Abstract
Chimeric antigen receptors (CARs) targeting CD19 have mediated dramatic anti-tumor responses in hematologic malignancies, but tumor regression has rarely occurred using CARs targeting other antigens. It remains unknown whether the impressive effects of CD19 CARs relate to greater susceptibility of hematologic malignancies to CAR therapies, or superior functionality of the CD19 CAR itself. We discovered that tonic CAR CD3ζ phosphorylation, triggered by antigen-independent clustering of CAR scFvs, can induce early exhaustion of CAR T cells that limits anti-tumor efficacy. Such activation is present to varying degrees in all CARs studied, with the exception of the highly effective CD19 CAR. We further identify that CD28 costimulation augments, while 4-1BB costimulation ameliorates, exhaustion induced by persistent CAR signaling. Our results provide biological explanations for the dramatic anti-tumor effects of CD19 CARs and for the observations that CD19.BBz CAR T cells are more persistent than CD19.28z CAR T cells in clinical trials.
Introduction
Genetic engineering of T cells to express chimeric antigen receptors (CARs) is a promising new approach for adoptive immunotherapy of cancer. CARs are synthetic immune receptors that link antigen binding domains, commonly a single chain variable fragment (scFv), with T cell signaling domains to endow T cells with non-MHC restricted specificity for cell surface antigens1, 2. Recent clinical trials have demonstrated impressive activity of CD19 CAR T cells against B cell malignancies3–10. However, CARs targeting other antigens have thus far shown limited anti-tumor efficacy11–16. It remains unknown whether this reflects increased susceptibility of hematologic malignancies to this therapeutic approach, or superior functionality of the CD19 CAR constructs compared to CARs targeting other antigens.
It is well accepted that anti-tumor efficacy of adoptively transferred T cells requires efficient expansion and persistence in vivo17–20, but there is currently little understanding of how CAR structure impacts these properties. We have observed that human T cells expressing CARs not infrequently mediate potent in vitro cytolysis, but show limited expansion, persistence, and anti-tumor efficacy in immunodeficient xenograft models. T cell exhaustion is a major factor limiting anti-viral and anti-tumor responses in the setting of chronic antigen exposure21–28. Exhausted T cells have low proliferative and cytokine producing capacities, high rates of apoptosis, and express high levels of inhibitory receptors such as PD-1, TIM-3, and LAG-327, 28. Whether exhaustion plays a significant role in limiting CAR efficacy, and how CAR structural design impacts the development of exhaustion, has not been previously studied.
Here, we demonstrate that early T cell exhaustion is a primary factor limiting anti-tumor efficacy of CAR expressing T cells, and that CAR structure plays a central role in predisposing CAR T cells to chronic activation and exhaustion. By dissecting the basis for differential in vivo activity between CD19 vs. GD2 CARs that show equivalent in vitro cytotoxicity, we discovered that antigen-independent signaling can drive early exhaustion in CAR T cells and limit anti-tumor efficacy in vivo. Furthermore, we demonstrate that tonic activation and exhaustion is not unique to the GD2 CAR, but is present to varying degrees in all CARs tested, with the exception of the highly effective CD19 CAR.
Recent reports have demonstrated significantly enhanced persistence of CD19 CARs incorporating the 4-1BB costimulatory domain compared to those including the CD28 domain3, 4. Using the GD2 CAR as a model for chronic CAR signaling, we additionally identify that the CD28 endodomain augments, while the 4-1BB endodomain ameliorates, key aspects of exhaustion, whether induced by antigen-independent signaling or induced by persistent exposure to antigen. Therefore, this data provides a biological basis for the differential persistence observed using CD19 CAR T cells with CD28 versus 4-1BB costimulatory endodomains in clinical trials3, 4, and is the first to identify anti-exhaustion effects associated with 4-1BB signaling.
Results
GD2.28z CAR T cells have poor efficacy in vivo despite strong activity in vitro
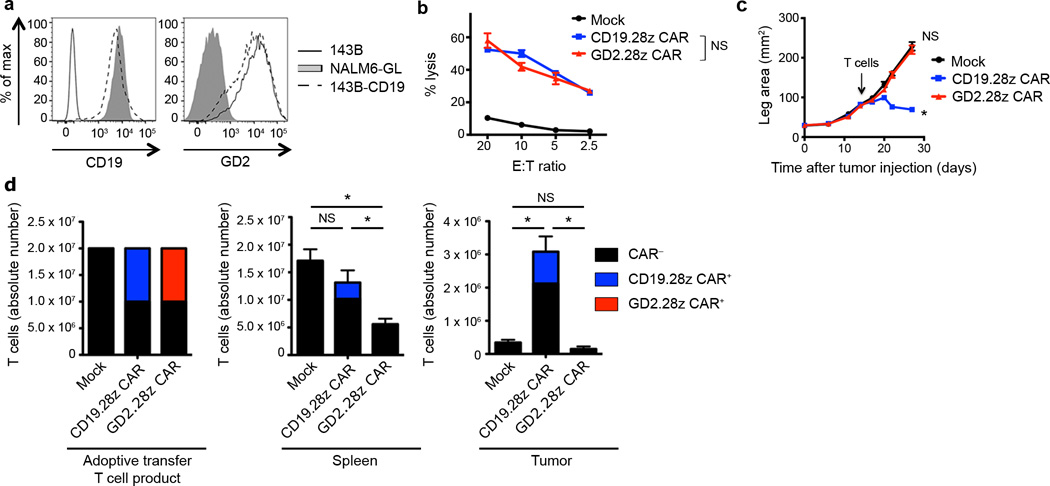
In an attempt to develop CAR therapies for GD2-expressing solid tumors, we designed a second-generation, GD2-specific CAR incorporating the scFv derived from the 14g2a antibody29, 30 plus the CD28 and CD3ζ signaling domains (Supplementary Fig. 1a). T cells expressing the GD2.28z CAR efficiently lysed the GD2+ osteosarcoma line 143B in vitro, but failed to mediate anti-tumor effects in vivo in a xenograft mouse model (Supplementary Fig. 2a–b). Poor in vivo activity could not be attributed to the incorporation of an IgG1 CH2CH3 spacer domain as recently reported31, because mice treated with a GD2.28z CAR without the spacer domain (GD2.sh.28z CAR) also showed no anti-tumor efficacy in vivo (Supplementary Fig. 2c). Interestingly, a CD19 specific CAR with identical signaling domains5, 32 (Supplementary Fig. 1a) and comparable in vitro activity produced rapid and complete eradication of the CD19+ NALM6-GL leukemia in vivo (Supplementary Fig. 2d–e). This discrepancy led us to explore whether differences between in vivo efficacy of GD2 vs. CD19.28z CARs, despite similar in vitro cytotoxic capacities, were related to differential potencies of the CARs themselves or differential susceptibility of these tumors to T cell therapies.
To control for tumor-associated differences, we stably expressed CD19 on the 143B osteosarcoma cell line (143B-CD19; Fig. 1a). In vitro cytolytic assays demonstrated that GD2.28z CAR and CD19.28z CAR T cells mediated comparable in vitro lysis of 143B-CD19 (Fig. 1b). However, notable differences in anti-tumor efficacy were observed in vivo. While GD2.28z CAR T cells had no effect on 143B-CD19 tumor growth rates, CD19.28z CAR T cells induced regression of established 143B-CD19 tumors (Fig. 1c). Tumors in CD19.28z CAR treated mice that eventually outgrew lacked CD19 expression (Supplementary Fig. 3a), whereas, tumors that outgrew in GD2.28z CAR T cell treated mice maintained GD2 expression (Supplementary Fig. 3b). Furthermore, anti-tumor efficacy correlated with CAR T cell expansion and persistence in vivo. While we readily identified CD19.28z CAR T cells in the spleen and tumor 14 days following adoptive transfer, GD2.28z CAR T cells could not be found (Fig. 1d). In summary, despite equivalent in vitro cytolytic efficacy, CD19.28z CAR T cells persist and eradicate all CD19+ tumor disease in vivo, while GD2.28z CAR T cells do not persist and do not impact tumor growth.
Figure 1.
GD2.28z CAR T cells have discrepant in vitro and in in vivo activity. (a) CD19 and GD2 antigen expression on the 143B-CD19 osteosarcoma line. Representative of n=5. (b) In vitro 51chromium release assay evaluating cytolytic activity of mock-transduced, CD19.28z CAR, and GD2.28z CAR T cells against 143B-CD19. Assay performed 9 days after initial activation. n=3 replicates/point; representative of 4 donors. (c) Growth curves of 143B-CD19 tumors in NSG mice. Mice were inoculated with 106 143B-CD19 periosteally on day 0 followed by adoptive transfer of 107 CAR transduced or mock-transduced T cells on day 14. Mock n=5, CD19.28z CAR n=7, GD2.28z CAR n=7. (d) Left: composition of T cell product adoptively transferred into mice in (c). Middle and right: quantification of T cells within the spleen and tumor 14 days following adoptive T cell transfer into mice from (c). n=8 mice/group. * = p<0.05 by Student’s T-test. Bar graphs represent mean ± SEM. In vivo results are representative of four experiments.
GD2.28z CAR T cells become exhausted during ex vivo expansion
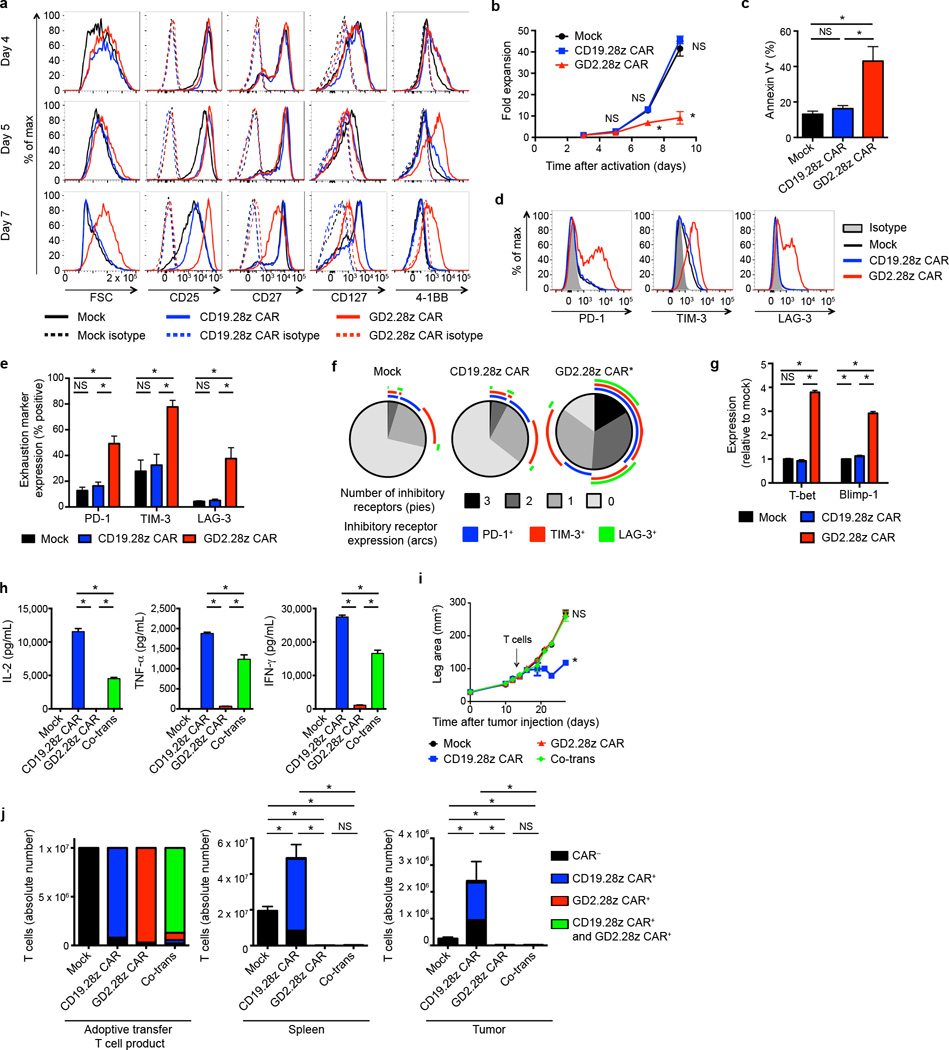
We next sought to characterize GD2.28z versus CD19.28z CAR T cells during ex vivo expansion (Supplementary Fig. 4). While T cell activation levels were indistinguishable on day 4, GD2.28z CAR T cells began to show increased size, higher CD25 and 4-1BB expression, and lower CD27 and CD127 expression, compared to CD19.28z CAR T cells or mock-transduced controls on days 5–7 (Fig. 2a). Despite increased activation, GD2.28z CAR T cells expanded less efficiently ex vivo (Fig. 2b) and showed higher rates of apoptosis (Fig. 2c). By day 9, GD2.28z CAR T cells showed a cell surface and transcriptional profile consistent with exhaustion, including higher expression of PD-1, TIM-3, and LAG-3 (Fig. 2d–f) and of exhaustion-associated transcription factors T-bet and Blimp-1 (Fig. 2g)28, 33–35. Furthermore, GD2.28z CAR T cells produce less than 100× the levels of IL-2, TNF-α , and IFN-γ compared to CD19.28z CAR T cells following exposure to 143B-CD19 (Fig. 2h). We could find no evidence that IL-2 added to the culture media substantially impacted exhaustion, and this phenotype could not be rescued with the addition of IL-7 into the culture media (Supplementary Fig. 5). Together, these phenotypic, functional, and transcriptional studies demonstrate that GD2.28z CAR T cells rapidly become exhausted during ex vivo culture, whereas similar effects do not occur in mock-transduced and CD19.28z CAR T cells stimulated in the same manner.
Figure 2.
GD2.28z CAR T cells become exhausted during ex vivo expansion. (a) Activation marker expression 4–7 days after initial activation. Representative of 3 donors. (b) Expansion during ex vivo culture. n=3 replicates/point; representative from 3 donors. (c) Quantification of apoptosis of CAR T cells generated from 4 unique donors evaluated 9–10 days following initial activation. (d) Representative exhaustion marker expression 9 days following initial activation. (e) Quantification of exhaustion marker expression pooled from 4 donors, 9–11 days following activation. (f) Exhaustion marker SPICE55 analysis from (e). (g) ΔΔCT q-RT-PCR expression levels of exhaustion transcription factors relative to mock, 9–11 days following initial activation. n=4 technical replicates; representative of 3 donors. (h) Cytokine production of flow-sorted single-transduced (CD19.28z CAR+ or GD2.28z CAR+) and co-transduced (Co-trans; CD19.28z CAR+ and GD2.28z CAR+) T cells, co-incubated with 143B-CD19 at 9 days after initial activation. n=3 replicates/group; representative of 3 donors. T cells with media: <5 pg/mL IL-2, TNF-α and IFN-γ. (i) Tumor growth curves of NSG mice inoculated with 106 143B-CD19 periosteally on day 0 followed by adoptive transfer of 107 transduced CAR T cells on day 14. n=5 mice/group. (j) Left: composition of T cell product adoptively transferred into mice in (i). Middle and right: quantification of T cells within the spleen and tumor 14 days following adoptive T cell transfer into mice from (i). n=6 mice/group. SPICE analysis: * = p<0.05 by Wilcoxon signed rank test. All other data: * = p<0.05 by Student’s T-test. Bar graphs represent mean ± SEM.
GD2.28z CAR has a dominant inhibitory effect on CAR T cell efficacy
To confirm that poor cytokine production by the GD2.28z CAR T cells was a result of T cell exhaustion rather than differences in the potency of CAR-antigen interactions, we co-transduced T cells to express both CD19.28z and GD2.28z CARs. Flow-sorted CD19.28z CAR+ GD2.28z CAR+ T cell populations (Supplementary Fig. 6a) produced lower amounts of IL-2, TNF-α and IFN-γ compared to T cells expressing the CD19.28z CAR alone when exposed to 143B-CD19 (Fig. 2h) and NALM6-GL (Supplementary Fig. 6b). Therefore, the GD2.28z CAR exerts a dominant, inhibitory effect on the ability of T cells to produce cytokines in response to antigen. In vivo experiments from Fig. 1c–d were also repeated to include T cells co-transduced with both the CD19.28z CAR and GD2.28z CAR (Supplementary Fig. 6c). A dominant, inhibitory effect of the GD2.28z CAR was also observed in vivo (Fig. 2i–j and Supplementary Fig. 3a). These data demonstrate that the inferior activity of the GD2.28z CAR is not related to poor CAR-antigen interactions or impaired signaling upon exposure to antigen, but rather reflects exhaustion associated with expression of the GD2.28z CAR.
CAR signaling during ex vivo expansion leads to exhaustion
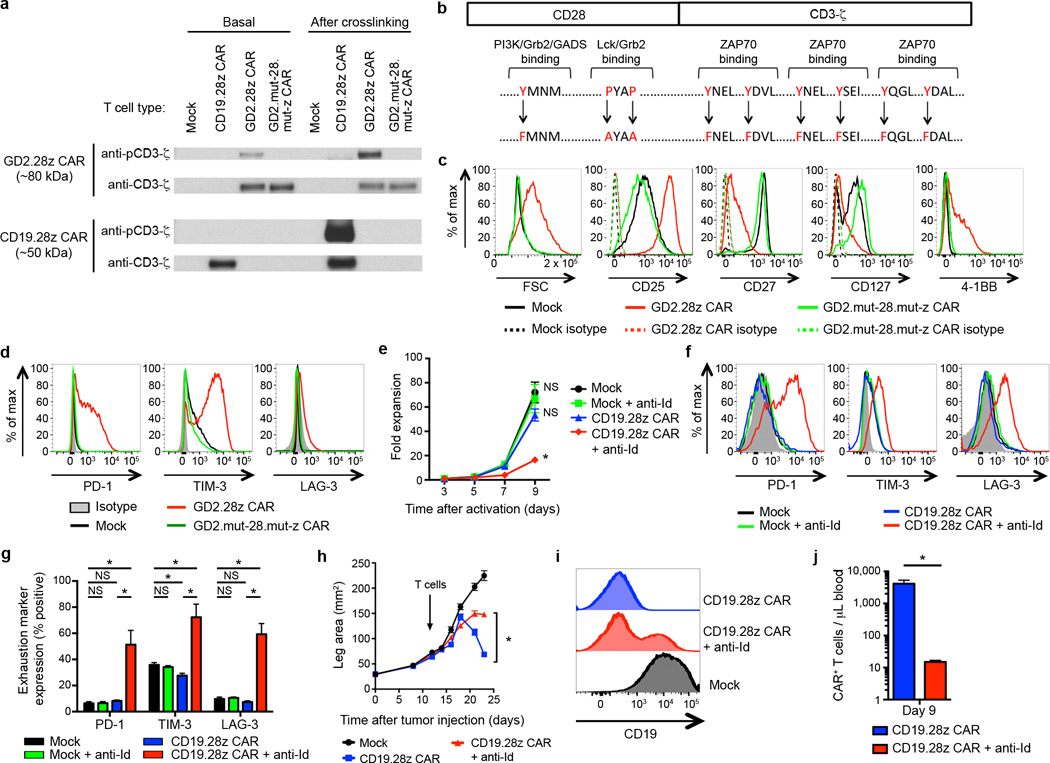
Both T cells bearing the CD19.28z CAR or the GD2.28z CAR were expanded ex vivo using anti-CD3/CD28 beads, yet only the GD2.28z CAR T cells showed early exhaustion. We found no evidence that exposure to CD19+ B cells in the initial PBMC culture could protect CD19.28z CAR T cells from exhaustion (Supplementary Fig. 7), leading us to hypothesize that signaling through the GD2.28z CAR itself played an essential role in inducing early exhaustion. To evaluate signaling of the GD2.28z and CD19.28z CARs, we probed CAR phosphorylation status via western blots with phospho-CD3ζ specific antibodies. The GD2.28z CAR demonstrated basal levels of zeta phosphorylation (Fig. 3a), while the CD19.28z CAR showed zeta phosphorylation only following crosslinking with an anti-idiotype antibody. These data demonstrate tonic signaling via the GD2.28z CAR during ex vivo expansion, and an absence of such signaling via the CD19.28z CAR.
Figure 3.
Tonic CAR signaling during ex vivo expansion leads to early exhaustion. (a) Western blot evaluating phosphorylation levels of CAR signaling domains versus total CAR signaling domains, using an anti-phospho-CD3ζ and anti-CD3ζ antibody, respectively. Evaluated day 5 after initial activation. “Basal” phosphorylation evaluated without further stimulation. “After crosslinking” evaluated following incubation with OKT3, anti-CD19 CAR idiotype, or anti-GD2 CAR idiotype antibodies. Representative of 3 donors. (b) Nine amino acid point mutations were introduced to eliminate signaling via the GD2.28z CAR (GD2.mut-28.mut-z CAR). (c) Activation and (d) exhaustion marker expression of GD2.mut-28.mut-z CAR T cells 10 days after initial activation. Representative of 3 donors. (e) Ex vivo expansion and (f-g) exhaustion marker expression of CD19.28z CAR T cells cultured ± anti-idiotype antibody (anti-Id; 0.25 ug/ml) and a crosslinking F(ab’)2 (2.5 ug/ml). n=4 replicates/group; representative of 3 donors. (h) Tumor growth curves of NSG mice inoculated with 106 143B-CD19 periosteally on day 0 followed by adoptive transfer of 107 transduced CAR T cells on day 14. Mice received mock-transduced or CD19.28z CAR cultured with or without anti-idiotype antibody. No anti-idiotype antibody was given to mice. n=8 mice/group. (i) CD19 expression on tumors 10 days following adoptive transfer into mice from (h). (j) Quantification of T cells within the blood 9 days following adoptive T cell transfer into mice from (h). n=3 mice/group. In vivo results are representative of two experiments. * = p<0.05 by Student’s T-test. Bar graphs represent mean ± SEM.
To confirm an essential role for GD2.28z CAR signaling in the development of exhaustion, we knocked out GD2.28z CAR signaling via amino acid point mutations in the SH2/SH3 binding sites on CD28 that facilitate PI3K and LCK binding36, 37, and the three ITAMs required for CD3ζ signaling38 (GD2.mut-28.mut-z CAR; Fig. 3b and Supplementary Fig. 1a). T cells bearing the GD2.mut-28.mut-z CAR showed no signs of over-activation (Fig. 3c) and did not develop exhaustion (Fig. 3d), despite equivalent cell surface expression levels compared to the wild type (WT) GD2.28z CAR (Supplementary Fig. 8a). Furthermore, tonic CD19.28z CAR signaling induced by culturing T cells in the presence of a cross-linking anti-idiotype antibody replicated the exhaustion phenotype in CD19.28z CAR T cells in vitro (Fig. 3e–g) and in vivo (Fig. 3h–j). Together, these data led us to propose that tonic signaling via the GD2.28z CAR results in early exhaustion.
Tonic GD2.28z CAR signaling is antigen independent
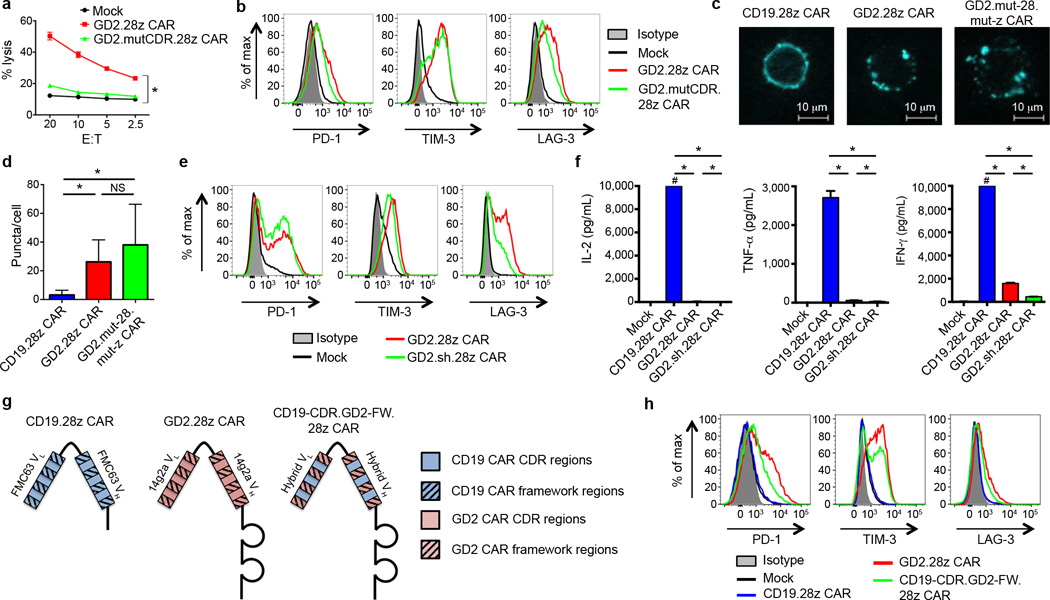
To identify factors responsible for tonic GD2.28z CAR signaling, we looked for the presence of GD2 in the culture system. Activated T cells expressed very low levels of the enzymes responsible for GD2 synthesis (36 fold less GD3 synthase and 205 fold less GD2 synthase than GD2+ tumor lines) and we could find no evidence for surface GD2 expression on activated T cells (Supplementary Fig. 9a–b). However to rule out the possibility that low levels of surface GD2, soluble GD2, or another antigen that cross-reacted with the complementarity determining (CDR) regions were responsible for tonic GD2.28z CAR signaling, we introduced seven point mutations into the antigen binding domain of the GD2.28z CAR to abrogate antigen binding (GD2.mutCDR.28z CAR; Supplementary Fig. 8b). The GD2.mutCDR.28z CAR expressed efficiently on the T cell surface, but did not lyse GD2 positive tumors (Fig. 4a), nor bind the 14g2a anti-idiotype antibody (1A7; Supplementary Fig. 8c). Despite the inability to bind antigen, GD2.mutCDR.28z CAR expressing T cells continued to demonstrate early exhaustion (Fig. 4b). We also synthesized a soluble version of the GD2 and CD19 scFvs to assess binding to cross-reactive antigens on the surface of activated T cells, but none were identified (Supplementary Fig. 9c). We conclude that tonic signaling via the GD2.28z CAR was not due to GD2 or a cross-reactive antigen in the culture system.
Figure 4.
Tonic GD2.28z CAR signaling is antigen independent. (a) In vitro 51Cr release assay against the 143B-CD19 osteosarcoma cell line. Both GD2.28z CAR and GD2.mutCDR.28z CAR had comparable transduction efficiencies (85 and 81%, respectively). Assay performed 9 days after initial T cell activation. n=3 replicates/point; representative of 3 donors. (b) Exhaustion marker expression on GD2.mutCDR.28z CAR vs. GD2.28z CAR T cells. Representative of 3 donors. (c) Fluorescence microscopy of T cells transduced with CAR-Cerulean fusion proteins (blue). (d) Quantification of the number of puncta per cell from (c). 30 cells per group; repeated for 3 donors. (e) Exhaustion marker expression levels of a version of the GD2.28z CAR designed to be structurally more like the CD19.28z CAR, based upon removal of the IgG1 hinge and substitution of the GD2 linker with the CD19 linker (GD2.sh.28z CAR). (f) Cytokine release levels upon co-incubation with 143B-CD19 on day 10 of ex vivo culture. # designates values >10,000 pg/mL. n=3 replicates/group; representative of 3 donors. (g) Structure and (h) exhaustion marker expression of a hybrid CAR combining the CDR regions of the CD19.28z CAR and the framework regions of the GD2.28z CAR (CD19-CDR.GD2-FW.28z CAR), 9 days post activation during ex vivo culture. Representative of 3 donors. For cell puncta analysis, * = p<0.0001 by Kolmogorov-Smirnov test. All other data, * = p<0.05 by Student’s T-test. Bar graphs represent mean ± SEM.
Tonic GD2.28z CAR signaling is due to CAR clustering
The antigen independence of tonic GD2.28z CAR signaling suggested that structural components of the GD2.28z CAR, separate from the antigen-binding site, were responsible for the constitutive signaling. Given numerous studies describing scFv oligomerization in vitro39–41, we hypothesized that GD2.28z CAR scFv self-interactions may lead to receptor clustering and tonic signaling. Using CAR-fluorescent protein fusion constructs that retained functionality (Supplementary Fig. 10a), we observed that the CD19.28z CARs uniformly distribute across the T cell membrane, whereas GD2.28z CARs aggregate in punctae on the surface (Fig. 4c–d, Supplementary Fig. 10b). Clustering was not due to an active capping process secondary to signaling through the CAR receptor, as GD2.mut-28.mut-z CAR T cells also demonstrated a punctated phenotype. Consistent with this, T cells co-transduced with both CAR-Cerulean and CAR-Venus proteins showed a higher intensity of Förster resonance energy transfer (FRET) signaling in GD2.28z CAR T cells compared to CD19.28z CAR T cells (Supplementary Fig. 10c). Together, this data is consistent with a model where tonic signaling of the GD2.28z CAR results from antigen-independent physical interactions between CAR receptors, leading to self-association and aggregation on the cell surface.
Two structural elements, the linker and the spacer domain, were distinct between the CD19.28z and GD2.28z CARs, and thus we substituted the CD19 versions into the GD2.28z CAR to evaluate the impact on early exhaustion. Neither incorporation of the CD19 linker nor removal of the IgG1 spacer domain (GD2.sh.28z CAR; Supplementary Fig. 1a and Supplementary Fig. 8d) reduced CAR exhaustion (Fig. 4e–f). In contrast, replacing the CD19.28z CAR scFv framework regions with those of the GD2.28z CAR scFv induced early exhaustion in the CD19.28z CAR (CD19-CDR.GD2-FW.28z CAR; Fig. 4g–h). Although attempts were made to prevent exhaustion in the GD2.28z CAR by substitution of the CD19 scFv framework regions, CARs with this structure were not expressed on the T cell surface and hence could not be adequately evaluated. Together, we conclude that framework regions within the 14g2a scFv are sufficient to induce tonic signaling, over-activation, and early exhaustion in CAR T cells, which substantially limits anti-tumor efficacy.
Exhaustion occurs to variable degrees across several CARs
To evaluate whether ex vivo exhaustion is a phenomenon unique to the GD2.28z CAR, we evaluated activation and exhaustion in T cells expressing three other CARs: two CARs specific for CD2220 (HA22 or m971 scFvs) and one CAR specific to ErbB238 (4D5 scFv), all incorporating CD28-CD3ζ signaling domains identical to the GD2.28z CAR (Supplementary Fig. 1b). We observed varying degrees of activation following ex vivo expansion, with the CD22-HA22 28z CAR T cells showing lower level activation, while CD22-m971.28z and ErbB2.28z CAR T cells showed higher levels of activation (Supplementary Fig. 11a). The degree to which these cells showed increased activation during ex vivo expansion correlated with an increase in exhaustion marker expression (Supplementary Fig. 11b–d) and exhaustion-association transcription factor expression (Supplementary Fig. 11e). We conclude that tonic activation leading to early exhaustion is likely present at varying degrees in most scFv based CARs, with the exception of the CD19 CAR.
4-1BB endodomains ameliorate exhaustion of CAR T cells
Recent clinical trials have demonstrated differing persistence rates of CD19 CAR T cells incorporating CD28 versus 4-1BB costimulatory domains3, 4. Because exhausted T cells are known to persist poorly in vivo, we sought to evaluate whether the costimulatory domains impact the development of exhaustion in the setting of chronic CAR signaling. Using the GD2 CAR as a model for chronic CAR signaling, we compared results in cells bearing the WT GD2.28z CAR (with both CD3ζ and CD28) versus CARs capable of signaling via CD3ζ alone and CD28 alone (Supplementary Fig. 1a and Supplementary Fig. 8e). These studies confirmed the essential role for CD3ζ signaling in exhaustion (Supplementary Fig. 12a–b). Importantly however, signaling through the WT receptor (which incorporates both CD3ζ and CD28) led to higher expression of exhaustion markers and exhaustion-related transcription factors compared to signaling via CD3ζ alone. Thus, CD28 endodomains incorporated into the CAR receptor augment the development of exhaustion in this model system.
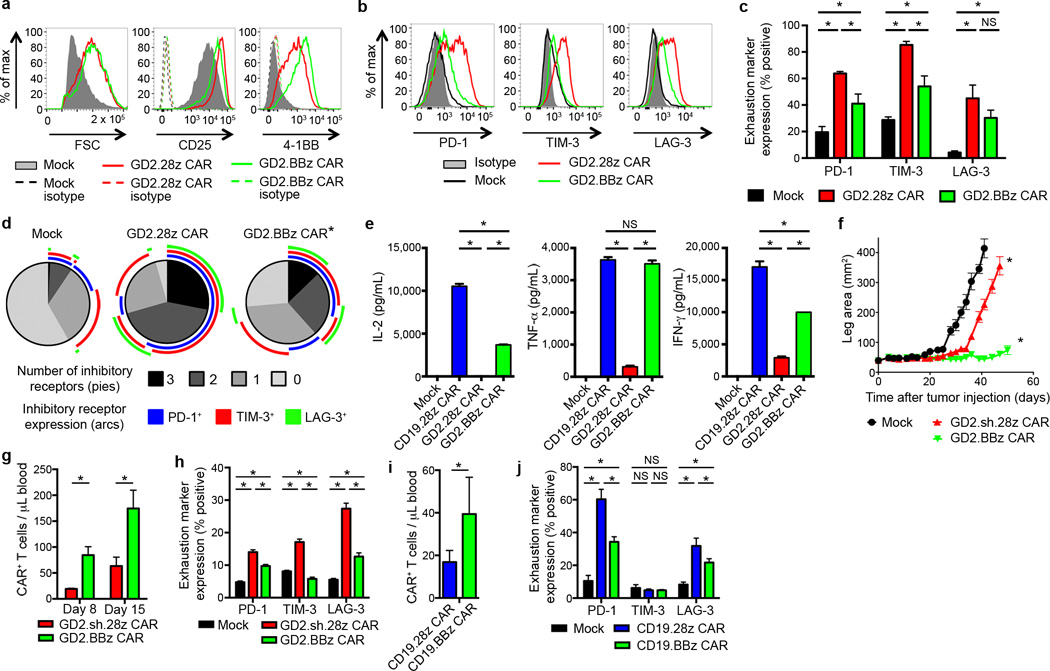
We next sought to investigate whether 4-1BB endodomains impacted the development of exhaustion in this system by developing a GD2 CAR incorporating 4-1BB rather than CD28 (GD2.BBz CAR; Supplementary Fig. 1a and Supplementary Fig. 8f). While GD2.BBz CAR T cells showed similar levels of over-activation compared to GD2.CD28z CAR T cells (Fig. 5a), GD2.BBz CAR T cells expressed lower levels of exhaustion markers (Fig. 5b–d), produced higher levels of cytokines (Fig. 5e) and mediated improved anti-tumor effects in vivo (Fig. 5f). GD2.BBz CAR T cells also demonstrated increased persistence in vivo (Fig. 5g) and decreased exhaustion marker expression in vivo (Fig. 5h). Similarly, we observed improved persistence and anti-tumor efficacy with CD22-m971.BBz CARs versus CD22-m971.28z CARs (Supplementary Fig. 1b and Supplementary Fig. 13), which also show evidence of tonic activation during ex vivo expansion. Furthermore, 4-1BB has ameliorating effects on exhaustion in the CD19 CAR, where we find improved persistence and decreased exhaustion marker expression of T cells in vivo following treatment of leukemic mice (Fig. 5i–j and Supplementary Fig. 12c). We conclude that CD28 endodomains augment exhaustion whereas 4-1BB endodomains provide an anti-exhaustion signal that can mitigate adverse effects induced by chronic CAR signaling. Anti-exhaustion effects of 4-1BB endodomains can diminish exhaustion induced by antigen-independent signaling or that induced by high tumor burdens.
Figure 5.
4-1BB endodomain ameliorates exhaustion in CAR T cells. (a) Activation marker expression 7 days following initial activation and (b) exhaustion marker expression 9 days after initial activation of GD2 CARs with the CD28 or 4-1BB co-stimulatory domains. (Representative of 4 donors). (c) Quantification of exhaustion marker expression pooled from 4 donors, 9–11 days following initial activation. (d) Exhaustion marker SPICE55 analysis of (c). (e) Cytokine release upon co-incubation with 143B-CD19 for 24 hours starting on day 10 following initial activation. n=3 replicates/group; representative of 3 donors. (f) Tumor growth curves of NSG mice inoculated with 5 × 105 143B-CD19 periosteally on day 0, then treated with 3 mg cyclophosphamide intraperitoneally on day 2 and with adoptive transfer of 107 transduced CAR T cells on day 4. n=10 mice/group. (g) Quantification of T cells within the blood 8 and 15 days following adoptive transfer into mice from (f). n=8 mice/group. (h) Exhaustion marker expression of CAR T cells from (f) on day 14 following adoptive transfer. (i) Quantification of T cells within the blood of NSG mice inoculated with 106 NALM6-GL followed by adoptive transfer of 5×106 CAR T cells three days later. T cells were quantified on day 8 following adoptive transfer. n=5 mice/group. (j) Exhaustion marker expression of CAR T cells from (i). In vivo results are representative of three experiments. SPICE analysis: * = p<0.05 by Wilcoxon signed rank test. All other data: * = p<0.05 by Student’s T-test. Bar graphs represent mean ± SEM.
4-1BB amelioration of exhaustion is associated with a unique transcriptional profile
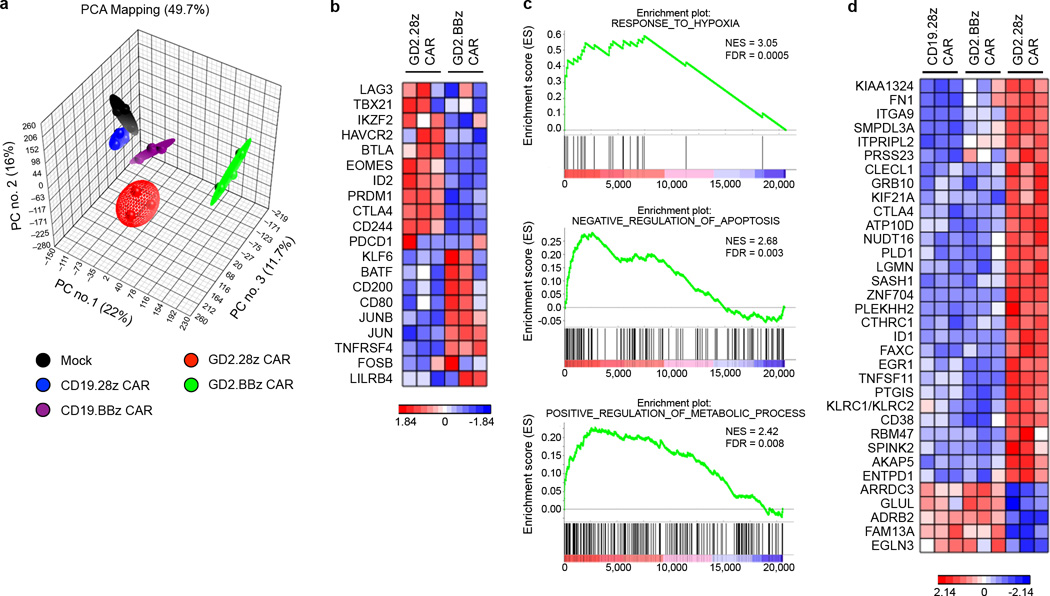
We next sought to explore molecular pathways that contribute to the ameliorating effect of 4-1BB signaling on CAR T cell exhaustion. Global transcriptional profiles of non-transduced and CAR T cells from three donors were analyzed 9 days after initial activation (Supplementary Table 1). Principal component analysis demonstrated that despite donor variability, transcriptional profiles clustered based on the CAR expressed (Fig. 6a). T cells expressing CD19.28z and CD19.BBz CARs had global transcriptional profiles most similar to untransduced T cells, while T cells expressing the GD2.28z or GD2.BBz CARs had profiles distinct from the untransduced/CD19 CAR T cells, and distinct from one another.
Figure 6.
Ameliorating effect of 4-1BB signaling is associated with a unique transcriptional profile.
(a) Principal component analysis (PCA) of global transcriptional profiles of CAR T cells generated from 3 unique healthy donors, evaluated 9 days following initial activation. Transduction efficiency was >90% for all samples. (b) Heatmap of genes previously reported to impact T cells exhaustion43. Exhaustion related transcription factors (TBX21, EOMES, PRDM1, IKZF2), inhibitory receptors (LAG3, HAVCR2, CTLA4, BTLA, CD244), and transcription factors reported to be preferentially expressed in memory vs. exhausted cells (KLF6, JUN, JUNB) were compared. (c) Representative GSEA results from running the unfiltered GD2.BBz CAR vs. GD2.28z CAR T cell rank list against the MSigDB C5 gene ontology sets. (d) Heatmap of genes uniquely dysregulated in exhausted GD2.28z CAR T cells. Genes are those up/downregulated > 2 fold in GD2.28z CAR vs. CD19.28z CAR T cells, up/downregulated > 2 fold in GD2.28z CAR vs. GD2.BBz CAR T cells, and < 2 fold difference in GD2.BBz CAR vs. CD19.28z CAR T cells. Genes associated with response to hypoxia (EGLN3, EGR, PTGIS, ID1), apoptosis (ID1), and metabolism (GLUL, ATP10D, SMPDL3A), or T cell suppressive pathways (CTLA4, CD38, LGMN, CLECL1, ENTPD1, KLRC1/2) were identified.
Gene set enrichment analysis (GSEA) demonstrated that GD2.28z CAR T cells showed enrichment of genes previously identified as up-regulated in exhausted T cells in mouse models42 (Supplementary Fig. 14a), confirming that GD2.28z CAR T cells are biologically similar to previously published models of T cell exhaustion. While some exhaustion genes were also dysregulated in GD2.BBz CAR T cells (Supplementary Fig. 14b), several key genes previously described to contribute to exhaustion43 were differentially expressed between the GD2.28z and GD2.BBz CAR T cells (Fig. 6b), and thus may contribute to the improved functionality of GD2.BBz CAR T cells. In particular, GD2.28z CAR T cells showed higher expression of genes encoding for inhibitory receptors such as LAG3, HAVCR2 (TIM-3), CTLA4, BTLA, and CD244 (2B4), and exhaustion-related transcription factors TBX21 (T-bet), EOMES, PRDM1 (Blimp-1), and IKZF2 (Helios). GD2.BBz CAR T cells on the other hand expressed higher levels of transcription factors reported to be associated with memory, such as KLF6, JUN and JUNB43. Together, these results suggest quantitative reductions in some exhaustion-associated molecules may contribute to the improved functionality observed in GD2.BBz CAR T cells.
Using GSEA with the Molecular Signatures Database to conduct unbiased comparisons of GD2.28z versus GD2.BBz CAR T cells, we also identified several novel gene sets enriched in GD2.BBz T cells which could also contribute to the improved functionality observed. Many of the enriched gene sets were associated with response to hypoxia, including 32 gene sets from 16 publications related to hypoxia responses (Fig. 6c and Supplementary Fig. 14c). Other enriched gene sets included negative regulation of apoptosis/programmed cell death and genes involved in regulation of cellular metabolism (Fig. 6c). Further, a three-way comparison of the transcriptional profiles of GD2.28z, GD2.BBz and CD19.28z CAR T cells was employed to identify those genes specifically “dysregulated” in GD2.28z CAR T cells, but “normalized” in GD2.BBz CAR T cells (Fig. 6d). Similar to the GSEA analyses, this analysis identified a set of 34 genes, many of which have been previously associated with the response to hypoxia (EGLN3, EGR1, PTGIS, ID1), apoptosis (ID1), metabolism (GLUL, ATP10D, SMPDL3A), or T cell suppressive pathways (CTLA4, CD38, LGMN, CLECL1, ENTPD1, KLRC1/2). Taken together, our data suggest that 4-1BB ameliorates exhaustion in part by diminishing expression of known exhaustion related genes, but may also modulate metabolism, apoptosis and/or response to hypoxia pathways.
Discussion
Impressive early clinical results reported following administration of CD19 CAR T cells demonstrate the promise that CAR therapies hold as an immune-based treatment of cancer3–10. However thus far, this success has not been translated beyond CD19+ hematologic malignancies and it remains unclear to what extent this relates to variable potencies of the chimeric receptors themselves. Here, we provide the unexpected observation that GD2 CARs incorporating the scFv derived from the well-studied 14g2a antibody tonically signal during anti-CD3/CD28 based ex vivo expansion, which induces rapid exhaustion in CAR T cells. This exhaustion appears indistinguishable from exhaustion occurring in non-engineered T cells in the setting of cancer and chronic viral infection21–25, 27, 28 and is characterized by altered transcriptional profiles, high expression of exhaustion markers, poor proliferative capacity, poor cytokine production, and poor anti-tumor efficacy in vivo.
The observation that some CARs tonically signal, leading to impaired T cell function and exhaustion, illustrates the importance of optimizing CAR engineering and a need for enhanced understanding of how receptor structure impacts function. Our work contributes to a growing literature demonstrating that structural characteristics of CAR ectodomains can critically impact functionality of these novel receptors31, 44, 45. Additionally, a recent report by Frigault et al. identified that some CARs constitutively signal, which is associated with inferior in vivo efficacy46. Our results likely explain this phenomenon by directly demonstrating CAR aggregation, CAR CD3ζ domain phosphorylation, tonic T cell activation, and ultimately exhaustion. We further localize the cause of GD2 CAR clustering to interactions within the scFv framework regions. This is consistent with a sizable literature reporting that scFvs and other antibody fragments frequently oligomerize39–41, and raises the possibility that tonic signaling impacts the efficacy of many CARs incorporating other scFvs. Indeed, we observed substantial levels of antigen-independent activation and exhaustion across several different CARs.
Our results demonstrate that common in vitro cytotoxicity assays are likely insufficient to predict in vivo efficacy of CARs, since exhausted GD2.28z CAR T cells maintain cytolytic capacity in 51chromium release assays, despite poor cytokine production and poor in vivo efficacy. This “split functionality” has been previously reported with exhausted T cells28, 33, 47. By CD107a degranulation assays, we observed preserved, yet notably diminished, degranulation capacity of the GD2 CAR T cells compared to CD19 CAR T cells (Supplementary Fig. 10a). Our results suggest that the ability of CAR T cells to be polyfunctional cytokine producers in response to antigen exposure may be a better predictor of CAR T cell anti-tumor efficacy in vivo.
Our evaluation of CAR T cell exhaustion was in the context of a tonically signaling CAR, though our results also have broader implications regarding potential mechanisms of CAR treatment failure. Exhaustion of tumor reactive T cells is an increasingly well-established mechanism of immune evasion in cancer21–26. CAR extrinsic factors such as high tumor burden are also likely to induce exhaustion and limit the efficacy of these effectors in vivo. Our data demonstrate that incorporation of 4-1BB in CAR design ameliorates the development of exhaustion in this setting, and are consistent with reports that 4-1BB signaling can enhance T cell function in the setting of chronic viral infection48, 49. They also provide an explanation for recent reports demonstrating that CAR T cells incorporating the 4-1BB costimulatory domain have more prolonged persistence compared to those incorporating the CD28 costimulatory domain in clinical trials3–5, 7, 9, 10, 50.
The mechanisms that lead to exhaustion in the setting of tonic T cell signaling are complex and remain poorly understood. In an effort to better understand how 4-1BB signaling can ameliorate CAR T cell exhaustion, we compared the transcriptional profiles of GD2 CARs containing CD28 and 4-1BB costimulatory domains. In addition to diminished expression of known exhaustion-associated molecules in GD2.BBz compared to GD2.28z CAR T cells, we identified three pathways that are upregulated in GD2.BBz CAR T cells: response to hypoxia, cellular metabolism, and negative regulation of apoptosis. Hypoxia inducible factors have been recently reported to enhance effector T cell functions in the setting of chronic LCMV infection51, and the ability of metabolism to influence T cell function and memory formation is becoming increasingly apparent52–54. To what extent these pathways contribute to the exhaustion-ameliorating effects of 4-1BB in CAR T cells is a focus of our ongoing studies.
In conclusion, our work implicates T cell exhaustion as a major factor limiting the efficacy of CAR therapies. The identification that CAR ectodomains can drive CAR T cell exhaustion, and that antigen-independent activation and exhaustion occur to varying degrees across a spectrum of CARs, highlights the importance of optimizing CAR engineering. This report also identifies 4-1BB as a potent mitigator of exhaustion in chronically stimulated CAR T cells, thus providing a basis for differential CAR T cell persistence observed in clinical trials using CARs with various costimulatory endodomains, and providing new insights into the biology of human T cell exhaustion.
Online Methods
Cells and culture conditions
All cell lines used in this report were STR fingerprinted within the past year, and validated to be mycoplasma free by PCR. The GD2+ human osteosarcoma cell line 143B was transfected with CD19 (NM_001770.3) in the pCMV6-XL5 vector (Origene) using Lipofectamine 2000 (Life Technologies) to create a new, CD19+GD2+ cell line (143B-CD19). To isolate a stable CD19 expressing line, transfected cells underwent two rounds of flow sorting (FACSAria II, BD Bioscience) and single cell cloning. The following cell lines were cultured in DMEM (Life Technologies): 143B, 143B-CD19, 293GP (retroviral vector packaging line), 293T, and H3 (PG13-based, stable producer line of MSGV-FMC63-28z CD19 CAR retroviral vector)32. The following cell lines were cultured in RPMI-1640 (Life Technologies): K562 (chronic myelogenous leukemia), NALM6-GL (acute lymphoblastic leukemia line, stably transfected with GFP and luciferase), and LAN5 (neuroblastoma). DMEM and RPMI-1640 media were supplemented with 10% heat inactivated fetal bovine serum (FBS, Gemini Bioproducts), 10 mM HEPES, 100 U/mL penicillin, 100 ug/mL streptomycin, and 2 mM L-glutamine (Life Technologies).
Human PBMCs from healthy donors were obtained from the Department of Transfusion Medicine, National Institutes of Health (NIH) Clinical Center, under an NIH institutional review board (IRB) approved protocol, after informed consent and in accordance with the Declaration of Helsinki. PBMCs from 24 donors were used. PBMCs were depleted of monocytes by plastic adherence. PBMCs were cultured in AIM-V (Life Technologies), supplemented with 5% FBS, 10 mM HEPES, 100 U/mL penicillin, 100 ug/mL streptomycin, 2 mM L-glutamine and r-human IL-2 (aldesleukin, Prometheus).
Construction of chimeric antigen receptors (CAR) genes
The GD2.28z CAR (MSGV-14g2a-28z) was constructed by linking sequences from a signal peptide derived from the human immunoglobulin heavy chain (GenBank: AAA58735.1, aa 1–19) to a GD2-specific single chain variable fragment (scFv) derived from the 14g2a antibody29, followed by a spacer containing the CH2CH3 domains from human IgG1 (GenBank: AAC82527.1, aa 98–329). This sequence was reverse translated, codon optimized, and synthesized (Mr. Gene.) The resulting product was subcloned into an MSGV-1 based retroviral backbone plasmid encoding the transmembrane and intracellular domains of CD28 and the intracellular domain of CD3ζ (derived from MSGV-139-28z, as previously described)56.
All modifications made in the ecto- or endodomain of the GD2 CAR were generated through sequence manipulation, reverse translation, codon optimization, synthesis (Life Technologies), and subcloning into the MSGV-14g2a-28z vector. To generate the GD2.mut-28.mut-z CAR, nine amino acid point mutations were introduced within the CAR signaling domains. Within the CD28 motif, the following mutations were made (numbering based on GenBank: AAA51944.1): Y191F, P208A and P211A. Within CD3ζ, tyrosines of the three ITAMs were mutated to phenylalanines (numbering based on GenBank: AAH25703.1): Y72F, Y83F, Y111F, Y123F, Y142F and Y153F. Versions of the GD2 CAR containing only CD3ζ signaling or CD28 signaling were generated by deleting either the intracellular component of CD28 (V179 to S220) or the CD3ζ motif, respectively. GD2 CARs containing the 4-1BB endodomain (GD2.BBz CAR) were generated using sequences from the CD19-BBz transgene, as previously described19. The GD2.mutCDR.28z CAR was synthesized by introducing mutations in the complementary determining regions (CDR) of the 14g2a scFv. The CDR regions of the heavy and light chains of the 14g2a scFv were identified using the International Immunogenetics Information System (iMGT) V-QUEry and Standardization (V-QUEST) tool57. Seven residues were selected and mutated (Supplementary Fig. 8b). A hybrid CAR combining the scFv framework regions of the GD2 CAR with the CDR regions of the CD19 CAR (CD19-CDR.GD2-FW.28z CAR) was also synthesized based on CDR identification using V-QUEST. CAR-fluorescent protein fusion constructs were synthesized by subcloning the sequences for the Cerulean or Venus protein immediately 3’ to the CD3ζ domain of the CARs. Soluble versions of the GD2 and CD19 scFvs were constructed by linking the scFv sequences to the CH2CH3 domain of rat IgG2a (GenBank: ADX94416.1, aa 114–321).
The second generation ErbB2.28z CAR construct was synthesized by subcloning the 4D5 scFv from the MSGV-4D5–28BBz vector38 into the MSGV-14g2a-28z vector. Second generation CD22.28z and BBz CAR constructs (based off of the HA22 scFv and m971 scFv) have been previously described20.
Retroviral vector production and transduction
GD2 CAR, CD22 CAR and ErbB2 CAR-encoding retroviral supernatants were produced via transient transfection of the 293GP cell line, as previously described20. Briefly, 293GP cells were transfected via Lipofectamine 2000 (Life Technologies) with the plasmids encoding the CARs and the RD114 envelope protein. Supernatants were collected 48 and 72 hours post-transfection. CD19 CAR-encoding retroviral supernatant was harvested from the H3 producer cell clone.
Monocyte depeleted PBMCs were activated with anti-CD3/CD28 beads (Life Technologies) in a 3:1 bead:cell ratio with 40 IU/mL IL-2 for 3 days. Activated T cells were then retrovirally transduced on days 3 and 4 as previously described20 using Retronectin (Takara) coated plates, and cultured in 300 IU/mL IL-2. Anti-CD3/CD28 beads were removed on day 5. Media and IL-2 were changed every two days (Supplementary Fig. 4). Transduction efficiencies were routinely 80–90% for all CARs. T cells co-expressing the CD19 CAR and GD2 CAR were generated by transducing with a 50:50 mixture of both supernatants simultaneously.
Flow cytometry
All samples were analyzed with an LSR Fortessa or FACSAria II (BD Bioscience) and data were analyzed using FlowJo and SPICE55 software. GD2 CARs were detected with the 14g2a anti-idiotype antibody 1A758. CD19 CARs were detected with the FMC63 anti-idiotype antibody 136.20.159. The GD2.mutCDR.28z CAR and CD19-CDR.GD2-FW.28z CAR were detected via the IgG1 CH2CH3 domain using a goat anti-human IgG H+L (polyclonal, catalog number: A-21091, Life Technologies). All other CARs were detected with biotinylated protein L (Pierce Protein Biology.) T cell phenotype was evaluated via: CD25 (clone BC96, eBioscience), CD27 (clone O323, eBioscience), CD127 (clone eBioRDR5, eBioscience), 4-1BB (clone 4B4, eBioscience), PD-1 (clone eBioJ105, eBioscience), TIM-3 (clone F35-2E2, eBioscience), LAG-3 (polyclonal goat anti-human, catalog number FAB2319P, R&D Systems and clone 3DS223H, eBioscience), annexin V (BD Bioscience) and 7-AAD (BD Bioscience). All FACS plots presenting CAR T cell phenotype data were conducted on gated CAR+ cells. For mock-transduced T cells, whole T cell populations were used for analysis.
Tumor antigen expression was assessed by anti-CD19 (clone SJ25C1, BD Bioscience) and anti-GD2 (clone 14g2a, BD Bioscience). Following xenograft experiments, human cells were identified via human-HLA-A,B,C (clone G46-2.6, BD Bioscience) and human T cells were identified via CD45 (clone HI30, eBioscience). All samples from in vivo experiments were stained in the presence of mouse Fc block (anti-mouse CD16/CD32, clone 2.4G2, BD Bioscience).
To detect binding of soluble scFvs to tumor and T cells by flow cytometry, supernatants were harvested from 293T cells transiently transfected with constructs encoding for the soluble GD2 and CD19 scFvs. Target cells were incubated with harvested supernatants, washed, and then stained with an anti-Rat IgG F(ab’)2 secondary (polyclonal, catalog number 12–4822-87, eBioscience).
Chromium release, CD107a degranulation, and cytokine assays
Standard 51Cr release assays were performed to evaluate CAR T cell cytolytic ability. Target tumor cells were loaded with 100 µCi 51Cr for 1 hour, and then 10,000 tumor cells were co-incubated with CAR T cells for 6 hours at effector-to-target (E:T) ratios ranging from 40:1 to 2.5:1. Supernatants were harvested and 51Cr release quantified using a TopCount Reader (Packard). Percent lysis was calculated as: % lysis = (experimental lysis – spontaneous lysis)/(maximal lysis – spontaneous lysis) × 100%, where maximal lysis was induced by incubation in a 2% Triton X solution. CD107a degranulation assays were conducted by coincubating 5×10 5T cells with 106 tumor cells for 4 hours in the presence of 2 µM monensin and a CD107a antibody (clone eBioH4A3, eBioscience), and then evaluated by flow cytometry. Cytokine production by CAR T cells was evaluated by co-incubation with target tumor cells at a 1:1 ratio (104 cells each) for 24 hours. Supernatants were harvested and cytokine levels measured using a MesoScale Discovery TH1/TH2 multi-array 96 well system. Prior to each assay, T cells were purified using a Pan T cell Isolation Kit II (Miltenyi Biotec). Calculated E:T ratios incorporated the transduction efficiency of CAR T cell cultures. In cases of unequal transduction efficiency, untransduced T cells were supplemented to ensure that both the number of CAR+ T cells and total number of T cells remained consistent across CAR T cell groups.
Western Blots
Whole cell lysates of CAR T cells were generated by lysing 5 × 106 washed cells in 150 µl of RIPA buffer (PBS, 1% NP40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) with 1× cOmplete EDTA free protease inhibitor (Roche) and 0.5 mM sodium vanadate (New England BioLabs), and incubating for 30 minutes on ice. Samples were sonicated at 4°C for 5 minutes to shear DNA. Western blots were then performed on supernatants of centrifuged samples, using an anti-CD3ζ antibody (clone E-3, Santa Cruz Biotech) or an anti-pY142 CD3ζ antibody (clone K25–407.69, BD Bioscience) primary and CleanBlot-IP Detection Reagent (Thermo Scientific) secondary. For analysis of T cells following receptor crosslinking, Mock, CD19.28z CAR and GD2.28z CAR T cells were pre-incubated on ice for 10 minutes with OKT3, the 135.20.1 CD19 CAR anti-idiotype antibody, or the 1A7 GD2 CAR anti-idiotype antibody, respectively (10 µg/mL in PBS). Samples were washed and incubated for 5 minutes on ice with a crosslinking goat anti-mouse F(ab’)2 (catalog number 115-006–071, Jackson ImmunoResearch; 25 µg/ml). Samples were then stimulated at 37°C for 10 minutes, centrifuged and immediately lysed as above.
In vitro idiotype-mediated activation of CD19.28z CAR
The 135.20.1 CD19 CAR anti-idiotype antibody (0.25 µg/ml) and a crosslinking goat anti-mouse F(ab’)2 (catalog number 115-006–071, Jackson ImmunoResearch; 2.5 µg/ml) were added directly to the culture media of CD19.28z CAR T cells, starting at the time of initial exposure to CD19.28z CAR retroviral supernatant (day 3). Idiotype and secondary were replenished every two days. Prior to flow cytometry, T cells were blocked in 10% normal mouse serum (Life Technologies), and then stained in the presence of 2% normal mouse serum.
Fluorescence microscopy imaging and analysis
T cells were transduced to express CAR-Cerulean fusion proteins. On day 9, CAR T cells were stained with the DiD membrane dye (Life Technologies) and with the LIVE/DEAD Fixable Blue Dead Cell (Life Technologies) in PBS. Cells were washed, wet-mounted, and images acquired with a Zeiss Apotome fitted with an AxioCam MRm camera, using a Zeiss plan apochromat 20× objective. Exposure settings were fixed at the beginning of acquisition and were unchanged for the duration of each experiment. Image analysis was performed using ImageJ. Dead cells were eliminated from analysis. The region of interest (ROI) for each cell, identified using the DiD membrane stain, was applied to the Ceruelan (CFP) channel within which the dimensions of each region of interest and the maximum intensity projections were counted. Only a maximum intensity projection of greater than one was considered punctate (maximum intensity less than or equal to one was considered background). The threshold for the DiD stain was set at 10% of maximum pixel intensity. The threshold for the Cerulean channel was set at 20% of maximum pixel intensity.
Förster resonance energy transfer (FRET) analysis
T cells were transduced to express either only the Cerulean or Venus tagged versions, or co-transduced to express both the Cerulean and Venus tagged versions. T cells were then flow sorted on day 7 (FACSAria II, BD Bioscience) to obtain pure CAR+ populations with equivalent MFIs to normalize for CAR surface expression. CAR T cells were rested overnight in culture, then adhered to glass coverslips using CellTak (Corning), fixed in 4% paraformaldehyde for 5 minutes, and mounted using ProLong Gold (Life Technologies).
Images for FRET analyses were collected on a Yokogawa CSU-10 spinning disk confocal microscope coupled to a Nikon TE-2000 using a Nikon 60× 1.4 NA objective. The illumination source for donor excitation was a 100 mW 445 nm laser (Oxxius, Lannion, France; part number LBX-445-100-CIR-PP; 442 nm wavelength selected). The illumination source for acceptor excitation was a 150 mW 514 nm Sapphire laser (Coherent, Inc., Santa Clara, CA; part number 1176578). Excitation power and wavelength control were provided by a Polychromatic acousto-optic modulator (Neos Technologies, Melbourne, FL; Model number 64040-75-.1–8CH-16B). The laser light was coupled into a single mode optical fiber and coupled to the Yokogawa spinning disk unit. The excitation light was passed through a 445/514/561 multiband Yokogawa dichroic beamsplitter (Semrock, Lake Forest, IL; part number Di01-T445/515/561-13×15×0.5) to the objective. Emission light was reflected from the dichroic and passed through a 483/32 bandpass filter for donor emission (Semrock, Lake Forest, IL; part number FF01-483/32-25) and a 535/22 bandpass filter for acceptor emission (Semrock, Lake Forest, IL; part number FF01-535/22-25) mounted in a Lambda 10-2 optical filter changer (Sutter Instrument, Novato, CA). The fluorescence was detected using a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ). The microscope and components were controlled using the open source software60.
Three images were collected for each field of view: DD (donor-donor) images using 442 nm excitation while collecting 483/32 fluorescence, DA (donor-acceptor) image using 442 nm excitation while collecting 535/22 fluorescence, and AA (acceptor-acceptor) images using 514 nm excitation while collecting 535/22 fluorescence. Sensitized acceptor emission FRET analysis was performed with the pFRET plugin61 in Fiji62, using quantum yields of 0.57 for Venus and 0.61 for Cerulean63 and a 60% detector sensitivity in all channels. The FRET signal was measured in 100 to 150 cells, in one region of interest per cell obtained by thresholding the uncorrected FRET images with the same value for all measurements.
Real time PCR
mRNA was extracted from cells using the RNeasy Mini Kit (Qiagen) and reverse transcribed into cDNA using the SuperScript First-Strand Synthesis System (Life Technologies). All reactions were performed with TaqMan Fast Universal PCR Master Mix (Applied Biosystems) on an Applied Biosystems StepOnePlus Real-Time PCR machine, using the following TaqMan primers (Applied Biosystems) with noted amplification factors (mean ± SEM): GM2/GD2 synthase (B4GALNT1, Hs00155195_m1, 1.92 ± 0.01); GM1/GD1b/GA1 synthase (B3GALT4, Hs00534104_s1, 1.94 ± 0.04); GD3 synthase (ST8S1A1, Hs00268157_m1, 1.90 ± 0.02); T-bet (TBX21, Hs00203436_m1, 1.90 ± 0.02); Blimp-1 (PRDM1, Hs00153357_m1, 1.92 ± 0.01); β-actin (ACTB, Hs99999903_m1, 1.89 ± 0.02). Delta CT calculations were relative to β-actin and were corrected for PCR efficiencies.
Microarray, gene expression, and GSEA analysis
RNA was extracted from cells using the RNeasy Mini Plus Kit (Qiagen). RNA was in vitro transcribed, fragmented, and hybridized and applied to an Affymetrix Human Genome U133 Plus 2.0 array according to the manufacturer’s instructions (Affymetrix) and in accordance to the microarray core facility NCI Frederick. Microarray data analyses were performed using Partek Genomics Suite Version 6.6. Data were uploaded using the original CEL z-scoring and normalized using the robust multichip average (RMA) algorithm. Differentially expressed genes were selected at ≥ 2-fold difference and p < 0.05. Cluster analyses and principal component analysis were conducted with Partek default settings. Heatmaps were generated with expression data normalized to mean of zero and standard deviation of one. GSEA analysis (http://www.broadinstitute.org/gsea/index.jsp) was performed using default parameter settings. Published gene sets of gene ontology (MSigDB C5) and curated gene sets from the biomedical literature (MSigDB C2) were used for analysis. Significance was defined as having FDR < 0.05.
In vivo studies
All animal studies were carried out under protocols approved by the National Cancer Institute (NCI) Bethesda Animal Care and Use Committee. Xenograft studies were performed using NSG mice (NOD.Cg-Prkdcscid ILrgtm1Wjl/SzJ, Jackson Laboratory) 6–12 weeks of age. Equivalent number of male and female mice were used. The 143B and 143B-CD19 osteosarcoma lines were inoculated at 5 × 105 or 106 periosteal to the tibia, and the NALM6-GL leukemia line was inoculated at 5 × 105 or 5 × 106 intravenously. Mice then received adoptive transfer of 107 transduced CAR T cells intravenously 3–14 days later, as indicated in individual experiments. Where indicated, mice also received 3 mg of cyclophosphamide (Millipore) intraperitoneally on day 2. To control for differences in transduction efficiency, untransduced T cells were supplemented to ensure that both the number of CAR+ T cells and total number of T cells remained constant across CAR T cell groups. In osteosarcoma experiments, mice received cytokine support of 1 µg IL-7 (Cytheris Inc.) and 5 µg M25 antibody (anti-IL-7; Immunex) intraperitoneally three times per week following T cell transfer64. Osteosarcoma burden was quantified via two-dimensional leg area measurements. NALM6-GL leukemia burden was evaluated using the Xenogen IVIS Lumina (Caliper Life Sciences). Mice were injected intraperitoneally with 3 mg D-luciferin (Caliper Life Sciences), and then imaged 4 minutes later with an exposure time of 30 seconds. Luminescence images were analyzed using Living Image software (Caliper Life Sciences). Spleens were mechanically disaggregated by passage through a 70 µM filter. Tumors were mechanically dissociated using a GentleMacs (Miltenyi) and passed through 70 µM filters.
Study design
Characterization of the activation, exhaustion, and anti-tumor efficacy of CAR T cells was replicated across at least 3 unique healthy donors for all experiments reported. For all osteosarcoma in vivo xenograft studies, the primary endpoint was tumor size and mice were humanely euthanized when the tumor size reached 20 mm. For all leukemia in vivo xenograft studies, leukemic burden was monitored by bioluminescence imaging. Animals were humanely euthanized at 21 days. Group sizes were determined by an a priori power analysis for a two-tailed, two-sample t test with an α of 0.05 and power of 0.8, in order to detect a 10% difference in tumor size at endpoint. No blinding or randomization was used during the experiments. No samples or animals were excluded from analysis.
Statistics
All figures are representative of at least three experiments unless otherwise noted. All graphs report mean ± SEM values of biological replicates. For the SPICE analysis of exhaustion markers, statistical significance was calculated using a Wilcoxon signed rank test. Quantification of CAR punctae was compared by a Kolmogorov-Smirnov test. Statistical significance of all other data was calculated using a Student’s T-test. P<0.05 was considered significant and is designated with an asterisk in all figures.
Supplementary Material
Acknowledgements
We kindly thank S. Gottschalk (Texas Children’s Hospital) for providing sequences for the 14g2a scFv and IgG1 CH2CH3; J. Kochenderfer (National Cancer Institute, NCI, US National Institutes of Health, NIH) for the H3 MSGV-FMC63-28z retroviral vector producer line; S. Grupp (Children’s Hospital of Philadelphia) for the NALM6-GL cell line; R. Morgan (BlueBird Bio, previously NCI, NIH) for the MSGV-139-28z and MSGV-4D5-28BBz vectors; and C. June (University of Pennsylvania) for sequences to the 4-1BB CAR endodomain. We also thank L. Cooper (MD Anderson) for providing the 136.20.1, FMC63 anti-idiotype antibody; the Biological Research Branch of NCI for providing the 1A7, 14g2a anti-idiotype antibody; the Clinical Support Laboratory of the Frederick National Laboratory for Cancer Research (FNLCR) for assisting in MesoScale cytokine release assays; and the Laboratory of Molecular Technology (LMT) Microarray Group of FNLCR for assisting in microarray assays. We thank N. Restifo and M. Roederer for careful review of this manuscript.
This work was supported by the Intramural Research Program of the NIH, including the NCI and the National Institute of Biomedical Imaging and Bioengineering (NIBIB): ZIA BC 011073 (A.H.L, K.M.W., J.P.S., A.J.W., M.E.K., V.R.V., R.J.O., and C.L.M); ZIA BC 011565 (W.M.H. and T.J.F.); ZIA BC 011332 (M.M and R.N.K); and ZIA EB 000071-06 (M.I. and G.H.P.) This research was also supported by a Stand Up To Cancer – St. Baldrick’s – NCI Pediatric Dream Team Translational Cancer Research Grant. Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
Accession codes
All generated microarray data are available at the Gene Expression Omnibus (Geo) with accession code GSE65856.
Author Contributions
A.H.L., W.M.H., J.F.S., M.M., M.I., M.E.K., R.N.K., G.H.P., T.J.F., R.J.O. and C.L.M. designed the research; A.H.L., W.M.H., K.M.W., M.M., M.I., J.P.S., A.J.W, M.E.K., V.R.V., G.H.P. conducted experiments; A.H.L., W.M.H., J.F.S., M.M., M.I., G.H.P., T.J.F., R.J.O. and C.L.M. analyzed data; and A.H.L. and C.L.M. wrote the paper.
References
- 1.Lee DW, et al. The Future Is Now: Chimeric Antigen Receptors as New Targeted Therapies for Childhood Cancer. Clinical Cancer Research. 2012;18(10):2780–2790. doi: 10.1158/1078-0432.CCR-11-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadelain M, Brentjens R, Rivière I. The Basic Principles of Chimeric Antigen Receptor Design. Cancer Discovery. 2013;3(4):388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee DW, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. The Lancet. 2014 doi: 10.1016/S0140-6736(14)61403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maude SL, et al. Chimeric Antigen Receptor T Cells for Sustained Remissions in Leukemia. New England Journal of Medicine. 2014;371(16):1507–1517. doi: 10.1056/NEJMoa1407222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochenderfer JN, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119(12):2709–2720. doi: 10.1182/blood-2011-10-384388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter DL, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. New England Journal of Medicine. 2011;365(8):725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grupp SA, et al. Chimeric Antigen Receptor-Modified T Cells for Acute Lymphoid Leukemia. New England Journal of Medicine. 2013;368(16):1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savoldo B, et al. CD28 costimulation improves expansion and persistence of chimeric antigen receptor-modified T cells in lymphoma patients. The Journal of Clinical Investigation. 2011;121(5):1822–1826. doi: 10.1172/JCI46110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brentjens RJ, et al. CD19-Targeted T Cells Rapidly Induce Molecular Remissions in Adults with Chemotherapy-Refractory Acute Lymphoblastic Leukemia. Science Translational Medicine. 2013;5(177):177ra138–177ra138. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davila ML, et al. Efficacy and Toxicity Management of 19-28z CAR T Cell Therapy in B Cell Acute Lymphoblastic Leukemia. Science Translational Medicine. 2014;6(224) doi: 10.1126/scitranslmed.3008226. 224ra225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kershaw MH, et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clinical Cancer Research. 2006;12(20):6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JR, et al. Adoptive transfer of chimeric antigen receptor re-directed cytolytic T lymphocyte clones in patients with neuroblastoma. Molecular therapy. 2007;15(4):825–833. doi: 10.1038/sj.mt.6300104. [DOI] [PubMed] [Google Scholar]
- 13.Pule MA, et al. Virus-specific T cells engineered to coexpress tumor-specific receptors: persistence and antitumor activity in individuals with neuroblastoma. Nature medicine. 2008;14(11):1264–1270. doi: 10.1038/nm.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis CU, et al. Antitumor activity and long-term fate of chimeric antigen receptor-positive T cells in patients with neuroblastoma. Blood. 2011;118(23):6050–6056. doi: 10.1182/blood-2011-05-354449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Till BG, et al. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012;119(17):3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamers CHJ, et al. Treatment of Metastatic Renal Cell Carcinoma With CAIX CAR-engineered T cells: Clinical Evaluation and Management of On-target Toxicity. Mol Ther. 2013;21(4):904–912. doi: 10.1038/mt.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins PF, et al. Cutting edge: persistence of transferred lymphocyte clonotypes correlates with cancer regression in patients receiving cell transfer therapy. The Journal of Immunology. 2004;173(12):7125–7130. doi: 10.4049/jimmunol.173.12.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowolik CM, et al. CD28 Costimulation Provided through a CD19-Specific Chimeric Antigen Receptor Enhances In vivo Persistence and Antitumor Efficacy of Adoptively Transferred T Cells. Cancer Research. 2006;66(22):10995–11004. doi: 10.1158/0008-5472.CAN-06-0160. [DOI] [PubMed] [Google Scholar]
- 19.Milone MC, et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Molecular Therapy. 2009;17(8):1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haso W, et al. Anti-CD22-chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia. Blood. 2013;121(7):1165–1174. doi: 10.1182/blood-2012-06-438002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmadzadeh M, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 2009;114(8):1537–1544. doi: 10.1182/blood-2008-12-195792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakuishi K, et al. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. The Journal of experimental medicine. 2010;207(10):2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baitsch L, et al. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. The Journal of clinical investigation. 2011;121(6):2350. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Q, et al. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117(17):4501–4510. doi: 10.1182/blood-2010-10-310425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo S-R, et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer research. 2012;72(4):917–927. doi: 10.1158/0008-5472.CAN-11-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New England Journal of Medicine. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138(1):30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 28.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 29.Rossig C, et al. Targeting of GD2-positive tumor cells by human T lymphocytes engineered to express chimeric T-cell receptor genes. International journal of cancer. 2001;94(2):228–236. doi: 10.1002/ijc.1457. [DOI] [PubMed] [Google Scholar]
- 30.Pulè MA, et al. A chimeric T cell antigen receptor that augments cytokine release and supports clonal expansion of primary human T cells. Molecular Therapy. 2005;12(5):933–941. doi: 10.1016/j.ymthe.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Hudecek M, et al. The non-signaling extracellular spacer domain of chimeric antigen receptors is decisive for in vivo antitumor activity. Cancer immunology research. 2014 doi: 10.1158/2326-6066.CIR-14-0127. canimm. 0127.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochenderfer JN, et al. Construction and Pre-clinical Evaluation of an Anti-CD19 Chimeric Antigen Receptor. Journal of immunotherapy (Hagerstown, Md: 1997) 2009;32(7):689. doi: 10.1097/CJI.0b013e3181ac6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin H, et al. A Role for the Transcriptional Repressor Blimp-1 in CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2009;31(2):309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doering TA, et al. Network Analysis Reveals Centrally Connected Genes and Pathways Involved in CD8< sup>+</sup> T Cell Exhaustion versus Memory. Immunity. 2012;37(6):1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338(6111):1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friend LD, et al. A dose-dependent requirement for the proline motif of CD28 in cellular and humoral immunity revealed by a targeted knockin mutant. The Journal of experimental medicine. 2006;203(9):2121–2133. doi: 10.1084/jem.20052230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodson LF, et al. Targeted knock-in mice expressing mutations of CD28 reveal an essential pathway for costimulation. Molecular and cellular biology. 2009;29(13):3710–3721. doi: 10.1128/MCB.01869-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. A herceptin-based chimeric antigen receptor with modified signaling domains leads to enhanced survival of transduced T lymphocytes and antitumor activity. The Journal of Immunology. 2009;183(9):5563–5574. doi: 10.4049/jimmunol.0900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieba L, Honegger A, Krebber C, Plückthun A. Disrupting the hydrophobic patches at the antibody variable/constant domain interface: improved in vivo folding and physical characterization of an engineered scFv fragment. Protein Engineering. 1997;10(4):435–444. doi: 10.1093/protein/10.4.435. [DOI] [PubMed] [Google Scholar]
- 40.Dolezal O, et al. Single-chain Fv multimers of the anti-neuraminidase antibody NC10: the residue at position 15 in the VL domain of the scFv-0 (VL− VH) molecule is primarily responsible for formation of a tetramer-trimer equilibrium. Protein engineering. 2003;16(1):47–56. doi: 10.1093/proeng/gzg006. [DOI] [PubMed] [Google Scholar]
- 41.Whitlow M, et al. Multivalent Fvs: characterization of single-chain Fv oligomers and preparation of a bispecific Fv. Protein Engineering. 1994;7(8):1017–1026. doi: 10.1093/protein/7.8.1017. [DOI] [PubMed] [Google Scholar]
- 42.Wherry EJ, et al. Molecular Signature of CD8+ T Cell Exhaustion during Chronic Viral Infection. Immunity. 2007;27(4):670–684. doi: 10.1016/j.immuni.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 43.Crawford A, et al. Molecular and Transcriptional Basis of CD4+ T Cell Dysfunction during Chronic Infection. Immunity. 40(2):289–302. doi: 10.1016/j.immuni.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guest RD, et al. The role of extracellular spacer regions in the optimal design of chimeric immune receptors: evaluation of four different scFvs and antigens. Journal of Immunotherapy. 2005;28(3):203. doi: 10.1097/01.cji.0000161397.96582.59. [DOI] [PubMed] [Google Scholar]
- 45.James SE, et al. Antigen sensitivity of CD22-specific chimeric TCR is modulated by target epitope distance from the cell membrane. The Journal of Immunology. 2008;180(10):7028–7038. doi: 10.4049/jimmunol.180.10.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frigault MJ, et al. Identification of chimeric antigen receptors that mediate constitutive or inducible proliferation of T cells. Cancer immunology research. doi: 10.1158/2326-6066.CIR-14-0186. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agnellini P, et al. Impaired NFAT nuclear translocation results in split exhaustion of virus-specific CD8+ T cell functions during chronic viral infection. Proceedings of the National Academy of Sciences. 2007;104(11):4565–4570. doi: 10.1073/pnas.0610335104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vezys V, et al. 4-1BB signaling synergizes with programmed death ligand 1 blockade to augment CD8 T cell responses during chronic viral infection. The Journal of Immunology. 2011;187(4):1634–1642. doi: 10.4049/jimmunol.1100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, et al. 4-1BBL induces TNF receptor-associated factor 1-dependent Bim modulation in human T cells and is a critical component in the costimulation-dependent rescue of functionally impaired HIV-specific CD8 T cells. The Journal of Immunology. 2007;179(12):8252–8263. doi: 10.4049/jimmunol.179.12.8252. [DOI] [PubMed] [Google Scholar]
- 50.Porter D, et al. Chimeric Antigen Receptor Modified T Cells Directed Against CD19 (CTL019 cells) Have Long-Term Persistence and Induce Durable Responses In Relapsed, Refractory CLL. Blood. 2013;122(21):4162–4162. [Google Scholar]
- 51.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat Immunol. 2013;14(11):1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460(7251):103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Windt Gerritje JW, et al. Mitochondrial Respiratory Capacity Is a Critical Regulator of CD8+ T Cell Memory Development. Immunity. 2012;36(1):68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460(7251):108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roederer M, Nozzi JL, Nason MC. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry Part A. 2011;79(2):167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 56.Morgan RA, et al. Recognition of glioma stem cells by genetically modified T cells targeting EGFRvIII and development of adoptive cell therapy for glioma. Human gene therapy. 2012;23(10):1043–1053. doi: 10.1089/hum.2012.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Research. 2008;36(suppl 2):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sen G, et al. Induction of IgG antibodies by an anti-idiotype antibody mimicking disialoganglioside GD2. Journal of Immunotherapy. 1998;21(1):75–83. doi: 10.1097/00002371-199801000-00010. [DOI] [PubMed] [Google Scholar]
- 59.Jena B, et al. Chimeric Antigen Receptor (CAR)-Specific Monoclonal Antibody to Detect CD19-Specific T Cells in Clinical Trials. PLoS ONE. 2013;8(3):e57838. doi: 10.1371/journal.pone.0057838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edelstein A, et al. Computer control of microscopes using µManager. Current protocols in molecular biology. 2010 doi: 10.1002/0471142727.mb1420s92. 14.20. 11-14.20. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elangovan M, et al. Characterization of one-and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29(1):58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Olenych SG, Claxton NS, Ottenberg GK, Davidson MW. The fluorescent protein color palette. Current Protocols in Cell Biology. 2007 doi: 10.1002/0471143030.cb2105s36. [DOI] [PubMed] [Google Scholar]
- 64.Martin CE, et al. IL-7/anti-IL-7 mAb complexes augment cytokine potency in mice through association with IgG-Fc and by competition with IL-7R. Blood. 2013;121(22):4484–4492. doi: 10.1182/blood-2012-08-449215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.