Abstract
T regulatory (Treg) cells are central to the maintenance of immune homeostasis. The transcription factor Foxp3 is essential for specifying the Treg cell lineage during development, and continued expression of Foxp3 in mature Treg cells is necessary for suppressive function. Treg cells can lose Foxp3 expression under certain conditions, and this is associated with autoimmune pathology. Here we review recent insights into the mechanisms that maintain Treg cell stability and function, and place these findings within the broader understanding of mechanisms that establish Treg cell identity during development. We integrate emerging principles in Treg cell lineage maintenance with the mechanisms that allow Treg cells to sense and respond to varied inflammatory environments, and outline important areas of future inquiry in this context.
Introduction
Regulatory T (Treg) cells play an indispensable role in homeostasis of the immune system. Perturbations of Treg cell differentiation and function lead to autoimmune diseases and immunopathology (1). Foxp3, a member of the forkhead transcription factor family, is an essential regulator of both the establishment of the Treg cell lineage and the suppressor function of these cells (2-4). Although recent studies have shown that Foxp3 is temporarily expressed in non-Treg cells and that epigenetic modifications unrelated to Foxp3 function play critical role in Treg cell lineage establishment (5, 6), sustained expression of Foxp3 is an essential feature of Treg cells. Whereas effector T cells can differentiate into different T helper subsets (Th1, Th2, Th17, etc.) in response to a wide range of pathogens and cytokines in the inflammatory environment, Treg cells do not further differentiate into stable subsets (7). However, they display a certain level of functional plasticity that involves the ability to sense cytokines in their milieu and adjust the expression of a subset of genes accordingly; this functional plasticity is essential for the appropriate regulation of the surrounding immune response.
While, the flexibility of Treg cells to acclimate to their microenvironment is vital to their suppressive function, it also poses a potential threat to immune homeostasis. Most Treg cells recognize self-antigens, and thus loss of Foxp3 expression and the concomitant loss of suppressive function can result in auto-reactive cells that promote autoimmune disease. Recent studies have provided insight into the Treg cell-intrinsic programs in place to maintain Foxp3 expression and safeguard Treg cell identity, revealing a central role for a Foxp3 intronic enhancer that serves as a sensor of both TCR and cytokine signals and translates these inputs into increased Foxp3 transcription during Treg cell activation (8, 9). We discuss these findings here, and place them in the context of the broader understanding of the cellular and molecular mechanisms that regulate Foxp3 expression during Treg cell lineage establishment and maintenance.
What constitutes Treg cell identity?
A prerequisite for studying the regulation of Treg cell lineage formation and stability is the identification of key characteristics and molecular markers defining Treg cell identity. The central feature of Treg cells is their immune suppressor function, mediated through a set of diverse mechanisms (10, 11). Other important characteristics of Treg cells include their dependence on IL-2, absence of expression of effector cytokines associated with other T helper cell lineages such as IFN-γ, IL-4, and IL-17, and distinct regulation of their intracellular metabolism (12). Among several cellular markers that have been associated with Treg cell fate and function, expression of the transcriptional regulator Foxp3 is the most specific feature that distinguishes Treg cells from other T helper lineages. First, as the Treg cell lineage specification transcription factor, Foxp3 expression is required for the Treg cell differentiation. Germline deletion of the Foxp3 gene leads to Treg cell deficiency and the development of lethal autoimmune syndrome (2-4). Second, beyond its role in Treg differentiation, continuous Foxp3 expression is also required in mature Treg cells for their suppressive function and the full manifestation of the aforementioned key features of Treg cells. Deletion of Foxp3 in fully differentiated mature Treg cells results in the deregulation of its target genes and the loss of suppression function (13). Last but not least, Foxp3 helps to prevent Treg cells from acquiring alternative fates since the ablation or severe attenuation of Foxp3 expression leads to the expression of effector cytokine genes that are characteristic of other CD4 helper lineages. (13-15). In mice containing a Foxp3 GFP reporter null allele (Foxp3GFPKO), GFP+ T cells, which are “wannabe” Treg cells but are unable to express Foxp3 protein, produce Th2 and Th17 cytokines IL-4 and IL-17 (14). The production of proinflammatory cytokines IL-2, TNFα, IFN-γ, IL-17, and IL-4 is also increased in mature Treg cells when Foxp3 is acutely ablated using retroviral expression of Cre (13). Foxp3 can suppress Th17 differentiation by inhibiting the function of Th17 lineage specifying transcription factor RORγt (16). Therefore, Foxp3 expression is a central contributor to Treg cell identity.
However, despite its importance and specificity, Foxp3 expression cannot be considered an unambiguous marker for Treg cells. Comparison of the transcriptional profile of Treg cells with that of conventional CD4+ T cells made to express Foxp3 through retroviral transduction showed that Foxp3 expression alone is not sufficient to confer T cells with the expression of many Treg cell signature genes, despite the ability of Foxp3 to regulate the expression of many of its cofactors (17, 18). Thus, it appears that the formation of Treg cell identity, characterized by its unique transcription program, requires the activation of additional genes. Indeed, enforced expression of Eos, IRF4, Satb1, Lef1, or GATA-3 cooperates with Foxp3 to activate the expression of most, but still not all of the Treg signature genes (19). DNA methylation analysis revealed that Treg cells establish a specific CpG hypomethylation pattern associated with Treg-cell-specific gene expression (6, 20). Importantly, this Treg-cell-specific CpG hypomethylation pattern can be established in Foxp3-null Treg cells but is absent in Foxp3+ iTreg cells (6). Therefore, it is possible that a Foxp3+ cell is not a bona fide Treg cell due to the lack of a Treg-cell-specific epigenetic landscape and the expression of other genes that are required to establish the full Treg cell transcription program. The potential disconnect between Foxp3 expression and Treg cell identity warrants caution in using Foxp3 as a sole marker of Treg cell identity. Moreover, these findings highlight the question of what mechanisms, beyond Foxp3 expression, establish Treg cell identity. We discuss these below.
Establishment of the regulatory T cell identity
Regulatory T cells are generated in the thymus (tTreg) and the periphery (pTreg). The majority of the Treg cells emerge in the thymus at the CD4 single positive stage when Foxp3 expression is induced (21). TCR signal strength, IL-2, and TGF-β are required for Foxp3 upregulation during Treg cell differentiation in thymus, and these are discussed in turn.
It has been postulated that TCRs from Treg cells have intermediate affinity to self-antigens – that is, stronger affinity than that of naïve CD4 T cell TCRs and weaker affinity than the threshold for negative selection. Several lines of evidence support this notion. In a double transgenic mouse carrying a specific TCR expressed in T cells and its cognitive antigen expressed ubiquitously driven by SV40 promoter (TS1xHA28), tTreg generation increased significantly (22, 23). TCR sequencing studies showed that Treg cell and naïve T cell TCR repertoires are mostly non-overlapping although a small percentage of TCRs are shared by both T cell populations (24, 25). To test the self-reactivity of TCRs derived from Treg or naïve T cells, Rag1 knockout Tcli-TCR transgenic T cells were transduced by retroviral vectors carrying individual TCRs and adoptively transferred into lymphopenic recipients. T cells expressing Treg TCRs, not naïve T cell TCRs, undergo increased homeostatic expansion after adoptive transfer, suggesting these TCRs have higher affinity to self-antigens (24). Recently, a mouse was generated that carries a GFP reporter driven by the promoter of Nur77 transcription factor (Nur77GFP); GFP fluorescence intensity of T cells in this mouse is a direct reflection of TCR signal strength (26). In this model, Foxp3+ Treg cells were shown to express higher levels of GFP in the thymus and in the periphery as compared to non-Treg CD4 T cells. Interestingly, by using a series of TCRs that react to an ovalbumin peptide at increased affinity, Hsieh et al showed Tregs can be generated at a surprisingly wide range of TCR strength, which could contribute to the partial overlap of the Treg and naïve T cell TCR repertoires (27). Future study is needed to illuminate how the timing and duration of antigen stimulation in combination with TCR strength can provide the optimum signal for Treg development.
Although the self-antigens that select Treg cells are not identified so far, several studies have demonstrated that Autoimmune Regulator (Aire)-mediated thymic expression of self-antigens drive development of a subpopulation of tTregs. Aire is expressed in the medulla thymic epithelial cells (mTECs) and drives promiscuous expression of peripheral tissue antigens in these cells for T cell selection (28). Malchow and colleagues showed that thymic generation of a Treg cell subset recognizing a prostate specific antigen was dependent on Aire expression (29). Through TCR repertoire sequencing study, Hsieh’s group showed that Aire-dependent expression of self-antigens preferably select Tregs carrying lower frequency TCRs (30). Recently, Yang and colleagues discovered that tTreg cells generated at perinatal stage in an Aire-dependent manner are a distinct population compared to adult generated tTregs in terms of TCR repertoire and gene expression profile, adding an age-related layer to Treg cell selection and function (31).
In addition to TCR stimulation, signals downstream of IL-2 are also critical for thymic Treg cell differentiation. An early study showed that thymic transgenic expression of IL-2Rβ driven by thymic-specific proximal lck promoter is sufficient to restore Treg cell generation and rescue the lethal autoimmunity associated with IL-2Rβ-deficient mice (32)(33). The numbers of thymic Tregs, as defined by FoxP3 expression, in IL-2Rα deficient mice were reduced by approximately 50% as compared to wild-type (WT) mice, and ablation of IL-2Rγ completely block Treg cell development (34). Furthermore, a population of thymic Treg precursors (CD4+CD25+Foxp3−) can up-regulate Foxp3 expression in response to IL-2 and, to a lesser extent, IL-15, in the absence of TCR signal (35). IL-2 signals downstream of the IL-2R are transduced via the activity of STAT5. Mice engineered to express a constitutively active STAT5 (CA-STAT5) in lymphocytes displayed a dramatic increase in the numbers of Treg cells, as compared to WT mice (36, 37). Interesting, TCR signal strength defined by Nur77GFP reporter expression did not increase in the Tregs from the CA-STAT5 mice, suggesting that TCR and IL-2 signals can work independently in turning on Foxp3 expression (26).
The role of TGF-β in tTreg generation was explored by previous studies showing that ablation of TGF-βRI lead to defective tTreg generation in neonatal mice, although this deficiency could be compensated later on by homeostatic expansion of Tregs (38). TGF-β signal can enhance Treg survival during negative selection, as TGF-β receptor deficient thymic Tregs undergo enhanced apoptosis due to reduced expression of Bcl2 and increased expression of Bim, Bax, and Bak (39). Interestingly, mice deficient of both TGF-βRI and IL-2Rα show a complete absence of tTreg cells, suggesting compensatory roles for IL-2 and TGF-β during tTreg differentiation (38).
Generation of Tregs also occurs in the periphery through the conversion of naïve CD4 T cells. Early studies showed that TGF-β treatment of naive CD4 T cells can induce Foxp3 expression and convert them into induced Treg cells (iTreg) (40). Although iTreg cells acquire certain property of tTregs, such as immune suppression function, they don’t carry the epigenetic markers of tTregs and rapidly lose Foxp3 expression in vivo (41). In contrast to in vitro generated iTregs, in vivo generated pTregs are very similar to tTregs in terms of gene expression, suppression function, and lineage stability. Currently, the most well studied cases of in vivo pTreg cell generation were on Treg cells residing in the gut induced by microbiota. Round and colleagues showed colonization of germ-free mice with B. fragilis increases colonic Treg cell numbers. Induction of Treg cells is dependent on an immunomodulatory molecule, polysaccharide A (PSA), produced by B. fragilis (42). Honda’s group showed similar Treg cell induction by colonizing gut with Closditrium species. In this study, Treg cell induction is dependent on TGF-β signaling pathway as TGF-β neutralizing antibody treatment abolishes Treg induction (43). Treg cells induced by gut microbes in both studies express IL-10 and are protective against colitis. By sequencing TCR repertoire of colonic Treg cells, Lanthrop and colleagues showed that TCRs of these Treg cells were different from Treg cells in other tissues. Using a GFP-NFAT reporter cell line, they identified individual TCRs that recognize a number of colonic bacterial isolates. Naïve T cells expressing these colonic microbe-reactive TCRs convert to Foxp3+ Helios− pTreg cells specifically in the colon lamina propria, suggesting TCR stimulation is required for pTreg generation (44).
Due to a large number of commensal bacteria residing in the gut, generation of pTreg cells is also influenced by metabolites present in the gut microenvironment. Early studies showed that vitamin A and its derivative retinoid acid (RA) could promote TGF-β dependent conversion of naive CD4 T into Foxp3+ Treg cells. CD103+ dendritic cells from gut and mesenteric lymph node facilitate this conversion process (45, 46). Recently, three groups reported that short-chain fatty acids (SCFAs) including propionate and butyrate, products from metabolism of gut microbiota, could promote naive T cell to Treg cell conversion in vitro and in vivo (47-49). One mechanism proposed by these studies was that SCFAs could inhibit histone deacetylase (HDAC) activity, and promote histone acetylation at the Foxp3 locus, which in turn leads to Foxp3 transcription. Future studies will elucidate if other classes of gut microbe metabolites are involved in tuning of gut immune cells and maintenance host-microbe homeostasis.
Differentiations of tTreg and pTreg occur at different locations and start from different precursor cells, yet similar factors are involved in both processes. TCR stimulation is required for both tTreg and pTreg cell differentiation, but it is not likely that same TCR signal strength leads to Foxp3 induction in both tTreg and pTreg cells. Differentiation of tTreg requires intermediate to high TCR signal, while pTreg generation prefers weaker TCR stimulation in the absence of pro-inflammatory cytokines. TGF-β is also involved in generation of both Treg subsets. While it exerts anti-apoptotic effect on tTreg cells (39), TGF-β induces Smad binding to Foxp3 locus and directly promotes Foxp3 transcription in pTreg (50-52). The similarity and differences between establishement of tTreg and pTreg cell identities are likely due to different kind of antigens they recognize (self for tTreg cell and environmental for pTreg cell) despite their shared gene expression profile and suppressive functionality.
Epigenetic regulation during Treg cell identity formation
Although the molecular mechanism of Foxp3 induction was the focus of most studies on Treg differentiation over the past decade, it has been increasingly clear that epigenetic changes also play an indispensable role in the formation of the Treg cell lineage. A recent study showed that only a very small portion (300 out of ~160,000) of DNA methylated regions in conventional T cells are demethylated and become accessible in Treg cells (20). These Treg cell - specific demethylated regions (TSDRs) are enriched in Treg signature genes such as Foxp3, Ctla4, and Il2ra, suggesting that highly specific epigenetic modifications occur during Treg development (6, 20). Genome-wide mapping of accessible chromatin regions by DNase I hypersensitivity assay (DHS) also revealed similar global open chromatin regions in Treg cells and activated conventional T cells except for a small number of loci with Treg specific expression pattern (53). Expression of Foxp3 is not required for setting up the vast majority of the open chromatin regions in Treg cells, instead, Foxp3 selectively binds to these regions to shape the expression of key Treg cell signature genes (53). Similar observations were reported for the emergence of active enhancers defined by p300 binding regions during Th1 and Th2 cell differentiation (54). Lineage specification factors T-bet and GATA3 for Th1 and Th2 cells, respectively, are not required for the formation of most of the active enhancers. Instead, STAT5 plays a major role in defining the enhancer landscape in these cells (54). Surprisingly, TSDRs and Foxp3-bound DHS sites occupy two separate sets of loci, with the exception for the Foxp3 locus itself, implicating two different programs are involved in regulating gene expression in Tregs (20). The contributions of Foxp3 dependent and independent mechanisms in influencing epigenetic landscape and controlling gene expression of Treg cells remains to be determined.
Lineage stability of mature Treg cells
A number of groups have examined the lineage stability of Treg cells in a variety of contexts. Treg cells showed considerable instability when stimulated in vitro in the presence of pro-inflammatory cytokines such as IL-6 or anti-OX40 antibody (55-57). Although in vitro culture does not recapitulate physiological conditions, these studies raised important questions as to whether pro-inflammatory conditions influence Treg cell stability, and whether subsets of Foxp3+ cells (may or may not be fully committed Treg cells, as discussed later) exist that are prone to losing Foxp3 expression.
Treg cell stability can also be examined by using an adoptive transfer approach, which entails transferring congenically marked Treg cells isolated from Foxp3 reporter mice into recipient mice, and measuring Foxp3 expression in these cells. Additional cell surface markers may be used in combination with the Foxp3 reporter to isolate a more defined subset of Treg cells. Cells can also be labeled with a cell division dye prior to transfer into recipients to facilitate the measurement of cell proliferation. In addition, recipient mice can be manipulated to examine the effects of specific conditions or treatments on the stability of transferred Treg cells. These experiments present the advantage of in vivo physiological conditions, although it should be noted that the stress associated with cell isolation and transfer may still have an impact on outcome. Results from one such study showed that over 90% of transferred Foxp3+ cells can maintain Foxp3 expression after being transferred together with conventional T cells into lymphopenic mice (58). Similar stability of Foxp3 expression was observed when Treg cells were transferred into lymphoreplete recipients. Interestingly, cells that have unstable Foxp3 expression are mostly limited to the Foxp3+CD25− subset, indicating the heterogeneity of Foxp3+ cells and raising the possibility that some Foxp3+ cells may not be fully committed Treg cells, or alternatively, that some Treg cells are inherently less stable than others.
To overcome potential drawbacks associated with methods employing in vitro culture and adoptive transfer approaches, more recent studies have utilized genetic fate mapping approaches to study lineage stability of Treg cells. One group used Cre recombinase driven by a constitutive Foxp3 BAC transgene, together with a reporter allele that express YFP upon Cre-mediated recombination, thus rendering YFP positive all cells that have expressed FoxP3 at some point in their lifetime; 10-20% of peripheral YFP+ cells were shown to be Foxp3− (59). Furthermore, using this approach in nonobese diabetic (NOD) mice revealed that ~20 and 30% of YFP+ cells were Foxp3− in the pancreatic lymph nodes and pancreas, respectively. Thus, autoimmune conditions appear to exacerbate instability of Foxp3 expression. Because Cre recombinase is constitutively expressed, it is possible that non-Treg cells that transiently expressed Foxp3 were marked. Although the frequency of these transient Foxp3 expressing cells may be low, gradual accumulation of these marked cells as mice age may account for the relatively large proportion of ex-Foxp3 cells observed in this study.
One way to minimize the continuous accumulation of cells that are labeled due to transient Foxp3 expression in non-Treg cells is to pulse-label Foxp3 expressing cells. Using an inducible Cre allele driven by the Foxp3eGFP-CreERT2 locus in combination with a Cre-activated YFP reporter, Rubtsov et al (60) showed that over 95% of YFP+ cells are still GFP+ at either 2 weeks or 5 months after Cre activation. Similar results were obtained when mice were subjected to sub-lethal irradiation-induced lymphopenia, bacterial infection, CD40 ligation-induced Th1 type inflammation and autoimmune inflammation. These results support the notion that the great majority of Treg cells can maintain stable Foxp3 expression in vivo, and raise the possibility that unstable Foxp3 expression observed in some of the previous studies might have resulted from the presence of Foxp3+ cells that have not fully committed to the Treg cell fate.
Recent studies have characterized these unstable Foxp3+ cells by distinguishing cells that have only started to express Foxp3 from those that have expressed Foxp3 for longer periods by examining the expression levels of RFP in Foxp3+ CD4 T cells from Foxp3GFPCre × ROSA26-RFP mice (5). High RFP expression levels indicate a longer history of Foxp3 expression whereas low or negative RFP expression indicates more recent initiation of Foxp3 expression. When GFP+ RFPhi or GFP+ RFP−/lo cells were cultured in vitro, over 98% of GFP+ RFPhi cells maintained GFP expression (marker of Foxp3 expression), but over 10% of GFP+ RFP−/lo cells lost GFP expression. Furthermore, addition of IL-6 or TGF-β blocking antibody to the culture or transferring cells into Rag1-deficient hosts did not substantially increase the loss of GFP expression in GFP+ RFPhi cells, but resulted in ~50% of GFP+ RFP−/lo cells losing GFP expression. Notably, GFP+ RFP−/lo cells have high levels of CpG methylation of CNS2, which is demethylated in committed Treg cells (5, 41). These findings confirm that fully committed Treg cells are a stable population, and show that a small population of Foxp3+ cells that have only recently started expressing Foxp3 present the main source of the ex-Foxp3+ cells observed in inflammatory or lymphopenic environment (5).
Most of the studies examining the stability of Treg cell lineage have focused on Foxp3 expression due to the central role of Foxp3 in specifying Treg cell lineage. However, because Foxp3 alone is insufficient to drive the full Treg cell transcription program and to define Treg cell identity, Treg cells can conceivably be destabilized when the expression or activity of essential cofactors is compromised despite stable Foxp3 expression. For example, deletion of Foxo1 in Treg cells severely impairs Treg cell function and results in a lethal inflammatory disorder in mice without reducing the frequency of Foxp3+ cells and the Foxp3 expression levels in individual cells (61). Instead, Foxo1 regulates the expression of many genes, including inhibiting the expression of IFN-γ, the deletion of which partially restored the function of Foxo1 deficient Treg cells. Thus, Foxo1 expression seems to be essential for Treg cell identity considering its critical contribution to Treg-cell-specific transcription program and function. Treg-cell-specific epigenetic landscape, including CpG hypomethylation may also regulate Treg cell identity beyond its role in regulating Foxp3 expression since a number of the hypomethylated regions lie close to or within loci of genes with known functions in Treg cells (6). Further insights on the stability of Treg cell identity beyond Foxp3 expression may be gained from a more comprehensive understanding of the identity and regulation of key factors collaborating with Foxp3 to specify the core Treg-cell transcription program that support Treg function and suppress alternative cell fates.
Nonetheless, these studies point to the multifaceted characteristics of Treg cell identity, the complex and temporal nature of the Treg lineage commitment process, as well as limitation of Foxp3 as a marker of commitment of Treg cell fate.
Plasticity and heterogeneity of Treg cells
It is increasingly clear that Treg cells are not a homogenous population with a rigid transcriptional program. To be able to optimally control diverse types of immune responses in potentially drastically different and rapidly changing local environments, Treg cells have to possess a certain degree of functional plasticity to tailor their suppressive function and homeostatic properties to fulfill specific regulatory roles in diverse contexts. To achieve functional plasticity, Treg cells alter their transcriptional program to meet specific regulatory needs while preserving their core immune suppressive features. Treg cells co-opt a growing list of transcription factors that promote specific types of effector T cell differentiation, to control the same types of effector T cell response. For example, Treg cell specific deletion of an essential Th2 differentiation factor, Irf4, specifically abolished the ability of Treg cells to control Th2 responses (62). T-bet expression allows Treg cells to adopt features of Th1 cells such as the expression of CXCR3, which is a key chemokine receptor mediating the accumulation of Th1 cells at local inflammation sites (63). Deletion of T-bet impaired the ability of Treg cells to control Th1 type inflammation. Similarly, expression of Stat3, a key transcription factor for Th17 cells, in Treg cells is essential for control of Th17 responses and expression of Bcl6, a crucial transcription factor in follicular helper T cells, endows Treg cells the ability to control germinal center responses (64-66). Expression of other transcription factors and certain microRNAs has been shown to bestow Treg cells with capabilities to regulate specific subsets of immune responses (67-71). In addition, there is an emerging recognition and appreciation of the existence of Treg cells residing in non-lymphoid tissues and their unique functions and phenotypes (72). These tissue-residing Treg cells often have distinct transcription programs as compared to Treg cells in lymphoid organs. For example, Treg cells in the visceral adipose tissue (VAT) express high levels of PPARγ, which is the master transcription factor regulating adipocyte differentiation (73). PPARγ expression is essential for establishing the unique VAT Treg transcription program, their phenotype, and homeostasis. Similarly, Treg cells accumulate in skeletal muscles following acute injury to promote muscle repair (74). These muscle-resident Treg cells also exhibit distinct transcription program that may support their repair function. Therefore, the function of Tregs partially depends on a certain degree of plasticity they exhibit in response to the microenvironment.
Foxp3 intronic enhancers and Treg cell fate
The ability of Treg cells to preserve their core identity while exhibiting flexibility in their function, phenotype, and associated transcription program suggests that powerful intrinsic mechanisms exist to protect their identity. Given the central role of Foxp3 in safeguarding Treg cell fate, it is imperative to understand the mechanisms of stable Foxp3 expression in Treg cells. The Foxp3 gene contains multiple evolutionally conserved non-coding sequence (CNS) elements, which are usually identified as enhancers that regulate gene expression. Indeed, three intronic Foxp3 CNS regions (named CNS1, 2 and 3) are implicated in different aspects of regulation of Foxp3 expression and Treg fate decision (Box 1). CNS3 is involved in the initial induction of Foxp3 in tTregs by recruiting cRel to the Foxp3 locus (75). CNS1 contains a Smad binding motif downstream of TGF-β signal, and is implicated in the generation of pTregs (75, 76). CNS1 deficient mice exhibit allergic-type Th2 inflammation at mucosal interfaces like gut and lung, due to defect in pTreg induction at these sites (51, 52, 75). Recent studies examining CNS2 deficient mice have revealed that, unlike CNS1 and CNS3, CNS2 serves a pivotal role in maintaining Foxp3 expression in mature Tregs (8, 9, 75).
Box1. By controlling Foxp3 transcription, cis-regulatory elements on the Foxp3 gene locus play central roles in the differentiation and stability of Treg cells.
Foxp3 promoter
Although the basal Foxp3 promoter contains sites for the binding of several transcription factors, including NF-κB, Foxo1/3a, NFAT, AP-1, SP1, STAT5, and Runx, its transactivation activity appears to be weak. On the one hand, a weak transactivation activity may help prevent promiscuous Foxp3 induction. On the other hand, other cis-regulatory elements are needed for the induction and maintenance of Foxp3 expression in Treg cells.
CNS1
CNS1 is critical for Foxp3 induction during peripheral differentiation of Treg cells, but not for the thymic Treg differentiation. The binding of NFAT, Smad3, and RAR/RXR likely facilitates the induction of Foxp3 expression in naïve CD4 T cells in the presence of TGF-β and retinoic acid. Enhancement of peripheral Treg generation by commensal bacteria-derived butyrate also requires CNS1.
CNS2
CNS2 contains a CpG island, which is methylated in non-Treg cells, including Treg precursors, and is demethylated in committed Treg cells. CpG methylation on CNS2 inhibits the binding of transcription factors. Consistent with this, CNS2 is not required for Foxp3 induction during Treg differentiation in the thymus and periphery. Instead, it is required for maintaining Foxp3 expression in activated and dividing mature Treg cells, especially in the presence of proinflammatory cytokines including IL-4 and IL-6, or when IL-2 is limited. Upon TCR activation, CNS2 promotes Foxp3 expression by interacting with Foxp3 promoter in a Calcineurin/NFAT-dependent manner, which may allow CNS2-bound factors including STAT5 to access Foxp3 promoter.
CNS3
This conserved non-coding DNA sequence element exhibits histone marks indicative of regulatory function even in thymic and peripheral Treg cell precursors. Its deletion leads to impaired differentiation of both thymic and peripheral Treg cells. It is bound by c-Rel, which is also required for efficient Foxp3 induction. Thus, CNS3 appears to be a pioneering element facilitating Foxp3 induction during Treg differentiation.
CNS2 and preservation of Treg cell identity
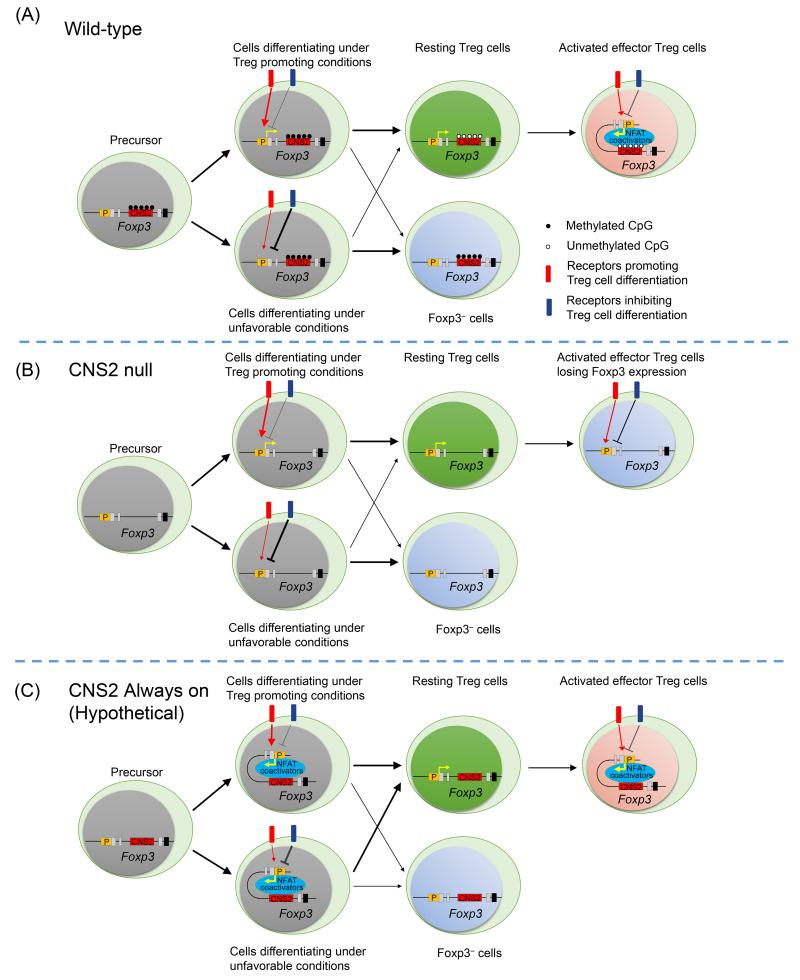
CNS2 is unique among all Foxp3 CNSs in that it contains the only CpG-rich region within the Foxp3 locus. CNS2 is heavily methylated in Treg precursors in the thymus and conventional T cells in the periphery (41, 77, 78). CD4 T cells with unstable Foxp3 expression such as Treg cells induced by TGF-β treatment in vitro (iTregs) also retain methylated CNS2 (41, 78). Demethylation of CNS2 starts during Treg development in the thymus concurrently with induction of Foxp3 expression, and completes soon after mature Tregs emigrate into the periphery (Figure 1A)(41, 79). Therefore, demethylation of CNS2 is only associated with mature Tregs with sustained Foxp3 expression, suggesting a role of CNS2 in promoting stable Foxp3 expression. Indeed, deletion of CNS2 resulted in specific impairment in Foxp3 expression in mature Treg cells, whereas Treg cell development in the thymus and Foxp3 induction during iTreg cell differentiation in vitro appeared normal (Figure 1B)(75). Two recent studies examined the physiological role and mechanisms of CNS2-mediated maintenance of Treg cell identity (8, 9). Unexpectedly, CNS2 deletion did not result in early onset severe autoimmune diseases observed in Foxp3 null mice. Instead, these mice showed mild lymphoproliferative diseases and increased inflammation in multiple tissues as they age. Elevation of both Th1 and Th2 type inflammation in CNS2 deficient mice suggests that CNS2 confers Treg cell function in controlling multiple types of immune responses. Surprisingly, CNS2 deficient and wild type Treg cells had comparable overall Foxp3 expression, suggesting that only a subset of Tregs require CNS2 for maintaining Foxp3 expression in steady state. Indeed, CNS2 deficient Treg cells that express low levels of Foxp3 exhibit gene expression signatures indicative of high levels of TCR activation, cell proliferation, responsiveness to chemotactic and proinflammatory cytokines, and suppressor function (9). Furthermore, CNS2 deficient Treg cells are prone to losing Foxp3 expression in response to strong TCR activation, stimulation with proinflammatory cytokines including IL-4 and IL-6, and deprivation of IL-2 (8, 9). Interestingly, CNS2 deficient Treg cells appeared to be particularly incapable of maintaining Foxp3 expression in certain tissues such as small intestine, liver, and lung, where increased inflammation was observed in CNS2 KO mice (8, 9). These results suggest that CNS2 is especially important for the lineage stability of Treg cells at the environmental barriers, where increased stimulation by proinflammatory cytokines and abundance of foreign antigens can lead to Treg cell activation (Figure 1B). Therefore, CNS2 is critical for maintaining Foxp3 expression when committed Treg cells get activated and further differentiate into effector Treg cells. Conversely, the powerful stabilizing effects on Foxp3 expression may also explain the timing of its demethylation, as premature activation of CNS2 through demethylation may lead to excessive induction of Foxp3 expression during Treg cell differentiation (Figure 1C). Indeed, CNS2 is required for Foxp3 induction in naïve CD4+ T cells following treatment of cells with a DNA methyltransferase inhibitor (9).
Figure 1.
Role of CNS2 in Treg cell differentiation and Treg cell lineage stability. (A) During Treg cell differentiation in wild-type mice, the probability of a precursor cell turning on Foxp3 expression is influenced by the relative strength of extrinsic signals promoting or inhibiting Foxp3 expression. Methylated CpG on CNS2 in precursor cells prevents CNS2 from promoting inappropriate Foxp3 induction, thus permitting proper regulation of Treg cell differentiation by extrinsic signals. CpG on CNS2 becomes demethylated during Treg differentiation and is fully demethylated in committed Treg cells. Although CNS2 is dispensable for the stability of Foxp3 expression in resting Treg cells. When committed resting Treg cells further differentiate into highly activated effector Treg cells, CNS2 plays a critical role in preventing Foxp3 expression from being destabilized by extrinsic environmental cues such as the presence of proinflammatory cytokines IL-4 and IL-6 or the absence of sufficient amounts of IL-2. These extrinsic cues often also influence Foxp3 induction during Treg cell differentiation. Mechanistically, upon TCR activation, CNS2 interacts with Foxp3 promoter in a NFAT-dependent manner to help maintain Foxp3 expression, probably by facilitating the access of CNS2-bound positive factors such as STAT5 to Foxp3 promoter. (B) CNS2 deletion does not affect Treg cell differentiation or the stability of Foxp3 expression in resting Treg cells. Instead, CNS2 is indispensable for maintaining Foxp3 expression in activated effector Treg cells. (C) In a hypothetic scenario, if CNS2 is not methylated in Treg precursors, its activity may lead to inappropriate Foxp3 induction and dysregulation of Treg cell differentiation.
TCR signal controls Foxp3 expression in activating Treg cells
Although TCR signal plays a critical role in Treg development, two recent studies characterizing mice with mature Treg-specific TCR ablation showed that TCR expression is not required for normal homeostasis of mature resting Tregs in the periphery. However, these TCR-less Tregs are defective in expression of genes associated with activated Tregs and their suppressor capacity (80, 81). Similar to these observations, resting Treg cells from CNS2 deficient mice can maintain their Foxp3 expression fairly well. When they are activated and enter cell cycle, a large proportion of these Tregs lose Foxp3 expression (8, 9). This suggests that TCR activation, which is the main driver of Treg cell activation and proliferation, might provide two signals: one can destabilize Foxp3 expression; the other is sensed by CNS2 and stabilizes Foxp3 expression. This notion was further supported by previous observation that CNS2 can respond to TCR activation in reporter assays in vitro (75, 82).
Indeed, CREB and NF-κB, two transcription factors activated by TCR signal, contributed to CNS2-dependent stabilization of Foxp3 expression (9, 82, 83). Furthermore, the calcineurin-NFAT signaling pathway downstream of TCR also plays an essential role in mediating CNS2-dependent maintenance of Foxp3 expression (9). Although NFAT was previously shown to play a critical role in mediating Foxp3 induction by binding to Foxp3 promoter and CNS1, its role in maintenance of Treg cell identity was not clear (76, 84). NFAT binds to both CNS2 and Foxp3 promoter to mediate a specific looping interaction in Treg cells upon TCR stimulation (9). In addition, factors known to be important for mediating enhancer-promoter looping interactions, Med12 and Nipbl, could bind to CNS2 (85). Knocking down these two factors by shRNAs resulted in increased loss of Foxp3 expression in wild type Treg cells, but not in CNS2 deficient Treg cells. Thus, TCR activation triggers a specific calcineurin/NFAT-dependent interaction between CNS2 and Foxp3 promoter, leading to the stabilization of Foxp3 expression, possibly through recruiting CNS2-associated transcriptional activators to the Foxp3 promoter (Figure 1A). For example, Stat5 binding to the Foxp3 promoter may benefit from the interaction between CNS2 and Foxp3 promoter, providing a potential mechanistic link between TCR and IL-2 signaling for their cooperation to stabilize Foxp3 expression (8, 9, 34, 86, 87). The exact mechanism of how these CNS2-binding factors collaborate remains to be further elucidated. Nonetheless, both TCR activation and IL-2 signaling are central players in many types of immune responses. Thus, sensing these signals by CNS2 may be a robust way to protect the identity of Treg cells in diverse immune contexts.
Signals that down-regulate Foxp3 expression in Treg cells
Although CNS2 can largely negate the destabilizing effects of aforementioned signals on wild type Treg cells, it is conceivable that under chronic inflammatory conditions, stronger destabilizing signals and/or weaker stabilizing signals afforded by CNS2 may tip the balance, leading to loss of Treg cell identity (88, 89). Thus, it is of great importance to understand how these signals including T-cell activation, pro-inflammatory cytokine stimulation, and limited amounts of IL-2 destabilize Foxp3 expression in CNS2 deficient Treg cells. Curiously, increased loss of Foxp3 expression in CNS2 deficient Treg cells was only observed in dividing cells in vivo and in vitro (8, 9). The close association between loss of Foxp3 expression and cell proliferation raised two non-mutually exclusive possibilities. First, cell division and cell cycle progression per se may contribute to unstable Foxp3 expression. Second, signals driving Treg cell proliferation may destabilize Foxp3 expression. A recent study found that cis-elements on the Foxp3 locus were partially re-methylated during Treg cell division and suggested that Stat6 and Stat3, which are activated by IL-4 and IL-6, respectively, may destabilize Foxp3 expression by recruiting Dnmt1 to the Foxp3 locus (8). This is consistent with previously observed requirement of Stat3 and Stat6 in inhibition of Foxp3 expression by IL-6 and IL-4, respectively (56, 90-93). Future studies will shed light on how the interplay among cell proliferation, TCR and cytokine derived signaling molecules, and epigenetic regulations affect the stability of Foxp3 expression.
Role of IL-2 in maintenance of Foxp3 expression
In addition to its important role in both tTreg and pTreg differentiation, IL-2 signaling also promotes stable Foxp3 expression in mature Tregs. In a Treg lineage-tracing study, among several experimental conditions tested, IL-2 antibody neutralization was the only condition that led to loss of Foxp3 expression in a sizable (>15%) fraction of Treg cells (60). Thus, it appears that CNS2 is not sufficient to completely prevent Treg cells from losing Foxp3 expression when IL-2 signaling is low. Indeed, recent studies showed that in the presence of limited amount of IL-2 or when IL-2 signal is blocked by a neutralizing antibody, CNS2 appeared to be even more important for maintaining Foxp3 expression, suggesting that sensing TCR activation through CNS2 and sensing IL-2 signaling are two key non-redundant mechanisms employed by mature Treg cells to maintain stable Foxp3 expression (8, 9). CNS2 is not required for IL-2 to promote Foxp3 expression, but may help to increase the effectiveness of low levels of IL-2 in stabilizing Foxp3 expression, possibly through facilitating Stat5 binding to the Foxp3 promoter and other cis-regulatory elements on the Foxp3 locus (8). The important role of IL-2 signaling in stabilizing Foxp3 expression may partially explain the previously observed correlation between CD25 expression levels and the stability of Foxp3 expression in Treg cells (58). More recently, reduced CD25 expression was also observed in PTEN deficient Treg cells which appeared to have unstable Foxp3 expression (94, 95). Thus, IL-2 signaling mediated stabilization of Foxp3 expression is a critical mechanism for maintaining Treg cell lineage stability.
Concluding remarks
Accumulating experimental evidences showed that keeping a clear division between the regulatory and effector T cell lineages is essential for immune system homeostasis. Regulatory T cell utilizes multiple molecular mechanisms to maintain its lineage stability in steady state or under a variety of inflammatory conditions. It is not surprising that the signals initiate Treg differentiation in thymus, including TCR and IL-2, also play a pivotal role in protecting mature Treg identity. This might be the simplest solution for Tregs to maintain their lineage stability when they also have to afford a certain degree of functional plasticity in adaption to their specific microenvironment. In this context, the Foxp3 CNS2 region serves as a gatekeeper between Tregs and conventional T cells. CNS2 is methylated in naïve T cells, which ensures the transient nature of Foxp3 induction by T cell activation or homeostatic expansion, so that naïve T cells cannot be accidentally converted to Tregs en masse. Demethylation of CNS2 in mature Treg cells turns it from dormant state to an active sensor of TCR and cytokine signals. During Treg activation, CNS2 brings multiple transcriptional activators to the proximity of the Foxp3 promoter through a looping mechanism, resulting in stabilized Foxp3 expression and Treg identity protection.
Despite recent progresses, important questions on how Treg lineage is preserved are still not fully answered (Box 2). The signaling pathways leading to CNS2 demethylation during Treg development are not clear, despite recent findings demonstrated that the TET proteins may be involved in the demethylation process (79). Although it is now recognized that Treg cell identity is established by building a unique epigenetic landscape and induction of Foxp3 expression, very little efforts were spent so far on investigating how Treg specific epigenetic modifications are maintained in mature Treg cells. Finally, CNS2-promoter looping and subsequent stabilization of Foxp3 transcription is simply one example on how DNA looping can regulate gene expression in Treg cell. Whether DNA looping is a common mechanism to protect the expression of Treg signature genes need to be explored in the future.
Box2. Outstanding questions.
What signal induces CNS2 demethylation during Treg cell development? Are the signaling pathways similar for tTreg and pTreg cells?
How IL-4 and IL-6 destabilize Foxp3 expression in activated Treg cells? Do they recruit repressors or remove transcriptional activators at the Foxp3 locus?
Why does cell cycle entry make Treg cell more vulnerable to loss of Foxp3 expression?
Among the Treg cell specific demethylated regions and DHS sites, which ones are essential for Treg cell function?
How genome-wide epigenetic landscape is maintained in Treg cell in a pro-inflammatory microenvironment?
In addition to TCR and IL-2, what other signals can CNS2 sense?
Which transcription factors are driving promoter-CNS2 looping? Which ones are simply passengers recruited to the promoter by looping to increase Foxp3 transcription?
Highlights.
Foxp3 CNS2 is a gatekeeper between regulatory and conventional T cell lineages
TCR and IL-2 signals dictate both differentiation and stability of Treg cells
Dynamic enhancer-promoter interaction is required for stable Foxp3 expression
Acknowledgement
We thank members of the Zheng laboratory for helpful discussions. X. L. was supported by a postdoc fellowship from the Nomis Foundation. Y. Z. was funded by the Nomis Foundation, the Rita Allen Foundation, the Hearst Foundation, the National Multiple Sclerosis Society, and National Institute of Health (AI099295, AI107027).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–87. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–61. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 3.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 4.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4:337–42. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 5.Miyao T, Floess S, Setoguchi R, Luche H, Fehling HJ, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3(+) T cells reflects promiscuous Foxp3 expression in conventional T cells but not reprogramming of regulatory T cells. Immunity. 2012;36:262–75. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, Huehn J, Fehling HJ, Sparwasser T, Nakai K, Sakaguchi S. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–99. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Yu F, Sharma S, Edwards J, Feigenbaum L, Zhu J. Dynamic expression of transcription factors T-bet and GATA-3 by regulatory T cells maintains immunotolerance. Nat Immunol. 2015;16:197–206. doi: 10.1038/ni.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 2014;158:749–63. doi: 10.1016/j.cell.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Liang Y, LeBlanc M, Benner C, Zheng Y. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 2014;158:734–48. doi: 10.1016/j.cell.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–45. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 11.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng H, Chi H. Metabolic control of regulatory T cell development and function. Trends Immunol. 2015;36:3–12. doi: 10.1016/j.it.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8:277–84. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 14.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–5. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 15.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–70. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 16.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, Shaffer SA, Goodlett DR, Rudensky AY. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13:1010–9. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu W, Ergun A, Lu T, Hill JA, Haxhinasto S, Fassett MS, Gazit R, Adoro S, Glimcher L, Chan S, Kastner P, Rossi D, Collins JJ, Mathis D, Benoist C. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–80. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa H, Ohkura N, Vandenbon A, Itoh M, Nagao-Sato S, Kawaji H, Lassmann T, Carninci P, Hayashizaki Y, Forrest AR, Standley DM, Date H, Sakaguchi S, Consortium F. Differential roles of epigenetic changes and Foxp3 expression in regulatory T cell-specific transcriptional regulation. Proc Natl Acad Sci U S A. 2014;111:5289–94. doi: 10.1073/pnas.1312717110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HM, Hsieh CS. Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. J Immunol. 2009;183:2261–6. doi: 10.4049/jimmunol.0901304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–6. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 23.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3:756–63. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–77. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 25.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–59. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–89. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 2012;37:475–86. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 29.Malchow S, Leventhal DS, Nishi S, Fischer BI, Shen L, Paner GP, Amit AS, Kang C, Geddes JE, Allison JP, Socci ND, Savage PA. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339:1219–24. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perry JS, Lio CW, Kau AL, Nutsch K, Yang Z, Gordon JI, Murphy KM, Hsieh CS. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 2014;41:414–26. doi: 10.1016/j.immuni.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S, Fujikado N, Kolodin D, Benoist C, Mathis D. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015 doi: 10.1126/science.aaa7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malek TR, Porter BO, Codias EK, Scibelli P, Yu A. Normal lymphoid homeostasis and lack of lethal autoimmunity in mice containing mature T cells with severely impaired IL-2 receptors. J Immunol. 2000;164:2905–14. doi: 10.4049/jimmunol.164.6.2905. [DOI] [PubMed] [Google Scholar]
- 33.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–78. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 34.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol. 2005;6:1142–51. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 35.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–11. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–21. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burchill MA, Goetz CA, Prlic M, O’Neil JJ, Harmon IR, Bensinger SJ, Turka LA, Brennan P, Jameson SC, Farrar MA. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J Immunol. 2003;171:5853–64. doi: 10.4049/jimmunol.171.11.5853. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9:632–40. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 39.Ouyang W, Beckett O, Ma Q, Li MO. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 2010;32:642–53. doi: 10.1016/j.immuni.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–5. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 50.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, von Boehmer H. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–27. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 51.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–9. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J Exp Med. 2012;209:1529–35. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178:6725–9. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 56.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, Shah B, Chang SH, Schluns KS, Watowich SS, Feng XH, Jetten AM, Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vu MD, Xiao X, Gao W, Degauque N, Chen M, Kroemer A, Killeen N, Ishii N, Li XC. OX40 costimulation turns off Foxp3+ Tregs. Blood. 2007;110:2501–10. doi: 10.1182/blood-2007-01-070748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc Natl Acad Sci U S A. 2009;106:1903–8. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X, Bailey-Bucktrout SL, Jeker LT, Penaranda C, Martinez-Llordella M, Ashby M, Nakayama M, Rosenthal W, Bluestone JA. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat Immunol. 2009;10:1000–7. doi: 10.1038/ni.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rubtsov YP, Niec RE, Josefowicz S, Li L, Darce J, Mathis D, Benoist C, Rudensky AY. cell. 2010;329:1667–71. doi: 10.1126/science.1191996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, Peng M, Chan P, Ma Q, Mo Y, Meijer D, Zhao K, Rudensky AY, Atwal G, Zhang MQ, Li MO. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;491:554–9. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–8. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–82. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35:337–48. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, Paul WE, Bosselut R, Wei G, Zhao K, Oukka M, Zhu J, Belkaid Y. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121:4503–15. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–11. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 71.Ulges A, Klein M, Reuter S, Gerlitzki B, Hoffmann M, Grebe N, Staudt V, Stergiou N, Bohn T, Bruhl TJ, Muth S, Yurugi H, Rajalingam K, Bellinghausen I, Tuettenberg A, Hahn S, Reissig S, Haben I, Zipp F, Waisman A, Probst HC, Beilhack A, Buchou T, Filhol-Cochet O, Boldyreff B, Breloer M, Jonuleit H, Schild H, Schmitt E, Bopp T. Protein kinase CK2 enables regulatory T cells to suppress excessive TH2 responses in vivo. Nat Immunol. 2015;16:267–75. doi: 10.1038/ni.3083. [DOI] [PubMed] [Google Scholar]
- 72.Burzyn D, Benoist C, Mathis D. Regulatory T cells in nonlymphoid tissues. Nat Immunol. 2013;14:1007–13. doi: 10.1038/ni.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–53. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burzyn D, Kuswanto W, Kolodin D, Shadrach JL, Cerletti M, Jang Y, Sefik E, Tan TG, Wagers AJ, Benoist C, Mathis D. A special population of regulatory T cells potentiates muscle repair. Cell. 2013;155:1282–95. doi: 10.1016/j.cell.2013.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–12. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 77.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–63. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 78.Baron U, Floess S, Wieczorek G, Baumann K, Grutzkau A, Dong J, Thiel A, Boeld TJ, Hoffmann P, Edinger M, Turbachova I, Hamann A, Olek S, Huehn J. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37:2378–89. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 79.Toker A, Engelbert D, Garg G, Polansky JK, Floess S, Miyao T, Baron U, Duber S, Geffers R, Giehr P, Schallenberg S, Kretschmer K, Olek S, Walter J, Weiss S, Hori S, Hamann A, Huehn J. Active demethylation of the Foxp3 locus leads to the generation of stable regulatory T cells within the thymus. J Immunol. 2013;190:3180–8. doi: 10.4049/jimmunol.1203473. [DOI] [PubMed] [Google Scholar]
- 80.Levine AG, Arvey A, Jin W, Rudensky AY. Continuous requirement for the TCR in regulatory T cell function. Nat Immunol. 2014;15:1070–8. doi: 10.1038/ni.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vahl JC, Drees C, Heger K, Heink S, Fischer JC, Nedjic J, Ohkura N, Morikawa H, Poeck H, Schallenberg S, Riess D, Hein MY, Buch T, Polic B, Schonle A, Zeiser R, Schmitt-Graff A, Kretschmer K, Klein L, Korn T, Sakaguchi S, Schmidt-Supprian M. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity. 2014;41:722–36. doi: 10.1016/j.immuni.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 82.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–51. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–31. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 84.Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, Schmidt-Weber CB. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 85.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–90. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 87.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, Hennighausen L, Wu C, O’Shea JJ. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–75. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bailey-Bucktrout SL, Martinez-Llordella M, Zhou X, Anthony B, Rosenthal W, Luche H, Fehling HJ, Bluestone JA. Self-antigen-driven activation induces instability of regulatory T cells during an inflammatory autoimmune response. Immunity. 2013;39:949–62. doi: 10.1016/j.immuni.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Komatsu N, Okamoto K, Sawa S, Nakashima T, Oh-hora M, Kodama T, Tanaka S, Bluestone JA, Takayanagi H. Pathogenic conversion of Foxp3+ T cells into TH17 cells in autoimmune arthritis. Nat Med. 2014;20:62–8. doi: 10.1038/nm.3432. [DOI] [PubMed] [Google Scholar]
- 90.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 91.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, Reid SP, Levy DE, Bromberg JS. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–73. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fu S, Zhang N, Yopp AC, Chen D, Mao M, Chen D, Zhang H, Ding Y, Bromberg JS. TGF-beta induces Foxp3 + T-regulatory cells from CD4 + CD25 - precursors. Am J Transplant. 2004;4:1614–27. doi: 10.1111/j.1600-6143.2004.00566.x. [DOI] [PubMed] [Google Scholar]
- 93.Takaki H, Ichiyama K, Koga K, Chinen T, Takaesu G, Sugiyama Y, Kato S, Yoshimura A, Kobayashi T. STAT6 Inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem. 2008;283:14955–62. doi: 10.1074/jbc.M801123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. 2015;16:178–87. doi: 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, Townamchai N, Gerriets VA, Rathmell JC, Sharpe AH, Bluestone JA, Turka LA. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015;16:188–96. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]