Summary
The PIDDosome—PIDD-RAIDD-caspase-2 complex—is a proapoptotic caspase-activation platform of elusive significance. DNA damage can initiate complex assembly via ATM phosphorylation of the PIDD death domain (DD), which enables RAIDD recruitment to PIDD. In contrast, the mechanisms limiting PIDDosome formation have remained unclear. We identify the mitotic checkpoint factor, BubR1, as a direct PIDDosome inhibitor, acting in a noncanonical role independent of Mad2. Following its phosphorylation by ATM at DNA breaks, ‘primed’ PIDD relocates to kinetochores via a direct interaction with BubR1. BubR1 binds the PIDD DD, competes with RAIDD recruitment, and negates PIDDosome-mediated apoptosis after ionizing radiation. The PIDDosome thus sequentially integrates DNA damage and mitotic checkpoint signals to decide cell fate in response to genotoxic stress. We further show that by sequestering PIDD at the kinetochore, BubR1 acts to delay PIDDosome formation until the next cycle, defining a new mechanism by which cells evade apoptosis during mitosis.
Introduction
The PIDDosome is a caspase-activation platform whose significance remains unclear more than a decade after its biochemical isolation by Tschopp and colleagues (Bock et al., 2012; Janssens and Tinel, 2012; Kumar, 2009; Tinel and Tschopp, 2004). Initial views of the complex as a stress-inducible, proapoptotic device have been supported by studies implicating the PIDDosome in cell death responses to DNA damage and other stimuli (Ando et al., 2012; Berube et al., 2005; Jelinek et al., 2013; Niizuma et al., 2008). However, there are experimental settings in which one or more PIDDosome components show inconsistent phenotypes (Kim et al., 2009; Manzl et al., 2009; Manzl et al., 2012; Ribe et al., 2012). Further impeding the functional elucidation of the complex, the identities of the PIDDosome’s upstream regulators and downstream substrates remain essentially unknown.
The PIDDosome comprises the death domain (DD) proteins, PIDD (p53-inducible protein with a DD; LRDD) and RAIDD (RIP-associated Ich-1/CED homologous protein with DD; CRADD), and their client caspase, caspase-2 (Duan and Dixit, 1997; Lin et al., 2000; Telliez et al., 2000; Wang et al., 1994). PIDD and RAIDD serve as core scaffold and caspase adaptor, respectively, which mobilize caspase-2 to the complex and enable its activation by induced proximity (Berube et al., 2005; Bouchier-Hayes et al., 2009; Tinel and Tschopp, 2004). Biochemical analyses have identified the PIDD-RAIDD interaction, rather than the RAIDD-caspase-2 interaction, as the rate-limiting step in PIDDosome assembly (Jang and Park, 2013). The interaction occurs at the PIDD DD through homotypic PIDD DD:RAIDD DD interactions analogous to the FAS-FADD interactions underlying death-inducing signaling complex (FAS-FADD-caspase-8) assembly (Park et al., 2007). How the PIDD-RAIDD interaction, and thus PIDDosome formation, is initiated in live cells is a central focus in the field. Recent reports have converged on DNA damage as primary trigger. First, DNA lesions induced by a range of genotoxins have been shown to favor the autocatalytic processing of full-length PIDD into the C-terminal PIDD-CC fragment, which retains the putative oligomerization domain and DD and exhibits maximal affinity for RAIDD (Tinel et al., 2007). More directly, ionizing radiation (IR)-activated ATM phosphorylates threonine 788 within the PIDD DD, triggering a conformational change that is necessary and sufficient for RAIDD binding under overexpression conditions and is required for PIDDosome formation at the endogenous level (Ando et al., 2012; Terry et al., 2014)
In addition to priming PIDD for RAIDD recruitment and complex assembly, DNA damage also acts to limit PIDDosome activity. This occurs at least in part via checkpoint kinase 1 (Chk1) signaling, though the underlying mechanism is unknown (Ando et al., 2012; Manzl et al., 2013; Myers et al., 2009; Pan et al., 2009; Sidi et al., 2008). Here we identify the mitotic checkpoint effector, BubR1, as a direct PIDDosome inhibitor in response to IR-induced DNA damage. BubR1 is a core component of the mitotic checkpoint complex (MCC), a Mad2-BubR1-Bub3-Cdc20 tetramer that inhibits the anaphase-promoting complex/cyclosome (APC/C) in response to dysfunctional kinetochore (KT)-microtubule attachments (Karess et al., 2013; Suijkerbuijk et al., 2012). As a PIDDosome inhibitor, however, BubR1 acts at the KT but independently of the MCC. Mechanistically, BubR1 binds the PIDD DD to outcompete RAIDD recruitment, thus effectively preventing PIDDosome assembly and apoptosis induction until completion of mitosis.
Results
BubR1, but not Mad2, suppresses PIDDosome signaling after DNA damage
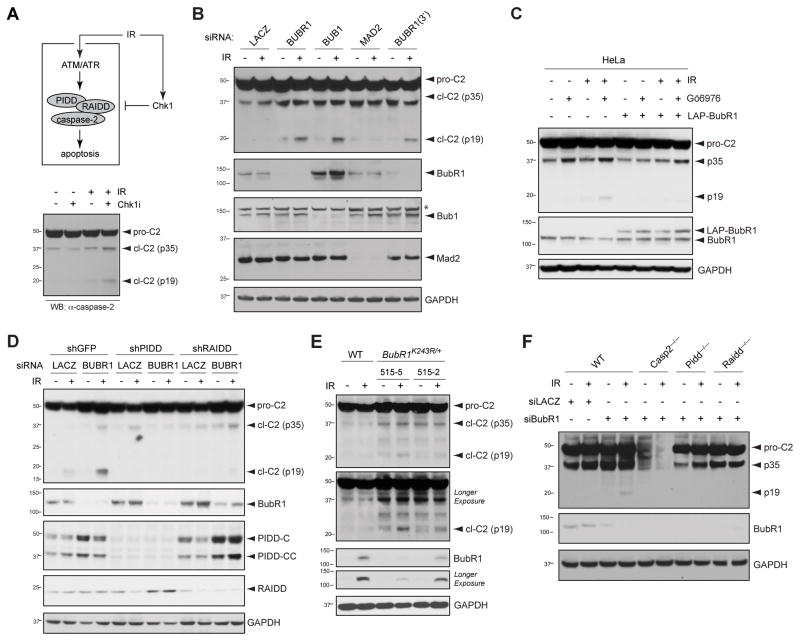
PIDDosome inhibition by Chk1 after DNA damage can be readily assessed by the inability of IR to trigger caspase-2 cleavage unless Chk1 is simultaneously inhibited via siRNA or Chk1 inhibitor such as Gö6976 (Figure 1A and S1A; note the appearance of the p19 mature cleavage product in lane 4) (Ando et al., 2012; Myers et al., 2009; Sidi et al., 2008). To identify novel negative regulators of the PIDDosome, we initiated screens for genes whose knockdown phenocopies Chk1i after IR. As shown in Figure 1B, independent siRNAs to BubR1 triggered robust caspase-2 cleavage specifically after IR. Overexpression of BubR1 was sufficient to restore PIDDosome suppression in Chk1-inhibited cells, indicating that BubR1 acts downstream or in parallel of Chk1 (Figure 1C). Bub1, which recruits BubR1 to KTs to initiate the mitotic checkpoint (Johnson et al., 2004), was also required to suppress caspase-2 activation after IR (Figure 1B) and so was the upstream mitotic checkpoint kinase, Aurora B (Figure S1B). Notably, strong knockdown of Mad2, which like BubR1 is essential for MCC function, failed to enable caspase-2 processing under these conditions (Figure 1B).
Figure 1. BubR1, but not Mad2, suppresses PIDDosome signaling after IR.
(A) Diagram of the PIDDosome signaling pathway triggered by DNA damage. Pathway activation after IR by ATM phosphorylation of PIDD is restrained by Chk1. Combined delivery of IR and Chk1 inhibitor (Chk1i, such as Gö6976) activates the PIDDosome and results in apoptosis. pro-C2, procaspase-2; cl-C2 (p35), putative caspase 2 cleavage intermediate; cl-C2 (p19), p19 fragment (mature cleavage product).
(B) HeLa cells transfected with the indicated siRNAs were treated with or without 10 Gy IR and harvested 24 hr after IR. Lysates were analyzed by western blot. *non-specific band.
(C) HeLa cells were transfected with or without LAP-BubR1WT and treated with DMSO or Go6976 (1 μM) with or without 10 Gy IR and harvested 24 hr after IR. Lysates were analyzed by western blot.
(D) HeLa cells stably expressing the indicated shRNAs transfected with siLacZ or siBubR1 were treated with or without IR (10 Gy) and harvested 24 hours after IR. Lysates were analyzed by western blot.
(E) SV40 transformed MEF cells of indicated genotypes were treated or not treated with IR and harvested 24 hours after IR. Lysates were analyzed by western blot.
(F) SV40 transformed MEF cells of indicated genotypes and transfected with siLacZ or siBubR1 were treated with or without IR (10 Gy) and harvested 24 hours after IR. Lysates were analyzed by western blot.
See also Figure S1
To ascertain that caspase-2 cleavage triggered by depletion of BubR1 was dependent on the PIDDosome, we analyzed HeLa cells with stable PIDD or RAIDD knockdowns (Ando et al., 2012). As shown in Figure 1D, PIDD and RAIDD were indeed essential for caspase-2 cleavage after IR+BubR1 knockdown. Similar experiments also placed Bub1 and Aurora B in the PIDDosome pathway (Figure S1C, D).
To further validate BubR1 as a suppressor of PIDDosome signaling, we analyzed two BubR1K243R heterozygous MEF lines in which mutationally impaired BubR1 acetylation reduces total BubR1 levels to variable degrees (Park et al., 2013). Reduction of BubR1 was sufficient to trigger caspase-2 cleavage after IR, the extent of which correlated with the severity of BubR1 reduction (Figure 1E, compare lanes 4 and 6). To assess the PIDDosome-dependence of these effects, we depleted BubR1 from Pidd−/− and Raidd−/− MEFs (Berube et al., 2005; Manzl et al., 2009). Knockout of either gene blocked caspase-2 cleavage, showing that mouse BubR1, like human BubR1, acts to suppress PIDDosome activity (Figure 1F). Together, these experiments identified an evolutionarily conserved role for BubR1 in PIDDosome inhibition. The additional involvements of Aurora B and Bub1 (which in the mitotic checkpoint act upstream of BubR1), but not Mad2 (which acts at the level of BubR1), provided first evidence for a novel, MCC-independent function of BubR1 in PIDDosome regulation.
PIDDosome control by mitotic kinases dictates cell fate after IR
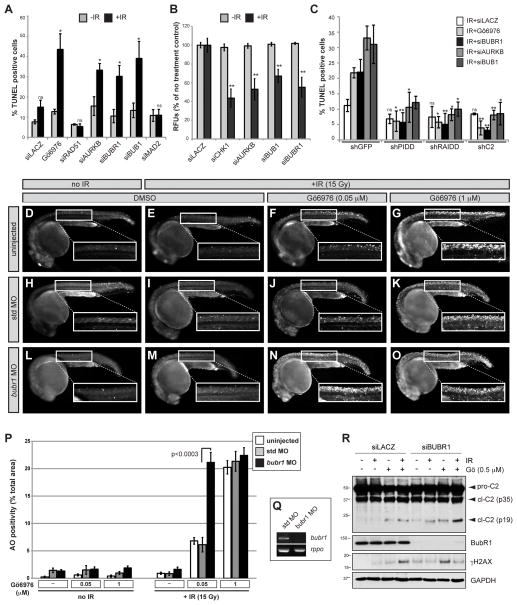
The levels of active caspase-2 enzyme produced by the PIDDosome in response to IR+Chk1i are necessary and sufficient for apoptotic cell death in most settings (Ando et al., 2012; Ho et al., 2009; Manzl et al., 2013; Myers et al., 2009; Pan et al., 2009; Sidi et al., 2008). To test whether suppression of PIDDosome signaling by BubR1, Bub1 and Aurora B was similarly significant to cell fate, we analyzed radiation responses in vitro and in vivo. Caspase-2-dependent apoptosis after IR is most reliably studied in cells devoid of p53, which prevents the activation of additional caspases via p53-dependent pathways. Thus, in HPV-harboring HeLa cells or p53 mutant zebrafish embryos, all apoptosis induced by IR+Chk1i depends on caspase-2 (Figure 2C, compare bars 2 and 17) (Sidi et al., 2008).
Figure 2. BubR1 suppresses PIDDosome-mediated apoptosis.
(A) HeLa cells transfected with the indicated siRNAs or Go6976 (1 μM) were treated with or without IR (10 Gy) (black and grey bars respectively), harvested 24 hr post IR stained for TUNEL and analyzed by flow cytometry. Data are means +/− SD of 3 independent experiments. *p < 0.05, ns, non-significant; two-tailed Student’s t-test.
(B) HeLa cells were transfected with the indicated siRNAs, treated with or without 10 Gy IR (black and grey bars respectively), and stained with alamar Blue at 72 hr post IR. Data are means +/− SD of 3 independent experiments. **p < 0.01, two-tailed Student’s t-test.
(C) HeLa cells stably expressing the indicated shRNAs were transfected with the indicated siRNAs or Go6976 (1 μM), treated with IR(10 Gy), harvested 24 hr post IR and stained for TUNEL. Data are means +/− SD of 3 independent experiments. Significance vs. corresponding shGFP controls: *p < 0.05, **p < 0.01, ns, non-significant; two-tailed Student’s t-test.
(D–O) p53M214K/M214K zebrafish embryos were non-injected or injected at the one-cell stage with standard control (std) or bubr1 MOs, incubated 17 hr later with or without Gö6976 at indicated concentrations (μM), treated with or without 15 Gy IR and stained with the cell death marker acridine orange (AO) after 7 hours. All embryos imaged live at 24hpf.
(P) Quantification of AO stains shown in (D–O). White bars, uninjected; gray bars, std MO; black bars, bubr1 MO. Data collected from 3 independent experiments (≥ 10 embryos per condition). All data are reported as means ± SEM (two-tailed Student’s t-test).
(Q) RT-PCR of bubr1 and rppo transcripts from embryos injected with std MO or bubr1 MO. Note the nonsense-mediated decay of bubr1 transcript in the bubr1 MO injected embryos.
(R) HeLa cells transfected with the indicated siRNAs were treated with DMSO or Go6976 (0.5μM) with or without IR (10 Gy) and harvested 24 hr post IR. Lysates were analyzed by western blot.
See also Figure S2
Similar to Chk1i, siRNA depletions of BubR1, Bub1 and Aurora B triggered a robust, PIDDosome-dependent apoptotic response to IR in otherwise radioresistant HPV+ HeLa cells or SV-40 MEFs (Figures 2A–C and S2A). In contrast, knockdowns of Mad2 or Rad51, which have no effect on caspase-2 cleavage (Figures 1B and S1A), failed to trigger apoptosis after IR (Figure 2A). These results indicated that PIDDosome control by BubR1, Bub1 and Aurora B is biologically significant and, again, independent of their canonical MCC signaling function.
We next tested the in vivo relevance of these observations in the zebrafish system, in which the caspase-2 apoptotic response to IR+Chk1i was originally identified (Sidi et al., 2008). As expected from this study, 18-hour post-fertilization (hpf) p53 mutant embryos failed to respond to IR unless Chk1 was simultaneously inhibited (Figures 2E, G; quantification of all acridine orange stains is shown in Figure 2P). While morpholino (MO) knockdown of the zebrafish bubr1 orthologue, bub1bb, failed to individually phenocopy Chk1i (Figure 2G, M), it strongly potentiated the effects of a low, individually insufficient dose of Chk1i (50 nM) to trigger a marked apoptotic response (Figure 2F, M, N; knockdown levels shown in 2Q). This potentiated response closely matched that observed in embryos treated with a 20-fold greater dose of Chk1i (Figure 2N, G, P). Such a cooperative genetic interaction between Chk1 and BubR1 was also observed in human cells, in which reduced doses of Chk1i and BubR1 siRNA synergistically triggered caspase-2 activation after IR (Figure 2R). When tested in zebrafish, the Aurora B inhibitor, AZD1152, and the Bub1 inhibitor, 2OH-BNPP1 (Kang et al., 2008), mirrored the effects of BubR1 knockdown (Figure S2B). Together, the human and zebrafish data confirmed that BubR1, Bub1 and Aurora B exert key roles in PIDDosome control after DNA damage. Additionally, the cooperative/synergistic interactions observed between Chk1 and these factors suggested that the latter control the PIDDosome in parallel with, rather than downstream of, Chk1.
ATM-phosphorylated PIDD relocates to kinetochores during early mitosis
The involvement of mitotic checkpoint factors in PIDDosome regulation led us to consider that the complex might be present in dividing cells. Notably, because none of the available antibodies to PIDD, RAIDD or caspase-2 have been validated for immunofluorescence (IF), the precise cellular location of the PIDDosome has remained unsolved to date.
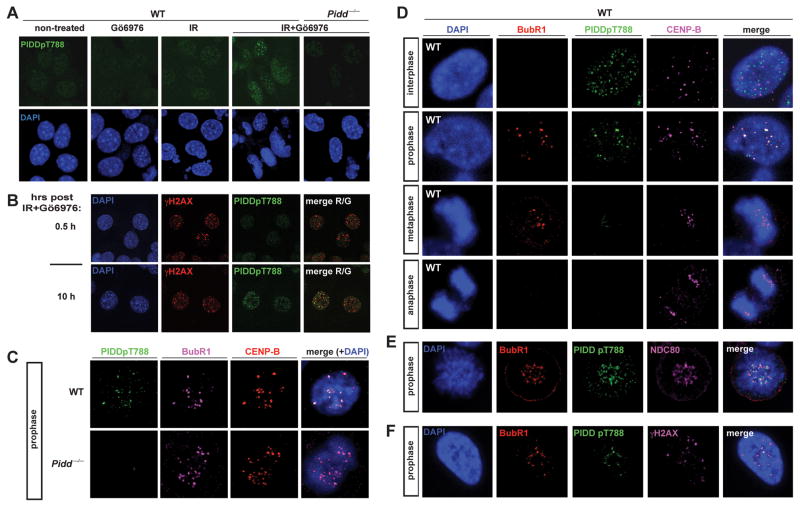
We tested the recently generated α-PIDDpT788 antibody, which specifically detects ATM-phosphorylated PIDD by immunoblot (Ando et al., 2012). This phosphoryation event primes PIDD for RAIDD recruitment and, like caspase-2 cleavage, is primarily detected in cells treated with IR+Chk1i (Ando et al., 2012). As shown in Figure 3A, α-PIDDpT788 reacted to nuclear foci-like structures in MEFs treated with IR+Chk1i, but did not in untreated or single-treated cells, altogether mirroring PIDDpT788 western blots (Ando et al., 2012). As would be expected from a protein phosphorylated by ATM, phospho-PIDD colocalized with γH2AX and ATMpS1981 foci (Figures 3B and S3B). PIDDpT788 foci were undetectable in Pidd−/− MEFs, validating signal specificity (Figure 3A; see also Figure 3C for mitosis-specific IF signals). Therefore, α-PIDDpT788 enables the visualization of endogenous, primed PIDD in cells.
Figure 3. ATM-phosphorylated, ‘primed’ PIDD localizes at kinetochores during early mitosis.
(A) SV40 transformed MEFs of indicated genotypes grown on coverslips were treated with or without Go6976 (1 μM) or IR (10 Gy), harvested 10 hr post IR, stained with the indicated antibodies and visualized by confocal microscopy (single 0.8 μm sections are shown).
(B) SV40 transformed WT MEFs grown on coverslips were treated with Gö6976 (1 μM) and IR (10 Gy), harvested 0.5 or 10 hr post IR, stained using the indicated antibodies and visualized by confocal microscopy (single 0.8 μm sections are shown).
(C) SV40 transformed MEFs of indicated genotypes grown on coverslips were treated with Gö6976 (1 μM) and IR (10 Gy) and harvested 10 hr post IR. Coverslips were stained using the indicated antibodies and visualized by confocal microscopy (single 0.8 μm sections are shown).
(D–F) SV40 transformed WT MEFs grown on coverslips were treated with Gö6976 (1 μM) and IR (10 Gy), harvested 10 hr post IR, stained using the indicated andibodies and visualized by confocal microscopy (single 0.8 μm sections are shown).
See also Figure S3
While phospho-PIDD foci were detectable on both interphase and mitotic chromosomes (as assessed by DNA morphology and detection of KT markers; see below), γH2AX and ATMpS1981 foci could not be readily detected during mitosis (Figures S3A, B). This suggested two possibilities: 1) phospho-PIDD remains at sites of DNA damage that no longer contain phosphorylated H2A and active ATM; or, 2) following its phosphorylation by ATM at the breaks, PIDD relocates to a distinct nuclear compartment during mitosis. Strikingly, we found that in both mouse and human cells, 70–75% of phospho-PIDD foci were localized at KTs during prophase (total of 383 foci scored in 0.8 μm sections of 54 cells over 7 experiments, Figures 3C–F; PC3 cells shown in Figure S3C). KT localization of phospho-PIDD was demonstrated by staining overlaps with the centromere protein CENP-B (Figure 3C, D), the KT protein NDC80 (Figure 3E), and importantly BubR1 itself, which is recruited to KTs during prophase to initiate the spindle assembly checkpoint (Figure 3C–F and Figure S3C). The above numbers translated into approximately half of KTs, on average, being occupied by phospho-PIDD (46.6+/−19.4 %, total of 1203 KTs scored). The presence of phospho-PIDD at KTs was restricted to prophase, after which the signal sharply declined during metaphase and was no longer detectable during anaphase (Figure 3D). In prophase nuclei, the 25–30% of phospho-PIDD foci that did not reside at KTs were also not reliably located at DNA breaks, as observed in the occasional prophase nuclei with detectable γH2AX foci (5/21 prophases over three experiments, Figure 3F; note that the only two phospho-PIDD foci that do overlap with γH2AX also overlap with BubR1, likely reflecting coincidental IR-induced DNA breaks at centromeres). We conclude that subsequent to its DNA damage-induced phosphorylation by ATM, PIDD relocates from DNA breaks to KTs, where it colocalizes with its negative regulator, BubR1. To our knowledge, KTs and DNA breaks define the first reliably identified cellular localizations of the endogenous PIDD protein.
PIDD localization at kinetochores requires BubR1 and is necessary for PIDDosome inhibition
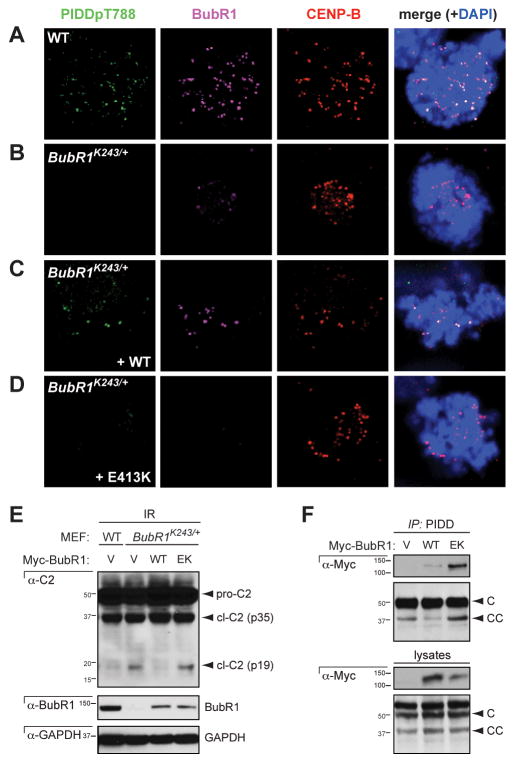
Notably, the KT localization of phospho-PIDD was lost in BubR1K243R MEFs in which BubR1 localization at KTs is substantially decreased (Figure 4A, B). Wild-type BubR1, but not the KT-deficient BubR1E413K mutant (Elowe et al., 2010), restored phospho-PIDD recruitment to KTs in these cells (Figure 4C, D). These observations showed that BubR1 is required for PIDD localization at KTs. Consistent with this finding, silencing of Bub1 or Aurora B also compromised PIDDpT788 recruitment to KTs (Figure S4).
Figure 4. Localization of PIDD at the kinetochore depends on BubR1 and is required for PIDDosome control.
(A–D) SV40 transformed MEFs of the indicated genotypes were transfected with the indicated cDNAs, treated with Go6976 (1 μM) and IR (10 Gy) and harvested 10 hr post IR. Coverslips were stained using the indicated antibodies and visualized by confocal microscopy (single 0.8 μm sections are shown).
(E) SV40 transformed MEFs of the indicated genotypes were transfected with the indicated cDNAs, treated with or without IR (10 Gy) and harvested 24 hr post IR. Lysates were analyzed by western blot.
(F) HeLa cells transfected with the indicated cDNAs were harvested 24 hr post transfection, lysed and immunoprecipitated using a polyclonal (AL233) PIDD antibody. Immunoprecipitates were analyzed by western blot using the indicted antibodies.
See also Figure S4
We then asked whether the requirement for BubR1 in PIDD recruitment to KTs was relevant to BubR1-mediated PIDDosome control. Whereas WT BubR1 restored PIDDosome suppression in BubR1K243R MEFs, BubR1E413K failed to do so (Figure 4E). The inability of BubR1E413K to rescue PIDDosome inhibition was not due to a failure to physically bind PIDD (Figure 4F), which as will be shown below is central to BubR1-mediated inhibition of the PIDDosome (see Figure 5). Therefore, the presence of PIDD at KTs, while dependent on BubR1 function, is also necessary for PIDDosome inhibition by BubR1.
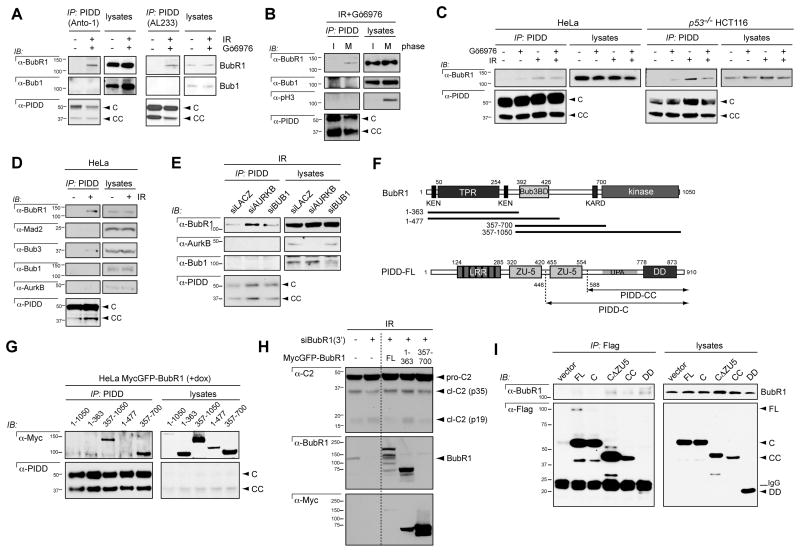
Figure 5. BubR1 interacts with PIDD.
(A) HeLa cells treated with or without Gö6976 (1 μM) plus IR (10 Gy) were harvested 24 hr post IR, lysed and immunoprecipitated with monoclonal (Anto-1) or polyclonal (AL233) PIDD antibodies. Immunoprecipitates were analyzed by western blot using the indicated antibodies.
(B) HeLa cells treated with Gö6976 (1 μM) plus IR (10 Gy) were separated into mitotic and interphase fractions by mitotic shake-off and harvested 24 hr post IR. (AL233) PIDD immunoprecipitates were analyzed by western blot.
(C) HeLa cells and TP53−/− HCT116 cells were treated with DMSO or Gö6976 (1 μM) with or without IR (10 Gy) and harvested 24 hr post IR. (AL233) PIDD immunoprecipitates were analyzed by western blot.
(D) HeLa cells treated with or without IR (10 Gy) were harvested 24 hr post IR. (Al233) PIDD immunoprecipitates were analyzed by western blot.
(E) HeLa cells were transfected with the indicated siRNAs, treated with IR (10 Gy) and harvested 24 hr post IR. (AL233) PIDD immunoprecipitates were analyzed by western blot.
(F) Diagrams of BubR1 highlighting the three major functional domains and the constructs used in (G) (TPR, tetratricopeptide) and of PIDD-FL highlighting the autoproteolytic cleavage sites and the constructs used in (I). LRR, leucine-rich repeats; ZU-5, ZO-1 and UNC5-like; DD, death domain.
(G) HeLa cells stably expressing inducible Myc-tagged forms of the constructs shown in (F) were treated with Doxycyline (1 μM) and harvested 24 hr post Dox. (AL233) PIDD immunoprecipitates were analyzed by western blot.
(H) HeLa cells stably expressing inducible Myc-tagged forms of the constructs shown in (F) were transfected with siRNAs to LacZ or the 3’UTR of BubR1. 24 hr later the cells were treated with Doxycyline (1 μM) then 24 hr post-Dox were treated with IR (10Gy). Cells were harvested 24 hr post IR. (AL233) PIDD immunoprecipitates were analyzed by western blot. Dashed line indicates deletion of irrelevant lanes from the original blot.
(I) HeLa cells transfected with the indicated Flag-tagged PIDD deletion constructs were harvested 24 hr post transfection. Flag immunoprecipitates were analyzed by western blot.
See also Figures S5 and S6
BubR1 directly interacts with PIDD after DNA damage
Our observations that BubR1 is required for PIDD localization and inhibition at KTs led us to ask whether these proteins physically interact. We readily detected BubR1, but not Bub1, in PIDD or PIDDpT788 pulldowns from mitotic or even unsynchronized HeLa cells exposed to IR+Chk1i (Figures 5A, B and S5A). The PIDD-BubR1 interaction was not observed in interphase cells, nor was it detected in untreated cells regardless of cell cycle. These results were indicative of a strong, mitosis-specific and stimulus-dependent interaction between PIDD and BubR1, consistent with their colocalization at KTs.
In both HeLa and TP53−/− HCT116 cells, the affinity of BubR1 for PIDD was maximal after IR alone and minimal in untreated cells or cells treated with Chk1i alone (Figure 5C). The stimulatory effect of IR was not due to enrichment in mitotic cells (Figure S5B, compare bars 1 and 2). The strong IR-induced interaction between BubR1 and PIDD was only moderately affected by Chk1i (Figure 5C), consistent with Chk1 and BubR1 controlling the PIDDosome in parallel (Figure 2P, R). In contrast with BubR1, Bub1 and Aurora B failed to interact with PIDD after IR, whereas the obligate BubR1 cofactor, Bub3, could be detected in the pulldowns, albeit at low levels (Figure 5D). Finally, despite participating in the MCC with BubR1 and Bub3, Mad2 failed to bind PIDD at detectable levels, even after IR (Figure 5D). These results further agreed with a Mad2- and MCC-independent role for BubR1 in PIDDosome control.
Similar to BubR1, Bub1 and Aurora B act to suppress PIDDosome activity after IR (Figures 1, 2 and S1). We considered that they may be required upstream of the BubR1-PIDD interaction, similar to their upstream requirements in BubR1-mediated mitotic checkpoint control (Johnson et al., 2004; Morrow et al., 2005). Surprisingly, depletions of Bub1 or Aurora B did not compromise the ability of BubR1 to bind PIDD, and if anything stimulated the interaction (Figure 5E). These results indicated that Bub1 and Aurora B regulate the PIDDosome either downstream of the PIDD-BubR1 interaction or in a separate pathway.
To define the PIDD interaction domain on BubR1, we analyzed PIDD pulldowns from HeLa cells stably expressing BubR1 deletion constructs (Figure 5F) (Han et al., 2013). This refined the minimal PIDD binding domain to an internal region of BubR1 spanning amino acids 357–700 (Figure 5G). Importantly, this segment was sufficient to restore PIDDosome inhibition in BubR1-depleted cells (Figure 5H; note that the 357–700 fragment does retain KT localization in Figure S6). Amino acids 357–700 exclude the kinase domain and the N-terminal Mad3 homology domain involved in Cdc20 sequestration and APC/C inhibition (Karess et al., 2013). While the segment does contain the Bub3 binding domain (amino acids 393–426), the Bub3BD is also unlikely to contribute to the interaction (Figure 1G, note that the 1–477 fragment fails to interact). Thus the PIDD interaction domain, which we designate PIDDID, likely maps to the non-characterized, 426–700 internal region of BubR1.
We next determined the BubR1 binding site on PIDD. BubR1 bound transfected Flag-PIDD-CC, with the DD therein being sufficient for binding (Figure 5I). Surprisingly, BubR1 failed to interact with PIDD-C even though this fragment also contains the DD (Figure 5F, I). This could be imputed to the additional presence within PIDD-C of a ZO-1 and UNC5-like (ZU-5) domain, whose deletion was sufficient to restore BubR1 binding (Figure 5I). Thus, BubR1 interacts with PIDD via the DD but only when supported by PIDD-CC and not PIDD-C. These binding properties are identical to that described for RAIDD (Tinel et al., 2007). Altogether, these data demonstrate a direct PIDD-BubR1 interaction that involves the PIDD DD and a previously uncharacterized internal region in BubR1, designated PIDDID. At the endogenous level, the interaction is triggered by DNA damage in a Chk1-independent manner and occurs specifically during mitosis, when PIDDosome inhibition takes place at the KT.
BubR1 outcompetes RAIDD at the PIDD DD
We next asked whether binding of BubR1 to the PIDD DD might interfere with RAIDD recruitment to the complex, thus accounting for the mechanism by which BubR1 inhibits the PIDDosome. We first performed competition assays with transiently transfected GFP-BubR1 and VSV-RAIDD constructs. Consistent with our hypothesis, increasing doses of BubR1 displaced RAIDD from Flag-PIDD (Figure 6A). Reciprocally, increasing doses of RAIDD gradually displaced BubR1 from PIDD. Interestingly, the third PIDD DD client protein known to date, receptor-interacting protein 1 (RIP1), was also displaced from PIDD by RAIDD, as previously shown (Janssens et al., 2005), as well as by BubR1 (Figure 6A).
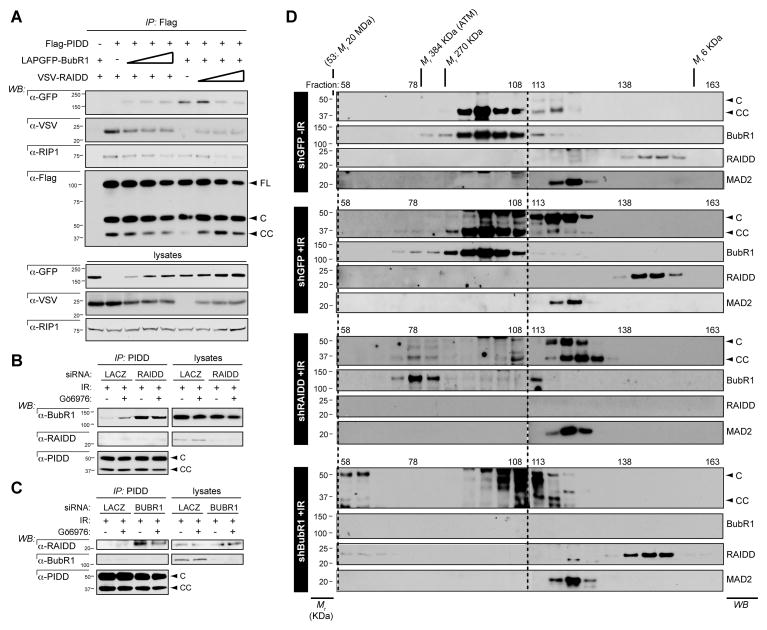
Figure 6. BubR1 competes with RAIDD for docking onto the PIDD DD.
(A) HeLa cells were transiently transfected with fixed amounts of expression vectors encoding Flag-PIDD and LapGFP-BubR1 and increasing amounts of VSV-RAIDD or vice versa. Lysates were harvested 24 hr post transfection and Flag immunoprecipitates were analyzed by western blot.
(B) HeLa cells transfected with the indicated siRNAs were treated with DMSO or Gö6976 plus IR (10Gy) and harvested 24 hours post IR. PIDD immunoprecipitates were analyzed by western blot.
(C) HeLa cells transfected with the indicated siRNAs were treated with DMSO or Gö6976 plus IR (10Gy) and harvested 24 hours post IR. (Al233) PIDD immunoprecipitates were analyzed by western blot.
(D) HeLa cells stably expressing the indicated shRNAs were treated with or without IR (10 Gy), lysed 24 hr after IR and run on a S400 HiPrep 16/60 Sephacryl column (1 ml/min). An aliquot of each fraction was concentrated and analyzed by western blot with the indicated antibodies. Blots for high fractions (left of dashed line) are longer exposures than blots for low fractions.
To validate BubR1 as a competitive inhibitor of RAIDD recruitment to PIDD, we asked whether reduction of the endogenous protein would increase the affinity of the reciprocal partner (Figures 6B, C). Consistent with the competition assays, depletion of BubR1 triggered a marked increase in RAIDD recruitment to PIDD after DNA damage (Figure 6C). Reciprocally, reduction of RAIDD exacerbated the BubR1-PIDD interaction (Figure 6B). None of these effects were due to changes in PIDD-CC levels, increases in which could have accounted for the hyperaffinities of either protein for PIDD in response to removal of the other (Figures 6B, C).
To test whether the BubR1 vs. RAIDD competition effectively impacts PIDDosome formation, we monitored complex formation by size exclusion chromatography (Figure 6D). Reduction of BubR1 after IR was sufficient to shift PIDD and RAIDD to fractions >384 kDa, consistent with their inclusion in the PIDDosome (~660 kDa) (Figure 6D, shBubR1 panel, fractions 58–63). Reciprocally, depletion of RAIDD after IR shifted PIDD-CC and BubR1 to fractions >270 kDa, reflecting their physical interaction (Figure 6D, shRAIDD panel, fractions 73–83; note that these fractions do not contain Mad2, ruling out that the observed shift in BubR1 reflected its recruitment to the MCC). Importantly, the shifts of PIDD-CC and BubR1 observed in shRAIDD cells were also observed in the presence of physiological levels of RAIDD after IR, albeit to a lesser extent (Figure 6D, compare shGFP - and +IR panels). This was to be expected from the ability of BubR1 to bind PIDD and suppress PIDDosome signaling in cells with intact RAIDD (Figures 1, 2 and 5). Altogether, the data identify BubR1 as the first described direct inhibitor of PIDDosome assembly after DNA damage.
PIDDosome assembly requires mitotic exit
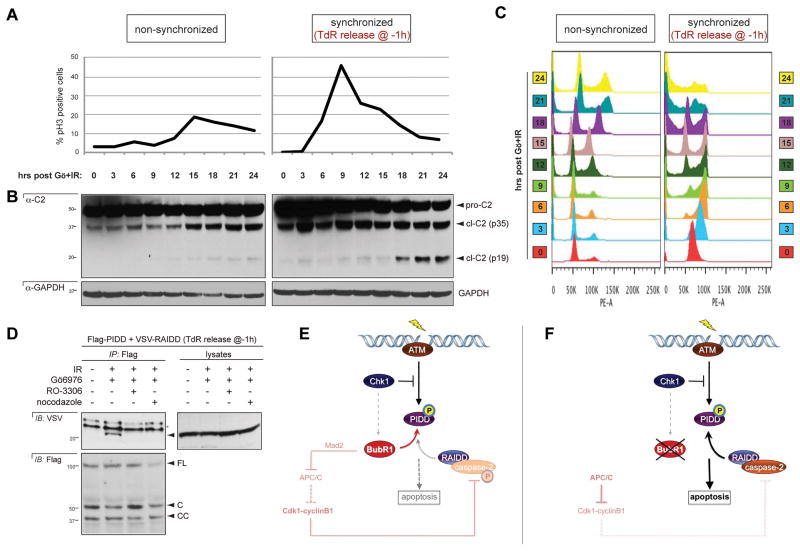
PIDDosome inhibition by BubR1 occurs at the KT (Figure 4), suggesting that PIDDosome assembly might be linked to mitotic progression. To determine the timing of PIDDosome activation during the cell cycle, we first monitored caspase-2 cleavage in synchronized cells. Caspase-2 cleavage as triggered by IR+Chk1i was first detectable 18 hr after release from TdR, coinciding with mitotic exit and cell accumulation in the next G1 (Figures 7A–C, and Figure S7 for no-treatment control). To directly determine whether PIDDosome formation occurs before, during or after mitosis, we probed the PIDD-RAIDD interaction in cells in which mitotic entry or exit was blocked via the CDK inhibitor RO-336 and the microtubule poison nocodazole, respectively. Strikingly, both drugs completely abolished RAIDD recruitment to PIDD after IR+Chk1i (Figure 7D). Therefore, PIDDosome assembly after DNA damage requires exit from mitosis. These results corroborated the identification of a mitotic effector, BubR1, as a key regulator of PIDDosome formation.
Figure 7. Exit from mitosis is required for PIDDosome formation after IR.
(A) HeLa cells were synchronized or not synchronized using the thymidine double block method then released, treated with Gö6976 plus IR (10Gy) and harvested every 3 hr post IR. Half of each harvest was stained for pHH3 and analyzed by flow cytometry.
(B) HeLa cells used in (A). Half of each harvest was lyzed and analyzed by western blot.
(C) Cell cycle profiles of the cells from (A, B) analyzed for DNA content by propidium iodide staining and analyzed by flow cytometry.
(D) HeLa cells transfected with Flag-PIDD and VSV-RAIDD were treated with Go6976 (1 μM) plus IR (10Gy) with or without RO-3306 (10μM) or Nocodazole (200ng/ ml) to prevent mitotic entry/ exit respectively. Cells were lysed 24 hr post IR. Flag immunoprecipitates were analyzed by western blot.
(E–F) Model for PIDDosome control after DNA damage, in the presence (E) or absence (F) of BubR1. Mitotic checkpoint inputs indicated in black (DNA damage-dependent events) or pink (spindle assembly checkpoint-dependent events). See the Discussion for details.
See also Figure S7
Discussion
While the stimuli and molecules that trigger PIDDosome formation have begun to emerge, the factors that act to restrain platform assembly have remained elusive. Here we identify BubR1, a mitotic surveillance protein, as a direct PIDDosome inhibitor. BubR1 outcompetes RAIDD at the PIDD docking site, thus targeting the rate-limiting step in PIDDosome formation. The inhibition occurs at KTs during early mitosis, after PIDD has been primed for complex assembly via ATM-mediated phosphorylation during the interphase. The PIDDosome thus sequentially integrates DNA-damage and mitotic checkpoint functionalities —via ATM and BubR1, respectively— to decide whether to kill a compromised cell as it threatens to divide (Figure 7E). The data also shed new light on the mechanisms by which apoptosis is actively repressed during mitosis.
PIDDosome inhibition defines a new role for BubR1 in the DNA damage response
There is evidence, however limited, that BubR1 plays a role in the DNA damage response (DDR) (reviewed in Karess et al., 2013). For example, BubR1 and Mad2, both core members of the MCC, have been shown to delay anaphase onset in response to excessive levels of DNA damage, thus acting as a last resort to arrest the cell cycle especially in situations where interphase DDR checkpoints are compromised (Fang et al., 2006; Kim and Burke, 2008; Royou et al., 2005). In contrast to this co-opted role, we here identify BubR1 as a bona fide DDR effector, acting independently of the MCC to suppress PIDDosome-mediated apoptosis.
How does the DDR engage BubR1 and mobilize it to PIDD? Like BubR1, the DDR effector Chk1 antagonizes PIDDosome signaling from zebrafish to mammals (Ando et al., 2012; Ho et al., 2009; Manzl et al., 2013; Myers et al., 2009; Pan et al., 2009; Sidi et al., 2008). A Chk1-BubR1 pathway has also been reported to operate in the cellular response to taxol (Zachos et al., 2007). Thus Chk1 might relay DDR signals to BubR1. Consistent with this view, BubR1 is sufficient to restore PIDDosome inhibition in the absence of Chk1. However, we also found that Chk1 activity was only partially required for the PIDD-BubR1 interaction and that PIDDosome control by Chk1 and BubR1 is synergistic rather than epistatic. Collectively, these observations argue in favor of predominantly independent requirements for Chk1 and BubR1 in PIDDosome control, similar to their reported independent requirements in IR-induced cell cycle arrest in yeast and Drosophila (Fang et al., 2006; Kim and Burke, 2008; Royou et al., 2005).
Role of the kinetochore
Elegant studies in yeast indicate that BubR1-mediated mitotic arrest in response to DNA damage can occur independently of KTs (Kim and Burke, 2008). In the case of PIDDosome control by BubR1, however, proper recruitment of BubR1 and PIDD to KTs is critical for inhibition of the complex. Paradoxically, while Aurora B and Bub1 are required for BubR1 and PIDD localization at KTs (Figure S4) (Johnson et al., 2004; Morrow et al., 2005), neither is necessary for the BubR1-PIDD interaction (Figure 5E). This indicates that BubR1 initially binds PIDD prior to their recruitment at KTs. Because BubR1 itself is required for PIDD localization at KTs, it is likely that BubR1-PIDD dimers/complexes assembled away from the KT then shuttle together to the KT.
If not at the KT, where might BubR1 and PIDD initially interact? Lee and colleagues identified the DNA repair factor, PARP, as a BubR1 interactor (Fang et al., 2006). Thus BubR1 may initially bind PIDD when the latter localizes at DNA breaks during the interphase (site of its phosphorylation by ATM). While we were not able to detect the presence of BubR1 foci in interphase cells, let alone at DNA breaks, this possibility remains to be explored further. We note however that the PIDD-BubR1 interaction is primarily a mitotic event (Figure 5B), arguing that PIDD and BubR1 first meet in the early prophase nucleus prior to their recruitment at KTs.
While not required for the PIDD-BubR1 interaction, Aurora B and Bub1 are nonetheless efficient PIDDosome suppressors after IR. It seems likely that their requirement for BubR1 and PIDD localization at KTs, where PIDDosome inhibition occurs, underlies their functions as PIDDosome antagonists. Such a requirement for Bub1 downstream of the BubR1-PIDD interaction is supported by their epistatic, rather than synergistic, relationship in PIDDosome control (Figure S5C). In contrast, we found that Aurora B acts synergistically with BubR1 (Figure S5D). This suggests additional independent roles for Aurora B in the regulation of the complex.
As discussed above, the PIDD-BubR1 interaction as detected by co-IP is necessary but not sufficient for PIDDosome inhibition: subsequent recruitment of PIDD-BubR1 dimers/complexes to KTs is also required. KT proteins might act to stabilize the interaction or might be necessary for fully preventing the docking of RAIDD onto the BubR1-bound PIDD DD. Alternatively, BubR1-mediated recruitment of PIDD to the KT might act to sequester PIDD from nuclear or cytoplasmic RAIDD. The development of IF-compatible antibodies should illuminate the localization of RAIDD in prophase cells.
Multistep control of PIDDosome formation by the mitotic machinery
The tight connection between the PIDDosome and the mitotic checkpoint machinery unveiled by our data is further supported by several observations. First, deletion of caspase-2 suffices to drive aneuploidy in mouse cancer models (Ho et al., 2009; Parsons et al., 2013; Puccini et al., 2013; Ren et al., 2012). Second, Kornbluth and colleagues recently reported that cdk1-cyclinB1, a key target for destruction by APC/CCdc20, phosphorylates caspase-2 on S340 during mitosis to inhibit its activity until the checkpoint is satisfied (Andersen et al., 2009). S340 phosphorylation either impairs caspase-2 recruitment to RAIDD or locks the zymogen into a non-cleavable state, thus constitutively preventing its activation regardless of whether it is mobilized by PIDD-RAIDD (Andersen et al., 2009). Thus the mitotic checkpoint apparatus controls PIDDosome assembly at least at two levels. The first, upstream of APC/CCdc20 and in response to DNA damage, limits the PIDD-RAIDD interaction via the competitive binding of BubR1 onto the PIDD DD at KTs. The second, downstream of APC/CCdc20 and in response to aberrant microtubule-KT attachments, prevents the RAIDD-caspase-2 interaction or the maturation of the caspase (Figure 7E). It follows that completion of mitosis, when APC/CCdc20 degrades both BubR1 and cdk1-cyclinB1, or mitotic checkpoint dysfunction at the level or upstream of BubR1 such as seen in human cancers (Karess et al., 2013), might act as one-two punches for PIDDosome activation, by enabling both RAIDD recruitment to, and caspase-2 activation by, the complex (Figure 7F). Indeed, we found that formation of the PIDDosome after IR coincides with mitotic exit.
Multilevel control of PIDDosome formation by the mitotic machinery is consistent with the long known propensity of mitotic cells to evade apoptosis and may contribute, alongside cdk1-mediated inhibitory phosphorylation of caspase-9 and other pathways, to a conserved mitotic program of apoptosis inhibition (Allan and Clarke, 2007; Andersen et al., 2009; Marash et al., 2008; Wan et al., 2014).
Experimental Procedures
Cell Culture and Reagents, RNAi and DNA transfections
HPV-HeLa, HEK293T, TP53−/− HCT116 and PC3 cells were cultured in DMEM medium (GIBCO) supplemented with 10% fetal bovine serum (Sigma- Aldrich) and 1% penicillin /streptomycin (GIBCO). Caspase-2−/−, Raidd−/−, Pidd−/−, BubR1K243R/+ and corresponding WT, SV40-transformed MEFs, kindly provided by Andreas Villunger, Douglas Green and Hyunsook Lee, were cultured as previously described (Manzl et al., 2009). Unless otherwise specified, cells seeded in 10cm plates were grown to 50–80% confluence for treatment with DMSO or Go6976 (1 μM final; Calbiochem) 1 hr before IR (10 Gy, Gammacell 1000 137Cs irradiator). Cells were synchronized using TdR: cells were incubated for 16 hours with 200 μM thymidine, washed twice and allowed to recover in media for 9 hours then incubated for a further 15 hours in 200 μM thymidine (Sigma-Aldrich), washed twice, given one hour to recover and treated as specified. Mitotic and interphase cells were separated by mitotic shake-off. Plates were vortexed and washed twice with PBS to remove rounded cells (mitotic) then the remaining cells (interphase) were removed using cell scrapers. Nocodazole, AZD1152 and RO-3306 were purchased from Sigma-Aldrich, 2-OH-BNPP1 was a kind gift of Hongtao Yu (Kang et al., 2008). RNAi protocols and sequences and DNA transfection protocols can be found in Supplemental Experimental Procedures.
Morpholino Injections, Drug Treatment and Acridine Orange Labeling in Zebrafish Embryos
Live p53M214K/M214K zebrafish embryos (Berghmans et al., 2005) were MO-injected at the one-cell stage or treated with kinase inhibitors at 17 hpf, irradiated with a 137Cs-irradiator at 18 hpf, labeled live with acridine orange 6hpIR, and analyzed with ImageJ as previously described (Sidi et al., 2008). See Supplemental Experimental Procedures for details including MO sequences.
Immunoblotting, coimmunoprecipitation and antibodies
Unless otherwise specified (see Supplemental Experimental Procedures), western blotting and coimmunoprecipitation protocols were as previously described (Ando et al., 2012). A full list of antibodies and protocols is provided in the Supplemental Experimental Procedures.
Confocal microscopy
MEFs and PC3 cells (5 × 104) were seeded directly onto coverslips coated with collagen (EMD Millipore), fixed in 1% paraformaldehyde, permeabilized in 0.5% Triton-X100 and stained as described. Confocal microscopy was performed using a Leica TCS SP5 II Confocal over an inverted microscope. Images were acquired using LAS software. See Supplemental Experimental Procedures for a table of antibodies used and specific staining methods.
Size Exclusion Chromatography (Gel Filtration)
SEC was performed using an AKTApurifier FPLC System according to the manufacturer’s instructions (GE). Cells were washed twice with PBS and resuspended in hypotonic buffer (20 mM Hepes- KOH, 10 mM KCl, 1 mM MgCl2, 1mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (PhosSTOP, Roche). Resuspended cells were subjected to three rounds of freeze thawing in liquid nitrogen. Debris was removed by centrifugation at 10,000×g for 20 min at 4 °C, followed by filtration at 0.2 μm. The column was equilibrated with gel filtration buffer (150 mM NaCl, 20 mM Hepes- KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, pH 7.5). Whole cell lysates (5 mg) were applied to a S400 (HiPrep 16/60 Sephacryl) gel filtration column (Amersham Biosciences). Samples were eluted at 1 ml/min collected at every 0.75ml and monitored with an online detector at 280 nm. Every fifth sample was then concentrated using Amicon® Ultracel® 3K centrifugal filter units (Millipore) and analyzed by western blot.
Statistics
Statistical significance was analyzed by two-tailed Student’s t tests. Data are represented as mean ± SD or SEM, as indicated.
Supplementary Material
Acknowledgments
We thank Evan Closser and Patrick Bradley for zebrafish husbandry, Don Cleveland, Geert Kops, Hongtao Yu, Hyunsook Lee and Sabine Elowe for sharing reagents, Ruth Rodriguez-Barrueco and Jose M. Silva for technical advice, and Andreas Villunger, Julio A. Aguirre-Ghiso, Robert Maki and Emily Arias-Foley for helpful discussions. This work was supported by NIH grants 5R01CA178162 (S.S), 1F30CA186448 (P.H.L), and awards from the Searle Scholars Program and JJR Foundation to S.S.
Footnotes
Supplemental Data include Supplemental Experimental Procedures and nine figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allan LA, Clarke PR. Phosphorylation of caspase-9 by CDK1/cyclin B1 protects mitotic cells against apoptosis. Mol Cell. 2007;26:301–310. doi: 10.1016/j.molcel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Andersen JL, Johnson CE, Freel CD, Parrish AB, Day JL, Buchakjian MR, Nutt LK, Thompson JW, Moseley MA, Kornbluth S. Restraint of apoptosis during mitosis through interdomain phosphorylation of caspase-2. EMBO J. 2009;28:3216–3227. doi: 10.1038/emboj.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, Look AT, Wang J, Bouchier-Hayes L, Sidi S. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol Cell. 2012;47:681–693. doi: 10.1016/j.molcel.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, Yeh WC, Mak TW, Benchimol S. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci USA. 2005;102:14314–14320. doi: 10.1073/pnas.0506475102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock FJ, Peintner L, Tanzer M, Manzl C, Villunger A. P53-induced protein with a death domain (PIDD): master of puppets? Oncogene. 2012;31:4733–4739. doi: 10.1038/onc.2011.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchier-Hayes L, Oberst A, McStay GP, Connell S, Tait SW, Dillon CP, Flanagan JM, Beere HM, Green DR. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol Cell. 2009;35:830–840. doi: 10.1016/j.molcel.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Elowe S, Dulla K, Uldschmid A, Li X, Dou Z, Nigg EA. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J Cell Sci. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- Fang Y, Liu T, Wang X, Yang YM, Deng H, Kunicki J, Traganos F, Darzynkiewicz Z, Lu L, Dai W. BubR1 is involved in regulation of DNA damage responses. Oncogene. 2006;25:3598–3605. doi: 10.1038/sj.onc.1209392. [DOI] [PubMed] [Google Scholar]
- Han JS, Holland AJ, Fachinetti D, Kulukian A, Cetin B, Cleveland DW. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LH, Taylor R, Dorstyn L, Cakouros D, Bouillet P, Kumar S. A tumor suppressor function for caspase-2. Proc Natl Acad Sci USA. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang TH, Park HH. PIDD mediates and stabilizes the interaction between RAIDD and caspase-2 for the PIDDosome assembly. BMB Rep. 2013;46:471–476. doi: 10.5483/BMBRep.2013.46.9.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Tinel A. The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ. 2012;19:13–20. doi: 10.1038/cdd.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Jelinek M, Balusikova K, Kopperova D, Nemcova-Furstova V, Sramek J, Fidlerova J, Zanardi I, Ojima I, Kovar J. Caspase-2 is involved in cell death induction by taxanes in breast cancer cells. Cancer Cell Int. 2013;13:42. doi: 10.1186/1475-2867-13-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–1589. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess RE, Wassmann K, Rahmani Z. New insights into the role of BubR1 in mitosis and beyond. Int Rev Cell Mol Biol. 2013;306:223–273. doi: 10.1016/B978-0-12-407694-5.00006-7. [DOI] [PubMed] [Google Scholar]
- Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IR, Murakami K, Chen NJ, Saibil SD, Matysiak-Zablocki E, Elford AR, Bonnard M, Benchimol S, Jurisicova A, Yeh WC, et al. DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis. 2009;14:1039–1049. doi: 10.1007/s10495-009-0375-1. [DOI] [PubMed] [Google Scholar]
- Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat Rev Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Manzl C, Fava LL, Krumschnabel G, Peintner L, Tanzer MC, Soratroi C, Bock FJ, Schuler F, Luef B, Geley S, et al. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis. 2013;4:e942. doi: 10.1038/cddis.2013.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Krumschnabel G, Bock F, Sohm B, Labi V, Baumgartner F, Logette E, Tschopp J, Villunger A. Caspase-2 activation in the absence of PIDDosome formation. J Cell Biol. 2009;185:291–303. doi: 10.1083/jcb.200811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzl C, Peintner L, Krumschnabel G, Bock F, Labi V, Drach M, Newbold A, Johnstone R, Villunger A. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Diff. 2012;19:1722–1732. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell. 2008;30:447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Myers K, Gagou ME, Zuazua-Villar P, Rodriguez R, Meuth M. ATR and Chk1 suppress a caspase-3-dependent apoptotic response following DNA replication stress. PLoS Genet. 2009;5:e1000324. doi: 10.1371/journal.pgen.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niizuma K, Endo H, Nito C, Myer DJ, Kim GS, Chan PH. The PIDDosome mediates delayed death of hippocampal CA1 neurons after transient global cerebral ischemia in rats. Proc Natl Acad Sci USA. 2008;105:16368–16373. doi: 10.1073/pnas.0806222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ren KH, He HW, Shao RG. Knockdown of Chk1 sensitizes human colon carcinoma HCT116 cells in a p53-dependent manner to lidamycin through abrogation of a G2/M checkpoint and induction of apoptosis. Cancer Biol Ther. 2009;8:1559–1566. doi: 10.4161/cbt.8.16.8955. [DOI] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Lee HO, Choi E, Lee YK, Kwon MS, Min J, Park PG, Lee S, Kong YY, Gong G, et al. Loss of BubR1 acetylation causes defects in spindle assembly checkpoint signaling and promotes tumor formation. J Cell Biol. 2013;202:295–309. doi: 10.1083/jcb.201210099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons MJ, McCormick L, Janke L, Howard A, Bouchier-Hayes L, Green DR. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Diff. 2013;20:1174–1182. doi: 10.1038/cdd.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccini J, Shalini S, Voss AK, Gatei M, Wilson CH, Hiwase DK, Lavin MF, Dorstyn L, Kumar S. Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc Natl Acad Sci USA. 2013;110:19920–19925. doi: 10.1073/pnas.1311947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren K, Lu J, Porollo A, Du C. Tumor-suppressing function of caspase-2 requires catalytic site Cys-320 and site Ser-139 in mice. J Biol Chem. 2012;287:14792–14802. doi: 10.1074/jbc.M112.347625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribe EM, Jean YY, Goldstein RL, Manzl C, Stefanis L, Villunger A, Troy CM. Neuronal caspase 2 activity and function requires RAIDD, but not PIDD. Biochem J. 2012;444:591–599. doi: 10.1042/BJ20111588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Barrueco R, Marshall N, Silva JM. Pooled shRNA screenings: experimental approach. Methods Mol Biol. 2013;980:353–370. doi: 10.1007/978-1-62703-287-2_21. [DOI] [PubMed] [Google Scholar]
- Royou A, Macias H, Sullivan W. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr Biol. 2005;15:334–339. doi: 10.1016/j.cub.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, Hoffmans R, Pascual J, Imamura S, Kishi S, Amatruda JF, et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell. 2008;133:864–877. doi: 10.1016/j.cell.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23:745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, Lin LL. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- Terry MR, Arya R, Mukhopadhyay A, Berrett KC, Clair PM, Witt B, Salama ME, Bhutkar A, Oliver TG. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J. 2007;26:197–208. doi: 10.1038/sj.emboj.7601473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, Liu J, Shaik S, Chen G, Deng J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell. 2014;29:377–391. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, Gillespie DA. Chk1 is required for spindle checkpoint function. Dev Cell. 2007;12:247–260. doi: 10.1016/j.devcel.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.