SUMMARY
The circadian clock orchestrates global changes in transcriptional regulation on a daily basis via the bHLH-PAS transcription factor CLOCK:BMAL1. Pathways driven by other bHLH-PAS transcription factors have a homologous repressor that modulates activity on a tissue-specific basis, but none have been identified for CLOCK:BMAL1. We show here that the cancer/testis antigen PASD1 fulfills this role to suppress circadian rhythms. PASD1 is evolutionarily related to CLOCK and interacts with the CLOCK:BMAL1 complex to repress transcriptional activation. Expression of PASD1 is restricted to germline tissues in healthy individuals, but can be induced in cells of somatic origin upon oncogenic transformation. Reducing PASD1 in human cancer cells significantly increases the amplitude of transcriptional oscillations to generate more robust circadian rhythms. Our results describe a function for a germline-specific protein in regulation of the circadian clock and provide a molecular link from oncogenic transformation to suppression of circadian rhythms.
INTRODUCTION
The circadian clock coordinates temporal control of physiology by regulating the expression of at least 40% of the genome on a daily basis (Zhang et al., 2014). Disruption of circadian rhythms through environmental stimuli (e.g. light at night) or genetic means can lead to the onset of diseases such as: diabetes, cardiovascular disease, premature aging and cancer (Filipski and Lévi, 2009; Jeyaraj et al., 2012; Kondratov et al., 2006; Marcheva et al., 2010). Understanding the molecular basis of circadian transcriptional regulation in health and disease states offers the opportunity to control vast transcriptional programs that promote health and well-being (Takahashi et al., 2008).
The heterodimeric basic helix-loop-helix PER-ARNT-SIM (bHLH-PAS) transcription factor CLOCK:BMAL1 sits at the core of the molecular circadian clock in mammals. CLOCK:BMAL1 drives expression of core clock factors Period (Per) and Cryptochrome (Cry), as well as thousands of additional clock-controlled output genes (Koike et al., 2012; Zhang et al., 2014). PER and CRY complexes interact with CLOCK:BMAL1 in the nucleus to inhibit transcriptional activation and close the feedback loop, generating intrinsic ~24-hour timing (Gustafson and Partch, 2014; Koike et al., 2012; Lee et al., 2001) This cell-autonomous molecular oscillator is present in nearly every mammalian tissue (Nagoshi et al., 2004; Welsh et al., 2004; Yoo et al., 2004) and is regulated by tissue-specific factors to fine-tune circadian output genes according to cell type (Panda et al., 2002; Storch et al., 2002). While the molecular circadian clock is broadly recognized as a systemic transcriptional regulator, factors that provide tissue-specific regulation of the clock and its outputs remain to be elucidated.
CLOCK and BMAL1 belong to the bHLH-PAS family of transcription factors, which share similar domain architecture but regulate diverse processes including adaptation to hypoxia, xenobiotic metabolism and neuronal development (Crews, 1998; Ema et al., 1996; Gu et al., 1998; Mimura et al., 1999). Homotypic interactions between N-terminal DNA-binding bHLH domains and tandem PAS domains guide formation of specific heterodimeric transcription factor complexes (Huang et al., 2012; Scheuermann et al., 2009; Wang et al., 2013). By contrast, bHLH-PAS C-termini interact with regulatory factors that modulate activity of the complexes (Freedman et al., 2002; Kobayashi et al., 1997). In CLOCK:BMAL1, the BMAL1 C-terminus harbors the essential transactivation domain (TAD) that recruits coactivators CBP/p300 and cryptochrome repressors (Czarna et al., 2011; Kiyohara et al., 2006; Takahata et al., 2000) and a short helical region encoded by CLOCK Exon 19 interacts with the histone methyltransferase MLL1 and a vertebrate-specific repressor CIPC (Katada and Sassone-Corsi, 2010; Zhao et al., 2007). Deletion of Exon 19 prevents proper chromatin targeting of CLOCK:BMAL1 to interfere with circadian transcriptional regulation (Gekakis et al., 1998; Vitaterna et al., 2006).
One interesting feature shared by bHLH-PAS transcription factors is their regulation by paralogous PAS domain-containing repressors. By definition, each paralog repressor shares significant homology with an activator subunit, but either possesses a repressive domain and/or lacks a domain(s) necessary for activation (Ema et al., 1996; Evans et al., 2008; Makino et al., 2001; Teh et al., 2006). These repressors are often expressed in a highly restricted manner to control the tissue-specificity of transcriptional outputs (Fan et al., 1996; Yamamoto et al., 2004). However, the mechanisms by which the pathway-specific paralogs repress transcriptional activation by their cognate heterodimers, and importantly, how their homology to activator subunits is used to impinge on transcriptional regulation are not well understood.
The dedicated bHLH-PAS family repressor for circadian rhythms has not yet been identified. Here we show that the protein PAS domain containing 1 (PASD1) is evolutionarily related to the circadian transcription factor subunit CLOCK and interacts with the CLOCK:BMAL1 complex to inhibit transcriptional activation and suppress circadian timekeeping. Furthermore, deletion of one region, highly conserved with CLOCK Exon 19, alleviates repression by PASD1 to suggest that it utilizes molecular mimicry to interfere with CLOCK:BMAL1 function. As a cancer/testis antigen, expression of PASD1 is natively restricted to gametogenic tissues but can be upregulated in somatic tissues as a consequence of oncogenic transformation. Our work suggests that mechanisms to suppress circadian cycling can be hard-wired in a tissue-specific manner and we show here that they can be co-opted in cancer cells to attenuate clock function.
RESULTS
Identification of a CLOCK paralog in humans
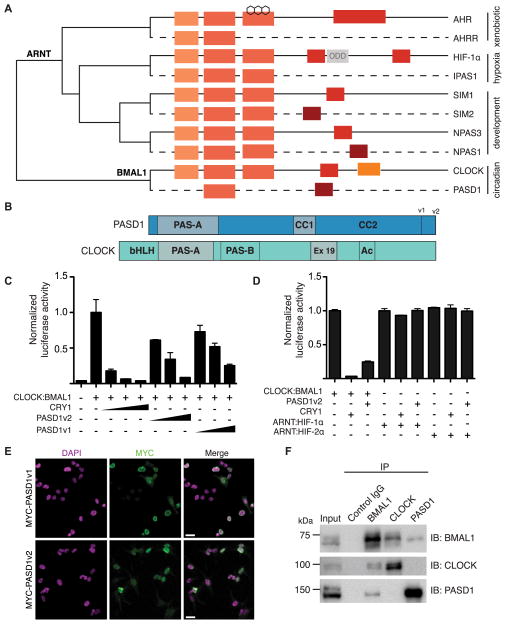
The absence of a paralogous repressor for CLOCK:BMAL1 prompted us to search for human clock protein paralogs that might serve this purpose. Using a PAS domain-based query with the SMART algorithm (Letunic et al., 2014; Schultz et al., 1998) we discovered an uncharacterized PAS domain-containing protein in humans, PAS domain containing protein 1 (PASD1), that is homologous to CLOCK (Figures 1A and S1A). In contrast to CLOCK, PASD1 lacks the DNA-binding bHLH domain and the PAS-B domain, both of which are needed to interact with BMAL1 to form a functional heterodimeric transcription factor (Huang et al., 2012). Similarity between PASD1 and CLOCK is restricted to the PAS-A domain and a helical region in the C-terminus (Figures 1B and S1A) defined by CLOCK Exon 19, which is essential for CLOCK:BMAL1 function (Gekakis et al., 1998; Katada and Sassone-Corsi, 2010; King et al., 1997). PASD1 has two splice isoforms (PASD1v1 and PASD1v2) that differ only by extension of the C-terminus (Figure 1B) (Liggins et al., 2004). The C-terminus of PASD1 has more predicted secondary structure than CLOCK, with two conserved helical regions designated coiled coil 1 and 2 (CC1 and CC2, Figure 1B). Based on these sequence analyses, PASD1 demonstrates two key properties of a pathway-specific bHLH-PAS repressor—homology to a bHLH-PAS activator subunit with the loss of domain(s) critical for transcriptional activation (Makino et al., 2001; Mimura et al., 1999; Moffett et al., 1997).
Figure 1. Identification of a novel circadian repressor that is homologous to CLOCK.
(A) Cladogram of bHLH-PAS transcription factors showing their evolutionary relationship. Each branch of transcription factors within the bHLH-PAS family has a truncated transcriptional repressor that clusters with its related activator subunit and shares similar domain architecture. bHLH: basic helix-loop-helix domain; PAS: PER-ARNT-SIM domain; TAD: Transactivation domain; RPD, repression domain; ODD: Oxygen-dependent degradation domain; MYST, histone acetyltransferase motif from the MYST family (Doi et al., 2006). The cladogram was generated using EMBL-EBI Clustal Omega. Activator subunits have solid lines; repressors have dashed lines. (B) Comparison of human PASD1 and CLOCK domain organization. Gray shading highlights regions of high sequence identity with PASD1. CC1: Coiled-coil domain 1, CC2: Coiled-coil domain 2, Ex 19: Exon 19, Ac: histone acetyltransferase motif. v1 and v2 refer to splice isoforms. See also Figure S1A. (C) Both isoforms of PASD1 inhibit CLOCK:BMAL1 activity in Per1:luc reporter gene assays. HEK293T cells were transfected with Per1:luc, CLOCK, BMAL1 and increasing amounts of CRY1 or PASD1 plasmid as indicated. (n = 3 experiments, mean ± SD) (D) CRY1 and PASD1 do not inhibit transactivation of a VEGF:luc reporter by the bHLH-PAS homologs HIF-1α:ARNT or HIF-2α:ARNT. HA-tagged P1P2N mutants stabilize expression of HIF-1α or HIF-2α under normoxic conditions (Dioum et al., 2009). (n = 3) (E) HEK293T cells were transiently transfected with MYC-PASD1v1 and MYC-PASD1v2 and subcellular localization was determined by immunofluorescence. Scale bar, 20 μm. (F) Co-immunoprecipitation of endogenous BMAL1, CLOCK with PASD1-GFP from U2OS Per2:dluc PASD1-GFP cells. See also Figure S1.
PASD1 is an X-linked gene that is broadly conserved in mammals but absent in murine lineages (Figure S1B), a property common to X-linked genes involved in spermatogenesis (Mueller et al., 2013). In healthy individuals, PASD1 is only expressed in germline tissues such as the testis (Djureinovic et al., 2014); however, PASD1 can be found in somatic tissues upon oncogenic transformation (Ait-Tahar et al., 2009; Cooper et al., 2006; Liggins et al., 2004). PASD1 is therefore designated as a cancer/testis antigen, a classification shared with a large family of proteins that are normally expressed only in the germline and whose expression can provoke immune responses when aberrantly upregulated in neoplastic somatic cells (Ait-Tahar et al., 2009; Liggins et al., 2004; Whitehurst, 2014). While the immunogenicity and expression of PASD1 in a diverse array of human cancers have been well characterized (Joseph-Pietras et al., 2010; Liggins et al., 2010), the cellular function of this protein remains unknown. These data prompted us to investigate whether PASD1 could represent the dedicated bHLH-PAS family repressor that negatively regulates CLOCK:BMAL1.
PASD1 is a nuclear protein that represses transcriptional activation by CLOCK:BMAL1
To determine if PASD1 regulates CLOCK:BMAL1 activity, we conducted a reporter gene assay in human embryonic kidney 293T (HEK293T) cells using the Per1:luc luciferase reporter (Gekakis et al., 1998). PASD1 had no effect on the Per1:luc reporter by itself or in combination with either CLOCK or BMAL1 alone, indicating that it cannot drive transcriptional activation (Figure S2). Titration of either splice isoform of PASD1 led to dose-dependent repression of CLOCK:BMAL1 activity, similar to the core clock repressor Cryptochrome 1 (Figure 1C). Furthermore, both CRY1 and PASD1 demonstrated specificity for CLOCK:BMAL1 as co-expression of either repressor with bHLH-PAS homologs HIF-1α:ARNT or HIF-2α:ARNT had no effect on their transcriptional activation of a hypoxia response element from VEGF (Figure 1D).
Regulation of CLOCK:BMAL1 transcriptional activity is likely to occur in the nucleus where the complex is localized. To determine the subcellular localization of PASD1, we transfected HEK293T cells with MYC-tagged versions of both splice isoforms of PASD1 and used immunofluorescence to visualize PASD1. Both MYC-tagged PASD1 splice isoforms are localized exclusively in the nucleus, predominantly at the periphery where heterochromatin is generally compartmentalized in metazoan cells (Padeken and Heun, 2014) (Figure 1E). To determine if PASD1 interacts with CLOCK:BMAL1, we performed co-immunoprecipitations of each protein from nuclear lysate of a U2OS cell line stably expressing PASD1-GFP. Endogenous BMAL1 precipitated both CLOCK and PASD1-GFP, while CLOCK precipitated low levels of BMAL1 but no detectable PASD1-GFP (Figure 1F). PASD1-GFP precipitated BMAL1 but not CLOCK, suggesting that PASD1 may target the CLOCK:BMAL1 complex through interaction with BMAL1. Taken together, these data show that PASD1 is a potent and specific, nuclear repressor of the CLOCK:BMAL1 complex.
Identification of the PASD1 repressive domain
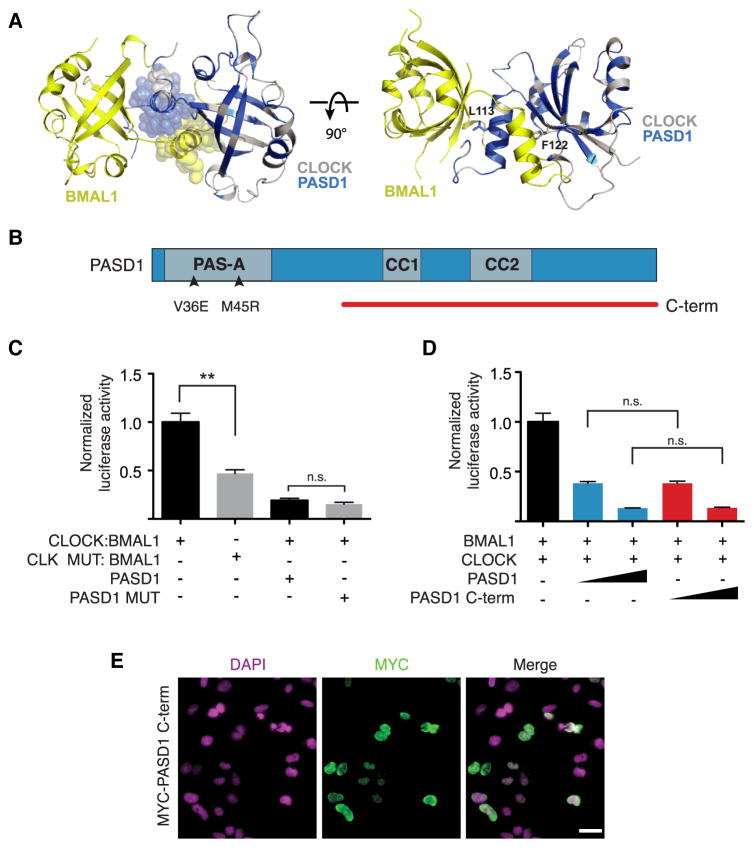
To understand how PASD1 regulates CLOCK:BMAL1 activity, we set out to identify the repressive domain on PASD1. We initially focused our attention on the PASD1 PAS-A domain because it shares a high degree of conservation with CLOCK (Figure S1A). Moreover, PASD1 residues conserved with CLOCK are predominantly localized at the PAS-A interface of the CLOCK:BMAL1 heterodimer (Figure 2A). Mutations in CLOCK PAS-A (L113E/F122R) at this interface diminish interaction with BMAL1 and decrease transactivation by the complex in the Per1:luc reporter assay (Huang et al., 2012). We reasoned that if PASD1 represses CLOCK:BMAL1 by sequestration of the BMAL1 PAS-A domain, then mutation of homologous residues in PASD1 (V36E/M45R) should reduce inhibition of CLOCK:BMAL1 by disrupting the interaction (Figure 2B). The CLOCK PAS-A L113E/F122R mutant caused a significant decrease in Per1:luc activation with BMAL1 as previously reported (Huang et al., 2012), but we saw no effect of the PAS-A V36E/M45R mutation on the ability of PASD1 to repress CLOCK:BMAL1 (Figure 2C). Moreover, we determined that both full-length PASD1 and the isolated C-terminus repressed CLOCK:BMAL1 activity to the same degree (Figure 2D) and localized to the nucleus (Figure 2E), demonstrating that the PASD1 C-terminus is sufficient to repress CLOCK:BMAL1 activity.
Figure 2. The C-terminus of PASD1 is sufficient to repress CLOCK:BMAL1.
(A) Mapping of residues conserved between PASD1 and CLOCK onto the CLOCK:BMAL1 PAS-A domain interface (PDB: 4F3L). BMAL1 (yellow), CLOCK residues conserved with PASD1 (blue) or non-conserved (white). Location of the CLOCK PAS-A mutations (L113E/F122R) that disrupt the CLOCK:BMAL1 heterodimer and reduce transactivation are shown (Huang et al., 2012). (B) Schematic of PASD1 expression constructs. Arrowheads indicate point mutations in the PAS-A domain. (C) Mutation of the PASD1 PAS-A β-sheet interface (V36E/M45R) does not affect repression of CLOCK:BMAL1, while mutation of analogous residues in the PAS-A domain of CLOCK (L113E/F122R) reduces activation of the Per1:luc gene (Huang et al., 2012) (n = 3 experiments, mean ± SD). Significance was determined by Student’s t-test: **, p < 0.01, n.s., not significant. (D) The PASD1 C-terminus is sufficient to repress CLOCK:BMAL1 Per1:luc luciferase expression (n = 3). Significance was determined by Student’s t-test: n.s., not significant. (E) HEK293T cells were transiently transfected with MYC-PASD1v1 C-term and subcellular localization was determined by immunofluorescence. Scale bar, 20 μm. See also Figure S2.
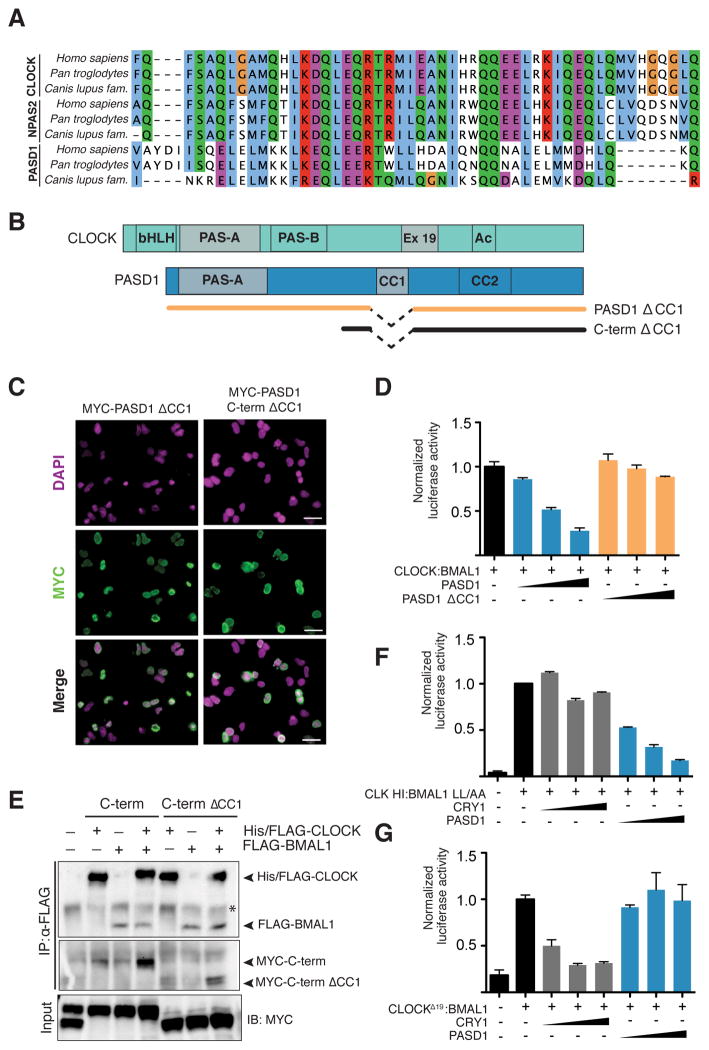
The other major region of conservation between PASD1 and CLOCK exists within the coiled-coil domain 1 (CC1) in the C-terminus of PASD1, which exhibits significant homology with Exon 19 of CLOCK (Figure 3A). This short helical region of CLOCK is important for transcriptional activation and necessary to sustain a robust amplitude of cycling (Gekakis et al., 1998; Katada and Sassone-Corsi, 2010; Vitaterna et al., 2006), suggesting that its conservation within PASD1 might play a role in its regulation of CLOCK:BMAL1 activity. We deleted the CC1 region from the full-length protein (PASD1ΔCC1) or the isolated C-terminus (C-term ΔCC1) to probe its role in CLOCK:BMAL1 regulation (Figure 3B). Although CC1-truncated forms of PASD1 retained nuclear localization and were expressed to the degree as full-length protein (Figures 3C and S3A), repression of CLOCK:BMAL1-driven luciferase activity was significantly impaired (Figure 3D). Co-expression of CLOCK and BMAL1 drives their nuclear localization (Kondratov et al., 2003; Kwon et al., 2006). Full-length PASD1 and the C-terminus both interacted with co-expressed CLOCK and BMAL1, and complex formation was visibly reduced with deletion of CC1 (Figures 3E and S3B). Therefore, the CC1 domain of PASD1 is important for interaction with its cognate transcription factor and transcriptional repression.
Figure 3. PASD1 requires its CC1 domain and Exon 19 of CLOCK to repress CLOCK:BMAL1.
(A) TCoffee alignment of CLOCK and NPAS2 Exon 19 with PASD1 CC1 domain. (B) Schematic of PASD1 expression constructs used in luciferase assays. (C) Deletion of CC1 domain in the full-length protein or C-terminus does not affect nuclear localization. HEK293T cells were transfected with MYC-tagged constructs and subcellular localization was visualized by immunofluorescence. Scale bar, 20 μm. (D) Deletion of the PASD1 CC1 domain (residues 365–415) relieves repression of CLOCK:BMAL1 activation of the Per1:luc gene (n = 3 experiments, mean ± SD). See also Figure S3A. (E) PASD1-MYC tagged C-term ΔCC1 shows decreased interaction when both CLOCK and BMAL1 are precipitated compared to the MYC tagged C-terminus. Asterisk, non-specific protein. See also Figure S3B. (F) Mutation of two residues in the PAS-B domain HI loop of CLOCK (Q361P/W362R; HI) and BMAL1 (L606A/L607A; LL/AA) that reduce repression by CRY1 do not affect PASD1-mediated repression in Per1:luc luciferase assays (n = 3). (G) CRY1 can inhibit transactivation by CLOCKΔ19:BMAL1 but PASD1 cannot (n = 3). See also Figure S3.
Regulation of CLOCK:BMAL1 by PASD1 CC1 and CLOCK Exon 19 are functionally linked
To probe how PASD1 impinges on transcriptional activation by CLOCK:BMAL1, we tested two transcription factor mutants that interfere discretely with two key regulatory domains on CLOCK:BMAL1: the BMAL1 TAD or CLOCK Exon 19. Both CLOCK:BMAL1 mutants had reduced activity relative to wild-type CLOCK:BMAL1, but overall activity of the heterodimer was sufficiently robust (~3–4 fold activation of the Per1:luc reporter over background) to probe regulation of the complexes (Figure S3C). We first tested the CLOCK HI:BMAL1 LL/AA mutant, which interferes with sequestration of the BMAL1 TAD by CRY1 (unpublished data). Although the CLOCK HI:BMAL1 LL/AA mutant abolished repression by CRY1, PASD1 still potently repressed transcriptional activation to suggest that PASD1 does not inhibit CLOCK:BMAL1 through sequestration of the BMAL1 TAD (Figure 3F). We then tested the ability of PASD1 to repress CLOCKΔ19:BMAL1, which lacks the 51 amino acids encoded by Exon 19 (Gekakis et al., 1998; King et al., 1997). CRY1 could still potently repress the residual transactivation potential in the CLOCKΔ19:BMAL1 mutant (Figure 3G); however, PASD1 could no longer exhibited repression of CLOCKΔ19:BMAL1 activity. We interpret these data to mean that PASD1 interferes with CLOCK:BMAL1 function in a manner that depends on the activating potential of Exon 19; once disrupted in the CLOCKΔ19:BMAL1 mutant, PASD1 can no longer further repress the heterodimer.
PASD1 suppresses circadian cycling
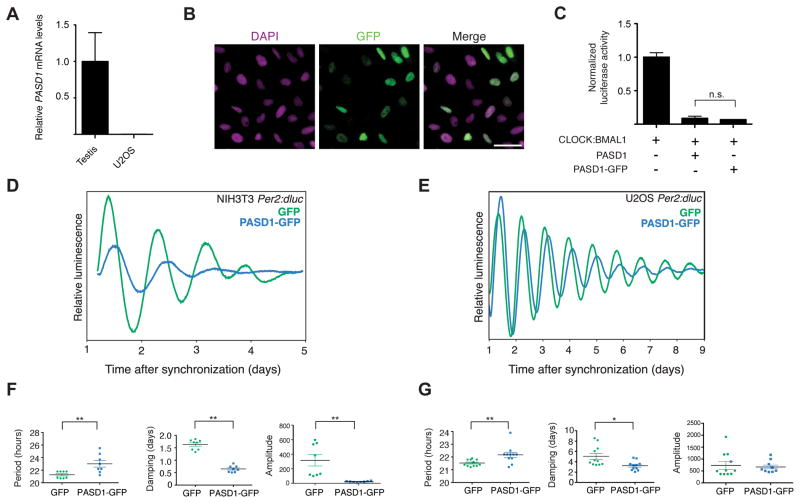
To determine the effect of PASD1 expression on intact molecular circadian oscillators, we examined circadian cycling in mouse NIH3T3 fibroblast cells, which completely lack the PASD1 gene, and human U2OS osteosarcoma cells that cycle with high amplitude (Hirota et al., 2008; Vollmers et al., 2008) but do not express PASD1 as determined by RT-qPCR (Figures 4A and S4A). We generated Per2:dluc reporter cell lines stably expressing PASD1-GFP or a GFP control after determining that GFP fusion does not alter PASD1 subcellular localization or attenuate PASD1 activity towards CLOCK:BMAL1 (Figures 4B–C and S4B). In both lines, expression of PASD1-GFP led to significant alterations in the molecular oscillator (Figures 4D–E) marked by an increase in the period (~1 h) and rate of damping (Figures 4F–G), which indicates defects in cell-autonomous clocks that lead to desynchronization of the population (Izumo et al., 2006). In mouse NIH3T3 fibroblasts, we also noted a significant decrease in amplitude upon PASD1-GFP expression that was not as pronounced in U2OS cells, which could be attributable to the higher basal amplitude of cycling in the U2OS cell line. (Figures 4F–G). Collectively, these findings demonstrate that introduction of PASD1 into naïve cells attenuates the robustness of the molecular circadian oscillator.
Figure 4. Overexpression of PASD1 lengthens period and increases damping of circadian cycling in cell culture.
(A) Comparison of PASD1 expression in human testis and U2OS cells by RT-qPCR. See also Figure S4A. (B) Immunofluorescence of PASD1-GFP in a U2OS Per2:dluc cell line stably expressing PASD1-GFP. Scale bar, 20 μm. See also Figure S4B. (C) Fusion of GFP to PASD1 does not affect repression of CLOCK:BMAL1. Significance was determined by Student’s t-test: n.s., not significant. (D) Representative bioluminescence records from mouse NIH3T3 Per2:dluc cells expressing GFP or PASD1-GFP (n = 8). (E) Representative bioluminescence records from human U2OS Per2:dluc cells expressing GFP or PASD1-GFP (n = 11). (F) PASD1-GFP expression in NIH-3T3 Per2:dluc cells significantly lengthens period, increases damping rate and decreases amplitude. Period (hours) ± SEM: GFP 21.3 ± 0.2, PASD1-GFP 23.0 ± 0.53. Damping (days) ± SEM: GFP 1.29 ± 0.3, PASD1-GFP 0.65 ± 0.046. (G) PASD1-GFP expression in U2OS Per2:dluc cells significantly lengthens period and increases damping rate. Period (hours) ± SEM: GFP 21.55 ± 0.07, PASD1-GFP 22.2 ± 0.2. Damping (days) ± SEM: GFP 5.05 ± 0.6, PASD1-GFP 3.24 ± 0.27, n =11. Significance was determined by Student’s t-test: *, p < 0.05; **, p < 0.01. See also Figure S4.
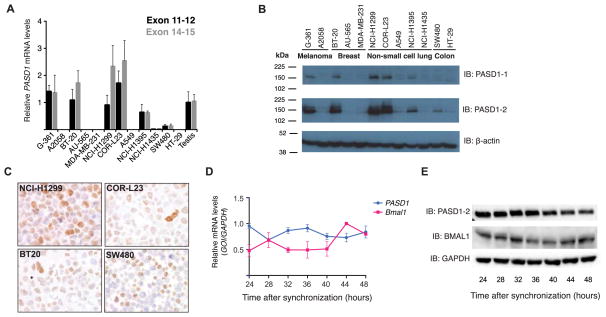
Identification of PASD1-positive cancer cell lines
PASD1 is not expressed in cells of somatic origin unless it has been de-repressed due to malignant transformation (Kim et al., 2013). The circadian clock is frequently disrupted in cancer, allowing cells to escape its daily temporal control of regulated processes and facilitate tumor growth (Filipski and Lévi, 2009; Sahar and Sassone-Corsi, 2009; Takahashi et al., 2008). To determine if upregulation of PASD1 influences the robustness of the circadian clock in human cancer, we screened a panel of cancer cell lines by examining mRNA and protein expression of the two PASD1 splice isoforms. TaqMan probes common to both splice isoforms (Exon 11–12) or specific for the longer PASD1v2 isoform (Exon 14–15) reported similar levels of expression, indicating that the longer isoform is predominantly expressed in human cancer cells (Figure 5A) (Cooper et al., 2006). Among cancer cell lines with the highest PASD1 mRNA expression, we found that G-361 melanoma, NCI-H1299 non-small cell lung carcinoma, COR-L23 large cell lung carcinoma, and the BT-20 breast cancer line had PASD1 transcript levels comparable to human testis (Figure 5A). Relative levels of PASD1 protein correlated with differences between mRNA transcripts when analyzed by western blotting and expression of the longer isoform was confirmed by use of isoform-specific antibodies (Figure 5B) (Cooper et al., 2006). Immunohistochemical analysis of PASD1 expression in several cancer cell lines revealed cell-to-cell heterogeneity in nuclear PASD1 expression, particularly within SW480 colon cancer cells (Figure 5C). Heterogeneous expression of cancer biomarkers is often seen in tumors and can drastically affect the efficacy of cancer therapeutics (Marusyk et al., 2012). Moreover, in the context of circadian regulation, heterogeneous expression of PASD1 could lead to differences in period among individual cells that would serve to desynchronize cell-autonomous molecular oscillators to diminish overall clock function in the tumor microenvironment.
Figure 5. PASD1 mRNA and protein is expressed in a diverse array of human cancers.
(A) Examination of PASD1 transcripts in a panel of cancer cell lines by RT-qPCR. Exon 11–12 TaqMan probe (Hs01098424_m1) recognizes both splice isoforms, while the Exon 14–15 (Hs00542871_m1) TaqMan probe recognizes only the long isoform. PASD1 expression was normalized to TBP (TATA Box Binding Protein), 18S RNA and HPRT1 (Hypoxanthine phosphoribosyltransferase 1) and all samples are presented relative to PASD1 expression in human testis, normalized to 1. Error bars indicate standard deviation from n = 3 measurements. (B) Western blot analysis of PASD1 protein expression in the same cell lines as in panel A. The PASD1-1 monoclonal antibody recognizes an epitope between residues 195–474, common to both splice isoforms. The PASD1-2 monoclonal antibody recognizes an epitope between residues 640–773 that is specific to the longer isoform (Cooper et al., 2006). (C) Immunohistochemistry of PASD1 positive cancer cell lines. Hematoxylin and eosin staining (blue), nuclei; PASD1 staining (brown). (D) RT-qPCR of PASD1 and Bmal1 in NCI-H1299 cells after circadian synchronization with 100 nM dexamethasone. Relative mRNA values are normalized to GAPDH and error bars represent the mean ± SEM of two independent experiments. (E) Western blot of PASD1 and BMAL1 protein expression in circadian synchronized NCI-H1299 cells. Data are representative of two independent experiments.
Other repressors of CLOCK:BMAL1 are transcriptionally regulated by the clock, giving rise to a circadian peak in mRNA abundance (Albrecht et al., 1997; Anafi et al., 2014; Annayev et al., 2014; Honma et al., 2002; Miyamoto and Sancar, 1999; Shearman et al., 1997; Zhao et al., 2007). We examined PASD1 and Bmal1 mRNA and protein expression over a circadian period in NCI-H1299 cells after synchronization of cellular clocks by dexamethasone. We found that expression of Bmal1 mRNA was rhythmic on the circadian timescale, but exhibited low amplitude in its oscillation (ANOVA, p = 0.05) (Figure 5D). PASD1 mRNA expression was antiphasic to Bmal1 with an even lower amplitude of oscillation that did not reach criteria for significance (ANOVA, p = 0.12). Protein levels of BMAL1 followed the same trend, with cyclical yet low amplitude circadian rhythms while PASD1 levels did not appear to cycle after synchronization (Figure 5E). Taken together, these data suggest that PASD1 may be a CLOCK:BMAL1 target; however, we were unable to detect a circadian oscillation of PASD1 mRNA or protein levels in NCI-H1299 cells.
Downregulation of PASD1 improves amplitude of cycling in human cancer cells
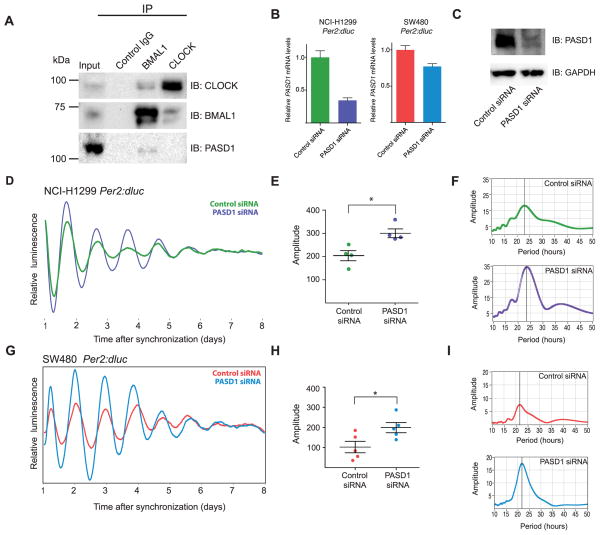
To assess the effect of endogenous PASD1 expression on circadian rhythms in human cancer cells, we chose to study the NCI-H1299 cell line (high expression) and the SW480 cell line (lower levels with more heterogeneous expression) by stably incorporating the Per2:dluc reporter gene (Zhang et al., 2009). Co-immunoprecipitation experiments in NCI-H1299 Per2:dluc cells showed that a pool of endogenous PASD1 and BMAL1 interact (Figure 6A). Transfection of the Per2:dluc lines with PASD1 siRNA achieved knockdown of PASD1 in NCI-H1299 cells as assessed by RT-qPCR, although to a lesser degree in SW480 cells (Figure 6B). siRNA treatment of NCI-H1299 Per2:dluc cells reduced PASD1 protein levels (Figure 6C) and resulted in a significant increase in the amplitude of circadian cycling (Figures 6D–E). Fast Fourier Transform (FFT) power spectra of cycling data from NCI-H1299 Per2:dluc cells also demonstrated that knockdown of PASD1 improved amplitude (Figure 6F). This waveform analysis converts time domain cycling data into the frequency domain, illustrating both the period of the oscillation and its amplitude, demonstrated by the height of the strongest spectral peak that defines the circadian period. The amplitude of circadian cycling was also significantly improved upon PASD1 knockdown in SW480 Per2:dluc cells (Figures 6G–I). We detected a modest but non-significant trend towards a longer period in both lines with PASD1 knockdown (Figure S5). The marked improvement of cycling amplitude in two distinct cancer cell lines upon knockdown of PASD1 demonstrates that it can suppress circadian clock function when upregulated in human cancer.
Figure 6. Reducing expression of PASD1 in cancer cells increases robustness of circadian rhythms.
(A) Co-immunoprecipitation of endogenous CLOCK, BMAL1 and PASD1 from NCI-H1299 nuclear extract. (B) Reduction of PASD1 mRNA in NCI-H1299 Per2:dluc and SW480 Per2:dluc cells after siRNA by RT-qPCR. (n = 3 experiments, mean ± SD) (C) Knockdown of PASD1 protein in NCI-H1299 Per2:dluc cells with siRNA. Due to the lower level of heterogeneous expression of PASD1 in SW480 cells, protein levels were at the limit of detection by western blot using our polyclonal rPASD1 antibody; therefore, knockdown was assessed only using RT-qPCR. (D) Representative bioluminescence records from NCI-H1299 Per2:dluc lung cancer cells transfected with control scramble or PASD1 siRNA (n = 4). (E) Mean amplitude values from independent experiments. Data are represented as mean ± SEM. Significance was assessed by Student’s t-test, *, p < 0.05. (F) FFT analysis spectrum of cycling traces shown in part D. G to I) Same as in panels D to F for SW480 Per2:dluc colon cancer cells. Student’s t-test, *, p < 0.05. See also Figure S5.
DISCUSSION
Our findings establish that PASD1 is the bHLH-PAS paralog repressor for the circadian transcription factor CLOCK:BMAL1. As such, PASD1 fulfills a role analogous to the aryl hydrocarbon receptor repressor (AhRR) (Mimura et al., 1999), inhibitory PAS protein (IPAS) (Makino et al., 2001), and neuronal PAS domain protein 1 (NPAS1) (Teh et al., 2006), each dedicated to repression of a specific bHLH-PAS signaling pathway. Several bHLH-PAS paralog repressors require their homologous PAS domains to interfere with the function of cognate transcription factors (Makino et al., 2001; Mimura et al., 1999), while others such as NPAS1 and SIM2 possess homologous PAS domains but utilize repressive domains in their C-termini (Moffett and Pelletier, 2000; Moffett et al., 1997; Teh et al., 2006). We discovered PASD1 by searching for PAS domain-containing homologs to CLOCK and BMAL1 in humans. However, our studies addressing the biochemical mechanism of PASD1 regulation highlighted an essential role for the C-terminal CC1 domain that bears homology to the essential regulatory region encoded by CLOCK Exon 19. The Id protein family also shares structural homology with CLOCK and BMAL1 in the helix-loop-helix DNA-binding domain that it uses to repress the complex (Duffield et al., 2009; Ward et al., 2010). However, by targeting a prevalent DNA-binding motif, these proteins also repress many other transcriptional networks. Because PASD1 invokes sequence similarity with CLOCK to regulate CLOCK:BMAL1 activity, it appears to be more specific for the circadian pathway. We showed that PASD1 does not repress CLOCK:BMAL1 activity like cryptochromes, which sequester the BMAL1 transcriptional activation domain from coactivators (unpublished data). Instead, PASD1 requires the activating potential of CLOCK Exon 19 to repress transcriptional activation by CLOCK:BMAL1. Collectively, these data suggest that PASD1 utilizes molecular mimicry of CLOCK Exon 19 to interfere with CLOCK:BMAL1 function.
CLOCK Exon 19 is essential for CLOCK:BMAL1 function and its deletion generates a dominant negative ClockΔ19 mutant that suppresses circadian rhythms (Gekakis et al., 1998; King et al., 1997). The 51 amino acids encoded by CLOCK Exon 19 are needed to interact with the coactivator histone methyltransferase MLL1 (Katada and Sassone-Corsi, 2010) and a vertebrate-specific repressor, CLOCK-Interacting Protein Circadian, CIPC (Zhao et al., 2007). Even though MLL1 interacts with both CLOCK and BMAL1 by co-immunoprecipitation, it requires CLOCK Exon 19 to coordinate rhythmic changes in Histone H3 Lysine4 trimethylation (Katada and Sassone-Corsi, 2010). Unlike MLL1 and CIPC, we did not detect interaction of PASD1 with CLOCK; instead, our data show that PASD1 requires its CC1 domain to interfere with CLOCK:BMAL1 function that is mediated by Exon 19. The reciprocal relationship between PASD1 CC1 and CLOCK Exon 19 is further supported by similarities between PASD1 overexpression in 3T3 fibroblasts and heterozygous expression of the dominant negative ClockΔ19 mutant, both of which exhibit decreased amplitude, long period and rapid damping (Vitaterna et al., 2006). However, the exact mechanism by which PASD1 impinges on transcriptional activation by CLOCK:BMAL1 remains to be determined.
The limited distribution of PASD1 across tissues is analogous to other bHLH-PAS repressors that are expressed selectively to control developmental or tissue-specific programs of transcriptional activation (Fan et al., 1996; Makino et al., 2001; Michael and Partch, 2013; Yamamoto et al., 2004). One powerful example of this is inhibition of the hypoxia inducible factor, HIF (HIF-1α:ARNT), in the hypoxic cornea by its paralog repressor IPAS, which prevents neovascularization in the cornea that would interfere with vision (Makino et al., 2001). By virtue of its limited tissue distribution, PASD1 is poised to suppress circadian rhythms in the germline and, as a consequence of its demethylation and upregulation, in somatic cancers (Cooper et al., 2006; Whitehurst, 2014). Notably, mouse and hamster testis do not exhibit molecular circadian rhythms (Alvarez and Sehgal, 2005; Miyamoto and Sancar, 1999; Morse et al., 2003). It has yet to be demonstrated that testis from other mammals, including humans, do not have circadian rhythms; however, based on the data presented here, we speculate that high levels of PASD1 in human testis could lead to suppression of circadian rhythms in the germline (Cooper et al., 2006). Connections between the lack of circadian cycling in undifferentiated embryonic stem cells and the germline are just coming to light (Paulose et al., 2012; Umemura et al., 2014; Yagita et al., 2010), making PASD1 an interesting link that could be explored further.
To date, cancer/testis antigens have largely been explored as targets for cancer immunotherapy (Ait-Tahar et al., 2009; Joseph-Pietras et al., 2010; Whitehurst, 2014). It is still unclear whether they simply serve as cancer biomarkers or whether their upregulation in somatic cancer has consequences for tumor progression by promoting return to a germ-like state (Simpson et al., 2005). Recent studies show that some cancer/testis antigens possess activities consistent with the latter hypothesis, from promoting destabilization of tumor suppressors to regulating genomic stability (Cappell et al., 2012; Doyle et al., 2010). Here we describe a role for a cancer/testis antigen in suppression of the circadian clock, showing that PASD1 can attenuate clock function even when heterogeneously expressed in cancer cells. Circadian disruption has been connected to increased incidence of diabetes, cardiovascular disease, and cancer (Filipski and Lévi, 2009; Jeyaraj et al., 2012; Marcheva et al., 2010) but there is a growing appreciation for reciprocal regulation of circadian rhythms by disease or altered metabolic states. In particular, consumption of a high fat diet alters the metabolic state to suppress CLOCK:BMAL1-driven transcriptional activation and dampen circadian amplitudes, allowing for the rewiring of vast transcriptional programs (Eckel-Mahan et al., 2013; Hatori et al., 2012). These studies demonstrate the importance of maintaining robust circadian amplitudes to promote proper temporal regulation of physiology. Our discovery of PASD1 as a circadian bHLH-PAS paralog repressor that is only expressed in somatic tissues after oncogenic transformation suggests that it may represent a molecular link from oncogenesis to circadian disruption.
EXPERIMENTAL PROCEDURES
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 10 minutes, then permeabilized and blocked in phosphate buffered saline (PBS) containing 1% horse serum and 0.05% Triton X-100 for 15 minutes. Primary incubation was carried out for 1 hour at room temperature (RT) in blocking solution with chicken anti-GFP (1:1,500; Aves labs) or mouse anti-MYC (1:1,000; Abcam). Cells were washed 3x with PBS, then incubated with secondary Alexa Fluor conjugated antibodies (1:2,000; Molecular Probes) and DAPI (Life Technologies) for 30 minutes. Slides were mounted in Fluoromount-G (Southern Biotech) and analyzed on a Keyence BZ-9000 Fluorescence Microscope.
Antibodies
Polyclonal antibodies were generated against human PASD1 (rPASD1) (epitope: DQMRSAEQTRLMPAEQRDS, residues 751–770) and human BMAL1 (Rey et al., 2011) (epitope: LEADAGLGGPVDFSDLPWPL, residues 607–626) by immunization of KLH-conjugated peptides in rabbits using standard protocols (Pierce Biotechnology). Serum was affinity purified using the SulfoLink Immobiliation Kit using the manufacturer’s instructions (Thermo) after conjugation of the antigenic peptide to the immobile phase. Purified antibodies were dialyzed into 0.15M glycine, 50mM Tris-HCl, pH 7.0 with 50% glycerol and aliquoted for storage at -80°C. Hybridoma supernatants containing anti-PASD1 monoclonal antibodies (PASD1-1 and PASD1-2) were as previously described (Cooper et al., 2006).
Supplementary Material
Acknowledgments
We appreciate comments from A. Sancar, A.C. Liu, and L. Hinck on the manuscript. This work was supported by grants from the University of California Cancer Research Coordinating Committee and R01 GM107069 (to C.L.P), NIH Ruth Kirschstein fellowship F31 CA189660 and a Paul & Anne Irwin Cancer Research Award (to A.K.M.), and Cancer Research UK Programme grant A10702 (to A.B.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ait-Tahar K, Liggins AP, Collins GP, Campbell A, Barnardo M, Lawrie C, Moir D, Hatton C, Banham AH, Pulford K. Cytolytic T-cell response to the PASD1 cancer testis antigen in patients with diffuse large B-cell lymphoma. Br J Haematol. 2009;146:396–407. doi: 10.1111/j.1365-2141.2009.07761.x. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- Alvarez JD, Sehgal A. The thymus is similar to the testis in its pattern of circadian clock gene expression. J Biol Rhythms. 2005;20:111–121. doi: 10.1177/0748730404274078. [DOI] [PubMed] [Google Scholar]
- Anafi RC, Lee Y, Sato TK, Venkataraman A, Ramanathan C, Kavakli IH, Hughes ME, Baggs JE, Growe J, Liu AC, et al. Machine Learning Helps Identify CHRONO as a Circadian Clock Component. PLoS Biol. 2014:12. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annayev Y, Adar S, Chiou YY, Lieb JD, Sancar A, Ye R. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem. 2014;289:5013–5024. doi: 10.1074/jbc.M113.534651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappell KM, Sinnott R, Taus P, Maxfield K, Scarbrough M, Whitehurst AW. Multiple Cancer Testis Antigens Function To Support Tumor Cell Mitotic Fidelity. Mol Cell Biol. 2012;32:4131–4140. doi: 10.1128/MCB.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CDO, Liggins AP, Ait-Tahar K, Roncador G, Banham AH, Pulford K. PASD1, a DLBCL-associated cancer testis antigen and candidate for lymphoma immunotherapy. Leukemia. 2006;20:2172–2174. doi: 10.1038/sj.leu.2404424. [DOI] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH- PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Czarna A, Breitkreuz H, Mahrenholz CC, Arens J, Strauss HM, Wolf E. Quantitative analyses of cryptochrome-mBMAL1 interactions: mechanistic insights into the transcriptional regulation of the mammalian circadian clock. J Biol Chem. 2011;286:22414–22425. doi: 10.1074/jbc.M111.244749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- Djureinovic D, Fagerberg L, Hallström B, Danielsson A, Lindskog C, Uhlén M, Pontén F. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 2014;20:476–488. doi: 10.1093/molehr/gau018. [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–974. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield GE, Watson NP, Mantani A, Peirson SN, Robles-Murguia M, Loros JJ, Israel MA, Dunlap JC. A Role for Id2 in Regulating Photic Entrainment of the Mammalian Circadian System. Curr Biol. 2009;19:297–304. doi: 10.1016/j.cub.2008.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, De Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Morita M, Ikawa S, Tanaka M, Matsuda Y, Gotoh O, Saijoh Y, Fujii H, Hamada H, Kikuchi Y, et al. Two new members of the murine Sim gene family are transcriptional repressors and show different expression patterns during mouse embryogenesis. Mol Cell Biol. 1996;16:5865–5875. doi: 10.1128/mcb.16.10.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BR, Karchner SI, Allan LL, Pollenz RS, Tanguay RL, Jenny MJ, Sherr DH, Hahn ME. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol Pharmacol. 2008;73:387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan CM, Kuwana E, Bulfone A, Fletcher CF, Copeland NG, Jenkins NA, Crews S, Martinez S, Puelles L, Rubenstein JL, et al. Expression patterns of two murine homologs of Drosophila single-minded suggest possible roles in embryonic patterning and in the pathogenesis of Down syndrome. Mol Cell Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- Filipski E, Lévi F. Circadian disruption in experimental cancer processes. Integr Cancer Ther. 2009;8:298–302. doi: 10.1177/1534735409352085. [DOI] [PubMed] [Google Scholar]
- Freedman SJ, Sun ZYJ, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit, HIF3alpha. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- Gustafson CL, Partch CL. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry. 2014 doi: 10.1021/bi500731f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JAJ, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Wook J, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. PNAS. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS. Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex. Science (80-) 2012;337:189–194. doi: 10.1126/science.1222804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo M, Sato TR, Straume M, Johnson CH. Quantitative Analyses of Circadian Gene Expression in Mammalian Cell Cultures. PLoS Comput Biol. 2006;2:e136. doi: 10.1371/journal.pcbi.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, et al. Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature. 2012;483:96–99. doi: 10.1038/nature10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Pietras D, Gao Y, Zojer N, Ait-Tahar K, Banham AH, Pulford K, Rice J, Savelyeva N, Sahota SS. DNA vaccines to target the cancer testis antigen PASD1 in human multiple myeloma. Leuk Off J Leuk Soc Am Leuk Res Fund, UK. 2010;24:1951–1959. doi: 10.1038/leu.2010.196. [DOI] [PubMed] [Google Scholar]
- Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Kulkarni P, Hannenhalli S. Derepression of Cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation. BMC Cancer. 2013;13:144. doi: 10.1186/1471-2407-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara YB, Tagao S, Tamanini F, Morita A, Sugisawa Y, Yasuda M, Yamanaka I, Ueda HR, van der Horst GTJ, Kondo T, et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc Natl Acad Sci U S A. 2006;103:10074–10079. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Numayama-Tsuruta K, Sogawa K, Fujii-Kuriyama Y. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt) J Biochem. 1997;122:703–710. doi: 10.1093/oxfordjournals.jbchem.a021812. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science (80-) 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 2006;26:7318–7330. doi: 10.1128/MCB.00337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggins AP, Brown PJ, Asker K, Pulford K, Banham AH. A novel diffuse large B-cell lymphoma-associated cancer testis antigen encoding a PAS domain protein. Br J Cancer. 2004;91:141–149. doi: 10.1038/sj.bjc.6601875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liggins AP, Lim SH, Soilleux EJ, Pulford K, Banham AH. A panel of cancer-testis genes exhibiting broad-spectrum expression in haematological malignancies. Cancer Immun a J Acad Cancer Immunol. 2010;10:8. [PMC free article] [PubMed] [Google Scholar]
- Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- Michael AK, Partch CL. bHLH-PAS proteins: functional specification through modular domain architecture. OA Biochem. 2013;1:16. [Google Scholar]
- Mimura J, Ema M, Sogawa K, Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 1999;13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Sancar A. Circadian regulation of cryptochrome genes in the mouse. Mol Brain Res. 1999;71:238–243. doi: 10.1016/s0169-328x(99)00192-8. [DOI] [PubMed] [Google Scholar]
- Moffett P, Pelletier J. Different transcriptional properties of mSim-1 and mSim-2. FEBS Lett. 2000;466:80–86. doi: 10.1016/s0014-5793(99)01750-0. [DOI] [PubMed] [Google Scholar]
- Moffett P, Reece M, Pelletier J. The murine Sim-2 gene product inhibits transcription by active repression and functional interference. Mol Cell Biol. 1997;17:4933–4947. doi: 10.1128/mcb.17.9.4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D, Cermakian N, Brancorsini S, Parvinen M, Sassone-Corsi P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol Endocrinol. 2003;17:141–151. doi: 10.1210/me.2002-0184. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr Opin Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Paulose JK, Rucker EB, Cassone VM. Toward the Beginning of Time: Circadian Rhythms in Metabolism Precede Rhythms in Clock Gene Expression in Mouse Embryonic Stem Cells. PLoS One. 2012:7. doi: 10.1371/journal.pone.0049555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol. 2011:9. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Reppert SM. Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Simpson AJG, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Ozaki T, Mimura J, Kikuchi Y, Sogawa K, Fujii-Kuriyama Y. Transactivation mechanisms of mouse clock transcription factors, mClock and mArnt3. Genes to Cells. 2000;5:739–747. doi: 10.1046/j.1365-2443.2000.00363.x. [DOI] [PubMed] [Google Scholar]
- Teh CHL, Lam KKY, Loh CC, Loo JM, Yan T, Lim TM. Neuronal PAS domain protein 1 is a transcriptional repressor and requires arylhydrocarbon nuclear translocator for its nuclear localization. J Biol Chem. 2006;281:34617–34629. doi: 10.1074/jbc.M604409200. [DOI] [PubMed] [Google Scholar]
- Umemura Y, Koike N, Matsumoto T, Yoo S-H, Chen Z, Yasuhara N, Takahashi JS, Yagita K. Transcriptional program of Kpna2/Importin-α2 regulates cellular differentiation-coupled circadian clock development in mammalian cells. Proc Natl Acad Sci. 2014 doi: 10.1073/pnas.1419272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Panda S, DiTacchio L. A high-throughput assay for siRNA-based circadian screens in human U20S cells. PLoS One. 2008:3. doi: 10.1371/journal.pone.0003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wu Y, Li L, Su XD. Intermolecular recognition revealed by the complex structure of human CLOCK-BMAL1 basic helix-loop-helix domains with E-box DNA. Cell Res. 2013;23:213–224. doi: 10.1038/cr.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward SM, Fernando SJ, Hou TY, Duffield GE. The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression. J Biol Chem. 2010;285:38987–39000. doi: 10.1074/jbc.M110.175182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annu Rev Pharmacol Toxicol. 2014;54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J, et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci U S A. 2010;107:3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto J, Ihara K, Nakayama H, Hikino S, Satoh K, Kubo N, Iida T, Fujii Y, Hara T. Characteristic expression of aryl hydrocarbon receptor repressor gene in human tissues: organ-specific distribution and variable induction patterns in mononuclear cells. Life Sci. 2004;74:1039–1049. doi: 10.1016/j.lfs.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, Janes J, et al. A Genome-wide RNAi Screen for Modifiers of the Circadian Clock in Human Cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WN, Malinin N, Yang FC, Staknis D, Gekakis N, Maier B, Reischl S, Kramer A, Weitz CJ. CIPC is a mammalian circadian clock protein without invertebrate homologues. Nat Cell Biol. 2007;9:268–275. doi: 10.1038/ncb1539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.