Abstract
Background
Biofilm formation is associated with various aspects of bacterial and fungal infection. This study was designed to assess the impact of diverse natural polyelectrolytes, such as DNA, F-actin, neurofilaments (NFs), vimentin and purified Pf1 bacteriophage on biofilm formation and swarming motility of select pathogens including Pseudomonas aeruginosa associated with lung infections in CF patients.
Results
The bacteriophage Pf1 (1 mg/ml) significantly increased biofilm mass produced by Pseudomonas aeruginosa P14, Escherichia coli RS218 and Bacillus subtilis ATCC6051. DNA, F-actin, NFs and Pf1 also increased biofilm mass of the fungal C. albicans 1409 strain. Addition of F-actin, DNA or Pf1 bacteriophage to 0.5% agar plates increased swarming motility of Pseudomonas aeruginosa Xen5.
Conclusions
The presence of polyelectrolytes at infection sites is likely to promote biofilm growth and bacterial swarming.
Keywords: Bacteria, C. albicans, Filamentous bacteriophages, DNA, F-actin, Biofilm mass
Background
Bacterial biofilm, a complex community of surface-associated cells surrounded by a self-produced polymer matrix consisting mostly of polysaccharides and DNA can form at infection sites and promote chronic infection [1–3]. Biofilm increases bacterial resistance to the host immune system, provides a place for development of resistant microorganisms and is associated with increased resistance to exogenous antibiotic treatment [4–6]. Therefore, once established, biofilm infections are nearly impossible to eradicate [7]. Biofilm formation involves the production of an extracellular matrix, but the composition of this matrix is not well defined. In Pseudomonas aeruginosa PA14, decreased production of extracellular polymers reduces static biofilm development [8]. An increasing number of studies support the hypothesis that DNA, F-actin, and possibly other filamentous polymers released from host inflammatory cells, as well as products of bacterial autolysis, trigger genetic changes in planktonic bacteria that initiate the formation of biofilm [9, 10]. Extracellular DNA and F-actin have been shown to function as a structural support for bacterial biofilm architecture [4, 11, 12] and have been proposed as therapeutic targets to prevent biofilm growth [5, 13]. Culturing Pseudomonas aeruginosa (PA) in settings that include neutrophils enhances formation of biofilms by stimulating the expression of alginate [5]. Recently, extracellular DNA was found to smooth the progress of efficient traffic flow of PA cells throughout the complex matrix of biofilm by maintaining coherent cell alignment, thereby avoiding traffic jams and ensuring an efficient supply of cells to the migrating front [14]. However, biofilm formation and swarming by clinical isolates of PA were found to be negatively associated [15]. Another consequence of negatively-charged polymers such as DNA and F-actin present at infection sites results from their ability to bind and inactivate host-derived cationic antibacterial molecules [16, 17]. Therefore, poly-anionic filamentous viruses produced by Pseudomonas, which are important for the virulence of these pathogens in humans, might inactivate host antibacterial molecules by similar mechanisms. The PA strain PAO1 strongly up-regulates genes encoding Pf1-like phages at the start of biofilm formation, and Pf1-like virions are present at 100 to 1000 fold higher levels in biofilms when compared with planktonic PA cultures [18]. The Pf1 phage exists largely as bundles of long filamentous phages extending from the bacterial surface [19]. Bundle formation was previously found to be mediated by polyvalent counterions, including cationic host defense molecules [20]. A repeatable pattern of cell death and lysis that occurs in biofilms during the normal course of development is also phage-dependent, and phage-mediated cell death is an important mechanism of differentiation inside microcolonies that facilitates dispersal of a subpopulation of surviving cells [21]. This report indicates that addition of diverse natural polyelectrolytes, including purified Pf1 bacteriophage, to different bacteria or Candida albicans cultures results in increased biofilm formation. The effects of Pf1 on biofilm formation by bacterial strains that it cannot infect suggests that the physicochemical properties of Pf1 and similar polyelectrolytes, rather than any specific biological effect of the bacteriophage on the bacteria is required for it to support biofilm formation by different microorganisms.
Results and discussion
Polyelectrolyte-induced formation of microorganisms’ biofilm
In agreement with previous reports [5, 13] F-actin increased biofilm formation by PA PAO1 on glass (Fig. 1a and b). Increased chemiluminescence of PA Xen5 bacteria culture after 48 h growth in the presence of F-actin or DNA was observed (Fig. 1b: rows 1-control, 2,3,4- increasing concentration of F-actin; 5,6,7—increasing concentration of DNA). DNA staining with YOYO-1 revealed an increased concentration of DNA in S. aureus Xen29 biofilm formed in the presence of exogenous DNA (Fig. 1d). This observation suggests that bacteria might use their own and likely other species’ extracellular DNA to form a biofilm matrix. Increased attachment of PA PAO1 to the cytoskeleton fraction of A549 cells attached to the coverglass surface after Triton x-100 treatment indicates that bacteria are able to bind the F-actin network (Fig. 1e and f). Compared to DNA and Pf1, F-actin has a stronger stimulatory effect on biofilm mass of PA Xen5 and S. aureus Xen29 (Fig. 2). Among all tested bacteria strains, the largest increase in biofilm production was observed after 48 h for B. subtilis growth in the presence of 0.25 and 1 mg/ml Pf1 (p < 0.001) (Fig. 3b). A statistically significant increase in biofilm formation was also observed at 48 h in PA P14 culture after addition of 0.25 and 1 mg/ml Pf1 (p < 0.05 and p < 0.001 respectively), in E coli MG1655 culture with addition of 1 mg/ml Pf1 (p < 0.001) and E.coli RS218 culture with addition of 0.25 and 1 mg/ml Pf1 (p < 0.05 and p < 0.001 respectively) (Fig. 3). The observed differences between the amounts of biofilm formed in the presence of Pf1 bacteriophage for different bacterial strains might be a result of growth rate differences.
Fig. 1.
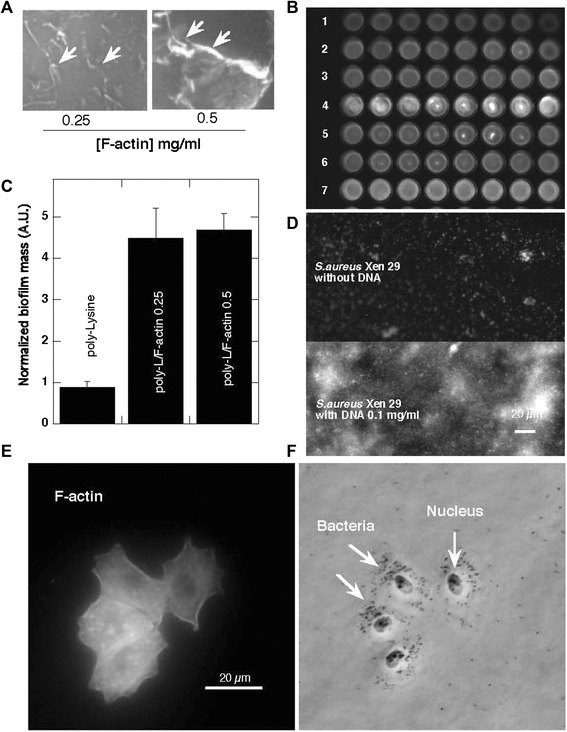
Bacterial biofilm formation in the presence of F-actin and DNA. a Arrows indicate F-actin filaments stained with rhodamine-phalloidin and attached to a glass slide coated with poly-Lysine (0.1% water solution). b Biofilm mass of Pseudomonas aeruginosa PAO1 formed in the presence of F-actin. c Chemiluminescence of Pseudomonas aeruginosa Xen5 48 h after bacteria addition to LB broth (row 1) containing F-actin (rows 2, 3, 4) or DNA (rows 5, 6, 7) at concentrations of 0.01, 0.05 and 0.1 mg ml −1 respectively. d DNA staining (YOYO-1) of Staphylococcus aureus Xen29 biofilm after 48 h growth with and without the presence of DNA. e F-actin filaments of lung epithelial A549 cytoskeleton stained with rhodamine-phalloidin (cells were treated with 0.01% Triton X-100 for 10 min and washed 3x with PBS before staining). f Pseudomonas aeruginosa PAO1 cells preferentially attached to the cytoskeleton of A549 cell remnants
Fig. 2.

F-actin, DNA and Pf1 bacteriophages increase Staphylococcus aureus Xen29 and Pseudomonas aeruginosa Xen5 biofilm formation. Panels a and b indicate biofilm mass assessed with crystal violet staining at 24 and 48 h time points. *Significantly increased compared to control
Fig. 3.

Pf1-mediated increase of biofilm formation. Biofilm formation of Pseudomonas aeruginosa P14 (panel a), Bacillus subtilis ATCC 6051 (panel b), Escherichia coli MG1655 (panel c) and Escherichia coli RS218 (panel d) in the presence of Pf1. Biofilm mass was assessed using crystal violet staining at 24 and 48 h time points. *Significantly increased compared to control
DNA, F-actin, neurofilaments and vimentin are all anionic polyelectrolyte filaments present within the cellular and extracellular space, that have large net negative charge densities distributed over their surfaces and all promote biofilm formation (Figs. 1, 2 and 3 and data not shown). However, there are some differences between their activity that may result from filament properties or the growth of different bacteria strains. Neurofilaments have little effect on PA Xen5 biofilm formation compared to their effect on S. aureus Xen29, which is more than doubled in the presence of neurofilaments at a concentration of 0.1 mg/ml. Vimentin slightly increases biofilm formation of both PA Xen5 and S. aureus Xen29 (data not shown). Polyelectrolyte-induced biofilm formation was also observed for the fungus C. albicans 1409, which is a frequent cause of fungal infection in humans. The fungal biofilm matrix is mostly produced by the fungal cells themselves and is composed of different types of biopolymers [22]. As shown in Fig. 4, DNA, Pf1, F-actin and NFs all increase the formation of C. albicans biofilm. Among these polyelectrolytes, the strongest activity was observed for DNA, Pf1, F-actin, and NFs (Fig. 4). At the tested concentration range (0.1-1 mg/ml) NFs reveal the potential to promote C. albicans biofilm formation only at low concentrations. At a concentration greater than 0.5 mg/ml, NFs decrease the amount of biofilm formed, an effect that might be related to their high negative charge density. At a high concentration NFs might function as neutrophil extracellular traps (NETs).
Fig. 4.

Biofilm formation of C. albicans. Increase of C. albicans 1409 biofilm formation in the presence of DNA (panel a), F-actin (panel b), Pf1 (panel c) and NFs (panel d) (n = 6). *Significantly different compared to control
Increase of P. aeruginosa swarming activity in the presence of polyelectrolytes
Bacterial swarming motility and production of exopolysaccharides both contribute to biofilm formation, yet it is unclear how bacteria coordinate these two processes [23]. For some bacteria, the ability to swarm positively correlates with production of exopolysaccharides [24]. In contrast, alginate oligomers (OligoG) were found to bind to the bacterial surface, modulate surface charge, induce microbial aggregation, and inhibit swarming motility [25]. Figure 5 shows the morphology of a P. aeruginosa Xen5 biofilm that develops in the presence of different biopolymers. At the macroscopic level, a more fragmented pattern of growth was observed in the presence of F-actin, compared to uniform growth in DNA- or Pf1-containing samples. As indicated by the arrow in Fig. 5, on the edges of the biofilm where the swarming assures spreading of bacteria, more rounded structures less than 10 μm in size are observed in substrates containing biopolymers compared to control substrates. Quantitative analysis of biofilm indicates that all tested biopolymers increase swarming activity of PA Xen5. Among these biopolymers, there is a higher average increase of swarming area for substrates containing DNA than F-actin or Pf1 at the higher concentrations (0.1 mg/ml). In agreement with our study, recent results indicate that among the tested swarming mutants, a large range of biofilm formation levels was observed [26].
Fig. 5.
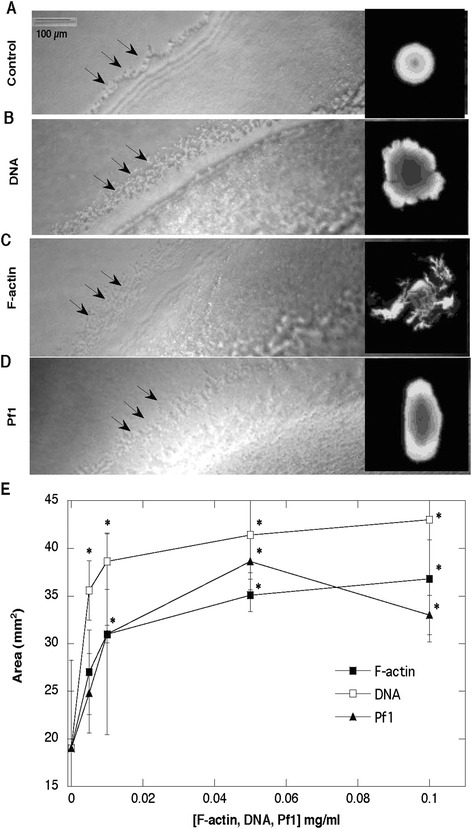
Morphology of Pseudomonas aeruginosa Xen5 biofilm growth on agar with addition of DNA, F-actin and Pf1 bacteriophage. In the presence of different biopolymers (0.1 mg/ml), after 16 h of bacterial growth, the swarming edges show some similar features (they are more diffuse and less defined; panel a-d). Developing area of spreading PA biofilm due to swarming activity was assessed using Image Gauge software (panel e). *Significantly different compared to control
Lower mass of biofilm formed in the presence of DNase I
Two clinically important biofilm-forming bacterial pathogens, PA and S. aureus were used to assess the effects of DNAse I and cationic antibacterial agents on biofilm mass. Addition of Pulmozyme (DNAse I) at 0.5 μg/ml significantly decreases the biofilm mass of both pathogens grown for 48 h with and without DNA (p < 0.01). In an earlier study, the addition of poly(aspartic acid) was found to reduce adhered microflora and decrease dental plaque formation [27]. The combination of DNase I and poly(aspartic acid) decreased PA biofilm formed in the presence of activated neutrophils by 79.2% on hydrogel contact lenses [13], and disrupted mature PA biofilm [4]. In our experimental settings, poly(aspartic acid) decreased biofilm formation but the effect did not reach a statistically significant level (data not shown). This difference might potentially be associated with the length of polymer used. On the other hand, in accordance with previous observations that the polycationic antimicrobial peptide LL-37 blocks biofilm formation at concentrations below its MIC, [28] decreased biofilm mass of both pathogens in the presence of LL-37 peptide (p < 0.001) was observed. A similar decrease was found in the presence of the cationic antibacterial steroid ceragenin CSA-13 (Fig. 6).
Fig. 6.
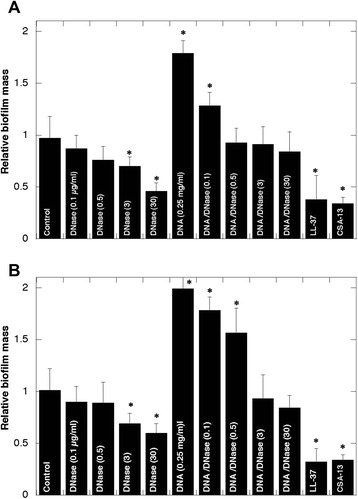
Biofilm formation in the presence of DNA and DNase I. Pseudomonas aeruginosa Xen 5 (panel a) and Staphylococcus aureus Xen 29 (panel b) biofilm at 48 h in 50% LB with DNA 0.25 mg/ml in the absence/presence of DNase I (0.1–30 μg/ml) and in the presence of LL-37 or CSA-13 (20 μM each). Inhibitory effect of DNase was compared to the effect of CSA-13 and LL-37 (n = 4). *Significantly different compared to control
Discussion
Accumulation and death of neutrophils recruited in response to pathogen invasion and tissue destruction release various negatively-charged biopolymers including DNA, vimentin, and F-actin. Destruction of neuronal cells can be expected to release NFs in addition to F-actin and DNA, whereas damaged muscle cells might predominantly release actin. Bacteria also support the accumulation of extracellular anionic biopolymers since they can release exopolysaccharides, bacterial DNA, as well as different filamentous phage. A growing body of evidence indicates that these biopolymers can interfere with host immunoresponse to infections. Extracellular DNA (eDNA) is a major structural component in many biofilms of pathogenic bacteria [1, 29]. Additionally, the functional DNA binding-uptake system that can be inhibited by the presence of DNase l [9], DNA-induced changes in gene expression [30], and sphingomyelinase-mediated cross-linking of eDNA in the biofilm skeletal framework [31] are all involved in biofilm formation by different bacterial pathogens. Results of our study indicate that the ability to stimulate biofilm formation might be a common feature of negatively charged biopolymers, including Pf1-like phages expressed by PA strains preferentially infecting CF patients. However, there are conflicting interpretations as to whether production of the filamentous phage virions encoded within P. aeruginosa as they colonize the airway increase the virulence of P. aeruginosa strains. The increased production of biofilm in the presence of Pf1 suggests that phage expression should be considered as a factor that increases the virulence of Pseudomonas.
Determining the relevant concentration range of phage and other polyelectrolytes is guided by several considerations. Infected, purulent body fluids such as cystic fibrosis sputum can contain as much as 50 mg ml−1 DNA and 10 mg ml−1 F-actin. Since Pf1 is not lytic and diffuses very slowly from its site of production, the relevant virus concentration is that adjacent to the bacteria, as seen in [19]. Quantitative studies of the rate or extent of Pf1, Pf4, or Pf5 production by Pseudomonas strains have not yet been reported, but E. coli can produce 830 copies of the structurally similar 800 nm long filamentous phage M13 per hour [32]. Making the conservative estimate that a 0.5 μm x 2 μm cylindrical Pseudomonas produces 800 viruses extending from the bacterial surface, and using the fact that a polymer of length 2 μm and persistence length 2 μm has an end-to-end distance of 1.7 μm [33], the local concentration of Pf1 within the zone 1.7 μm away from the bacterial surface is approximately 1 mg ml−1. This concentration is within the range at which Pf1 promotes biofilm growth. Therefore [Pf1] between 0.1 and 5 mg ml−1 is likely to be relevant to a purulent setting. These findings also indicate that the physicochemical properties of filamentous bacteriophage, and its concentration, rather than the ability of Pf1 to infect and propagate in bacteria, are sufficient to promote biofilm growth. Pf1 has a very similar size, shape, and charge density as F-actin and aggregates similarly with multivalent cations. These features might explain the strong ability of both polymers to induce biofilm formation [5, 34], as well as that of neurofilaments and vimentin [35]. In agreement with recent studies showing an increase of C. albicans biofilm formation induced by its own DNA as well as DNA released from bacteria [12, 36], we observed that DNA and other natural polyelectrolytes such as F-actin, Pf1 and NFs can all stimulate this process. The dose-dependent effect recorded for NFs (concentrations above 0.5 mg/ml resulted in decreased biofilm formation) was previously reported for DNA [36].
The effect of anionic biopolymers to promote biofilm formation may depend on their effect on swarming mobility. Many bacteria move in groups, in a mode described as swarming to colonize surfaces and form a biofilm to survive external stresses, including exposure to antibiotics [37]. Considering the possibility of charge-driven interactions, antibiotic resistance might result from the ability to sequester positively-charged antibiotics by negatively-charged polymeric components [17, 38] of the biofilm network before the antibiotics reach and insert into bacterial cells. This mode of bactericidal agent inactivation affects natural host defense cationic peptides such as cathelicidin LL-37.
Conclusion
The presence of polyelectrolytes at infection sites such as DNA, F-actin, neurofilaments and Pf1 bacteriophage is likely to promote biofilm formation. However, the mass of developing biofilm in the presence of the same kind of polyelectrolytes varies among different microorganism strains.
Methods
Materials
Cetrimide Agar (Pseudomonas selective agar base) and deoxyribonucleic acid from herring sperm were from Sigma. Luria-Bertani broth (LB) and tryptic soy broth (TSB) were from DIFCO (Sparks, MD). Dulbecco’s Modified Eagle media (DMEM) was from GIBCO, (Grand Island, NY). Fetal bovine serum was from Hyclone, (Logan, UT). PA Xen 5 and S. aureus Xen29 strains engineered through conjugation and transposition of a plasmid carrying transposon Tn5 luxCDABE were purchased from Caliper Life Science Inc. (CA, USA). B. subtilis ATCC6051, E. coli MG1655, E. coli RS218 and C. albicans 1409 was purchased from ATTC collection and Polish Collection of Microorganism (PCM). High affinity nucleic acid stain YOYO®-1 was from Life Technologies (Grand Island, NY). Monomeric G-actin was prepared from acetone powder of rabbit skeletal muscle [39] and kept in non-polymerizing solution contained 2 mM TRIS, 0.2 mM CaCl2, 0.5 mM ATP, 0.2 mM DTT pH 7.4. Actin polymerization was induced by adding 150 mM KCl and 2 mM MgCl2. Neurofilaments (NFs) were isolated from bovine spinal cord according to procedures described previously [40]. Protease free, Pf1 bacteriophage strain LP11-92 that was propagated in P. aeruginosa LA23-99 was from ASLA Biotech Ltd. (Riga, Latvia).
Cell Culture
Human lung epithelial carcinoma cells, line A549, were cultured in DMEM supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. Cells were seeded and allowed to spread for 24–72 h before experiments were performed. A549 cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 min at room temperature and subjected to 0.1% solution of TRITON X-100 treatment to expose the cytoskeleton F-actin network. When required, staining with 1:100 rhodamine-labeled phalloidin (Invitrogen) in PBS for 30 min was performed. To assess bacterial ability to interact with the cytoskeleton, a suspension of PA PAO1 (105 CFU/ml) was added to A549 cell remnants for five minutes. Fluorescence and bacterial attachment were evaluated after 3 washes with PBS, using a LEICA DM IRBE. Images were captured with a Cool SNAP (HQ) camera (Trenton, NJ, USA).
Biofilm formation
The mass of biofilms was assessed using Crystal Violet (CV) staining (0.1%) [41] and chemiluminescence intensity measurements as an additional determinant of biofilm viability [42]. In each experiment an overnight culture of P. aeruginosa Xen 5, P. aeruginosa PAO1, P. aeruginosa P14 in TSB or S. aureus Xen 29, B. subtilis ATCC6051, E. coli MG1655, E. coli RS218 and C. albicans 1409 in LB was diluted to ~ 105 CFU/ml. When required, bacterial suspensions were placed on glass slides coated with poly-Lysine/F-actin or in flat bottom PVC microplates (MP Biomedicals; Solon, OH) sealed with a gas permeable membrane (USA Scientific; Ocala, FL). Bacteria or C. albicans were grown in 50% LB broth with or without F-actin, DNA, NFs, vimentin or Pf1 bacteriophage. A stationary biofilm was allowed to form for 24 or 48 h. PA Xen5 and S. aureus Xen29 chemiluminescence signals were evaluated using a Fuji Film LAS-300 system. Densitometry analysis was performed using Image Gauge (version 4.22) software.
Pseudomonas aeruginosa Xen5 swarming motility
A swarming motility assay was assessed as described previously [43]. Briefly, 3 μl of bacterial inoculum containing ~108 CFU/ml were placed in the center of 0.5% agar containing modified M9 medium with different concentrations of F-actin, DNA or Pf1 bacteriophage (0.005–0.1 mg/ml) in 6 well plates. Swarming area was evaluated from images captured with a Fuji Film LAS-300 system analyzed with Image Gauge (version 4.22) software.
Statistical analysis
Data are reported as mean ± SD from 3–8 repeats. Differences between means were evaluated using the unpaired Student’s t-test and differences p < 0.05 were considered statistically significant.
Acknowledgements
This work was supported by the National Science Center, Poland under Grant: UMO-2012/07/B/NZ6/03504 (to RB), Medical University of Białystok grant: 143-62679 L (to R.B.) and the: GM096971 grant (to PJ).
Footnotes
Competing interests
In 2012, Dr. P. Janmey and Dr. R. Bucki were involved in a sponsored research agreement with Genentech in a project directed at evaluating the potential clinical use of p-ASP to improve Pulmozyme activity, but not otherwise related to the present study. Other authors: none to declare. None of the research reported in this paper was supported by any corporate entity.
Authors’ contributions
RB: study design, carried out evaluation of LL-37 activity in the presence of polyelectrolytes and helped to draft the manuscript KN: participated in study design, carried out biofilm studies; UW: biofilm studies, helped to draft the manuscript; MW: participated in study design and helped to draft the manuscript; FB: F-actin studies, participated in study design, helped to draft the manuscript; KC: helped to draft the manuscript, evaluation of LL-37 bactericidal activity in different settings; MWr: participated in study design, helped to draft the manuscript; PAJ: participated in study design, helped to draft the manuscript; All authors read and approved the final manuscript.
Contributor Information
Robert Bucki, Phone: 00-48-85-748-5483, Email: buckirobert@gmail.com.
Katarzyna Niemirowicz, Phone: 00-48-85-748-5483, Email: katia146@wp.pl.
Urszula Wnorowska, Phone: 00-48-85-748-5483, Email: u.wnorowska@gmail.com.
Marzena Wątek, Phone: 00-48-41 367-42-08, Email: marzena.watek@wp.pl.
Fitzroy J. Byfield, Phone: 001-215-573-9787, Email: fbyfield@mail.med.upenn.edu
Katrina Cruz, Phone: 001-215-573-9787, Email: katcruz@mail.med.upenn.edu.
Marta Wróblewska, Phone: 00-48-22-628-27-39, Email: martamwroblewska@gmail.com.
Paul A. Janmey, Phone: 00-1-215-573-9787, Email: janmey@mail.med.upenn.edu
References
- 1.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295(5559):1487. [DOI] [PubMed]
- 2.Moreau-Marquis S, Stanton BA, O’Toole GA. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther. 2008;21(4):595–9. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garde C, Welch M, Ferkinghoff -Borg J, Sams T. Microbial Biofilm as a Smart Material. Sensors (Basel). 2015;15(2):4229–41. [DOI] [PMC free article] [PubMed]
- 4.Parks QM, Young RL, Poch KR, Malcolm KC, Vasil ML, Nick JA. Neutrophil enhancement of Pseudomonas aeruginosa biofilm development: human F-actin and DNA as targets for therapy. J Med Microbiol. 2009;58(Pt 4):492–502. [DOI] [PMC free article] [PubMed]
- 5.Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, et al. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun. 2005;73(6):3693–701. [DOI] [PMC free article] [PubMed]
- 6.Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis. 2015;34(5):877-86. [DOI] [PubMed]
- 7.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J. Novel entries in a fungal biofilm matrix encyclopedia. MBio. 2014;5(4):e01333–14. [DOI] [PMC free article] [PubMed]
- 8.Kim HS, Park HD. Ginger extract inhibits biofilm formation by Pseudomonas aeruginosa PA14. PLoS One. 2013;8(9):e76106. doi: 10.1371/journal.pone.0076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen FC, Tao L, Scheie AA. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J Bacteriol. 2005;187(13):4392–400. doi: 10.1128/JB.187.13.4392-4400.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas VC, Thurlow LR, Boyle D, Hancock LE. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J Bacteriol. 2008;190(16):5690–8. [DOI] [PMC free article] [PubMed]
- 11.Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol. 2011;193(23):6552–9. [DOI] [PMC free article] [PubMed]
- 12.Pammi M, Liang R, Hicks J, Mistretta T-A, Versalovic J. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol. 2013;13:257. [DOI] [PMC free article] [PubMed]
- 13.Robertson DM, Parks QM, Young RL, Kret J, Poch KR, Malcolm KC, et al. Disruption of contact lens-associated Pseudomonas aeruginosa biofilms formed in the presence of neutrophils. Invest Ophthalmol Vis Sci. 2011;52(5):2844–50. [DOI] [PMC free article] [PubMed]
- 14.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, et al. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A. 2013;110(28):11541–6. [DOI] [PMC free article] [PubMed]
- 15.Murray TS, Ledizet M, Kazmierczak BI. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J Med Microbiol. 2010;59(Pt 5):511–20. doi: 10.1099/jmm.0.017715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bucki R, Sostarecz AG, Byfield FJ, Savage PB, Janmey PA. Resistance of the antibacterial agent ceragenin CSA-13 to inactivation by DNA or F-actin and its activity in cystic fibrosis sputum. J Antimicrob Chemother. 2007;60(3):535–45. [DOI] [PubMed]
- 17.Bucki R, Byfield FJ, Janmey PA. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur Respir J. 2007;29(4):624–32. doi: 10.1183/09031936.00080806. [DOI] [PubMed] [Google Scholar]
- 18.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413(6858):860–4. [DOI] [PubMed]
- 19.Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186(23):8066–73. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang JX, Wen Q, Bennett A, Kim B, Sheils CA, Bucki R, Janmey PA. Anionic poly(amino acid)s dissolve F-actin and DNA bundles, enhance DNase activity, and reduce the viscosity of cystic fibrosis sputum. Am J Physiol Lung Cell Mol Physiol. 2005;289(4):L599–605. [DOI] [PubMed]
- 21.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, et al. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185(15):4585–92. [DOI] [PMC free article] [PubMed]
- 22.Lopez-Ribot JL. Large-Scale Biochemical Profiling of the Candida albicans Biofilm Matrix: New Compositional, Structural, and Functional Insights. MBio. 2014;5(5):e01781-14. doi: 10.1128/mBio.01781-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, et al. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharides production in Pseudomonas aeruginosa. Appl Environ Microbiol. 2014, doi:10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed]
- 24.Kalai Chelvam K, Chai LC, Thong KL. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog. 2014;6(1):2. doi: 10.1186/1757-4749-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Powell LC, Pritchard MF, Emanuel C, Onsøyen E, Rye PD, Wright CJ, et al. A nanoscale characterization of the interaction of a novel alginate oligomer with the cell surface and motility of Pseudomonas aeruginosa. Am J Respir Cell Mol Biol. 2014;50(3):483–92. [DOI] [PubMed]
- 26.Mireles JR, Toguchi A, Harshey RM. Salmonella enterica serovar typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J Bacteriol. 2001;183(20):5848–54. doi: 10.1128/JB.183.20.5848-5854.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan YH, Lath DL, Graaf T, Lilley TH, Brook AH. Moderation of oral bacterial adhesion on saliva-coated hydroxyapatite by polyaspartate. J Appl Microbiol. 2003;94(3):456–61. [DOI] [PubMed]
- 28.de la Fuente-Nunez C, Korolik V, Bains M, Nguyen U, Breidenstein EB, Horsman S, et al. Inhibition of bacterial biofilm formation and swarming motility by a small synthetic cationic peptide. Antimicrob Agents Chemother. 2012;56(5):2696–704. [DOI] [PMC free article] [PubMed]
- 29.Vlassov VV, Laktionov PP, Rykova EY. Extracellular nucleic acids. Bioessays. 2007;29(7):654–67. doi: 10.1002/bies.20604. [DOI] [PubMed] [Google Scholar]
- 30.Caceres SM, Malcolm KC, Taylor-Cousar JL, Nichols DP, Saavedra MT, Bratton DL, et al. Enhanced In Vitro Formation and Antibiotic Resistance of Non-attached Pseudomonas aeruginosa Aggregates through Incorporation of Neutrophil Products. Antimicrob Agents Chemother. 2014;58(11):6851-60. [DOI] [PMC free article] [PubMed]
- 31.Huseby MJ, Kruse AC, Digre J, Kohler PL, Vocke JA, Mann EE, et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc Natl Acad Sci U S A. 2010;107(32):14407–12. [DOI] [PMC free article] [PubMed]
- 32.Ploss M, Kuhn A. Kinetics of filamentous phage assembly. Phys Biol. 2010;7(4):045002. doi: 10.1088/1478-3975/7/4/045002. [DOI] [PubMed] [Google Scholar]
- 33.Hill DF, Short NJ, Perham RN, Petersen GB. DNA sequence of the filamentous bacteriophage Pf1. J Mol Biol. 1991;218(2):349–64. [DOI] [PubMed]
- 34.Schwab UE, Ribeiro CM, Neubauer H, Boucher RC. Role of actin filament network in Burkholderia multivorans invasion in well-differentiated human airway epithelia. Infect Immun. 2003;71(11):6607–9. [DOI] [PMC free article] [PubMed]
- 35.Hansma HG, Pietrasanta LI, Auerbach ID, Sorenson C, Golan R, Holden PA. Probing biopolymers with the atomic force microscope: a review. J Biomater Sci Polym Ed. 2000;11(7):675–83. [DOI] [PubMed]
- 36.Sapaar B, Nur A, Hirota K, Yumoto H, Murakami K, Amoh T, et al. Effects of extracellular DNA from Candida albicans and pneumonia-related pathogens on Candida biofilm formation and hyphal transformation. J Appl Microbiol. 2014;116(6):1531–42. [DOI] [PubMed]
- 37.Du H, Xu Z, Anyan M, Kim O, Leevy WM, Shrout JD, Alber M. High density waves of the bacterium Pseudomonas aeruginosa in propagating swarms result in efficient colonization of surfaces. Biophys J. 2012;103(3):601–9. [DOI] [PMC free article] [PubMed]
- 38.Bucki R, Namiot DB, Namiot Z, Savage PB, Janmey PA. Salivary mucins inhibit antibacterial activity of the cathelicidin-derived LL-37 peptide but not the cationic steroid CSA-13. J Antimicrob Chemother. 2008;62(2):329–35. [DOI] [PMC free article] [PubMed]
- 39.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–71. [PubMed] [Google Scholar]
- 40.Leterrier JF, Käs J, Hartwig J, Vegners R, Janmey PA. Mechanical effects of neurofilament cross-bridges. Modulation by phosphorylation, lipids, and interactions with F-actin. J Biol Chem. 1996;271(26):15687–94. [DOI] [PubMed]
- 41.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72(2):157–65. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Leszczynska K, Namiot A, Cruz K, Byfield FJ, Won E, Mendez G. Potential of ceragenin CSA-13 and its mixture with pluronic F-127 as treatment of topical bacterial infections. J Appl Microbiol. 2011;110(1):229–38. [DOI] [PMC free article] [PubMed]
- 43.Tremblay J, Deziel E. Improving the reproducibility of Pseudomonas aeruginosa swarming motility assays. J Basic Microbiol. 2008;48(6):509–15. doi: 10.1002/jobm.200800030. [DOI] [PubMed] [Google Scholar]


