Abstract
Cell cycle checkpoints provide surveillance mechanisms to activate the DNA damage response, thus preserving genomic integrity. The heterotrimeric Rad9-Rad1-Hus1 (9-1-1) clamp is a DNA damage response sensor and can be loaded onto DNA. 9-1-1 is involved in base excision repair (BER) by interacting with nearly every enzyme in BER. Here, we show that individual 9-1-1 components play distinct roles in BER directed by MYH DNA glycosylase. Analyses of Hus1 deletion mutants revealed that the interdomain connecting loop (residues 134–155) is a key determinant of MYH binding. Both the N- (residues 1–146) and C-terminal (residues 147–280) halves of Hus1, which share structural similarity, can interact with and stimulate MYH. The Hus1K136A mutant retains physical interaction with MYH but cannot stimulate MYH glycosylase activity. The N-terminal domain, but not the C-terminal half of Hus1 can also bind DNA with moderate affinity. Intact Rad9 expressed in bacteria binds to and stimulates MYH weakly. However, Rad91-266 (C-terminal truncated Rad9) can stimulate MYH activity and bind DNA with high affinity, close to that displayed by heterotrimeric 91-266-1-1 complexes. Conversely, Rad1 has minimal roles in stimulating MYH activity or binding to DNA. Finally, we show that preferential recruitment of 91-266-1-1 to 5′-recessed DNA substrates is an intrinsic property of this complex and is dependent of complex formation. Together, our findings provide a mechanistic rationale for unique contributions by individual 9-1-1 subunits to MYH-directed BER based on subunit asymmetry in protein-protein interactions and DNA binding events.
Keywords: DNA repair, DNA binding protein, DNA damage response, DNA glycosylase, genomic instability
Graphical abstract

1. Introduction
Cell cycle checkpoints provide surveillance mechanisms to activate the DNA damage response (DDR), thus preserving genomic integrity [1,2]. Activation of DDR leads to cell cycle arrest (which allows time for DNA repair) and enhances DNA repair. When DNA damage is extreme, apoptosis is triggered. The checkpoint system includes an array of proteins that function as sensors, transducers, and effectors [3–6]. The heterotrimeric Rad9/Rad1/Hus1 (9-1-1) complex is a DDR sensor [7,8] and is loaded onto DNA by the Rad17-RFC2–5 clamp loader [9–12]. 9-1-1 is essential for embryonic development, genomic stability, and telomere integrity [13–16]. Besides serving as a damage sensor [17], 9-1-1 is involved in many DNA metabolisms [14] including base excision repair (BER) (reviewed in [18]). Remarkably, 9-1-1 interacts with nearly every enzyme in BER and is proposed to constitute a platform to coordinate BER. Many BER proteins interact selectively with specific subunit(s) of 9-1-1 [19–21].
The first step in BER is carried out by a DNA glycosylase, which cleaves damaged or mismatched bases. MYH (also called MUTYH) DNA glycosylase excises adenine when it is mispaired with 8-oxo-7,8-dihydroguanine (Go) or guanine and thus reduces G:C to T:A mutations [22–24]. The resulting apurinic/apyrimidinic (AP) site is processed by AP-endonuclease 1 (APE1), allowing the downstream BER enzymes to complete the DNA repair process. Mutations in the human MYH (hMYH) gene can lead to colorectal cancer (as in MYH-associated polyposis or MAP) [25], while APE1 is essential for cell viability [26]. MYH contains unique motifs that mediate interactions with partner proteins involved in DNA replication, mismatch repair, and DDR (reviewed in [22,23]). We have shown that the interdomain connector (IDC) located between the N- and C-terminal domains of hMYH is uniquely oriented [27] to interact with Hus1 [21] and APE1 [28].
The ring structure of 9-1-1 [29–31] is remarkably similar to that of the proliferating cell nuclear antigen (PCNA) [32–34]. Each 9-1-1 subunit folds into two globular domains linked by an interdomain connecting loop (IDCL) (Fig. 1A). According to the PCNA-DNA structure [35], the 9-1-1 ring is supposed to encircle double-stranded DNA [30]. Although the three subunits of the 9-1-1 complex are structurally similar, they exhibit key differences. These differences are most pronounced in the IDCLs between their N- and C-terminal domains [29–31]. These structural distinctions between 9-1-1 components have been suggested to dictate protein-binding specificity for individual subunits. For example, hMYH and hAPE1 bind preferentially to the Hus1 subunit [20,21]. Functionally, the Hus1 subunit alone can stimulate MYH activity [21]. In this paper, we constructed a panel of Hus1 mutant proteins to identify domains required for binding MYH, stimulating MYH glycosylase activity, and binding DNA. Subsequently we tested the roles of other 9-1-1 components in MYH activation and DNA binding. This systematic and quantitative biochemical strategy revealed key differences in the ability of 9-1-1 subunits to bind DNA and functionally interact with MYH, thus supporting a model whereby each subunit of 9-1-1 plays a distinct and directed role in BER.
Fig. 1.

(A) Structure of the human 9-1-1 complex. The crystal structure 91-272-1-1 is shown (RCSB codes:3A1J) [30]. The structure of the extreme C-terminal domain of Rad9 (Rad9Cter) has not been determined. The key differences among the three subunits are circled in red. The DNA in the channel of the 9-1-1 ring is tilted toward Rad9 and the N-terminal domain of Hus1 based on the data from this paper. (B) Structure of Hus1 showing the locations of K136 and V137 based on the structure of 91-272-Rad1-Hus1 (RCSB codes:3A1J) [30]. N and C indicate N- and C-termini, respectively.
2. Materials and methods
2.1. Glutathione-S-transferase (GST)-tagged Hus1 protein constructs
The plasmid pGEX-3X-hHus1 containing GST-tagged hHus1 was obtained from Dr. A. E. Tomkinson at the University of New Mexico. GST fusions incorporating hHus1 deletion constructs were made by polymerase chain reaction (PCR) using primers listed in Table S1 in the supplementary material. The PCR products were digested with BamHI and SalI and ligated into the BamHI-XhoI-digested pGEX-4T-2 vector (GE Healthcare). The K136A and V137A mutants of the hHus1 gene were constructed by Quick Change site-directed mutagenesis (Stratagene) using pGEX-3X-hHus1 plasmid as a template and primers listed in Table S1 in the supplementary material.
2.2. Cloning and purification of His-tagged hHus1 and deletion constructs
Plasmid pET21a-hHus1 containing full-length wild-type hHus1 cDNA has been described [21]. PCR products containing truncated hHus1 cDNA fragments were digested with BamHI and SalI (as described in GST cloning) and ligated into BamHI-XhoI-digested pET21a. The K136A and V137A mutants of the hHus1 gene were constructed by Quick Change site-directed mutagenesis (Stratagene) using pET21-hHus1 plasmid as a template as described for GST clones.
E. coli Rosetta cells (Novagen) harboring the hHus1-His expression plasmid were grown and induced as described [21]. The hHus1-His proteins were purified over a Ni-NTA resin (Qiagen) and a 1 ml Hitrap Heparin column (GE Healthcare) as described [21]. Fractions containing the majority of hHus1-His proteins were pooled, divided into small aliquots, and stored at −80°C. The purified proteins were analyzed by polyacrylamide gel electrophoresis containing SDS (SDS-PAGE) (Fig. 2), with protein concentrations determined by the Bradford method [36].
Fig. 2.
Purified His-tagged Hus1 constructs, Rad1, Rad9, Rad91-266, 91-266-1-1, and mMyh (as indicated) were analyzed by SDS-PAGE and stained by Coomassie blue. Lanes 1, 8, and 10 serve as molecular weight markers for lanes 2–7, 9, and 11–15, respectively. Lanes 1–7 and lanes 8–15 are run on 15% and 12% gels, respectively.
2.3. Cloning and purification of human hRad91-266-Rad1-Hus1
The cDNAs of hRad9 and hRad1 were amplified by PCR from pGEX-4T3-hRad9 and pGEX-3X-hRad1 plasmids, respectively (both from Dr. A. E. Tomkinson) using primers listed in Table S1 in the supplementary material. The hRad1 gene was cloned between the BamHI and SalI sites of pACYCDuet-1 (EMD Biosciences) to obtain the clone pACYCD-hRad1. The hHus1 cDNA was then inserted into the second cassette of the pACYCD-hRad1 plasmid using BglII and XhoI sites to obtain pACYCDuet-hRad1-hHus1. A hRad9 cDNA fragment encoding amino acid residues 1–266 was cloned between the BamHI and SalI sites of pETDuet-1 to obtain pETD-hRad91-266. Both hRad1 and hRad91-266 proteins contained an N-terminal His tag while hHus1 was tagged with a C-terminal S-tag.
E. coli BL21 Star/DE3 harboring both pACYCDuet-hRad1-hHus1 and pETD-hRad91-266, were grown and induced as described [21]. The 91-266-1-1 complex was first purified over a Ni-NTA resin (Qiagen). Proteins eluted from the Ni column were then diluted with buffer S [20 mM Tris-HCl (pH 7.8), 50 mM NaCl, 0.5 mM DTT and 0.1 mM phenylmethylsulfonyl fluoride] and further purified over a 1 ml Hitrap Q column (GE Healthcare). Fractions containing 91-266-1-1 were collected and further purified using a Superose 12 column (in Buffer S) (GE Healthcare). The size of the 91-266-1-1 complex was estimated by comparing its elution position on the Superose 12 column with those of marker proteins (bovine thyroglobulin, apoferritin, β-amylase, and bovine serum albumin). Fractions containing 91-266-1-1 were collected and further purified and concentrated over a 1 ml Hitrap Heparin column as described [21]. The eluted 91-266-1-1 complex was divided into small aliquots and stored at −80 °C. The purified complex was analyzed by SDS-PAGE (Fig. 2, lane 14).
2.4. Purification of hRad91-266, hRad9, and hRad1
PCR products containing hRad91-266 or hRad1 cDNAs were cleaved by BamHI and SalI and then cloned into BamHI-XhoI-digested pETDuet-1 and pET21a to obtain clones pETDuet-hRad91-266 and pET21a-hRad1, respectively. Full length Rad9 cDNA was cloned by inserting a BglII and XhoI fragment from pACYC-Rad9 [37] into BamHI-XhoI-digested pET21a to obtain clones pET21-hRad9. His-tagged hRad91-266, hRad9, and hRad1 were purified over a Ni-NTA resin similar to hHus1 (above). hRad91-266 was further purified using 1 ml Hitrap SP (GE Healthcare) while hRad9 and hRad1 were purified using Hitrap Q (GE Healthcare) columns. The fractions containing respective proteins were pooled, divided into small aliquots, and stored at −80°C. The purified proteins were analyzed by SDS-PAGE (Fig. 2, lanes 11–13).
2.5. Purification of mouse Myh (mMyh)
Full length mMyh cDNA cloned in pcDNA3.1 (kindly provided by Dr. Yusaku Nakabeppu at Kyushu University, Japan) [38] was subcloned into pET21a (EMD Bioscience) by PCR amplification using primers listed in Table S1 in the supplementary material. The PCR product was digested with Nhe1 and XhoI and ligated into the digested pET21a. mMyh was purified as described for hMYH [27] and was >85% pure as analyzed by SDS-PAGE (Fig. 2, lane 15).
2.6. GST pull-down assay
GST-pull-down assays were performed similarly as described [28]. To eliminate the effect of nucleic acid on protein–protein interactions, 50 μg/ml of ethidium bromide was added to the immobilized proteins for 30 min prior adding their interacting partners.
2.7. Assay of mMyh glycosylase activities
The mMyh substrate is a 44-mer duplex DNA with sticky ends filled-in by Klenow fragment from 40-mer duplex DNA containing an A/Go base mismatch (Table S1 in the supplementary material). The 5′-end of the A-containing strand was labeled with [γ-32P] ATP as described [39]. The MYH glycosylase assay was performed as described previously [27]. The products were then treated with 0.1 M NaOH at 90°C for 30 min to cleave the phosphodiester bonds at AP sites. Reaction mixtures were loaded onto 14% sequencing gels containing 7 M urea. The gel was exposed to a Phosphor Imager screen allowing the image to be detected with a Typhoon FLA9500 and quantified using Image Quant Software (GE Healthcare).
2.8. Electrophoresis mobility shift assay (EMSA)
The DNA binding activities of 9-1-1 subunits were assayed as previously described by Lu et al [40]. The binding reactions (20 μl)contained 20 mM Tris-HCl, pH 7.6, 80 mM NaCl, 1 mM DTT, 1 mM EDTA, 1.5% glycerol, 200 ng/ml poly dI/dC, 50 μg/ml bovine serum albumin, 0.09 nM DNA, and various concentrations of protein. After electrophoresis on a nondenaturing 6% polyacrylamide gel in 50 mM Tri-borate, pH 8.3, and 1 mM EDTA(TBE) buffer, the gel was dried and exposed to a Phosphor Imager screen. In some cases, the gel buffer contained 150 mM imidazole, pH 9 and 1 mM EDTA. Images were detected using a Typhoon FLA9500 and enzyme-bound and free DNA bands were quantified by Image Quant Software (GE Healthcare).
2.9. Fluorescence anisotropy assay
For fluorescence anisotropy assays, one of the DNA strands was 5′-end labeled with a fluorescein (Fl) tag (6-carboxyfluorescein) and annealed to the complementary strand (Table S1 in the supplementary material). Fluorescence anisotropy was measured as described [41] using the Beacon 2000 variable temperature fluorescence polarization system (Panvera, Madison, WI) equipped with fluorescein excitation (490 nm) and emission (535 nm) filters. Binding reactions were performed as described for EMSA assays, except that no glycerol was added, the DNA concentration was 2 nM, and the reaction volume was 100 μl. After adding Fl-DNA, samples were incubated for 1 min before measurement of anisotropy (Ac) and total fluorescence emission (F535). Each data point represents the mean of 10 measurements for each binding reaction. Association of the target protein with Fl-DNA substrates was detected by an increase in the anisotropy of the Fl-DNA emission because of restricted segmental motion and retarded rotational correlation time in the protein:DNA complexes relative to the free DNA substrate [42,43].
9-1-1 binding to DNA substrates was considered in terms of a binding model in which a single Fl-DNA may be sequentially targeted by multiple protein molecules (P) to yield a saturated protein:DNA complex (Equation 1).
Although it was not possible to assess binding constants describing individual steps of oligomeric protein assembly on Fl-DNA substrates by this model, indices of binding affinity and cooperativity were estimated by Equation 1,
where Ac-DNA and Ac-complex are the intrinsic anisotropy values of the unbound and protein-saturated Fl-DNA substrates, respectively; [P]½ is the protein concentration yielding half-maximal DNA saturation; and h is the Hill (cooperative) coefficient. Nonlinear regression analyses of Ac data sets taken across at least 15 protein concentrations were performed using PRISM version 3.0 (GraphPad, San Diego, CA).
3. RESULTS
3.1. MYH binds to the interdomain connecting loop of Hus1
We have shown that hMYH physically interacts with 9-1-1 mainly via the Hus1 subunit [21]. The Hus1 binding site is located within the IDC region (residues 295–350) of hMYH [21]. However, the regions of hHus1 protein engaged in the physical interaction with MYH have not been determined. Thus, we generated a panel of hHus1 deletion constructs fused to GST (Figs. 3A and 3B) to map the MYH interacting region. Because MYH binds preferentially to the Hus1 subunit over Rad1 and Rad9 [21] and the IDCL (residues 134–155) of Hus1 is strikingly different from those of Rad1 and Rad9 [29–31] (Fig. 1A), we focused on examining this IDCL region. In the GST pull-down experiments, we used mMyh [44], which is 77% identical and 90% similar to hMYH, due to technical difficulties for purifying hMYH. First, we compared the ability of hHus1N (residues 1–146) and hHus1C (residues 147–280) to bind mMyh. Immobilized GST-hHus1N and GST-hHus1C (Fig. 3B, lane 6 and 8, respectively) both precipitated mMyh in a similar manner as the wild-type GST-hHus1 (Fig. 3C, lanes 3, 7, and 9) whereas bead-bound GST did not (Fig. 3C, lane 2). Because GST-hHus1C was easily degraded during purification (Fig. 3B, lane 8), we further analyzed variants of GST-hHus1N. Adding the missing IDCL residues to hHus1N gave the hHus1N(+) construct (residues 1–155) which did not change mMyh binding (Fig. 3C, compare lanes 6 and 7). In contrast, deletion of the remaining IDCL residues from hHus1N in the hHus1N(−) construct (residues 1–133) resulted in a reduced interaction with mMyh (Fig. 3C, lanes 7 and 8). Further deletion into the N-terminal domain in hHus1N1 (residues 1–90) resulted in a total loss of mMyh binding (Fig. 3C, lane 10). Thus, our results indicate that both the N- and C-terminal halves of Hus1 can interact with MYH and residues 134–146 of the N-terminal domain of hHus1 are critical for Hus1N-MYH interaction, which may be supplemented by a minor role for residues 91–133.
Fig. 3.
The interdomain connecting loop (IDCL) of hHus1 is required for its interaction with mMyh. (A) The GST-tagged hHus1 constructs used in the pull-down analyses are depicted. Numbers represent residues within the truncated Hus1. The IDCL between the N- and C-terminal domains of hHus1 consists of residues 134–155. (B) GST and the GST-hHus1 fusion proteins (as indicated on top of each lane) used in the mMyh pull-down experiments were analyzed by SDS-PAGE. The membrane was stained with Coomassie Blue. (C) Immobilized GST and GST-hHus1 constructs were used to pull-down His-mMyh. Lane 1 contains 10% of input mMyh protein. Lanes 2–10 used GST and GST-hHus1 constructs as indicated in lanes 1–9 of (B) in the His-mMyh pull-down experiments. His-mMyh was detected by Western blotting with an anti-His antibody.
3.2. Both N- and C-terminal domains of hHus1 can stimulate MYH glycosylase activity
We have shown that the hHus1 subunit alone can stimulate Schizosaccharomyces pombe (Sp) Myh1 glycosylase activity slightly less effectively than the intact Sp9-1-1 complex [21]. Stimulation of SpMyh1 glycosylase activity requires a substantial molar excess of hHus1 over SpMyh1 protein. Since we had demonstrated that only discrete domains of hHus1 were required for binding to mMyh, we next tested whether truncated hHus1 proteins could stimulate mMyh glycosylase activity. We kept the ratio of mMyh to DNA near 1 in order to observe better stimulation effect. As shown in Fig. 4, both Hus1N and Hus1C could stimulate mMyh glycosylase activity but required higher ratios of protein over mMyh to achieve the same degree of stimulation observed with intact Hus1. Interestingly, inclusion of the complete IDCL (in Hus1N+) conferred weaker Myh-stimulating activity versus Hus1N, which only contains half of the IDCL. In contrast, Hus1N1 (residues 1–90) did not detectably stimulate mMyh glycosylase activity (Fig. 4A, lanes 19–21), consistent with its inability to bind mMyh (Fig. 3C).
Fig. 4.
mMyh glycosylase activity can be enhanced by Hus1 deletion constructs. (A) Assay of mMyh glycosylase activity in the presence of Hus1 constructs. Lane 1, 5′-32P -labeled A/Go-containing DNA. Lane 2, 0.18 nM A/Go-DNA was incubated with 0.2 nM mMyh. Lanes 3–5 are similar to lane 2 but included 0.4, 1.6, and 6.4 nM hHus1, respectively. Lane 6, A/Go-DNA was incubated with 6.4 nM hHus1 without mMyh. Lanes 7–22 are similar to lanes 3–6 except with Hus1N+, Hus1N, Hus1C, or Hus1N1 as indicated. The concentrations of Hus1 constructs are based on monomers. Arrows mark the intact DNA substrate (S) and the cleavage product (P). (B) Quantitative analyses of mMyh glycosylase stimulation by each hHus1 construct tested in (A). The percentage (%) of cleavage was calculated as P/(P+S). Fold stimulation was calculated using % of cleavage in Myh alone in lane 2 as standard. Error bars indicate SD; n = 3.
To further identify residues within the hHus1 IDCL region that are important for interacting with and stimulating mMyh activity, we inspected the Hus1 structure and selected two residues in IDCL region for mutagenesis (Fig. 1B). K136 and V137 precede the α-helix in the IDCL region of Hus1 and are conserved between hHus1 and SpHus1. Both K136A and V137A mutants of hHus1 exhibited similar physical interactions with mMyh as observed with wild-type Hus1 (Fig. 3C, lanes 3–5). However, Hus1V137A retained (Fig. 5A), whereas Hus1K136A lost (Fig. 5B), the ability to stimulate mMyh glycosylase activity. These data demonstrate that physical interaction between Hus1 and MYH is not sufficient for Hus1 to enhance the MYH glycosylase.
Fig. 5.
mMyh glycosylase activity can be enhanced by Hus1V137A but not by Hus1K136A. (A) Assay of mMyh glycosylase activity in the presence of Hus1 and Hus1V137A. Lane 1, 5′-32P-labeled A/Go-containing DNA. Lane 2, 0.18 nM A/Go-DNA was incubated with 0.2 nM mMyh. Lanes 3–7 are similar to lane 2 but included with 0.6, 1.2, 2.4, 4.8, and 9.6 nM hHus1, respectively. Lane 8, A/Go-DNA was incubated with 9.6 nM hHus1 without mMyh. Lanes 9–14 are similar to lanes 3–8 except with Hus1V137A. The reaction products were separated on large denaturing gels (38 cm × 43 cm). Arrows mark the intact DNA substrate (S) and the cleavage product (P). (B) Assay of mMyh glycosylase activity in the presence of Hus1 and Hus1K136A. Reactions are similar to (A) except that lanes 9–14 are with Hus1K136A and products were separated on small gels (8 cm × 8 cm). (C) Quantitative analyses of mMyh glycosylase stimulation by Hus1 constructs from results in (A) and (B). Fold stimulation was calculated as in Fig. 4B. Error bars indicate SD; n = 3.
3.3. Functional interaction of 9-1-1 with MYH
Our previous results showed that the Sp9-1-1 complex can stimulate SpMyh1 glycosylase activity [21]. To determine whether a similar functional relationship exists among the homologous mammalian components, we expressed and purified the human Rad91-266-Rad1-Hus1 (91-266-1-1) complex. The purified 91-266-1-1 complex was >90% pure and contained all three subunits in equal molar ratios (Fig. 2, lane 13). In comparison to size markers, the 91-266-1-1 complex eluted from a gel filtration column at the position indicating a molecular weight of about 100 kDa (data not shown), which is consistent with the theoretical value of a trimeric complex. We then compared the ability of human 91-266-1-1 and its substituent subunits (hRad91-266, Rad1, and Hus1) to stimulate mMyh glycosylase activity. As shown in Fig. 6, Rad91-266 and Hus1 (based on monomer concentrations) each stimulated mMyh glycosylase activity to a similar degree but slightly weaker than the 91-266-1-1 complex. Addition of Rad1 also resulted in a detectable enhancement of mMyh glycosylase activity but to a significantly lesser degree when compared to Rad91-266, Hus1, or the 91-266-1-1 complex. Thus, individual subunits of the 9-1-1 complex exert different functional consequences upon binding MYH.
Fig. 6.
mMyh glycosylase activity can be enhanced by 9-1-1 and its subunits. (A) Assay of mMyh glycosylase activity in the presence of human 91-266-1-1, Rad91-266, Rad1, Hus1, and Rad9 (labeled as Rad9full). Lane 1, 5′-32P -labeled A/Go-containing DNA. Lane 2, 0.18 nM A/Go-DNA was incubated with 0.2 nM mMyh. Lanes 3–5 are similar to lane 2 but included 0.4, 1.6, and 6.4 nM 91-266-1-1, respectively. Lane 6, A/Go-DNA was incubated with 6.4 nM 91-266-1-1 without mMyh. Lanes 7–22 are similar to lanes 3–6 except with Rad91-266, Rad1, Hus1, or Rad9full as indicated. The concentrations of Rad91-266, Rad1, Hus1, and Rad9full are based on monomers. Arrows mark the intact DNA substrate (S) and the cleavage product (P). (B) Quantitative analyses of mMyh glycosylase stimulation by 91-266-1-1 and its subunits from results in (A). Fold stimulation was calculated as in Fig. 4B. Error bars indicate SD; n = 3. (C) Rad9 lacking the C-terminal domain has higher affinity for MYH than does intact Rad9. Immobilized GST, GST-hRad9full, and GST-hRad91-266 constructs were used to pull-down His-mMyh. GST pull-down experiments were performed similar to Fig. 3C.
We have shown that hMYH interacts with 9-1-1 mainly via the Hus1 subunit and that Rad9 is the weakest binding partner [21]. However, Fig. 6B shows that Rad91-266 has the same stimulatory capability of Hus1 on MYH activity. To investigate whether the C-terminal domain of Rad9 causes this disparity in MYH interaction, we tested the effect of Rad9fullon MYH stimulation. As shown in Fig. 6 (lanes 19–21), retention of the C-terminal domain of Rad9 weakened MYH stimulation. At a Rad9/Myh ratio of 16, Rad9full enhanced mMyh activity by 2.4-fold while Rad91-266 stimulated mMyh activity by 4.2-fold (p≤0.01). In addition, deletion of the C-terminal domain of Rad9 increases MYH binding (Fig. 6C). Thus, the C-terminal domain of Rad9 plays a role in MYH physical and functional interaction.
3.4. DNA binding of 9-1-1 and its subunits
It has been shown that human 91-272-1-1 can bind to DNA [30]. The 9-1-1 ring is supposed to encircle double-stranded DNA [30]. To determine the roles of individual 9-1-1 subunits in DNA recognition, we first measured their ability to bind the linear blunt-ended HC40:HG40 DNA substrate by electrophoresis mobility shift assay (EMSA). 91-266-1-1, Rad91-266, Hus1, and Rad1 all bound HC40:HG40 DNA but generated complexes migrating at different positions in the native gel (Fig. 7). 91-266-1-1 and Rad1 formed discrete bands with DNA (Fig. 7, lanes 1–13). The 91-266-1-1 complex reached 30% shift at 1120 nM protein (Fig. 7, lane 7). Rad1 had the weakest DNA binding activity compared to the other subunits, with only 5% of the DNA substrate shifted in the presence of 1120 nM protein (Fig. 7, lane 13). Rad91-266 shifted the DNA probe without forming a consistently sized DNA-protein complex which gradually migrated towards the gel origin with increasing protein concentration. At 560 nM, all DNA labeled probe was shifted by Rad91-266 toward the gel origin. By contrast, the Hus1:DNA complex barely entered the gel. By EMSA, Hus1 appeared to show relatively high affinity for the HC40:HG40 DNA substrate with about 50% of the DNA substrate bound at 560 nM protein concentration.
Fig. 7.

DNA binding activity of 9-1-1 and its subunits. Binding of human 91-266-1-1, Rad91-266, Rad1, and Hus1 with HC40:HG40-DNA was performed by EMSA. The protein:DNA complexes were fractionated with 6% polyacrylamide gels containing TBE buffer. Lanes 1, 14, and 20: 5′-32P -labeled DNA. Lanes 2–7, DNA was incubated with 0.035, 0.07, 0.14, 0.28, 0.56, or 1.12 μM 91-266-1-1, respectively. Lanes 8–13 are similar to lanes 2–7 except with Rad1. Lanes 15–19 and lanes 20–25 are similar to lanes 3–7 except with Rad91-266 and Hus1, respectively, as indicated. Arrows mark the positions of free DNA substrate and the bound complexes.
Using anisotropy assays [42,43], we then quantified and compared the DNA binding affinities of these proteins with different DNA substrates in solution. Rad91-266 bound to A/Go-20 DNA with a [P]1/2 (protein concentration yielding half-maximal DNA binding) value of 258± 13 nM (Fig. 8A and Table 1), similar to the [P]1/2 of 91-266-1-1 (208± 13 nM). By contrast, the [P]1/2 of Hus1 binding to A/Go-20 DNA was 2170 ± 210 nM. Rad1 binding to A/Go-20 was very weak and saturation could not even be approached at concentrations up to 10 μM (data not shown). By comparing [P]1/2 values, we conclude that the affinities of the 91-266-1-1 complex and its components for A/Go-20 DNA is in the order: 91-266-1-1 ≈ Rad91-266> Hus1 ≫ Rad1. To test whether 9-1-1 has a preference to bind A/Go-DNA, we compared its binding with homoduplex C:G-DNA. 91-266-1-1 bound to HC20:HG20 DNA with a [P]1/2 value of 229± 15 nM (Fig. 8B and Table 1). Thus, DNA binding by 91-266-1-1 is independent of a base/base mismatch.
Fig. 8.
DNA binding curves of 9-1-1 and its subunits by fluorescence anisotropy assay. (A), (C), and (D) represent DNA binding of 91-266-1-1 (circles), Rad91-266 (squares), and Hus1 (triangles) with FAM-labeled A/GO-20, HC40:HG40 (blunt-ended), and HC40:HG20-DNA (5′-recessed), respectively. (B) shows the 91-266-1-1 binding with A/GO-20 (close circles) and homoduplex HC20:HG20 DNA (open circles). The data of the 91-266-1-1 binding with A/GO-20 are the same shown in (A). (E) represents the 91-266-1-1 binding with 3′-recessed HC20:HG40-DNA. The concentrations of Rad91-266 and Hus1 are based on monomers. Nonlinear regression analyses of anisotropy values taken across at least 15 protein concentrations were performed by PRISM version 3.0 using Equation 2. Rad1 DNA binding curves are not shown because saturation could not be approached even at concentrations up to 10 μM.
Table 1.
Protein concentration yielding half-maximal DNA binding ([P]1/2) for 91-266-1-1 and its subunits determined by anisotropy.
| DNA | Enzyme | [P]1/2 (nM) |
|---|---|---|
| A/Go-20 | 91-266-1-1 | 208 ± 13 |
| A/Go-20 | Rad91-266 | 258 ± 13 |
| A/Go-20 | Hus1 | 2170 ± 210 |
|
| ||
| HC20:HG20 | 91-266-1-1 | 229 ± 15 |
|
| ||
| HC40:HG40 | 91-266-1-1 | 185 ± 14 |
| HC40:HG40 | Rad91-266 | 233 ± 4 |
| HC40:HG40 | Hus1 | 582 ± 47 |
|
| ||
| HC40:HG20 | 91-266-1-1 | 18.2 ± 0.9 |
| HC40:HG20 | Rad91-266 | 246 ± 11 |
| HC40:HG20 | Hus1 | 512 ± 38 |
|
| ||
| HC20:HG40 | 91-266-1-1 | 118 ± 10 |
When using longer blunt-ended DNA (HC40:HG40 DNA) without a base/base mismatch in the anisotropy assay (Fig. 8C), the [P]1/2 values of 91-266-1-1 and Rad91-266 were 185± 14 and 233± 4 nM, respectively. These [P]1/2 values are similar to those with A/Go-20 or HC20:HG20 DNA (Table 1). The [P]1/2 value of Hus1 binding to HC40:HG40-DNA was 582± 47 nM which is about 4-fold stronger than to A/Go-20 DNA. There is a discrepancy between the apparent DNA binding affinities of 91-266-1-1, Rad91-266 and Hus1 when measured by anisotropy as compared to EMSA methods. In particular, 91-266-1-1 showed higher affinity for HC40:HG40-DNA than Rad91-266 or Hus1 when measured using the anisotropy assay (Table 1) but lower DNA binding affinity than these individual subunits when compared by EMSA (Fig. 7). The substantial differences in apparent binding affinities determined by these two methods suggest that the DNA binding by the 91-266-1-1 clamp may be more sensitive to gel sieving or exhibit faster dynamics than complexes formed using Rad91-266 alone. Because the anisotropy-based assay measures binding under true equilibrium conditions, the data obtained by this method provide a better index of solution binding affinity.
It has been shown that 9-1-1 is preferentially loaded by Rad17-RFC2–5 onto DNA substrates possessing 5′ recessed ends [9,10,12][10,12]. However, it has not been reported that 91-266-1-1 prefers to associate with 5′-recessed DNA substrates in the absence of its clamp loader. To examine whether 91-266-1-1, Rad91-266 and Hus1 have this property, we measured their binding affinities for HC40:HG20-DNA containing a 5′ recessed end. The [P]1/2 value for 91-266-1-1 binding to HC40:HG20-DNA was 18.2± 0.9 nM (Fig. 8D and Table 1) which is 10-fold lower than its [P]1/2 (185± 14 nM) for HC40:HG40-DNA. Conversely, the binding affinities of Rad91-266 and Hus1 for HC40:HG20-DNA were similar to those for HC40:HG40-DNA (Table 1). Because Rad1 binds DNA very weakly, its binding to 5′ recessed DNA substrates was not tested. To confirm the preferential binding of 5′ recessed DNA by 91-266-1-1, we measured its affinity for the HC20:HG40-DNA substrate which contains a 3′ recessed end. The [P]1/2 value for 91-266-1-1 binding to HC20:HG40-DNA was 118± 10 nM (Fig. 8E and Table 1) which is close to the [P]1/2 (185± 14 nM) for HC40:HG40-DNA but is 6.5-fold higher than the [P]1/2 (18.2± 0.9 nM) for HC40:HG20-DNA. Given that these observations were made in the absence of the Rad17-RFC2–5 clamp loader, we suggest that this preference for 5′ recessed DNA substrate is an intrinsic property of the 9-1-1 complex. However, the binding preference for 5′ recessed DNA ends was not observed with Rad91-266 and Hus1 indicating that enhanced recognition of these DNA structures by 9-1-1 proteins only occurs in the heterotrimeric context.
3.5. DNA binding activity of Hus1 deletion constructs and mutants
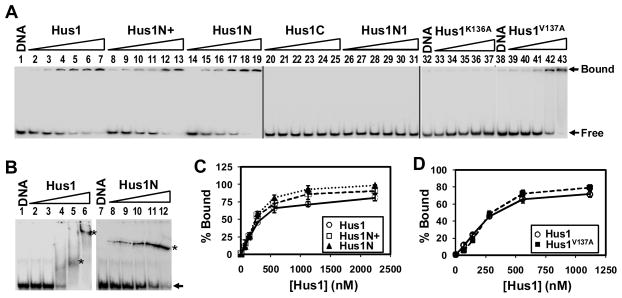
To determine which domain(s) of Hus1 is responsible for DNA binding, we analyzed Hus1 deletion constructs and mutants described in Fig. 3A. Hus1 shows relatively similar affinity for DNA using both EMSA and anisotropy assays (Figs. 7 and 8). We elected to use EMS As to qualitatively assess Hus1 domain binding to DNA because this assay is convenient and requires far smaller quantities and lower concentrations of enzymes. One complication with these EMSA experiments was that DNA complexes with Hus1, Hus1N+, Hus1N, and the two Hus1 point mutants barely entered the gels when run using TBE buffer. Because similar patterns were observed even at low protein concentrations and with different Hus1 deletion constructs, it is unlikely that the DNA is simply precipitated by Hus1 in the binding buffer. To investigate whether Hus1:DNA complex is precipitated after loading onto the polyacrylamide gel containing TBE buffer, we loaded the binding reactions on gels containing different buffers. As shown in Fig. 9B, Hus1:DNA complexes migrated well into gels containing 150 mM imidazole/1 mM EDTA (pH 9.3) buffer, revealing multiple Hus1:DNA complex species. However, gel patterns were usually distorted, likely due to the low buffering capacity of imidazole. As shown in Fig. 9B (lanes 8–12), the Hus1N:DNA complex was also well resolved using gels containing 150 mM imidazole. By comparison, the percentages of shifted Hus1:DNA and Hus1N:DNA complexes in gels containing imidazole were similar to those retained in the wells of TBE gels (compare lanes 2–6 of Fig. 9B with lanes 2–6 of Fig. 9A and lanes 8–12 of Fig. 9B with lanes 14–18 of Fig. 9A). From these data, we conclude that the bands retained near the wells of gels containing TBE buffer are likely specific protein-DNA complexes. Based on this premise, TBE-buffered EMSA analyses of the remaining Hus1 mutants revealed that intact Hus1, Hus1N, and Hus1N+ could bind HC40:HG40-DNA (Fig. 9C) while Hus1C and Hus1N1 did not detectably bind DNA (Fig. 9A, lanes 20–31). Hus1V137A had similar DNA binding activity as wild-type Hus1 while Hus1K136A did not bind DNA (Fig. 9A, lanes 32–43 and Fig. 9D). Together, these data demonstrate that a domain between residues 91–146 of Hus1 that involves K136 is required for DNA binding.
Fig. 9.
DNA binding activity of Hus1 deletion constructs and mutants. (A) Binding of Hus1 variants with HC40:HG40-DNA was performed by EMSA in gels containing TBE buffer. Lanes 1, 32, and 38 contained 5′-32P -labeled DNA. Lanes 2–7, DNA was incubated with 0.07, 0.14, 0.28, 0.56, 1.12, or 2.24 μM hHus1, respectively. Lanes 8–13, 14–19, and 26–31 are similar to lanes 2–7 except with Hus1N+, Hus1N, or Hus1N1 as indicated. Lanes 20–25, DNA was incubated with 0.035, 0.07, 0.14, 0.28, 0.56, or 1.12 μM Hus1C, respectively. Lanes 33–37 and 39–43 are similar to lanes 2–6 except with Hus1V137A and Hus1K136A, respectively. Arrows mark the positions free DNA substrate and the bound complex. (B) Binding of Hus1 and Hus1N with HC40:HG40-DNA was performed by EMSA in gels containing 150 mM imidazole/1 mM EDTA buffer. Lanes 1–6 are similar to lanes 1–6 in (A) and lanes 8–12 are similar to lanes 14–18 in (A) except the gels were run with different buffers. The position of free DNA substrate is marked by an arrow and the bound complexes are marked by stars on the right of the bands. (C) Quantitative analyses of the percentage of DNA bound by each Hus1 deletion construct from results in (A). The percentage (%) of DNA bound was calculated as bound/(bound+free). Error bars indicate SD; n = 3. (D) Quantitative analyses of the percentage of DNA bound by Hus1V137A as compared to wild-type Hus1. Analyses are similar to (C).
4. DISCUSSION
In the present study, we reveal the differential roles played by the 9-1-1 subunits in physical and functional interactions with the MYH glycosylase and DNA binding. The three subunits of 9-1-1 are structurally similar, but exhibit key differences [29–31] (Fig. 1A). This structural asymmetry correlates with an asymmetry in protein-protein interactions (reviewed in [18]). In particular, hMYH and hAPE1 bind preferentially to the Hus1 subunit [20,21]. The IDCL domain of Hus1 is substantially different from those of Rad9 and Rad1 [29–31] (Fig. 1A). Both hHus1N (residues 1–146) and hHus1C (residues 147–280) share structural symmetry and contain half of IDCL. However, the half IDCL of hHus1N contains a α-helix while that of hHus1C is disordered [29–31] (Fig. 1B). Here, we show that MYH binds to both Hus1 halves and that residues 134–155 in the IDCL of hHus1 are critical for MYH interaction. Both Hus1N and Hus1C can also enhance MYH activity but to a lower extent than intact Hus1. Curiously, Hus1N+ (residues 1–155), which contains the complete IDCL, was less effective in stimulating Myh activity than Hus1N, which only contains one half of the IDCL. The cause of the different Myh-activating potentials of each mutant Hus1 requires further investigation.
In our initial attempt to identify MYH interacting motif(s) on Hus1, we examined two Hus1 mutants. Although both Hus1V137A and Hus1K136A mutants physically interact with mMyh similar to wild-type Hus1, Hus1V137 retains, whereas Hus1K136A lacks, the ability to stimulate mMyh glycosylase activity. The K136A mutation, constructed in the intact Hus1 protein, appears to inactivate the ability of Hus1C to enhance the MYH activity. We offer two possibilities. It is possible that the K136A mutation may alter the conformation of Hus1C. Another possibility is that there may be some exposed surfaces that exist in truncated Hus1-C domain that are hidden in the intact Hus1K136A protein. In addition, Hus1V137A has DNA binding activity similar to wild-type Hus1 while Hus1K136A cannot bind DNA. It has been shown that positively charged residues in the center of the PCNA clamp make contact with DNA [35]. Because K136 of Hus1 is not located near the center of the 9-1-1 clamp [29–31] (Fig. 1B), the reason why Hus1K136A is unable to bind DNA remains unclear. It also remains to be confirmed whether the DNA binding ability of Hus1 is associated with its stimulation of MYH activity. However, such a mechanical connection might be not necessary because Hus1C can both physically and functionally interact with MYH, but it cannot bind DNA.
Among the 9-1-1 subunits, we have shown that Hus1 is the preferred partner, while intact Rad9 is the poorest interacting partner for hMYH [21] and SpMyh1 [19]. The Hus1 binding site is located within the IDC region (residues 295–350) of hMYH [21]. Recently, Agustina et al. [45] showed that hRad9 interacts with N-terminal region (residues 1–74) of hMYH. Thus, the physical interactions of MYH with Hus1 and Rad9 appear different. Here we show that truncated Rad91-266 and Hus1 can stimulate mMyh activity to a similar extent, however, intact Rad9 has a weaker MYH stimulation than Rad91-266(Fig. 6). In addition, intact Rad9 has a weaker physical interaction with MYH than Rad91-266 (Fig. 6C). Thus, the C-terminal domain of Rad9 has a negative effect on MYH physical and functional interaction. It has been shown that 91-272-1-1, but not intact 9-1-1 (purified from bacteria, thus without post-transcriptional modification), binds DNA. This suggests that the C-terminal domain of Rad9 may block DNA binding in the intact 9-1-1 clamp. It will be interesting to learn whether phosphorylation of the C-terminal domain of Rad9 can alter MYH interaction or DNA binding because this domain is phosphorylated upon DNA damage and is required for checkpoint function [46,47].
Our results show that 91-266-1-1 binding to DNA is independent of a base/base mismatch suggesting it is not involved in DNA damage recognition. Our DNA binding data also show that 91-266-1-1 prefers binding to 5′ recessed DNA. This is consistent with the previous finding that 9-1-1 has a preference to be loaded on DNA substrates possessing 5′ recessed ends by Rad17-RFC2–5 clamp loader [10,12,48]. Our results suggest that this preference for 5′ recessed DNA substrate is an intrinsic property of the 9-1-1 complex even in the absence of the clamp loader. We believe that 91-266-1-1 is loaded onto the 5′-recessed DNA without the clamp loader for two reasons. First, the oligonucleotides used in our experiments are relatively short and they can run through the channel of the ring and contact the positively charged residues in the channel of the clamp. Second, we observed high binding affinity ([P]1/2 is 18.2 nM) with 91-266-1-1 and 5′-recessed DNA (Table 1). Such high binding affinity indicates that the clamp stays at the single-stranded and double-stranded DNA junction and does not slide away. The preference for binding DNA containing 5′ recessed ends is not observed with Rad91-266 and Hus1. Thus, preferential recruitment of 9-1-1 to 5′-recessed DNA is dependent of complex formation. The DNA binding modes of Rad91-266 and Hus1 are also distinct from that of 91-266-1-1 as indicated by their different gel mobility patterns in EMSA analyses (Fig. 7).
Through this study we demonstrated, for the first time, that the structural asymmetry of the 9-1-1 subunits contributes to differential DNA binding (Table 1). Our data show that Rad91-266 has significantly stronger DNA binding affinity than the other two subunits (Fig. 8 and Table 1). In fact, hRad91-266 binds to A/Go-20 and C40/G40 with similar affinity as 91-266-1-1. The N-terminal half of Hus1, which is adjacent to Rad9 in the 9-1-1 clamp [29–31], also has moderate DNA binding activity. A domain between residues 91–146 of Hus1 is required for DNA binding. Within this region, K93 of Hus1 is located in the inner ring of the 9-1-1 complex [29–31] and is predicted to interact with DNA [49]. However, the C-terminal half of Hus1 does not bind DNA and Rad1 plays a minimal role in DNA binding. Based on these data, we suggest that DNA encircled by the 9-1-1 clamp may be tilted toward Rad9 and the N-terminal domain of Hus1 (Fig. 1A). Our results are in agreement with the E. coli β-clamp-DNA complex [50] and the simulated PCNA/FEN1/DNA model [49]. However, our data do not support the simulated 91-270-1-1/FEN1/DNA model in which all three subunits of 91-270-1-1 make contacts with DNA [49].
In conclusion, our results demonstrate that each subunit of the 9-1-1 complex plays a distinct role in MYH-directed BER and supports a model whereby Hus1 binds to and stimulates MYH [21] and APE1 [20], Rad9 interacts with DNA, and Rad1 may recruit other BER factors such as FEN1 [31,49]. We have shown previously that deletion of fission yeast S. pombe SpMyh1 strongly abrogated the association of SpHus1 with telomeres [51]. hMYH knockdown in human cells decreases Chk1 and Cdc25C phosphorylation in response to H2O2 treatment [52] and decreases p-Chk1 and p-Cdk2 after hydroxyurea treatment [45]. Thus, we suggest that MYH first recognizes a mismatch then recruits 9-1-1 to sites of DNA damage through MYH-Hus1 interaction. 9-1-1 recruitment enhances MYH activity and recruits/activates APE1. Since 9-1-1 has no preference for mismatch and prefers to bind 5′ recessed end, one possibility is that after APE1 function, the exposure of single-stranded DNA may secure 9-1-1 at the site where Rad9 and the N-terminal domain of Hus1 bind to DNA and prevent the clamp to slide away. This model may provide a biochemical rationale for the 9-1-1-dependent enhancement of BER via increasing local repair enzyme concentrations.
Supplementary Material
Highlight.
Individual Rad9-Rad1-Hus1 (9-1-1) subunits play distinct roles in BER.
The interdomain connecting loop of Hus1 is a key determinant of MYH binding.
The K136A mutant demonstrated that Hus1 binding is uncoupled from MYH activation.
The order of DNA binding affinity is:91-266-1-1 ≥ Rad91-266> Hus1 ≫ Rad1.
Preferential binding of 9-1-1 to 5′-recessed DNA is independent of clamp loader.
Acknowledgments
We thank Dr. Yusaku Nakabeppu (Kyushu University, Japan) for the mouse Myh clone and Dr. Alan Tomkinson (University of New Mexico) for the cDNA of hRad9 and hRad1. We thank Dr. Eric Toth (University of Maryland Medical School) for critical reading of this manuscript and constructing Fig. 1B. This work was supported by the National Cancer Institute of the National Institute of Health grants R01-CA78391 and S10-OD011969 to A.L. and R01-CA102428 to G.M.W.
The abbreviations used are
- 9-1-1
Rad9, Rad1, and Hus1 heterotrimer complex
- AP
apurinic/apyrimidinic
- APE1
AP endonuclease
- BER
base excision repair
- DDR
DNA damage response
- DTT
dithiothreitol
- ECL
Enhanced Chemiluminescence
- EMSA
electrophoresis mobility shift assay
- Go or 8-oxoG
7,8-dihydro-8-oxo-guanine
- GST
glutathione-S-transferase
- h
human
- h coefficient
Hill (cooperative) coefficient
- IDC
interdomain connector
- IDCL
interdomain connecting loop
- m
mouse
- MYH
MutY homolog
- [P]1/2
protein concentration yielding half-maximal DNA binding
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- Rad91-266
hRad9 containing residues 1–266 of 391
- RFC
replication factor C
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Sp
Schizosaccharomyces pombe
- TBE
Tri-borate-EDTA buffer
Footnotes
Conflict of interest statement
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 4.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 5.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 6.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 7.Hang H, Lieberman HB. Physical interactions among human checkpoint control proteins HUS1p, RAD1p, and RAD9p, and implications for the regulation of cell cycle progression. Genomics. 2000;65:24–33. doi: 10.1006/geno.2000.6142. [DOI] [PubMed] [Google Scholar]
- 8.St Onge RP, Udell CM, Casselman R, Davey S. The human G2 checkpoint control protein hRAD9 is a nuclear phosphoprotein that forms complexes with hRAD1 and hHUS1. Mol Biol Cell. 1999;10:1985–1995. doi: 10.1091/mbc.10.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci U S A. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- 13.Francia S, Weiss RS, Hande MP, Freire R, d’Adda dF. Telomere and telomerase modulation by the mammalian Rad9/Rad1/Hus1 DNA-damage-checkpoint complex. Curr Biol. 2006;16:1551–1558. doi: 10.1016/j.cub.2006.06.066. [DOI] [PubMed] [Google Scholar]
- 14.Francia S, Weiss RS, d’Adda dF. Need telomere maintenance? Call 911. Cell Div. 2007;2:3–8. doi: 10.1186/1747-1028-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–7248. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss RS, Enoch T, Leder P. Inactivation of mouse Hus1 results in genomic instability and impaired responses to genotoxic stress. Genes Dev. 2000;14:1886–1898. [PMC free article] [PubMed] [Google Scholar]
- 17.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madabushi A, Lu A-L. The novel role of cell cycle checkpoint clamp Rad9-Hus1-Rad1 (the 9-1-1 complex) in DNA repair. In: Berhardt Leon V., editor. Advances in Medicine and Biology. Nova Publishers; Hauppauge NY: 2011. pp. 41–74. [Google Scholar]
- 19.Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2005;280:408–417. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- 20.Gembka A, Toueille M, Smirnova E, Poltz R, Ferrari E, Villani G, Hubscher U. The checkpoint clamp, Rad9-Rad1-Hus1 complex, preferentially stimulates the activity of apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta in long patch base excision repair. Nucleic Acids Res. 2007;35:2596–2608. doi: 10.1093/nar/gkl1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY homolog (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400:53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu AL, Bai H, Shi G, Chang DY. MutY and MutY homologs (MYH) in genome maintenance. Front Biosci. 2006;11:3062–3080. doi: 10.2741/2033. [DOI] [PubMed] [Google Scholar]
- 24.Markkanen E, Dorn J, Hubscher U. MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA. Front Genet. 2013;4:18. doi: 10.3389/fgene.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 26.Wilson DM, III, Thompson LH. Life without DNA repair. Proc Natl Acad Sci U S A. 1997;94:12754–12757. doi: 10.1073/pnas.94.24.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luncsford PJ, Chang DY, Shi G, Bernstein J, Madabushi A, Patterson DN, Lu AL, Toth EA. A structural hinge in eukaryotic MutY homologues mediates catalytic activity and Rad9-Rad1-Hus1 checkpoint complex interactions. J Mol Biol. 2010;403:351–370. doi: 10.1016/j.jmb.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY protein (hMYH) physically interacts with protein involved in long-patch DNA base excision repair. J Biol Chem. 2001;276:5547–5555. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- 29.Dore AS, Kilkenny ML, Rzechorzek NJ, Pearl LH. Crystal structure of the Rad9-Rad1-Hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 30.Sohn SY, Cho Y. Crystal structure of the human Rad9-Hus1-Rad1 clamp. J Mol Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Xu M, Bai L, Gong Y, Xie W, Hang H, Jiang T. Structure and functional implications of the human Rad9-Hus1-Rad1 cell cycle checkpoint complex. J Biol Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burtelow MA, Roos-Mattjus PM, Rauen M, Babendure JR, Karnitz LM. Reconstitution and molecular analysis of the hRad9-hHus1-hRad1 (9-1-1) DNA damage responsive checkpoint complex. J Biol Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 33.Shiomi Y, Shinozaki A, Nakada D, Sugimoto K, Usukura J, Obuse C, Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, Rad9-Rad1-Hus1 and Rad17-RFC. Genes to Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 34.Venclovas C, Thelen MP. Structure-based predictions of Rad1, Rad9, Hus1 and Rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Guan X, Bai H, Shi G, Theriot CA, Hazra TK, Mitra S, Lu AL. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res. 2007;35:2463–2472. doi: 10.1093/nar/gkm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano S, Tominaga Y, Ichinoe A, Ushijima Y, Tsuchimoto D, Honda-Ohnishi Y, Ohtsubo T, Sakumi K, Nakabeppu Y. Mutator phenotype of MUTYH-null mouse embryonic stem cells. J Biol Chem. 2003;278:38121–38124. doi: 10.1074/jbc.C300316200. [DOI] [PubMed] [Google Scholar]
- 39.Lu AL. Repair of A/G and A/8-oxoG mismatches by MutY adenine DNA glycosylase. In: Vaughan P, editor. DNA Repair Protocols: Prokaryotic Systems. Humana Press Inc; Totowa, NJ: 2000. pp. 3–16. [DOI] [PubMed] [Google Scholar]
- 40.Lu AL. Isolation and analyses of MutY and MutY homologs (MYH) Methods in Enzymology. 2006;408:64–78. doi: 10.1016/S0076-6879(06)08005-0. [DOI] [PubMed] [Google Scholar]
- 41.Lee CY, Bai H, Houle R, Wilson GM, Lu AL. An Escherichia coli MutY mutant without the six-helix barrel domain is a dimer in solution and assembles cooperatively into multisubunit complexes with DNA. J Biol Chem. 2004;279:52653–52663. doi: 10.1074/jbc.M405271200. [DOI] [PubMed] [Google Scholar]
- 42.Jameson DM, Sawyer WH. Fluorescence anisotropy applied to biomolecular interactions. Methods Enzymol. 1995;246:283–300. doi: 10.1016/0076-6879(95)46014-4. [DOI] [PubMed] [Google Scholar]
- 43.Kishor A, Brewer G, Wilson GM. Analyses of RNA-ligand interactions by fluorescence anisotropy. In: Dinman JD, editor. Biophysical Approaches to Translational Control of Gene Expression. Springer Science+Business Media, Inc; New York: 2012. pp. 173–198. [Google Scholar]
- 44.Ushijima Y, Tominaga Y, Miura T, Tsuchimoto D, Sakumi K, Nakabeppu Y. A functional analysis of the DNA glycosylase activity of mouse MUTYH protein excising 2-hydroxyadenine opposite guanine in DNA. Nucleic Acids Res. 2005;33:672–682. doi: 10.1093/nar/gki214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agustina L, Hahm SH, Han SH, Tran AH, Chung JH, Park JH, Park JW, Han YS. Visualization of the physical and functional interaction between hMYH and hRad9 by Dronpa bimolecular fluorescence complementation. BMC Mol Biol. 2014;15:17. doi: 10.1186/1471-2199-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kai M. Role of the checkpoint clamp in DNA damage response. Biomolecules. 2013;3:75–84. doi: 10.3390/biom3010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roos-Mattjus P, Hopkins KM, Oestreich AJ, Vroman BT, Johnson KL, Naylor S, Lieberman HB, Karnitz LM. Phosphorylation of human Rad9 is required for genotoxin-activated checkpoint signaling. J Biol Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- 48.Bermudez VP, Lindsey-Boltz LA, Cesare AJ, Maniwa Y, Griffith JD, Hurwitz J, Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRad17-replication factor C complex in vitro. Proc Natl Acad Sci U S A. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Querol-Audi J, Yan C, Xu X, Tsutakawa SE, Tsai MS, Tainer JA, Cooper PK, Nogales E, Ivanov I. Repair complexes of FEN1 endonuclease, DNA, and Rad9-Hus1-Rad1 are distinguished from their PCNA counterparts by functionally important stability. Proc Natl Acad Sci U S A. 2012;109:8528–8533. doi: 10.1073/pnas.1121116109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang DY, Shi G, Durand-Dubief M, Ekwall K, Lu AL. The role of MutY homolog (Myh1) in controlling the histone deacetylase Hst4 in the fission yeast Schizosaccharomyces pombe. J Mol Biol. 2011;405:653–665. doi: 10.1016/j.jmb.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang BJ, Shi G, Lu AL. Mammalian MutY homolog (MYH or MUTYH) protects cells from oxidative DNA damage. DNA Repair (Amst) 2014;13:10–21. doi: 10.1016/j.dnarep.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.