Abstract
L-Asparaginase (ASNase) is a front-line chemotherapy for acute lymphoblastic leukemia (ALL), which acts by deaminating asparagine and glutamine. To evaluate the importance of glutaminase activity, we exploited a recently developed mutant of Helicobacter pylori ASNase (dm HpA), with amino acid substitutions M121C/T169M. The mutant form has the same asparaginase activity as wild-type but lacks glutaminase activity. Wild-type and dm HpA were compared with the clinically used ASNases from E. coli (L-ASP) and Erwinia chrysanthemi (ERWase). Asparaginase activity was similar for all isoforms, while glutaminase activity followed the rank order: ERWase > L-ASP > wild-type HpA > dm HpA. Cytotoxic efficacy of ASNases was tested on 11 human leukemia cell lines and two patient-derived ALL samples. Two cell lines which we had previously shown to be asparagine-dependent were equally sensitive to the asparaginase isoforms. The other nine lines and the two patient-derived samples were more sensitive to isoforms with higher glutaminase activities. ERWase was overall the most effective ASNase on all cell lines tested whereas dm HpA, having the lowest glutaminase activity, was the least effective. These data demonstrate that asparaginase activity alone may not be sufficient for ASNase cytotoxicity, and that glutaminase activity may be required for full anti-leukemic efficacy.
Keywords: Asparaginase, Glutaminase, ALL, Leukemia
Introduction
L-Asparaginases (ASNases) are naturally occurring bacterial enzymes currently used as a universal component of treatment of childhood acute lymphoblastic leukemia (ALL), especially for remission induction 1–3. The drug acts through the hydrolysis of asparagine and glutamine, two vital amino acids for leukemia cell growth, to aspartic acid and glutamic acid, respectively, releasing ammonia in the process 4. Leukemic lymphoblasts are highly sensitive to asparagine depletion due to low levels of asparagine synthetase (ASNS) and thus generally depend on extracellular supply of asparagine. Up-regulation of ASNS and glutamine synthetase, as well as glutamine transporters, are associated with resistance in vitro 5,6. Level of ASNS mRNA, protein and activity, however, vary widely in pediatric ALL samples 7–12 and are not necessarily associated with in vitro resistance to L-asparaginase, thus indicating additional mechanisms of resistance to ASNases besides modulation of ASNS 13. Lymphoblasts and lymphocytes, especially B lymphoblastoid cell lines 14,15, are highly dependent on glutamine when compared to other cell types 16. Glutamine is an essential co-substrate of ASNS, as it donates the amino group required for asparagine synthesis 17, indicating a critical dependency on glutamine supply for asparagine neo-synthesis. Strategies targeting glutamine 18,19 in addition to asparagine may therefore improve treatment outcomes in newly diagnosed, and in relapsed, ALL patients.
Novel ASNases besides E. coli ASNase (L-ASP) with different activities have been developed in an attempt to decrease side effects and increase efficacy20. Erwinia chrysanthemi ASNase or Erwinase (ERWase) has similar asparaginase but more than 10-fold higher glutaminase activity in comparison to E. coli ASNase, with favorable Km and faster catalysis (higher kcat) 21. Alternatively, other investigators seek to reduce the glutaminase activity of ASNase to reduce toxicity 22–24. Glutaminase activity of L-ASP was recently shown to be necessary for its anticancer activity against ASNS positive cell types but not ASNS-negative cell types 24. Recently, we have shown on a panel of 9 human leukemia cell lines that ALL cells have greater dependence on glutamine than asparagine 25. It is therefore critical to establish whether glutaminase activity is necessary for ALL chemotherapy. In this study, we test whether glutaminase activity is important for cytotoxicity of L-asparaginases on human leukemia cells, using a recently characterized mutant H. pylori ASNase lacking glutaminase activity.
Materials and Methods
Cell lines
Human leukemia cell lines were obtained from ATCC (Manassas, VA) and the German Collection of Cell Lines (DSMZ, Braunschweig, Germany; Table 1). We also used patient-derived human leukemia samples, BLQ1 and TxL2 (Ph-positive ALL), which had been passaged through NOD.Cg-PrkdcscidIl2rgtm1Wjll/SzJ mice and cultured as described previously 25,26.
Table 1.
Human leukemia cell lines
| CELLS | ASN ase | PHENOTYPE | TISSUE | LEUKEMIA | MOLECULAR GENETIC |
|---|---|---|---|---|---|
| K-562 | R | Erythromyeloblast | Bone marrow | CML | t(9;22) Ph1 BCR/ABL |
| SUP-B15 | S | Pre-B lymphoblast | Bone marrow | ALL rel. | t(9;22) Ph1 BCR/ABL |
| BV173 | I/R | Pre-B lymphoblast/ myeloblast | Peripheral Blood | ALL/CML | t(9;22) Ph1 BCR/ABL |
| SD-1 | R | Pre-B lymphoblast | Peripheral blood | ALL | t(9;22) Ph1 BCR/ABL |
| HL-60 | R | Pro-myeloblastoid | Peripheral blood | APL | c-myc overexpress. |
| MOLT4 | S | T lymphoblast | Peripheral Blood | ALL | low p53, high MYB |
| RS4;11 | S | Pro-B lymphoblast | Bone marrow | ALL rel. | t(4;11) MLL/AF4 |
| SEM | I | Pro-B lymphoblast | Peripheral blood | ALL | t(4;11) MLL/AF4 |
| RCH-ACV | R | Pre-B lymphoblast | Bone marrow | ALL | t(1;19) E2A/PBX1 |
| NALM6 | R | Pre-B lymphoblast | Peripheral blood | ALL rel. | t(5;12) |
| REH | R | Pro-B lymphoblast | Peripheral blood | ALL rel. | t(12;21) TEL/AML1 |
|
| |||||
| BLQ1 | * | Pre-B lymphoblast | Bone Marrow | ALL | t(9;22) Ph1 BCR/ABL |
| TxL2 | * | Pre-B lymphoblast | Bone Marrow | ALL | t(9;22) Ph1 BCR/ABL |
Adapted and updated from 7. S: sensitive; I: intermediate; R: resistant; rel.: relapse.
BLQ1 and TxL2 were cultured with OP9 feeder layer during treatment with ASNase.
Double mutant Helicobacter pylori construct
The expression and biochemical characterization of recombinant Helicobacter pylori L-asparaginase (wt HpA) has been previously described 27. The enzyme showed a strong preference for asparagine (Km: 0.290 mM) over glutamine (Km: 46.4 mM) 21,27. A novel glutamine-inactive form of Helicobacter pylori ASNase (M121C/T169M mutant, dm HpA) was recently generated by site-directed mutagenesis28. The M121C mutant was designed to have the same asparaginase activity as wild type (wt), but lack glutaminase activity. However, only with the addition of the serendipitous mutation T169M was wild-type asparaginase activity maintained in the glutaminase-free mutant. The biochemical characterization of the single and double mutant and a preliminary analysis of the cytotoxicity of the latter on HL-60 cells at 24 hours has been described elsewhere28.
Measurement of asparaginase and glutaminase activities
L-asparaginase derived from Escherichia coli (L-ASP) was obtained from Amatheon (Miami, FL). Erwinase investigational drug was kindly provided for experimental evaluations by Dr. Paul Plourde (Jazz Pharmaceuticals, Langhorne, Pennsylvania). Quantitative detection of ammonia was performed with Nessler’s reagent as previously described 25,29 and adapted to 96-well plates for measurement of asparaginase and glutaminase activities with multiple dilutions. This method allows for direct comparison of asparaginase and glutaminase activities between different ASNase isoforms. The production of ammonia by L-asparaginase over time was expressed relative to the slope of known ammonia standards. The resulting value represented the activity of the enzyme in international units (IUs), in which one IU equaled the amount of enzyme that catalyzed the formation of 1 μmol of ammonia per min. Briefly, 10 mM asparagine or 40 mM glutamine, pH 8.6, were incubated at 37° C for 15 min with different dilutions of ASNases (0.001 IU to 20 IU) in a final volume of 100 μl/well using 96-well plates. The reaction was stopped on ice by adding 5 μl of 1.5 M TCA and microplates were centrifuged at 3700 x g. 10 μl of each ammonium sulfate standard or reaction mix were mixed in duplicate with 190 μL of Nessler’s Working Reagent, and absorbance was measured at 436 nm. While each ASNase isoform has different optimal pH for their respective asparaginase and glutaminase activities which may differ from the physiological or culture medium pH, we did not observe any significant differences in asparaginase and glutaminase activities measured at pH 8.6 and 6.5. In addition, the ranking order of the isoforms was identical (data not shown).
Data were fitted with a non-linear regression 5-parameter model.
Determination of cell viability
Cells were plated onto 96-well plates at a density of 100,000–120,000 cells/well in RPMI medium supplemented with 10% FBS, Na Pyruvate, GlutaMax and gentamicin. L-ASNases were tested in a final concentration range from 0.03 to 3 IU/ml for 72 hours. Experiments with primary human leukemia cells were conducted over OP9 feeder layers, with leukemia cells collected at 72 hours by vigorous trituration25. Viable cells were determined by trypan blue exclusion in triplicate wells, counted by a blinded observer. Each dose-response experiment was repeated for n = 4 to 5 for each cell line. In additional experiments, cell death was measured by flow cytometry. Cells plated at 500,000 cells/well in 24-well plates were incubated for 48 hours with ASNases. Cells were then washed twice in ice-cold PBS. DAPI was added and the samples were run on LSR II Analyzer (Becton Dickinson).
Statistical analysis
Two-way ANOVA with Bonferroni post-hoc test was used to analyze the percentage of remaining viable leukemia cells or percent inhibition. Inferences for concentration analysis were determined with Wilcoxon signed-rank test. A p-value of less than 0.05 was considered significant. IC50 and percentage inhibition at 3 IU/ml were calculated from normalized data using non-linear regression with GraphPad Prism. Correlation was determined with Pearson’s test.
Results
ASNases have similar maximal asparaginase activity with different glutaminase activity
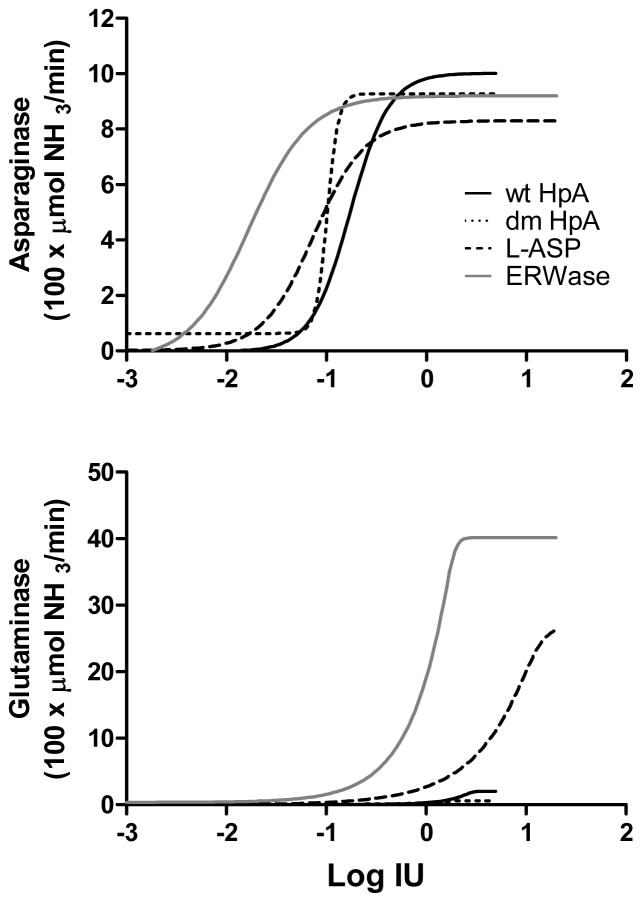
To test the dual activity of each ASNase isoform simultaneously, asparaginase and glutaminase activities were determined as described in Material and Methods. Under our assay conditions, asparaginase activity reached a plateau for all isoforms between 0.1 and 1 IU (Fig 1A), and was not significantly different between ASNase isoforms. Glutaminase activity also plateaued, but at higher enzyme concentrations (Fig 1B). Glutaminase activity of ERWase was higher than for L-ASP, whereas for wt HpA it was small compared to other isoforms, and for dm HpA it was close to background activity even at 5 IU. This is summarized in Table 2 where asparaginase and glutaminase activities are expressed as a percentage of ERWase (arbitrarily fixed at 100%) at a dose of 3 IU. Glutaminase activity showed the following ranking order: ERWase > L-ASP > wt HpA > dm HpA.
Figure 1.
Dose response fit of asparaginase activity (A) and glutaminase activity (B) of four ASNases (n =2). The X-axis represents the Log IU for each isoforms calculated from 10 to 12 different concentrations ranging from 0.001 to 20 IU. Graphs shown are a non-linear regression 5-parameter model.
Table 2.
Activity ratio of ASNases to ERWase
| % ERWase activity at 3 IU | dm HpA | wt HpA | L-ASP | ERWase |
|---|---|---|---|---|
| Asparaginase | 101% | 109% | 90% | 100% |
| Glutaminase | 1.4% | 4.0% | 19% | 100% |
The asparaginase and glutaminase activities were calculated from Fig 1. Data are expressed as percentage of ERWase interpolated at 3 IU with ERWase arbitrarily set at 100% for both relative asparaginase and relative glutaminase activity.
Cytotoxicity of ASNases on human leukemia cell lines
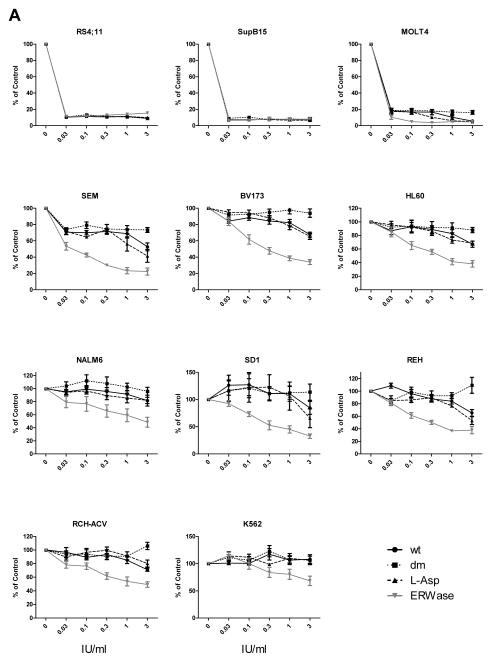
L-Asparaginases ranging from 0.01 to 3 IU/ml were tested on 11 human leukemia cell lines (Table 1, Fig 2A). Two asparagine-dependent B-ALL cell lines 25, SupB15 and RS4;11, were equally sensitive to all four ASNase isoforms, including the asparaginase-only dm HpA. The T lymphoblast cell line MOLT-4, which was previously shown to be sensitive to asparagine depletion 25, showed inhibition of cell growth to all 4 isoforms, though dm HpA was slightly less cytotoxic than the other 3 isoforms (p<0.05 at 3 IU/ml). All other cell lines were more sensitive to isoforms with higher glutaminase activities, with several showing little or no sensitivity to dm HpA. ERWase with the highest glutaminase activity was the most cytotoxic overall. wt HpA and L-ASP showed a similar pattern for all 11 cell lines tested. Interestingly there was a divergence in cytotoxicity between wt and dm HpAses, particularly at the highest dose of 3 IU/ml, in MOLT-4, SEM, BV173, REH and RCH-ACV (Table 3). This is supported by the determination of IC50 for each cell line, which differentiated the leukemia cell lines into two distinct groups (Table 4). The first group were asparaginase-sensitive (Sup-B15, RS4;11 and MOLT-4), with equivalent and very low IC50 for all ASNase isoforms. The remaining eight cell lines were overall more resistant to ASNases and showed differential sensitivities to the four isoforms. All eight cell lines in this latter group showed some sensitivity to ERWase, and little to no sensitivity to dm HpA.
Figure 2.
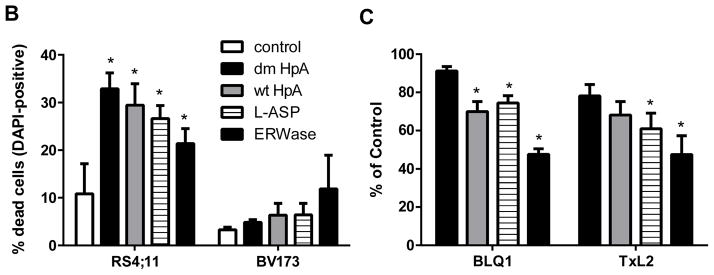
Cytotoxic efficacy of ASNases on human leukemia cells. A, Cells were cultured for 72 hours with a range of 0.03 IU/ml to 3 IU/ml ASNase isoforms. After 72 hours, cell viability was determined in triplicate by trypan blue exclusion (n = 4 or 5 per cell line). Percentage of live cells at 5 concentrations compared to untreated control (100%) is plotted in each graph. Statistical analysis was performed with two way ANOVA with Bonferroni post-tests for each ASNase isoforms vs. control and also dm HpA vs. other isoforms however for clarity, symbols are not indicated on the graphs. B, RS4;11 and BV173 cells were cultured for 48 hours with 3 IU/ml ASNase isoforms (n = 3). Cell death was quantified with DAPI-positive cells using flow cytometry, and expressed as a percentage of total measured cells. dm: double-mutant H. pylori, wt: wild-type H. pylori, L-Asp: E. coli, ERWase: Erwinia chrysanthemi. *p<0.05 vs. control was determined by t-test. C, Primary human leukemia cells in direct co-culture with OP9 feeder layer were incubated for 72 hours with 3 IU/ml ASNase isoforms (n = 4). Percentage of live cells compared to untreated control (100%) is indicated;. *p<0.05 vs. dm HpAse was determined by two way ANOVA with Bonferroni post-tests.
Table 3.
Percent inhibition at 3 IU/ml ASNase
| Cell line | dm HpA | wt HpA | L-ASP | ERWase |
|---|---|---|---|---|
| SupB15 | 92 | 92 | 93 | 92 |
| RS4;11 | 91 | 91 | 90 | 85 |
| MOLT-4 | 84 | 94* | 95* | 95* |
| SEM | 27 | 47** | 58** | 78** |
| BV173 | 6 | 32** | 35** | 66** |
| HL-60 | 12 | 33 | 31 | 62** |
| SD-1 | 0 | 15 | 34 | 67** |
| REH | 0 | 35** | 47** | 62** |
| RCH-ACV | 0 | 29** | 20** | 51** |
| NALM6 | 4 | 18 | 18 | 51** |
| K-562 | 0 | 0 | 0 | 32** |
The percentage of inhibition at 3 IU/ml was calculated from normalized data (Fig 2). Each ASNase isoforms was compared to untreated cells control using 2 way ANOVA with Bonferroni post-tests.
p<.05,
p<.01 vs. dm HpA; n = 4–5 for each cell line.
Table 4.
IC50 in IU/ml
| Cell line | dm HpA | wt HpA | L-ASP | ERWase |
|---|---|---|---|---|
| SupB15 | 0.0043 | 0.0033 | 0.0032 | 0.0030 |
| RS4;11 | 0.0055 | 0.0052 | 0.0054 | 0.0057 |
| MOLT-4 | 0.0108 | 0.0097 | 0.0088 | 0.0039 |
| SEM | nd | 2.05 | 1.14 | 0.07 |
| BV173 | nd | 5.21* | 4.72* | 0.34 |
| HL-60 | nd | 5.39* | 4.62* | 0.51 |
| SD-1 | nd | nd | 10.54* | 0.55 |
| REH | nd | 5.26* | 3.13 | 0.35 |
| RCH-ACV | nd | 6.67* | 11.82* | 1.13 |
| NALM6 | nd | 12.62* | 10.56* | 1.40 |
| K-562 | nd | nd | nd | 5.37* |
IC50s were calculated from normalized data (Fig 2) using non-linear regression with GraphPad Prism.
extrapolated for values > 3 IU/ml, n=4–5 for each cell line, nd: not determined >100 IU/ml.
Cytotoxicity was also assessed at 48 hours by flow cytometry in BV173 (asparagine independent) and RS4;11 (asparagine dependent) cells using DAPI (Fig 2B). All ASNases isoforms significantly increased cell death in RS4;11 cells. The percentage of cell death in BV173 cells did not reach statistical significance; however, the rank order among ASNase isoforms was relative to glutaminase activity, similar to Fig 2A for BV173 cells. Cell viability estimates performed at 48 hours with trypan blue parallel the 72 hours data (not shown).
Cytotoxicity of ASNases on primary leukemia cells
To confirm our findings in a more clinically relevant model, patient-derived leukemia cells BLQ1 and TxL2 were co-cultured with OP9 stromal cell line and incubated with a single dose of 3 IU/ml ASNase isoforms for 72 hours. The pattern of inhibition of these two cell types was similar to the majority of the cell lines, with no or little inhibition from dm HpA, slightly more cytotoxicity from wt HpA and L-ASP, and the highest cytotoxicity from ERWase (Fig 2C). The overall level of inhibition was lower than with cell line monoculture, possibly reflecting protection by the feeder layer and/or a higher intrinsic level of resistance.
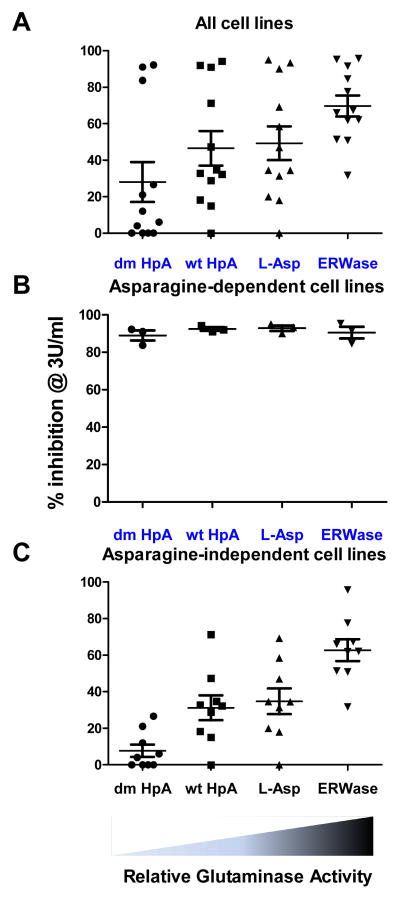
Glutaminase activity determines ASNase cytotoxic efficacy
Data from percentage inhibition at 3 IU/ml (Table 3) were plotted for each individual ASNase isoform (Fig. 3). Overall, the percentage inhibition at 3 IU/ml increased with increased relative glutaminase activity (p=0.042, R2=0.92; Fig 3A). Correlations could not be determined for asparaginase activity since all four isoforms had similar asparaginase activities. When the three asparaginase-sensitive cell lines (Sup-B15, RS4;11 and MOLT-4) were analyzed separately, the percentage inhibition was the same with all four isoforms, with no correlation with glutaminase activity as determined by Pearson’s correlation test (p=0.74, R2=0.07; Fig 3B). The remaining eight cell lines showed a very strong correlation between percentage inhibition and glutaminase activity (p=0.019, R2=0.96; Fig 3C).
Figure 3.
Glutaminase activity vs. cytotoxic efficacy. A, Each cell line data for percentage inhibition at 3 IU/ml was plotted and mean ± SD was calculated for the four ASNase isoforms (n=11 cell lines per ASNase). B, Data for 3 asparaginase-sensitive cell lines (Sup-B15, RS4;11 and MOLT-4) without other 8 cell lines are shown for % inhibition at 3 IU/ml. C, Data for 3 asparaginase-sensitive cell lines (Sup-B15, RS4;11 and MOLT-4) were excluded and the remaining 8 cell lines are shown for percentage inhibition at 3 IU/ml. Relative glutaminase activity for each isoform is indicated below graphs.
Discussion
Use of an asparaginase-only, glutaminase-inactive mutant of Helicobacter pylori ASNase allowed us to confirm the importance of glutaminase activity on ASNase cytotoxicity. Asparaginase activity alone may not be sufficient to explain ASNase cytotoxicity in many cases of ALL. ERWase, which had the highest glutaminase activity of all ASNase isoforms tested, was also systematically the most effective in preventing cell growth for all cell lines, whereas asparaginase-only dm HpA, without significant glutaminase activity, was the least effective, with measurable cytotoxicity on only three out of 11 cell lines (Sup-B15, RS4;11 and MOLT-4). These observations are supported by our data showing an increase in cytotoxic efficacy as measured by percentage inhibition and IC50 correlating with higher glutaminase activity. While some cell lines are clearly very asparagine dependent (Sup-B15, RS4;11 and MOLT-4) the majority of lines exhibited sensitivity primarily related to glutaminase activity.
The individual cell line pattern of sensitivity to asparaginase and glutaminase activities shown in this study was consistent with our previous report showing human cell lines growth in asparagine-free, glutamine-free or in asparagine- & glutamine-free media 25. Cell lines previously established to be asparagine-dependent25 were the most and equally sensitive to all four ASNases tested in this study, including asparaginase-only dm HpA isoform. Other cell lines which were sensitive primarily to glutamine depletion were sensitive to ASNase isoforms with glutaminase activities but not to asparaginase-only dm HpA isoform. Interestingly, cell lines NALM6 and K-562, previously shown to be insensitive to both asparagine and glutamine depletion, were the most resistant to all ASNases in the present study, but did show some modest sensitivity to ERWase. Thus it seems clear that ALL cell sensitivity to various ASNase isoforms is determined by the intrinsic sensitivity to the two amino acids they deplete.
Because the four ASNase isoforms we used have similar asparaginase activities, we were not able to test for synergistic effects between asparagine and glutamine depleting activities of these enzymes. Indeed, it is probable that asparagine and glutamine depletion have additive or even synergistic effects. We have previously reported in several human cell lines that depletion of asparagine yields further leukemia cytotoxicity beyond that caused by depletion of glutamine alone 25. Yet it is clear from the present study that the glutaminase activity is critical for cytotoxicity against most human ALL cell lines.
Despite the demonstrated benefit of ASNase on ALL survival, these drugs can have significant side effects. Allergic reaction and antibody formation can limit dosing and efficacy, and often lead to change in the ASNase form used. Several side effects, such as immunosuppression, pancreatitis and intestinal damage, are believed to be related primarily to glutamine starvation 20. Use of an ASNase from Acinetobacter with very high glutaminase activity (Km = 5.8 μM) led to increased neurotoxicity in clinical trial 30. Unfortunately, if both cytotoxicity and side effects are related to glutamine depletion, this makes it difficult to develop strategies to reduce side effects without impairing efficacy. For example, glutamine supplementation has been advised to provide protection to intestinal and immune cells, and to maintain acid-base balance in the kidney, for children with critical illness 31. However, in cancer patients, glutamine supplementation could promote the growth of neoplastic cells 32, particularly cancer cells of lymphoid origin, which are highly dependent on glutamine compared to other cells 14–16. Recently, ovarian cancer cell sensitivity to ASNase has also been shown to be related more closely to glutamine than to asparagine depletion 33.
While cancer cells which lack ASNS may be extremely sensitive to asparagine depletion in monoculture in vitro, Iwamoto et al. has shown that mesenchymal cells in the leukemia microenvironment can secrete asparagine, which protects these cells from ASNase 34. We have observed a similar phenomenon with adipocytes, which are also highly prevalent in the bone marrow environment 25. Since glutamine is an essential substrate of ASNS for asparagine synthesis 17, ASNase dual activity is important for cancer cell cytotoxicity 23,35. This dual activity could also reduce microenvironmental production of asparagine, thereby improving overall treatment efficacy.
In summary, our data show that ASNase isoforms with less glutaminase activity should be used with caution. Even in cases where ALL cells at diagnosis are clearly and totally asparagine-dependent, the existence of subclones with ASNS expression cannot be completely ruled out. Furthermore, B-cell evolution of different leukemic subclones during treatment (i.e. clonal shift), could promote development of ALL cells with higher ASNS expression 36. Finally, ASNS negative leukemia cells can still potentially obtain asparagine from cells in their microenvironment which do express ASNS34. Therefore, any benefit in side effect profile from using an asparaginase-only enzyme must be weighed against the potential cost in terms of decrease in efficacy and a potential danger of relapse.
Highlights.
We tested activity of a novel glutaminase-inactive asparaginase on leukemia cells
Sensitivity to different asparaginase isoforms varies between ALL cell lines
Cytotoxicity of asparaginase is determined primarily by its glutaminase activity
Acknowledgments
This work was supported by the Gabrielle’s Angel Foundation, the charitable support of the Nautica Malibu Triathlon event produced by MESP, Inc., The T.J. Martell Foundation, and the NIH/NCI (CA139060).
Footnotes
Contribution: J.H.P., V.I.A. and S.D.M. designed the experiments; C.S., M.M., and E.T. produced and provided the HpA asparaginases; J.H.P. performed the experiments, analyzed results and made the figures; J.H.P., C.S., V.I.A. and S.D.M. wrote the manuscript. All authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. Int J Nanomedicine. 2006;1(3):241–254. [PMC free article] [PubMed] [Google Scholar]
- 2.Pieters R, Hunger SP, Boos J, et al. L-asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer. 2011;117(2):238–249. doi: 10.1002/cncr.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asselin BL, Whitin JC, Coppola DJ, et al. Comparative pharmacokinetic studies of three asparaginase preparations. J Clin Oncol Off J Am Soc Clin Oncol. 1993;11(9):1780–1786. doi: 10.1200/JCO.1993.11.9.1780. [DOI] [PubMed] [Google Scholar]
- 4.Oettgen HF, Old LJ, Boyse EA, et al. Inhibition of leukemias in man by L-asparaginase. Cancer Res. 1967;27(12):2619–2631. [PubMed] [Google Scholar]
- 5.Aslanian AM, Kilberg MS. Multiple adaptive mechanisms affect asparagine synthetase substrate availability in asparaginase-resistant MOLT-4 human leukaemia cells. Biochem J. 2001;358(Pt 1):59–67. doi: 10.1042/0264-6021:3580059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, Li Z, Broome JD. Epigenetic changes in the repression and induction of asparagine synthetase in human leukemic cell lines. Leukemia. 2005;19(3):420–426. doi: 10.1038/sj.leu.2403639. [DOI] [PubMed] [Google Scholar]
- 7.Fine BM, Kaspers GJL, Ho M, Loonen AH, Boxer LM. A genome-wide view of the in vitro response to l-asparaginase in acute lymphoblastic leukemia. Cancer Res. 2005;65(1):291–299. [PubMed] [Google Scholar]
- 8.Abbatiello SE, Pan Y-X, Zhou M, et al. Mass spectrometric quantification of asparagine synthetase in circulating leukemia cells from acute lymphoblastic leukemia patients. J Proteomics. 2008;71(1):61–70. doi: 10.1016/j.jprot.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanian MN, Butterworth EA, Kilberg MS. Asparagine synthetase: regulation by cell stress and involvement in tumor biology. Am J Physiol Endocrinol Metab. 2013;304(8):E789–799. doi: 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermanova I, Zaliova M, Trka J, Starkova J. Low expression of asparagine synthetase in lymphoid blasts precludes its role in sensitivity to L-asparaginase. Exp Hematol. 2012;40(8):657–665. doi: 10.1016/j.exphem.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Su N, Pan Y-X, Zhou M, et al. Correlation between asparaginase sensitivity and asparagine synthetase protein content, but not mRNA, in acute lymphoblastic leukemia cell lines. Pediatr Blood Cancer. 2008;50(2):274–279. doi: 10.1002/pbc.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimitriou H, Choulaki C, Perdikogianni C, Stiakaki E, Kalmanti M. Expression levels of ASNS in mesenchymal stromal cells in childhood acute lymphoblastic leukemia. Int J Hematol. 2014;99(3):305–310. doi: 10.1007/s12185-014-1509-y. [DOI] [PubMed] [Google Scholar]
- 13.Appel IM, den Boer ML, Meijerink JPP, et al. Up-regulation of asparagine synthetase expression is not linked to the clinical response L-asparaginase in pediatric acute lymphoblastic leukemia. Blood. 2006;107(11):4244–4249. doi: 10.1182/blood-2005-06-2597. [DOI] [PubMed] [Google Scholar]
- 14.Kitoh T, Kubota M, Takimoto T, et al. Metabolic basis for differential glutamine requirements of human leukemia cell lines. J Cell Physiol. 1990;143(1):150–153. doi: 10.1002/jcp.1041430120. [DOI] [PubMed] [Google Scholar]
- 15.Le A, Lane AN, Hamaker M, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15(1):110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newsholme P. Why is L-glutamine metabolism important to cells of the immune system in health, postinjury, surgery or infection? J Nutr. 2001;131(9 Suppl):2515S–22S. doi: 10.1093/jn/131.9.2515S. discussion 2523S–4S. [DOI] [PubMed] [Google Scholar]
- 17.Richards NGJ, Kilberg MS. Asparagine synthetase chemotherapy. Annu Rev Biochem. 2006;75:629–654. doi: 10.1146/annurev.biochem.75.103004.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willems L, Jacque N, Jacquel A, et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood. 2013;122(20):3521–3532. doi: 10.1182/blood-2013-03-493163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samudio I, Konopleva M. Asparaginase unveils glutamine-addicted AML. Blood. 2013;122(20):3398–3400. doi: 10.1182/blood-2013-09-526392. [DOI] [PubMed] [Google Scholar]
- 20.Avramis VI, Panosyan EH. Pharmacokinetic/pharmacodynamic relationships of asparaginase formulations: the past, the present and recommendations for the future. Clin Pharmacokinet. 2005;44(4):367–393. doi: 10.2165/00003088-200544040-00003. [DOI] [PubMed] [Google Scholar]
- 21.Covini D, Tardito S, Bussolati O, et al. Expanding targets for a metabolic therapy of cancer: L-asparaginase. Recent Patents Anticancer Drug Discov. 2012;7(1):4–13. doi: 10.2174/157489212798358001. [DOI] [PubMed] [Google Scholar]
- 22.Avramis VI. Asparaginases: biochemical pharmacology and modes of drug resistance. Anticancer Res. 2012;32(7):2423–2437. [PubMed] [Google Scholar]
- 23.Reinert RB, Oberle LM, Wek SA, et al. Role of glutamine depletion in directing tissue-specific nutrient stress responses to L-asparaginase. J Biol Chem. 2006;281(42):31222–31233. doi: 10.1074/jbc.M604511200. [DOI] [PubMed] [Google Scholar]
- 24.Chan WK, Lorenzi PL, Anishkin A, et al. The glutaminase activity of L-asparaginase is not required for anticancer activity against ASNS-negative cells. Blood. 2014;123(23):3596–3606. doi: 10.1182/blood-2013-10-535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehsanipour EA, Sheng X, Behan JW, et al. Adipocytes cause leukemia cell resistance to L-asparaginase via release of glutamine. Cancer Res. 2013;73(10):2998–3006. doi: 10.1158/0008-5472.CAN-12-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fei F, Stoddart S, Groffen J, Heisterkamp N. Activity of the Aurora kinase inhibitor VX-680 against Bcr/Abl-positive acute lymphoblastic leukemias. Mol Cancer Ther. 2010;9(5):1318–1327. doi: 10.1158/1535-7163.MCT-10-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cappelletti D, Chiarelli LR, Pasquetto MV, et al. Helicobacter pyloril-asparaginase: a promising chemotherapeutic agent. Biochem Biophys Res Commun. 2008;377(4):1222–1226. doi: 10.1016/j.bbrc.2008.10.118. [DOI] [PubMed] [Google Scholar]
- 28.Maggi M, Chiarelli LR, Valentini G, Scotti C. Engineering of Helicobacter pylori L-asparaginase: characterization of two functionally distinct groups of mutants. PloS One. 2015;10(2):e0117025. doi: 10.1371/journal.pone.0117025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Distasio JA, Niederman RA, Kafkewitz D, Goodman D. Purification and characterization of L-asparaginase with anti-lymphoma activity from Vibrio succinogenes. J Biol Chem. 1976;251(22):6929–6933. [PubMed] [Google Scholar]
- 30.Warrell RP, Arlin ZA, Gee TS, et al. Clinical evaluation of succinylated Acinetobacter glutaminase-asparaginase in adult leukemia. Cancer Treat Rep. 1982;66(7):1479–1485. [PubMed] [Google Scholar]
- 31.Mok E, Hankard R. Glutamine supplementation in sick children: is it beneficial? J Nutr Metab. 2011;2011 doi: 10.1155/2011/617597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purwaha P, Lorenzi PL, Silva LP, Hawke DH, Weinstein JN. Targeted metabolomic analysis of amino acid response to L-asparaginase in adherent cells. Metabolomics. 2014:1–11. doi: 10.1007/s11306-014-0634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwamoto S, Mihara K, Downing JR, Pui C-H, Campana D. Mesenchymal cells regulate the response of acute lymphoblastic leukemia cells to asparaginase. J Clin Invest. 2007;117(4):1049–1057. doi: 10.1172/JCI30235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Offman MN, Krol M, Patel N, et al. Rational engineering of L-asparaginase reveals importance of dual activity for cancer cell toxicity. Blood. 2011;117(5):1614–1621. doi: 10.1182/blood-2010-07-298422. [DOI] [PubMed] [Google Scholar]
- 36.Zweidler-McKay PA. Clone wars: IgH subclones in preB-ALL. Blood. 2012;120(22):4280–4281. doi: 10.1182/blood-2012-09-455402. [DOI] [PubMed] [Google Scholar]