in other mouse models lacking lysosomal proteins, and with alterations characteristic for advanced chronic liver injury.
Introduction
Glycosylated lysosomal membrane protein (GLMP, formerly known as NCU-G1) was first described as a nuclear protein, capable of regulating gene transcription [1]. Later studies identified GLMP as a bona fide lysosomal membrane protein [2–4]. The biological function of GLMP is unknown, but a high degree of conservation of the amino acid sequence indicates an important function [1]. Recently, we created and characterized a novel mouse model with no detectable expression of GLMP (Glmp gt/gt mouse, formerly known as Ncu-g1 gt/gt mouse), and showed that the predominant phenotype is chronic liver injury which had developed into a well-established fibrosis by the age of 6 months [5]. Further analyses identified accumulation of iron in Kupffer cells [5], which has been shown to be associated with metabolic dysregulation [6]. Specific accumulation of iron in Kupffer cells has been shown to correlate with the severity of metabolic liver injury [7].
Lysosomal disorders are a group of congenital metabolic disorders caused by malfunctioning of a protein related to normal lysosomal function [8–13]. Most lysosomal disorders are characterized by impaired turnover of certain metabolites, which accumulate intralysosomally and affect normal lysosome and lysosome-related processes, such as autophagy [9–11]. Defective recycling of macromolecules through lysosomal pathways may cause insufficient recycling of metabolites and metabolic stress [11]. In fact, metabolic irregularities have been demonstrated in several mouse models with lysosomal disorders [14].
Liver fibrosis is a result of chronic liver injury, and is characterized by capillarization of sinusoids and redirection of blood directly from the portal tract to the hepatic vein [15–17]. The impaired perfusion of the liver and loss of fenestrations across the endothelial cell layer contribute to the imbalance in metabolite exchange between plasma and the liver [15]. The liver is also an important organ for regulation of glucose, lipid and protein metabolism, and chronic liver injury is known to cause alterations to glucose and lipid homeostasis [18].
Using the Glmp gt/gt mouse, a mouse model lacking this lysosomal membrane protein, and characterized by chronic liver injury and liver fibrosis, we assessed the metabolic consequences of GLMP ablation, focusing on liver.
Materials and Methods
Materials
[1-14C]oleic acid (58.2 mCi/mmol), D-[14C(U)]glucose (2.9 mCi/mmol) and [1-14C]acetic acid (56.0 mCi/mmol) were from PerkinElmer NEN (Boston, MA, US). Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), Penicillin-Streptomycin (Pen-Strep), Fungizone and Dulbecco's phosphate-buffered saline (DPBS, with Mg2+ and Ca2+) were purchased from Life-Technologies (Carlsbad, CA, US). HEPES, L-carnitine, oleic acid (OA), glucose, bovine serum albumin (BSA), Triton X-100, sodium dodecyl sulfate (SDS) and protease inhibitor cocktail were from Sigma-Aldrich (St. Louis, MO, US). Sodium chloride and Tris Base were from VWR (Radnor, PA, US). Corning CellBIND tissue culture plates (12-well and 96-well plates) were obtained from Corning Life-Sciences (Schiphol-Rijk, The Netherlands). OptiPhase Supermix and UniFilter-96 GF/B were obtained from PerkinElmer (Shelton, CT, US). Thin layer chromatography plates were purchased from Merck (Darmstadt, Germany). Accu-Chek Aviva Nano Blood Glucose Meter System was purchased from Roche Applied Science (Mannheim, Germany). RNeasy Plus kit was obtained from Qiagen (Hilden, Germany). Brilliant III Ultra Fast SYBR Green qPCR Master Mix was obtained from Agilent Technologies (Santa Clara, CA, US) and LightCycler 480 SYBR Green I Master Mix was purchased from Roche Applied Science. Primers were designed and purchased from DNA Technology (Risskov, Denmark) and Life Technologies. Hypnorm was from VetaPharma (Leeds, UK). Midazolam was from Actavis (Parsippany, NJ, US). Animal diets were purchased from Scanbur (Karlslunde, Denmark). Protein assay reagent was purchased from Bio-Rad (Bio-Rad, Hercules, CA, US) and the content of proteins was determined using the Coomassie reagent [19]. All other chemicals used in this study were of standard commercial high-purity quality.
Animal experiments
All animal experiments were reviewed and approved by the Norwegian Animal Research Authority, and performed according to national laws and regulations. Wild type (WT) and Glmp gt/gt mice [5] were housed in an approved animal facility with access to standard rodent chow and water ad libitum unless otherwise stated. Mice were 3 weeks old at the beginning of the feeding experiment. The body weight and food intake of a total of 27 male WT and 31 male Glmp gt/gt mice, housed in 12 different cages, and a total of 13 female WT and 20 female Glmp gt/gt mice, housed in 7 different cages were monitored at intervals of 7 days for 12 weeks, and then at 2, 3.5, 4.5, 6, 7.5 and 9 months of age. Feed efficiency was calculated as the ratio between body weight gain/week for each individual mouse and the average feed intake/mouse. Biological samples were collected at designated age points (1, 2, 3.5, 6, 7.5, and 9 months). Blood was obtained from randomly fed mice through cardiac puncture, and sera were collected after coagulation and centrifugation at 1500 x g for 20 minutes. Liver, spleen and epididymal fat pads (representing visceral adipose tissue) were dissected at selected time points, weighed and frozen in liquid nitrogen. The axillary and inguinal fat depots (representing subcutaneous adipose tissue) and interscapular brown adipose tissue were inspected at 1 and 4.5 months of age. All biological samples were stored at -80°C until further analysis.
Resting blood glucose and serum and liver lipid levels
Resting blood glucose levels were measured in male WT and Glmp gt/gt mice (age = 5 months) fed ad libitum using Accu-Chek Aviva Nano Blood Glucose Meter System. Serum levels of non-esterified fatty acids (NEFA) were analyzed using NEFA-kit according to the supplier´s protocol (ILS Laboratories Scandinavia AS, Oslo, Norway). Triacylglycerol (TAG) was quantified in serum and whole liver homogenates using TG PAP 150-kit (BioMerieux, Marcy l’Etoile, France) according to the supplier’s protocol.
Isolation of primary hepatocytes
Isolation of primary hepatocytes from WT and Glmp gt/gt mice (age = 4–5 weeks) was carried out by a two-step perfusion method as described [20]. Liver parenchymal and non-parenchymal cells were separated by differential centrifugation as described elsewhere [21]. Primary hepatocyte preparations with high viability (> 95%) were used in further studies.
Uptake and oxidation of oleic acid and glucose
Determination of [14C]oleic acid (OA) or [14C]glucose uptake and oxidation has been described previously [22]. Primary hepatocytes were isolated as described above, and cultured in hepatocyte growth medium (DMEM high glucose, enriched with 20% FBS, Pen-Strep and Fungizone) on a 96-well microplate (80.000 cells/well) (Corning CellBIND). For measurements of OA or glucose uptake and oxidation, the growth medium was removed after 24 h and the cells were exposed to [14C]OA (0.5 μCi/mL, 100 μM) bound to BSA (40 μM) at a ratio of 2.5:1 in DPBS (with Mg2+ and Ca2+) with HEPES (10 mM), L-carnitine (1 mM) for 4 h or [14C]glucose (0.6 μCi/mL, 200 μM) in DPBS (with Mg2+ and Ca2+) with HEPES (10 mM), L-carnitine (1 mM) and BSA (7.2 μM) for 4 h. [14C]CO2 production and total uptake of OA or glucose (sum of CO2 and cell-associated radioactivity) were calculated using cell protein levels for standardization as described previously [22].
Thin layer chromatography
Primary hepatocytes were isolated as described above, and cultured in hepatocyte growth medium in 12-well microplates (200.000 cells/well) (Corning CellBIND). The growth medium was removed after 24 h and the cells were exposed either to fatty acid medium, consisting of [14C]OA (0.5 μCi/mL, 100 μM) bound to BSA (40 μM) at a ratio of 2.5:1 in DPBS with HEPES (10 mM) and L-carnitine (1 mM) for 4 h, or to lipogenesis medium, consisting of [14C]acetate (0.5 μCi/mL, 100 μM) in DMEM enriched with glucose (5.5 mM) and BSA (10 μM) for 4 h. The media were collected and stored at -20°C until further analysis, and the remaining cells washed 3x with PBS before adding 250 μL H2O for cell lysis and solubilized cells were stored at -20°C. Cell-associated lipids were extracted with chloroform:methanol according to Folch et al. [23] and separated by thin layer chromatography as previously described [23]. The content of radiolabelled lipids was normalized against cell protein content.
Analysis of gene expression
RNA extractions from mouse liver were carried out according to the manufacturer using RNeasy Plus kit. The expression of selected mRNA transcripts (S1 Table) was analyzed by qPCR using a LightCycler 480 (Roche Diagnostics, Manheim, Germany). PCR efficiencies were experimentally determined for each primer pair. Relative gene expression was calculated using the ΔΔCt-method, with Beta-actin and Eukaryotic translation elongation factor 2 as reference genes.
Western blotting
Tritosomes from liver and kidneys from WT and Glmp gt/gt mice were isolated as described elsewhere [3]. Tritosome preparations (6 μg protein) were electrophoresed on NuPAGE 4–12% Bis-Tris Mini Gels (Life Technologies), and transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA). After blocking, membranes were incubated overnight at 4°C with rabbit anti-GLMP serum [3] or rabbit anti-LAMP1 (1:1000, C54H11, Cell Signaling, Beverly, MA, US). This was followed by 1 h incubation with goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (1:4000, 65–6120, Life Technologies).
Statistical methods
All results are expressed as mean ± SEM. Linear mixed models (LMM) were used to analyze overall differences in liver/body weight ratio, spleen/body weight ratio, serum TAG and serum NEFA levels, using IBM SPSS software (SPSS Inc. Chicago, IL, US). Other data were analyzed using two-tailed T-test (SigmaPlot, Systat Software Inc, Chicago, IL, US).
Results
Confirmation of GLMP ablation in Glmp gt/gt mice
In agreement with our previous report, analysis of lysosome-enriched fractions from mouse liver and kidneys after tyloxapol treatment [3] confirmed the lack of GLMP expression in Glmp gt/gt mice (S1 Fig).
Similar body weight gain and feed intake in WT and Glmp gt/gt mice
Development of body weight in wild type (WT) and Glmp gt/gt mice was monitored from 3 weeks of age. As shown in Fig 1A, the body weight gains were similar for male WT and Glmp gt/gt mice up to the age of 7.5 months, but the Glmp gt/gt siblings were outgrown by WT mice at the age of 9 months. The feed efficiency was indistinguishable between the genotypes up to the age of 14 weeks (Fig 1B). Similar data were also obtained for female mice (data not shown).
Fig 1. Loss of GLMP does not affect body weight gain and feed efficiency in young mice.
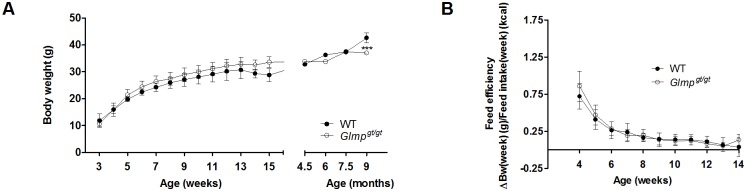
(A) Body weight gain for male WT and Glmp gt/gt mice were monitored every week from 3 to 15 weeks of age, and then at 4.5, 6, 7.5 and 9 months of age. (B) The feed efficiencies of WT and Glmp gt/gt mice were monitored from 4 to 14 weeks of age (n = 27–31) (*** p < 0.005 vs. WT). Values are presented as mean ± SEM.
Smaller epididymal fat pads and overall hepatosplenomegaly in Glmp gt/gt mice
In spite of similar body weight, the Glmp gt/gt mice appeared leaner compared to the WT. Male WT and Glmp gt/gt mice were sacrificed at the age of one and 4.5 months, and their epididymal fat pads/body weight ratios were determined. As shown in Fig 2A and 2B, Glmp gt/gt mice had significantly smaller epididymal fat pads compared to WT at the age of one month, with an increased difference at 4.5 months of age. In contrast, the axillary and inguinal fat depots and the interscapular brown adipose tissue were comparable betweent he genotypes (data not shown). Next, we assessed the liver/body weight and spleen/body weight ratios in WT and Glmp gt/gt mice. The Glmp gt/gt liver was significantly enlarged at 0.8, 1, 2, 4.5 and 7.5 months of age compared to age-matched WT (Fig 2C), while Glmp gt/gt spleen was significantly enlarged at 1, 2, 3.5, 4.5, 6, 7.5 and 9 months of age compared to age-matched WT animals (Fig 2D).
Fig 2. GLMP ablation causes reduced size of epididymal fat pads and hepatosplenomegaly.
(A) Representative pictures of epididymal fat pads and (B) epididymal fat pads/body weight ratios in 1 (n = 4) and 4.5 months (n = 10–12) old WT and Glmp gt/gt mice. (C) Spleen/body weight ratios and (D) liver/body weight ratios in WT and Glmp gt/gt mice were examined at designated time-points between 2 weeks and 9 months of age (n = 10–12) (* p < 0.05, ** p < 0.01, *** p < 0.005 vs. WT). Values are presented as mean ± SEM.
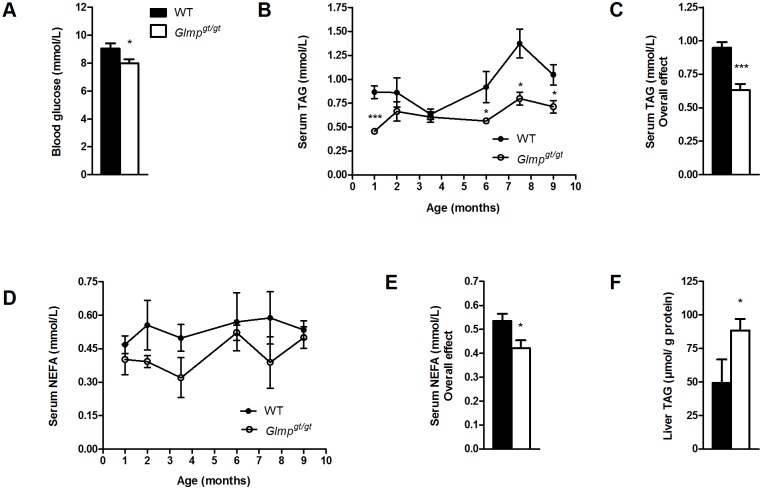
Decreased blood glucose, serum lipids and increased liver triacylglycerol (TAG) in Glmp gt/gt mice
Five months old Glmp gt/gt mice had significantly reduced levels of blood glucose compared to WT mice when fed ad libitum (Fig 3A). Analysis of serum concentrations of triacylglycerol (TAG) in WT and Glmp gt/gt mice aged 1–9 months, showed an overall level that was significantly lower in the Glmp gt/gt mice (p < 0.001) (Fig 3B and 3C). A significant decrease was also found for serum non-esterified fatty acids (NEFA) concentrations in Glmp gt/gt compared to WT mice aged 1–9 months (p < 0.001) (Fig 3D and 3E). Liver content of TAG was measured in 6 months old WT and Glmp gt/gt mice. As shown in Fig 3F, there were significantly higher levels of liver TAG in Glmp gt/gt compared to WT mice.
Fig 3. Glmp gt/gt mice have reduced blood glucose and circulating lipids, but increased liver triacylglycerol accumulation.
(A) Blood glucose was analyzed in ad libitum fed 5 months old WT and Glmp gt/gt mice (n = 5). (B, C) The levels of circulating triacylglycerol (TAG) and (D, E) non-esterified fatty acids (NEFA) in WT and Glmp gt/gt mice were analyzed at designated time-points between 1 and 9 months of age (n = 4–8). (F) Liver TAG content was analyzed in 6 months old WT and Glmp gt/gt liver homogenates, and normalized to protein content (n = 6–7) (* p < 0.05, ** p < 0.01, *** p < 0.005 vs. WT). Values are presented as mean ± SEM.
Changes in expression of metabolic genes in Glmp gt/gt liver
Metabolic homeostasis is regulated by peroxisome proliferator-activated receptors (PPARs) [24]. In livers from 3.5 months old mice, GLMP ablation resulted in altered mRNA expression of the three PPAR isoforms. Fig 4A shows a decreased expression of Ppara and increased expression of Ppard and Pparg in Glmp gt/gt liver relative to WT animals. In contrast, the PPAR co-regulator Pgc1a was not differently expressed between the two genotypes. Genes involved in de novo lipogenesis (Fasn, Scd1 and Scd2) were found to be significantly upregulated in Glmp gt/gt relative to WT liver (Fig 4B). The fatty acid transporter, Cd36 showed a 4.6-fold increased expression in Glmp gt/gt liver, while cytoplasmic fatty acid binding protein Fabp1 was significantly decreased compared to WT (Fig 4C). The expression of the initiator of peroxisomal fatty acid beta-oxidation, Acox1 was significantly decreased in Glmp gt/gt liver, while no differences were found for the expression of Acox2 (Fig 4D). Genes involved in mitochondrial fatty acid beta-oxidation (Cpt1a and Acadl) were not differently expressed between the two genotypes (Fig 4E). Furthermore, the expression of the lipid droplet associated proteins perilipin 2 and 5 (Plin2 and Plin5) was significantly decreased in Glmp gt/gt liver (Fig 4F). A decreased expression of a lipoprotein lipase regulator, Angptl4 was also detected in Glmp gt/gt liver, while the expression of Apoc3, a very low-density lipoprotein (VLDL) regulatory protein, was increased compared to WT (Fig 4G). With regard to genes involved in glucose metabolism, a decreased expression of the glucose transporter, Glut2 was detected in Glmp gt/gt liver compared to WT. No differences in expression were detected for liver specific hexokinase (Gck), while the expression of Pdk4, an inhibitor of pyruvate dehydrogenase complex, was significantly upregulated in Glmp gt/gt liver compared to WT (Fig 4H).
Fig 4. Altered expression of genes involved in metabolism in Glmp gt/gt liver.
(A) qPCR analyses of 3.5 months old mouse livers from wild type (WT) and Glmp gt/gt mice showed altered expression of genes involved in metabolic regulation (Pgc1a, Ppara, Ppard, Pparg), (B) lipogenesis (Fasn, Scd1, Scd2), (C, D, E) lipid metabolism (Cd36, Fabp1, Acox1, Acox2, Cpt1a, Acadl) (F) lipid droplets (Plin2, Plin5), (G) very low-density lipoproteins (Angptl4, Apoc3) and (H) glucose metabolism (Glut2, Gck, Pdk4) in Glmp gt/gt liver (n = 4–8, * p < 0.05, ** p < 0.01, *** p < 0.005 vs. WT). Values are presented as mean ± SEM.
Altered glucose and fatty acid uptake and oxidation in Glmp gt/gt hepatocytes
To assess whether GLMP ablation affects glucose and fatty acid metabolism in liver cells, hepatocytes were isolated from 4–6 weeks old Glmp gt/gt and WT mouse liver and exposed to either [14C]glucose or [14C]oleic acid (OA) for 4 h. Ablation of GLMP resulted in increased glucose uptake (Fig 5A) and oxidation (Fig 5B) in Glmp gt/gt hepatocytes. Exposure to [14C]OA revealed a significantly higher uptake of [14C]OA in Glmp gt/gt hepatocytes compared to WT cells (Fig 5C), while the oxidation capacity for OA was comparable between the two genotypes (Fig 5D).
Fig 5. Glucose and oleic acid oxidation in WT and Glmp gt/gt hepatocytes.
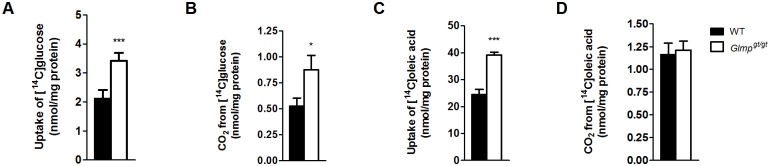
Overnight cultured primary hepatocytes from WT and Glmp gt/gt mice were incubated with [14C]glucose (0.6 μCi/mL, 200 μM) or [14C]oleic acid (OA) (0.5 μCi/mL, 100 μM) for 4 h. (A) Total [14C]glucose uptake, (B) [14C]glucose oxidation, (C) [14C]OA uptake and (D) [14C]OA oxidation were measured. Substrate uptake was calculated as the sum of cell-associated radioactivity and [14C]CO2 production. Substrate oxidation was measured as [14C]CO2 production. Results were normalized to protein content (n = 20 experiments, representing 4 individual mice/genotype, with 5 replicates each, * p < 0.05, *** p < 0.005 vs. WT). Values are presented as mean ± SEM.
To further explore metabolism of OA, we performed thin layer chromatography on cell culture media and cell homogenates after 4 h exposure to [14C]OA. No significant difference in secretion of triacylglycerol (TAG) into the media could be detected (data not shown), but there was a significant increase of accumulated total lipids in Glmp gt/gt hepatocytes compared to WT hepatocytes (Fig 6A). This difference was primarily contributed to by a significant increase in TAG accumulation (Fig 6B). Incorporation of labelled OA in other lipids assayed, diacylglycerol (DAG) (Fig 6C), non-esterified fatty acids (NEFA) (Fig 6D), phospholipids (Fig 6E), and cholesterol esters (CE) (Fig 6F) did not significantly differ between Glmp gt/gt and WT hepatocytes.
Fig 6. Glmp gt/gt hepatocytes incorporate more [14C]oleic acid into triacylglycerol.
Overnight cultured primary hepatocytes from WT and Glmp gt/gt mice were incubated with [14C]oleic acid (0.5 μCi/mL, 100 μM) for 4 h. (A) The total amount of cell accumulated lipids, (B) triacylglycerol (TAG), (C) diacylglycerol (DAG), (D) non-esterified fatty acids (NEFA), (E) phospholipids and (E) cholesterol esters (CE) were analyzed using thin layer chromatography (n = 9 experiments, representing 3 individual mice/genotype, with 3 replicates each, ** p < 0.01 vs. WT). Values are presented as mean ± SEM.
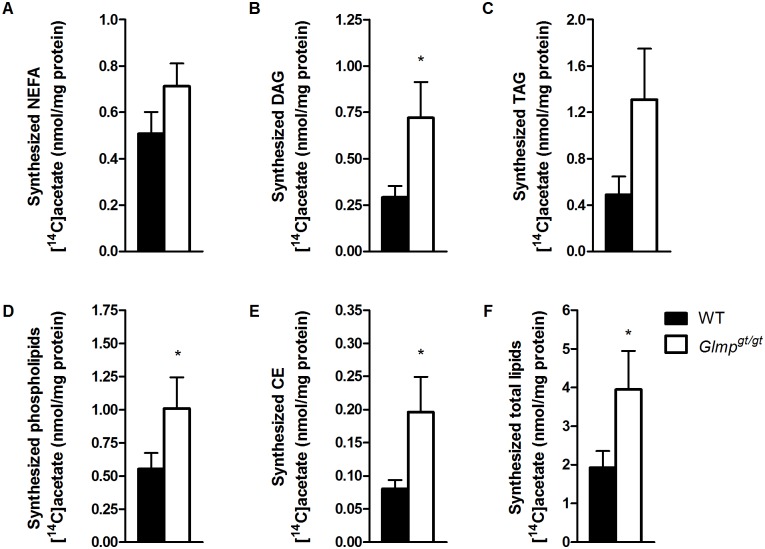
Increased de novo lipogenesis in Glmp gt/gt hepatocytes
Primary hepatocytes from WT and Glmp gt/gt mice were exposed to [14C]acetate for 4 h, and thin layer chromatography was performed on cell homogenates in order to assess de novo lipogenesis. The levels of newly synthesized NEFA showed no significant difference between the two genotypes (Fig 7A), but the incorporation into DAG was found to be significantly elevated (Fig 7B). A clear tendency for increased synthesis of TAG was also observed for Glmp gt/gt hepatocytes (Fig 7C), which, however, did not reach statistical significance. Higher incorporation of labelled acetate into PL (Fig 7D) and CE (Fig 7E) was also detected in Glmp gt/gt hepatocytes. Taken together, these results indicate increased lipogenesis in Glmp gt/gt hepatocytes compared to WT cells, which was also reflected in the total amount of synthesized lipids (Fig 7F).
Fig 7. Increased de novo lipogenesis in Glmp gt/gt hepatocytes.
Overnight cultured primary hepatocytes from WT and Glmp gt/gt mice were incubated with [14C]acetate (0.5 μCi/mL, 100 μM) for 4 h. (A) The amount of synthesized non-esterified fatty acids (NEFA), (B) diacylglycerol (DAG), (C) triacylglycerol (TAG), (D) phospholipids, (E) cholesterol esters (CE) and (F) total lipids were analyzed using thin layer chromatography (n = 8 experiments, representing 4 individual mice/genotype, with 2 replicates each, * p < 0.05 vs. WT). Values are presented as mean ± SEM.
Discussion
In this study, we showed that wild type (WT) and Glmp gt/gt mice initially had similar growth rates. The feed efficiency was also comparable for the two genotypes up to the age of 14 weeks. Yet the epididymal fat pads were significantly smaller in Glmp gt/gt siblings compared to WT mice at both one and 4.5 months of age. Other fat depots such as the axillary and inguinal white adipose tissue and the interscapular brown adipose tissue, however, were comparable between the genotypes. Hepatosplenomegaly contributed to the similar body weights until the age of 9 months when the Glmp gt/gt siblings had significantly reduced body weight. The reduced sizes of the epididymal fat pads were reflected in lower levels of circulating triacylglycerol (TAG) and non-esterified fatty acids (NEFA) in Glmp gt/gt mice. In addition, the Glmp gt/gt mice had lower blood glucose concentration. Gene expression analysis using liver mRNA from WT and Glmp gt/gt mice indicated changes in both glucose and lipid metabolic pathways, and biochemical analysis of whole liver homogenates detected higher TAG levels in Glmp gt/gt liver. Exposing isolated primary hepatocytes from WT and Glmp gt/gt mice to radiolabelled oleic acid (OA) showed that the Glmp gt/gt hepatocytes accumulated significantly more OA than WT hepatocytes, and that the increased OA taken up was stored as TAG. Exposure to radiolabelled glucose and acetate revealed a higher oxidation of glucose and increased de novo lipogenesis, respectively, in Glmp gt/gt hepatocytes compared to WT cells.
In most lysosomal disorders, the primary lysosomal dysfunction induces secondary alterations in other metabolic pathways [25], some of which may be adaptive changes in order to provide metabolites for de novo synthesis of macromolecules, because lysosomal dysfunction might inhibit adequate recycling of metabolites [14, 26]. As a consequence, reduced adiposity and body weight, possibly caused by higher tissue demand for metabolites, has been observed in several mouse models with lysosomal disorders [14, 26]. However, the body weight of younger Glmp gt/gt mice was comparable to that of their WT siblings since the reduced adiposity was compensated by hepatosplenomegaly. Similar observations have been reported for the mouse models for mucopolysaccharidosis (MPS) type I and IIIB [14]. Interestingly, as seen in the Glmp gt/gt mouse, reduced adiposity, reduced blood glucose, reduced lipid levels and hepatomegaly were also reported in these mice [14]. A similar feed efficiency between WT and Glmp gt/gt mice suggested that the Glmp gt/gt mice did not suffer from malabsorption as was also shown for mouse models with other lysosome deficiencies [14].
We have recently established that the Glmp gt/gt mice suffer from a mild chronic liver injury, as indicated by the modest increase in serum transaminase levels. However, by the age of 6 months, the sustained liver injury led to a well-established fibrosis with increased inflammation and oxidative stress [5], conditions known to disturb the liver's capacity for regulating glucose, lipid and protein homeostasis [18]. As the liver is an important organ for glucose homeostasis [27], the small decrease in resting blood glucose may be caused by increased glucose flow to the liver. Somewhat conflicting studies have been reported regarding glucose uptake capacity in fibrotic liver, which has been ascribed to differences in experimental setup [28–32]. A recent study using primary hepatocytes isolated from early, and advanced fibrotic rat livers, demonstrated a gradual decrease in mitochondrial function [33], an established secondary effect of advanced liver fibrosis [34]. Hepatocytes from early fibrotic livers increased the glycolytic flux in order to compensate for impaired ATP-production by oxidative phosphorylation [33]. Although the ATP-production capacity of Glmp gt/gt mitochondria needs to be further elucidated, we showed by using radiolabelled glucose that the Glmp gt/gt hepatocytes consumed significantly higher levels of glucose compared to WT hepatocytes. Increased flow of glucose to the liver is known to stimulate the expression of lipogenic genes [35]. By using radiolabelled acetate, we showed that de novo lipogenesis was induced in Glmp gt/gt hepatocytes, an expected result given the increased expression of lipogenic genes in Glmp gt/gt liver.
Unexpectedly, the expression of Pdk4 was significantly increased in Glmp gt/gt liver, suggesting an inhibition of the pyruvate dehydrogenase complex, an event usually associated with inhibition of de novo lipogenesis and activation of gluconeogenesis [36]. An increase in Pdk4 expression has been reported in two independent mouse models of liver fibrosis, resulting from chemical-induced liver damage [37] and chronic biliary injury [38]. Interestingly, the increased Pdk4 expression in the chemical-induced model was not accompanied by increased expression of gluconeogenic genes [37]. Similarily, the Mdr2 -/- mouse liver (model for chronic biliary injury [39]) had increased expression of lipogenic genes, and developed steatotic nodules [38]. In both studies, the increased Pdk4 expression was explained as preventive adaptations to the hepatic injury [37, 38]. Increased Pdk4 expression in the Glmp gt/gt liver is likely to be an adaptive response to the chronic liver injury, although increased glucose flux through the pentose phosphate pathway, which contributes to the antioxidant defense, and NADPH production required for de novo lipogenesis [40] is also a possibility.
Abnormal accumulation of lipids in the liver is a known consequence of dysregulation of metabolism [41]. However, in our initial report, histological analysis failed to reveal obvious steatosis in Glmp gt/gt liver [5]. Here, using a biochemical approach, we showed that Glmp gt/gt mice accumulated slightly more liver TAG than WT animals. The small, relative difference might be undetectable using histological methods. In support of the elevated TAG accumulation in Glmp gt/gt liver, the increased expression of PPARγ as seen in the Glmp gt/gt liver is a hallmark of hepatic steatosis [42], as it stimulates expression of lipogenic genes [18, 43] and the fatty acid transporter, CD36 [44]. Increased Cd36 mRNA expression has been found to correlate with increased hepatic TAG content in different models of hepatic steatosis [45–47]. After exposure to radiolabelled OA, an increased fatty acid uptake concomitant with increased storage as TAG in Glmp gt/gt hepatocytes was observed, supporting the observed TAG accumulation in Glmp gt/gt liver. Apart from increased fatty acid uptake by hepatocytes and de novo lipogenesis, compromised assembly and secretion of VLDL particles can contribute to lipid accumulation [18, 41, 48, 49]. In our experimental setup, no significant decrease of TAG secretion was detected for Glmp gt/gt hepatocytes. The increased expression of Apoc3, as detected in Glmp gt/gt liver is probably a result of decreased PPARα expression and increased influx of glucose [50]. Apoc3 expression is usually positively correlated with plasma TAG levels [51, 52]. However, ApoC3-containing lipoprotein secretion is stimulated by elevated levels of plasma NEFA [53], which were decreased in Glmp gt/gt mice.
The detected liver steatosis in Glmp gt/gt mice may have been promoted by chronic inflammation [54, 55]. In our previous study, increased expression of inflammatory cytokines, including tumor necrosis factor α (TNFα) and interleukin 1β (IL-1β) were detected in Glmp gt/gt liver [5]. Kupffer cells, activated by liver injury, have been shown to be essential for the development of diet-induced hepatic steatosis in rats, as they alter metabolic pathways in hepatocytes through TNFα secretion [56]. TNFα promotes the lipogenic inducer sterol regulatory element binding protein-1c (SREBP1c) [57] and the protein expression of CD36 [58]. In addition, TNFα stimulates lipolysis in peripheral adipose tissue to release free fatty acids into the circulation [59]. The reduced adipose tissue size and reduced levels of circulating fatty acids observed in Glmp gt/gt mice may be due to prolonged stimulation of adipose tissue lipolysis. Finally, activated Kupffer cells secrete IL-1β, which downregulates the expression of PPARα [60], a positive regulator of lipid catabolism [61].
The recent identification of a lysosomal nutrient sensing machinery (LYNUS) has provided a direct link between intralysosomal nutrient levels and the expression of metabolic genes [62, 63]. In periods of nutrient scarcity, LYNUS releases the transcription factor EB into the nucleus where it promotes the expression of genes involved in autophagy and lysosomal biogenesis, in addition to genes involved in fatty acid β-oxidation through induced expression of PPARα and PGC1α [63–65]. A significant decrease in Ppara expression was detected in Glmp gt/gt liver, and we might hypothesize that ablation of the lysosomal membrane protein, GLMP, influences normal LYNUS function, a secondary effect observed in many lysosomal disorders [11], thus affecting normal Ppara expression and lipid turnover. Dysregulation of metabolism in Glmp gt/gt liver might in turn affect adipose tissue. However, one of the limitations of the present study is the inability to dismiss the metabolic alterations in Glmp gt/gt liver as secondary effects. We are currently establishing a cell-type-specific transgenic mouse model in order to address this question in future studies.
In summary, we showed that Glmp gt/gt mice have a similar body weight gain and feed efficiency as WT mice until 9 months of age. Similar to some other mouse models with a dysfunctional lysosomal protein, a reduced adiposity and an liver fibrosis were also detected in Glmp gt/gt mice [5]. Analysis of serum glucose and lipid levels showed a significant decrease in Glmp gt/gt mice. In addition to increased hepatic TAG content and changes in the expression of metabolic genes in Glmp gt/gt liver, our data strongly indicate metabolic dysregulation in Glmp gt/gt mice. Analysis using primary hepatocytes supported the findings in Glmp gt/gt liver, and showed enhanced uptake of fatty acids, increased de novo lipogenesis, and higher consumption of glucose in Glmp gt/gt hepatocytes. Ablation of the lysosomal membrane protein GLMP has recently been shown to cause chronic liver injury in mice [5]. In this study, we demonstrated that metabolic dysregulation is a likely contributor to the pathophysiology following GLMP ablation.
Supporting Information
Lysosome-enriched fractions from mouse kidney and liver after tyloxapol treatment were used to verify the ablation of GLMP expression in Glmp gt/gt mice. LAMP1 served as loading control.
(TIF)
(DOC)
Acknowledgments
We thank Ruomei Li, Karen Kristine Sørensen and Trond Berg for sharing their expertise in mouse hepatocyte isolation, Mads Bengtsen for his help with setting up qPCR analysis, and Hilde Letnes and Hilde Hyldmo for excellent technical assistance.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors have no support or funding to report.
References
- 1. Steffensen KR, Bouzga M, Skjeldal F, Kasi C, Karahasan A, Matre V, et al. Human NCU-G1 can function as a transcription factor and as a nuclear receptor co-activator. BMC Mol Biol. 2007;8:106–121. Epub 2007/11/21. 10.1186/1471-2199-8-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. 10.1126/science.1174447 . [DOI] [PubMed] [Google Scholar]
- 3. Schieweck O, Damme M, Schröder B, Hasilik A, Schmidt B, Lubke T. NCU-G1 is a highly glycosylated integral membrane protein of the lysosome. Biochemical Journal. 2009;422:83–90. 10.1042/bj20090567 . [DOI] [PubMed] [Google Scholar]
- 4. Schröder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10(22):4053–4076. Epub 2010/10/20. 10.1002/pmic.201000196 . [DOI] [PubMed] [Google Scholar]
- 5. Kong XY, Nesset CK, Damme M, Løberg E-M, Lübke T, Mæhlen J, et al. Loss of lysosomal membrane protein NCU-G1 in mice results in spontaneous liver fibrosis with accumulation of lipofuscin and iron in Kupffer cells. Disease Models & Mechanisms. 2014;7(3):351–362. 10.1242/dmm.014050 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dongiovanni P, Fracanzani AL, Fargion S, Valenti L. Iron in fatty liver and in the metabolic syndrome: A promising therapeutic target. Journal of Hepatology. 2011;55(4):920–932. 10.1016/j.jhep.2011.05.008 . [DOI] [PubMed] [Google Scholar]
- 7. Nelson JE, Wilson L, Brunt EM, Yeh MM, Kleiner DE, Unalp-Arida A, et al. Relationship between the pattern of hepatic iron deposition and histological severity in nonalcoholic fatty liver disease. Hepatology. 2011;53(2):448–457. 10.1002/hep.24038 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parkinson-Lawrence EJ, Shandala T, Prodoehl M, Plew R, Borlace GN, Brooks DA. Lysosomal storage disease: revealing lysosomal function and physiology. Physiology. 2010;25(2):102–115. 10.1152/physiol.00041.2009 . [DOI] [PubMed] [Google Scholar]
- 9. Schultz ML, Tecedor L, Chang M, Davidson BL. Clarifying lysosomal storage diseases. Trends in Neurosciences. 2011;34(8):401–410. 10.1016/j.tins.2011.05.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cox TM, Cachón-González MB. The cellular pathology of lysosomal diseases. The Journal of Pathology. 2012;226(2):241–254. 10.1002/path.3021 . [DOI] [PubMed] [Google Scholar]
- 11. Platt FM, Boland B, van der Spoel AC. Lysosomal storage disorders: The cellular impact of lysosomal dysfunction. The Journal of Cell Biology. 2012;199(5):723–734. 10.1083/jcb.201208152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boustany R-MN. Lysosomal storage diseases-the horizon expands. Nat Rev Neurol. 2013;9(10):583–598. 10.1038/nrneurol.2013.163 . [DOI] [PubMed] [Google Scholar]
- 13. Alroy J, Lyons JA. Lysosomal Storage Diseases. Journal of Inborn Errors of Metabolism & Screening. 2014;2:1–20. 10.1177/2326409813517663 [DOI] [Google Scholar]
- 14. Woloszynek JC, Coleman T, Semenkovich CF, Sands MS. Lysosomal Dysfunction Results in Altered Energy Balance. Journal of Biological Chemistry. 2007;282(49):35765–35771. 10.1074/jbc.M705124200 . [DOI] [PubMed] [Google Scholar]
- 15. Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis In: Abbas AKGSJHPM, editor. Annual Review of Pathology: Mechanisms of Disease, Vol 6. Annual Review of Pathology-Mechanisms of Disease. 6 2011. p. 425–456. 10.1146/annurev-pathol-011110-130246 [DOI] [PubMed] [Google Scholar]
- 16. Pinzani M, Rosselli M, Zuckermann M. Liver cirrhosis. Best Practice & Research Clinical Gastroenterology. 2011;25(2):281–290. 10.1016/j.bpg.2011.02.009 [DOI] [PubMed] [Google Scholar]
- 17. Tsochatzis EA, Bosch J, Burroughs AK. Liver cirrhosis. The Lancet. 2014;383(9930):1749–1761. 10.1016/S0140-6736(14)60121-5 [DOI] [PubMed] [Google Scholar]
- 18. Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. Journal of Hepatology. 2012;56(4):952–964. 10.1016/j.jhep.2011.08.025 [DOI] [PubMed] [Google Scholar]
- 19. Bradford MM. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Analytical Biochemistry. 1976;72(1–2):248–254. 10.1006/abio.1976.9999 . [DOI] [PubMed] [Google Scholar]
- 20. Hansen B, Arteta B, Smedsrød B. The physiological scavenger receptor function of hepatic sinusoidal endothelial and Kupffer cells is independent of scavenger receptor class A type I and II. Mol Cell Biochem. 2002;240(1–2):1–8. 10.1023/A:1020660303855 . [DOI] [PubMed] [Google Scholar]
- 21. Berg T, Boman D. Distribution of lysosomal enzymes between parenchymal and Kupffer cells of rat liver. Biochimica et Biophysica Acta (BBA)—Enzymology. 1973;321(2):585–596. 10.1016/0005-2744(73)90201-5 . [DOI] [PubMed] [Google Scholar]
- 22. Wensaas AJ, Rustan AC, Lövstedt K, Kull B, Wikström S, Drevon CA, et al. Cell-based multiwell assays for the detection of substrate accumulation and oxidation. Journal of Lipid Research. 2007;48(4):961–967. 10.1194/jlr.D600047-JLR200 . [DOI] [PubMed] [Google Scholar]
- 23. Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry. 1957;226(1):497–509. . [PubMed] [Google Scholar]
- 24. Wang Y-X. PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res. 2010;20(2):124–137. 10.1038/cr.2010.13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ballabio A, Gieselmann V. Lysosomal disorders: From storage to cellular damage. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2009;1793(4):684–696. 10.1016/j.bbamcr.2008.12.001 . [DOI] [PubMed] [Google Scholar]
- 26. Woloszynek JC, Kovacs A, Ohlemiller KK, Roberts M, Sands MS. Metabolic Adaptations to Interrupted Glycosaminoglycan Recycling. Journal of Biological Chemistry. 2009;284(43):29684–29691. 10.1074/jbc.M109.020818 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moore MC, Coate KC, Winnick JJ, An Z, Cherrington AD. Regulation of Hepatic Glucose Uptake and Storage In Vivo. Advances in Nutrition: An International Review Journal. 2012;3(3):286–294. 10.3945/an.112.002089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leatherdale BA, Chase RA, Rogers J, Alberti KGMM, Davies P, Record CO. Forearm glucose uptake in cirrhosis and its relationship to glucose tolerance. Clinical Science. 1980;59(3):191–198. . [DOI] [PubMed] [Google Scholar]
- 29. Shmueli E, Walker M, Alberti G, Record CO. Normal splanchnic but impaired peripheral insulin-stimulated glucose uptake in cirrhosis. Hepatology. 1993;18(1):86–95. 10.1002/hep.1840180115 . [DOI] [PubMed] [Google Scholar]
- 30. Kruszynska YT, Meyer-Alber A, Darakhshan F, Home PD, McIntyre N. Metabolic handling of orally administered glucose in cirrhosis. The Journal of Clinical Investigation. 1993;91(3):1057–1066. 10.1172/JCI116263 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johansson U, Wahren J, Eriksson SL. Splanchnic and peripheral glucose metabolism in cirrhosis. Journal of Hepatology. 1994;20(6):760–767. 10.1016/S0168-8278(05)80147-3 . [DOI] [PubMed] [Google Scholar]
- 32. Imano E, Kanda T, Nakatani Y, Motomura M, Arai K, Matsuhisa M, et al. Impaired splanchnic and peripheral glucose uptake in liver cirrhosis. Journal of Hepatology. 1999;31(3):469–473. 10.1016/S0168-8278(99)80039-7 . [DOI] [PubMed] [Google Scholar]
- 33. Nishikawa T, Bellance N, Damm A, Bing H, Zhu Z, Handa K, et al. A switch in the source of ATP production and a loss in capacity to perform glycolysis are hallmarks of hepatocyte failure in advance liver disease. Journal of Hepatology. 2014;60(6):1203–1211. 10.1016/j.jhep.2014.02.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krahenbuhl S, Reichen J. Adaptation of mitochondrial metabolism in liver cirrhosis. Different strategies to maintain a vital function. Scandinavian journal of gastroenterology. 1992;193:90–96. . [DOI] [PubMed] [Google Scholar]
- 35. Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metabolism. 2006;4(2):107–110. 10.1016/j.cmet.2006.06.008 . [DOI] [PubMed] [Google Scholar]
- 36. Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. American Journal of Physiology-Endocrinology and Metabolism. 2003;284(5):E855–E862. 10.1152/ajpendo.00526.2002 . [DOI] [PubMed] [Google Scholar]
- 37. Dateki M, Kunitomo M, Yoshioka K, Yanai K, Nakasono S, Negishi T. Adaptive gene regulation of pyruvate dehydrogenase kinase isoenzyme 4 in hepatotoxic chemical-induced liver injury and its stimulatory potential for DNA repair and cell proliferation. Journal of Receptors and Signal Transduction. 2011;31(1):85–95. 10.3109/10799893.2010.538405 . [DOI] [PubMed] [Google Scholar]
- 38. Katzenellenbogen M, Pappo O, Barash H, Klopstock N, Mizrahi L, Olam D, et al. Multiple Adaptive Mechanisms to Chronic Liver Disease Revealed at Early Stages of Liver Carcinogenesis in the Mdr2-Knockout Mice. Cancer Research. 2006;66(8):4001–4010. 10.1158/0008-5472.can-05-2937 . [DOI] [PubMed] [Google Scholar]
- 39. Popov Y, Patsenker E, Fickert P, Trauner M, Schuppan D. Mdr2 (Abcb4)-/- mice spontaneously develop severe biliary fibrosis via massive dysregulation of pro- and antifibrogenic genes. Journal of Hepatology. 2005;43(6):1045–1054. 10.1016/j.jhep.2005.06.025 . [DOI] [PubMed] [Google Scholar]
- 40. Riganti C, Gazzano E, Polimeni M, Aldieri E, Ghigo D. The pentose phosphate pathway: An antioxidant defense and a crossroad in tumor cell fate. Free Radical Biology and Medicine. 2012;53(3):421–436. 10.1016/j.freeradbiomed.2012.05.006 . [DOI] [PubMed] [Google Scholar]
- 41. Musso G, Gambino R, Cassader M. Non-alcoholic fatty liver disease from pathogenesis to management: an update. Obesity Reviews. 2010;11(6):430–445. 10.1111/j.1467-789X.2009.00657.x . [DOI] [PubMed] [Google Scholar]
- 42. Inoue M, Ohtake T, Motomura W, Takahashi N, Hosoki Y, Miyoshi S, et al. Increased expression of PPARγ in high fat diet-induced liver steatosis in mice. Biochemical and Biophysical Research Communications. 2005;336(1):215–222. 10.1016/j.bbrc.2005.08.070 . [DOI] [PubMed] [Google Scholar]
- 43. Sahini N, Borlak J. Recent insights into the molecular pathophysiology of lipid droplet formation in hepatocytes. Progress in Lipid Research. 2014;54(0):86–112. 10.1016/j.plipres.2014.02.002 . [DOI] [PubMed] [Google Scholar]
- 44. Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, et al. Hepatic Fatty Acid Transporter Cd36 Is a Common Target of LXR, PXR, and PPARγ in Promoting Steatosis. Gastroenterology. 2008;134(2):556–567. 10.1053/j.gastro.2007.11.037 [DOI] [PubMed] [Google Scholar]
- 45. Degrace P, Moindrot B, Mohamed I, Gresti J, Du Z-Y, Chardigny J-M, et al. Upregulation of liver VLDL receptor and FAT/CD36 expression in LDLR-/- apoB100/100 mice fed trans-10,cis-12 conjugated linoleic acid. Journal of Lipid Research. 2006;47(12):2647–2655. 10.1194/jlr.M600140-JLR200 . [DOI] [PubMed] [Google Scholar]
- 46. Buqué X, Martínez MJ, Cano A, Miquilena-Colina ME, García-Monzón C, Aspichueta P, et al. A subset of dysregulated metabolic and survival genes is associated with severity of hepatic steatosis in obese Zucker rats. Journal of Lipid Research. 2010;51(3):500–513. 10.1194/jlr.M001966 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. García-Monzón C, Lo Iacono O, Crespo J, Romero-Gómez M, García-Samaniego J, Fernández-Bermejo M, et al. Increased soluble CD36 is linked to advanced steatosis in nonalcoholic fatty liver disease. European Journal of Clinical Investigation. 2014;44(1):65–73. 10.1111/eci.12192 . [DOI] [PubMed] [Google Scholar]
- 48. Feldstein AE. Novel Insights into the Pathophysiology of Nonalcoholic Fatty Liver Disease. Seminars in Liver Disease. 2010;30(4):391–401. 10.1055/s-0030-1267539 . [DOI] [PubMed] [Google Scholar]
- 49. Musso G, Gambino R, Cassader M. Recent insights into hepatic lipid metabolism in non-alcoholic fatty liver disease (NAFLD). Progress in Lipid Research. 2009;48(1):1–26. 10.1016/j.plipres.2008.08.001 . [DOI] [PubMed] [Google Scholar]
- 50. Caron S, Verrijken A, Mertens I, Samanez CH, Mautino G, Haas JT, et al. Transcriptional Activation of Apolipoprotein CIII Expression by Glucose May Contribute to Diabetic Dyslipidemia. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31(3):513–519. 10.1161/atvbaha.110.220723 . [DOI] [PubMed] [Google Scholar]
- 51. Schonfeld G, George PK, Miller J, Reilly P, Witztum J. Apolipoprotein C-II and C-III levels in hyperlipoproteinemia. Metabolism. 1979;28(10):1001–1010. 10.1016/0026-0495(79)90004-0 . [DOI] [PubMed] [Google Scholar]
- 52. Fredenrich A, Giroux LM, Tremblay M, Krimbou L, Davignon J, Cohn JS. Plasma lipoprotein distribution of apoC-III in normolipidemic and hypertriglyceridemic subjects: comparison of the apoC-III to apoE ratio in different lipoprotein fractions. Journal of Lipid Research. 1997;38(7):1421–1432. . [PubMed] [Google Scholar]
- 53. Pavlic M, Valéro R, Duez H, Xiao C, Szeto L, Patterson BW, et al. Triglyceride-Rich Lipoprotein-Associated Apolipoprotein C-III Production Is Stimulated by Plasma Free Fatty Acids in Humans. Arteriosclerosis, Thrombosis, and Vascular Biology. 2008;28(9):1660–1665. 10.1161/atvbaha.108.169383 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shiri-Sverdlov R, Wouters K, Gorp PJv, Gijbels MJ, Noel B, Buffat L, et al. Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. Journal of Hepatology. 2006;44(4):732–741. 10.1016/j.jhep.2005.10.033 . [DOI] [PubMed] [Google Scholar]
- 55. Ma KL, Ruan XZ, Powis SH, Chen Y, Moorhead JF, Varghese Z. Inflammatory stress exacerbates lipid accumulation in hepatic cells and fatty livers of apolipoprotein E knockout mice. Hepatology. 2008;48(3):770–781. 10.1002/hep.22423 . [DOI] [PubMed] [Google Scholar]
- 56. Huang W, Metlakunta A, Dedousis N, Zhang P, Sipula I, Dube JJ, et al. Depletion of Liver Kupffer Cells Prevents the Development of Diet-Induced Hepatic Steatosis and Insulin Resistance. Diabetes. 2010;59(2):347–357. 10.2337/db09-0016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Endo M, Masaki T, Seike M, Yoshimatsu H. TNF-α Induces Hepatic Steatosis in Mice by Enhancing Gene Expression of Sterol Regulatory Element Binding Protein-1c (SREBP-1c). Experimental Biology and Medicine. 2007;232(5):614–621. . [PubMed] [Google Scholar]
- 58. Wang C, Hu L, Zhao L, Yang P, Moorhead JF, Varghese Z, et al. Inflammatory Stress Increases Hepatic CD36 Translational Efficiency via Activation of the mTOR Signalling Pathway. PloS one. 2014;9(7):e103071 10.1371/journal.pone.0103071 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ruan H, Miles PDG, Ladd CM, Ross K, Golub TR, Olefsky JM, et al. Profiling Gene Transcription In Vivo Reveals Adipose Tissue as an Immediate Target of Tumor Necrosis Factor-α: Implications for Insulin Resistance. Diabetes. 2002;51(11):3176–3188. . [DOI] [PubMed] [Google Scholar]
- 60. Stienstra R, Saudale F, Duval C, Keshtkar S, Groener JEM, van Rooijen N, et al. Kupffer cells promote hepatic steatosis via interleukin-1β–dependent suppression of peroxisome proliferator-activated receptor α activity. Hepatology. 2010;51(2):511–522. 10.1002/hep.23337 [DOI] [PubMed] [Google Scholar]
- 61. Mandard S, Müller M, Kersten S. Peroxisome proliferator-activated receptor α target genes. CMLS, Cell Mol Life Sci. 2004;61(4):393–416. 10.1007/s00018-003-3216-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14(5):283–296. 10.1038/nrm3565 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. 2012;31(5):1095–1108. 10.1038/emboj.2012.32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15(6):647–658. 10.1038/ncb2718 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Settembre C, Ballabio A. Lysosome: regulator of lipid degradation pathways. Trends in Cell Biology. 2014;(0):743–750. 10.1016/j.tcb.2014.06.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lysosome-enriched fractions from mouse kidney and liver after tyloxapol treatment were used to verify the ablation of GLMP expression in Glmp gt/gt mice. LAMP1 served as loading control.
(TIF)
(DOC)
Data Availability Statement
All relevant data are within the paper.