SUMMARY
G protein-coupled receptors (GPCRs) comprise the largest family of cell-surface receptors, regulate a wide range of physiological processes, and are the major targets of pharmaceutical drugs. Canonical signaling from GPCRs is relayed to intracellular effector proteins by trimeric G proteins, composed of α, β, and γ subunits (Gαβγ). Here, we report that G-protein β subunits (Gβ) bind to DDB1 and that Gβ2 targets GRK2 for ubiquitylation by the DDB1-CUL4A-ROC1 ubiquitin ligase. Activation of GPCR results in PKA-mediated phosphorylation of DDB1 at Ser645 and its dissociation from Gβ2, leading to increase of GRK2 protein. Deletion of Cul4a results in cardiac hypertrophy in male mice that can be partially rescued by the deletion of one Grk2 allele. These results reveal a non-canonical function of the Gβ protein as a ubiquitin ligase component and a mechanism of feedback regulation of GPCR signaling.
INTRODUCTION
G-protein coupled receptors (GPCRs) comprise the largest known family of cell-surface receptors, regulate numerous physiological processes, and have a major impact on medicine with about 30% of current therapeutics targeting these seven transmembrane receptors (Rockman et al., 2002; Shenoy and Lefkowitz, 2005). The canonical GPCR signals are commonly relayed to intracellular effector proteins by trimeric G proteins, composed of an α, β, and γ subunit (Gαβγ) (Siderovski et al., 2007). The inactive GDP-bound Gα associates with Gβγ when the GPCRs are un-occupied and switches to active GTP-bound form and dissociates from Gβγ upon activation of GPCRs by their respective agonists. The GTP-bound Gα activates adenylyl cyclase, resulting in the increase of cAMP, activation of cAMP-dependent protein kinase A (PKA) and downstream effector molecules. Hydrolysis of GTP by the intrinsic GTPase activity of Gα returns it to its GDP-bound form to form a heterotrimeric Gαβγ complex and complete the G-protein cycle.
Negative regulation and termination of most agonist-activated GPCRs are described as desensitization. Classically, activated receptors are subsequently phosphorylated by a family of kinases called G protein-coupled receptor kinases (GRKs, also known as β-adrenergic receptor kinase or β-ARKs) (Premont and Gainetdinov, 2007). The phosphorylated receptor then recruits the tethering adaptor protein β-arrestin that uncouples the receptor and G protein and promotes desensitization, internalization and down-regulation of the GPCR. Although many proteins have been identified to interact with GPCRs, GRKs and β-arrestins are the only two families of proteins that have the ability to interact generally with the agonist-stimulated GPCRs to inhibit signaling and desensitize receptors (DeWire et al., 2007). The molecular mechanism by which GRK2-terminates β-AR signaling is relatively well understood (Lefkowitz and Shenoy, 2005). GRK2 distributes in the cytoplasm of unstressed cells but translocates, through binding with free Gβγ dimers, to the plasma membrane following agonist stimulation of the β-AR through direct interaction of its C-terminal PH-domain with a Gβγ dimer, and then phosphorylates the agonist-occupied β-AR (Lodowski et al., 2003). Abnormally elevated GRK2 protein level is linked with multiple pathological conditions in humans (Gurevich et al., 2012), including myocardial infarction (Yu et al., 2005), heart failure, portal hypertension (Liu et al., 2005), insulin resistance (Morisco et al., 2006), and Alzheimer’s disease (Leosco et al., 2007). Despite extensive studies demonstrating a critical role of GRK2 in the regulation of β-AR signaling and heart function, the regulation of GRK2 protein levels, as well as other members of the broader GRK family, remains poorly understood at present.
Previously, we and other groups reported that human cells express as many as ninety DDB1-binding WD40 proteins (DWD, also known as DCAF for DDB1- and CUL4-associated factors and CDW for CUL4 and DDB1-associated WD40 repeats) {Angers, 2006 #90; He, 2006 #24; Higa, 2006 #25; Jin, 2006 #117}. Among these estimated 90 human DWD proteins are the five members of the G-protein β subunits (Gβs) (Gβ1 – 5). Structurally, each Gβ protein contains seven WD40 repeats with a perfectly matched DWD box in the fourth WD40 repeat (Figure 1A). This raises the possibility that Gβ proteins could have a previously unrecognized function as a component of cullin-RING E3 ubiquitin ligases (CRLs) involved in GPCR regulation. The present study has established Gβ2 as a component of E3 targeting GRK2.
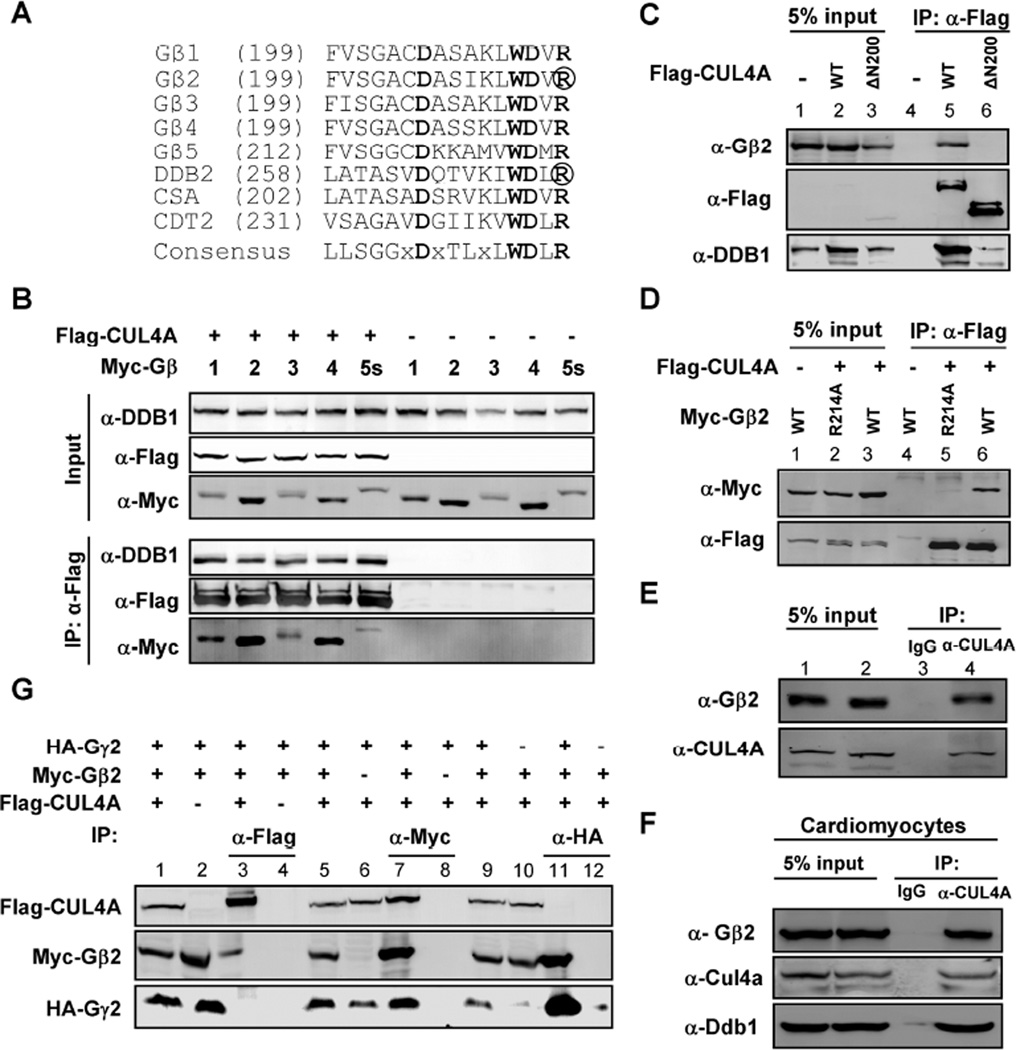
Figure 1. G protein β subunits bind to DDB1 and CUL4 independent of Gγ proteins.
(A) G protein β subunits contain the DDB1-binding WD40 (DWD) motif. The amino acid sequences spanning the DWD box from five human Gβ proteins are aligned (Gβ1, NCBI number: NP_002065.1; Gβ2: NP_005264.2; Gβ3: NP_002066.1; Gβ4: NP_067642.1; Gβ5: NP_006569.1). Also included are three well-characterized human DWD proteins, DDB2 (NP_000098.1), CSA (NP_000073.1), and CDT2 (NP_057532.3). Highly conserved residues are in bold, and residues essential for DDB1 binding, Arg273 in DDB2 and Arg214 in Gβ2, are circled.
(B) Gβ proteins bind with DDB1-CUL4A. 293T cells were co-transfected with plasmid expressing indicated proteins. Protein-protein bindings were determined by co-immunoprecipitation (co-IP). (‘α-Flag’ means anti-Flag antibody, the same below).
(C) The N-terminal domain of CUL4A is required for binding with Gβ2. 293T cells were co-transfected with plasmid expressing indicated proteins and protein-protein bindings were determined by co-IP (‘5% input’ means 5% total protein for IP experiments were loaded, the same below).
(D) The conserved Arg214 in the DWD box of Gβ2 is required for the binding with CUL4A. 293T cells were co-transfected with plasmids expressing indicated proteins and protein-protein bindings were determined by co-IP assay.
(E, F) Endogenous Gβ2 binds with CUL4A in HEK293 cells (E) and primary rat cardiomyocytes (F) as determined by the co-IP assay.
(G) Gβ2 binds with CUL4A independent of Gγ. 293T cells were co-transfected with plasmids expressing indicated proteins and protein-protein bindings were determined by co-IP.
RESULTS
G-protein β subunits bind with DDB1-CUL4A independent of Gγ
To test whether Gβ subunits bind with DDB1 and CUL4, five Myc-tagged Gβ proteins were expressed either individually or together with Flag-tagged CUL4A (epitope-tagging of DDB1 was avoided as it severely impairs its binding with DWD proteins and endogenous DDB1 is normally expressed at high levels sufficient for bridging DWD proteins to bind with CUL4). Co-immunoprecipitation assays demonstrated readily detectable binding of all five Gβ proteins with CUL4A (Figure 1B). CUL4A and CUL4B proteins use a N-terminal domain to bind with a linker subunit, DDB1, and through DDB1, bind with their DWD proteins (Angers et al., 2006; Hu et al., 2004). Deletion of this domain from CUL4A (ΔN200) completely disrupted its binding with Gβ2 (Figure 1C). A highly conserved signature Arginine (Arg) residue, known to be critical for binding with DDB1, follows the WD dipeptide of the DWD box. Mutation of this Arg in DDB2 (R273) is found in human xeroderma pigmentosum patients and disrupts DDB2–DDB1-CUL4 interaction (Rapic-Otrin et al., 2003). This Arg residue is invariably conserved in all Gβ subunits and conserved during evolution (Figure S1A). We found that mutation of this Arg in Gβ2 (R214) disrupted its association with DDB1-CUL4A (Figure 1D). The interaction of CUL4A and Gβ2 was also readily detected at the endogenous level in both HEK293 cells (Figure 1E) and rat primary cardiomyocytes (Figure 1F). Taken together, these results demonstrate that the Gβ subunits are bona fide DDB1-binding proteins, suggesting the possibility that multiple Gβ-DDB1-CUL4-ROC1 complexes may exist in vivo. Following commonly used nomenclature for cullin-RING E3 ubiquitin ligases (CRL), we have designated the Gβ-DDB1-CUL4-ROC1 complexes as CRL4Gβ where the substrate-recruiter DWD protein Gβ (see below) is superscripted.
Gβ subunits are present in cells either as Gαβγ heterotrimeric complexes, or as Gβγ dimers during GPCR activation, but rarely exist as monomers (Giguere et al., 2012; Wan et al., 2012). Gβ and Gγ subunits usually bind very tightly, and in most cases, a Gβγ dimer cannot be dissociated under nondenaturing conditions (Dupre et al., 2009). To determine whether Gβ-DDB1 binding is involved with or is independent of Gγ, we expressed differentially tagged Gβ2, Gγ2 and CUL4A and determined their interaction(s) by co-IP assay. This experiment demonstrated that, while Gβ2 could be easily detected in both Gγ2 and CUL4A immunocomplexes, no Gγ2 was detected in the CUL4A complex nor was CUL4A detected in the Gγ2 complex (Figure 1G), suggesting that Gβ2 interacts with DDB1-CUL4A independently of Gγ.
GRK2 is a substrate of the CRL4Gβ2 ubiquitin ligase
The main function of DWD proteins in CRL4 complexes is to recruit specific substrate(s) to the CRL4 ligase for ubiquitylation. To search for the substrate of CRL4Gβ2 ligase, we established stable cell pools expressing SBP (Streptavidin Binding Peptide Tag)-Flag-Gβ2 and SBP-Flag-Gβ2(R214A), performed tandem affinity purification (TAP) of Gβ2 complexes from cells treated with MG132, an inhibitor of the 26S proteosome, and subjected immune complexes to mass spectrometric analyses. These analyses identified multiple Gα and Gγ proteins in both the wild-type and R214A mutant Gβ2 immune complexes (Table S1), validating the IP-mass spec analysis and also indicating that R214 is not essential for the binding of Gβ2 with either Gα or Gγ. Consistent with the binding assay, CUL4A was identified in the wild-type, but not R214A mutant, Gβ2 immune complex. Notably, G-protein coupled receptor kinase 2 (GRK2, also known as β-adrenergic receptor kinase or βARK1) was identified in R214A mutant, but not wild-type, Gβ2 immune complexes. When assayed directly by expression and co-IP, GRK2 was able to bind to both the wild-type and R214A mutant of Gβ2 (Figure 2A). These results identify GRK2 as a binding protein for Gβ2 and also suggest that GRK2-Gβ2 association may be enhanced by the disruption of Gβ2’s association with DDB1.
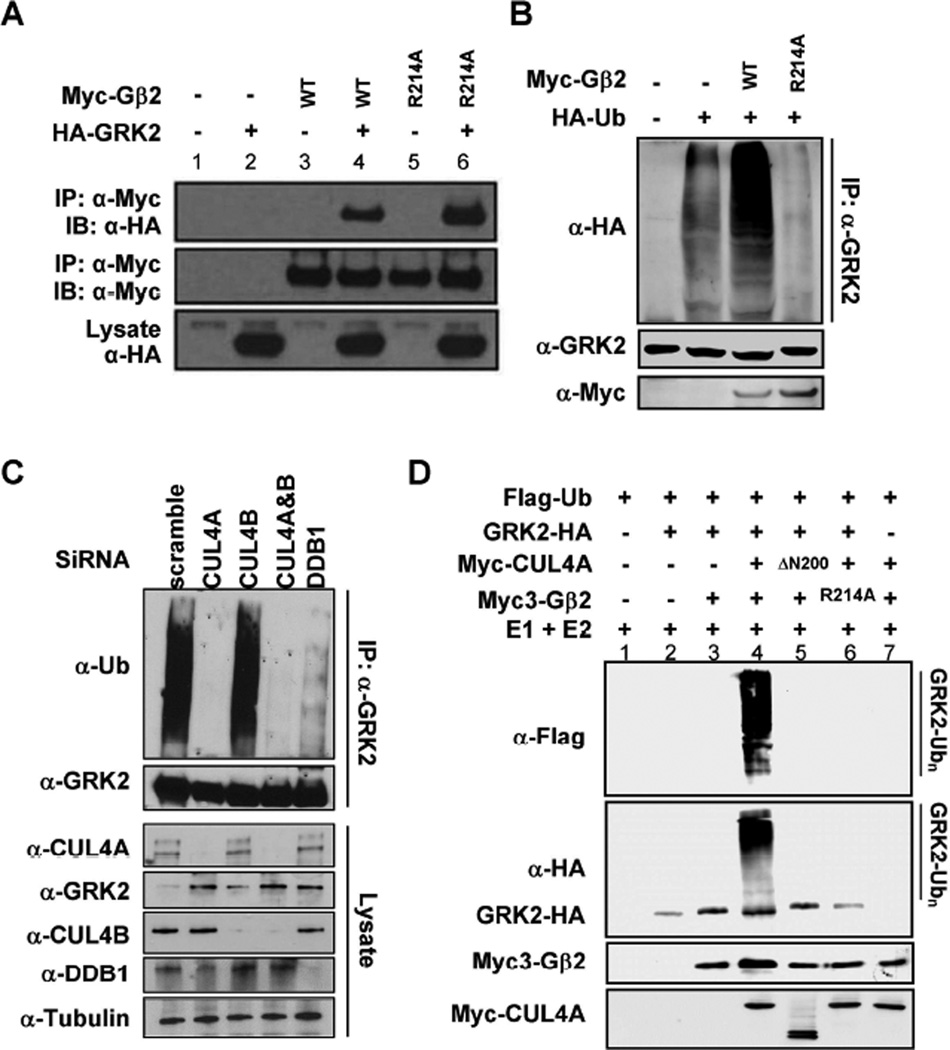
Figure 2. GRK2 is a substrate of CRL4Gβ2 E3 ubiquitin ligase.
(A) GRK2 binds to Gβ2. 293T cells were co-transfected with plasmids expressing indicated proteins and protein-protein interactions were determined by co-IP.
(B) Gβ2 promotes GRK2 ubiquitylation. HEK293 cells were transfected with plasmids expressing indicated proteins. Endogenous GRK2 was immunoprecipitated and analyzed for ubiquitylation by immunoblotting.
(C) Knocking down of CUL4A and DDB1 abolishes GRK2 ubiquitylation in vivo. HEK293 cells were transfected with siRNA oligonucleotides targeting indicated genes. The efficiency of knocking down was verified by immunoblotting. In vivo GRK2 ubiquitylation was determined by immunoprecipitation using an antibody specific to GRK2, followed by immunoblotting with an antibody specific to ubiquitin.
(D) In vitro ubiquitylation of GRK2 by CRL4Gβ2 E3 ligase. Purified GRK2 protein was incubated with CUL4A immunocomplex alone or with purified Gβ2 in the presence of E1, E2, ATP and ubiquitin. After termination, the reaction mixtures were resolved by SDS-PAGE, followed by immunoblotting with indicated antibodies.
To determine whether GRK2 is a substrate of CRL4Gβ2 E3 ligase, we over-expressed wild-type or R214A Gβ2 mutant in HEK293 cells and then detect GRK2 ubiquitylation level by IP and Western blot. The ubiquitylation of endogenous GRK2 protein was readily detected and was significantly enhanced by the expression of wild-type, but not the R214A mutant, Gβ2 (Figure 2B), providing evidence that GRK2 is ubiquitylated by a process involving Gβ2. The levels of ubiquitylated GRK2 in cells expressing the Gβ2(R214A) mutant were even lower than those observed in untransfected cells, suggesting a dominant negative inhibition of endogenous Gβ2 by the DDB1-binding deficient R214A mutant Gβ2.
To determine whether CUL4 and DDB1 promote GRK2 ubiquitylation, we transfected siRNA to HEK293 cells to knock down CUL4A, CUL4B and DDB1 expression, individually or in combination, and determined the ubiquitylation of endogenous GRK2. Knocking down either CUL4A or DDB1, but not CUL4B, substantially reduced the ubiquitylation of GRK2 and this reduction was associated with an increase in steady state levels of GRK2 by more than 50% (Figure 2C). This result supports the notion that CUL4A, which localizes predominantly in the cytoplasm, is the major ubiquitin ligase of GRK2 and that CUL4B, which shares 80% amino acid identity with CUL4A but is mostly nuclear (Nakagawa and Xiong, 2011), plays a very minor role in GRK2 regulation. An in vitro ubiquitylation assay showed that incubation of immunopurified GRK2 with immunopurified CUL4A and Gβ2 complexes resulted in robust GRK2 ubiquitylation in the presence of E1, E2, ATP and ubiquitin (Figure 2D). GRK2 ubiquitylation was not observed in the absence of the E3 CUL4A complex (lane 3), absence of the substrate GRK2 (lane 7), upon deletion of the N-terminal domain of CUL4A (lane 5), or upon mutation of R214 within Gβ2 that is required for DDB1 binding (lane 6). Collectively, these results demonstrate that GRK2 is a substrate of CRL4AGβ2 ubiquitin ligase.
CRL4Gβ2 regulates the stability and steady state levels of GRK2 protein
After determining that GRK2 is a substrate of CRL4Gβ2 ubiquitin ligase, we next examined whether CRL4Gβ2 regulates the steady state levels of GRK2. It was found that GRK2 is a relatively unstable protein with an estimated half-life (t1/2) of less than 3 hours (Figure S2A). Treatment of cells with MG132 significantly increased the half-life of GRK2 beyond the experimental duration (6 hours) (Figure 3A), suggesting that GRK2 is degraded by the 26S proteosome. We then determined the effect of expression of Gβ2 on endogenous GRK2. We found that overexpression of wild-type Gβ2 resulted in a decrease of GRK2 levels in a dose-dependent manner, while parallel overexpression of the R214A mutant of Gβ2 had little effect on GRK2 levels (Figure S2B). Transfection of cells with three different siRNA oligonucleotides targeting Gβ2 identified two, #1 and #2, that resulted in a significant reduction of Gβ2 and a commensurate increase of GRK2 by 50% (Figure 3B). Likewise, knocking down DDB1 or CUL4A, but not CUL1, also resulted in a similar increase in GRK2 protein levels by 50–60% (Figures 3C, S2C). This result further supports the notion that CUL4A is the major ubiquitin ligase of GRK2.
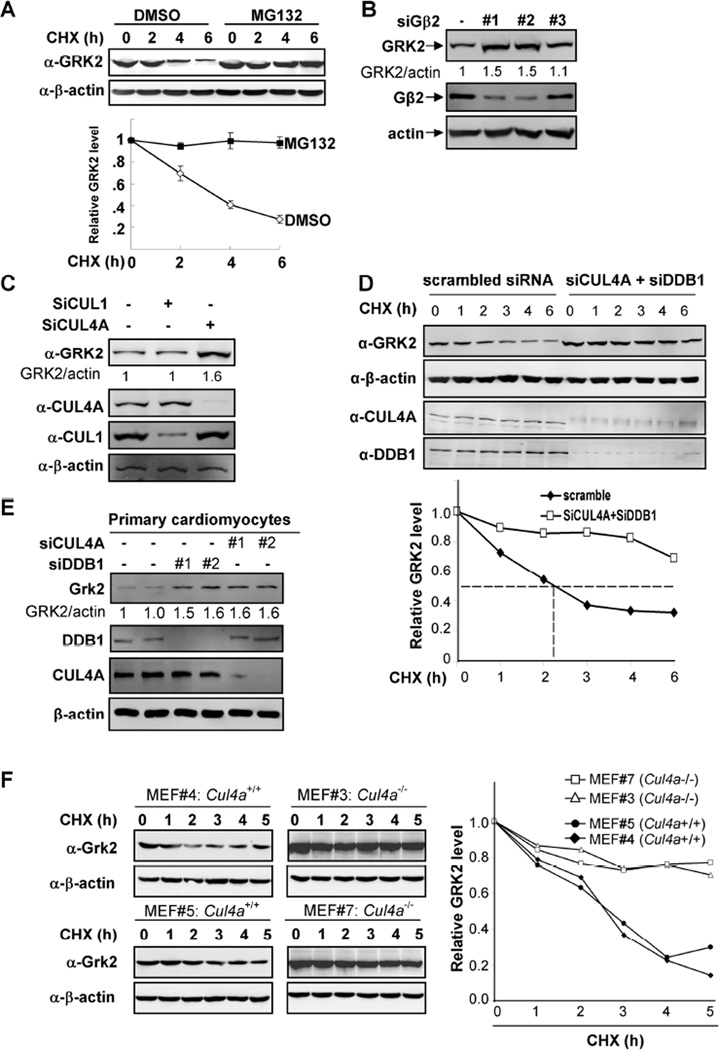
Figure 3. CRL4G|52 regulates the stability and steady state level of GRK2 protein.
(A) GRK2 is degraded by the 26S proteosome. HEK293 cells were treated wither either MG132 or solvent DMSO. The half-life of endogenous GRK2 protein was determined by cycloheximide (CHX)-chase.
(B, C) Knocking down of Gβ2 or CUL4A increases GRK2 protein level. HEK293 cells were transfected with three different siRNA oligo nucleotides targeting Gβ2 (B) or one targeting CUL4A (C). The GRK2 protein levels were determined by Western blotting and normalized against β-actin.
(D) GRK2 is stabilized by knocking down of both DDB1 and CUL4A. HEK293 cells were transfected with siRNA oligonucleotides targeting both CUL4A and DDB1. The half-life of GRK2 protein was determined by CHX treatment for different length of time as indicated and Western blotting with indicated antibodies.
(E) Knocking down Cul4a or Ddb1 increases Grk2 in rat primary cardiomyocyte cells. Two different siRNA oligos against either rat Cul4a or Ddb1 were transfected into rat cardiomyocyte cells.
(F) Deletion of Cul4a stabilizes GRK2 protein. The stability of GRK2 protein was determined in four littermate-matched MEFs by CHX treatment for different length of time as indicated and Western blotting with indicated antibodies.
Since DDB1 has also been reported to function as a subunit in a HECT ubiquitin ligase (Maddika and Chen, 2009), we next examined whether GRK2 regulation by DDB1 is mainly mediated by CUL4A. We found that co-depletion of both CUL4A and DDB1 did not result in any additional increase in GRK2 levels (Figure S2C), suggesting that DDB1-CUL4A is the ubiquitin ligase of GRK2. Supporting this conclusion, knocking down both CUL4A and DDB1 increased the half-life of GRK2 from 2.3 hours to more than 6 hours of experimental duration (Figure 3D). Likewise, when either Cul4a or Ddb1 was knocked down in rat primary cardiomyocytes, GRK2 protein level was also increased by about 50–60% (Figure 3E).
We then isolated four littermate-matched Cul4a+/+ and Cul4a−/− mouse embryonic fibroblast (MEF) lines and determined the half-life of Grk2 protein. We found that deletion of the Cul4a resulted in Grk2 stabilization from roughly 2.5 hours to longer than 5 hours (Figure 3F). Taken together, these results indicate that CRL4AGβ2 is the major ubiquitin ligase that controls the level of GRK2 protein in vivo.
β-AR activation disrupts Gβ2 binding to DDB1-CUL4A and up-regulates GRK2
Isoproterenol (ISO), a medication used clinically for its inotropic and chronotropic effects on the heart, is a sympathomimetic β-AR agonist. ISO treatment stabilized GRK2 in HEK293 cells, extending its half-life from about 2 hours to more than 6 hours (Figure 4A). To determine how ISO stabilizes GRK2, we examined the assembly of CRL4AGβ2 complex and found that ISO treatment reduced Gβ2’s association with DDB1-CUL4A as early as within 10 minutes and substantially (~ 80%) by 30 minutes of stimulation in a dose-dependent manner (Figures 4B and S3A). ISO treatment had little effect on either the steady state levels of CUL4A and DDB1 or CUL4A-DDB1 association. Similarly, treatment of rat cardiomyocytes with two different G-protein activating hormones, glucagon and epinephrine, also caused rapid (<10 min) reduction of Gβ2’s binding with DDB1-CUL4A (Figure 4C). After 30 minutes of treatment, DDB1-bound Gβ2 was reduced by 80% and 85% in glucagon and epinephrine treated cells, respectively, indicating that Gβ2-DDB1 association is regulated broadly by different GPCRs, most likely through the dissociation of Gβ2 from the DDB1-CUL4A ubiquitin ligase. We also detected the localization of endogenous GRK2, Gβ2, CUL4A and DDB1 under ISO treatment. Consistently, we found that ISO promoted GRK2 membrane localization (Figure3C). In addition, we found that CUL4A mainly localized in cytoplasm.
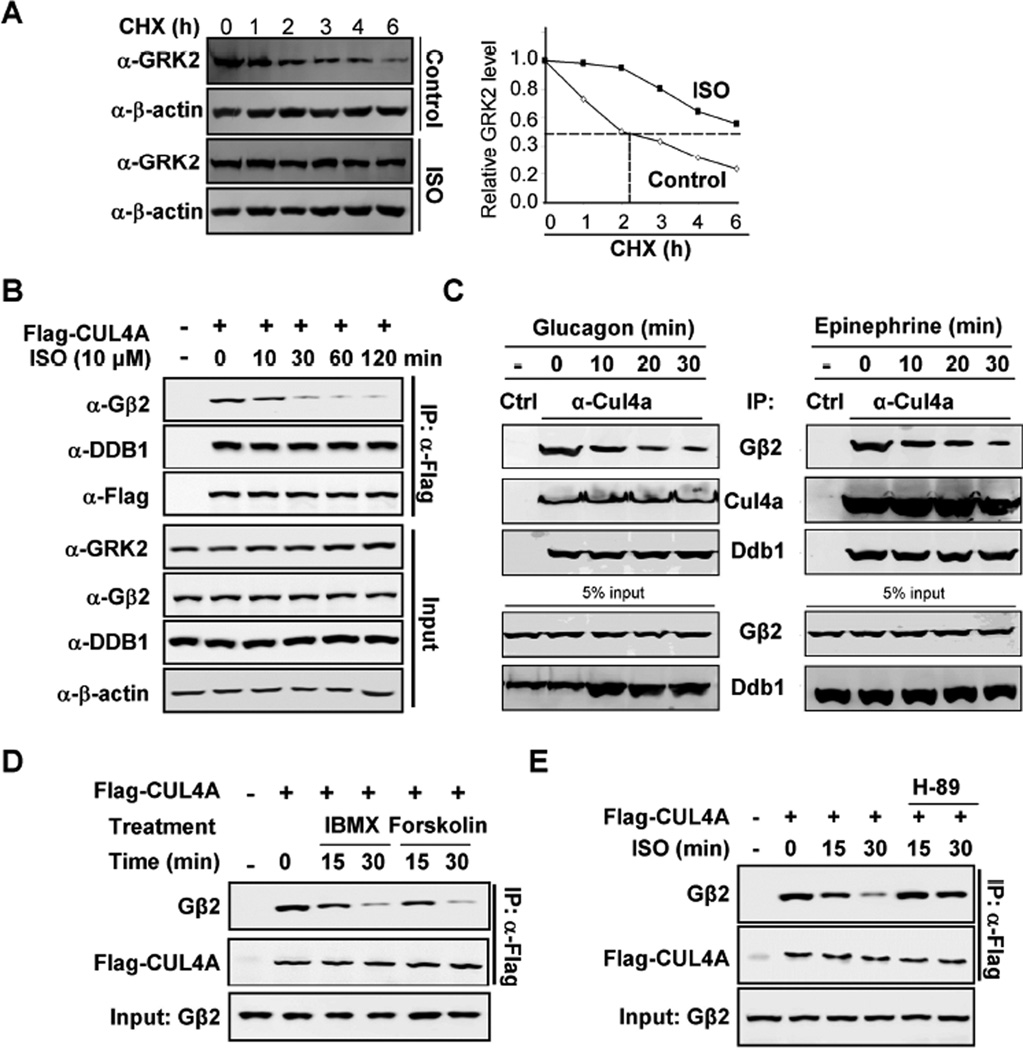
Figure 4. Activation of GPCR disrupts Gβ2 binding to DDB1.
(A) ISO stabilizes GRK2. HEK293 cells were treated with or without ISO, followed by CHX treatment as indicated time point. The protein levels of GRK2 were determined by Western blotting and quantified along with β-actin.
(B) Time dependent decrease of DDB1-CUL4A and Gβ2 binding by ISO. HEK293 cells were transfected with plasmids expressing Flag-CUL4A and then treated cells with ISO for indicated length of time. The levels of individual proteins and the protein-protein interactions were determined by Co-IP and Western analyses using indicated antibodies.
(C) Time dependent decrease of endogenous DDB1-CUL4A and Gβ2 binding by glucagon and epinephrine in cardiomyocytes.
(D) Dissociation of CUL4A and Gβ2 by IMBX/forskolin treatment. Flag-CUL4A was transfected into HEK293 cells and then treated the cells with IMBX/forskolin. The individual proteins were determined by Co-IP and Western blot analyses.
(E) PKA inhibitor H-89 blocks ISO effects on CUL4A-Gβ2 dissociation. Flag-CUL4A was transfected into HEK293 cells and then treated the cells with ISO/H-89. The individual proteins were determined by Co-IP and Western blot analyses.
To determine how GPCR activation leads to the dissociation of Gβ2 from CUL4A, potential candidate pathways downstream of β2AR were investigated. Treatment of cells with either adenylyl cyclase activator forskolin or phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX) disrupted CUL4A-Gβ2 binding (Figure 4D), indicating Gβ2-DDB1 binding is negatively regulated by cAMP which is the key second messenger downstream of β2AR and many other GPCRs. Furthermore, addition of H-89, a classical inhibitor of PKA, blocked the ISO effect on dissociating CUL4A-Gβ2 binding (Figures 4E and S3B). These results demonstrate that β2AR activation dissociates CUL4A-Gβ2 through a cAMP-PKA signaling pathway.
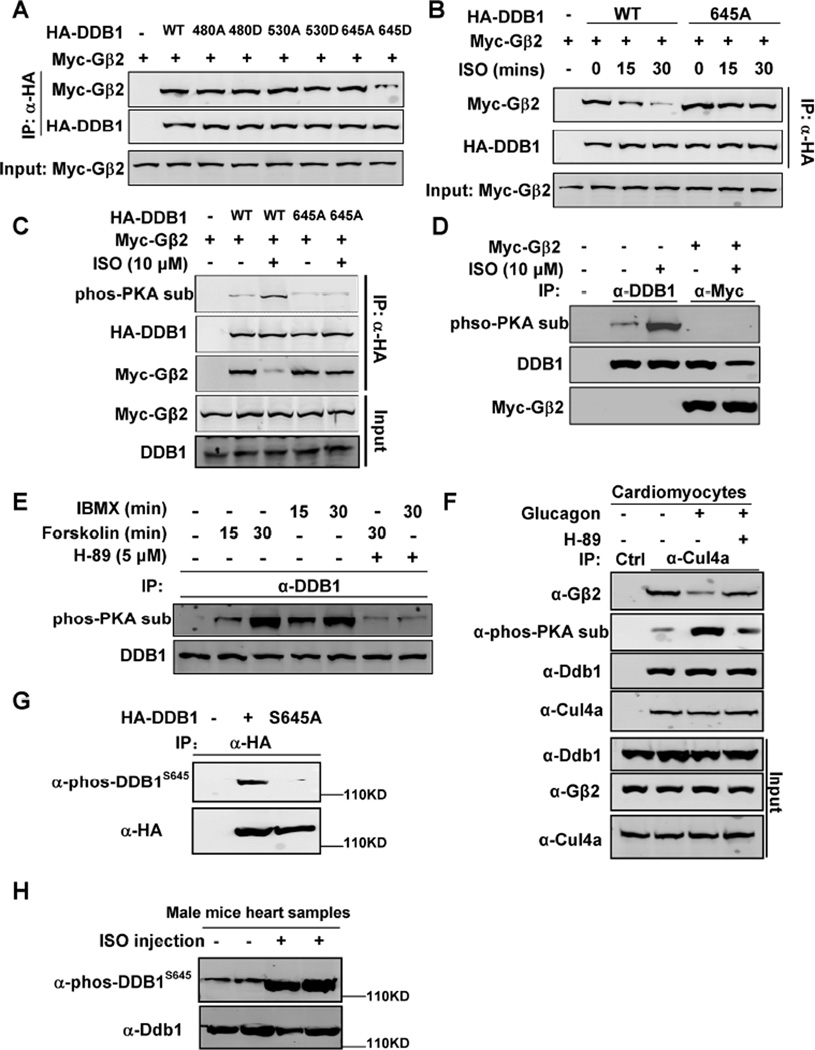
Gβ2-DDB1 complex is dissociated by PKA phosphorylation on DDB1 S645
To elucidate the mechanism by which PKA disassociates CUL4A-DDB1 from Gβ2, we inspected the protein sequences of both DDB1 and Gβ2 and found that there are 3 potential PKA phosphorylation sites (S480, S530 and S645) in DDB1, but none in Gβ2. Mutation analyses showed that while a phosphor-mimetic mutation of S645 (S645D) in DDB1 disrupted its binding to Gβ2, S450D or S530D did not (Figure 5A). DDB1S645A mutant is resistant to ISO-induced dissociation from Gβ2 (Figure 5B), supporting a critical role of DDB1 S645 phosphorylation in modulating DDB1-Gβ2 association. To confirm DDB1 phosphorylation by PKA, a monoclonal phospho-PKA substrate antibody was used to examine DDB1 phosphorylation level in cells treated with ISO. This experiment demonstrates that also associated with the ISO-induced disruption of DDB1-Gβ2 binding, there is a substantial increase of phosphorylation at a PKA site in the wild-type, but not S645A mutant, DDB1 (Figure 5C). We compared the level of ectopically and endogenously expressed DDB1 in this experiment and found that they were analogous. Notably, Gβ2 only bound un-phosphorylated, but not phosphorylated, DDB1 (Figure 5D). Inhibition of phosphodiesterase by IBMX or activation of adenylyl cyclase by forskolin both induced endogenous DDB1 phosphorylation, as detected by the phospho-PKA substrate antibody, which was blocked by the PKA inhibitor, H-89 (Figure 5E). Consistently, glucagon treatment in rat primary cardiomyocytes also induced PKA phosphorylation on DDB1 and disrupted DDB1- Gβ2 binding, and both effects were blocked by H-89 (Figure 5F). Furthermore, we made an anti-phosphorylated DDB1 at S645 antibody (anti-phos-DDB1S645) and characterized it (Figure 5G). This anti-phos-DDB1S645 antibody could recognize wild-type DDB1, but not S645A mutant. Four littermate-matched male mice were injected with ISO as described, and then using this anti-phos-DDB1S645 antibody, we found that ISO induced DDB1 phosphorylation at S645 in vivo (Figure 5H).
Figure 5. Gβ2-DDB1 complex is dissociated by PKA phosphorylation on DDB1 S645.
(A) DDB1645D mutant disrupts its binding to Gβ2. Myc-tagged Gβ2 and HA-tagged DDB1 or DDB1 mutant were transfected into HEK293 cells. The protein-protein interaction was determined by Co-IP and Western blot analyses.
(B) DDB1645A mutant blocks ISO effect on disrupting DDB1-Gβ2 binding. HEK293 cells were transfected with plasmids expressing Myc-Gβ2 and HA-DDB1/645A mutant, and then treated with ISO, followed by Co-IP and WB.
(C) ISO induces phosphorylation of the wild type DDB1 but not DDB1645A mutant. Myc-tagged Gβ2 and HA-tagged DDB1 or DDB1 mutant were transfected into HEK293 cells and then treated with ISO. The individual proteins were immunoprecipitated and subjected to Western blot with indicated antibodies.
(D) Gβ2 only binds to un-phosphorylated DDB1. Myc-tagged Gβ2 were transfected into HEK293 cells and then treated with ISO. The Myc-Gβ2 was immunoprecipitated and Western blot was performed to detect the co-precipitated DDB1.
(E) IBMX and Forskolin induce endogenous DDB1 phosphorylation. HEK293 cells were treated with IBMX, Forskolin, and H-89, as indicated. The individual proteins were precipitated with indicated antibodies and detected by Western blot analyses.
(F) Glucagon treatment in cardiomyocytes also induces DDB1 phosphorylation and disrupts DDB1- Gβ2 binding.
(G) Wild-type, but not S645A mutant, DDB1 was detected by anti-phos-DDB1S645 antibody.
(H) ISO induced DDB1 phosphorylation at S645 in vivo. 4 littermate-matched male mice were injected ISO as described, and their heart samples were harvested for Western blots analyses.
Together, these results establish that PKA phosphorylates DDB1 at S645 in response to agonist stimulation to disassociate DDB1 from Gβ2, thereby linking the regulation of GRK2 by CRL4AGβ2 ubiquitin ligase to GPCR signaling. Phosphorylation has been previously linked to the regulation of protein ubiquitylation by SCF/CRL1 E3 ligases where phosphorylation of a substrate often promotes its binding with substrate recognition factor (the F-box protein). Phosphorylation-mediated regulation of CRL4Gβ2 is distinctively different and occurs on the linker protein, DDB1.
Male Cul4a−/− mice develop cardiac hypertrophy which is partially rescued by lose of one Grk2 allele
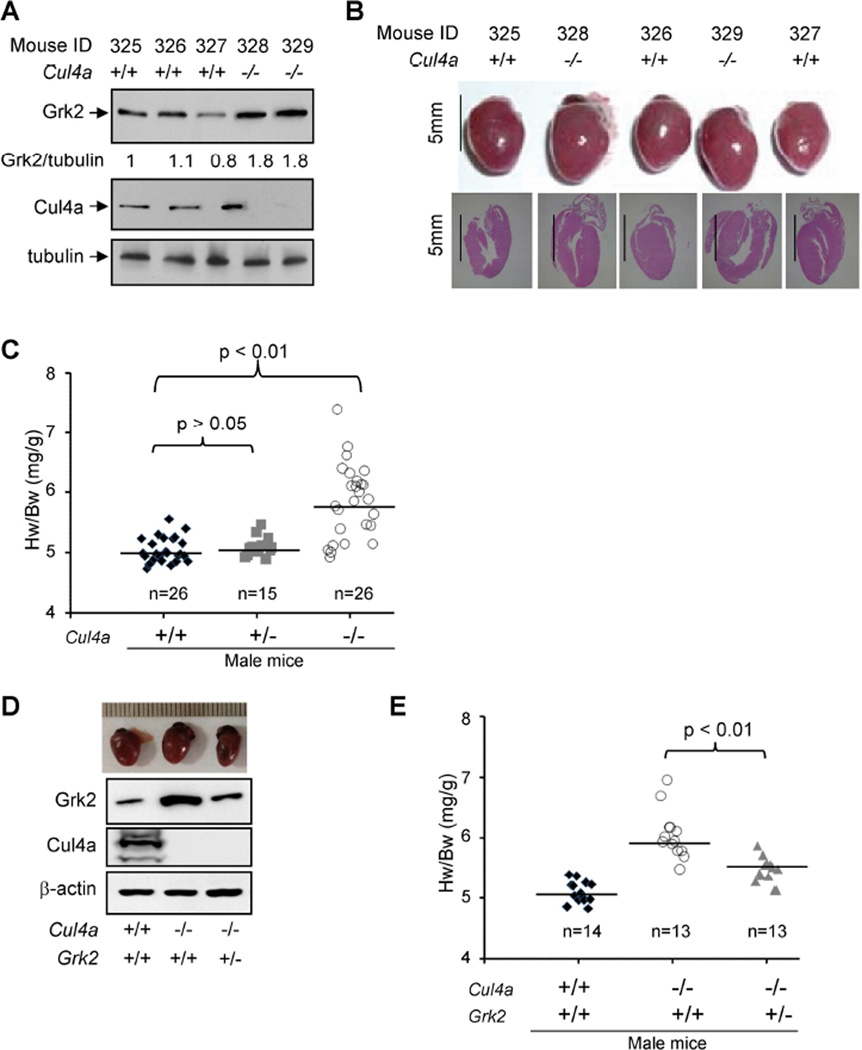
We next determined the function of CUL4A and DDB1 in the regulation of GRK2 in vivo. Whereas deletion of Ddb1 or Cul4b in mice results in embryonic lethality (Cang et al., 2006; Cox et al., 2010; Jiang et al., 2012; Liu et al., 2012), Cul4a null mice are viable and display no detrimental developmental defects throughout their life span with the exception of being sensitized to DNA damage and liver toxicity (Kopanja et al., 2009; Liu et al., 2009). We previously generated a Cul4a null strain with deletion of exons 4–8 encoding the DDB1 binding domain, which resulted in only a mild decrease in the proliferation of MEFs and viable mice (Kopanja et al., 2009). We first determined the level of Grk2 protein in the heart of wild-type and Cul4a−/− male mice. Similar to our findings in cultured MEF cells, deletion of the Cul4a gene resulted in an average 60% increase of the steady state Grk2 protein in the heart of Cul4a−/− male mice (Figure 6A, p<0.01, see Figure S4).
Figure 6. Male Cul4a−/− mice develop cardiac hypertrophy which is partially rescued by lose of one Grk2 allele.
(A) Deletion of Cul4a increases Grk2 protein in heart. The steady state levels of Grk2 protein were determined in five 10-week old littermate male mice by Western blotting.
(B) Male Cul4a−/− mice develop heart hypertrophy. 10-week-old littermate male mice were dissected and their hearts were analyzed by H&E staining.
(C) Male Cul4a−/− mice develop heart hypertrophy. The heart weights (HW) of 67 age-matched male mice of different genotypes were determined and normalized to the body weight (BW). The statistical significances of heart weight differences between different genotypes were determined by p value calculation as indicated.
(D, E) Cul4a−/−, but not Cul4a−/−;Grk2+/− male mice develop heart hypertrophy. The heart weights (HW) of age-matched 40 male mice of different genotypes were determined and normalized to the body weight (BW). The statistical significances of heart weight differences between different genotypes were determined by p value calculation as indicated.
Abnormally elevated GRK2 protein level is linked with multiple pathological conditions in humans, including myocardial infarction, heart failure and hypertension. We therefore further examined the heart phenotype of Cul4a−/− mice. Gross examination revealed prominent cardiac hypertrophy in male (Figure 6B), but not female Cul4a−/− mice (data not shown) when compared to wild-type littermates. To confirm this phenotype, we dissected 67 two-month old male mice (26 wild-type, 15 Cul4a+/−, and 26 Cul4a−/−) and determined their heart-to-body weight ratio (Hw/Bw). This study demonstrated a significant cardiac hypertrophy in Cul4a−/−(p<0.01), but not in Cul4a+/− (p>0.05) heterozygous, male mice (Figure 6C).
Finally, to further establish a functional link of Grk2 and Cul4a in heart protection, we crossed Grk2+/− mice with Cul4a+/− mice and characterized Cul4;Grk2 double mutant mice. We found that while Grk2 protein level was increased by 70% in Cul4−/− heart, it was reduced almost back to normal (10% increase) by the loss of one Grk2 allele in Cul4−/−;Grk2+/− heart when compared with the wild-type heart (Figure 6D). We dissected 40 two-month old male mice (14 wild-type, 13 Cul4a−/−, and 13 Cul4a−/− ;Grk2+/−) and determined the heart-to-body weight ratio. Associated with the restoration of Grk2 protein level, deletion of one Grk2 allele partially reduced the heart hypertrophy phenotype of Cul4a null male mice (Figure 6E). Collectively, these molecular, cellular and physiological analyses establish that Gβ2 functions as a component of the CRL4Gβ2 E3 ubiquitin ligase to regulate the level of GRK2 protein.
DISCUSSION
G protein β subunit functions as a substrate recruiter for E3 ubiquitin ligase
Gβ proteins have been extensively investigated since their initial discovery more than 30 years ago (Northup et al., 1980). The well-established function of Gβ protein is to participate in GPCR signaling either as a subunit of Gαβγ heterotrimeric complex that couples to GPCRs or as a subunit of the Gβγ heterodimer upon receptor activation. In this study, we reported a non-canonical role of Gβ—as a substrate recognition factor to recruit a specific substrate to an E3 ubiquitin ligase. Specifically, we have shown that a member of the Gβ family, Gβ2, targets a substrate, GRK2, for ubiquitylation and degradation by the DDB1-CUL4A-ROC1 (CRL4A) E3 ligase. These evidences include demonstration of the physical association of Gβ2 with DDB1-CRL4A and the regulation of this association by a β2AR agonist and cAMP-PKA pathway, in vivo and in vitro ubiquitylation of GRK2 by the CRL4AGβ2 E3 ligases and Grk2 accumulation in Cul4a null male mice. Genetically, we showed that Cul4a null male mice develop heart hypertrophy and that deletion of one allele of Grk2 restored the Grk2 protein back to near normal level and partially rescued heart defects in Cul4a null mice.
Five Gβ proteins share a high degree of sequence homology, including, in particular, the DWD box region and the critical Arg residue to which mutation in Gβ2 disrupts the association with DDB1-CRL4A and the regulation of GRK2 by CRL4AGβ2 E3 ligase. We have demonstrated that all five Gβ proteins can interact with CUL4A. We speculate that the function of substrate targeting for CRL4 E3 ligase is not only specific to Gβ2, and rather, that the other members of the Gβ family may also function in targeting protein ubiquitylation.
Gβ and DDB1 are key components of a PKA regulated E3 ubiquitin ligase for GRK2
Heart stress leads to the release of epinephrine and norepinephrine to activate β-ARs in cardiomyocytes, resulting in the activation of adenylyl cyclase, which increases cAMP and, ultimately, increases heart output. Activation of β-ARs also initiates a GRK-dependent desensitization process, leading to signal shutoff. This activation and desensitization system ensures acute response to heart stress and prevents prolonged heart stimulation. Disruption of this balance has long been linked to various heart diseases. In fact, it was reported over two decades ago that marked desensitization of β-ARs in the failing heart is accompanied by up-regulation of GRK2/βARK1 level and activity (Ungerer et al., 1993). Transgenic expression of Grk2 in mouse hearts resulted in attenuation of ISO-stimulated contractility, reduced cAMP production, and impaired cardiac function (Chen et al., 1998; Koch et al., 1995). These findings underscore the critical importance of regulating the GRK2 level for proper heart function.
Nearly all studies on GRK2 regulation have been focused on its mRNA expression. Although the degradation of GRK2 by the proteosome pathway has been reported (Penela et al., 1998), little is known about the identity of the GRK2 E3 ligase. The only reported E3 for GRK2 ubiquitylation is MDM2 (Salcedo et al., 2006). Considering the well-established function of MDM2 in p53 regulation and lack of significant defects in GPCR signaling and heart function in p53-Mdm2 double mutant mice (Mdm2 deletion causes embryonic lethality which can be rescued by co-deletion of p53), it appears that MDM2 does not play a major role in GRK2 regulation.
Five lines of evidence provided in this study collectively identify CRL4A as a major and physiologically significant GRK2 E3 ligase. First, GRK2 is ubiquitylated by CRL4AGβ2 and CRL4AGβ3 E3 ligase complexes in vivo and in vitro. Second, depletion or deletion of either CUL4A or DDB1 in established human cell lines or primary MEFs stabilized GRK2 and increased the steady-state levels of GRK2. Third, GRK2 protein is stabilized by agonist stimulation of β2AR that dissociates Gβ2 from DDB1-CUL4A. Fourth, deletion of Cul4a in mice resulted in elevated Grk2 level and cardiac hypertrophy and impaired heart function. Lastly, deletion of one allele of Grk2 in Cul4a−/− mice restored the level of Grk2 back to near normal and rescued the cardiac hypertrophy phenotype of Cul4a null male mice.
PKA-medicated feedback regulation of GPCR signaling
GPCR signaling, broadly involved in a plethora of cellular processes, is terminated by a common two-step mechanism. First, GRK phosphorylates the active receptor and converts it into a target for high affinity binding with arrestin. Second, bound arrestin shields the surface of the receptor to preclude G protein binding and/or promotes receptor internalization, thereby deactivating the GPCR. In this report, we revealed a feedback mechanism—PKA-regulated GRK2 stabilization—in the negative regulation of β2AR signaling. This regulation is directly coupled to β2AR signaling. Upon stimulation of β2AR by agonists, activation of PKA by elevated intracellular cAMP results in DDB1 phosphorylation and disruption of Gβ and DDB1 interaction, leading to GRK2 stabilization and eventual suppression of further β2AR signaling.
Utilizing ubiquitylation to regulate GRK2 offers a major advantage over the transcriptional regulation to regulate β2AR, as it enables cells to rapidly accumulate or terminate the accumulation of GRK2. Within an hour after ISO treatment, there was a noticeable GRK2 accumulation (Figure 4B). Such a rapid increase in GRK2 levels would facilitate acute desensitization. Likewise, cells could also quickly terminate GRK2 accumulation through simply re-associating DDB1 with Gβ once cAMP and PKA return to basal levels. There are two possible regulatory steps where cells could initiate and terminate CRL4Gβ-mediated GRK2 ubiquitylation: One controlling the association between DDB1 and Gβ and the other between Gβ and substrate GRK2. Our studies reveal a regulatory step on the DDB1-Gβ2 association as PKA-mediated phosphorylation in DDB1 disrupts its binding with Gβ2 after ISO treatment.
How broadly could CRL4A ubiquitin ligases regulate GPRC signaling? Aside from its traditional role in phosphorylating and desensitizing β-ARs in the regulation of heart function and protection, emerging evidence has substantially expanded the role of GRK2, including the regulation of GPCR trafficking in a phosphorylation-independent manner, phosphorylation of non-receptor proteins, and even interaction directly with signaling molecules (Evron et al., 2012). We speculate that many of these GRK2-regulated cellular processes may also be regulated by the CRL4Gβ2 E3 ligases. Furthermore, given that all five Gβ proteins can bind with DDB1-CUL4A, it is tempting to speculate that besides GRK2, additional proteins involved in GPCR signaling could be targeted for the ubiquitylation by the CRL4A E3 ligases.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293 and HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% newborn Calf Serum, 100 units/ml penicillin, and streptomycin (Gibco). MEF (mouse embryonic fibroblast) cells were maintained in DMEM medium supplemented with 10% fetal calf serum (Gibco), 1% L-glutamine, 100 units/ml penicillin, and streptomycin. Cell transfection was performed using Lipofectamine 2000 (Life Technologies) or calcium phosphate method. Cells were harvested at 48–60 hours post-transfection for protein analyses. To establish stable -expressing cells, wild type and R214A mutant pBabe-SBP-Flag-Gβ2 retroviruses were generated and used to infect HEK293 cells and stable pools were selected in puromycin (1 µg/ml)-containing media for 7 days.
Antibodies and immunological procedures
Protein lysates were prepared by lysing HEK293 cells in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM PMSF, 25 mM NaF, and a mixture of protease inhibitors. Cell lysate (20 µg) was resolved by SDS-PAGE, followed by Western blotting analysis. Antibodies recognizing Flag (Sigma), GRK2 (Santa Cruz), HA (Santa Cruz), Myc (Santa Cruz), phospho-PKA substrates (Cell Signaling) and β-actin (Cell Signaling) were purchased commercially. Antibodies to DDB1 and CUL4A have been described before (Hu et al., 2004).
For immunoprecipitation experiments, 800 µg total protein in cell lysate was incubated with anti-Flag M2-agarose (Sigma) or anti-GRK2 beads (Santa Cruz) for 3h at 4 °C. Beads were washed three times with lysis buffer and centrifuged at 2,000 × g for 3 min between each wash. Protein was eluted from beads with 50 µl of SDS sample buffer. Lysates were resolved on 8–15% SDS-PAGE gels and transferred onto nitrocellulose (Bio-Rad) for Western blotting.
In Vitro Ubiquitin Ligation Assays
Plasmids expressing Myc-CUL4A, HA-GRK2, Myc3-Gβ2 or Myc3-Gβ2R214A were individually transfected into 293T cells by Lipofectamine 2000. 48 hours after transfection, cells were lysated into a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% SDS, and a cocktail of protease inhibitors, followed by immunoprecipitation using Myc or HA sepharose (Santa Cruz). Immunocomplexes were washed with the lysis buffer and eluted by Myc or HA antigen peptides. Immunopurified HA-GRK2 protein was mixed with Myc-CUL4A and Myc3-Gβ2 in a ubiquitin ligation buffer (50 mM Tris-HCl/pH 7.4, 5 mM MgCl2, 2 mM NaF, 2 mM ATP, 10 nM okadaic acid, 0.6 mM DTT, 12 µg of bovine ubiquitin, 1 µg of FLAG-tagged ubiquitin (Sigma), 60 ng of E1 (E301, Boston Biochem), 500 ng of E2 (human Ubc5c), final volume = 30 µl). The reaction was incubated at 37 °C for 1 h on a rotator with slow shaking and then terminated by boiling at 95 °C with SDS sample buffer for 10 min prior to SDS-PAGE. GRK2 ubiquitylation was examined by immunoblotting with either anti-FLAG or anti-HA antibody.
Primary rat cardiomyocytes isolation, culture and transfection
Primary rat cardiomyocytes were freshly isolated form newborn rats (Wistar rats) and cultured for removing the adherent cells with fibroblastoid morphology. Primary rat cardiomyocytes were incubated in DMEM medium supplemented with 10% FBS, 8 mM glutamine, 25 mM glucose, penicillin/streptomycin and 100 µM Brdu. Primary cardiomyocytes contract when grown at the required density. Amaxa® Rat Cardiomyocyte –Neonatal Nucleofector® Kit was used for transfection as manufactory’s protocol. Briefly, the required number of cells (2 × 106 cells per well/sample) was centrifuged at 340 × g for 1 min at room temperature and the cell pellet resuspended carefully in 100 µl room temperature Nucleofector® Solution per sample, combining 100 µl of cell suspension with 200 nM siRNA targeting either rat Cul4a or Ddb1. The cell/RNA suspension was then transferred into a certified cuvette for the appropriate Nucleofector® Program G-009, followed by adding 500 µl of the pre-equilibrated culture media to the cuvette and gently transferring the sample immediately into the prepared gelatin coated 6-well plate (final volume 2 ml media per well), using the supplied pipettes and avoiding repeated aspiration of the sample. Cells were incubated in a humidified 37 °C/ 5% CO2 incubator until analysis.
ISO injection
Isoproterenol (Sigma) dissolved in 150 mM NaCl and 1 mM acetic acid was delivered chronically, at a rate of 8.7 mg per kilogram of body weight per day to 2-month-old littermate-matched male mice (n = 4) by using an implanted miniosmotic pump (ALZET model 2001) as described. Seven days after implantation of isoproterenol-loaded pumps, hearts were harvested, and protein from these heart samples were detected by Western blots.
Statistical analysis
Comparisons between the two groups were performed with unpaired, 2-tailed Student’s t-test (Excel software). P values < 0.05 were considered statistically significant. Data are presented as the mean ± SD.
Supplementary Material
Highlights.
Gβ has a non-canonical role as a substrate recruiter of E3 ubiquitin ligase
Gβ2-DDB1-CUL4-ROC1 is a ubiquitin ligase targeting GRK2
β-AR signaling regulates GRK2 via PKA-mediated DDB1 phosphorylation
Deleting one Grk2 allele partially rescued the heart hypertrophy in Cul4a null mice
ACKNOWLEDGEMENTS
We thank the members of the Fudan MCB and Xiong laboratories for discussions throughout this study, Brenda Temple of UNC R. L. Juliano Structural Bioinformatics Core facility for the molecular modeling, Bev Koller, Howard Rockman, Dennis Abraham, Kathleen Caron and John Sondek for discussions, and Cam Patterson for discussion and critical reading of the paper. This work was supported by Chinese Ministry of Sciences and Technology 973 (Grant No. 2015CB910401, 2011CB910600), NSFC (Grant No. 31271454, 81225016, 81430057), Shanghai Key basic research program (12JC1401100), Shanghai Outstanding Academic Leader (Grant No.13XD1400600) and the Youth Science and Technology Leading Talent by MOST to Q.Y.L, NIH grants EY022611 and CA 132809 (to K.L.G.) and GM067113 (to Y.X.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- Cang Y, Zhang J, Nicholas SA, Bastien J, Li B, Zhou P, Goff SP. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell. 2006;127:929–940. doi: 10.1016/j.cell.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Chen L-C, Manjeshwar S, Lu Y, Moore D, Ljung B-M, Kuo W-L, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Floss T, Gertsenstein M, van Campenhout C, Kuhn R, Wurst W, et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010;20:1154–1184. doi: 10.1101/gr.105106.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Rebois RV, Hebert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evron T, Daigle TL, Caron MG. GRK2: multiple roles beyond G protein-coupled receptor desensitization. Trends Pharmacol Sci. 2012;33:154–164. doi: 10.1016/j.tips.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere PM, Laroche G, Oestreich EA, Siderovski DP. G-protein signaling modulator-3 regulates heterotrimeric G-protein dynamics through dual association with Gbeta and Galphai protein subunits. J Biol Chem. 2012;287:4863–4874. doi: 10.1074/jbc.M111.311712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: more than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, McCall CM, Ohta T, Xiong Y. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat Cell Biol. 2004;6:1003–1009. doi: 10.1038/ncb1172. [DOI] [PubMed] [Google Scholar]
- Jiang B, Zhao W, Yuan J, Qian Y, Sun W, Zou Y, Guo C, Chen B, Shao C, Gong Y. Lack of Cul4b, an E3 ubiquitin ligase component, leads to embryonic lethality and abnormal placental development. PLoS One. 2012;7:e37070. doi: 10.1371/journal.pone.0037070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch WJ, Rockman HA, Samama P, Hamilton RA, Bond RA, Milano CA, Lefkowitz RJ. Cardiac function in mice overexpressing the beta-adrenergic receptor kinase or a beta ARK inhibitor. Science. 1995;268:1350–1353. doi: 10.1126/science.7761854. [DOI] [PubMed] [Google Scholar]
- Kopanja D, Stoyanova T, Okur MN, Huang E, Bagchi S, Raychaudhuri P. Proliferation defects and genome instability in cells lacking Cul4A. Oncogene. 2009;28:2456–2465. doi: 10.1038/onc.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Leosco D, Fortunato F, Rengo G, Iaccarino G, Sanzari E, Golino L, Zincarelli C, Canonico V, Marchese M, Koch WJ, Rengo F. Lymphocyte G-protein-coupled receptor kinase-2 is upregulated in patients with Alzheimer’s disease. Neurosci Lett. 2007;415:279–282. doi: 10.1016/j.neulet.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yin Y, Li Y, Prevedel L, Lacy EH, Ma L, Zhou P. Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 2012;22:1258–1269. doi: 10.1038/cr.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Premont RT, Kontos CD, Zhu S, Rockey DC. A crucial role for GRK2 in regulation of endothelial cell nitric oxide synthase function in portal hypertension. Nat Med. 2005;11:952–958. doi: 10.1038/nm1289. [DOI] [PubMed] [Google Scholar]
- Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Keeping G proteins at bay: a complex between G protein-coupled receptor kinase 2 and Gbetagamma. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- Maddika S, Chen J. Protein kinase DYRK2 is a scaffold that facilitates assembly of an E3 ligase. Nat Cell Biol. 2009;11:409–419. doi: 10.1038/ncb1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisco C, Lembo G, Trimarco B. Insulin resistance and cardiovascular risk: New insights from molecular and cellular biology. Trends Cardiovasc Med. 2006;16:183–188. doi: 10.1016/j.tcm.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Xiong Y. X-linked mental retardation gene CUL4B targets ubiquitylation of H3K4 methyltransferase component WDR5 and regulates neuronal gene expression. Mol Cell. 2011;43:381–391. doi: 10.1016/j.molcel.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northup JK, Sternweis PC, Smigel MD, Schleifer LS, Ross EM, Gilman AG. Purification of the regulatory component of adenylate cyclase. Proc Natl Acad Sci U S A. 1980;77:6516–6520. doi: 10.1073/pnas.77.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penela P, Ruiz-Gomez A, Castano JG, Mayor F., Jr Degradation of the G protein-coupled receptor kinase 2 by the proteasome pathway. J Biol Chem. 1998;273:35238–35244. doi: 10.1074/jbc.273.52.35238. [DOI] [PubMed] [Google Scholar]
- Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- Rapic-Otrin V, Navazza V, Nardo T, Botta E, McLenigan M, Bisi DC, Levine AS, Stefanini M. True XP group E patients have a defective UV-damaged DNA binding protein complex and mutations in DDB2 which reveal the functional domains of its p48 product. Hum Mol Genet. 2003;12:1507–1522. doi: 10.1093/hmg/ddg174. [DOI] [PubMed] [Google Scholar]
- Rockman HA, Koch WJ, Lefkowitz RJ. Seven-transmembrane-spanning receptors and heart function. Nature. 2002;415:206–212. doi: 10.1038/415206a. [DOI] [PubMed] [Google Scholar]
- Salcedo A, Mayor F, Jr, Penela P. Mdm2 is involved in the ubiquitination and degradation of G-protein-coupled receptor kinase 2. EMBO J. 2006;25:4752–4762. doi: 10.1038/sj.emboj.7601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK, Lefkowitz RJ. Seven-transmembrane receptor signaling through beta-arrestin. Sci STKE. 2005;2005 doi: 10.1126/stke.2005/308/cm10. cm10. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Kimple AJ, Willard FS, Begley TP. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc.; 2007. Large G-Proteins. [Google Scholar]
- Ungerer M, Bohm M, Elce JS, Erdmann E, Lohse MJ. Altered expression of beta-adrenergic receptor kinase and beta 1-adrenergic receptors in the failing human heart. Circulation. 1993;87:454–463. doi: 10.1161/01.cir.87.2.454. [DOI] [PubMed] [Google Scholar]
- Wan Y, Yang Z, Guo J, Zhang Q, Zeng L, Song W, Xiao Y, Zhu X. Misfolded Gbeta is recruited to cytoplasmic dynein by Nudel for efficient clearance. Cell Res. 2012;22:1140–1154. doi: 10.1038/cr.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Huang S, Patterson E, Garrett MW, Kaufman KM, Metcalf JP, Zhu M, Dunn ST, Kem DC. Proteasome degradation of GRK2 during ischemia and ventricular tachyarrhythmias in a canine model of myocardial infarction. Am J Physiol Heart Circ Physiol. 2005;289:H1960–H1967. doi: 10.1152/ajpheart.00328.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.