Abstract
Microscopy and micro-irradiation imaging techniques have significantly advanced our knowledge of DNA damage tolerance and the assembly of DNA repair proteins at the sites of damage. While these tools have been extensively applied to the study of nucleotide excision repair and double-strand break repair, their application to the repair of oxidatively-induced base lesions and single-strand breaks is just beginning to yield new insights. This review will focus on examining micro-irradiation techniques reported to create base lesions and single-strand breaks; these lesions are considered to be primarily addressed by proteins involved in the base excision repair (BER) pathway. By examining conditions for generating these DNA lesions and reviewing information on the assembly and dissociation of repair complexes at the induced lesion sites, we hope to promote further investigations into BER and to stimulate further development and enhancement of these techniques for the study of BER.
Keywords: DNA repair, DNA damage, base excision repair, single-strand break repair, micro-irradiation, PARP-1, XRCC1, PARP inhibitors, reactive oxygen species
Graphical Abstract
1. Introduction
Genomic DNA is attacked by a number of endogenous and exogenous agents that create base damage and both single- and double-strand breaks. These heterogeneous DNA damaged substrates require highly coordinated repair mechanisms to ensure genomic integrity and cell survival. DNA repair proteins sense damage sites, signal the recruitment of specific repair proteins, and initiate repair. Numerous biochemical and biophysical studies over the past 50 years have provided insight into the mechanism by which DNA damage is sensed and repaired; however, a temporal description of the assembly of DNA repair proteins at the sites of damage is lacking. The diversity of DNA lesions and DNA repair pathways, some with overlapping substrate specificities, has left a number of open questions about how the repair proteins assemble/disassemble to coordinate their activities to efficiently repair disparate DNA lesions.
Fluorescent microscopy has emerged as a powerful technique to visualize the response of DNA repair proteins to damage. Detection of damaged-induced DNA repair foci from globally applied agents, such as ultraviolet light (UV-A, 320-400 nm), ionizing radiation, hydrogen peroxide (H2O2), or chemotherapeutics, has been reported using immunofluorescent detection or fluorescently-tagged proteins of interest. These techniques have identified key players in DNA repair pathways, and the responses of repair proteins after damage to be quantified. While these techniques have provided detailed information about the kinetics of recruitment or the order of assembly for nucleotide excision repair (NER) and double strand break (DSB) repair (1, 2), information is still lacking for single strand break (SSB) and base excision repair (BER) pathways. Also, the global nature of the damaging agent does not allow for a synchronized start to damage initiation or for cellular compartment specific applications.
Fortunately, the past 20 years has seen significant advancement of laser micro-irradiation techniques. Since 1993 when Limoli and Ward demonstrated that UV light can induce strand breaks on bromodeoxyuridine (BrdU)-sensitized DNA (3), a number of groups have leveraged this technique and expanded the use of microlaser technology to examine the recruitment, kinetics, and specificity of DNA repair proteins at sites of laser-induced DNA damage (4-9). The coupling of laser scanning confocal microscopes and damage-inducing laser wavelengths has created improved tools for inducing base lesions and strand breaks, while allowing the immediate visualization of the repair complex response (4, 6, 9).
The fields most impacted by the advances in micro-irradiation have been NER and DSB repair (4-7, 10, 11). However, a growing number of studies have been examining the response of DNA repair factors to the sites of SSBs and oxidatively-induced base lesions (12-27). These types of laser-induced DNA damage allow the response of BER proteins to be examined.
BER is primarily responsible for the repair of endogenously or exogenously damaged DNA bases, such as alkylated or oxidized bases. The large number of substrates recognized by BER requires a diverse set of proteins that are responsible for the recognition, removal, and repair of base lesions. A number of DNA glycosylases are involved in damage recognition, as are scaffold proteins, like X-ray repair cross-complementing protein 1 (XRCC1) and poly(ADP-ribose) polymerase 1 (PARP-1), and end-tailoring enzymes like AP-endonuclease (APE1), polynucleotide kinase phosphatase (PNKP) (28, 29) and tyrosyl-DNA phosphodiesterase 1 (TDP1). BER is also responsible for repairing SSBs (30), and a number of distinct sub-pathways have been described (e.g., single-nucleotide and long-patch BER) (29-31).
The diversity and complexity of BER make it a rich target for micro-irradiation studies; however, a major limitation to the application of micro-irradiation to the study of BER has been the induction of specific DNA lesions for the study of specific sub-pathways. Recently, a more concerted effort has been focused on examining the dynamics of DNA repair proteins involved in BER. A number of interesting findings have been reported about the recruitment kinetics and persistence of BER repair proteins at sites of DNA damage, and several reports have indicated the possibility of novel sub-pathways that are independent of key scaffold proteins, like XRCC1 or PARP-1 (12, 16, 19, 32).
These studies offer intriguing insights into the complexity of the BER pathway; however, a fundamental limitation to the general acceptance of these results is that the micro-irradiation conditions differ widely in the various reports, and detailed information about the generation and characterization of the induced lesions is often not adequately described. Nevertheless, comparison of a number of published studies reveals some consistencies, despite the use of different techniques to induce DNA damage. This consistency argues that further investigation of BER by micro-irradiation, with improved standardization in reporting of the generation and characterization of damaging conditions, could significantly advance the detailed mechanistic understanding of the BER pathway and its response to SSBs and oxidatively-induced base lesions. A wide variety of techniques have been reported to generate these types of lesions, though only a handful of papers have investigated the behaviors of the BER proteins. To help promote further investigation, we have specifically reviewed the application of micro-irradiation techniques to the generation of SSBs and base lesions.
2. UV wavelengths to induce of DNA damage
A variety of UVA wavelengths have been utilized to create strand breaks (4, 6). Most reports of UVA applications are in the fields of NER (6, 11) and DSB repair (4, 6, 9), though some studies have utilized these wavelengths to examine SSBs and base lesions.
2.1 337 nm
Application of nanosecond (ns) 337 nm pulses at 0.04 μJ have been reported to recruit PARP-1, XRCC1, and flap endonuclease 1 (FEN1) to sites of damage (6). These proteins were detected by immunofluorescence, so no kinetic information about the recruitment was obtained. Analysis of the DNA damage caused by this wavelength revealed the production of 8-oxo-2′-deoxyguanosine (8-oxodG) lesions at sites of damage, along with 6,4-photoproducts (6,4PPs) and cyclobutane pyrimidine dimers (CPD) (6).
2.2 351 nm
Micro-irradiation by 351 nm (0.44 J/m2) has also been reported to induce oxidative DNA lesions (33). No characterization of the induced damage was presented in this report beyond a substantial induction of phosphorylated gammaH2AX (γH2AX), and the authors acknowledge this wavelength results in a mixture of base lesions, SSBs, and DSBs. Recruitment of RNA-binding protein fused-in-sarcoma/translocated-in-sarcoma (FUS/TLS) was shown to be PARP-1 dependent (33). However, the authors claim that this recruitment was specific to oxidatively-induced DNA lesions was not thoroughly demonstrated.
2.3 364/365 nm
Using a longer wavelength (365 nm) generates SSBs and simple base lesions without the accompanying generation of UV photoproducts thereby allowing analysis of BER or SSB repair proteins (19). Lan et al. (19) utilized a ns pulsed 365 nm laser to create a ~1 μm damage site in a variety of cell lines. Protein recruitment was visualized by immunofluorescent detection at specific time points post-damage and by real-time imaging of the recruitment of green fluorescence protein (GFP)-tagged DNA repair proteins. Recruitment of XRCC1, DNA polymerase β (Pol β), DNA Ligase III (LIGIII), proliferating cell nuclear antigen (PCNA), and the p150 subunit of chromatin assembly factor I (CAF1-p150) were observed at 0.19 μJ peak pulse power, and poly(ADP-ribose) (PAR) synthesis by PARP-1 was also detected. Recruitment kinetics for XRCC1, Pol β, and LIGIII were consistent with their recruitment to SSBs, as signaled by PARP-1 (19, 34). The PARP inhibitor 1,5- dihydroxyisoquinoline (DIQ) significantly reduced the recruit of XRCC1, Pol β, and LIGIII to the DNA damage sites. GFP-tagged XRCC1 persisted at damage sites for more than an hour, while Pol β and LIGIII dissociated within 30 min (19).
Application of greater laser power, 0.49 – 1 μJ, resulted in the production of oxidative damage (19). The production of oxidatively-induced DNA damage at these higher laser powers was confirmed by immunofluorescent staining with an 8-oxodG specific antibody and by the recruitment of GFP-tagged oxidative damage DNA glycosylases, endonuclease VIII-like 1 (NEIL1), endonuclease VIII-like 2 (NEIL2), endonuclease III-like protein 1 (NTH1), and 8-oxoguanine glycosylase 1 (OGG1) (19). This higher laser power also resulted in the recruitment of DSB repair proteins, such as Nijmegen breakage syndrome 1 (NBS1), breast cancer gene 1 (BRCA1), and RAD52, although no data to further analyze this recruitment was provided (19).
Utilization of two laser energies (0.19 and 0.49 μJ) allowed Lan et al. to examine different sub-pathways involved in BER and SSB repair. SSBs generated by low laser energy were repaired by PARP-1-dependent recruitment of XRCC1, Pol β, and LIGIII (19), consistent with previous reports (34). Recruitment of PCNA and CAF1 was less robust under these conditions and proceeded more slowly than that for Pol β and LIGIII, consistent with PCNA and CAF1 involvement in later stages of long-patch BER and chromatin assembly at the end of SSB repair, respectively (19). Inhibition of PARP-1 by DIQ mildly suppressed the recruitment of PCNA and CAF1-p150 to sites of DNA damage under low laser energy, suggesting independence from PARP-1 activation (19). Finally, while loss of XRCC1 significantly reduced recruitment of PCNA to DNA damage sites, loss of Pol β did not, suggesting a PCNA-dependent long-patch repair pathway utilizing replicative DNA polymerases δ and/or ε (19, 35).
Another 365 nm pulsed laser system, firing ns pulses at 0.7 nW, was reported by Muniandy et al. for the study of repair of interstrand cross-links (ICLs) generated by the addition of psoralen (36). Della-Maria et al. (14) utilized this system, without the addition of psoralen, to study the recruitment of XRCC1 and PNKP to the sites of laser-induced damage. Without psoralen to create cross-linked adducts, the authors proposed that this system generated free radical-induced SSBs in the irradiated region with no γH2AX foci detected (14). Recruitment kinetics of enhanced yellow fluorescent protein (YFP) fused XRCC1 showed a longer time to peak recruitment (~18 min) and retention of XRCC1 at the damage site for more than an hour (14). While the retention appears consistent with a previous report (19), the slow accumulation of XRCC1 at the damage site is not. A mutation in XRCC1 (A482T) that alters phosphorylation by casein kinase II and weakens affinity for PNKP was examined as well, and recruitment of this mutant peaked at ~4 min, more consistent with previous work, and was released from the damage site more rapidly (~25 min) (14). Recruitment of PNKP to DNA damage sites was found to require XRCC1, and no defect in the recruitment of PNKP was observed with the A482T mutant; the retention time of PNKP at the damage site was also reduced (~7 min to ~4 min (14)). Though the presence of oxidatively-induced DNA damage and UV photoproducts were not tested in this study, unique kinetics of SSB repair were obtained using this methodology.
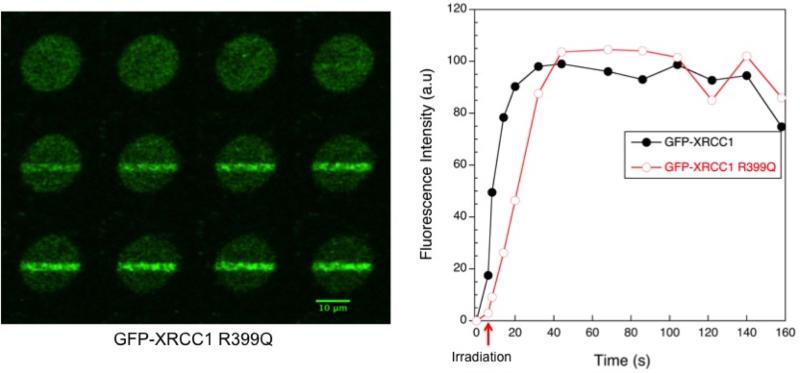
Our laboratory modified the initial conditions reported by Lan et al. (19) to include the sensitizer, BrdU. Using a continuous wave 364 nm laser and an applied energy of 0.17 μJ/pixel, we were able to monitor recruitment of XRCC1 and Pol β to sites of damage in mouse embryonic fibroblasts (MEFs), demonstrated in Figure 1 and 2 (26). We were also able to monitor the activation of PARP-1 at the damage sites by immunofluorescence detection of PAR. Consistent with Lan et al. (19), no recruitment of DSB proteins was observed at this energy, and it is expected that oxidatively-induced DNA lesions are generated by this exposure, though specific recruitment of glycosylases that remove oxidatively-induced base lesions was not observed.
Figure 1. Recruitment of GFP-XRCC1 and GFP-XRCC1 R399Q to DNA damage sites induced by 365 nm laser irradiation.
Cell deficient in XRCC1 (82) were transfected with GFP-XRCC1 or GFP-XRCC1 R399Q and sensitized with 10 μM BrdU for 24 h. Cells were irradiated with a 365 nm laser at 0.17 μJ/pixel. A single experiment is presented, though the recruitment trend observed and the images presented are representative of many experiments [43].
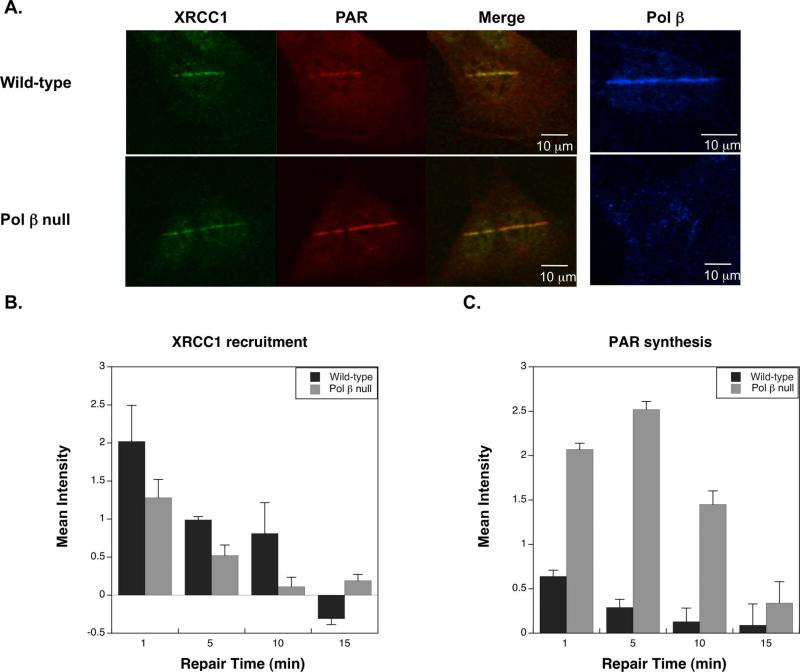
Figure 2. Recruitment of BER proteins to the sites of induced DNA damage by 365 nm laser irradiation (26).
A. Wild-type and Pol β null cells one min after irradiation. B. Time course of XRCC1 recruitment to the site of damage. C. PAR synthesis at the site of induced damage (error bars reflect SEM). Recruitment of XRCC1 is significantly different between the two cell lines at the 5 min time point (p < 0.05, Student's t-test), and the PAR levels are significantly different for the 1, 5, and 10 min time points (p < 0.01, Student's t-test).
Using these conditions in a number of BER repair proficient and deficient cell lines, we have been able to quantify the levels of XRCC1 and Pol β recruited to induced damage sites and the amount of PAR produced. We have also observed characteristic differences in these values based on the BER capacity of the cells (17, 18, 21, 26, 27, 37). Wild-type cells typically have rapid recruitment of XRCC1 and Pol β to sites of damage and rapid production of PAR by PARP-1 (peaking within 1 min). Time courses of recruitment show that XRCC1 and Pol β rapidly dissociate with only a small amount of XRCC1 or Pol β persisting at the damage site after 10 min, and the PAR response observed is also quickly resolved, as the repair of the induced-damage is conducted (Figure 2). Pol β deficient cells show a hyperactivation of PARP-1 over an extended time. This had been previously observed using other techniques (26, 38), along with a reduced recruitment of XRCC1 to the DNA damage site (26). XRCC1 deficiency also resulted in the hyperactivation of PARP-1, though not to the extent observed in Pol β deficient cells (18). Mutation of mouse XRCC1 at Val88 impairs the ability of XRCC1 to bind Pol β. Stably-transfected cell lines expressing XRCC1 V88R showed a characteristic increase in PAR, consistent with the XRCC1 deficient cell lines, and no recruitment of Pol β to the site of DNA damage (18). Interestingly, this mutant also showed reduced recruitment to the site of DNA damage, indicating an alteration in kinetics due to loss of the interaction with Pol β.
Examination of cells deficient in high mobility group N1 (HMGN1), a protein that affects the interaction of DNA repair factors with chromatin and controls their access to DNA damage, revealed that loss of HMGN1 reduced production of PAR by PARP-1 after laser-induced damage compared to wild-type cells (27). In addition to the reduction in PAR production, recruitment XRCC1 was also reduced compared to wild-type in HMGN1-deficient cells. This was consistent with previous reports that showed a reduction in XRCC1 recruitment after inhibition of PARP-1 activity by the PARP-1 inhibitor DIQ (19). These results are consistent with HMGN1 playing a regulatory role in PARP-1 activation.
Taken together, micro-irradiation by a 364/365 nm laser can create SSBs, oxidatively-induced DNA base damage, and DSBs in a dose-dependent manner, allowing a comprehensive picture of recruitment and kinetics to be assembled using immunofluorescent staining or live cell imaging of fluorescently-tagged proteins. An important caveat is that careful evaluation of applied energy levels is essential to ensure that the desired DNA damage mixtures are being created and that undesirable photoproducts (e.g., DSBs) are being avoided. Despite the promise of this powerful tool for the examination of SSB and BER proteins at the sites of DNA damage, there are some drawbacks to this approach that should be noted.
2.4 Caveats for the use of UV lasers to induce DNA damage
One major limitation to the application of the technique is the requirement for specialized lasers. Several manufacturers offer 351 nm, 355 nm or 365 nm lasers, but the requirements for UV transmissive filters and objectives still make this a specialized laser system, not accessible to a large number of researchers. Another consideration is the use of photosensitizers, like halogenated nucleotide analogues (e.g., BrdU) or DNA binding dyes (e.g., Hoechst), to lower the energy required to induce DNA strand breaks. Another consideration is that these agents can cause undesirable effects in chromatin structure that could alter the kinetics of DNA damage response, and may have additional damaging effects during long term imaging with visible light (5, 6, 11, 39). These effects are most often observed in the kinetics of DSB repair followed by live cell imaging, yet they are important to consider when evaluating the application of a laser technique.
Additionally, there are inconsistencies in the kinetics observed by this technique. As described above, our laboratory observed the rapid recruitment of XRCC1 and Pol β to sites of damage using immunofluorescent staining of MEFs, and this rapid recruitment was also seen in the other reports using immunofluorescent staining of HeLa cells (6, 19). However, our UV conditions also showed a rapid dissociation of XRCC1 and Pol β with only a small population of either protein persisting after 10 min (Figure 2). This result differs significantly from reports where fluorescently-tagged XRCC1 persisted at damage sites in HeLa and EM-9 cells for more than an hour (14, 19). Immunofluorescent staining for LIGIII after pulsed 365 nm damage showed a rapid recruitment and a time-dependent dissociation with the majority of the protein released after 20 min (19). The same conditions with GFP-tagged LIGIII showed a rapid recruitment, but significantly slower release of the GFP-LIGIII, although the authors do not seem to think this difference was significant (19). Recruitment kinetics of XRCC1 after uranium ion irradiation are consistent with the rapid recruitment and dissociation of XRCC1, with the majority of XRCC1 released by 10 min (40).
There are a number of issues in trying to compare immunofluorescent staining trends with direct imaging results of fluorescently-tagged proteins; however, the different results between these two methods are intriguing and argue for a more comprehensive study using fluorescent recovery after photobleaching.
3. DNA damage induced with visible wavelengths
3.1 405 nm with sensitizers
A more widely accessibly application of micro-irradiation utilizes visible laser wavelengths with laser scanning confocal imaging to create DNA damage and the recruitment of repair factors is monitored. Several reports have followed SSB and BER proteins after 405 nm micro-irradiation employing sensitizers like BrdU or Hoechst (12, 13, 15, 22-24, 41, 42). Using this technique, the recruitment of fluorescently-tagged PARP-1 (13, 15, 41), poly(ADP-ribose) glycohydrolase (PARG) (23), PCNA (13, 22-24), XRCC1 (12, 13, 15, 22, 24, 41), and DNA ligases I and III (24) to sites of induced-damage have been examined. While variations in the applied methodologies exist, most reports utilize a 405 nm diode laser set at 50-100% transmission to irradiate a ~1 μm nuclear spot through a 63x oil immersion objective. Under these conditions, γH2AX and PAR have been observed by immunofluorescence, indicating that a mixture of SSBs and DSBs are being generated by the laser irradiation (24).
Most studies utilizing these conditions have focused on the generation and repair of SSBs, and the recruitment dynamics for a number of proteins have been reported. Two interesting findings have come out of these reports. First, the kinetics of PARP-1 and XRCC1 recruitment are mediated by PARP-1 catalytic activity. GFP-PARP-1 was found to rapidly accumulate at sites of DNA damage (<1 min) and then undergo gradual release (15, 41), with most of PARP-1 dissociating by 30 min (15). Inhibition of PARP-1 with PARP inhibitors or through mutation of the catalytic domain resulted in a delay in the accumulation of PARP-1 at damaged sites and caused persistence of PARP-1 at these sites with maximum intensity persisting to 30 min (15, 41). In addition to altered PARP-1 recruitment, XRCC1 is not recruited to DNA damage sites in the absence of PARP-1 protein (parp1−/− MEFs (41) or in the presence of PARP inhibitors (15, 41); this was consistent with a report utilizing 365 nm irradiation (19). Second, as discussed above, SSB repair by BER proteins can be separated into single-nucleotide and long-patch sub-pathways (35). Single-nucleotide BER is typically carried out by XRCC1, Pol β, and LIGIII or DNA ligase I (LIGI), while long-patch BER can involve PCNA. PCNA stimulates the activity of FEN1 to remove the DNA flaps generated during long-patch DNA synthesis and may load replicative DNA polymerases δ or ε for gap-filling DNA synthesis, and also recruits LIGI for the final ligation step.
Recruitment of both PARP-1 and PCNA driven sub-pathways of BER was also observed using the 405 nm conditions described. A slow, constant recruitment of LIGI, identical to PCNA recruitment, was observed after DNA damage. PARP-1, XRCC1 and LIGIII all showed rapid recruitment to sites of damage with a maximal accumulation at between 1-2 min, followed by dissociation over the next 10-30 min (15, 22, 24). Inactivation of the single-nucleotide sub-pathway by inhibition of PARP-1 did not eliminate SSB repair by the PCNA-driven long-patch BER pathway (15, 19).
Induction of oxidative damage by 405 nm sensitized by Hoechst dye (60% laser power for 30 iterations) was reported recently (12). A significant amount of oxidatively-induced DNA damage was detected by immunostaining for 8-oxodG and validated by the recruitment of GFP-OGG1 to sites of laser-induced DNA damage (12). Induction of oxidative damage was not described in the previous reports discussed above.
In addition to oxidative damage, irradiation with 405 nm, coupled with Hoechst, has been shown to generate CPDs, but not the severely helix distorting 6-4 photoproducts (11). This effect is thought to occur through modulation of the DNA and chromatin structure by the intercalating dye (6, 11). Again, like the oxidatively-induced DNA base lesions seen after irradiation with 405 nm, the presence of CPDs or the response of NER proteins was not reported.
3.2 405 nm without sensitizers
Using 405 nm irradiation without sensitization at a low dose (~0.4 - 2.5 μJ/pixel) and a high dose (~5 - 23 μJ/pixel), Hanssen-Bauer et al. found recruitment of PCNA and FEN1 to sites of DNA damage at energies 10-15 times higher than those required for the recruitment of Pol β and PNKP (16). Application of PARP inhibitors did not affect the recruitment of PCNA, as previously reported (15), or FEN1. In this report, application of PARP inhibitors (4- amino-1,8-naphthalimide (4-AN) and PJ-34) only slightly altered the recruitment of XRCC1. This finding is different than those that employ the 405 nm laser with sensitization.
Another interesting finding was the identification of two distinct recruitment kinetic features for PARP-1 observed with two different PARP inhibitors, 4-AN and PJ-34. 4-AN resulted in increased accumulation of PARP-1, consistent with previous reports using the same inhibitor (15) and NU1025 (41); although the kinetics of accumulation were not reported and the persistence at DNA damage site was not confirmed. In contrast, application of PJ-34 reduced the accumulation of PARP-1 at sites of DNA damage, and equivalent accumulation of PARP-1 was not achieved in the presence of this inhibitor until 15 times more energy was applied. Since both inhibitors block catalytic activity of PARP, the authors attributed this difference in inhibitor action to alterations in the affinity of PARP-1 for DNA (16); however, other instances of this phenomenon with PJ-34 cannot be found.
In a separate study, XRCC1 recruitment to DNA damage sites was also examined. This study determined that the BRCA1 C-terminus 1 (BRCT1) domain of XRCC1 was essential for recruitment to damage sites and mediated the DNA repair functions of the scaffold protein (43). Additionally, the recruitment of three common single nucleotide polymorphisms in XRCC1 (R196W, R280H, and R399Q) was examined. No effect in the recruitment to damaged sites or in the co-recruitment of Pol β and PNKP was observed for these variants was noted, though the retention of R280H and R399Q at damaged sites was decreased compared to wild-type (43).
In addition to these studies, it was reported that a single 1.6 μW scan from a 405 nm pulse laser without sensitizers could induce SSBs without significant detection of DSBs (44). Increasing the power to 16-800 μW produced a mixture of DSBs and oxidatively-induced base lesions in addition to SSBs at the irradiated sites; no recruitment of BER factors was reported.
3.3 435 nm
Application of other visible wavelengths to the study of SSBs or oxidatively-induced DNA damage has been less common. Irradiation by 435 nm was reported by Berquist et al. to examine the recruitment of wild-type and mutant XRCC1 to sites of DNA damage (45). Using ~10 nW of laser power, SSBs were generated with no observation of γH2AX foci at the damage site indicating the lack of DSBs. The authors report specific generation of SSBs at this wavelength, though controls with other DNA damage markers were not reported. In this study recruitment of YFP-tagged wild-type XRCC1 and XRCC1 containing point mutations that interrupt critical partner protein interactions or alter protein stability (V86R, Pol β interaction; E98K, DNA binding; R109A, Pol β interaction and DNA binding; C398Y, protein stability) to sites of 435 nm induced DNA damage were measured (45). Additionally, the recruitment of YFP-tagged XRCC1 single-nucleotide polymorphisms (SNPs) (E98K, P161L, R194W, R280H, C389Y, R399Q, and Y576S) were also examined (45). All of the mutant proteins, except E98K and C398Y, showed rapid wild-type like recruitment to the site of DNA damage (within 10-15 s). E98K displayed abnormal localization to the nucleolus, which was thought to contribute to its lack of recruitment, and the C398Y mutation affected protein stability, which was reflected in a lower expression pattern in cells and lack of recruitment (45). The C398Y mutation could also reduce affinity for PARP-1, which could also explain its recruitment defect (45). In addition to recruitment, retention of XRCC1 at sites of DNA damage was also examined. Again, most of the mutant proteins showed a slow, wild-type like release from sites of damage over the course of 45-60 min (45), similar to the results reported by Lan et al. (19). The exception was R280H, which was released more quickly from the damaged site, within 15-30 min (45). This finding is consistent with kinetics reported by 405 nm-induced damage, although XRCC1 R399Q was also observed to dissociate more rapidly (43).
3.4 488 nm
A recent report by Solarczyk et al. utilized 488 nm visible light to induce DNA damage (46). With an applied laser energy of 17 mJ, recruitment of XRCC1 and LIGIII were seen at sites of induced DNA damage, as were phosphorylated ataxia telangiectasia-mutated gene (ATM), γH2AX, and replication protein A (RPA), indicating that 488 nm light created a mixture of SSBs and DSBs. The authors also evaluated the presence of oxidative DNA damage by immunostaining for 8-oxodG; no detectable levels of oxidative damage were observed by the 488 nm laser light; similarly, UV photoproducts were not detected. These findings are consistent with previous reports that UV photoproducts and oxidative DNA damage are produced at wavelengths less than 450 nm (47-49).
3.5 514 nm with sensitizer
Recruitment kinetics of the heterochromatin protein 1 (HP1) to sites of oxidative DNA damage were reported by Zarebski et al. using 514 nm irradiation of ethidium bromide (EtBr) sensitized cells (50). The presence of oxidatively-induced lesions and strand breaks was confirmed by immunofluorescent staining of 8-oxodG, XRCC1, and γH2AX. 8-oxodG was observed 1 min after the induction of damage, XRCC1 was visualized 30 min after damage, and γH2AX was observed from 15 min to 1.5 h after damage (50). The focus of the study was recruitment kinetics of the HP1 protein, and GFP-HP1 was shown to slowly accumulate over ~ 30 min following damage.
3.6 Caveats for the use of visible wavelengths for the induction of DNA damage
While visible laser wavelengths are more accessible to researchers because they are standard on most confocal microscopes, specific applications of these techniques to the study of SSB and BER proteins can be challenging due the creation of complex DNA lesion heterogeneity. More studies using 435, 488, and 514 nm would be required to truly assess their applicability for the study of SSBs and simple base lesions. The compatibility of these wavelengths, especially 488 and 514 nm, to most commercial confocal systems makes them attractive alternatives to 405 nm, that has been extensively utilized but with significant issues. As noted above, two contradictory studies about the influence of PARP-1 inhibitors on XRCC1 recruitment were reported with 405 nm micro-irradiation (16, 22). Since the type of DNA damage induced by 405 nm with and without sensitization was not fully characterized, it is unclear what the differences in the recruitment of XRCC1 in the presence of PARP-1 inhibitors could mean regarding PARP-1 independent sub-pathways for repair. However, it is clear that micro-irradiation with 405 nm can reproducibly create SSBs. Similar to reports with UV irradiation, there are differences in the reported DNA lesion dissociation kinetics of XRCC1, but most reports using 405 nm irradiation show rapid accumulation and dissociation of XRCC1 within 10-20 min of damage induction (13, 15, 22, 24, 41). Validation of the types of DNA damage created with 405 nm irradiation, and other visible wavelengths, including the presence of UV photoproducts and oxidatively-induced base lesions would make this specific approach more attractive.
4. Near Infrared (NIR) wavelengths induces SSBs and oxidatively-induced DNA lesions
Multiphoton excitation by a pulsed NIR laser has several advantages over the UV and visible damaging schemes. Nonlinear absorption by two or three lower energy photons results in higher energy deposition in a femtoliter confined volume, without the requirement of a confocal microscope (51, 52), thereby providing highly localized excitation. Multiphoton excitation also offers an improved signal to noise ratio and a reduction in photobleaching and photodamage. A number of multiphoton applications for DNA repair have been reviewed (5-8), and depending on the wavelength used to generate damage, a mixture of DNA lesions (base lesions, SSBs, UV photoproducts, and DSBs) can be created (6, 11, 53-55). It has also been established that NIR lasers used at powers above 7 mW generate reactive oxygen species (ROS) that produce oxidatively-induced base damage similar to that observed with H2O2 treatment of cells (56, 57).
Though it has proven difficult to precisely assay the lesion composition produced by multiphoton applications (39, 58, 59), a mixture of damage would allow the repair of a number of pathways to be examined, though the most frequently reported applications are for DSB repair (7, 10). Three specific applications of multiphoton excitation to BER and SSB repair are reviewed here, but several studies using wavelength from 775-1050 nm have reported recruitment of XRCC1 to sites of damage, although this was most often utilized as a marker of SSBs (57, 60).
4.1 750 nm with sensitization
A specific application of multiphoton excitation to examine the recruitment kinetics for SSB repair was reported by Abdou et al. (12). DNA damage was induced into Hoechst sensitized cells by a 750 nm NIR titanium sapphire laser and SSBs were introduced in a ~1 μm nuclear spot with 10 iterations of 10% laser power. GFP-XRCC1 was rapidly recruited to sites of induced DNA damage, while OGG1 was weakly recruited. The authors reported that these conditions allowed the separation of SSB repair from oxidative DNA damage repair by the BER machinery. Rapid accumulation of GFP-tagged PARP-1, XRCC1, LIGIII and PNKP were observed after damage. Rapid dissociation of complexes was not observed (>100 s), and these results seem consistent with the reports of retention of DNA repair factors at sites of damage for 5-10 min. Interestingly, loss of PARP-1 protein or inhibition of catalytic activity by PARP-1 inhibitors did not significantly affect the recruitment of XRCC1, LIGIII and PNKP to sites of damage, though a brief delay in accumulation was observed (12).
The authors identified LIGIII as the PARP-independent damage sensor that recruits XRCC1 and PNKP to sites of laser-damage. Partial loss of LIGIII reduced recruitment of XRCC1 and PNKP to sites of damage, and the role of LIGIII was further supported by the identification of the N-terminal zinc finger domain of LIGIII being required for damage sensing, and not the XRCC1 interaction (BRCT) domain. The identification of this PARP-independent sub-pathway may have interesting implications for the DNA damage response in cells with defects in SSB repair, and the observed recruitment of XRCC1 and PNKP after loss of PARP-1. The partial loss of LIGIII may indicate that another sub-pathway can accomplish SSB repair in these cells.
4.2 768 nm
In another application of multiphoton excitation that exploited the induction of ROS, Zielinska et al. examine recruitment of the BER enzyme OGG1 to sites of induced damage (25). Using a 768 nm NIR titanium sapphire laser at 10 mW for 250 ms, GFP-OGG1 was recruited to sites of laser-induced damage. Rapid accumulation of the protein was observed (<2 min) and dissociation was reported to occur within ~20 min (25), though this is a longer retention than was observed at 365 nm (19). Examination of a mutant, GFP-OGG1 (S326C), that has reduced 8-oxodG repair rates, revealed similar accumulation rates to wild-type, but much slower dissociation rates, with ~ 80% of the protein still retained at the site of damage 15 min after damage (25). Examination of other DNA repair proteins was not conducted in this study, and the DNA damage induced by this method has not been fully characterized, although the observed recruitment OGG1 is consistent with the 365 nm micro-irradiation studies (19).
4.3 800 nm
The recruitment of FEN1 to sites of laser-induced damage was examined by micro-irradiation of a ~2 μm spot with an 800 nm laser set at 10% power (61). Multiphoton excitation with 800 nm has been described to create UV photoproducts and DSBs at high power (11, 53, 55), so the contribution of other lesion types cannot be excluded even at this longer wavelength and lower power. The authors minimized the contributions of other pathways by excluding cells with replicative foci and avoiding nucleoli (61). They also assumed the number of DSBs was too low to impact FEN1 accumulation (61). Using these conditions, rapid recruitment of FEN1-YFP was observed after irradiation. FEN1 recruitment reached a maximum at 2-3 min, and FEN1 dissociation occurred over 15-20 min, consistent with reports on SSB repair kinetics (24, 25, 45, 62, 63). Accumulation of mCherry-PCNA and FEN1-YFP were also observed and this was consistent with long-patch repair of SSBs (61), as discussed in the previous sections (15, 22, 24).
4.4 Caveats for the use of NIR wavelengths for the induction of DNA damage
Multiphoton excitation for micro-irradiation offers a number of advantages over UV and visible wavelength; however, as with other techniques it also suffers from drawbacks. Two and three photon excitation lasers are not standard on most confocal systems and often require tuneable lasers that need to be coupled to microscopes and kept in good alignment. The induced DNA damage by nonlinear absorption appears to have a number of advantages in tight confinement, but the high power required to induce damage creates ROS as a by-product, and DNA damage heterogeneity results from this excitation as well.
Although this technique is most commonly utilized for DSB repair, these reports show it provides insight into SSB repair and BER; however, the DNA damage generated needs to be carefully characterized. The observation of a PARP-1 independent function of XRCC1 in SSB repair (12) is highly dependent on oxidatively-induced DNA damage not being created by the multiphoton excitation. Recruitment of XRCC1 independent of PARP-1 activity has been previously reported for oxidatively-induced DNA damage (discussed below) (32), but this finding is inconsistent with a number of other reports (15, 19, 41, 64).
Further, Kleppa et al. reported a reduction in accumulation of FEN1 at damage sites in the presence of PARP-1 inhibitors DPQ and NU1025 (61). This reduction in FEN1 recruitment contrasts with previous reports where FEN1 plays a role in long-patch repair of SSBs that is PARP-1 independent (15, 16). Different behaviors by PARP-1 inhibitors, 4-AN and PJ-34, were noted by Hanssen et al. (16), and since two different PARP inhibitors were utilized in this study, this may offer a partial explanation for these observations. Another possible explanation is that FEN1 is required to address the increase in oxidatively-induced DNA lesions occurring from the multiphoton micro-irradiation used (56, 57).
5. Induction of specific oxidative DNA damage by photosensitizers
As noted in previous sections, the ability to distinguish base lesions, SSBs, and DSBs is critical toward understanding the recruitment kinetics and sub-pathway dynamics involved in different repair pathways. A major limitation of techniques described to this point is the requirement of high laser intensity to produce ROS and base lesions. It has been proposed that the presence of intracellular antioxidants scavenge the ROS generated by micro-irradiation until an oxidative stress threshold is reached that overwhelms the delicate antioxidant balance (65). Träutlein et al. demonstrated this effect by adding N-acetyl-cysteine (NAC) in their multiphoton irradiation and quenching the production of ROS by the multiphoton excitation (57).
Halogenated sensitizers (BrdU) and DNA intercalating dyes (Hoechst and EtBr) allow the incident laser energy to be reduced, but how DNA damage is produced by these chemicals is not well understood, and their presence can have undesirable effects on chromatin structure and DNA mobility (5, 6, 11, 39). Photosensitizers that generate more specific types of oxidative damage would be desirable additions to micro-irradiation studies focused on the BER pathway. Chromophore-assisted laser or light inactivation (CALI) has been used in a number of fields to generate ROS to eliminate targets of interest (reviewed in (66)). Dyes (malachite green and fluorescein arsenical hairpin, FlAsH) and fluorescent proteins (KillerRed and miSOG) have been utilized to produce ROS site-specifically in cells (66). Combinations of these dyes or fluorescent proteins with micro-irradiation allow the generation of ROS within a defined area with temporal synchronization. Several examples of the application of CALI are reviewed below.
5.1 Ro19-8022
Excitation of the polar sensitizer Ro19-8022 has been shown to produce a DNA damage profile similar to singlet oxygen with 8-oxodG being the predominant base lesion observed (67). Micro-irradiation of Ro19-8022 by 365 nm revealed increased accumulation of oxidative damage DNA glycosylases OGG1, NEIL1, NEIL2, and NTH1 to sites of DNA damage (19). The addition of this sensitizer did not alter the recruitment of XRCC1, although an increase in Pol β was observed (19). This indicated an XRCC1-independent role for Pol β in the repair of oxidatively-induced DNA damage that was found to be mediated through the 8-kDa deoxyribose phosphate (dRP)-lyase domain (68).
Similarly, micro-irradiation of Ro19-8022 by 405 nm (10% laser power) was utilized by Campalans et al. to induce formation of oxidatively-induced DNA lesions (32). Without the sensitizer, fluorescently-tagged XRCC1, but not OGG1, was recruited to the sites of 405 nm induced DNA damage, and PARP-1 inhibitors DIQ or olaparib eliminated the recruitment of XRCC1 to these sites of damages, consistent with some previous reports (15, 19, 41). Addition of the sensitizer under the same damaging conditions resulted in the recruitment of XRCC1 and OGG1 to sites of induced damage, even in the presence of PARP-1 inhibitors (32). This suggested that recruitment of XRCC1 to sites of SSBs is dependent on PARP-1 activation, while recruitment of XRCC1 to sites of oxidatively-induced DNA base damage is independent of PARP-1 activity.
Finally, Menoni et al. used Ro19-8022 with low power 405 nm excitation to specifically introduce oxidatively-induced base lesions and to monitor the repair of these lesions by NER proteins, XPC-GFP and GFP-CSB (69). The presence of oxidatively-induced base lesions was confirmed by the recruitment of OGG1 to the sites of induced damage and by immunofluorescent staining with an 8-oxodG antibody. Further, no CPDs or 6-4PPs were observed using these damaging conditions, and no recruitment of other classical NER proteins was observed to sites of induced damage (69). Similar to Lan et al. (19), weak recruitment of XPC was observed without the photosensitizer, indicating a minor role for XPC in the repair of SSBs, and no recruitment of CSB was observed without the sensitizer (69). Interestingly, both XPC and CSB were strongly recruited to the sites of induced oxidative damage, though CSB recruitment was strongly linked to active transcription (69). Together, these data indicate a novel role for XPC and CSB in the repair of oxidatively-induced lesions.
5.2 Psoralen
Another recent report utilized psoralen to probe the role of BER in the resolution of ICLs (70). Psoralen or trioxsalen have been more widely used with micro-irradiation to investigate repair of ICLs by NER (36, 70-72). Psoralen treated cells were micro-irradiated with a 365 nm pulse laser set at 1.7 % power (described in the UV section (36)), and GFP-NEIL1 was found to accumulate to sites of induced damage (70). The damage induced by this system included oxidatively-induced base damage at higher laser powers. Specific recruitment of NEIL1 to ICLs, rather than oxidatively-induced DNA damage, was validated by addition of NAC prior to irradiation. Addition of NAC eliminated the production of oxidatively-induced DNA damage, similar to (57), but did not alter the accumulation of NEIL1 to induced damage sites .
5.3 Fluorescent protein, KillerRed
A more targeted ROS-generation approach utilizing the fluorescent protein KillerRed was recently reported (20). KillerRed is excited by 559 nm (~6 mJ/μm2) to generate superoxide (20, 73). To site-specifically introduce ROS, KillerRed was fused to the tetracycline repressor (tetR) and the transcription activator tetR+VP16 (TA), and these fusions were targeted to defined genomic regions containing a 90-kilobase (kb) tetracycline response element (TRE) binding cassette within the fusion proteins (20). Localization of the KillerRed-tetR allowed ROS to be introduced into regions of heterochromatin. The VP16 activator resulted in opening of condensed chromatin for transcription, and the KillerRed-TA fusion allowed the induction of ROS to euchromatin. Using this system, GFP-mediator complex subunit 1 (MED1), NEIL1, NEIL2, and NTH1 were found to accumulate at sites of induced damage in both types of chromatin structures, while PARP-1 was more efficiently recruited to heterochromatin structures (20). Accumulation of the long-patch repair proteins PCNA and FEN1 was observed in euchromatin regions, with FEN1 recruitment being dependent on PCNA (20). One other interesting finding in this study was a reduction of FEN1 and PCNA recruitment in the presence of RNA polymerase II inhibitor, which implies a role for FEN1 and PCNA in transcription coupled repair of oxidatively-induced DNA damage (20).
5.4 Caveats for the use of photosensitizers for the induction of DNA damage
Coupling micro-irradiation with photosensitizers allows for better control over the types of DNA damage being generated thereby providing better resolution and characterization of BER specific pathways. Additionally, the DNA lesions are more comparable to natural ROS damage. As before, there are limitations to these applications. The use of the Ro19-8022 photosensitizer is limited because it is not commercially available. Other photosensitizers (e.g., toluidine blue) are commercially available, but they have not been used with micro-irradiation yet.
Improvements in genome-editing tools now allow for the site-specific introduction of fluorophores like KillerRed, yet this protein also has drawbacks. KillerRed expression must be tightly regulated to avoid induction of ROS by white light (20). KillerRed has been shown to dimerize in cells (74) and to emit a weak green fluorescent signal that may interfere with recruitment monitoring for GFP-tagged proteins (75). An improved version of KillerRed, Supernova, has been generated with specific point mutations to prevent dimerization (76). These tools may allow for better generation of BER lesions and improved localization of site-specific DNA damage when utilized with micro-irradiation. The visible excitation of these agents also makes them more accessible to researchers. Yet, as with the other methods discussed here, assessment of the damage generated is still required to maximize the impact of the findings.
6. Conclusions and Future Directions
A common theme in the application of micro-irradiation techniques for the examination of DNA repair is the need for improved reporting of laser conditions and better characterization of lesions induced by the damage. While specific antibodies exist for UV photoproducts and 8-oxodG, researchers reporting micro-irradiation results do not often employ them (Table 1). Defining induced DNA lesion population heterogeneity can help researchers evaluate reported results, compare across techniques, and provide insight into cross-talk between DNA repair pathways. While no micro-irradiation technique is perfect, these methods have provided intriguing insights into NER and DSB repair, and appear to be poised to do the same for SSB repair and BER.
Table 1.
Summary of wavelengths reported to generate SSBs and base lesions.
| Wavelength | Damage Characterization | Proteins observed | References |
|---|---|---|---|
| 337 nm | 8-oxodG, 6,4PPs, CPDs γH2AX positive |
FEN1, PARP-1, XRCC1 | (6) |
| 351 nm | γH2AX positive | FUS/TLS | (33) |
| 364 nm | γH2AX negative | BRCA1, HMGN1, Pol β, XRCC1 | (17, 18, 21, 26, 27, 37) |
| 365 nm (low power) | γH2AX negative | CAF1-p150, LIGIII, PCNA, PNKP, Pol β XRCC1 | (14, 19) |
| 365 nm (high power) | γH2AX positive 8-oxodG |
BRCA1, CAF1-p150, NBS1, NEIL1, NEIL2, NTH1, OGG1, PCNA, RAD52 | (19) |
| 365 nm with Psoralen | γH2AX positive | NEIL1 | (70) |
| 365 nm with Ro19-8022 | 8-oxodG | NEIL1, NEIL2, NTH1, OGG1, Pol β, XRCC1 | (19) |
| 405 nm | Low power, γH2AX negative 16-800 μW, 8-oxodG and γH2AX positive |
FEN1, PCNA, Pol β, PNKP, XRCC1 | (16, 43, 44) |
| 405 nm with BrdU or Hoechst | 8-oxodG, CPDs γH2AX positive |
LIGI, LIGIII, OGG1, PARP-1, PARG, XRCC1 | (12, 13, 15, 22-24, 41, 42) |
| 405 nm with Ro19-8022 | 8-oxodG Negative for 6,4PPs, and CPDs |
CSB, OGG1, XPC, XRCC1 | (32, 69) |
| 435 nm | γH2AX negative | XRCC1 | (45) |
| 488 nm | γH2AX positive Negative for 8-oxodG, 6,4PPs, and CPDs |
ATM, LIGIII, PCNA, RPA XRCC1 | (46) |
| 514 nm with EtBr | 8-oxodG γH2AX positive |
XRCC1, HP1, OGG1 | (50) |
| 559 nm with Killer Red | γH2AX positive 8-oxodG |
FEN1, HP1, MED1, NEIL1, NEIL2, NTH1, PARP-1, PCNA, XRCC1 | (20) |
| 750 nm with Hoechst | γH2AX positive Negative for 8-oxodG |
XRCC1, OGG1, PARP-1, LIGIII, PNKP | (12) |
| 768 nm | None reported | OGG1 | (25) |
| 800 nm | None reported | FEN1, PCNA | (61) |
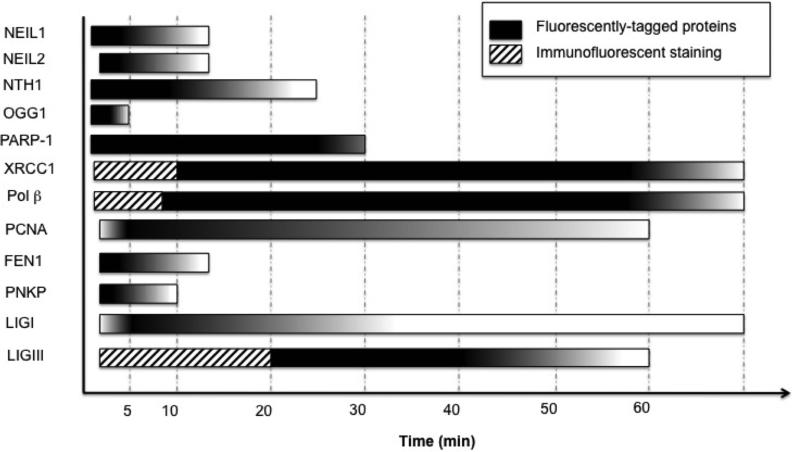
By examining the micro-irradiation studies together, a timeline of repair events can be observed (Fig. 3). The initial steps in lesion recognition involve proteins that arrive quickly and dissociated rapidly as well. Scaffold proteins like XRCC1 and PARP-1 appear to be retained at the damage, and this retention coincides with the final gap filling and ligation steps. Interestingly, FEN1, PCNA, and LIGI have different recruitment and retention profiles that with further study and better separation from single-nucleotide repair may reveal coordination of steps during long-patch BER.
Figure 3. Coordination of BER proteins responding to sites of induced damage.
Time scales were estimated from all of the reports reviewed with peak recruitment shaded darkly and gradual dissociation of proteins illustrated by the color gradient. For PCNA and LIGI a slower recruitment was observed (19, 24), so a gradient reflecting this accumulation was included. The endpoints reflect the longest reported time monitored. Hashed blocks indicate recruitment detected by immunofluorescent staining.
Some of the most intriguing results from micro-irradiation studies reviewed here come from the apparent inconsistencies between biochemical and biophysical reports. The role of PARP-1 in the recruitment of XRCC1 to sites of oxidatively-induced lesions is one example. While the results using 405 nm irradiation with the sensitizer Ro19-8022 are supported by the results without a sensitizer (16, 32), there is still a question of what lesions are being generated by the 405 nm irradiation (Table 1). If UV photoproducts are generated, then XRCC1 could be responding as part of NER rather than part of oxidative repair (77). The argument for a PARP-1 independent role in oxidative DNA lesion repair relies on the inhibition of catalytic activity by PARP-1 inhibitors (12, 32). Yet, the large number of PARP-1 inhibitors used in these studies makes comparisons difficult. PARP-1 inhibitors, NU1025 and 4-AN, still allow PARP-1 to be recruited to sites of DNA damage, though the accumulation kinetics are slightly altered and the persistence of PARP-1 at damage sites is extended (15, 41). Numerous reports have indicated the PARP inhibitors used in these studies do not prevent PARP-1 DNA binding (15, 41, 78-80). So, if PARP-1 is truly dispensable for the recruitment of XRCC1 to sites of DNA damage, then PARP-1 should not be observed at oxidatively-induced lesions. None of the reported studies indicating PARP-1 independent sub-pathways monitor the recruitment of PARP-1 to sites of induced damage, so future work should examine the recruitment of PARP-1 to sites of oxidatively-induced base lesions.
The lack of damage characterization and mixed use of inhibitors also affects the assessments about FEN1 and PCNA. In order to more effectively understand the use of long-patch repair proteins, the addition of chemicals like methoxyamine (MX), which blocks single-nucleotide BER , could help resolve competing repair pathways and allow better elucidation of sub-pathway specific behavior.
Micro-irradiation is a powerful technique that may allow DNA repair sub-pathways to be studied and dissected; however, it does have a fundamental limitation that must always be kept in mind: An individual repair process cannot be dynamically monitored in a population-based system. Despite the advantage of localized damage induction, this is not a single molecule technique that would allow a single discrete lesion to be generated and its repair monitored. Objectives used for most micro-irradiation applications generate pixels containing kb of DNA. That means DNA damage is occurring in highly concentrated regions and that a large number of repair proteins are responding to the sites of damage. This recruitment of a large number of proteins is a strategic advantage when using fluorescently-tagged proteins in wild-type backgrounds, yet it limits the precise monitoring of individual proteins to an individual damage site and insights into the stoichiometry of individual proteins at that damage site. While targeting techniques employed in the use of KillerRed limits the region of interest to ~90 kb (20), improvement in imaging techniques are still required to improve the resolution of individual proteins to induced damage sites. The increasing accessibility of super resolution microscopy techniques along with the growing number of compatible fluorophores for these systems holds significant promise for the study of DNA repair. This was recently demonstrated for ionizing radiation induced DNA damage sites, where individual DSBs were imaged (81).
Overall, micro-irradiation techniques have provided remarkable new insight into DNA damage repair pathways and allowed for the cross-talk between repair pathways to be investigated more precisely. The studies reviewed here demonstrate the power of this technique for examining BER and SSB repair, yet there is considerable room for improvement. Advances in genomic lesion quantification and in super resolution microscopy will allow for significant improvements in the detection and resolution of DNA repair complexes induced by micro-irradiation, and this will provide better insight into overlapping DNA repair pathways.
Highlights.
Micro-irradiation allows study of single strand break and base lesion repair
A wide range of wavelengths can be used to induce site-specific lesions
Novel recruitment dynamics can be observed by micro-irradiation techniques
Acknowledgments
The authors are supported by the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health (NIH), Z01-ES050158 and Z01-ES050159, and NRG is also funded by 1K99ES023813-01. We thank C. Jeff Tucker at the NIEHS Fluorescence Microscopy and Imaging Center for his expert assistance with the micro-irradiation studies and thoughtful comments on the manuscript. We also thank Bill Beard for helpful discussions.
Abbreviations
- DIQ
1,5- dihydroxyisoquinoline
- 4-AN
4- amino-1,8-naphthalimide
- 6,4PPs
6,4-photoproducts
- 8-oxodG
8-oxo-2′-deoxyguanosine
- OGG1
8-oxoguanine glycosylase 1
- APE1
AP-endonuclease
- ATM
ataxia telangiectasia-mutated gene
- BER
base excision repair
- BRCA1
breast cancer gene 1
- BRCT1
BRCA1 C-terminus 1
- BrdU
bromodeoxyuridine
- CALI
Chromophore-assisted laser or light inactivation
- CPD
cyclobutane pyrimidine dimers
- dRP
deoxyribose phosphate
- DSB
double-strand break
- LIGI
DNA ligase I
- LIGIII
DNA Ligase III
- Pol β
DNA polymerase β
- NEIL1
endonuclease VIII-like 1
- NEIL2
endonuclease VIII-like 2
- NTH1
endonuclease III-like protein 1
- EtBr
ethidium bromide
- FEN1
flap endonuclease 1
- FlAsH
fluorescein arsenical hairpin
- FUS/TLS
fused-in-sarcoma/translocated-in-sarcoma
- γH2AX
gammaH2AX
- GFP
green fluorescence protein
- HP1
heterochromatin protein 1
- HMGN1
high mobility group N1
- H2O2
hydrogen peroxide
- ICLs
interstrand cross-links
- MED1
mediator complex subunit 1
- MEFs
mouse embryonic fibroblasts
- NAC
N-acetyl-cysteine
- ns
nanosecond
- NIR
Near Infrared
- NBS1
Nijmegen breakage syndrome 1
- NER
nucleotide excision repair
- CAF1-p150
p150 subunit of chromatin assembly factor I
- PAR
poly(ADP-ribose)
- PARG
poly(ADP-ribose) glycohydrolase
- PARP-1
poly(ADP-ribose) polymerase 1
- γH2AX
phosphorylated H2AX
- PNKP
polynucleotide kinase phosphatase
- PCNA
proliferating cell nuclear antigen
- RPA
replication protein A
- ROS
reactive oxygen species
- SSBs
single-strand breaks
- TRE
tetracycline response element
- tetR
tetracycline repressor
- TA
transcription activator tetR+VP16
- TDP1
tyrosyl-DNA phosphodiesterase 1
- UV
ultraviolet light
- XRCC1
X-ray repair cross-complementing protein 1
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author.
References
- 1.Luijsterburg MS, et al. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vermeulen W. Dynamics of mammalian NER proteins. DNA Repair (Amst) 2011;10:760–771. doi: 10.1016/j.dnarep.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 3.Limoli CL, Ward JF. A new method for introducing double-strand breaks into cellular DNA. Radiat Res. 1993;134:160–169. [PubMed] [Google Scholar]
- 4.Lukas C, Bartek J, Lukas J. Imaging of protein movement induced by chromosomal breakage: tiny 'local' lesions pose great 'global' challenges. Chromosoma. 2005;114:146–154. doi: 10.1007/s00412-005-0011-y. [DOI] [PubMed] [Google Scholar]
- 5.Kim JS, et al. In situ analysis of DNA damage response and repair using laser microirradiation. Methods Cell Biol. 2007;82:377–407. doi: 10.1016/S0091-679X(06)82013-3. [DOI] [PubMed] [Google Scholar]
- 6.Kong X, et al. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrando-May E, et al. Highlighting the DNA damage response with ultrashort laser pulses in the near infrared and kinetic modeling. Front Genet. 2013;4:135. doi: 10.3389/fgene.2013.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botchway SW, Reynolds P, Parker AW, O'Neill P. Use of near infrared femtosecond lasers as sub-micron radiation microbeam for cell DNA damage and repair studies. Mutat Res. 2010;704:38–44. doi: 10.1016/j.mrrev.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Nagy Z, Soutoglou E. DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol. 2009;19:617–629. doi: 10.1016/j.tcb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 10.Botchway SW, Reynolds P, Parker AW, O'Neill P. Laser-induced radiation microbeam technology and simultaneous real-time fluorescence imaging in live cells. Methods Enzymol. 2012;504:3–28. doi: 10.1016/B978-0-12-391857-4.00001-X. [DOI] [PubMed] [Google Scholar]
- 11.Dinant C, et al. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J Cell Sci. 2007;120:2731–2740. doi: 10.1242/jcs.004523. [DOI] [PubMed] [Google Scholar]
- 12.Abdou I, Poirier GG, Hendzel MJ, Weinfeld M. DNA ligase III acts as a DNA strand break sensor in the cellular orchestration of DNA strand break repair. Nucleic Acids Res. 2015;43:875–892. doi: 10.1093/nar/gku1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolin C, et al. The impact of cyclin-dependent kinase 5 depletion on poly(ADP-ribose) polymerase activity and responses to radiation. Cell Mol Life Sci. 2012;69:951–962. doi: 10.1007/s00018-011-0811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Della-Maria J, et al. The interaction between polynucleotide kinase phosphatase and the DNA repair protein XRCC1 is critical for repair of DNA alkylation damage and stable association at DNA damage sites. J Biol Chem. 2012;287:39233–39244. doi: 10.1074/jbc.M112.369975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godon C, et al. PARP inhibition versus PARP-1 silencing: different outcomes in terms of single-strand break repair and radiation susceptibility. Nucleic Acids Res. 2008;36:4454–4464. doi: 10.1093/nar/gkn403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanssen-Bauer A, et al. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen. 2011;52:623–635. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton JK, Gassman NR, Dunigan BD, Stefanick DF, Wilson SH. DNA polymerase β-dependent cell survival independent of XRCC1 expression. DNA Repair (Amst) 2015;26:23–29. doi: 10.1016/j.dnarep.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton JK, et al. Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses. DNA Repair (Amst) 2013;12:774–785. doi: 10.1016/j.dnarep.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan L, et al. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan L, et al. Novel method for site-specific induction of oxidative DNA damage reveals differences in recruitment of repair proteins to heterochromatin and euchromatin. Nucleic Acids Res. 2014;42:2330–2345. doi: 10.1093/nar/gkt1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masaoka A, et al. Interaction between DNA Polymerase β and BRCA1. PLoS One. 2013;8:e66801. doi: 10.1371/journal.pone.0066801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortusewicz O, Leonhardt H. XRCC1 and PCNA are loading platforms with distinct kinetic properties and different capacities to respond to multiple DNA lesions. BMC Mol Biol. 2007;8:81. doi: 10.1186/1471-2199-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortusewicz O, Fouquerel E, Amé JC, Leonhardt H, Schreiber V. PARG is recruited to DNA damage sites through poly(ADP-ribose)- and PCNA-dependent mechanisms. Nucleic Acids Res. 2011;39:5045–5056. doi: 10.1093/nar/gkr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. Differential recruitment of DNA Ligase I and III to DNA repair sites. Nucleic Acids Res. 2006;34:3523–3532. doi: 10.1093/nar/gkl492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zielinska A, Davies OT, Meldrum RA, Hodges NJ. Direct visualization of repair of oxidative damage by OGG1 in the nuclei of live cells. J Biochem Mol Toxicol. 2011;25:1–7. doi: 10.1002/jbt.20346. [DOI] [PubMed] [Google Scholar]
- 26.Gassman NR, Stefanick DF, Kedar PS, Horton JK, Wilson SH. Hyperactivation of PARP triggers nonhomologous end-joining in repair-deficient mouse fibroblasts. PLoS One. 2012;7:e49301. doi: 10.1371/journal.pone.0049301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masaoka A, et al. HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem. 2012;287:27648–27658. doi: 10.1074/jbc.M112.370759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad R, Shock DD, Beard WA, Wilson SH. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285:40479–40488. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 30.Caldecott KW. DNA single-strand break repair. Exp Cell Res. 2014;329:2–8. doi: 10.1016/j.yexcr.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Prasad R, et al. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol Biol (Mosk) 2011;45:586–600. [PMC free article] [PubMed] [Google Scholar]
- 32.Campalans A, et al. Distinct spatiotemporal patterns and PARP dependence of XRCC1 recruitment to single-strand break and base excision repair. Nucleic Acids Res. 2013;41:3115–3129. doi: 10.1093/nar/gkt025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rulten SL, et al. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 35.Frosina G, et al. Two pathways for base excision repair in mammalian cells. J Biol Chem. 1996;271:9573–9578. doi: 10.1074/jbc.271.16.9573. [DOI] [PubMed] [Google Scholar]
- 36.Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem. 2009;284:27908–27917. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gassman NR, et al. Bisphenol a promotes cell survival following oxidative DNA damage in mouse fibroblasts. PLoS One. 2015;10:e0118819. doi: 10.1371/journal.pone.0118819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jelezcova E, et al. Parp1 activation in mouse embryonic fibroblasts promotes Pol beta-dependent cellular hypersensitivity to alkylation damage. Mutat Res. 2010;686:57–67. doi: 10.1016/j.mrfmmm.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds P, Botchway SW, Parker AW, O'Neill P. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage - when is a DSB not a DSB? Mutat Res. 2013;756:14–20. doi: 10.1016/j.mrgentox.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakob B, et al. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mortusewicz O, Amé JC, Schreiber V, Leonhardt H. Feedback-regulated poly(ADP-ribosyl)ation by PARP-1 is required for rapid response to DNA damage in living cells. Nucleic Acids Res. 2007;35:7665–7675. doi: 10.1093/nar/gkm933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mortusewicz O, Leonhardt H, Cardoso MC. Spatiotemporal dynamics of regulatory protein recruitment at DNA damage sites. J Cell Biochem. 2008;104:1562–1569. doi: 10.1002/jcb.21751. [DOI] [PubMed] [Google Scholar]
- 43.Hanssen-Bauer A, et al. The region of XRCC1 which harbours the three most common nonsynonymous polymorphic variants, is essential for the scaffolding function of XRCC1. DNA Repair (Amst) 2012;11:357–366. doi: 10.1016/j.dnarep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan L, et al. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]
- 45.Berquist BR, et al. Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res. 2010;38:5023–5035. doi: 10.1093/nar/gkq193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solarczyk KJ, Zarębski M, Dobrucki JW. Inducing local DNA damage by visible light to study chromatin repair. DNA Repair (Amst) 2012;11:996–1002. doi: 10.1016/j.dnarep.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 48.Kielbassa C, Epe B. DNA damage induced by ultraviolet and visible light and its wavelength dependence. Methods Enzymol. 2000;319:436–445. doi: 10.1016/s0076-6879(00)19041-x. [DOI] [PubMed] [Google Scholar]
- 49.Hamada N, et al. Histone H2AX phosphorylation in normal human cells irradiated with focused ultrasoft X rays: evidence for chromatin movement during repair. Radiat Res. 2006;166:31–38. doi: 10.1667/RR3577.1. [DOI] [PubMed] [Google Scholar]
- 50.Zarebski M, Wiernasz E, Dobrucki JW. Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A. 2009;75:619–625. doi: 10.1002/cyto.a.20734. [DOI] [PubMed] [Google Scholar]
- 51.Goeppert-Mayer M. Elementary process with two quantum jumps. Ann. Phys. 1931;9:273–294. [Google Scholar]
- 52.Kaiser W, Garrett CGB. Two-Photon Excitation in CaF2 - Eu2+. Physical Review Letters. 1961;7:229–231. [Google Scholar]
- 53.Meldrum RA, Botchway SW, Wharton CW, Hirst GJ. Nanoscale spatial induction of ultraviolet photoproducts in cellular DNA by three-photon near-infrared absorption. EMBO Rep. 2003;4:1144–1149. doi: 10.1038/sj.embor.7400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tirlapur UK, König K. Femtosecond near-infrared laser pulse induced strand breaks in mammalian cells. Cell Mol Biol (Noisy-le-grand) 2001;47 Online Pub:OL131-134. [PubMed] [Google Scholar]
- 55.Mari PO, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirlapur UK, König K, Peuckert C, Krieg R, Halbhuber KJ. Femtosecond near-infrared laser pulses elicit generation of reactive oxygen species in mammalian cells leading to apoptosis-like death. Exp Cell Res. 2001;263:88–97. doi: 10.1006/excr.2000.5082. [DOI] [PubMed] [Google Scholar]
- 57.Träutlein D, Deibler M, Leitenstorfer A, Ferrando-May E. Specific local induction of DNA strand breaks by infrared multi-photon absorption. Nucleic Acids Res. 2010;38:e14. doi: 10.1093/nar/gkp932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harper JV, et al. Induction of persistent double strand breaks following multiphoton irradiation of cycling and G1-arrested mammalian cells-replication-induced double strand breaks. Photochem Photobiol. 2008;84:1506–1514. doi: 10.1111/j.1751-1097.2008.00378.x. [DOI] [PubMed] [Google Scholar]
- 59.Mohanty SK, Rapp A, Monajembashi S, Gupta PK, Greulich KO. Comet assay measurements of DNA damage in cells by laser microbeams and trapping beams with wavelengths spanning a range of 308 nm to 1064 nm. Radiat Res. 2002;157:378–385. doi: 10.1667/0033-7587(2002)157[0378:camodd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 60.Haince JF, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 61.Kleppa L, et al. Kinetics of endogenous mouse FEN1 in base excision repair. Nucleic Acids Res. 2012;40:9044–9059. doi: 10.1093/nar/gks673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankenberg-Schwager M. Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother Oncol. 1989;14:307–320. doi: 10.1016/0167-8140(89)90143-6. [DOI] [PubMed] [Google Scholar]
- 63.Frankenberg-Schwager M. Induction, repair and biological relevance of radiation-induced DNA lesions in eukaryotic cells. Radiat Environ Biophys. 1990;29:273–292. doi: 10.1007/BF01210408. [DOI] [PubMed] [Google Scholar]
- 64.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Green RM, Graham M, O'Donovan MR, Chipman JK, Hodges NJ. Subcellular compartmentalization of glutathione: correlations with parameters of oxidative stress related to genotoxicity. Mutagenesis. 2006;21:383–390. doi: 10.1093/mutage/gel043. [DOI] [PubMed] [Google Scholar]
- 66.Sano Y, Watanabe W, Matsunaga S. Chromophore-assisted laser inactivation--towards a spatiotemporal-functional analysis of proteins, and the ablation of chromatin, organelle and cell function. J Cell Sci. 2014;127:1621–1629. doi: 10.1242/jcs.144527. [DOI] [PubMed] [Google Scholar]
- 67.Will O, et al. Oxidative DNA damage and mutations induced by a polar photosensitizer, Ro19-8022. Mutat Res. 1999;435:89–101. doi: 10.1016/s0921-8777(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 68.Sobol RW, et al. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 69.Menoni H, Hoeijmakers JH, Vermeulen W. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J Cell Biol. 2012;199:1037–1046. doi: 10.1083/jcb.201205149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McNeill DR, et al. NEIL1 responds and binds to psoralen-induced DNA interstrand crosslinks. J Biol Chem. 2013;288:12426–12436. doi: 10.1074/jbc.M113.456087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cremer T, Peterson SP, Cremer C, Berns MW. Laser microirradiation of Chinese hamster cells at wavelength 365 nm: effects of psoralen and caffeine. Radiat Res. 1981;85:529–543. [PubMed] [Google Scholar]
- 72.Johnston BH, Johnson MA, Moore CB, Hearst JE. Psoralen-DNA photoreaction: controlled production of mono- and diadducts with nanosecond ultraviolet laser pulses. Science. 1977;197:906–908. doi: 10.1126/science.887929. [DOI] [PubMed] [Google Scholar]
- 73.Vegh RB, et al. Reactive oxygen species in photochemistry of the red fluorescent protein “Killer Red”. Chem Commun (Camb) 2011;47:4887–4889. doi: 10.1039/c0cc05713d. [DOI] [PubMed] [Google Scholar]
- 74.Shirmanova MV, et al. Phototoxic effects of fluorescent protein KillerRed on tumor cells in mice. J Biophotonics. 2013;6:283–290. doi: 10.1002/jbio.201200056. [DOI] [PubMed] [Google Scholar]
- 75.Nordgren M, et al. Potential limitations in the use of KillerRed for fluorescence microscopy. J Microsc. 2012;245:229–235. doi: 10.1111/j.1365-2818.2011.03564.x. [DOI] [PubMed] [Google Scholar]
- 76.Takemoto K, et al. SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation. Sci Rep. 2013;3:2629. doi: 10.1038/srep02629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moser J, et al. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner. Mol Cell. 2007;27:311–323. doi: 10.1016/j.molcel.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 78.Prasad R, et al. Suicidal cross-linking of PARP-1 to AP site intermediates in cells undergoing base excision repair. Nucleic Acids Res. 2014;42:6337–6351. doi: 10.1093/nar/gku288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murai J, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murai J, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Britton S, Coates J, Jackson SP. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol. 2013;202:579–595. doi: 10.1083/jcb.201303073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tebbs RS, et al. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]