SUMMARY
Vertebrate cells have evolved elaborate cell-autonomous defense programs to monitor subcellular compartments for infection and to evoke counter-responses. These programs are activated by pathogen-associated pattern molecules and by various strategies intracellular pathogens employ to alter cellular microenvironments. Here, we show that when uropathogenic E. coli (UPEC) infect bladder epithelial cells (BECs), they are targeted by autophagy but avoid degradation because of their capacity to neutralize lysosomal pH. This change is detected by mucolipin TRP channel 3 (TRPML3), a transient receptor potential cation channel localized to lysosomes. TRPML3 activation then spontaneously initiates lysosome exocytosis, resulting in expulsion of exosome-encased bacteria. These studies reveal a cellular default system for lysosome homeostasis that has been co-opted by the autonomous defense program to clear recalcitrant pathogens.
INTRODUCTION
The cell autonomous defense (CAD) program comprises a multilayered intracellular surveillance system to detect and counter the infections. The highly compartmentalized nature of host cells has resulted in the development of various organelle–specific sensors for pathogen-associated molecular patterns (PAMPs), directly noticing molecular signatures of pathogens (Akira et al., 2006). It has recently been proposed that additional sensors exist which detect specific hostile actions of pathogens (also referred to as Patterns of Pathogenesis), such as penetration of subcellular membrane and disruption of the actin cytoskeleton (Vance et al., 2009). Once these sensors are engaged, the CAD program elicits robust responses to clear the pathogens.
A common measure for clearing intracellular pathogens involves detection followed by sequestration of pathogen in an autophagosome, a de novo generated membrane-bound compartment, which is promptly shuttled to the lysosome where the pathogens are degraded (Levine et al., 2011). The lysosome is arguably the epicenter of the CAD. Its sterilizing power originates from the concerted actions of numerous factors in the lysosome lumen, such as antimicrobial peptides and proteases, as well as reactive oxygen and nitrogen species which are highly toxic to the bacteria (Goren, 1977). Importantly, the bactericidal actions of these agents are greatly enhanced by the low pH (4.5–5.0) generated inside lysosomes by proton pumping vacuolar (v)-ATPases (Goren, 1977). Lysosome function is also exquisitely sensitive to ion homeostasis and regulating ion flux across lysosomal membranes are several members of a subfamily of transient receptor potential (TRP) cation channels, referred to as mucolipin TRP channel 1–3 (TRPML1-3) (Xu, 2015). In view of the powerful degradative actions of lysosomes, several pathogens have evolved capacities to block the activity of v-ATPases, which markedly attenuates the lysosomes by impairing its acidification (Sturgill-Koszycki et al., 1994). At this time it is not known whether the CAD program has additional strategies in its arsenal to counter pathogen-mediated subversion of lysosomes.
A distinct cellular mechanism for the elimination of invading bacteria involving non-lytic expulsion of intracellular bacteria back to the extracellular milieu was recently described in bladder epithelial cells (BECs) (Bishop et al., 2007). Uropathogenic E. coli (UPEC) circumvent the normally impregnable bladder epithelium by binding avidly to the uroepithelium triggering focal exocytosis of specialized Rab27+ fusiform vesicles which serve as repositories for extra plasma membrane necessary for bladder expansion. When these extruded membranes are subsequently retracted into BECs, adherent UPEC gain entry and slip into Rab27+ vesicles (Bishop et al., 2007). Remarkably the BECs have the innate capacity to expel nearly 70% of the infecting bacteria (Bishop et al., 2007). Much of the underlying mechanisms of how intracellular UPEC are detected and shuttled from their intracellular location to the plasma membrane remain a mystery.
In this report we reveal that UPEC expulsion from BECs is initiated in lysosomes, and triggered by a TRP channel upon sensing pathogen-mediated neutralization of lysosomal pH.
RESULTS
Infected BECs expel membrane-encased UPEC
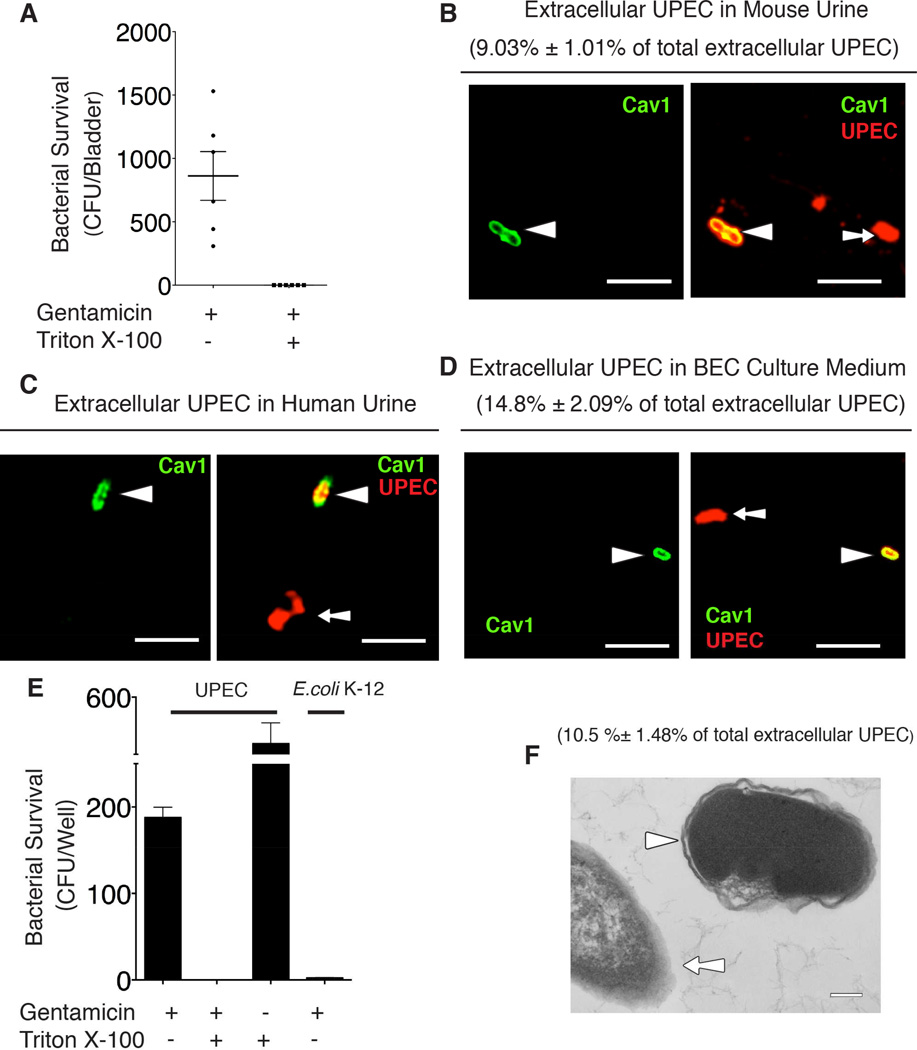
Impetus for this study came from the surprising finding that a significant portion of UPEC, when freshly isolated from urine of patients with urinary tract infections (UTIs), were consistently resistant to tobramycin. However, upon subculture, these isolates promptly lost their resistance. Given that tobramycin is not membrane permeable (Menashe et al., 2008), we speculated that this transient resistance was attributable to the presence of an encapsulating host membrane. To test this hypothesis, a suspension of bacteria collected from urine of infected mice was treated for 1 hr with gentamycin (an antibiotic more routinely used in antibiotic protection assays) with or without 0.1% Triton-X100, which selectively disrupts host membranes. We observed that, in contrast to the gentamicin-alone treated samples, Triton-X100 treated bacteria were no longer resistant to gentamycin (Figure 1A). We then found at least 10% of the bacteria in urine were stained positive for Caveolin-1, a host membrane marker (Figure 1B). The presence of membrane-encased bacteria in urine of human patients with acute UTIs was similarly observed (Figure 1C), thus, validating our notion.
Figure 1. Expelled UPEC are encased in membrane-bound vesicles.
(A) Cell-free urine from infected C57BL/6 mice was collected at 6 hours post infection (h.p.i.), and treated with gentamicin with/without 0.1% Triton X-100 for 1hr. The surviving CFU were quantified. Error bars, SEM. n=9.
(B – D) Immunofluorescence staining for Caveolin-1 (green) and UPEC (red) in cell-free urine collected from infected mouse at 6 h.p.i (B), UTI patients (C), or culture medium collected from infected BEC line at 8 h.p.i. (D). Arrow depicts naked bacteria and arrowhead depicts vesicle-encased UPEC. The membrane-encased UPEC were quantified and expressed as the percentage of total examined UPEC shown in the parenthesis. Scale bar: 5 µm. n=3 slides.
(E) Bacterial viability assay performed on cell-free medium collected from BECs infected with UPEC strain CI5 or E.coli K-12 strain MG1655 at 8 h.p.i., and treated with gentamicin or 0.1% Triton X-100 alone, or gentamicin plus 0.1% Triton X-100 for 1 hour. Error bars, SEM. n=18.
(F) TEM image of extracellular bacteria collected from the culture medium of infected BECs. Arrowhead depicts vesicle-encased bacterium and arrow depicts a naked bacterium. The membrane-encased ECU were quantified and expressed as the percentage of total examined UPEC shown in the parenthesis. Scale bar: 0.2 µm. n=3 grids
To elucidate the underlying basis for this phenomenon, we established an in vitro model of UTIs. The human BEC line 5637 was infected by UPEC, and after 8 hrs of incubation, the extracellular medium was assayed for the presence of membrane-encased bacteria. Data similar to that found from in vivo studies was obtained (Figure 1D, E). Interestingly, the presence of membrane-bound bacteria was not detectable when using a nonpathogenic E. coli K-12 strain to infect the BECs (Figure 1E). Shown in Figure 1F is a transmission electron microscope image of an extracellular UPEC revealing a distinct membrane around the bacteria. Cumulatively, our in vivo and in vitro data reveal that intracellular UPEC are exported within host cell-derived vesicles.
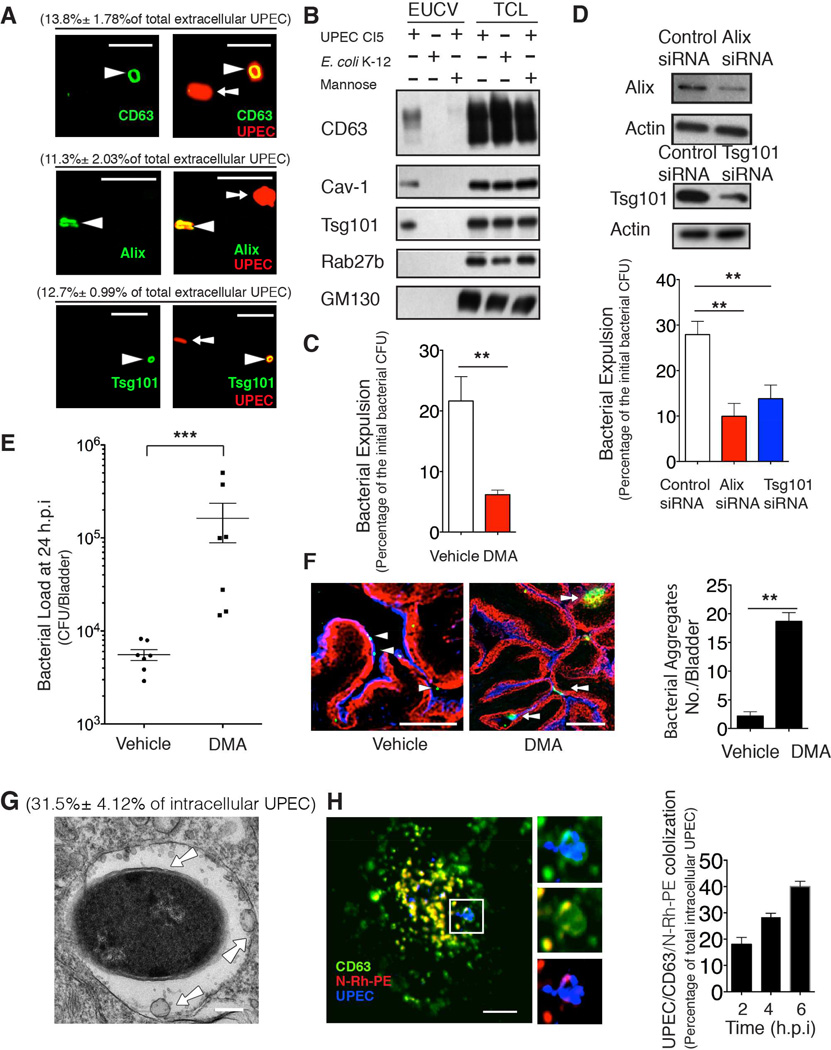
Expelled UPEC are contained within exosomes
Because several cellular products have been reported to be exported in microvesicles termed exosomes (Simons and Raposo, 2009), we explored the possibility that the extracellular UPEC containing vesicles (EUCVs) were exosomes. Exosomes are typically derived from the inward budding of multivesicular bodies (MVBs), and are of interest because they serve as highly efficient export vehicles (Thery et al., 2002). Immunofluorescent staining of EUCVs revealed colocalization of several proteins frequently found in exosomes, including the ESCRT proteins Alix and Tsg101, as well as the tetraspanin CD63 (Thery et al., 2006) (Figure 2A). To more quantitatively confirm the presence of exosome markers on EUCVs, the bacteria were first covalently linked to magnetic beads before exposure to BECs (Lonnbro et al., 2008), and EUCV-encased bacteria, were isolated from cell-free medium on a magnetic rack. We confirmed the presence of various exosome markers in the EUCV fractions (Figure 2B). As controls, we included 2% D-mannose in the medium to prevent UPEC adherence and entry (Abraham et al., 1988). We also infected BECs with nonpathogenic E. coli K12 MG1655. In both controls, we didn’t detect any exosome markers associating with extracellular bacteria (Figure 2B). As further controls to confirm the rigor of the EUCV purification procedure, and to exclude the possibility that EUCV are a result of cell lysis, in the EUCV fraction, we demonstrated the absence of intracellular organelle markers such as GM130, a marker of the Golgi, as well as the absence of Rab27b, a marker for the vacuole in which UPEC initially harbor (Bishop et al., 2007)(Figure 2B).
Figure 2. Vesicles encasing expelled bacteria are exosomes.
(A) Immunofluorescence staining of colocalization between exosome markers (green) and extracellular bacteria (red) collected from infected 5637 BEC line at 8 h.p.i.. The vesicle-encased UPEC were quantified and expressed as the percentage of total examined UPEC shown in the parenthesis. Scale bar: 5 µm. n=3 slides.
(B) Immunoblot analysis of lysates of EUCV purified at 12 h.p.i. from BECs infected with either UPEC, E. coli K12 strain, or UPEC with mannose in the medium. Total cell lysates (TCLs) were used to show similar number of cells were treated.
(C and D) Bacterial expulsion at 6 h.p.i. in infected BECs treated with DMSO (vehicle) or 15 nM dimethyl amiloride (DMA) (C), or transfected with control siRNA or Alix/Tsg101 siRNA. Knockdown efficiency is indicated by the western blot alongside. Error bar, SEM. n=18.
(E) Bacterial load at 24 h.p.i in infected bladder treated with either vehicle (DMSO) or 15 nM DMA. Error bar, SEM. n=9.
(F) Immunofluorescence staining of infected bladder treated with either vehicle or DMA for 24 hours. Collapsed bladder: superficial epithelium (blue), intermediate epithelium (red), UPEC (green). Arrowhead depicts single bacteria, and arrow depicts bacterial aggregates. The bacterial aggregates (> 10 µm) were quantified. Scale bar: 100 µm. n=3 mice.
(G) TEM image shows intracellular UPEC encased in MVBs at 4 h.p.i. The arrow depicts ILVs-encased in MVBs. Bacteria encased in MVBs were quantified and expressed as a percentage of total examined UPEC, shown in the parenthesis. Scale bar: 0.2 µm. n=3 grids
(H) Immunofluorescence staining of UPEC (blue) and MVB markers CD63 (green) and flourescent N-Rh-PE (red) at 4 h.p.i.. Scale bar: 5 µm. The CD63+ compartmenr-encased UPEC was quantified at 2, 4, 6 hours, expressed as the percentage of total examined UPEC, Scale bar: 5 µm. n=3 slides.
We then confirmed involvement of exosomes in bacterial export employing functional assays. To block exosome secretion pathway, we either treated UPEC-infected BECs with dimethyl amiloride (DMA), a known inhibitor of exosome release by disrupting calcium signaling (Savina et al., 2003), or knocked down expression of Alix or Tsg101, two key components implicated in exosome biogenesis (Baietti et al., 2012). We found that both DMA treatment (Figure 2 C) and Alix or Tsg101 KD (Figure 2D) markedly inhibited bacterial expulsion (See Experimental Procedure). To evaluate the relevance of exosome-mediated expulsion as a defense mechanism against UTI, DMA was transurethrally applied to the bladders at 2h post-infection. At 24h post-infection, we found the bacterial load in the DMA-treated mice was markedly more enhanced than controls (Figure 2E). Examination of these infected bladders revealed that when the BECs failed to expel bacteria, UPEC accumulated within the uroepithelium, forming large bacterial aggregates (Figure 2F). Cumulatively, our data point to exosomes as powerful vehicles for bacterial export following infection.
Intracellular UPEC are sequestered in MVBs prior to export
We next sought to identify the upstream activities leading to the packaging of UPEC in exosomes. Because exosomes originate from MVBs, we investigated whether UPEC could be found in MVBs. Shown in Figure 2G is a representative EM image of a compartment resembling the MVB which harbors multiple intraluminal vesicles (ILVs) and membrane-bound UPEC. CD63 is a marker of MVBs (Kobayashi et al., 2000), and fluorescent lipid phospholipid N-(Lissamine) rhodamine B sulfonyl dioleoylphosphatidylethanolamine (N-Rh-PE) preferentially localizes in MVBs (Vidal et al., 1997). Employing both probes, we verified that some of the intracellular compartments harboring UPEC were doubly positive for CD63 and N-Rh-PE (Figure 2H). Quantification revealed that the percentage of intracellular UPEC found in CD63+ vesicles gradually increased reaching a peak at 6 h.p.i. (Figure 2H).
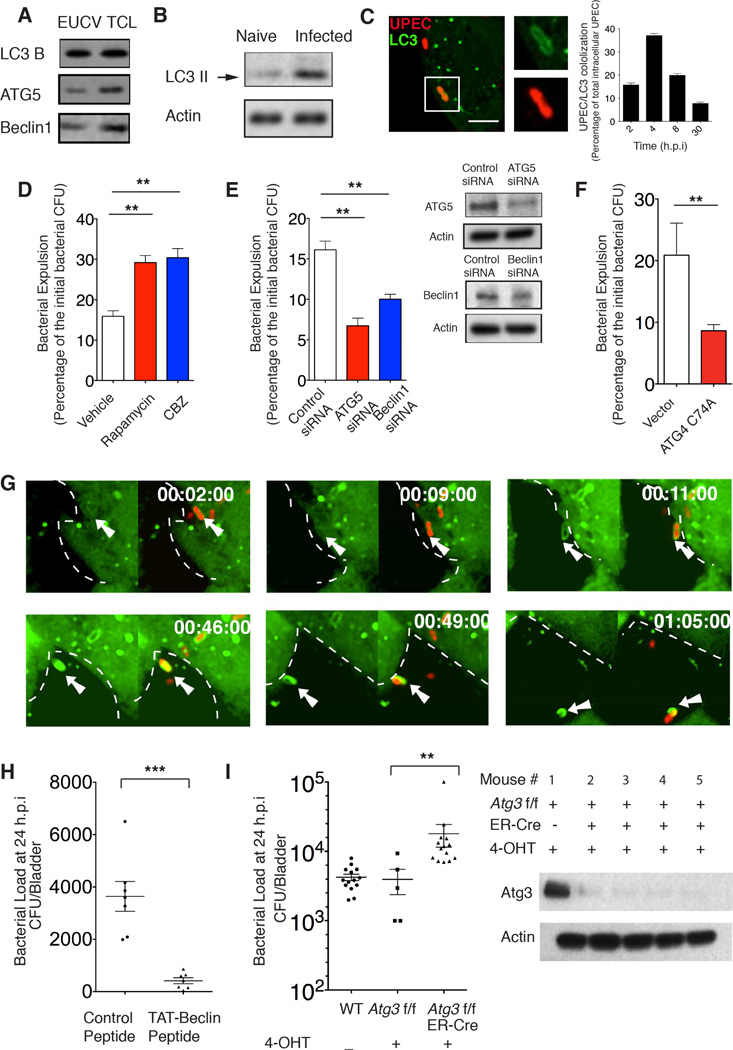
Bacterial expulsion involves autophagy machinery
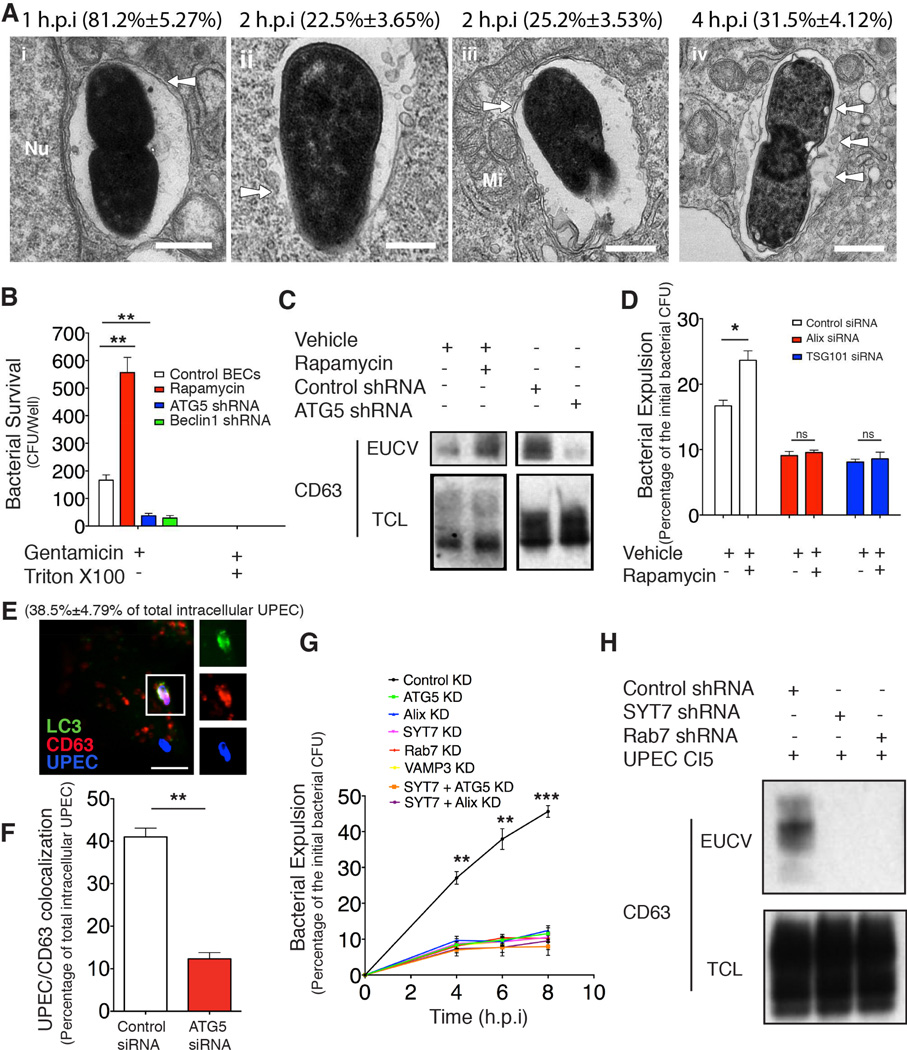
In view of the striking reduction in bacterial burden associated with expulsion activity, we hypothesized that this action was initiated by the cell autonomous defense system. To identify the putative intracellular surveillance mechanism involved in expulsion, we analyzed the molecular composition of the purified EUCVs. Interestingly, several components of the autophagy pathway (LC3, Beclin1 and ATG5) were detected in EUCV fractions (Figure 3A, Figure S1A). Whereas autophagy is best known as a cellular homeostasis process, there is also recognition of their involvement in the immune detection of intracellular pathogens (Levine et al., 2011). LC3 is a specific membrane marker of the autophagosome that, upon activation, becomes lipidated and converts to LC3II (Kabeya et al., 2000). We detected LC3 II in the BEC extract as early as 15 minutes after exposure to UPEC (Figure 3B). Correspondingly, in the same time frame, we observed intracellular UPEC being captured in the LC3+ autophagosomes (Figure 3C). Maximum association between autophagosome and UPEC was observed at 4 h.p.i. (Figure 3C). Most of the UPEC that were encased in autophagosomes also colocalized with ubiquitin and P62, markers that tag cargo for sequestration in autophagosomes (Levine et al., 2011)(Figure S1B, C). To investigate the involvement of the autophagy machinery in bacterial expulsion, we first exposed UPEC-infected BECs to chemical inducers of autophagy. We found that, both rapamycin and carbamazepine (CBZ) markedly enhanced bacterial expulsion (Figure 3D). The increased number of extracellular bacteria is not the result of autophagy-induced cell death, because neither terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) nor lactate dehydrogenase (LDH) release assays revealed any significantly enhanced signs of cell death following rapamycin treatment of infected BECs (Figure S1D, E). Conversely, when substrates involved in autophagosome formation, such as ATG5 or Beclin1 was knocked down, bacterial expulsion was significantly reduced, without affecting bacterial entry (Figure 3E, Figure S1F). Additionally, we overexpressed ATG4 C74A, a dominant negative mutant that has previously been shown to inhibit autophagosome formation (Fujita et al., 2008). While there was no difference in the bacterial uptake (Figure S1F), bacterial expulsion was markedly inhibited in these mutant BECs (Figure 3F).
Figure 3. Bacterial expulsion involves autophagy components.
(A) Immunoblot analysis of autophagy components in EUCVs collected from infected BECs at 12 h.p.i. .
(B) Immunoblot of naive BECs (lane1) and infected BECs (lane2) showing increase in LCII upon UPEC infection for 15 minutes.
(C) Immunofluorescence staining revealing co-association of autophagosome marker LC3 (green) with UPEC (red) in infected BECs at 2 h.p.i.. The number of the LC3-encased UPEC was quantified at 2, 4, 8, 30 h.p.i. and expressed as the percentage of total examined UPEC, Scale bar: 5 µm. n=3 slides.
(D to F) Bacterial expulsion levels at 6 h.p.i. in infected BECs treated with (D) DMSO (vehicle), 200nm Rapamycin or 30 µM CBZ, or (E) transfected with control siRNA, ATG5 siRNA or Beclin1 siRNA, or (F) transfected with empty vector or plasmid overexpressing ATG4 C74A. Knockdown efficiency in (E) is indicated by the western blot alongside. Error bars, SEM. (n=18).
(G) Still shots from a movie showing intracellular UPEC encapsulation in autophagosomes followed by export. Green, LC3; Red, UPEC. For each time point: left, LC3 only; right: LC3 & UPEC overlay. Dashed lines indicate the cell border assessed from background fluorescence, and the white arrows point to expelled-bacteria.
(H) Bacterial load at 24 h.p.i in infected bladder treated with either control or 30 µg TAT-Beclin peptide. Error bars, SEM. n=9.
(I) Bacterial load at 24 h.p.i in infected bladder of the wild type (WT), 4-OHT treated Control Atg3flox/flox or 4-OHT induced Atg3 KO in Atg3flox/flox ER-Cre mice. Western blot probing Atg3 in exfoliated superficial epithelium extract shows efficient knockout of Atg3 in the superficial epithelial cells of the bladder. Error bars, SEM. n=9.
Because the involvement of autophagy in bacterial expulsion was surprising, we attempted to visualize this process. We undertook live cell imaging of GFP-LC3-expressing BECs infected with RFP-labeled UPEC. In the video (Movie S1) and still images (Figure 3G), we observed UPEC encased in LC3-positive vesicles trafficking within a BEC before being expelled. Importantly, the autophagosome-encased bacteria rapidly moved towards the cell surface, and following ejection, the expelled bacterium was still encased in the LC3+ vesicles. (Movie S1).
To further implicate the autophagy pathway in reducing bacterial burden through bacterial expulsion in BECs, we administered a peptide, recently shown to trigger autophagy without harmful effects (Shoji-Kawata et al., 2013), into the infected bladder via a catheter. Compared to the control peptide-treated bladders, the autophagy-inducing peptide markedly reduced bacterial burden of infected bladders (Figure 3H). We then sought to specifically knock out Atg3, a key gene involved in autophagosome biogenesis in the superficial epithelial layer of the bladder. To achieve this, mice homologous for the floxed Atg3 allele were bred with ER-Cre mice to generate Atg3flox/flox ER-Cre mice, where the Atg3 could be inducibly deleted solely in the bladder epithelium by administering 4-hydroxytamoxin (4-OHT) locally in the bladder (see Experimental Procedure). As expected, in the exfoliated superficial epithelium extract (see Experimental procedure), we confirmed that this strategy ensured reproducible knockout of Atg3 in the superficial epithelium (Figure 3I). Using this animal model, we indeed observed significantly higher bacterial load in the autophagy-deficient bladder epithelium compared to the control (Figure 3I). Taken together, our in vitro and in vivo studies reveal the involvement of the autophagy pathway in the expulsion of bacteria by BECs.
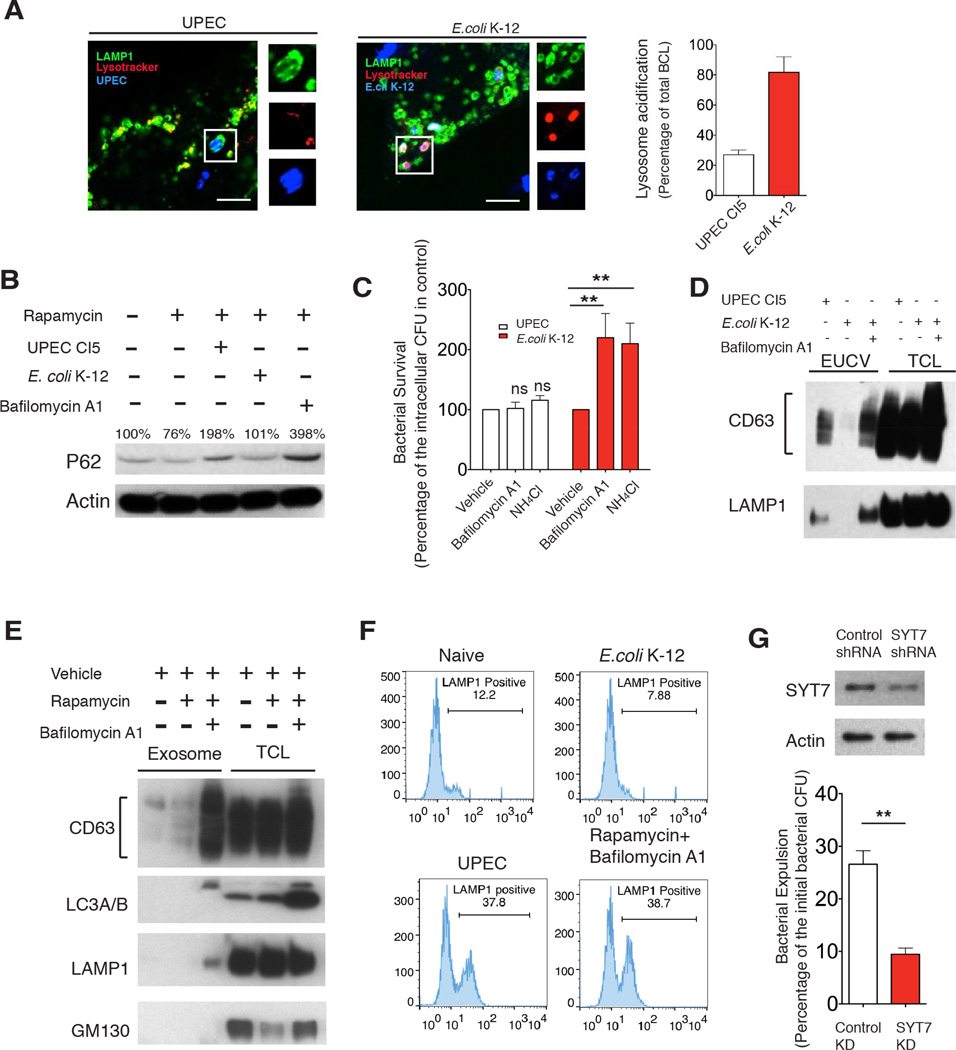
Bacterial neutralization of the lysosome triggers exocytosis
Since the ultimate fate of autophagic cargo is degradation within lysosomes, it was imperative to elucidate why this was replaced by expulsion in UPEC-infected BECs. We verified that autophagosomes still frequently fused with lysosomes in infected BECs (Figure S1G). However, the resulting autolysosomes failed to become acidified (Figure 4A). To test if this observation was specifically mediated by UPEC, we substituted the UPEC with the relatively innocuous E.coli K-12 strain MG1655 and in this case, bacteria-containing lysosomes were strongly acidified (Figure 4A).
Figure 4. Neutralization of bacteria-bearing lysosomes triggers lysosome exocytosis.
(A) Immunofluorescence staining of UPEC (blue) or E.coli K-12 (blue)-containing lysosomes (green) and lysotracker (red) at 6 h.p.i.. Lysotracker+ populations of bacteria-containing lysosomes were quantified, and expressed as the percentage of total examined lysosome-enclosed bacteria. Scale bar: 5 µm. n=3 slides.
(B) P62 level dynamics in naïve BECs (lane1), BECs treated with 200 nM rapamycin alone (lane 2) or rapamycin combined with UPEC (lane 3), E. coli K-12 (lane 4) or 1 µM bafilomycin A1 (lane 5).
(C) Intracellular bacteria CFU at 8 h.p.i in BECs infected with either UPEC or E.coli K-12 and treated with either vehicle, 1 µM bafilomycinA1 or 50 mM NH4 Cl. Error bars, SEM. n=18.
(D) Immunoblot quantification of CD63 or LAMP1 present in the EUCV purified from UPEC, or E.coli K-12 infected BECs, or E.coli K-12 infected BECs treated with bafilomycin A1. Comparable CD63 or LAMP1 in total cell lysate (TCL) suggest similar numbers of cells were used.
(E) Immunoblot quantification of CD63 in exosomes purified from naïve BECs, BECs treated with 200 nM rapamycin alone, or with 200 nM rapamycin for 4 hours followed by 1 µM bafilomycinA1 treatment for12 hours. Comparable CD63 level in total cell lysate suggest that similar number of cells were used.
(F) FACS analysis of cell surface LAMP1+ populations in naïve BECs, or BECs treated with rapamycin for 4 hours followed by bafilomycin A1 treatment for 12 hours, or infected with UPEC CI5 or E.coli K-12 for 12 hours.
(G) Bacterial expulsion levels at 6 h.p.i. in infected BECs expressing control shRNA, or SYT7 shRNA. Knockdown efficiency is indicated by the western blot alongside. Error bars, SEM. n=18.
Intracellular degradation of P62 is critically dependent on the pH of lysosomes (Klionsky et al., 2012). We found that upon induction of autophagy in BECs by rapamycin, there was a marked reduction in P62 levels (Figure 4B). However, UPEC infection, but not E.coli K-12 infection, caused dramatic accumulation of P62 in BECs, similar to the effect of bafilomycin A1, a well known lysosomal v-ATPase inhibitor (Figure 4B).
It is noteworthy that when the bafilomycin A1 or ammonium chloride was added to E. coli K-12 infected BECs, the number of intracellular bacteria increased compared to vehicle treated BECs, as these bacteria were no longer killed (Figure 4C). When these agents were added to UPEC-infected BECs, no change in intracellular bacterial numbers occurred, presumably because the lysosomes were already neutralized (Figure 4C).
These observations led us to hypothesize that the neutralization of the lysosome was a critical signal triggering the expulsion of bacteria-containing exosomes from lysosomes. In earlier experiments, when the E.coli K-12 was used to infect BECs, we failed to detect these bacteria in exosomes, as they were usually degraded in the lysosome (Figure 1E). However, when we infected BECs with E.coli K-12 followed by bafilomycin A1 treatment, we were able to detect significant amounts of extracellular E.coli associated with membranes enriched in CD63 (Figure 4D). To further test whether neutralization of lysosomal pH was sufficient to trigger exocytic events from lysosomes, we mimicked the infection condition by inducing autophagy with rapamycin in uninfected BECs for 4 hours followed by bafilomycin A1 treatment. We then isolated and examined regular exosomes shed from these cells. Remarkably, compared to untreated or rapamycin alone treated BECs, bafilomycin A1 treatment induced a striking increase in the amount of secreted exosomes (Figure 4E). Importantly, autophagic protein LC3 and the lysosomal marker LAMP1, were only present in exosomes after bafilomycin A1 treatment (Figure 4E). Based on these observations, we speculate that bacterial expulsion within exosomes is actually mediated by lysosome exocytosis, when the pH of lysosomes are neutralized. A hallmark for lysosome exocytosis is the appearance of lysosomal markers in the plasma membrane. Indeed, FACS analysis of bafilomycin A1 treated BECs, as well as UPEC, but not E.coli K-12 infected BECs showed striking increases in the cell surface LAMP1 (Figure 4F). Probing LAMP1 in isolated plasma membrane proteins fractions from BECs (Figure S2A), or assessing lysosomal enzyme activity in the medium of BECs treated in similar conditions, also confirmed this pattern of lysosome trafficking (Figure S2B). This movement is restricted to lysosomes as none of other organelles markers appeared on the plasma membrane of BECs treated with similar conditions.(Figure S2A). Figure 4G shows silencing synaptotagmin7 (SYT7), a key mediator of lysosome exocytosis (Reddy et al., 2001), led to significant abrogation of bacterial expulsion, further supporting the notion that bacterial expulsion is mediated by lysosome exocytosis. In sum, these results reveal that lysosome exocytosis resulting in expulsion of UPEC is actually a spontaneous cellular response to alteration of lysosomal conditions.
Sequence of bacterial trafficking through various subcellular compartments in BECs
So far, our studies have independently revealed the relevance of exosome formation, autophagy induction, and lysosome exocytosis in bacterial expulsion from infected BECs. However, the sequence of these events remains undefined. To better define the trafficking of intracellular UPEC, we undertook a morphological characterization of compartments (See Extended Experimental Procedures for identification criteria) housing the intracellular UPEC. At 1 h.p.i., which is relatively soon after bacterial invasion, most intracellular UPEC were contained within a single membrane vacuole (Figure 5Ai). This is consistent with earlier reports that UPEC initially reside in fusiform vesicles of BECs (Bishop et al., 2007). However, after further incubation (approximate 2 h.p.i.), UPEC started to partially or completely escape into the cytosol, as they did not appear to be completely encapsulated by a membrane (Figure 5A ii). At the same time point, some bacteria could also be found within autophagosomes, which are defined by their tightly fitting double-layered membranes (Figure 5A iii). At 4 h.p.i., intracellular UPEC were observed within MVBs which we defined as single membrane compartments which house membrane-bound bacteria as well as ILVs. (Figure 5A iv).
Figure 5. Sequential trafficking of intracellular UPEC before expulsion.
(A) TEM showing intracellular UPEC (i) encased in a single membrane (arrow) vacuole (ii) break their initial vacuole (arrow) or (iii) completely encased in an autophagosome with double-layered membrane (arrow) (iv) encased in a single membrane compartment containing membrane-bound bacteria (arrow) and ILVs (arrow), resembling a MVB. The bacteria associated with each structure is quantified and expressed as the percentage of total examined UPEC, provided in parenthesis. Scale bar: 0.2 µm. n=3 grids
(B) Extracellular UPEC surviving from gentamycin with/without 0.1% Triton X-100 treatment were quantified from BECs treated with rapamycin or ATG5 or Beclin1 shRNA. Error bars, SEM. n=18.
(C) Immunoblot quantification of CD63 associated with EUCV isolated at 12 h.p.i. from infected BECs treated with DMSO vehicle (lane1), 200 nM of rapamycin (lane2) or transfected with control shRNA (lane3) or ATG5 shRNA (lane4). The CD63 detected in the total cell lysate (TCL) was used to suggest similar number of cells were used.
(D) Bacterial expulsion levels at 6 h.p.i. in control or Alix/Tsg101 knockdown BECs, with/without 200 nM rapamycin treatment. Error bars, SEM. n=18.
(E) Immunofluorescence staining of colocalization between autophagosome marker LC3 (green) and MVB marker CD63 (red) when housing UPEC (blue) in infected BECs at 4 h.p.i.. The number of LC3+ and CD63+ compartment-encased UPEC was quantified and expressed as percentage of total ICU examined, shown in the parenthesis. Scale bar: 5 µm. n=3 slides.
(F) The number of CD63+ compartment-encased UPEC was quantified in control KD or ATG5 KD BECs and expressed as percentage of total ICU, n=3 slides.
(G) Bacterial expulsion level from infected control KD, or ATG5KD, Alix KD, VAMP3 KD, Rab7 KD, SYT7 KD BECs, or SYT7 & ATG5 double KD, or SYT7 & Alix double KD were quantified at 4, 6, and 8 h.p.i.. The time point immediately after 1h gentamycin treatment was considered as 0h. Error bars, SEM. n=12.
(H) Immunoblot quantification of CD63 associated with EUCV isolated at 12 h.p.i. from BECs transfected with control shRNA (lane1), SYT7 shRNA (lane2), or Rab7 shRNA (lane 3). CD63 detected in the total cell lysate (TCL) (lane 4, 5, 6) was used to suggest similar number of cells were used.
Next, we demonstrated autophagosome encapsulation precedes deposition of bacteria in ILVs within MVBs. When autophagy was enhanced, the number of exosome-encased UPEC increased correspondingly (Figure 5B, C), whereas this number was significantly reduced in ATG5 or Beclin1 KD BECs (Figure 5B, C). Additionally, when we examined Alix or Tsg101 KD BECs, we failed to detect rapamycin-enhanced expulsion in these cells (Figure 5D). We also observed that LC3 colocalizes with CD63+ compartments that house UPEC (Figure 5E), and when autophagy was abrogated, bacterial localization in CD63+ compartments was markedly reduced (Figure 5F).
We further sought to investigate if the lysosome is the site where bacterial expulsion occurred. We genetically blocked each key stage of the proposed trafficking pathway by selectively knocking down functionally relevant components, such as autophagosome formation (ATG5 KD), autophagosome and MVB fusion (VAMP3 KD) (Figure S2C) (Fader et al., 2009), MVB and lysosome fusion (Rab7 KD) (Figure S2C) (Vanlandingham and Ceresa, 2009), lysosome exocytosis (SYT7 KD), or simultaneously deficient in two of these processes. If parallel routes exist for bacterial export, then the impact of blocking traffic at an early stage (e.g. at autophagosome formation) or simultaneously inhibiting two of these processes will predictably have a larger inhibitory impact on exocytosis than any single blockage at the later stages, (e.g. lysosome exocytosis). However, we found that the inhibition with each of the KD was highly significant and importantly, comparable (Figure 5G). Furthermore, examination of the extracellular UPEC revealed that no detectable exosome associated with UPEC in SYT7 KD or Rab7 KD BECs (Figure 5H).
The above findings confirm that the exosome-encased UPEC are exclusively being exported from the lysosomes. This is surprising as under physiologic conditions, exosomes are routinely released from MVBs (Thery et al., 2002). We hypothesized that upon induction of autophagy, MVBs follow a distinct trafficking rout preferentially targeting their contents into lysosomes. We found that if we only induced autophagy in BECs, the expected exosome release is dramatically abrogated (Figure 4E). Additonally, although rapamycin plus bafilomycin A1 treatment led to a large scale release of exosomes (Figure 4E), this induced-exosome release was drastically reduced in Rab7 KD BECs where the fusion between MVBs and lysosomes was impaired, indicating that the exosomes released from lysosomes originated from MVBs (Figure S2D). To confirm this notion, we examined exosome release in SYT7 KD BECs. In these cells, only lysosome exocytosis is affected, but not MVB. We observed that whereas the induced large scale exosome release was markedly impaired in the SYT7KD cells (Figure S2E), the normal MVB-mediated exosomes secretion under physiological conditions was unaffected (Figure S2E). Cumulatively, our data support the model that UPEC are transported sequentially through multiple intracellular compartments, but they are not expelled from BECs until they reach the lysosomes and only when the lysosomal pH is neutralized by UPEC.
TRPML3 initiates bacterial expulsion from lysosomes
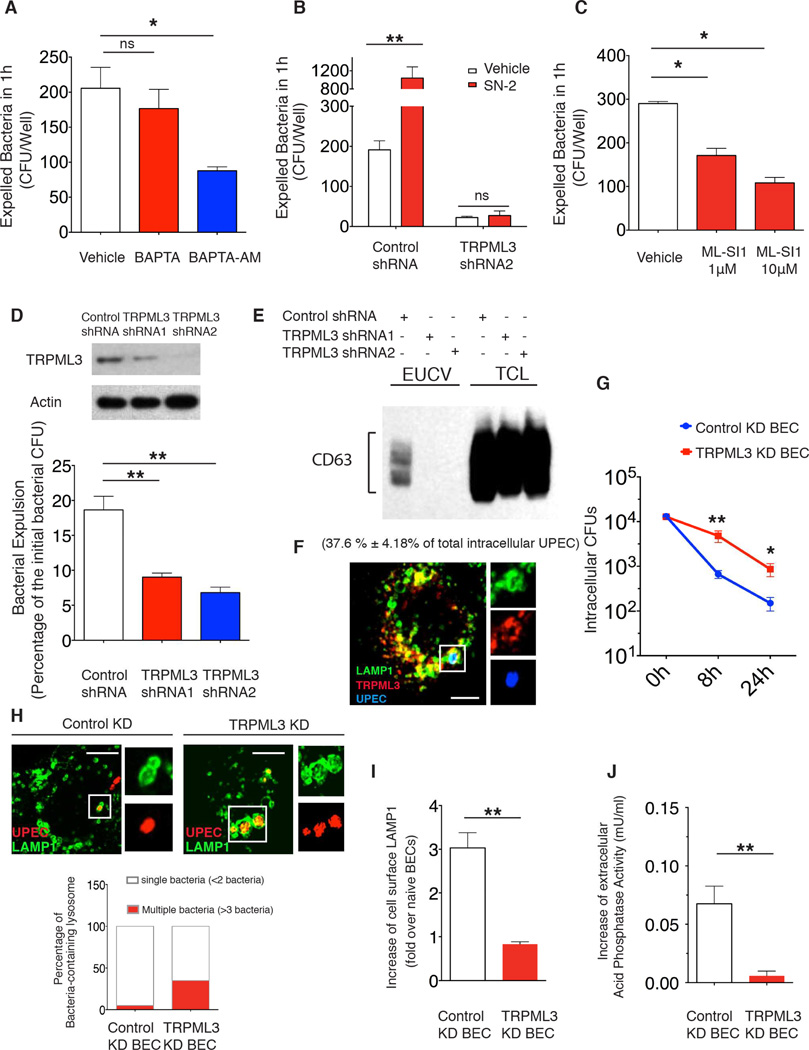
Next, we sought to identify the critical lysosomal sensor capable of detecting pH changes and regulating bacterial expulsion from infected BECs. Since calcium is essential for most exocytic events (Jahn and Sudhof, 1999), we pretreated BECs with BAPTA-AM, an intracellular calcium chelator, and found that BAPTA-AM, but not BAPTA which cannot penetrate BECs, significantly suppressed bacterial expulsion (Figure 6A), indicating that an intracellular calcium store was important for initiating bacterial expulsion. Several recent studies have revealed that the lysosome is actually a key calcium store and multiple calcium channels, including a subfamily of TRP cation channel known as mucolipin TRP channels (TRPML1-3), are located on lysosomes to regulate calcium efflux upon stimulation (Xu, 2015). Based on this knowledge, we decided to examine the involvement of any specific TRPMLs. We first tested the effect of SN2, a recently described agonist for the TRPMLs (Grimm et al., 2010), and found it drastically increased bacterial expulsion (Figure 6B), without accompanying cell lysis (Figure S4A). On the other hand, the ML-SI1(Samie et al., 2013), an antagonist of the TRPMLs markedly blocked bacterial expulsion (Figure 6C). To examine which of these TRPMLs is important for bacterial expulsion, we constructed BECs stably expressing shRNA targeting each of TRPMLs, and we found that TRPML3 KD, was most effective in abolishing bacterial expulsion activity, even in SN2-treated BECs (Figure 6D, 6B). In addition, we are able to detect CD63 associated with extracellular bacteria emanating from control but not from TRPML3 KD BECs, which further verified that TRPML3 is involved in the expulsion of exosome-encased UPEC (Figure 6E). TRPML3 is localized on the lysosome, including the ones encapsulating UPEC (Figure 6F). Interestingly, bacterial clearance is deficient in TRPML3 KD BECs which failed to expel intralysosomal UPEC, resulting in marked increase of the intracellular bacterial load (Figure 6G). Consistent with this observation, in the TRPML3 KD BECs, many bacteria in the lysosome appeared to have multiplied, forming large intracellular aggregates (Figure 6H).
Figure 6. TRPML3 is critical for neutralization-induced lysosome exocytosis.
(A to C) Quantification of expelled bacteria within 1h in infected BECs treated with DMSO vehicle, (A) 10 µM BAPTA or BAPTA-AM; (B) 25 µM SN-2; or (C) 1 µM or 10 µM ML-SI1. The drugs were applied at 6 h.p.i. Error bars, SEM. n=18.
(D) Bacterial expulsion levels at 6 h.p.i. in BECs expressing control shRNA or shRNA targeting two different regions of TRPML3. Knockdown efficiency is indicated by the western blot alongside. Error bars, SEM. n=18.
(E) Immunoblot quantification of CD63 in the purified EUCV from control or TRPML3 KD BECs at 12 h.p.i.. CD63 in the total cell lysate (TCL) was used to show similar number of cells were used.
(F) Immunofluorescence staining of the colocalization of UPEC (blue) containing lysosome (green) and TRPML3 (red) at 6 h.p.i.. The number of LAMP1+ TRPML3+ compartment-encased UPEC was quantified and expressed as percentage of total UPEC examined, shown in the parenthesis. Scale bar: 5 µm. n=3 slides
(G) Intracellular bacterial CFU quantified at 0h, 8h, and 24h.p. i. in control KD and TRPML3KD BECs. The time point immediately after 1h gentamycin treatment was considered as 0h. Error bars, SEM. n=12.
(H) Immunofluorescence staining of UPEC (red) in the lysosome (green) from control or TRPML3 KD BECs at 8 h.p.i.. The lysosomes containing either less than 2 or more than 3 bacteria were quantified and expressed as the percentage of total UPEC-containing lysosomes examined. n=3 slides.
(I) FACS analysis of the cell surface LAMP1+ population in naïve or 200nM rapamycin and 1 µM bafilymycinA1 treated controls or TRPML3 KD BECs. The fold increase of the cell surface LAMP1 over naïve BECs is shown. n=5.
(J) The increase of extracellular acid phosphatase activity in naïve or rapamycin and bafilomycinA1 treated control or TRPML3 KD BECs was compared. Error bars, SEM. n=15.
In view of the important role of TRPML3 in promoting bacterial expulsion, we investigated whether other pH neutralization-induced lysosomal responses also depended on TRPML3. FACS analysis of cell surface LAMP1, lysosomal enzyme release assay, as well as quantification of induced-exosome release in uninfected BECs all showed that bafilomycin A1 treatment failed to induce lysosome exocytosis in TRPML3 KD BECs (Figure 6I, J, and Figure S4B)
Although TRPML3 appears to be the principal modulator of UPEC exocytosis from lysosomes, it is noteworthy that in TRPML1 KD BECs, the number of expelled bacteria is also reduced, although not as striking as in the TRPML3 KD BECs (Figure S4C). However, overexpressing TRPML3 in TRPML1 KD BECs appeared to rescue the defect in lysosome exocytosis activity (Figure S4D), but transfecting TRPML1 failed to achieve a similar effect in the TRPML3 KD BECs, (Figure S4D). Therefore, it appears the bacterial expulsion activity is specifically associated with TRPML3 in BECs, and the function or localization of TRPML3 might somehow be affected in TRPML1 KD BECs.
TRPML3 senses abnormal lysosomal pH and triggers lysosome exocytosis
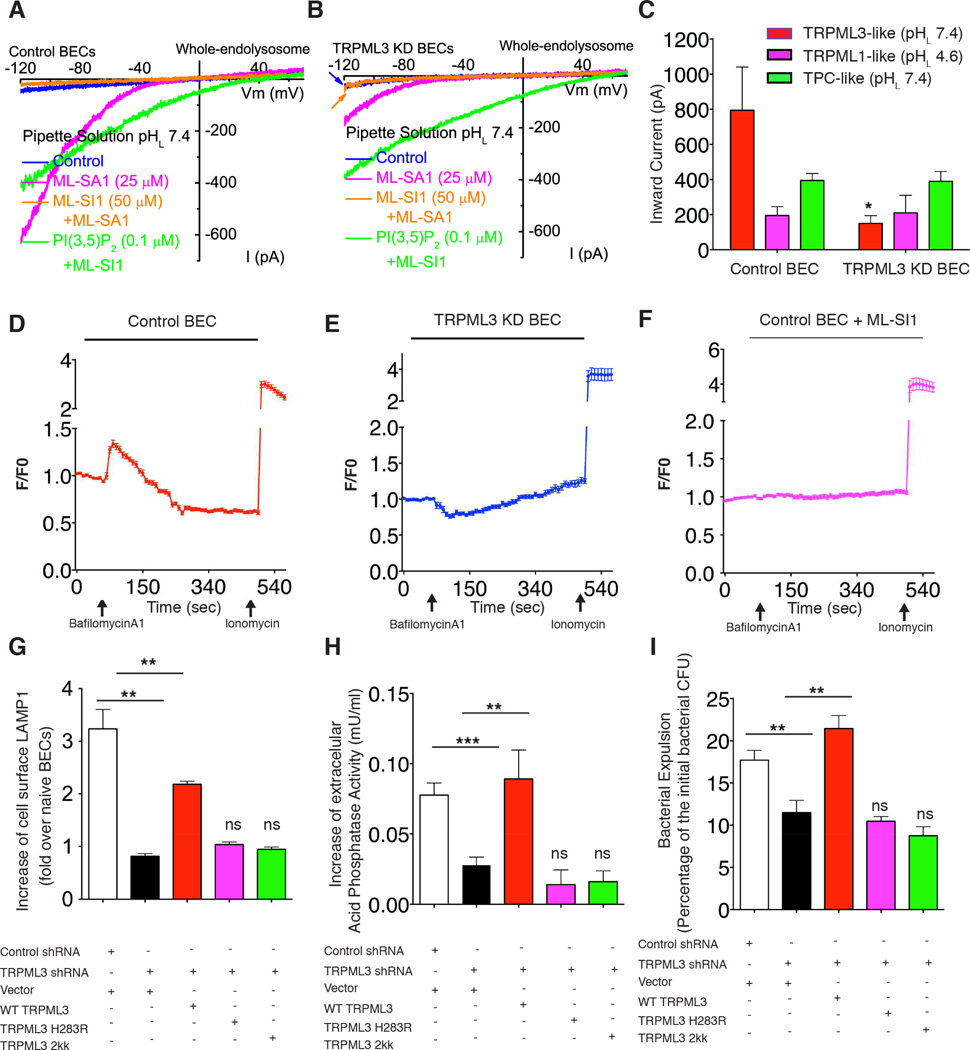
Even though TRPML3 is the critical determinant in lysosomes responsible for bacterial expulsion, it was still unclear how this channel promotes this process in response to lysosomal pH changes. We hypothesize that TRPML3 activation upon neutralization of lysosomal pH, mediates efflux of Ca2+ ions from lysosomes, which in turn induce lysosome exocytosis. To validate this notion, we undertook whole-endolysosome electrophysiology on isolated lysosomes from WT and TRPML3 KD BECs at neutral (pHL7.4) and acidic (pHL4.6) luminal conditions. TRPML-like currents were evoked by a TRPML synthetic agonist ML-SA1 (Shen et al., 2012) and suppressed by the addition of a TRPML antagonist ML-SI1 (Samie et al., 2013)(Figure 7A and 7B). At pHL7.4, TRPML-like currents (ITRPML-L) were greatly reduced by around 4 folds in lysosomes from TRPML3 KD BECs in comparison to those from WT, whereas no significant change was detected in ITRPML-L at pHL4.6 (Figure 7C). Since TRPML1 is known to be active at pHL4.6 (Dong et al., 2008), the ITRPML-L recorded at pHL7.4 is most likely mediated by TRPML3. Meanwhile, the conductance via the two pore channel (TPC) at pHL7.4 was unaffected by the absence of TRPML3 (Figure 7C), which is suggestive of the specificity of the TRPML3 KD. These studies reveal that the TRPML3 channel became active at neutral luminal pH, possibly mediating Ca2+ release from lysosomes. Consistent with this notion, calcium imaging revealed bafilomycinA1 treatment induced rapid calcium efflux from lysosome in WT (Figure 7D), but not in TRPML3 KD BECs (Figure 7E) or ML-SI1 treated BECs (Figure 7F). The inhibition of lysosomal calcium efflux by TRPML3 KD is not due to deficient calcium storage in the lysosomes, as glycyl-L-phenylalanine 2-naphthylamide (GPN), another pH-independent lysosome acting Ca2+ mobilization reagent (Haller et al., 1996), is still able to induce robust Ca2+ efflux from the lysosomes in TRPML3 KD BECs (Figure S4E).
Figure 7. TRPML3 senses lysosomal neutralization and triggers lysosome exocytosis.
(A to C) TRPML3 currents recorded from a whole endolysosome of (A) control BECs and (B) that of TRPML3 KD BECs, which is activated at pH 7.4 by TRPML agonist, ML-SA1 (magenta) and inhibited by TRPML antagonist, ML-SI1 (orange). (C) The quantification of the current. Error bar, SEM. n=5.
(D to F) Calcium efflux (measured as change of fluorescence F over basal fluorescence F0: F/F0) from lysosome upon bafilomycinA1 treatment in the (D) control BEC, (E) TRPML3 KD BECs, or (F) control BECs treated with 10 µM TRPML antagonist ML-SI 1; Error bars, SEM. (n=15).
(G to I) Increases in cell surface LAMP1 positive populations (G), acid phosphatase release (H), or bacterial expulsion (I) in either control BECs transfected with empty vector, or in TRPML3 KD BECs transfected with knockdown resistant form of WT, H283R, or a D458KD459K mutant. Error bars, SEM.
A recent study has shown that TRPML3 can be proton-inhibited, and substituting a histidine (H283) residue in TRPML3 into arginine could mimic this proton-inhibition (Kim et al., 2008). We sought to investigate whether a similar mutation would selectively abrogate the pH sensitivity of TRPML3, and block lysosome exocytosis in response to neutralization of pH. We constructed a similar mutation in the shRNA resistant form of TRPML3 and then overexpressed this mutant in TRPML3 KD BECs. As a positive control, we overexpressed a non-conducting pore mutant of TRPML3 (D458KD459K). Interestingly, while reconstitution of TRPML3 KD BECs with wild type TRPML3 rescued the deficiency in bacterial expulsion and lysosome exocytosis upon neutralization, overexpression of the pH-insensitive or non-conducting pore mutants failed to recapitulate any of these rescue effects (Figure 7 G, H, I). Thus, TRPML3 is normally inactive but when the pH in the lumen of lysosome is neutralized, this channel becomes activated releasing Ca2+ into the cytosol which in turn triggers spontaneous exocytosis of the lysosome and its contents.
DISCUSSION
Bacterial expulsion as a cell autonomous defense strategy
The intrinsic capacity of infected cells to expel intracellular pathogens has until recently been overlooked. Even when nonlytic ejection of bacteria was noticed, it was widely assumed to be a pathogen-driven mechanism for spreading (Hagedorn et al., 2009). Here, we reveal expulsion of UPEC from BECs to be a powerful cell autonomous defense mechanism to rapidly reduce intracellular bacterial load. Following infection, the UPEC-mediated lysosomal neutralization triggered a distinctive cellular response resulting in fusion of the malfunctioning lysosomes with plasma membrane. Blocking any stage of bacterial expulsion resulted in drastic increases in bacterial burden in the bladder. Therefore, these findings reveal a highly effective capacity of the host cell to sense defective lysosomes and to physically dispose of the potential threat. The discharge of bacteria from BECs while still encased in exosomes is intriguing. While it may be a byproduct of bacterial stopover in the MVB, the encircling host membranes may prevent the discharged UPEC, with their highly adhesive organelles (Abraham et al., 1988), from reattaching to the bladder epithelium ensuring their elimination when urine is voided. It is also noteworthy that a significant number of UPEC are still expelled from BECs in spite of deficiencies in our proposed pathway (Figure 5G), suggesting additional bacterial expulsion pathway exist. Indeed, previous studies have reported the relevance of Toll-like Receptor 4 (TLR4) in bacterial expulsion (Song et al., 2009).
Intracellular Trafficking of UPEC
This study has demonstrated the mobilization of the exocytic machinery by the CAD system to combat infection. By systematic backtracking the route of expulsion, we determined that UPEC were initially snared by autophagy, a degradative activity integral to the CAD program. Subsequently, autophagosome-encased bacteria are shuttled to MVBs to form amphisomes before ending up in lysosomes. The relevance of ILV formation within MVBs is noteworthy. Although currently, the autophagosomes are believed to directly fuse with lysosomes for cargo degradation, several recent reports point to a more complicated trafficking route (Lee et al., 2009). When various ESCRT complex components, which are involved in ILV formation, are deficient, the degradation of autophagy substrates within lysosomes is significantly impaired. (Filimonenko et al., 2007). Conceivably encapsulation of cargo in ILVs of MVBs may be necessary for optimal targeting of cargo into lysosomes. At this time, it is also unclear how exactly UPEC becomes encased in an exosome. Presumably, the outer membrane of the autophagosome fuses with the MVB membrane and the inner membrane surrounding the bacteria acquires exosomes markers when ILVs in the MVBs fuse with it.
Monitoring Lysosomal Homeostasis
The lysosome represents the cellular disposal depot and pathogens are also transported to lysosome for elimination. In view of the large pool of highly toxic compounds that they retain, the homeostasis of lysosomes has to be closely monitored. How this is achieved remains largely unclear. Here, we report that the fusion of malfunctioning lysosomes with the plasma membrane and ejection of their luminal contents is an effective mechanism for sustaining their homeostasis. This process is triggered when the luminal pH of lysosome is neutralized, a condition that greatly reduces its degradative powers. Sensing this pH change is a TRP family cation channel localized on the lysosome membrane. TRP channels are critical cellular sensors that respond to a wide range of stimuli and environmental conditions including temperature and osmotic pressure (Clapham, 2003). The TRP channels found on lysosomes belong to mucolipin subfamily (Xu, 2015). Upon activation, these TRPMLs mediate the release of Ca2+ and other cation, into the cytosol from the lysosomal lumen promoting various lysosomal trafficking events culminating in exocytosis (Samie et al., 2013). It is intriguing that the activity of specific TRPMLs is exquisitely sensitive to the ambient pH. While acidic pH potentiates TRPML1 (Dong et al., 2008), the activity of TRPML3 is activated only under neutral pH (Figure 7A–C). Since the lysosomal pH is usually acidic, TRMPL3 may have evolved specifically to counter lysosome-neutralizing pathogens. It is noteworthy that TRPML3 is only expressed in certain cell types, thus, additional mechanisms may exist for monitoring the lysosomal homeostasis.
The lysosomes have long been viewed as nondescript cauldrons where intracellular pathogens are deposited for killing. Our studies suggest that these organelles are highly dynamic with sensory capabilities and are able to perceive subversive actions of pathogens and then eliminate recalcitrant pathogens through exocytosis. However, in spite of the versatility of BEC lysosomes, UPEC infection remain highly prevalent. Presumably, UPEC may possess additional capabilities to subvert this defense strategy. Nevertheless, our studies raise the intriguing possibility of utilizing TRPMLs agonists as treatment for UTIs.
EXPERIMENTAL PROCEDURES
For bacterial strain, mice, antibody and reagent information, see Extended Experimental Procedure
Bacterial Expulsion Assay
The human 5637 BECs were infected at MOIs of 100:1. One hour later, the infected cells were washed and treated with 100µg/mL gentamicin for one hour to kill all extracellular bacteria. At this time, 6 wells with infected BECs from each cell line were permeabilized with 0.1% Triton X-100 to determine the initial bacterial CFU. To the remaining wells, fresh culture media (500 µL) plus methyl α-D-mannopyranoside and 25 µg/mL bacteriostatic reagent, trimethoprim (Sigma) was added and the mixture was incubated. At various time points, 50 µl of culture supernatant in each well was collected, and plated on MacConkey agar plates. Each assay was repeated three times, each time employed 6 wells.
Isolation of Extracellular Bacteria Containing Vesicles
BioMag carboxyl magnetite particles (BM570, Bangs Laboratories) were cross-linked to E.coli to generate “magnetic bacteria” (See more detail in Extended Experimental Procedure) which were added on dishes containing 2×107 BECs at an MOI 200:1. 12h later, the supernatant was collected and undergone serial centrifugation (1. 200g for 5mins; 2. 200g for 5min; 3. 310g for 5min) to remove the floating dead cells. The EUCVs in the supernatant was then purified and washed on a magnetic cell separation rack (BD IMagnet™, BD Biosciences, 552311) at 4°C. The molecular composition of the purified EUCVs was analyzed after SDS-PAGE and western blotting.
Extracellular Gentamicin Protection Assay
The culture medium from infected BECs or the urine from infected mice (see Extended Experimental Procedure) was collected and subjected to serial centrifugation (1. 200g for 5mins; 2. 200g for 5min; 3. 310g for 5min) to remove dead cells and any residual cell debris. Then 100 µg/mL gentamicin was added to the cell-free medium with or without 0.1% Triton-X100 for 1h. The bacteria from each treatment condition were washed in PBS for three times, and plated on MacConkey agar plates. Each assay was repeated three times, each time employed 6 dishes or three mice.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Y. He for Atg3f/f and Atg3f/f ER-Cre mice. We thank K. Hazen, and A. Crichton for collecting urine samples from UTI patients. We also thank Y. Gao, for advice in light microscopy imaging. M. Plue and J. Phillips are thanked for their assistance in TEM. We thank D. Tobin and R. Valdivia for manuscript review. The authors’ work is supported by the US National Institutes of Health grants R01 AI96305, R01 AI35678, R01 DK077159, R01 AI50021, R37 DK50814 and R21 AI056101.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary information includes three figures, one table and one movie, and can be found with this article on line.
AUTHOR CONTRIBUTIONS
Studies were designed by Y.M. and S.N.A. with help from H.X.. Y. M. carried out experiments, with G.L. contributing to the autophagy activation assay. X.Z. performed whole endolysosome electrophysiology studies. Data was analyzed by Y.M., with advice from S.N.A. and H.X. The manuscript was written by Y.M. and S.N.A. All authors contributed to manuscript review.
REFERENCE
- Abraham SN, Sun D, Dale JB, Beachey EH. Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature. 1988;336:682–684. doi: 10.1038/336682a0. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Bishop BL, Duncan MJ, Song J, Li G, Zaas D, Abraham SN. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med. 2007;13:625–630. doi: 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Sanchez DG, Mestre MB, Colombo MI. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochimica et biophysica acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren MB. Phagocyte lysosomes: interactions with infectious agents, phagosomes, and experimental perturbations in function. Annual review of microbiology. 1977;31:507–533. doi: 10.1146/annurev.mi.31.100177.002451. [DOI] [PubMed] [Google Scholar]
- Grimm C, Jors S, Saldanha SA, Obukhov AG, Pan B, Oshima K, Cuajungco MP, Chase P, Hodder P, Heller S. Small molecule activators of TRPML3. Chemistry & biology. 2010;17:135–148. doi: 10.1016/j.chembiol.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annual review of biochemistry. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Li Q, Tjon-Kon-Sang S, So I, Kiselyov K, Soyombo AA, Muallem S. A novel mode of TRPML3 regulation by extracytosolic pH absent in the varitint-waddler phenotype. EMBO J. 2008;27:1197–1205. doi: 10.1038/emboj.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Vischer UM, Rosnoblet C, Lebrand C, Lindsay M, Parton RG, Kruithof EK, Gruenberg J. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol Biol Cell. 2000;11:1829–1843. doi: 10.1091/mbc.11.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnbro P, Nordenfelt P, Tapper H. Isolation of bacteria-containing phagosomes by magnetic selection. BMC Cell Biol. 2008;9:35. doi: 10.1186/1471-2121-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menashe O, Kaganskaya E, Baasov T, Yaron S. Aminoglycosides affect intracellular Salmonella enterica serovars typhimurium and virchow. Antimicrobial agents and chemotherapy. 2008;52:920–926. doi: 10.1128/AAC.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- Samie M, Wang X, Zhang X, Goschka A, Li X, Cheng X, Gregg E, Azar M, Zhuo Y, Garrity AG, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Developmental cell. 2013;26:511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- Shen D, Wang X, Li X, Zhang X, Yao Z, Dibble S, Dong XP, Yu T, Lieberman AP, Showalter HD, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nature communications. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Song J, Bishop BL, Li G, Grady R, Stapleton A, Abraham SN. TLR4-mediated expulsion of bacteria from infected bladder epithelial cells. Proc Natl Acad Sci U S A. 2009;106:14966–14971. doi: 10.1073/pnas.0900527106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell host & microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlandingham PA, Ceresa BP. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem. 2009;284:12110–12124. doi: 10.1074/jbc.M809277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J Cell Sci. 1997;110(Pt 16):1867–1877. doi: 10.1242/jcs.110.16.1867. [DOI] [PubMed] [Google Scholar]
- Xu HR, D Lysosomal Physiology. Annu Rev Physiol. 2015;77:6.1–6.24. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.