Abstract
The IL-9R consists of the α-subunit and the γc-chain shared with other cytokine receptors, including IL-2R, an important regulator of T cells. We have previously shown that IL-2R is expressed in common clusters with MHC glycoproteins in lipid rafts of human T lymphoma cells raising the question what the relationship between clusters of IL-2R/MHC and IL-9R is. Confocal microscopic co-localization and FRET experiments capable of detecting membrane protein organization at different size scales revealed non-random association of IL-9R with IL-2R/MHC clusters at the surface of human T lymphoma cells. Accommodation of IL-9Rα in membrane areas segregated from the IL-2R/MHC domains could also be detected. The bipartite nature of IL-9R distribution was mirrored by STAT activation results. Our data indicate that co-compartmentation with MHC glycoproteins is a general property of γc receptors. Distribution of receptor chains between different membrane domains may regulate their function.
Keywords: membrane proteins, protein-protein interaction, FRET, confocal laser scanning microscopy
Introduction
Compartmentation of membrane proteins plays a crucial role in cell biological processes associated with the plasma membrane (e.g. signal transduction, protein sorting, etc.). These processes are usually accompanied by dynamic redistribution of protein patterns existing at the cell surface. The basic organization level of membrane proteins is defined by their molecular proximity (often manifested in their physical association as well). Accumulation of small-scale clusters into larger entities at the few tens of nanometer to micrometer scale can also be observed[1, 2].
The selective accumulation of proteins in (or their exclusion from) membrane areas with a specific lipid composition is a major tool for cells to manage distribution of membrane proteins[2]. Lipid rafts are specific microdomains characterized by high cholesterol and (glyco)sphingolipid content, the presence of GPI-anchored proteins and high microviscosity (“liquid ordered phase”), which enable the dynamic association and functional cooperation of proteins residing within the same raft. The dynamic exchange of components between different rafts (or between raft and non-raft regions) as well as the aggregation of smaller rafts into raft "macrodomains" also play a central role in the spatio-temporal organization of numerous cell biological processes[3–5].
There are several biophysical techniques capable of detecting co-localization of membrane proteins in intact or quasi-intact cells[2]. Since the rate of fluorescence resonance energy transfer (FRET) is inversely proportional to the sixth power of the donor–acceptor distance on the 1- to 10-nm scale, it enables us to detect molecular-scale interactions between fluorescently labeled proteins[6]. Organization of molecules at higher hierarchical levels can be studied by the wide range of microscopic techniques, whose resolution limit determines the scale at which organization of membrane species can be detected[7].
Confocal microscopy can give information on the lateral distribution of fluorescently labeled membrane species on the few-hundred-nanometer scale, which is usually sufficient when co-localization of membrane proteins (or other molecules) is investigated. In addition, by complementing co-expression data with FRET measurements we can get further insight into the details of cell surface clusters.
T cell function and homeostasis is regulated by a series of signals delivered by cytokines, a subset of which acts through receptor complexes sharing the common γc chain. These include IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21[8, 9]. Besides γc, IL-2 and -15 receptors share an additional signaling subunit (IL-2/15Rβ) and both have their own, “private” α-chains ensuring high affinity binding of the appropriate cytokine. In contrast, other members of the family exert their activities via heterodimeric receptor complexes consisting of a cytokine-specific α chain interacting with γc for signaling[10].
IL-9 is a multifunctional cytokine with pleiotropic effects on T cells[11, 12]. It is produced by several subsets of CD4+ T cells and its proliferative effect, as opposed to other growth factors (e.g. IL-2), is restricted to CD4+ T cells and it requires preliminary activation or tumoral transformation[13, 14]. Several observations suggest a potential role for IL-9 in T cell tumorigenesis[15]. Recently, IL-9 has been shown to influence TH17 and Treg cell development/function as well. IL-9 activates several signaling routes, including the Jak/STAT pathway[16, 17]. IL-9 itself induces phosphorylation of Jak1 and Jak3 tyrosine kinases pre-associated with IL-9Rα and γc, respectively, which is followed by recruitment and subsequent activation (i.e. tyrosine phosphorylation) of STAT1, STAT3 and STAT5 transcription factors. Despite the emerging number of reports on the biological activities of IL-9, the cell surface topology and molecular interactions of its receptor are still poorly characterized.
Using various approaches, previously we have revealed significant similarities in the molecular environment of IL-2 and IL-15 receptors in human T lymphoma cells[18]. We have shown preassembly of the heterotrimeric IL-2R and IL-15R, as well as co-localization of their α chains occurring at different size scales[7, 19–21] and the existence of supramolecular complexes of IL-2R, IL-15R and MHC glycoproteins in lipid rafts of T cells[21–23]. It can be hypothesized, that other members of the γc cytokine receptor family, such as the IL-9R complex, may also fulfill their tasks in a similar environment, maybe in the same clusters. In the present study, we used CLSM co-localization experiments combined with FRET measurements to investigate the cell surface organization of interleukin-9 receptor (IL-9R) and its relationship with IL-2R/MHC superclusters in different human T lymphoma cell lines.
The Pearson’s correlation coefficient is the most frequently applied measure of co-localization[24–26]. However, since its absolute value depends on the actual experimental setup and the biological system itself (e.g. expression levels of membrane species in question), co-localization data should be compared to carefully chosen biological controls and measurement-dependent confidence intervals in order to distinguish true co-localization (or segregation) from random localization. Here we implemented the method developed by Costes et al., by which one can construct a confidence interval for no correlation (C=0) by scrambling the images recorded for the pair of distributions in question[27]. Correlation coefficients above the upper limit of this interval can be considered as true co-localization, whereas values below the interval reflect true separation of the molecules.
Using this evaluation method, our CLSM and FRET experiments demonstrated interaction of IL-9Rα with IL-2R and MHC glycoproteins in MT-2 and Kit225/IL-9R human T lymphoma cells suggesting that IL-9Rα is another component of the aforesaid superclusters. Interestingly enough, our CLSM data revealed that IL-9Rα could also be detected in membrane areas segregated spatially from IL-2R/MHC-rich domains in addition to the superclusters mentioned above. This bipartite nature of IL-9R distribution was mirrored in its signaling capacity as suggested by flow cytometric detection of STAT1 phosphorylation.
These results along with our previous data for the IL-2/15R system indicate that compartmentation of the receptor complexes in common membrane microdomains with MHC glycoproteins may be a general property of γc cytokines in T cells. In addition, they hint at the possibility that expression of IL-9R in distinct membrane areas has a crucial role in the regulation of its function.
Results and Discussion
Whereas cell surface organization of IL-2R (and IL-15R) was extensively studied by using various biophysical techniques[18], the plasma membrane topology of other members of the γc family, especially that of the IL-9 receptor, is less well characterized. The expression of IL-9Rα is mainly restricted to T cell subsets also expressing the heterotrimeric IL-2 receptor (e.g. Treg cells, T lymphoma cells, etc.), in which IL-2 has a crucial regulatory role[28]. Considering that IL-9 shares the signaling γc subunit with IL-2, it is an intriguing question whether there is any correlation between cell surface localization of IL-9 and IL-2 receptors. Lipid raft-assisted co-assembly of IL-2R with MHC glycoproteins seems to be a hallmark shared by a variety of human T cells[21–23, 29], therefore spatial relationship of the two receptor kinds was tested in combination with MHC glycoproteins and lipid rafts.
Visual analysis of CLSM images suggests partial co-expression of IL-9Rα with IL-2Rα and MHC glycoproteins
For determining the localization of IL-9Rα with respect to IL-2R, MHC glycoproteins as well as to lipid rafts on the size scale of membrane microdomains, a few hundred nanometers, CLSM images of cells doubly or triply labeled with fluorescently tagged antibodies or cholera toxin B (a specific marker of lipid rafts) were recorded.
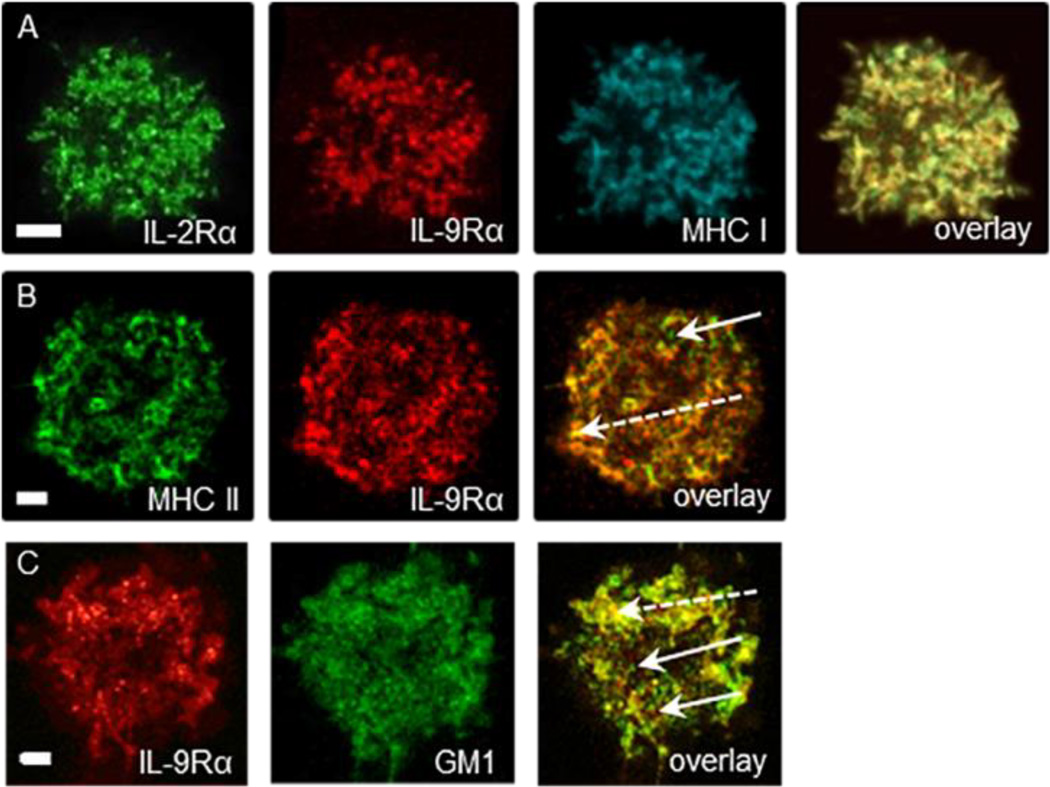
Visual analysis of CLSM images revealed a significant overlap between the distributions of IL-9Rα and IL-2Rα, or IL-9Rα and MHC glycoproteins in both cell types (Fig.1A,B). A less prominent, but still detectable overlay was found for the IL-9Rα/GM1 pair (Fig. 1C). CLSM images of cells triply labeled for IL-2Rα, IL-9Rα and MHC-I implied that co-localization of any pairwise combination of the above labels was mainly restricted to membrane areas where triple co-localization was present (Fig. 1A).
Figure 1. IL-9Rα is co-localized with IL-2Rα, MHC glycoproteins and lipid rafts.
(A) Triple co-localization of IL-2Rα, IL-9Rα, and MHC I glycoproteins in Kit225/IL-9R cells. CLSM images of the distribution of IL-2Rα labeled by Alexa Fluor 488-anti-Tac, IL-9Rα labeled by Alexa Fluor 546-AH9R7, and MHC I targeted by Alexa Fluor 647-W6/32 mAbs, recorded from a Kit225/IL-9R cell. Co-localization coefficients are C12=0.5 for IL-2Rα and IL-9Rα, C13=0.64 for IL-2Rα and MHC I and C23=0.26 for IL-9Rα and MHC I.
(B) Co-localization of IL-9Rα and MHC II glycoproteins in MT-2 cells. MHC II (HLA-DR) was labeled by Alexa Fluor 488-L243 and IL-9Rα by Alexa Fluor 546-AH9R7 mAbs, C12 = - 0.17.
(C) Co-localization of IL-9R and GM1-containing lipid rafts on MT-2 cells. IL-9Rα was tagged by Alexa Fluor 546-AH9R7 and GM1 with Alexa Fluor 647-CTX. C12 = 0.26.
In the overlay images mixed colors represent co-localization of the labeled proteins at the ≥200-nm scale. Pairwise correlation coefficients between the different channels are listed in Table 1. (Scale bar: 2 µm.)
The solid arrows indicate non-overlapping membrane regions, whereas dashed ones show the overlapping areas for the given pair of proteins.
In addition to the joint expression in common membrane regions, any pair of the above proteins also exhibited segregated distribution in certain membrane regions (Fig. 1B as an example for the MHC II – IL-9Rα pair). The relative amount of overlapping and non-overlapping areas for a given pair exhibited cell-to-cell variation as well (Fig. 1.).
Quantitative analysis of CLSM images
Although visual inspection allows us to disclose the presence or absence of overlay in selected membrane areas, it only gives qualitative information and lacks evidence regarding non-randomness of the observed patterns. In addition it may be biased by several subjective factors, including different expression levels, consequently visibility of the studied molecules.
Pairwise co-localization of membrane species was therefore characterized numerically by calculating the Pearson’s cross-correlation coefficient (C) from the confocal images. The numerical value of C depends on not only the amount of concrete overlap between the two molecules being examined, but is also influenced by their expression levels, the mixed occurrence of membrane regions with different co-localization properties as well as the experimental setup itself. Therefore, CLSM experiments and their evaluation should be complemented with appropriate controls.
In order to confirm the true co-localization or segregation of the investigated molecules (i.e. non-randomness of their spatial relationship), confidence intervals for no correlation were created for each pair of distributions on a cell-by-cell basis (see Materials and Methods)[27]. The correlation between distributions with a C value falling above, below or within the confidence interval was then categorized as “true positive”, “true negative” or “no correlation”, respectively. Average cross-correlation coefficients were determined separately for the “true negative” and “true positive” cases (Table 1).
Table 1.
Correlation between pairs of labeled molecules in CLSM images in Kit225/IL-9R and MT-2 cells.
| Labeled epitopes | Label 1 | Label 2 | Correlation (mean ± SD) | ||
|---|---|---|---|---|---|
| category | Kit225/IL-9Ra | MT-2b | |||
| IL-9Rα + IL-2Rα | AH9R7 | anti-Tac | − | − 0.33 ± 0.12 | − 0.36 ± 0.17 |
| + | 0.38 ± 0.13 | 0.38 ± 0.18 | |||
| IL-9Rα + MHC I HC | AH9R7 | W6/32 | − | − 0.39 ± 0.16 | − 0.39 ± 0.15 |
| + | 0.35 ± 0.22 | 0.46 ± 0.18 | |||
| IL-9Rα + MHC II | AH9R7 | L243 | − | − 0.39 ± 0.17 | − 0.30 ± 0.12 |
| + | 0.41 ± 0.21 | 0.43 ± 0.18 | |||
| IL-9Rα + GM1 | AH9R7 | CTX B | − | − 0.30 ± 0.22 | − 0.36 ± 0.16 |
| + | 0.42 ± 0.23 | 0.39 ± 0.19 | |||
| IL-2Rα + MHC I HC | anti-Tac | W6/32 | − | − 0.32 ± 0.16 | − 0.28 ± 0.19 |
| + | 0.43 ± 0.20 | 0.47 ± 0.21c | |||
| IL-2Rα + MHC II | anti-Tac | L243 | − | − 0.14d | no datae |
| + | 0.57 ± 0.17 | 0.48 ± 0.17 | |||
| TrfR + GM1 | MEM75 | CTX B | − | − 0.39 ± 0.16 | − 0.43 ± 0.17 |
| + | 0.28 ± 0.14 | 0.37d | |||
| MHC I HC + β2m | L368 | W6/32 | − | no datae | no datae |
| + | 0.74 ± 0.13 | 0.81 ± 0.09c | |||
Data show the average positive and negative correlation coefficients calculated in the fraction of cells of different correlation categories (“true positive” and “true negative”, indicated by ‘+’ and ‘−‘, respectively) as described in the Experimental Section.
Kit225/IL-9R cells, n≥30;
MT-2 cells, n≥20;
n≥12;
Since only one cell was detected in this category, no SD was calculated.
No cells were detected in this category.
Analysis of control samples with known co-localization properties
So as to prove the reliability of the evaluation method in our experimental system, as well as to determine the dynamic range of C values characterizing the actual experimental setup, we used positive and negative biological controls. As a negative control, the distributions of transferrin receptors (TrfR) and GM1 gangliosides were compared. TrfRs are localized in coated pits and are excluded from lipid rafts[30, 31]. As a positive control, the correlation between the light chain (β2m) and the heavy chain of MHC I was determined.
In accordance with expectation the correlation coefficient for each analyzed cell fell into the “true positive” category in the case of the positive biological control (Fig. 2) with a high average value (<C> ~0.7–0.8; Table 1). For the TrfR-GM1 pair ~80% of the cells had a C value in the “true negative” category (<C> ~ −0.4 for both cell types), proving spatially segregated distributions of these molecules. The theoretical minimum for the C values for totally segregated distributions is −1. However, in biological samples the size of the investigated objects (i.e. coated pits and lipid rafts in this particular case) can be comparable (or even smaller) than the overall resolution limit of the microscopic images, which can generate a non-zero random co-localization component, despite their segregated distributions; i.e. the resultant C value will be less negative than expected.
Figure 2. Cell fraction in the different pairwise correlation categories of positive and negative control.
The graph shows the fraction of cells in different correlation categories („true negative“, zero and „true positive“ fractions) of MT-2 (A and C) and Kit225/IL-9R cells (B and D). MHC I heavy and light chain were targeted by W6/32 and L368 mAbs tagged with spectrally different fluorophores in A and B, served as positive control. Transferrin receptor and GM1 ganglioside were targeted by cholera toxin B and MEM75 mAb tagged with spectrally different fluorophores in C and D, served as negative control.
A few percentage of cells with a C value in the “true positive” category could also be detected for the negative control, which is presumably due to the rather low expression and/or labeling accessibility of TrfR and GM1 molecules, i.e. the low signal-to-noise ratio which may increase uncertainties in determining the appropriate threshold; therefore the accuracy of calculations.
Non-random co-localization of IL-2Rα with MHC glycoproteins
Previously we have shown enrichment of IL-2R and MHC glycoproteins in common membrane areas in different human T lymphoma cell lines, including Kit225 cells[21–23]. Since the Kit225/IL-9R cell line was derived by stable transfection of IL-9Rα into Kit225 cells, it provides a good model system to explore the spatial relationship of IL-9Rα with the above-mentioned proteins. Firstly, we have checked whether transfection of IL-9Rα had any effect on the co-localization properties of IL-2R and MHC molecules. Correlation between the distributions of IL-2Rα and MHC glycoproteins were also tested in MT-2 cells exhibiting constitutive expression of the IL-9 receptor.
In accordance with our previous data both types of MHC glycoproteins showed substantial, non-random overlap with IL-2Rα. For the IL-2Rα/MHC II pair practically all the cells fell in the “true positive” category with an average C value ~0.6, confirming the significant non-random co-localization of the two proteins (Table 1). The majority of cells had a positive C value (~60%) for the IL-2Rα/MHC I pair as well, therefore our data provided evidence that IL-2R/MHC microdomains also existed in Kit225/IL-9R and MT-2 cells (Fig. 1A; Table 1). At the same time for the IL-2Rα/MHC I pair we could also detect cells in the “true negative” category (~30% and 10% for Kit225/IL-9R and MT-2 cells, respectively), indicating that spatially segregated domains of the two molecule types may also be present on the surface of these cells.
IL-9Rα enriched in separate membrane areas having distinct spatial correlation with IL-2Rα/MHC domains
Figure 3 shows the percentage of cells within the different correlation categories for the IL-9Rα/IL-2Rα, IL-9Rα/MHC I, IL-9Rα/MHC II and IL-9Rα/GM1 pairs.
Figure 3. Cell fraction in the different pairwise correlation categories between the distinct epitopes of interest.
The graph shows the fraction of cells in different correlation categories (“true negative”, zero and “true positive” fractions) of MT-2 (A, C, E and G) and Kit225/IL-9R cells (B, D, F and H). Pairwise co-localization coefficients between IL-9Rα and IL-2Rα (A and B), IL-9Rα and MHC I HC (C and D), IL-9Rα and MHC II (HLA-DR) (E and F) and IL-9Rα and GM1 ganglioside (G and H). IL-9Rα IL-2Rα, MHC I, MHC II and GM1 ganglioside were targeted with spectrally different fluorophores conjugated AH9R7, anti-Tac, W6/32, L243 mAbs and cholera toxin B, respectively.
A significant portion of cells exhibited an overall cross-correlation coefficient falling in the “true-positive” category in both cell types, independent of the pair being investigated; i.e. these molecules non-randomly co-localize with each other in common membrane domains, presumably in lipid rafts. As it was hinted by the visual observation of CLSM images, these cells could be characterized with an intermediate positive C value, indicating a strong, but not perfect co-localization between IL-9Rα and the other molecules (Table 1, Fig. 1).
Intermediate positive C values can simply be the consequence of large differences in the expression levels of the investigated molecules, which increases the chance for membrane regions containing exclusively the molecule present in more abundance. Indeed, with the exception of GM1, expression of IL-9Rα is significantly lower, than that of the other molecules (see Supplementary table). However, visual inspection of the images indicates that the existence of these membrane areas is not restricted to the more abundant molecule. Moreover, we could also detect cells with C values in the “true negative” category for all the examined pairs, implying that in these cases a significant amount of IL-9Rα is expressed in membrane areas spatially segregated from the IL-2Rα/MHC-rich domains.
The amount of cells in the “true negative” category was the highest in the case of the IL-9Rα/MHC I pair (~50% and ~70% for MT-2 and Kit225/K6 cells, respectively). Although MHC I can be accommodated by lipid rafts, numerous studies indicate that MHC I are associated with non-raft domains as well and their presence in these domains seems to be preferable to raft localization in many cases[21–23, 32–34]. This fact along with the high abundance of MHC I glycoproteins may contribute to the observed negative C values and may account for the fraction of cells in this category detected for the IL-2Rα/MHC I pair as well.
IL-9Rα is co-clustered with IL-2Rα and MHC glycoproteins
FRET measurements were also performed to test whether IL-9Rα was within molecular distance from the molecules it was found to share common membrane microdomains with. The median values of FRET efficiency distributions determined on a pixel-by-pixel basis for individual cells were averaged and used to characterize a given donor/acceptor pair.
The results of our acceptor photobleaching FRET experiments are listed in Table 2. We could detect FRET between IL-2Rα and MHC I or II glycoproteins confirming the existence of IL-2R/MHC-containing supramolecular clusters in these cells as well. Positive FRET efficiency was found to a varying degree between IL-9Rα and IL-2Rα, MHC class I or MHC class II, demonstrating association of these proteins on Kit225/IL-9R cells (Fig. 4). We got similar results with slight variations for MT-2 cells indicating the cell-type independent existence of the above association motifs.
Table 2.
FRET efficiencies measured by the acceptor photobleaching FRET technique in Kit225/IL-9R and MT-2 cells.
| Labeled epitopes | Donora | Acceptorb | E (%)c | |
|---|---|---|---|---|
| Kit225/IL-9R | MT-2 | |||
| IL-9Rα + IL-2Rα | AH9R4 | anti-Tac | 6.4 ± 2.3 | 5.2 ± 2.1 |
| AH9R7 | anti-Tac | 6.6 ± 2.1 | 6.1 ± 3.4 | |
| IL-9Rα + MHC I HC | AH9R7 | W6/32 | 17.2 ± 6.1 | 13.5 ± 7.0 |
| IL-9Rα + MHC II | AH9R7 | L243 | 15.9 ± 5.1 | 10.2 ± 5.2 |
| IL-2Rα + MHC I HC | anti-Tac | W6/32 | 21.7 ± 5.7 | 22.5 ± 5.2 |
| IL-2Rα + MHC II | anti-Tac | L243 | 21.9 ± 3.9 | 11.9 ± 2.9 |
| TrfR + GM1 | MEM75 | CTX B | 0.2 ± 1.5 | 1.1 ± 1.5 |
| β2m + MHC I HC | L368 | W6/32 | 30.3 ± 5.3 | 35.4 ± 5.4 |
Monoclonal antibodies tagged with Alexa Fluor 546 were used as donor, whereas Alexa Fluor 647-tagged mAbs or CTX B served as acceptor in our experiments.
FRET efficiencies were determined from the median values of frequency distribution histograms obtained for individual cells. Data are presented as mean ± SD. (n≥10) p < 0.001, p-values were calculated by one-tailed Student’s t test for each comparison with the negative control.
Figure 4. IL-9Rα is in the molecular proximity of IL-2Rα, MHC I and MHC II according to FRET imaging.
(A) Molecular scale association of IL-9Rα with IL-2Rα and MHC glycoproteins. Representative pixel-by-pixel FRET efficiency maps with the corresponding frequency distribution histograms measured on Kit225/IL-9R cells are depicted. IL-2Rα, MHC I and MHC II targeted with Alexa Fluor 647-conjugated mAbs (anti-Tac, W6/32 and L243, respectively) served as acceptor, whereas IL-9Rα labeled with Alexa Fluor 546-conjugated AH9R4 (for FRET with IL-2Rα) or AH9R7 mAbs (for FRET with MHC I or II) were used as donor in our experiments. Acceptors are indicated in the individual figures.
(B) Positive and negative controls. We measured the FRET efficiency between the light chain (β2m) and heavy chain of MHC I glycoproteins as a positive control, and between GM1 ganglioside and transferrin receptor as a negative control. The light and heavy chains of MHC I were targeted with Alexa Fluor 546-L368 and Alexa Fluor 647-W6/32 mAbs, respectively. GM1 ganglioside and TrfR were labeled with Alexa Fluor 555-CTX and Alexa Fluor 647-MEM75, respectively.
FRET efficiency maps were determined from the donor images taken before and after photobleaching of the acceptor. The color code ranges between FRET efficiencies of -100% (black) and 100% (white). Pixels with intensities lower than the background threshold appear in black. Frequency distribution histograms show FRET efficiencies in the individual pixels. Numerical values of FRET efficiencies are listed in Table 2. (Scale bar, 2 µm.)
In most cases frequency distributions of FRET efficiencies exhibited a significant amount of pixels lacking evidence for FRET between the examined pairs. For membrane proteins the observed FRET efficiency is influenced by several factors including steric relationship of the labeled epitopes as well as the stoichiometry/composition of protein clusters; therefore positive FRET efficiency is a strong indicator of molecular proximity/co-clustering, but the lack thereof does not necessarily prove the opposite. Still – based on the results of our CLSM experiments – it can be assumed that these pixels represent membrane regions where the two proteins are not co-localized with each other. Furthermore, our FRET results imply the combined occurrence of IL-9Rα domains with distinct co-localization properties even on the surface of the same cell.
Interestingly enough, the high incidence of Kit225/IL-9R cells in the “true negative” category observed for the IL-9Rα/MHC I and IL-9Rα/MHC II pairs in CLSM co-localization experiments was not reflected by the FRET data; i.e. FRET frequency distributions were shifted significantly toward the positive values for all cells in the experiments. At the same time C values are not only affected by the relative amount of membrane areas/blocks of pixels with different co-localization properties, but also influenced by the number of molecules residing in them; therefore even a few membrane regions containing exclusively IL-9Rα or MHC may shift the overall correlation coefficient toward the negative values if their abundance is high enough.
The light chain-heavy chain pair of MHC I displaying a high FRET efficiency (E~30-35%) was used as a positive control, whereas the TrfR-GM1 pair characterized by low FRET (E~1-2%) served as a negative control (Fig. 4B). According to our statistical analysis (see Table 2) all the FRET values detected between IL-9Rα and the other proteins of interest were significantly higher (p<0.001) than that of the negative control. It should be noted that contrary to co-localization experiments, the positive control used in FRET studies does not determine the maximum value of FRET; instead it is simply an indicator of the accuracy of the measurements.
As a rule-of-thumb, in a FRET experiment between proteins/epitopes with considerably different expression levels the more abundantly expressed epitope is labeled with the acceptor dye. Using this labeling strategy a donor is more likely to have a nearby acceptor yielding a higher apparent average FRET efficiency.
The proximity/random FRET increases when the acceptor concentration is increased, but at the expression level of the molecules (see Supplementary table) and the tendency of MHC glycoproteins for self-oligomerization[22], the contribution of random-FRET between functionally non-interacting proteins is negligible compared to the FRET observed in our experiments[35, 36].
STAT phosphorylation experiments support the existence of distinct IL-9Rα domains
We have tested whether the duality detectable in the localization of IL-9Rα has any effect on the signaling capacity of the receptor. Due to the practically direct link between IL-9 binding and STAT activation, observing the efficiency and time course of STAT phosphorylation may provide a good tool to assess functional consequences of membrane-related properties of the receptor (e.g. localization in specific membrane domains). Whereas STAT3 and STAT5 can be activated by both IL-2 and IL-9, IL-2-induced phosphorylation of STAT1, if any, is negligible[17]. Since Kit225/IL-9R cells require IL-2 for their growth, we followed phosphorylation of STAT1 using flow cytometry to measure the biological response to IL-9. In order to get rid of the remnants of IL-2 and its potential influence on IL-9 binding, we have also performed experiments with cells cultured for two days in IL-2-free medium and then treated with IL-9. According to our CLSM experiments these IL-2 deprived cells had similar co-localization properties than “normal” ones (data not shown).
Using IL-9 concentration just below the saturating level, phosphorylation of STAT1 usually took place in two phases (Fig. 5): first, there was a relatively fast response characterized by a low but still significant elevation of pY-STAT1 in all cells, which is followed by a slower second phase with a more prominent increase of the pY-STAT1 level. Treating cells with methyl-β-cyclodextrin (MβCD) before cytokine stimulation hardly affects the first phase, but delays the onset of the second phase.
Figure 5. IL-9 induced tyrosine phosphorylation of STAT1.
Flow cytometric intensity distribution histograms from a representative experiment demonstrating the time course of IL-9-induced tyrosine phosphorylation of STAT1 in control (upper panel) and MβCD-treated (lower panel) Kit225/IL-9R cells. STAT1 phosphorylation was monitored by flow cytometric detection of the binding of Alexa Fluor 647-conjugated antibodies specific for pY-STAT1. Cells were grown in IL-2-free medium for 48 hours, treated with 3 mM MβCD and then stimulated with IL-9 in fresh medium. Control cells were incubated in the buffer alone before cytokine stimulation. STAT1 phosphorylation was detected in nonstimulated cells (black line) as well as in cells stimulated with IL-9 for 5, 10 and 20 minutes (red, green and blue lines, respectively). Cells unstained with the anti-pTyr-STAT1 mAb served as a background sample (dashed line).
The latter finding is shown by the fact that the slower, more prominent, IL-9-evoked STAT1 phosphorylation in the presence of 1, or 3 mM MβCD was less substantial (Figure 5; lower panel) at 10 min, but was comparable to the control after 20-minute incubation with IL-9. This phenomenon implies that mainly the plasma membrane-related events of IL-9R signaling (e.g. recruitment of receptor subunits via diffusion) were disturbed by cholesterol depletion.
These data are in accordance with our CLSM results indicating that IL-9Rα is located in distinct membrane domains (presumably both in raft and non-raft regions) which influence the signaling properties of the receptor as well.
Conclusions
Our confocal microscopic co-localization and FRET experiments indicated that IL-9Rα forms supramolecular clusters with IL-2R and MHC molecules in lipid rafts of the studied human T lymphoma cells. Taking into account our current and previous results on IL-9, IL-15 and IL-2 receptors it is plausible to assume that superclusters of γc cytokine receptors with MHC glycoproteins may be generally present in the plasma membrane of activated or transformed T cells. The role of these association motifs needs to be clarified. Interactions within these clusters may affect the assembly and signaling capability of the receptors. Compartmentalization of IL-9 and IL-2 receptors in the same membrane area may provide a platform promoting subunit “switching,” and may ensure efficient and economical utilization of the common γc subunits by the two receptor kinds. MHC glycoproteins themselves may even directly participate in the signaling pathways activated by these cytokines using a similar regulatory cross-talk mechanism suggested previously between the insulin receptor and MHC I molecules[37]. Ligation of MHC I with specific mAbs may result in the activation of signaling pathways also stimulated by the γc cytokine receptors[38, 39]. Involvement of MHC I in the negative regulation of toll-like receptor-triggered inflammatory responses via reverse signaling has also been reported recently[40].
In addition to common membrane domains a considerable fraction of IL-9Rα is expressed in membrane areas spatially segregated from IL-2R/MHC-containing lipid rafts. The question arises whether the presence of the two subsets of IL-9Rα is mutually exclusive or they may co-exist on the same cell. Both qualitative inspection and quantitative evaluation of our CLSM images as well as our FRET and STAT phosphorylation experiments support the latter assumption; i.e. both types of IL-9Rα domains may be present on the same cell. It is plausible to assume that – similar to other members of the γc receptor family – distribution (and/or redistribution) of IL-9 receptor chains between different membrane domains has a crucial role in regulating IL-9 evoked responses[41–43]. However, further experiments are required to elucidate the details and the factors regulating distribution of IL-9R between these distinct membrane areas.
Finally, our results provide evidence that a combination of widely used FRET and co-localization approaches supplemented with the appropriate, quantitative evaluation methods may provide valuable information about localization and interactions of membrane proteins without using super resolution microscopy.
Experimental Section
Cell Culture and Cytokine Treatment
Kit225/IL-9R is a HTLV-nonexpressing, cytokine-dependent human adult T lymphoma cell line with a helper/inducer phenotype, derived by stable transfection of IL-9Rα into Kit225 cells[44]. MT-2 is a HTLV-I–immortalized human adult T-cell leukemia cell line showing physiological expression of IL-9Rα[45]. Both cell lines exhibit constitutive expression of γc as well as IL-2Rα and β chains and are responsive to both IL-2 and IL-9. The MT-2 cell line also produces IL-9.
Cells were cultured in RPMI 1640 medium (SIGMA-Aldrich) supplemented with 10% FBS, penicillin, and streptomycin. We also added 30 U/ml human recombinant IL-2 (NCI, BRB Preclinical Repository) to the medium of Kit225/IL-9R cells every 48 h. The medium of Kit225/IL-9R cells contained 800 µg/ml G418 (Calbiochem®) to suppress the growth of wild type cells.
In some experiments freshly harvested Kit225/IL-9R cells were washed twice in PBS and grown in IL-2-free medium for 48 h. Such cells were considered to be deprived of IL-2. Cells were then washed and incubated in fresh medium with IL-9 (0.25 µg/ml) at 37°C.
Monoclonal Abs
IL-2Rα was targeted with anti-Tac mAb (IgG2a; RepliGen Corporation) defining the cytokine-binding epitope. IL-9Rα was labeled with mAbs AH9R7 (IgG2bκ; BioLegend, San Diego, CA, USA) or AH9R4 (IgG2a, kindly provided by Jean-Christophe Renauld, Ludwig Institute for Cancer Research and Experimental Medicine Unit, Brussels, Belgium). AH9R7 strongly inhibits the action of IL-9 in a TS1H9RA3 assay[46], whereas AH9R4 shows little or no inhibition. The common γc chain was targeted with TugH4 mAb (IgG2b; BD Biosciences). Transferrin receptor (TrfR) was labeled by MEM75 (IgG1) mAb (EXBIO). Hybridomas producing W6/32 (IgG2aκ) mAb specific for the heavy chain of class I MHC molecules, L368 (IgG1κ) binding β2m, and L243 (IgG2a) binding MHC class II (HLA-DR) were kindly provided by F. Brodsky (University of California).
Aliquots of purified IgGs were conjugated with succinimidyl esters of Alexa Fluor 488, 546, 555 or 647 dyes (Molecular Probes-LifeTechnologies) or those of Cy3 or Cy5 (Amersham Pharmacia) as described in ref.[47].
Labeling Cells with Fluorescent Markers
Freshly harvested cells were washed twice in ice-cold PBS, pH 7.4. The cell pellet was suspended in PBS and incubated with saturating concentrations of fluorescent mAbs or fluorescent cholera toxin subunit B (CTX B; Molecular Probes-LifeTechnologies) for 40 min on ice. After washing cells were fixed with 1% formaldehyde/PBS. Special care was taken to keep the cells at ice-cold temperature to avoid induced aggregation of cell surface molecules or receptor internalization.
The number of antibody binding sites was determined on a FACSAria flow cytometer (Becton Dickinson) from the mean values of flow cytometric fluorescence intensity histograms. Mean fluorescence intensities (MFI) were background corrected, normalized by the dye-to-protein ratios of the mAbs and then calibrated with fluorescent microbeads carrying a known number of fluorophores (Quantum™ Alexa Fluor 488 MESF kit). Flow cytometric intensity histograms were evaluated using the REFLEX software[48].
Confocal Laser Scanning Microscopy
CLSM experiments performed on an Olympus FluoView 1000 Confocal microscope were used for co-localization and acceptor photobleaching FRET measurements. For the excitation of Alexa Fluor 488 the 488 nm line of an Argon ion laser; for Alexa Fluor 546 a 543-nm He-Ne laser; for Cy5 and Alexa Fluor 647 a 633-nm He-Ne laser was used. Fluorescence emissions of Alexa Fluor 488, Alexa Fluor 546 and Alexa Fluor 647 were detected through 500–530-nm, 555–625-nm and 655–755-nm band-pass filters, respectively. Signals from different channels were represented by green, red and cyan false colors. Images were taken in sequential mode to prevent cross-talk between the channels. Image stacks of 1 µm thick optical sections each containing 512×512-pixels were obtained with a 60× UPLSAPO oil immersion objective (NA 1.35).
Determining co-localization from image cross-correlation by CLSM
Co-localization of molecules with a resolution of ~200-300 nm (i.e. at the membrane domain level) was determined from CLSM images of doubly or triply labeled cells[21]. Fluorescently labeled and formaldehyde-fixed cells were mounted on poly-L-lysine-coated cover slips with Mowiol 4–88 (Calbiochem®) dissolved in glycerol to reduce unwanted photobleaching. Two to three optical sections were recorded from the “bottom” (touching the coverslip) or “top” segments (distal from the coverslip) of the cells. Before the quantitative analysis, images were low-pass filtered to reduce noise, and z projection images were created.
The co-localization of fluorescence labels recorded in two different fluorescence channels was analyzed according to a method adapted from Costes implemented in a custom-written Matlab (Mathworks, Natick, MA) program using DipImage functions (Delft University of Technology, Delft, The Netherlands). Briefly, a region of interest was manually drawn around the cells to be analyzed and the two images were segmented using the max-entropy algorithm[27]. Before analysis neighboring pixels were pooled so that the size of pixel blocks was approximately equal to the lateral resolution of the microscope. Co-localization was analyzed for blocks belonging to the foreground (clusters) in at least one of the images, i.e. a Boolean OR operation was carried out between the segmented images.
For a pair of images x and y, the cross-correlation coefficient between the intensity distributions of cell surface labeling was calculated as
| (1) |
where xij and yij are fluorescence intensities at block coordinates i, j in images x and y, and 〈x〉, 〈y〉 are the mean intensities. Determination of the correlation coefficient was followed by defining the 95% confidence interval of the correlation coefficient for no correlation between the fluorescence labels (C=0) by randomly scrambling pixel blocks in one of the images and measuring the correlation coefficient between this scrambled image and the other unscrambled image 500 times[27, 49]. Positive correlation between the fluorescence labels in the unscrambled images (i.e. true co-localization of molecules) was concluded if the correlation coefficient was above the 95% confidence interval. A correlation coefficient for the unscrambled images below the 95% confidence interval was regarded as negative (i.e. true separation of molecules of interest).
It should be noted, however, that a C value within the confidence interval does not necessarily mean the lack of correlation (or anti-correlation), but simply indicates that there is no statistical proof for any of them. E.g. a C value within the confidence interval may be simply the consequence of mixed occurrence of positive and negative correlation between the two distributions at the cell surface.
Pixel-by-Pixel FRET Measurements
Molecular proximity of cell surface proteins (i.e. molecular scale association) was investigated by acceptor photobleaching FRET technique. The method is based on detecting the enhanced emission of the donor after eliminating FRET by photobleaching the acceptor dye. Proteins of interest were labeled by monoclonal antibodies tagged with the donor (Alexa Fluor 546) or acceptor (Alexa Fluor 647 or Cy5) dyes. Single optical sections of the donor and acceptor intensity distributions from doubly labeled samples were recorded by using low excitation intensities of the 543- and 633-nm laser lines, respectively. Subsequently, acceptor molecules were bleached by repetitive scans with the 633-nm laser line at maximum laser power. After photobleaching, an image of the donor distribution was recorded again. Images were low-pass filtered to reduce noise and registered prior to analysis. Calculations were restricted to those pixels where the donor intensity was at least the double of the background measured in a cell-free area. Pixel-by-pixel FRET efficiencies were determined using the evaluation method described in Ref.[50]. The apparent FRET efficiency Eij at the pixel with coordinates i, j was calculated as
| (2) |
where and are donor fluorescence intensities detected before and after acceptor photobleaching, and B is the mean cellular autofluorescence in the donor channel measured from unlabeled cells. Donor intensities were corrected for the unwanted photobleaching of the donor dye. Other correction factors such as crosstalk of the acceptor or its photoproduct into the donor channel were negligible[50]. Results were presented as pseudocolor FRET efficiency maps and frequency distribution histograms of Eij values. For numerical representation, the median values of the frequency distribution histograms obtained for individual cells were averaged and presented in a tabulated form.
Cholesterol depletion by methyl-β-cyclodextrin (MβCD)
Cells were washed twice with Hank’s buffered salt solution (HBSS) and then suspended in HBSS (control) or in HBSS containing 1, or 3 mM of MβCD (CycloLab) at a concentration of 2×106 cells/ml. The cell suspension was incubated for 30 min at 37°C followed by washing with HBSS and then with PBS. The efficiency of cholesterol extraction by MβCD was assessed from the decrease of fluorescence intensity of Filipin III (Sigma; 0.1 mg/ml). Incubation (106 cells/ml) for 1 h at 37°C was followed by washing twice in PBS and cells were then analyzed on a FACSAria flow cytometer (Becton Dickinson). Filipin III was excited by a 375-nm laser line and detected at 450±20nm. Treatment with 1mM and 3 mM MβCD reduced the cholesterol content of Kit225/IL-9R cells by ~25% and ~50%, respectively.
Detection of cytokine-stimulated tyrosine phosphorylation of STATs
IL-9 induced tyrosine phosphorylation of STAT1 was followed by detecting the binding of phospho-STAT1 specific mAb (mouse IgG2a, BD Biosciences) using flow cytometry. This antibody detects a phosphorylated Tyr moiety (Y701) on STAT1 without appreciable cross-reaction with other Tyr-phosphorylated STATs. Cells were labeled as suggested by the supplier. Briefly, cells with or without cytokine treatment were fixed in 2% formaldehyde/PBS (10 min at 37°C), permeabilized in 90% methanol (30 min on ice) followed by washing twice and staining with Alexa Fluor 647-conjugated anti-phospho-STAT1 mAb in staining buffer (PBS + 2% FBS + 0.1% Sodium Azide). After staining cells were washed twice in PBS, fixed with 1% formaldehyde/PBS and analyzed on a FACSArray or FACSAria flow cytometer (Becton Dickinson) using their 635-nm and 633-nm laser lines, respectively, suitable for excitation of Alexa Fluor 647. Fluorescence emission was detected through a 553 to 679-nm band-pass or a >655-nm long-pass filter. Flow cytometric intensity histograms were evaluated using the REFLEX software[48]. Since the fluorescence intensity histogram of the isotype control and the unstained cells were not significantly different, cells unstained with the anti-pTyr-STAT1 mAb served as a background sample.
Supplementary Material
Acknowledgements
We thank Rita Szabó and Adrienn Bagosi for their excellent technical assistance. This work was supported by the following grants: TÁMOP 4.2.1./B-09/1/KONV-2010-0007, 4.2.2/B-10/1-2010-0024, 4.2.2.A-11/1/KONV-2012-0023 and 4.2.2.A-11/1/KONV-2012-0025 projects implemented through the New Hungary Development Plan and co-financed by the European Union and the European Social Fund; TÁMOP-4.2.4.A/2-11/1-2012-0001 ‘National Excellence Program’ co-financed by the European Union, the State of Hungary and the European Social Fund; Hungarian Scientific Research Fund Grants CK 78179, K 103965, K 103906 and NK 101337; Baross Gábor program (REG-EA-09-1-2009-0010). The work was also supported by the Internal Research Program of University of Debrecen and the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
References
- 1.Vereb G, Szollosi J, Matko J, Nagy P, Farkas T, Vigh L, Matyus L, Waldmann TA, Damjanovich S. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8053–8058. doi: 10.1073/pnas.1332550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodnar A, Vamosi G, Toth K, Jenei A, Matyus L, Damjanovich S. In: Biophysical Aspects of Transmembrane Signaling. Damjanovich S, editor. Vol. 8. Heidelberg; New York: Springer Verlag, Berlin; 2005. pp. 71–95. [Google Scholar]
- 3.Pike LJ. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 5.Nagy P, Matyus L, Jenei A, Panyi G, Varga S, Matko J, Szollosi J, Gaspar R, Jovin TM, Damjanovich S. J. Cell Sci. 2001;114:4063–4071. doi: 10.1242/jcs.114.22.4063. [DOI] [PubMed] [Google Scholar]
- 6.Edidin M. Curr. Protoc. Immunol. 2003;Chapter 18(Unit 18):10. doi: 10.1002/0471142735.im1810s52. [DOI] [PubMed] [Google Scholar]
- 7.Jenei A, Kormos J, Szentesi G, Veres AJ, Varga S, Bodnar A, Damjanovich S, Matyus L. Chemphyschem. 2009;10:1577–1585. doi: 10.1002/cphc.200900242. [DOI] [PubMed] [Google Scholar]
- 8.Rochman Y, Spolski R, Leonard WJ. Nat. Rev. Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugamura K, Asao H, Kondo M, Tanaka N, Ishii N, Nakamura M, Takeshita T. Adv. Immunol. 1995;59:225–277. doi: 10.1016/s0065-2776(08)60632-x. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Lupardus P, Laporte SL, Garcia KC. Annu. Rev. Immunol. 2009;27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renauld JC, Van Snick J. In: The Cytokine Handbook. Thomson AW, Lotze MT, editors. London: Academic Press; 2003. pp. 347–358. [Google Scholar]
- 12.Demoulin JB, Renauld JC. Int. Rev. Immunol. 1998;16:345–364. doi: 10.3109/08830189809043001. [DOI] [PubMed] [Google Scholar]
- 13.Noelle RJ, Nowak EC. Nat. Rev. Immunol. 2010;10:683–687. doi: 10.1038/nri2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowak EC, Noelle RJ. Cell Cycle. 2009;8:3798–3799. doi: 10.4161/cc.8.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoops L, Renauld JC. Growth Factors. 2004;22:207–215. doi: 10.1080/08977190410001720879. [DOI] [PubMed] [Google Scholar]
- 16.Bauer JH, Liu KD, You Y, Lai SY, Goldsmith MA. J. Biol. Chem. 1998;273:9255–9260. doi: 10.1074/jbc.273.15.9255. [DOI] [PubMed] [Google Scholar]
- 17.Demoulin JB, Renauld JC. Cytokines. Cell. Mol. Ther. 1998;4:243–256. [PubMed] [Google Scholar]
- 18.Bodnar A, Nizsaloczki E, Mocsar G, Szaloki N, Waldmann TA, Damjanovich S, Vamosi G. Immunol. Lett. 2008;116:117–125. doi: 10.1016/j.imlet.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Damjanovich S, Bene L, Matko J, Alileche A, Goldman CK, Sharrow S, Waldmann TA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13134–13139. doi: 10.1073/pnas.94.24.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Bakker BI, Bodnar A, van Dijk EM, Vamosi G, Damjanovich S, Waldmann TA, van Hulst NF, Jenei A, Garcia-Parajo MF. J. Cell Sci. 2008;121:627–633. doi: 10.1242/jcs.019513. [DOI] [PubMed] [Google Scholar]
- 21.Vamosi G, Bodnar A, Vereb G, Jenei A, Goldman CK, Langowski J, Toth K, Matyus L, Szollosi J, Waldmann TA, Damjanovich S. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11082–11087. doi: 10.1073/pnas.0403916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matko J, Bodnar A, Vereb G, Bene L, Vamosi G, Szentesi G, Szollosi J, Gaspar R, Horejsi V, Waldmann TA, Damjanovich S. Eur. J. Biochem. 2002;269:1199–1208. doi: 10.1046/j.0014-2956.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 23.Vereb G, Matko J, Vamosi G, Ibrahim SM, Magyar E, Varga S, Szollosi J, Jenei A, Gaspar R, Jr, Waldmann TA, Damjanovich S. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6013–6018. doi: 10.1073/pnas.97.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. J. Cell Sci. 1992;103(Pt 3):857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y, Zinchuk V, Grossenbacher-Zinchuk O, Stefani E. Interdiscip Sci. 2012;4:27–37. doi: 10.1007/s12539-012-0117-x. [DOI] [PubMed] [Google Scholar]
- 26.Zinchuk V, Grossenbacher-Zinchuk O. Prog. Histochem. Cytochem. 2009;44:125–172. doi: 10.1016/j.proghi.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao W, Lin JX, Leonard WJ. Curr. Opin. Immunol. 2011;23:598–604. doi: 10.1016/j.coi.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bene L, Kanyari Z, Bodnar A, Kappelmayer J, Waldmann TA, Vamosi G, Damjanovich L. Biochem. Biophys. Res. Commun. 2007;361:202–207. doi: 10.1016/j.bbrc.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Harder T, Simons K. Eur. J. Immunol. 1999;29:556–562. doi: 10.1002/(SICI)1521-4141(199902)29:02<556::AID-IMMU556>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Langlet C, Bernard AM, Drevot P, He HT. Curr. Opin. Immunol. 2000;12:250–255. doi: 10.1016/s0952-7915(00)00084-4. [DOI] [PubMed] [Google Scholar]
- 32.Matyus L, Bene L, Heiligen H, Rausch J, Damjanovich S. Immunol. Lett. 1995;44:203–208. doi: 10.1016/0165-2478(94)00215-d. [DOI] [PubMed] [Google Scholar]
- 33.Goebel J, Forrest K, Flynn D, Rao R, Roszman TL. Hum. Immunol. 2002;63:813–820. doi: 10.1016/s0198-8859(02)00458-5. [DOI] [PubMed] [Google Scholar]
- 34.Knorr R, Karacsonyi C, Lindner R. J. Cell Sci. 2009;122:1584–1594. doi: 10.1242/jcs.039727. [DOI] [PubMed] [Google Scholar]
- 35.King C, Sarabipour S, Byrne P, Leahy DJ, Hristova K. Biophys. J. 2014;106:1309–1317. doi: 10.1016/j.bpj.2014.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabian AI, Rente T, Szollosi J, Matyus L, Jenei A. Chemphyschem. 2010;11:3713–3721. doi: 10.1002/cphc.201000568. [DOI] [PubMed] [Google Scholar]
- 37.Ramalingam TS, Chakrabarti A, Edidin M. Mol. Biol. Cell. 1997;8:2463–2474. doi: 10.1091/mbc.8.12.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skov S. Tissue Antigens. 1998;51:215–223. [PubMed] [Google Scholar]
- 39.Skov S, Nielsen M, Bregenholt S, Odum N, Claesson MH. Blood. 1998;91:3566–3573. [PubMed] [Google Scholar]
- 40.Xu S, Liu X, Bao Y, Zhu X, Han C, Zhang P, Zhang X, Li W, Cao X. Nat. Immunol. 2012;13:551–559. doi: 10.1038/ni.2283. [DOI] [PubMed] [Google Scholar]
- 41.Pillet AH, Lavergne V, Pasquier V, Gesbert F, Theze J, Rose T. J Mol Biol. 2010;403:671–692. doi: 10.1016/j.jmb.2010.08.056. [DOI] [PubMed] [Google Scholar]
- 42.Rose T, Pillet AH, Lavergne V, Tamarit B, Lenormand P, Rousselle JC, Namane A, Theze J. J Biol Chem. 2010;285:14898–14908. doi: 10.1074/jbc.M110.104232. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Tamarit B, Bugault F, Pillet AH, Lavergne V, Bochet P, Garin N, Schwarz U, Theze J, Rose T. J Biol Chem. 2013;288:8691–8701. doi: 10.1074/jbc.M113.449918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hori T, Uchiyama T, Tsudo M, Umadome H, Ohno H, Fukuhara S, Kita K, Uchino H. Blood. 1987;70:1069–1072. [PubMed] [Google Scholar]
- 45.Miyoshi I, Kubonishi I, Yoshimoto S, Shiraishi Y. Gann. 1981;72:978–981. [PubMed] [Google Scholar]
- 46.De Smedt M, Verhasselt B, Kerre T, Vanhecke D, Naessens E, Leclercq G, Renauld JC, Van Snick J, Plum J. J Immunol. 2000;164:1761–1767. doi: 10.4049/jimmunol.164.4.1761. [DOI] [PubMed] [Google Scholar]
- 47.Sebestyen Z, Nagy P, Horvath G, Vamosi G, Debets R, Gratama JW, Alexander DR, Szollosi J. Cytometry. 2002;48:124–135. doi: 10.1002/cyto.10121. [DOI] [PubMed] [Google Scholar]
- 48.Szentesi G, Horvath G, Bori I, Vamosi G, Szollosi J, Gaspar R, Damjanovich S, Jenei A, Matyus L. Comput. Methods Programs Biomed. 2004;75:201–211. doi: 10.1016/j.cmpb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Lifshitz LM. IEEE Trans. Med. Imaging. 1998;17:299–303. doi: 10.1109/42.700742. [DOI] [PubMed] [Google Scholar]
- 50.Roszik J, Szollosi J, Vereb G. BMC Bioinformatics. 2008;9:346. doi: 10.1186/1471-2105-9-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.