SUMMARY
Axon regeneration in the central nervous system (CNS) requires reactivating injured neurons’ intrinsic growth state and enabling growth in an inhibitory environment. Using an inbred mouse neuronal phenotypic screen, we find that CAST/Ei mouse adult dorsal root ganglion neurons extend axons more on CNS myelin than the other eight strains tested, especially when pre-injured. Injury-primed CAST/Ei neurons also regenerate markedly in the spinal cord and optic nerve more than those from C57BL/6 mice and show greater spouting following ischemic stroke. Heritability estimates indicate that extended growth in CAST/Ei neurons on myelin is genetically determined, and two whole-genome expression screens yield the Activin transcript Inhba as most correlated with this ability. Inhibition of Activin signaling in CAST/Ei mice diminishes their CNS regenerative capacity whereas its activation in C57BL/6 animals boosts regeneration. This screen demonstrates that mammalian CNS regeneration can occur and reveals a molecular pathway that contributes to this ability.
INTRODUCTION
Axons do not regenerate after an injury to the adult mammalian central nervous system (CNS). Two conditions appear to be required for successful re-growth of injured CNS axons; an intrinsic capacity of the neurons to grow and an ability to overcome growth-inhibitory environments. Many factors inhibit growth in the CNS (Filbin, 2003; Fitch and Silver, 2008; Fournier et al., 2001; Silver et al., 2014) and eliminating some of these can promote regeneration (Alilain et al., 2011). In the mature CNS the intrinsic growth capacity of intact neurons is repressed to stabilize synaptic circuitry (Sun and He, 2010) and is not generally re-engaged after axonal injury (Cho et al., 2013). To experimentally increase intrinsic neuronal growth in the CNS, studies have genetically inhibited growth-suppressing genes (Sun et al., 2011), exposed CNS neurons to growth-promoting factors (Yin et al., 2006) and altered the neuronal growth state by a preconditioning axonal lesion (Kadoya et al., 2009; Mills et al., 2007; Neumann and Woolf, 1999). The data from many studies suggest that both the presence of extrinsic inhibition and the lack of an intrinsic growth capacity after injury are important in preventing axonal re-growth in adult CNS (Benowitz and Yin, 2007).
Neurons with axons in the peripheral nervous system retain an ability to regenerate following injury by inducing a robust intrinsic growth response; in addition, the PNS environment is growth-permissive (Hoffman, 2009). Preconditioning PNS neurons by a peripheral axonal injury primes them through massive transcriptional changes to regenerate more vigorously and faster in the face of a second injury because of an induction of networks of regeneration-associated genes (Tedeschi, 2012). Such pre-conditioned growth can be detected in vitro both on permissive substrates (Smith and Skene, 1997; Richardson, 1984) and to a more limited degree on myelin, the latter enabling the central axons of pre-conditioned neurons to begin to grow in the CNS (Kadoya et al., 2009; Mills et al., 2007; Neumann and Woolf, 1999).
Studies of the transcriptional response in preconditioned dorsal root ganglia (DRG) neurons have implicated regeneration-promoting roles for many transcription factors (c-Jun, Atf3, Klf family, Stat3, Sox11 and Smad1) and regeneration-associated genes (Gap43, Cap23, Arg1, Sprr1a, Hspb1 and tubulins: reviewed in Sun and He, 2010; Tedeschi, 2012). In retinal ganglion cells a growth-priming phenomenon analogous to preconditioning can be initiated by intraocular inflammation using agents such as zymosan, which activates expression of the neutrophil- and macrophage-derived growth factor Oncomodulin to increase neuronal expression of regeneration-associated genes (Benowitz and Popovich, 2011; Kurimoto et al., 2013).
Using an adult primary neuron cell-based assay in genetically diverse inbred mouse strains, we have investigated whether the capacity for adult sensory neurons to regenerate on an inhibitory substrate after a preconditioning lesion has a heritable component with the intention of then using this for identifying signaling pathways that may enable growth of injured CNS neurons. Our results identify a particular mouse strain with the capacity to regenerate injured axons in the CNS to a far greater extent than other inbred strains, and identify Activin signaling as a major component of this ability.
RESULTS
In vitro screening for heritability of DRG neuron axonal growth on central myelin
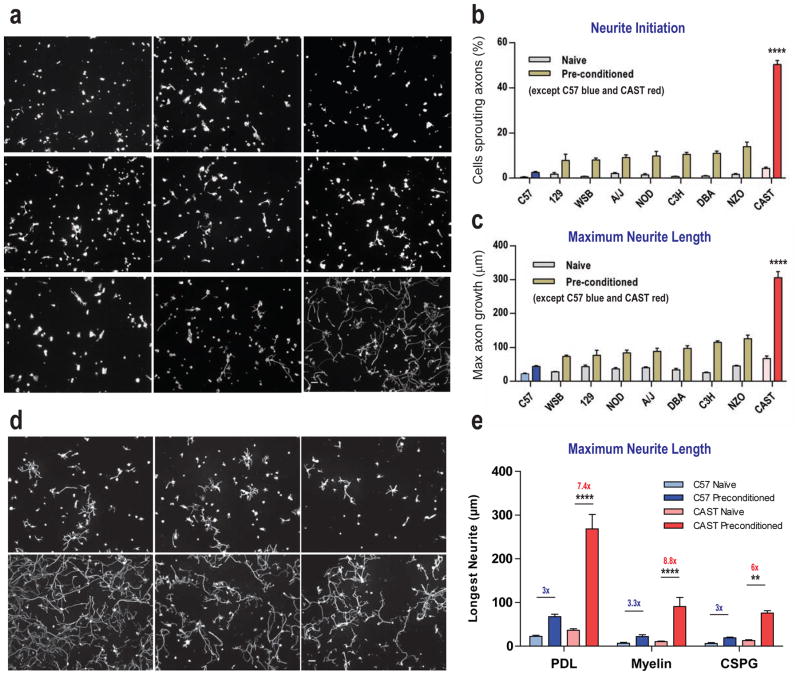
We examined nine diverse inbred mouse strains for a potential differential capacity to initiate adult axonal growth in a CNS-like environment. To accomplish this we screened adult primary sensory neurons obtained from the lumbar L4 and L5 DRGs of genetically distinct mouse strains (A/J, C3H/HeJ, C57BL/6J, DBA/2J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, and WSB/EiJ) for their ability to extend neurites when grown in culture on CNS myelin over 24 hours, as a model for axonal growth in the face of the inhibitory CNS environment. The genetic diversity of the nine strains, which are founder members of the Collaborative Cross has been well documented (Chesler et al., 2008). The uninjured central branch of DRG neurons can regenerate to some extent in the spinal cord in spite of the inhibitory growth environment when the peripheral axons of these cells are pre-injured (Kadoya et al., 2009; Mills et al., 2007; Neumann and Woolf, 1999). Consequently, the axonal outgrowth on myelin screens were performed using both naive (DRG neurons from un-injured mice) and preconditioned neurons (DRG neurons pre-injured by a sciatic nerve crush five days before culture).
These assays revealed that DRG neurons from CAST/Ei mice were capable of producing substantially higher levels of growth on a myelin substrate than all the other strains assayed, measured both as numbers of neurons with neurites and longest axonal process per neuron. The axonal growth in this strain surpassed the average growth of the other strains under naive conditions (2-fold increase, p<0.01, one-way ANOVA; post-hoc Sidak’s, for naive CAST/Ei against all other naïve strains) and markedly greater than the average growth of the other strains post pre-conditioning (3.5-fold increase, p<0.0001, one-way ANOVA; post-hoc Sidak’s, for conditioned CAST/Ei against all other pre-injured strains; Figure 1a–1c). Preconditioning increased growth on myelin in all the strains, but the relative increase in effect size was much greater for neurons from the CAST/Ei strain (Suppl Table 1). The low-power micrographs presented in Figure 1 show the extent of this growth in pre-injured CAST/Ei neurons, a finding that was replicated multiple times (see methods).
Figure 1. Screening for axonal growth in an inhibitory central growth environment.
A: Representative photomicrographs of pre-conditioned axonal growth for nine screened mouse strains (A/J, C3H/HeJ, C57BL/6J, DBA/2J, 129S1/SvImJ, NOD/LtJ, NZO/HlLtJ, CAST/EiJ, and WSB/EiJ) on Myelin. B: Maximal level of axonal sprouting achieved by naïve and pre-conditioned DRG neurons grown on myelin for 24 hours in these mouse strains. C: Maximal level of axonal outgrowth achieved in naïve and pre-conditioned DRG neurons by a sciatic nerve crush injury 5 days prior to growth on myelin for 24 hours. D: Representative photomicrographs of pre-conditioned DRG axonal growth for C57BL/6 and CAST/Ei DRG neurons on PDL, Myelin and CSPG respectively. E: Quantification of growth of naïve and pre-conditioned C57BL/6 and CAST/Ei DRG neurons on a neutral substrate, poly-d-lysine and on inhibitory substrates, myelin and CSPGs. Alternate image capture and counting schemes were used between data given A–C and D–E (see methods). Statistical analysis of B and C: One-way ANOVA; post-hoc Sidak’s corrected, p<0.0001 for pre-conditioned CAST/Ei against all other conditions. For E: Three-way ANOVA; post-hoc Sidak’s. ** p<0.01; **** p<0.0001. Scale, 100 μm. For growth of C57BL/6 and CAST/Ei DRG neurons in the PNS see Suppl Fig 1.
The sensory neurons from the CAST/Ei mice also exhibited elevated levels of growth compared to those from C57BL/6 mice when grown on chondroitin sulfate proteoglycans (CSPGs), an inhibitory substrate found in glial scars (Dickendesher et al., 2012; Fisher et al., 2011; Shen et al., 2009). Preconditioned CAST/Ei neurons grew far more robustly than preconditioned C57BL/6 neurons on both CSPGs (4-fold increase; p<0.05, 56.7 (17.0), CI: 8.3–105.1; three-way ANOVA; post-hoc Sidak’s) and on myelin (as an independent replicate of the earlier findings; 4.1-fold increase; p<0.001; 68.7 (17.0), CI: 20.3–117.1; three-way ANOVA; post-hoc Sidak’s; Figure 1e). These results show that injury-conditioned CAST/Ei DRG neurite growth is strikingly greater than that of conditioned C57BL/6 on two major CNS inhibitory substrates, myelin and CSPG, indicating a broad reduction in responsiveness of sensory neurons from this strain to potent CNS inhibitory cues.
In contrast, pre-conditioned growth on permissive substrates (dissociated DRG neurons on laminin, whole adult DRG explants on matrigel, and also within the PNS in vivo after a sciatic nerve crush) were broadly similar between CAST/Ei and C57BL/6 (1.3-fold CAST/Ei preconditioned vs. C57BL/6 preconditioned at 12hrs on laminin; p<0.001; one-way ANOVA; post-hoc Sidak’s; Suppl Fig 1b). These results imply that DRG neurons from CAST/Ei mice activate a growth program when injured that specifically enables extensive axonal regeneration on CNS inhibitory substrates, and that this differs from the generalized ‘preconditioning effect’ present across all the lines, which promotes regeneration strongly on permissive substrates or in the PNS but only to a much more limited extent on myelin or in the CNS.
Growth phenotype of injured CAST neurons in the CNS
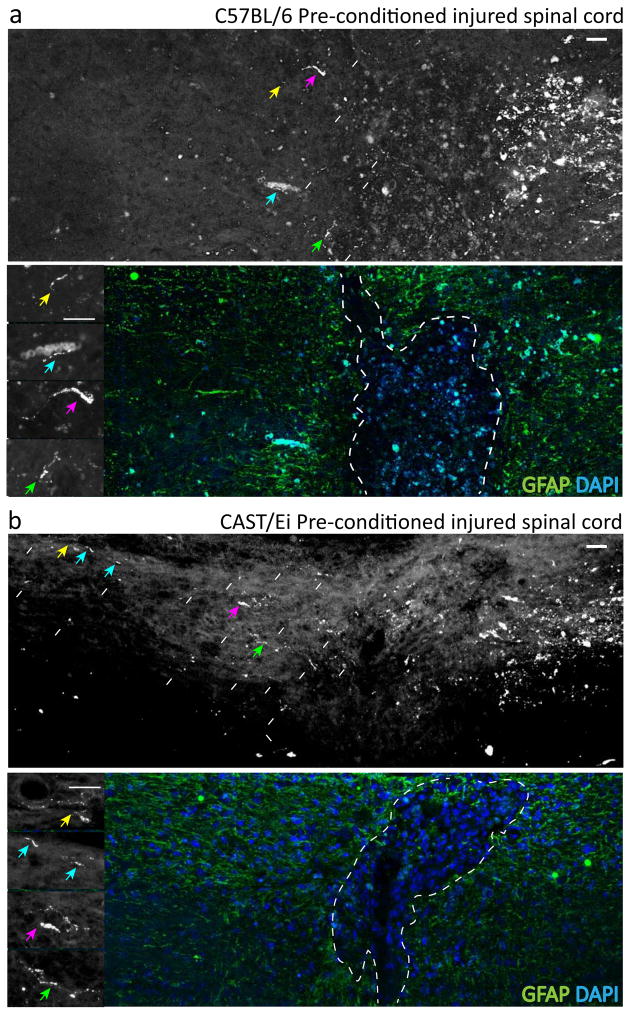
To determine whether the growth capacity of injured CAST/Ei DRG sensory neurons on myelin and CSPGs translated into enhanced growth in the CNS, we assayed three different animal models of CNS regeneration: dorsal column injury, optic nerve injury, and ischemic stroke. We first assayed regeneration of the centrally projecting DRG sensory axons in CAST/Ei or C57BL/6 mice after a dorsal column injury at T10, with and without a pre-conditioning lesion to the sciatic nerve. This is an established model of DRG axonal growth in the CNS, in which the pre-conditioning enables some regeneration of central axons of DRG neurons in the injured spinal cord to occur (Kadoya et al., 2009; Mills et al., 2007; Neumann and Woolf, 1999). Both strains received either a sham or sciatic nerve preconditioning injury a week prior to the dorsal column injury at T10. Six weeks after the dorsal column injury the sciatic nerve was re-exposed and injected with dextran 3000 (micro-Ruby) to retrogradely label ascending dorsal column fibers, and these were measured two weeks later.
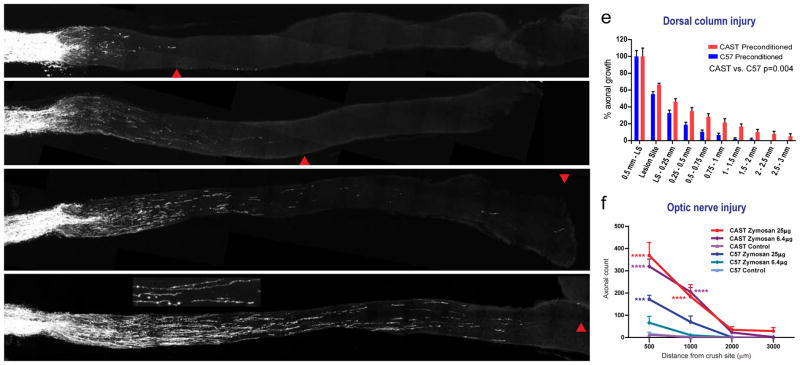
Axonal growth in the dorsal column after a lesion was very limited in both C57BL/6 and CAST/Ei mice that had not undergone a pre-conditioning lesion; however, the pre-conditioning of sensory neurons prior to the spinal cord injury resulted in growth into and beyond the lesion site in both strains (Figure 2a and b; Suppl Fig’s 2 & 3). With dorsal columns from conditioned CAST/Ei mice containing significantly more regenerating axonal trajectories quantified until 3mm rostral of the injury site (p=0.004; differences determined by permutation tests for CAST/Ei and C57BL/6 axonal growth profiles, Figure 3e). Overall pre-conditioned CAST/Ei sciatic dorsal column fibers regenerated 2,974±89μm rostral from the lesion site compared to 789±271μm in C57BL/6, (a 3.5-fold difference, p<0.01, 2184.9 (557.1) CI 826.3–3543.6; two-way ANOVA, post-hoc Sidak’s; Suppl Fig 4). In two out of seven CAST/Ei animals, axonal growth reached 5mm beyond the crush site. The limited extent of growth rostral to the injury site in C57BL/6 mice was consistent with previous results (Seijffers et al., 2007).
Figure 2. CAST vs. C57 axonal regeneration in the adult spinal cord.
Regeneration of central axons of pre-conditioned DRG neurons rostral to a dorsal column injury. A small level of growth is present in C57BL/6 mice (A) but it is significantly greater in CAST/Ei mice (B). Growth quantified in Figure 3. Arrows identifies retrogradely labeled axons from the PNS. The white dashed line indicates the border of the lesion. Asterisk indicates the lesion epicenter (R: rostral, C: caudal). Scale, 100 μm. See also Suppl Fig’s 3 & 2.
Figure 3. CAST vs. C57 axonal regeneration in the adult CNS.
A–D: Optic nerve injury treated with low dose (6.4 μg) zymosan in C57BL/6 results in minimal RGC re-growth (A), but the same dose initiates robust growth in CAST/Ei (C). Medium dose zymosan (25μg) stimulates growth in C57BL/6 optic nerves (B) however CAST/Ei RGC growth is much greater (D). Scale, 500 μm. Red arrow head signifies maximal axonal signal in that section. Inset; Magnified image of the regenerating fibers. Scale, 25μm. E: Quantification of percentage of labeled axons crossing the injury site rostral to the lesion site in preconditioned adult C57BL/6 and CAST/Ei spinal cords respectively. F: Quantification of numbers of axons reaching defined distances from the crush site in the optic nerve injury model of central regeneration. C57BL/6 and CAST/Ei 25μg or 6μg zymosan vs. control comparisons ***p<0.001, ****p<0.0001 (two-way ANOVA, post-hoc Sidak’s). For quantification of longest axonal growth in the injured spinal cord see Suppl Fig 4.
We next investigated whether an enhanced growth might also occur in CAST/Ei neurons which reside within the CNS. To do this, we assayed retinal ganglion cell (RGC) axon regeneration after optic nerve crush using cholera toxin B fragment (CTB) as an axonal tracer. Axon regeneration under baseline conditions was minimal in both strains 2 weeks after optic nerve (ON) crush. Zymosan is commonly used to condition these cells in a manner analogous to a priming nerve injury for DRG neurons (Yin et al., 2003), with a dose of 36 μg zymosan optimal for promoting RGC regeneration in C57BL/6 mice (Kurimoto et al., 2010; Yin et al., 2006). To compare relative regeneration between the two strains we tested the effects of two reduced doses of zymosan applied 5 days before and immediately after the optic nerve crush, in order to use zymosan conditioning doses that induce a lower than maximal growth in C57BL/6 mice.
In C57BL/6 animals 6.4μg and 25μg of zymosan produced modest and dose-dependent levels of axonal regeneration measured 14 days after the ON injury (Figure 3a–b), (Kurimoto et al., 2010; Yin et al., 2006). The same doses in CAST/Ei mice, however, produced substantially more extensive axonal regeneration, with axons observed as far as 4,000μm from the crush site reaching close to the optic chiasm. Differences determined by permutation tests for contrast based on average difference of regenerating axonal trajectories at 6.4μg zymosan CAST vs. C57, mean difference = 118.02, p=0.012; at 25μg zymosan CAST vs. C57, mean difference = 93.28, p=0.031 (Figure 3c–d, quantified in 3f). As robust growth occurred in CAST/Ei RGCs at a low dose of zymosan (6.4μg), one at which C57BL/6 RGCs respond minimally, these results suggest that CAST/Ei CNS neurons are either primed more easily for regrowth than those from C57BL/6 mice or overcome inhibitory substrates better.
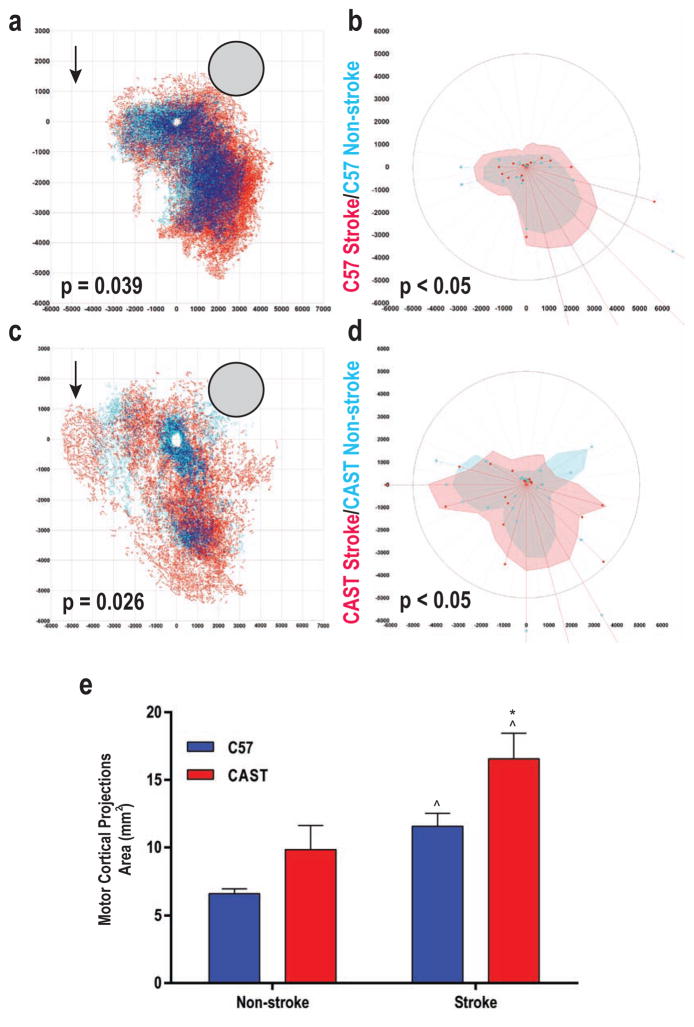
Next, we assayed whether CAST/Ei mice exhibit a greater axonal sprouting ability than C57BL/6 mice following an ischemic stroke produced by middle cerebral artery occlusion, where neuronal growth is impeded by myelin and CSPGs. To determine the extent of axonal sprouting we mapped cortical motor system connections in naive CAST/Ei and C57BL/6 mice one month after stroke, a time point that coincides with the formation of new connections (Clarkson et al., 2013; Li et al., 2010; Overman et al., 2012). In C57BL/6 animals, motor cortex projections showed only a limited degree of axonal sprouting (Figure 4a and 4b). However, sprouting in CAST/Ei mice extended from the motor cortex into the premotor cortex, as well as to posterior and lateral areas corresponding to the first and second somatosensory cortices (Figure 4c and 4d), with axonal projections >5mm from cell bodies (43% area increase in CAST/Ei stroke 16.55±1.9mm2 vs. C57BL/6 stroke 11.58±1.0mm2, p<0.05, Student’s t-test, two-tailed, Figure 4e) (Clarkson et al., 2013; Li et al., 2010; Overman et al., 2012). Pre-conditioning by axonal injury or zymosan administration was not necessary in this model to achieve enhanced axonal regeneration in the CAST/Ei mice, perhaps because the inflammation intrinsic to this model is sufficient to prime the cortical neurons for growth, and possibly, in part, because the axons do not have to traverse a physical lesion, as in the spinal cord and optic nerve CNS injury models.
Figure 4. CAST vs. C57 axonal regeneration post cortical stroke.
A: Connections of forelimb motor cortex in C57BL/6 with and without stroke. Light blue dots indicate the location of projections in non-stroke, red in stroke and dark blue dots are areas of dense overlap between the two conditions. Grey circle is site of tracer injection. X and Y axis represent micron distance from the center of the tracer injection. P-value: Hotellings inverse T-test. B: Polar distribution map in register with connectional plot in (a) showing localization of sprouting in stroke vs. control (Watson’s U2 test; P<0.05). Shaded polygons represent the 70th percentile of the distances of labeled projections from the injection site in each segment of the graph; weighted polar vectors represent normalized distribution of the quantity of points in a given segment of control (blue) or stroke (red) graphs. C: Same conventions as in panel A for CAST/Ei mice (n=5 per group). Note the connections in the far premotor cortex, 4 to 5.5mm anterior to the tracer injection site. These are not present in C57BL/6 after stroke (arrows). D: Same conventions as in Panel B. Overlay of control and post-stroke forelimb motor cortex projections. E: Area of motor cortex projections in C57BL/6 and CAST/Ei mice. Areas taken from 70th percentile of all distances in each radial segment from the injection site (B and D). Student’s t-test, two-tailed. ^ p<0.05, non-stroke vs. stroke C57BL/6 (n=5 for both stroke and non-stroke) and CAST/Ei (n=5 for both stroke and non-stroke) respectively; * p<0.05, CAST/Ei stroke vs. C57BL/6 stroke. See Supplemental Methods for further details.
Heritability estimates of axonal growth phenotype
We next wished to understand what molecular signaling pathways might operate in CAST/Ei mouse neurons to enable regeneration in a CNS environment and if these are genetically determined. To estimate the genetic contribution to the observed regenerative variance across strains we estimated narrow-sense heritability, defined as the additive contribution of genetic factors to the observed phenotype from normalized phenotypic data across all nine strains (Suppl Table 2). Pre-conditioned growth on myelin showed a high degree of heritability −72% (p = 2.2×10−14) for axonal length, suggesting that most of the inter-strain axonal growth variation can be attributed to genetic factors.
Genome-wide expression profiling of naïve and preconditioned DRGs across all nine strains
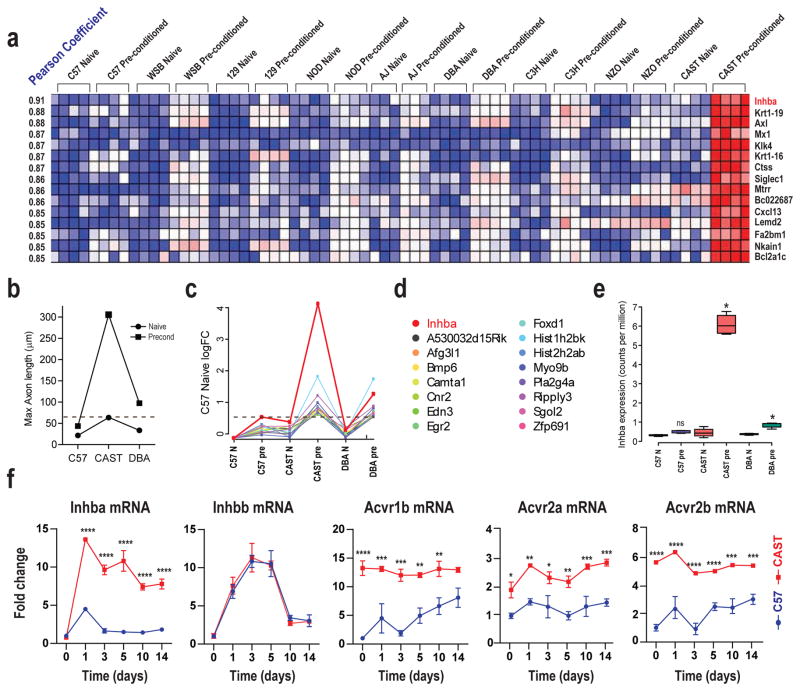
Given the high heritability, we undertook two bioinformatic strategies to identify candidate genes and pathways that may contribute specifically to the CAST/Ei pre-conditioned growth on myelin phenotype. In the first, we performed a genome-wide microarray expression profiling screen of naïve and pre-conditioned DRG tissue across all nine mouse strains (Suppl Table 3). We then correlated this with the degree of DRG axonal growth on myelin, and ranked the resulting genes by Pearson correlation coefficient (Suppl Table 4). In the second strategy, we assayed global transcript expression in naïve and pre-conditioned DRGs by RNAseq from three strains chosen for their differing regenerative capacity (C57BL/6 - low, DBA/2 – mid range and CAST/Ei - high). This global expression dataset was then compared to naïve and pre-conditioned axonal growth on myelin using an independent screening assay (see Methods) to determine the genes that most closely correlated to growth (for array and RNAseq data: GSE55424). Remarkably, in both bioinformatic analyses, Inhba was revealed to be the transcript most strongly correlated with ability of the pre-injured DRG neurons to grow on myelin (Figure 5a, 5c and 5d). The strain-dependent regulation of Inhba indicated by the profiling studies (Figure 5a and 5e) was confirmed in independent samples across time by QRT-PCR (Figure 5f). As two independent genome-wide screens identified Activin signaling as being most highly correlated to CAST/Ei active growth on myelin we chose to investigate this cascade further as detailed below. However, this is unlikely to be the only signaling system which may contribute to the CAST/Ei CNS growth phenotype, since we identified two sets of other highly correlated transcripts (Figure 5a and 5d) from the full screen data (GSE55424).
Figure 5. Genome-wide expression analysis of CAST CNS growth.
A: Naïve and preconditioned array data for the 15 genes whose DRG expression behavior most correlated with axonal growth on myelin. Each box represents data from a single array, each column a single DRG sample, each row a single gene, blue is less expressed, red most expressed. The transcripts are listed in order of Pearson correlation co-efficient between expression and axonal growth, the strains are ordered relative to preconditioned growth on myelin. B: The growth phenotype of each strain measured in a follow-up RNAseq screen determined by maximal axonal length. Horizontal red checked line represents the minimal growth level required in the screen for strong growth in the CNS. C: Expression data of the 16 genes identified by RNAseq screen. These data highlight the Inhba transcript expression (red line) as the most related to long distance regeneration in the mammalian CNS. D: Each of the transcripts identified by the screen with the color indicator for the expression profiles shown in (c). E: Actual expression of the Inhba transcript measured by RNAseq (units are counts per million reads); Inhba expression is significantly upregulated after pre-conditioning only in the DBA/2 and CAST/Ei strains (* p-value< 0.01, false discovery rate<0.01, EdgeR). Data from two independent screens then place Activin signaling at the apex of the strong central axonal regeneration phenotype, most clearly seen in the CAST/Ei strain. F: Time course of expression of Inhba, Inhbb and Activin receptor subtype transcripts in ipsilateral L4/5 DRG from naïve mice and 1, 3, 5, 10 and 14 days post-sciatic nerve crush for both C57BL/6 and CAST/Ei. QRT-PCR; Mean fold change ± SEM normalized to naïve C57BL/6 values For statistical analysis, two-way ANOVA was performed, post hoc Sidak’s (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). For Activin cascade member sites of expression in the DRG see Suppl Fig 5.
Overall the DRG gene expression levels in naïve mice and the transcriptional changes induced by sciatic nerve crush were very similar across the nine strains, with relatively few transcripts showing large strain-dependent expression differences across the dataset. To illustrate this, we performed a weighted gene co-expression network analysis (WGCNA) (Oldham et al., 2008) on the full mouse array data set. Briefly, absolute Pearson correlation coefficients between each transcript and every other gene in the expression analysis were computed, weighted and used to determine the topological overlap, a measure of connection strength, or ‘neighborhood sharing’ in the network. A set of ‘expression networks’ were then constructed where the members of each network have high topological overlap. Groups of genes with very similar expression patterns across large data sets are often functionally connected; part of the same tissue, cell type or biological pathway (Oldham et al., 2006; Oldham et al., 2008). Across the mouse array data set the largest of these expression networks (turquoise, n=1824 unique transcripts) represented genes commonly expressed and regulated after nerve injury between the strains (Suppl Table 5). This gene set also overlapped substantially with that group of transcripts that are regulated in the rat DRG following a sciatic nerve injury, and previously implicated as contributing to the conditioning response (p-value = 7.1×10−52 at the 7 day time point) (Chang and Jovanis, 1990; Kadoya et al., 2009). The turquoise ‘mouse preconditioning response’ network did not include Inhba, which is uniquely induced in CAST/Ei relative to marginal changes in expression in the other strains. In addition, Inhba is not upregulated in the rat DRG following sciatic nerve injury (Blesch et al., 2012; Costigan et al., 2010). Moreover, when the degree of regulation of annotated regeneration-associated genes (Sun and He, 2010; Tedeschi, 2012) between C57BL/6 and CAST/Ei was analyzed, we found virtually no difference (Suppl Fig 5). We interpret these data as indicating that those elements of the preconditioning transcriptional response that facilitate accelerated regeneration in the PNS are largely uniform across all nine mice strains, in keeping with the minimal difference we found in growth rates between C57BL/6 and CAST/Ei on permissive substrates or in the PNS (Suppl Fig 1). Our screen of pre-injured DRG neuronal growth on central myelin has assayed a completely different genomic program, one which enables primed CAST/Ei injured neurons to grow robustly on CNS growth inhibitory substrates.
Activin signaling components in C57 and CAST DRGs
Inhba mRNA encodes Activin-βA, and the Activin signaling cascade is a component of the TGF-beta superfamily. Activin comprises three functional protein forms, Activin-A (βAβA), Activin-B (βBβB) and Activin-AB (βAβB). The Activin-βB transcript Inhbb unlike Inhba was upregulated to a similar extent in CAST/Ei and C57BL/6 DRGs following sciatic nerve crush injury (Figure 5f). Activin is signaled by one Type I serine/threonine kinase receptor (Acvr1b, Alk4) and two Type II co-receptors (Acvr2a and Acv2b) (Harrison et al., 2003). These receptors form an active tetrameric complex composed of two of each receptor subtype. Acvr1b is expressed at higher levels in naïve CAST/Ei DRGs than in C57BL/6 DRGs (13.2-fold) but is not regulated by nerve injury. In C57BL/6 DRGs, levels of this transcript are low in the naïve state, but it slowly upregulates following injury. Both type II receptors are expressed at higher levels in naïve CAST/Ei than C57BL/6 DRGs and were minimally regulated by injury in either strain (Figure 5f). Using in situ hybridization and immunohistochemistry we demonstrate that Inhba mRNA, the Activin-βA peptide and all receptor sub-types are expressed in injured CAST/Ei DRG neurons. Furthermore, we confirm lack of Inhba mRNA induction and Activin-βA peptide expression in injured C57BL/6 DRG neurons (Suppl Fig 6).
Activin signaling promotes translocation of phosphorylated Smad2/3 from the cytoplasm to the nucleus to initiate downstream transcriptional signaling, and we found a large increase in nuclear phospho-Smads2/3 in pre-conditioned DRG neurons from CAST/Ei but not C57BL/6 mice (Suppl Fig 5). Thus pre-conditioned CAST/Ei, but not C57BL/6 neurons manifest activation of downstream signaling components of the Activin pathway.
Activin gain- and loss-of-function
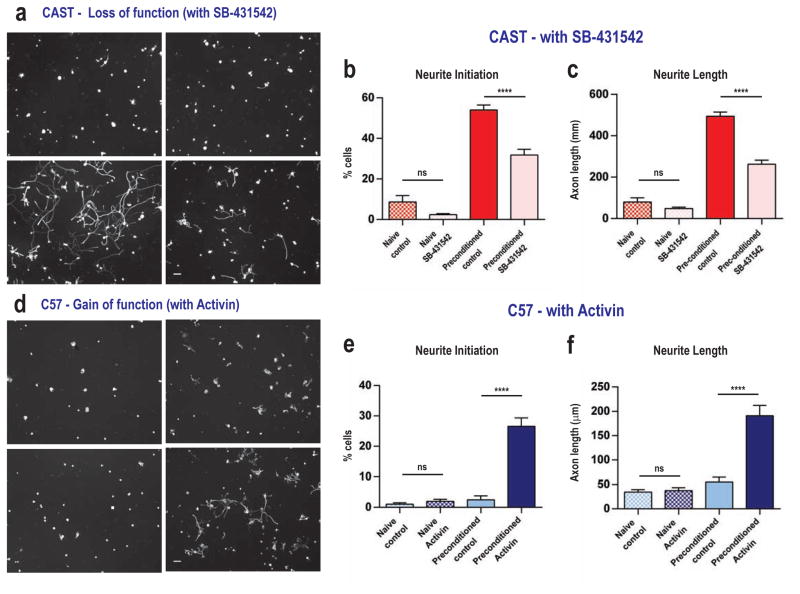
To evaluate whether Activin signaling is necessary for the elevated axonal growth of CAST/Ei DRG neurons on myelin, we cultured pre-conditioned CAST/Ei DRG neurons for 24 hours (the same protocol used for screening inbred strains) in the presence or absence of SB- 431542, a potent Activin-receptor antagonist (Laping et al., 2002). SB-431542 (100nM) significantly reduced neurite initiation (42% reduction, p<0.0001, 22.3 (2.6) CI: 15.6–28.9; two-way ANOVA, post-hoc Sidak’s) and maximal axon length (48% reduction, p<0.0001, 232.7 (21.9) CI: 176.6–288.9; two-way ANOVA, post-hoc Sidak’s), relative to vehicle controls (Figure 6a–6c).
Figure 6. Gain and loss of function assays of Activin action in promoting central axonal growth.
A–C: The activin receptor, type IB inhibitor SB-431542 (100nM) significantly decreases neuronal growth of pre-conditioned CAST/Ei DRG neurons on myelin, quantified in (b and c). D–F: A mixture of Activin-A, Activin-B and Activin-AB peptides (10 ng/ml) significantly increased axonal growth in pre-conditioned C57BL/6 DRG neurons, quantified in (e and f). Neuronal counts average of 4 wells per group, run 3 independent times as counted as in Figure 1a–c. For B, C, E, F: Mean ± SEM. ****p<0.0001 (two-way ANOVA, post-hoc Sidak’s). Scale, 100 μm.
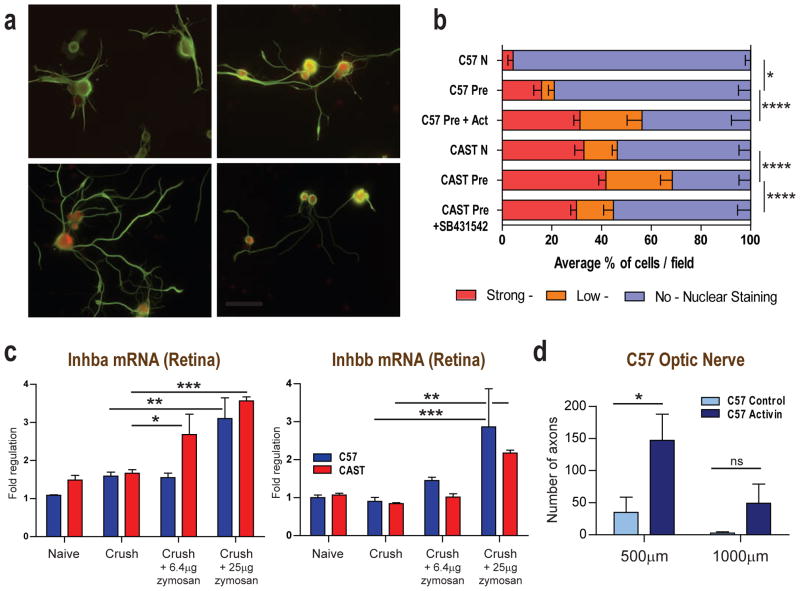
We also determined whether exogenous introduction of Activin ligands could promote axonal growth on myelin in C57BL/6 neurons. Dissociated pre-conditioned DRG neurons from C57BL/6 mice showed a 10.6-fold increase in sprouting (p<0.0001, 24.1 (2.2) CI: 18.8–29.4; two-way ANOVA, post-hoc Sidak’s) and a 3.6-fold increase in neurite elongation (p<0.0001, 135.6 (19.8) CI: 88.1–183.1; two-way ANOVA, post-hoc Sidak’s) on myelin when cultured in the presence of Activin A, B and AB peptides (10 ng/ml, Figure 6d–6f). To monitor Activin downstream signaling in these cells, we assayed nuclear pSmad2/3 localization, finding low overall levels in C57BL/6 naïve neurons (4.4% positive nuclei) with significantly more but still relatively low levels in pre-conditioned C57BL/6 neurons 21% (p<0.05, 0.19(0.05) CI: 0.03–0.35; Poisson Regression - rate estimate on a log scale, post-hoc Sidak’s) that increased robustly when treated with Activin to 56% nuclear staining (p<0.0001, 0.59(0.06) CI: 0.39–0.79; Poisson Regression - rate estimate on a log scale, post-hoc Sidak’s). Nuclear pSmad2/3 levels were relatively high in naïve dissociated CAST/Ei DRG neurons at 46%, but increased further after pre-conditioning to 69% (p<0.0001, 0.53(0.08) CI: 0.27–0.79; Poisson Regression - rate estimate on a log scale, post-hoc Sidak’s). In addition, the number of pSmad2/3-stained nuclei decreased from 69% to 45% when pre-conditioned CAST/Ei neurons were treated with SB-431542 (100nM) (p<0.0001, 0.56(0.08) CI: 0.31–0.81; Poisson Regression - rate estimate on a log scale, post-hoc Sidak’s; Figure 7a–b).
Figure 7. Molecular mechanisms of Activin function.
A: Representative cultured pre-conditioned C57BL/6 and CAST/Ei DRG neurons immunostained for pSmad2/3 and Tuj1 before and after Activin or SB-431542 treatment. Scale, 100 μm. B: Quantification of nuclear pSmad2/3 in naïve and preconditioned C57BL/6 and CAST/Ei DRG neurons treated with Activin or SB-431542. C: Q-PCR data showing that treating the mouse retina with 6.4μg intraocular zymosan increases Inhba mRNA expression only in CAST/Ei while 25μg induces regulation in both C57BL/6 and CAST/Ei animals. Introcular injection of 6.4μg and 25μg zymosan showing similar Inhbb regulation in C57BL/6 and CAST/Ei animals. D: A mixture of Activin-A, Activin-B and Activin-AB increases ON regeneration in C57BL/6 mice treated with 6.4μg of zymosan. Data quantified as numbers of optic nerve axons growing 500μm and 1000μm from crush site in presence of PBS or Activin peptide mix respectively For B: Mean ± SEM. Statistical analysis Poisson Regression, post-hoc Sidak’s. *p<0.05, ****p<0.0001. For C: Mean fold change ± SEM normalized to naïve C57BL/6 values, for comparisons shown *p<0.05, **p<0.01, ***p<0.001, (two-way ANOVA, post hoc Sidak’s). For D: Counts are average of 4 sections per animal, 5 animals. Average numbers of axons reaching defined distances from the crush site in each animal ± SEM; *p<0.05 (two-way ANOVA, post hoc Sidak’s). For in vitro gain of function assay using Activin peptides on rat retinal ganglion cells see Suppl Fig 7.
Zymosan at a dose (6.4μg) that produces extensive growth only in CAST/Ei mice, induced Inhba mRNA expression in the retina of this strain but not in retina from C57BL/6 mice, whereas a higher zymosan dose (25μg), which promotes regeneration in both strains, induced Inhba mRNA expression in both mouse lines (Figure 7c). To test if Activin signaling can boost RGC axonal regeneration in vivo we measured axonal growth in C57BL/6 optic nerves after crush where the retina had been treated with low dose (6.4μg) zymosan and then with 10 ng/ml of Activin immediately after optic nerve crush. This combination resulted in a 4.2-fold increase in the number of axons 500μm distal to the crush site relative to zymosan alone (p<0.05, 112.4 (39.5) CI: 14.9–209.9; two-way ANOVA, post-hoc Sidak’s; Figure 7d), showing that Activin enhanced CNS axonal regeneration in C57BL/6 mice after a low dose zymosan priming. Finally, we assayed if Activin promoted CNS neuron growth in another species, using rat RGCs in culture. Rat RGCs that had been injured and primed in vivo with low dose zymosan (6.4μg) showed a 2.8-fold increase in outgrowth when subsequently grown on PDL in vitro in response to the addition of Activin peptides (relative to control RGCs: p<0.01, Student’s t-test, two-tailed; Suppl Fig 7).
DISCUSSION
Neurons in the adult mammalian PNS have the capacity, also found in the CNS of some lower organisms, to mount a regenerative response following axonal damage. In stark contrast, this regenerative ability has been considered to be essentially lost in the mammalian adult CNS. Two factors have been considered cardinal to achieving long distance axonal growth in the adult mammalian CNS; removal of the growth inhibitory environment and initiation of intrinsic molecular growth programs that enable active axonal growth. Our data show that CAST/Ei mouse neurons, following a pre-conditioning injury, are not inhibited by extrinsic inhibitory cues from the CNS extracellular environment in the way that other mouse strains are. The extent of the CNS regeneration in primed RGCs from CAST/Ei mice (as determined by maximal axonal growth) is similar to that found when tonic growth suppressors are eliminated (Gaub et al., 2010; Sun et al., 2011). The discovery of this unexpected growth capacity of injured neurons in the CNS in this strain allow molecular mechanisms required to initiate and sustain regeneration in the adult mammalian CNS to be teased out and therapeutic implications studies without removal of tumor suppressor mechanisms, which although effective, carry an oncogenic risk (Sun et al., 2011).
We designed a cell-based phenotypic screen to look for heritable differences in neuronal growth capacity, using preconditioned DRG neurons on myelin to model one of the very few situations where axonal growth occurs in the CNS without genetic manipulation; namely when a prior peripheral nerve injury enables dorsal column fibers to regenerate into and beyond a spinal cord injury (Lu et al., 2004; Neumann et al., 2002; Neumann and Woolf, 1999). Our hypothesis was that differences in the growth of isolated DRG neurons across different mouse strains on a CNS inhibitory substrate might predict a differential capacity to grow within a CNS environment. Our screen revealed both that there is a large heritable component to the regeneration on myelin phenotype after preconditioning, and that CAST/Ei display a much higher level of growth in this circumstance than all the other strains. Furthermore, the growth of DRG neurons from this strain in vitro in this model accurately predicted increased growth in vivo of pre-conditioned dorsal column fibers after a spinal cord injury and RGCs axons after optic nerve crush when primed by zymosan, as well as an enhanced spatial reorganization of projections arising from cortical neurons after a stroke.
To successfully regrow after axonal injury, PNS neurons need to realign their genomic structure though epigenetic mechanisms (Cho et al., 2013; Finelli et al., 2013; Kiryu-Seo and Kiyama, 2011; Puttagunta et al., 2014) engage transcriptional programs with miRNAs (Li et al., 2012) and signaling pathways (Perry et al., 2012) to activate complex networks of transcription factors and repressors (Costigan et al., 2002; Costigan et al., 2010; Tedeschi, 2012). This injury response leads to the upregulation of the many genes required for initiating, guiding and sustaining axonal growth. Injuring the central axons of these DRG neurons, however, has no such effect (Ylera et al., 2009). Essentially peripheral nerve injury reprograms adult sensory and motor neurons from a quiescent to an active intrinsic axonal growth mode, but these changes are not a dedifferentiation to an immature state since most of the genes induced are not developmentally regulated. The increased intrinsic growth in DRG neurons after a peripheral axonal injury is accompanied by somewhat reduced responses to inhibitory cues, such as central myelin (Qiu et al., 2002). As such large scale transcriptional reprogramming does not occur after axonal injury in CNS neurons (Cho et al., 2013), this has been thought to an important contributor to the failure of CNS regeneration. We found that both C57BL/6 and CAST/Ei neurons display increased growth in a permissive environment after pre-conditioning, in agreement with multiple previous studies in rats and mice (Richardson and Issa, 1984; Smith and Skene, 1997) and that the extent of CAST/Ei pre-conditioned growth in the PNS was not markedly different from that in C57BL/6 mice. In keeping with this the major component of the transcriptional response to peripheral axonal injury (constituting preconditioning) was essentially identical across strains. Therefore an augmented or different transcriptional growth response after conditioning could not account for the superior regenerative capacity found for DRG neurons from CAST/Ei mice on myelin, opening up the possibility that the preconditioned growth on myelin phenotype results from a more discrete process, specifically activated in this strain by injury, which may add to the more generic pre-conditioned response that is uniform across strains. CAST/Ei mice seem to engage after preconditioning in the case of DRG neurons or zymosan priming for RGCs a specific transcriptional mechanism distinct from the standard induction of RAGs which enables them to grow well on myelin and CSPG, and within the CNS in vivo.
Our bioinformatic analysis for differential transcript expression correlates of the growth on myelin phenotype showed that induction of Activin-βA was a prime candidate. In naïve CAST DRG neurons, expression of transcripts that encode Activin-βA and Activin-βB peptides (Inhba and Inhbb) are low, but the expression of the Activin receptors are high. Following axonal injury, expression of Inhba and Inhbb mRNAs are upregulated, with the increase in Inhba being specific to this strain and leading to a markedly greater neuronal pSmad2/3 activation. How Activin signaling is selectively enhanced after injury in the CAST/Ei strain relative to the others, and the transcriptional cascades downstream of pSmad2/3 that increase axon growth and/or reduce responses to CNS inhibitory cues, will now need to be elucidated. Future work, for example by assaying growth variation on myelin in collaborative cross recombinant backcross strains (Threadgill et al., 2011), could help define the complete genomic pathway required for this growth phenotype.
To test if Activin-βA mechanistically contributed to the CNS regeneration phenotype, we used gain- and loss-of-function manipulations in vitro in DRG neurons and in vivo for RGCs. Antagonizing Activin/TGF-β signaling diminished CAST/Ei axonal growth after injury towards the level found in C57BL/6 mice, whereas conversely, Activin administered to preconditioned C57BL/6 neurons shifted regeneration capacity towards that present in CAST/Ei mice. The fact that Activin only induced active growth on myelin in pre-injured C57BL/6 DRG neurons means that other components induced by the conditioning injury need to be present for successful growth, components also provided by exposure to inflammation for RGCs, and we aim in the future to tease out what these are and how they interact with Activin.
A primordial function of Activin in the CNS is revealed by fly brain metamorphosis, a process that involves pruning of larval-specific dendritic and axonal processes and outgrowth of adult axonal structures. A genome-wide mutant screen for blockade of this growth plasticity resulted in only two genes, the drosophila Activin serine kinase receptor (Baboon) and dSmad2 (Zheng et al., 2003). In addition, the functional involvement of the two drosophila Activin co-receptors and dActivin in neuronal remodeling has since also been shown (Zheng et al., 2003; Zheng et al., 2006). Genome-wide studies have also found that Activin-A mediates tail regeneration in the leopard gecko (Gilbert et al., 2013) and fin re-growth in fish (Jazwinska et al., 2007). Activin can also promote embryonic chick DRG cell outgrowth (Fang et al., 2012) and its expression in the adult CNS, can be strongly regulated after injury (Tretter et al., 2000). In the injured CNS Activin promotes dendritic complexity (Ishikawa et al., 2010), modifying spine morphology and increasing the number of synaptic contacts on dendritic spines (Shoji-Kasai et al., 2007). Activin signaling through Smad2/3 is therefore a conserved neural/tissue regenerative pathway in flies, fish and birds; and as we now show, the pathway is also activated in the injured adult mammalian nervous system in a mouse strain in which, uniquely, substantial central axon regeneration can occur.
In conclusion, we have found that injured neurons in the mammalian central nervous system can unexpectedly mount a large regenerative response and show a central role for Activin signaling in mediating this, providing potential therapeutic opportunity for stroke and other forms of traumatic brain injury where injury and a priming inflammation co-exist. Identification of the regenerative potential of injured CNS neurons in the CAST/Ei strain provides an opportunity to begin to exhaustively interrogate molecular mechanisms that may enable CNS regeneration in mammals.
Methods
Additional information in supplemental materials.
Sciatic nerve crush
Sciatic nerve crush was performed by exposing the left sciatic nerve at the level of sciatic notch and crushed with smooth forceps for 30s.
Primary dissociated DRG cultures
For pre-conditioned DRG cultures, ipsilateral L4/L5 DRGs were collected 5 days after a sciatic nerve crush. Appropriate L4-5 DRGs from five mice (naïve or pre-conditioned) were pooled and processed in one tube per experiment as in (Ma et al., 2011; Seijffers et al., 2007). Once dissociated, 3,000 cells were grown per chamber in an 8-well slide, coated with myelin (100ng/well) or CSPGs (20ng/well, Millipore) and 1,500 cells were grown on laminin (5μg/well, Invitrogen).
Neurite growth assays
For quantification of sensory neuron culture experiments, neurons were plated in 8-well chamber slides. For data given in Figure 1a–1c: Nine non-overlapping low-power (4x) microscope images were taken per well, at least 4 wells were assayed per condition. All neurons in these images were analyzed by NeuroMath software. Neurons with neurites exceeding 50μm were defined as sprouting cells and the top 10% of the longest neurite lengths were quantified to obtain an average score, three independent experiments were performed. The values shown are the averages of these experiments and standard error of the mean (n=3). For data given in Figure 1d–1e: All neurons in at least 4 chambers of an 8-well slide were counted. Images were collected with ImageXpress micro High-Content Systems, neurite outgrowth was automatically quantified using the MetaXpress software.
Activin in-vitro gain and loss of function experiments
For gain of function experiment, either 10ng/ml of recombinant Activin A, B and AB (R&D, USA) or PBS containing 0.1% BSA was added into myelin coated eight chamber slides with 3,000 naïve or pre-conditioned C57BL/6 sensory neurons. For loss of function experiment, 3,000 naïve and pre-conditioned CAST/Ei sensory neurons were cultured on a myelin for 24 hours with 1% ethanol vehicle, 100nM SB 431542 (TOCRIS, USA).
Nuclear Smad2/3 counts: C57BL/6 and CAST/Ei DRG naïve and preconditioned DRG neurons cultured with or without the presence of either Activin or SB-431542 were assayed for nuclear pSmad2/3 expression by immunostaining (Cell Signaling, #8828). Cells were defined as either containing no, low or strong pSmad2/3 signal within the nucleus. Percentage of the cells lacking nuclear pSmad2/3 were compared. The values given are the averages of at least three independent experiments counted blind to treatment.
Spinal Cord Injury
Six-weeks-old mice were anesthetized with a mixture of ketamine/xylazine and received either sciatic nerve transection or sham operation. One week later, a T10 laminectomy was performed and the dorsal half of the spinal cord crushed with #5 modified forceps (Dumont, Fine Science Tools). The forceps were deliberately positioned to cut dorsal column axons completely. Six weeks after spinal cord injury, axons were traced by injecting 2 μl of 10% micro-Ruby tracer (3000 MW, Invitrogen) into the left sciatic nerve. Mice were kept for an additional two weeks before termination. For the quantification of dorsal column injury model, the retrogradely labeled axons in a series of regularly spaced sagittal sections (C57BL/6 n=5, CAST/Ei n=7) were imaged and the number of regenerating axons at different distances from the lesion epicenter was normalized to the number of labeled axons caudal (−0.5 mm/LS) to the lesion. The lesion site was defined according to GFAP immunostaining. Transverse sections of the spinal cord (3 mm caudal and 7–8 mm rostral to the lesion) were taken to confirm the completeness of the lesion and tracing efficiency among experimental groups. Mice with incomplete lesions were excluded from the analysis. Quantification was performed blinded to the treatment and the mouse strain.
Optic nerve injury
Optic nerve crush surgery and intraocular injections were performed under general anesthesia. The mice received either 6.4μg or 25μg of zymosan 5 days prior and immediately after optic nerve crush into the vitreous. Five days prior to sacrifice, CTB (3 μL, 1%; 103B, List Biological) was injected as an anterograde tracer. For C57BL/6 control n=5; 6.4μg n=4; 25μg n=5. For CAST/Ei control n=4; 6.4μg n=4; 25μg n=6. Quantification was performed blinded to the treatment and mouse strain.
Quantification of ON axon regeneration
Fourteen days after optic nerve injury, mice were perfused with saline and 4% PFA. Regenerating axons were visualized by staining with primary antibodies to CTB (1:500, GWB-7B96E4, GenWay). Axons were counted manually in at least eight longitudinal sections per case at pre-specified distances from the injury site, and these numbers were converted into the number of regenerating axons at various distances (Kurimoto et al., 2010).
Optic nerve gain of function experiment
C57BL/6 mice were injected with 6.4ug of zymosan 5 days prior and immediately after optic nerve crush to prime RGCs. The animals were then injected with either 0.1% BSA or 10ng/ml of Activin A, B and AB immediately after optic nerve crush. 5 mice were assayed for both control and activin treated groups. Animals were sacrificed 14 days after optic nerve injury and axonal regeneration quantified. Quantification as above was performed blinded to the treatment and mouse strain.
Stroke model
Stroke was produced in the forelimb motor cortex as described (Clarkson et al., 2013; Li et al., 2010; Overman et al., 2012) The neuroanatomical tracer biotinylated dextran amine (BDA) was injected into the motor cortex anterior to the stroke. The connections labeled by BDA in the ipsilateral hemisphere were quantified in tangential cortical sections and statistically tested between stroke and control in CAST/Ei and C57BL/6 as described (Clarkson et al., 2013; Li et al., 2010; Overman et al., 2012). For further details see supplemental materials.
Microarray Analysis
Total RNA was extracted from naïve and injured DRGs (6 DRGs per sample, 3 mice) across 9 strains and assessed with the Agilent Bioanalyzer (Agilent Technologies). Four biologically distinct replicates were run per sample for a total of 72 arrays. 200 ng of total RNA was amplified, biotinylated and hybridized to Illumina MouseRef-8 v2.0 Expression BeadChips. Raw data were analyzed using Bioconductor packages. Normalized expression values were used to compute Pearson correlations with phenotypic data.
RNAseq Analysis
Reads were quality controlled using FastQC, mapped to the reference genome using Bowtie2 (Langmead et al., 2009), counted using SummarizeOverlaps from the GenomicRanges package and genes were called differentially expressed using edgeR (Robinson et al., 2009). We searched for genes whose expression is significantly increased after pre-conditioning injury in the DBA/2 and CAST/Ei strain, to specifically select for genes responsible for the medium-low to strong growth phenotype using methods described in supplemental data. This resulted in a list of 16 genes for which we plotted the expression fold change (compared to the C57BL/6 naive expression level) across all 6 conditions.
Time course of expression of Inhba, Inhbb and Activin receptor subtype transcripts
Ipsilateral L4/5 DRGs were dissected from naïve mice and 1, 3, 5, 10 and 14 days post-sciatic nerve crush for both C57BL/6 and CAST/Ei. Each RNA sample was three animals pooled or 6 DRGs. Four distinct samples were collected for naïve (n=4) and three distinct samples for injured DRGs (n=3) for each time point were analyzed by QRT-PCR; Mean fold change ± SEM normalized to naïve C57BL/6 values. For the quantification of mRNA expression of the retina in naïve, post optic nerve crush, post optic nerve crush with 6.4μg zymosan and 25μg zymosan treated C57BL/6 and CAST/Ei, 3 retinas were pooled per RNA sample. Three independent RNA samples were analyzed by QRT-PCR (n=3); Mean fold change ± SEM normalized to naïve C57BL/6 values.
Supplementary Material
Acknowledgments
All: The Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. Members of the Kirby Neurobiology Center at BCH would like to thank the Intellectual and Developmental Disabilities Research Center (IDDRC) of Children’s Hospital (NIH P30 HD018655) for use of the Histology, Image Analysis, and Animal Behavior Cores. LIB: NIH (EY05690), Dept. of Defense (CDMRP DM102446). MC: NIH (NS074430), IRP-IFP Switzerland. CJW: NIH 2 R01 NS038153-15 and the Bertarelli Foundation. We thank Dr. Anthony Hill of the IDDRC Cellular Imaging Core for assistance with imaging and Dr. Nick Andrews for advice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alilain WJ, Horn KP, Hu H, Dick TE, Silver J. Functional regeneration of respiratory pathways after spinal cord injury. Nature. 2011;475:196–200. doi: 10.1038/nature10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benowitz LI, Popovich PG. Inflammation and axon regeneration. Curr Opin Neurol. 2011;24:577–583. doi: 10.1097/WCO.0b013e32834c208d. [DOI] [PubMed] [Google Scholar]
- Benowitz LI, Yin Y. Combinatorial treatments for promoting axon regeneration in the CNS: strategies for overcoming inhibitory signals and activating neurons’ intrinsic growth state. Dev Neurobiol. 2007;67:1148–1165. doi: 10.1002/dneu.20515. [DOI] [PubMed] [Google Scholar]
- Blesch A, Lu P, Tsukada S, Alto LT, Roet K, Coppola G, Geschwind D, Tuszynski MH. Conditioning lesions before or after spinal cord injury recruit broad genetic mechanisms that sustain axonal regeneration: superiority to camp-mediated effects. Exp Neurol. 2012;235:162–173. doi: 10.1016/j.expneurol.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HL, Jovanis PP. Formulating accident occurrence as a survival process. Accident; analysis and prevention. 1990;22:407–419. doi: 10.1016/0001-4575(90)90037-l. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mammalian genome : official journal of the International Mammalian Genome Society. 2008;19:382–389. doi: 10.1007/s00335-008-9135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155:894–908. doi: 10.1016/j.cell.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson AN, Lopez-Valdes HE, Overman JJ, Charles AC, Brennan KC, Thomas Carmichael S. Multimodal examination of structural and functional remapping in the mouse photothrombotic stroke model. J Cereb Blood Flow Metab. 2013;33:716–723. doi: 10.1038/jcbfm.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Befort K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan M, Belfer I, Griffin RS, Dai F, Barrett LB, Coppola G, Wu T, Kiselycznyk C, Poddar M, Lu Y, et al. Multiple chronic pain states are associated with a common amino acid-changing allele in KCNS1. Brain. 2010;133:2519–2527. doi: 10.1093/brain/awq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickendesher TL, Baldwin KT, Mironova YA, Koriyama Y, Raiker SJ, Askew KL, Wood A, Geoffroy CG, Zheng B, Liepmann CD, et al. NgR1 and NgR3 are receptors for chondroitin sulfate proteoglycans. Nat Neurosci. 2012;15:703–712. doi: 10.1038/nn.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Wang YN, Cui XL, Fang SY, Ge JY, Sun Y, Liu ZH. The role and mechanism of action of activin A in neurite outgrowth of chicken embryonic dorsal root ganglia. J Cell Sci. 2012;125:1500–1507. doi: 10.1242/jcs.094151. [DOI] [PubMed] [Google Scholar]
- Filbin MT. Myelin-associated inhibitors of axonal regeneration in the adult mammalian CNS. Nat Rev Neurosci. 2003;4:703–713. doi: 10.1038/nrn1195. [DOI] [PubMed] [Google Scholar]
- Finelli MJ, Wong JK, Zou H. Epigenetic regulation of sensory axon regeneration after spinal cord injury. J Neurosci. 2013;33:19664–19676. doi: 10.1523/JNEUROSCI.0589-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D, Xing B, Dill J, Li H, Hoang HH, Zhao Z, Yang XL, Bachoo R, Cannon S, Longo FM, et al. Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J Neurosci. 2011;31:14051–14066. doi: 10.1523/JNEUROSCI.1737-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol. 2008;209:294–301. doi: 10.1016/j.expneurol.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di Giovanni S. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell death and differentiation. 2010;17:1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- Gilbert RW, Vickaryous MK, Viloria-Petit AM. Characterization of TGFbeta signaling during tail regeneration in the leopard Gecko (Eublepharis macularius) Dev Dyn. 2013;242:886–896. doi: 10.1002/dvdy.23977. [DOI] [PubMed] [Google Scholar]
- Harrison CA, Gray PC, Koerber SC, Fischer W, Vale W. Identification of a functional binding site for activin on the type I receptor ALK4. J Biol Chem. 2003;278:21129–21135. doi: 10.1074/jbc.M302015200. [DOI] [PubMed] [Google Scholar]
- Hoffman PN. A conditioning lesion induces changes in gene expression and axonal transport that enhance regeneration by increasing the intrinsic growth state of axons. Exp Neurol. 2009;223:11–18. doi: 10.1016/j.expneurol.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Nishijima N, Shiota J, Sakagami H, Tsuchida K, Mizukoshi M, Fukuchi M, Tsuda M, Tabuchi A. Involvement of the serum response factor coactivator megakaryoblastic leukemia (MKL) in the activin-regulated dendritic complexity of rat cortical neurons. J Biol Chem. 2010;285:32734–32743. doi: 10.1074/jbc.M110.118745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Current biology : CB. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Kadoya K, Tsukada S, Lu P, Coppola G, Geschwind D, Filbin MT, Blesch A, Tuszynski MH. Combined intrinsic and extrinsic neuronal mechanisms facilitate bridging axonal regeneration one year after spinal cord injury. Neuron. 2009;64:165–172. doi: 10.1016/j.neuron.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryu-Seo S, Kiyama H. The nuclear events guiding successful nerve regeneration. Front Mol Neurosci. 2011;4:53. doi: 10.3389/fnmol.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto T, Yin Y, Habboub G, Gilbert HY, Li Y, Nakao S, Hafezi-Moghadam A, Benowitz LI. Neutrophils express oncomodulin and promote optic nerve regeneration. J Neurosci. 2013;33:14816–14824. doi: 10.1523/JNEUROSCI.5511-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto T, Yin Y, Omura K, Gilbert HY, Kim D, Cen LP, Moko L, Kugler S, Benowitz LI. Long-distance axon regeneration in the mature optic nerve: contributions of oncomodulin, cAMP, and pten gene deletion. J Neurosci. 2010;30:15654–15663. doi: 10.1523/JNEUROSCI.4340-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- Li S, Overman JJ, Katsman D, Kozlov SV, Donnelly CJ, Twiss JL, Giger RJ, Coppola G, Geschwind DH, Carmichael ST. An age-related sprouting transcriptome provides molecular control of axonal sprouting after stroke. Nat Neurosci. 2010;13:1496–1504. doi: 10.1038/nn.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Yu B, Wang S, Gu Y, Yao D, Wang Y, Qian T, Ding F, Gu X. Identification and functional analysis of novel micro-RNAs in rat dorsal root ganglia after sciatic nerve resection. J Neurosci Res. 2012;90:791–801. doi: 10.1002/jnr.22814. [DOI] [PubMed] [Google Scholar]
- Lu P, Yang H, Jones LL, Filbin MT, Tuszynski MH. Combinatorial therapy with neurotrophins and cAMP promotes axonal regeneration beyond sites of spinal cord injury. J Neurosci. 2004;24:6402–6409. doi: 10.1523/JNEUROSCI.1492-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CH, Omura T, Cobos EJ, Latremoliere A, Ghasemlou N, Brenner GJ, van Veen E, Barrett L, Sawada T, Gao F, et al. Accelerating axonal growth promotes motor recovery after peripheral nerve injury in mice. J Clin Invest. 2011;121:4332–4347. doi: 10.1172/JCI58675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills CD, Allchorne AJ, Griffin RS, Woolf CJ, Costigan M. GDNF selectively promotes regeneration of injury-primed sensory neurons in the lesioned spinal cord. Mol Cell Neurosci. 2007;36:185–194. doi: 10.1016/j.mcn.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann S, Bradke F, Tessier-Lavigne M, Basbaum AI. Regeneration of sensory axons within the injured spinal cord induced by intraganglionic cAMP elevation. Neuron. 2002;34:885–893. doi: 10.1016/s0896-6273(02)00702-x. [DOI] [PubMed] [Google Scholar]
- Neumann S, Woolf CJ. Regeneration of dorsal column fibers into and beyond the lesion site following adult spinal cord injury. Neuron. 1999;23:83–91. doi: 10.1016/s0896-6273(00)80755-2. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11:1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman JJ, Clarkson AN, Wanner IB, Overman WT, Eckstein I, Maguire JL, Dinov ID, Toga AW, Carmichael ST. A role for ephrin-A5 in axonal sprouting, recovery, and activity-dependent plasticity after stroke. Proc Natl Acad Sci U S A. 2012;109:E2230–2239. doi: 10.1073/pnas.1204386109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RB, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, Coppola G, McDonald MK, Gomes C, Geschwind DH, et al. Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttagunta R, Tedeschi A, Soria MG, Hervera A, Lindner R, Rathore KI, Gaub P, Joshi Y, Nguyen T, Schmandke A, et al. PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat Commun. 2014;5:3527. doi: 10.1038/ncomms4527. [DOI] [PubMed] [Google Scholar]
- Qiu J, Cai D, Dai H, McAtee M, Hoffman PN, Bregman BS, Filbin MT. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron. 2002;34:895–903. doi: 10.1016/s0896-6273(02)00730-4. [DOI] [PubMed] [Google Scholar]
- Richardson PM, Issa VM. Peripheral injury enhances central regeneration of primary sensory neurones. Nature. 1984;309:791–793. doi: 10.1038/309791a0. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, He Z, Silver J, Flanagan JG. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kasai Y, Ageta H, Hasegawa Y, Tsuchida K, Sugino H, Inokuchi K. Activin increases the number of synaptic contacts and the length of dendritic spine necks by modulating spinal actin dynamics. J Cell Sci. 2007;120:3830–3837. doi: 10.1242/jcs.012450. [DOI] [PubMed] [Google Scholar]
- Silver J, Schwab ME, Popovich PG. Central Nervous System Regenerative Failure: Role of Oligodendrocytes, Astrocytes, and Microglia. Cold Spring Harbor perspectives in biology. 2014 doi: 10.1101/cshperspect.a020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Skene JH. A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J Neurosci. 1997;17:646–658. doi: 10.1523/JNEUROSCI.17-02-00646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, He Z. Neuronal intrinsic barriers for axon regeneration in the adult CNS. Curr Opin Neurobiol. 2010;20:510–518. doi: 10.1016/j.conb.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi A. Tuning the orchestra: transcriptional pathways controlling axon regeneration. Front Mol Neurosci. 2012;4:60. doi: 10.3389/fnmol.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill DW, Miller DR, Churchill GA, de Villena FP. The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR journal / National Research Council, Institute of Laboratory Animal Resources. 2011;52:24–31. doi: 10.1093/ilar.52.1.24. [DOI] [PubMed] [Google Scholar]
- Tretter YP, Hertel M, Munz B, ten Bruggencate G, Werner S, Alzheimer C. Induction of activin A is essential for the neuroprotective action of basic fibroblast growth factor in vivo. Nat Med. 2000;6:812. doi: 10.1038/77548. [DOI] [PubMed] [Google Scholar]
- Yin Y, Cui Q, Li Y, Irwin N, Fischer D, Harvey AR, Benowitz LI. Macrophage-derived factors stimulate optic nerve regeneration. J Neurosci. 2003;23:2284–2293. doi: 10.1523/JNEUROSCI.23-06-02284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Henzl MT, Lorber B, Nakazawa T, Thomas TT, Jiang F, Langer R, Benowitz LI. Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat Neurosci. 2006;9:843–852. doi: 10.1038/nn1701. [DOI] [PubMed] [Google Scholar]
- Ylera B, Erturk A, Hellal F, Nadrigny F, Hurtado A, Tahirovic S, Oudega M, Kirchhoff F, Bradke F. Chronically CNS-injured adult sensory neurons gain regenerative competence upon a lesion of their peripheral axon. Current biology : CB. 2009;19:930–936. doi: 10.1016/j.cub.2009.04.017. [DOI] [PubMed] [Google Scholar]
- Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O’Connor MB, Lee CH, Lee T. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–315. doi: 10.1016/s0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zugates CT, Lu Z, Shi L, Bai JM, Lee T. Baboon/dSmad2 TGF-beta signaling is required during late larval stage for development of adult-specific neurons. EMBO J. 2006;25:615–627. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.