Abstract
The complex mechanisms that cells have evolved to meet the challenge of constant exposure to DNA-damaging stimuli, also serve to protect cancer cells from the cytotoxic effects of chemo- and radiotherapy. IGFBPs appear to be involved, directly or indirectly, in some of these protective mechanisms. Activation of p53 is an early response to genotoxic stress, and all six human IGFBP genes have predicted p53 response elements in their promoter and/or intronic regions, at least some of which are functional. IGFBP3 has been extensively characterized as a p53-inducible gene, but in some cases it is suppressed by mutant p53 forms. DNA double-strand breaks (DSBs), induced by radiotherapy and some chemotherapies, potentially lead to apoptotic cell death, senescence, or repair and recovery. DSB damage can be repaired by homologous recombination or non-homologous end-joining (NHEJ), depending on the cell cycle stage, availability of key repair proteins, and other factors. The epidermal growth factor receptor (EGFR) has been implicated in the NHEJ pathway, and EGFR inhibition may inhibit repair, promoting apoptosis and thus improving sensitivity to chemotherapy or radiotherapy. Both IGFBP-3 and IGFBP-6 interact with components of the NHEJ pathway, and IGFBP-3 can facilitate this process through direct interaction with both EGFR and the catalytic subunit of DNA-PK. Cell fate after DNA damage may in part be regulated by the balance between the sphingolipids ceramide and sphingosine-1-phosphate, and IGFBPs can influence the production of both lipids. A better understanding of the involvement of IGFBPs in the DNA damage response in cancer cells may lead to improved methods of sensitizing cancers to DNA-damaging therapies.
Keywords: Cancer, Chemotherapy, IGFBP, Non-homologous end-joining, p53, Radiotherapy
Introduction
The human genome is constantly challenged with DNA lesions originating from both internal and external influences (Jackson and Bartek 2009; Sirbu and Cortez 2013). Cells can respond to DNA damage by initiating DNA repair which, if successful, allows the cell to continue its normal function. Alternatively, severe DNA damage can lead to the induction of cell death by apoptosis, or to cell cycle arrest through senescence (d’Adda di Fagagna 2008). Both radio- and chemotherapy are designed to induce apoptosis – they generate DNA modifications and oxidative damage that eventually kill the rapidly dividing tumor cells. Among the various DNA lesions, double strand breaks (DSBs) are the most dangerous because they result in a direct cleavage of the DNA backbone. DSBs occur at an estimated rate of 10 per day per cell (Lieber 2010), their aberrant accumulation representing a major threat to genomic integrity (Jackson and Bartek 2009). DSBs may arise from the action of ionizing radiation, oxidative free radicals, mechanical stress, topoisomerase inhibition, or other damaging influences (Lieber 2010).
There is increasing evidence that members of the insulin-like growth factor (IGF) binding protein (IGFBP) family may influence cell responses to DNA damaging insults. Understanding the role that IGFBPs play in these processes is essential to increase knowledge both of the mechanisms by which genomic integrity is maintained in normal cell physiology, and also in the context of cancer cell sensitivity to DNA-damaging chemo- and radiotherapy. Recognition of how cell responsiveness to these therapies may be influenced by manipulating IGFBPs or their signaling pathways could lead to improved approaches to cancer treatment. The aim of this review is to provide an overview of DNA damage response pathways, with particular reference to non-homologous end-joining (NHEJ), and to discuss the involvement of IGFBPs in mediating or modulating these processes.
The IGFBPs
The insulin-like growth factors, IGF-I and IGF-II, associate in the circulation and the extracellular environment with members of the IGF binding protein (IGFBP) family, IGFBP-1 to IGFBP-6. The six high-affinity IGFBPs share cysteine-rich amino- and carboxyterminal domains, both of which contribute to the IGF-binding site (Baxter 2000; Forbes et al. 2012). In the circulation, IGFs have greatly extended half-lives when complexed with IGFBPs, in particular IGFBP-3 (by far the most abundant in adult human serum) and IGFBP-5, both of which form ternary complexes by interacting with a third protein, the acid-labile subunit (Baxter 2000). In the cellular environment, IGFBP-bound IGFs have restricted access to the cell-surface type 1 IGF receptor (IGF1R), since some IGF residues involved in receptor binding also contribute to IGFBP binding (Forbes et al. 2012; Sitar et al. 2006).
In addition to stabilizing IGFs in the bloodstream and regulating their activation of IGF1R, IGFBPs modulate cell function through interactions with other cell-surface and intracellular proteins that appear to be independent of their IGF-binding activity (Baxter 2014). Key interacting protein families include the integrins at the cell surface (Beattie, et al. 2010) and the nuclear hormone receptors within the nucleus (Yamada and Lee 2009).
IGFBPs, p53, and apoptosis
Activation of p53 is one of the key responses to DNA damage (Sperka et al. 2012), initiating both cell cycle arrest and cell death pathways. Its phosphorylation at Ser15 by ataxia-telangiectasia mutated protein (ATM) or ataxia-telangiectasia and Rad3-related (ATR) kinases, which is followed by phosphorylation at multiple other sites, is a crucial step in the transcriptional events that follow DNA damage (Loughery et al. 2014). In silico prediction of genes containing p53 response elements indicates that all six IGFBP genes include such sites, located in both intronic and promoter regions of the genes (Table 1) (p53FamTaG, http://p53famtag.ba.itb.cnr.it) (Sbisa et al. 2007). Induction of IGFBPs by p53 has been shown for IGFBP-1 (Leu and George 2007), IGFBP-2 (Grimberg et al. 2006), and IGFBP-3 (Buckbinder et al. 1995), and perhaps also IGFBP-5 (Kim et al. 2007).
Table 1.
Predicted p53 Response elements in human IGFBP genes
| Gene Name | RefSeqa | Chrb | REsc | Start | Size | Strand | Localization |
|---|---|---|---|---|---|---|---|
| IGFBP1 | NM_000596.2 | 7 | 3 | 45700053 | 43 | → | Promoter |
| 45700381 | 34 | → | Promoter | ||||
| 45701945 | 34 | → | Intron | ||||
| IGFBP2 | NM_000597.2 | 2 | 7 | 217328264 | 30 | → | Intron |
| 217335054 | 38 | → | Intron | ||||
| 217337336 | 38 | → | Intron | ||||
| 217338962 | 37 | → | Intron | ||||
| 217341622 | 42 | → | Intron | ||||
| 217342115 | 37 | → | Intron | ||||
| 217342527 | 37 | → | Intron | ||||
| IGFBP3 | NM_001013398.1 | 7 | 2 | 45727226 | 40 | ← | Intron |
| 45732422 | 41 | ← | Intron | ||||
| IGFBP4 | NM_001552.2 | 17 | 6 | 35850127 | 38 | → | Promoter |
| 35850210 | 43 | → | Promoter | ||||
| 35851306 | 37 | → | Promoter | ||||
| 35851380 | 34 | → | Promoter | ||||
| 35853307 | 37 | → | Promoter | ||||
| 35864072 | 44 | → | Intron | ||||
| IGFBP5 | NM_000599.3 | 2 | 3 | 217368891 | 43 | ← | Intron |
| 217370694 | 36 | ← | Intron | ||||
| 217388560 | 36 | ← | Promoter | ||||
| IGFBP6 | NM_002178.2 | 12 | 2 | 51775153 | 33 | → | Promoter |
| 51776111 | 36 | → | Promoter |
Data obtained from p53FamTaG (http://p53famtag.ba.itb.cnr.it)
aNCBI Reference Sequence Database
bChromosome
cNumber of response elements
The link between IGFBP-1 and p53 was demonstrated in the liver of irradiated mice, in which total and phospho-p53 increased markedly 1 h post-irradiation, followed by IGFBP-1 induction (Reynolds et al. 2004). The increased level of IGFBP-1 was reported to counteract p53-dependent apoptosis by binding to, and blocking the mitochondrial action of, the proapoptotic protein Bak (Leu and George 2007). IGFBP-2 was shown to be p53-dependent by the induction of IGFBP-2 mRNA in the thymus of irradiated wildtype mice, but not p53-null mice (Grimberg et al. 2006). p53 binding to 4 intronic consensus binding sites in the IGFBP2 gene has been demonstrated, and IGFBP-2 silencing by shRNA blocked the ability of p53 to inhibit IGF-I stimulated ERK activation in prostate cancer cells (Grimberg et al. 2006). In the case of IGFBP-3, potential p53 binding sites were first identified in IGFBP3 intronic regions and shown to be functional (Buckbinder et al. 1995), but hypermethylation of p53-responsive sequences in the IGFBP3 promoter region (not shown in Table 1) is able to selectively suppress p53-induced IGFBP-3 expression (Hanafusa et al. 2005). IGFBP-6, while not explicitly shown to be p53-dependent, is upregulated by DNA-damaging agents hydrogen peroxide, cisplatin, and doxorubicin (Xie et al. 2005). These findings suggest that members of the IGFBP family may be important mediators of p53 actions.
Among the p53-regulated IGFBPs, IGFBP-3 regulation has been most extensively studied. Because IGFBP-3 can induce apoptosis in many cell types, its p53-dependent upregulation in response to DNA-damaging stimuli such as ionizing radiation is seen as an important component of the DNA damage response (Grimberg et al. 2005; Williams et al. 2000). However, in some cell lines, IGFBP-3 is believed to be induced even in the absence of functional p53 (Butt et al. 2000; Grimberg et al. 2005). Interestingly, in a study of chemotherapy-induced hair loss, IGFBP-3 was downregulated after cyclophosphamide treatment in p53 null mice, although upregulated when in p53 wildtype mice (Botchkarev et al. 2000). Clearly the regulation of IGFBP-3 by p53 and its mutants is not as straightforward as originally thought, since different domains of p53 appear to be responsible for IGFBP-3 up- or downregulation (Harms and Chen 2005). Accordingly, IGFBP3 gene promoter activity is downregulated in cells expressing some “hot-spot” mutant p53 forms, a response interpreted as a p53 gain-of function (Vikhanskaya et al. 2007). The variant p53 family member, ΔNp63α, has also been shown to suppress IGFBP-3 expression, leading to the suggestion that p63 overexpression by some tumors might protect them from IGFBP-3-mediated apoptosis (Barbieri et al. 2005).
Detection and repair of double-strand breaks
Cells respond to DSB lesions by initiating the DNA damage response signaling cascade, which then activates cell cycle checkpoints and directs DNA repair. The rate and accuracy of repair depend on the clustering of the breaks and how capable the cells are of repairing the damage (Shikazono et al. 2009). Non-malignant cells generally have a slow turnover rate and are therefore proficient in reversing the DNA lesions and are spared from injurious damage when exposed to genotoxic insults. In malignant cells, many of which have hyperactivated DNA damage factors and rapid turnover rate, the cells’ repair machinery may attempt to rejoin the DSB ends at an accelerated rate that can lead to errors in the repair process.
The DNA damage response to double-strand breaks is typically initiated in a hierarchical fashion by recruitment of ATM or DNA-dependent protein kinase (DNA-PK) to the damage sites (Falck et al. 2005). ATR, in contrast, is recruited to single-stranded DNA. ATM and ATR activate the checkpoint kinases 1 or 2 (Chk1 or Chk2) (Abraham 2001), which in turn engage a myriad of downstream effectors that elicit an appropriate response (Smith et al. 2010; Zhou and Elledge 2000). The structurally-related transducer kinases heading the DNA damage response – ATM, ATR, and the catalytic subunit of DNA-PK (DNA-PKcs) – are all members of the PI3 kinase-related kinases (PIKK) family of Ser/Thr kinases (Abraham 2004).
DSB repair pathway choice
The mammalian genome has evolved two efficient, cell-cycle dependent DSB repair pathways – homologous recombination (HR) and non-homologous end-joining (NHEJ) (Thompson 2012). These pathways are complementary and operate under different circumstances. HR repair is a relatively slow, error-free process that occurs only during post-replicative S and G2 phases of the cell cycle during which a homologous DNA template (sister chromatid) is available (Moynahan and Jasin 2010; San Filippo et al. 2008). HR machinery copies the missing information from this DNA template into the break site, resulting in an exact reconstitution of the original sequence. In contrast, NHEJ repair rejoins two broken ends, with less stringency for homology, resulting in an intrinsically error-prone process. Repair errors can lead to gene mutations or wider chromosomal rearrangements. The lack of requirement for a homologous template (sister chromatid) means that NHEJ is not restricted to a certain phase of the cell cycle despite being assumed as the dominant repair pathway during the G1 and M phases (Weterings and Chen 2008). Malignant cells typically have fast turnover rates and, therefore, preferentially repair DSBs via the NHEJ pathway; if rapid re-joining does not occur, then resection may occur promoting HR repair (Shibata et al. 2011; Woods and Turchi 2013).
It appears that the two DSB repair pathways, HR and NHEJ are in direct competition – the HR (Mre11, Rad50 and Nbs1, or MRN) and NHEJ (Ku70/Ku80) sensors both bind independently yet almost simultaneously to the same DNA termini. During the late S/G2 phase where both NHEJ and HR repair mechanisms can co-exist, NHEJ is typically favored over HR because damage recognition by Ku70/80 precedes that of MRN (Kakarougkas and Jeggo 2014). Reduced activity of NHEJ proteins allows the process of DNA-end resection, in which the 5′-end of a damaged DNA strand is partially degraded, to occur, facilitating HR (Huertas 2010; Shibata et al. 2011). Recent studies have also shown that transient, PARP-dependent recruitment of RNA-binding proteins NONO and SFPQ to damaged sites can suppress HR repair whereas NHEJ is increased (Krietsch et al. 2012; Salton et al. 2010). Nevertheless if NHEJ progression is impeded, either due to high chromatin compaction or damage complexity, HR may still be activated to undertake the repair (Kakarougkas and Jeggo 2014).
Non-homologous end-joining
NHEJ begins with the recruitment and high-affinity binding of the abundant Ku heterodimer to DSB ends (Lieber 2010). Ku consists of Ku70 and Ku80 subunits that constitute the DNA-binding component of DNA-PK. High-affinity binding is conferred by the toroidal structure of Ku70/Ku80 that encircles about 20 bp of DNA (Grundy et al. 2014). By forming a synapse between broken DNA ends, the Ku heterodimer functions as a scaffold to support and align the DNA ends, while making the termini accessible to nucleases, polymerases, and ligases to promote the end-joining (Nick McElhinny et al. 2000).
A key binding partner of the Ku heterodimer is the ∼465 kDa kinase, DNA-PKcs (Meek et al. 2004; Uematsu et al. 2007). Autophosphorylation of DNA-PKcs upon binding to the DNA:Ku scaffold is required for the efficient repair of DSBs via the NHEJ pathway (Uematsu et al. 2007). In its dormant, unphosphorylated form, DNA-PKcs blocks the access of processing enzymes and ligases to the DNA ends, perhaps owing to its very large size. This block needs to be relieved before NHEJ repair can progress. Autophosphorylation of DNA-PKcs at SQ/TQ motifs – namely the 2609 (or ABCDE) cluster, the 2056 (PQR) cluster and C-terminal residues – releases the cap and allows processing and ligation of the broken ends to proceed (Davis et al. 2014; Dobbs et al. 2010). Phosphorylation at the Ser2056 cluster is mediated by DNA-PKcs itself (Chen et al. 2005), while phosphorylation at the Thr2609 cluster is mediated by ATM (Chen et al. 2007).
These phosphorylations enable DNA end-processing and recruitment of the ligation complex, which encompasses DNA ligase IV, X-ray repair cross-complementing protein 4 (XRCC4) and XRCC4-like factor (XLF; also termed Cernunnos) (Cottarel et al. 2013). The recruitment of this complex to the Ku-DSB scaffold is facilitated by DNA-PKcs. Most DNA termini are, however, incompatible – at least one of the strands will possess either a 3′ or 5′ single strand overhang – and, therefore, require additional processing to remove the non-ligatable ends. Candidate processing enzymes include the DNA polymerase μ and λ (polμ and polλ), terminal deoxynucleotidyltransferase, and the polynucleotide kinase (PNK) (Bernstein et al. 2009; Lieber 2010; Pawelczak et al. 2011). The endonuclease Artemis is also required for resecting a subset of DNA ends, especially the 3′-phosphoglycate ends commonly incurred by radiation. This Artemis endonuclease activity is facilitated by both DNA-PKcs and ATM (Pawelczak et al. 2011; Riballo et al. 2004).
Involvement of EGFR in the DNA damage response
DNA damage can trigger mitogenic growth factor signaling cascades, including activation of the epidermal growth factor receptor (EGFR). EGFR is a 170 kDa receptor tyrosine kinase involved in many physiological processes in both normal and malignant cells. EGFR overexpression occurs in many cancers, e.g. up to 50 % of triple negative breast cancers (Pintens et al. 2009) and may be associated with poor prognosis (Viale et al. 2009). These observations are the basis for a number of clinical trials that are exploring the efficacy of monoclonal anti-EGFR antibodies and EGFR tyrosine kinase inhibitors in breast cancer, as well as lung cancer and other forms of malignancy. In addition to activation of the EGFR by ligands such as EGF and TGFα, EGFR can be activated by DNA-damaging stimuli such as ionizing radiation and certain cytotoxic drugs (Kriegs et al. 2010), through mechanisms that are not fully established, but may involve the tyrosine kinase Src (Dittmann et al. 2008). This activation results in caveolin-1-dependent endocytosis and nuclear localization of EGFR (Dittmann et al. 2008; Khan et al. 2006). Nuclear EGFR has been shown to interact with both the catalytic and regulatory (Ku70/Ku80) subunits of DNA-PK, resulting in activation of the DNA-PK complex and favoring NHEJ repair (Dittmann et al. 2005). The phosphorylated DNA-PK clusters include 2056 and 2609, and in particular Thr2609 phosphorylation of DNA-PKcs is critical for EGFR-mediated radiation resistance (Javvadi et al. 2012). EGFR inhibition and/or downregulation by monoclonal antibodies abolishes EGFR nuclear translocation and, as a consequence, DNA-damage-induced DNA-PK activation (Dittmann et al. 2005). Similarly, the EGFR kinase inhibitor gefitinib was found to inhibit the formation of nuclear EGFR-DNA-PKcs complexes (Lin et al. 2014).
EGFR canonically triggers both the pro-survival PI3K/AKT pathway and the mitogen-activated protein kinases (MAPK)/extracellular signal-regulated kinase (ERK) pathway (Chen and Nirodi 2007). Both cytoplasmic signaling pathways appear to be involved in NHEJ and HR repair, potentially contributing to radioresistance (Golding et al. 2009). Akt has a role in phosphorylation of DNA-PKcs at the Ser2056 and Thr2609 clusters (Toulany et al. 2008) and, reciprocally, Akt can be activated by DNA-PKcs, providing a pro-survival feedback (Bozulic et al. 2008). This suggests that Akt and DNA-PKcs are inter-dependent for their maximal response to DNA damage. In parallel, in response to radiation, the EGFR/MEK/ERK signaling pathway is crucial for efficient HR and maximal activation of ATM (Golding et al. 2007). Taken together, either over-abundance of nuclear EGFR or activation of PI3K/AKT and/or MAPK/ERK signaling pathways induced by DNA-damaging therapies leads to an overdrive in DNA damage repair, which plays an important role in the resistance to chemo- and radiotherapy in breast and other cancer patients.
IGFBPs and the DNA damage response
There is limited literature on the involvement of IGFBPs in DNA DSB induction and signaling. Nevertheless evidence can be derived from studies investigating IGFBP effects on DNA-damaging agents, including chemotherapy, radiotherapy, UV irradiation, and reactive oxygen species. IGFBP-3 can directly mediate or accentuate apoptotic cell death by DNA-damage-associated agents such as ceramide (Gill et al. 1997), UV radiation (Hollowood et al. 2000), paclitaxel (Fowler et al. 2000), etoposide (Drivdahl et al. 2001; Lin et al. 2014) and doxorubicin (Granata et al. 2003). IGFBP-3 upregulation was shown to be associated with radiosensitive, compared to radioresistant, cervical cancer cell lines (Achary et al. 2000), and a similar observation has been reported in esophageal cancer cells (Yoshino et al. 2011). Similarly, increased expression of IGFBP-3 was associated with enhanced sensitivity to the cytotoxic effects of cisplatin in esophageal cancer, and this was reversed by silencing of IGFBP-3 (Zhao et al. 2012). These effects of IGFBP-3 may in some cases be dependent on its inhibition of prosurvival IGF1R signaling by sequestering IGFs, although the possibility of chemo- or radio-sensitizing effects without interference in IGF1R signaling cannot be excluded. Other IGFBPs may have opposite effects; for example, notwithstanding the reported p53-dependence of IGFBP-2, exogenous IGFBP-2 was found to decrease the glioma cell response to the DNA-damaging drug temozolomide (Han et al. 2014),
The involvement of sphingolipids
Ceramide accumulation, resulting from both ceramide synthase activation (de novo synthesis) and sphingomyelin breakdown, has been extensively documented as a response to cell irradiation leading to apoptotic cell death (Aureli et al. 2014). Cells isolated from mice that are deficient in acid sphingomyelinase, an important enzyme in sphingomyelin catabolism to ceramide, show resistance to radiation-induced apoptosis, and sensitivity is restored when the enzyme is reintroduced (Santana et al. 1996). IGFBP-3 potentiation of doxorubicin-induced apoptosis in human endothelial cell cultures was shown to be accompanied by an increase in ceramide levels, and inhibited by the ceramide synthase inhibitor fumonisin B1, suggesting that in these cells, IGFBP-3-dependent de novo synthesis of ceramide was an important contributor to doxorubicin-induced cell death (Granata et al. 2004).
Breast cancer cell death induced by exogenous C2 ceramide (a cell-permeable ceramide analogue) was found to be enhanced by exogenous recombinant IGFBP-3, but inhibited by recombinant IGFBP-4 or IGFBP-5 (Perks et al. 1999). IGFBP-1, -2 and -6 had no effect. The enhancing effect of exogenous IGFBP-3 on ceramide-dependent cell death appeared unrelated to its ability to sequester IGFs (which could activate IGF1R-mediated cell survival), since an IGFBP-3 analogue with greatly reduced IGF-binding affinity had a similar effect (Perks et al. 2002). The protective effect of IGFBP-5 was reversed by the addition of a hexapeptide containing the Arg-Gly-Asp integrin-binding motif, suggesting that the cellular environment is an important modulator of its response. In contrast to the protective effect of exogenous IGFBP-5 against ceramide-induced apoptosis, endogenous IGFBP-5 expressed by an adenoviral vector in breast cancer cells induced apoptosis alone, and enhanced radiation-induced loss of cell survival, but ceramide levels were not measured in this experiment (Butt et al. 2003).
While some studies suggest that IGFBP-3 may contribute to apoptosis by inducing pro-apoptotic ceramide under DNA-damaging conditions, there is also the potential for IGFBP-3 to promote resistance to chemotherapies via its upregulation of the pro-survival arm of this sphingolipid signaling system, the key components of which are sphingosine kinase (SphK) and the product of its enzymatic activity, sphingosine-1-phosphate (S1P). IGFBP-3 induction of SphK1 expression and activity was first reported in human endothelial cells (Granata et al. 2004) where it was shown to mediate cell-survival effects of IGFBP-3. Upregulation of this pathway by IGFBP-3 was also demonstrated in estrogen receptor-negative breast epithelial cells and several triple-negative breast cancer cell lines (Martin et al. 2014; Martin et al. 2009), in which IGFBP-3-induced S1P enhances EGFR signaling via receptor transactivation. In view of the fact that SphK1 and S1P promote chemo- and radio-resistance in a number of cell and animal models of cancer (Pchejetski et al. 2008; Truman et al. 2014), it is possible that through its induction of this pathway, and its interaction with EGFR to promote DNA damage repair, IGFBP-3 contributes to resistance to therapies that work by damaging DNA. This could be particularly relevant in cancer types with high IGFBP-3 expression, such as estrogen receptor-negative breast cancers (Shao et al. 1992), pancreatic cancer (Xue et al. 2008), and clear cell renal cell carcinoma (Chuang et al. 2008).
A role of IGFBP-3 in non-homologous end-joining
IGFBP-3 was shown several years ago to be a substrate for DNA-PKcs (Schedlich et al. 2003) which, as noted above, is a key component of the NHEJ pathway. Phosphorylation of IGFBP-3 by DNA-PKcs resulted in a loss of IGF-binding, but increased nuclear translocation and retention. A similar enhancement of nuclear import by DNA-PK phosphorylation was previously shown for SV40 large T-antigen (Xiao et al. 1997). The target site on IGFBP-3 for DNA-PK phosphorylation was inferred by mutagenesis to be Ser156, one of 3 clustered consensus sites for this family of kinases (Ser156, Ser165, and Thr170), and mutation of this residue prevented IGFBP-3-induced apoptosis in prostate cancer cells (Cobb et al. 2006). Conversely, IGFBP-3 phosphorylation by protein kinase CK2, while having little effect on the rate of nuclear uptake of IGFBP-3 (Schedlich et al. 2003), was shown to inhibit its apoptotic effect, and preventing CK2 activation enhanced apoptosis induced by IGFBP-3 (Cobb et al. 2009). These responses may be functionally related since CK2 downregulation has been shown to markedly increase DNA-PKcs activity (Olsen et al. 2010). However, the dependence of IGFBP-3 on DNA-PKcs phosphorylation of Ser156 for its proapoptotic effect in prostate cancer cells contrasts with an observation made in retinal endothelial cells, in which mutation of this residue to prevent its phosphorylation by DNA-PKcs increased the apoptotic response (Zhao et al. 2012). In this experimental system, DNA-PK was shown to contribute to the ability of IGFBP-3 to enhance cell survival, rather than death.
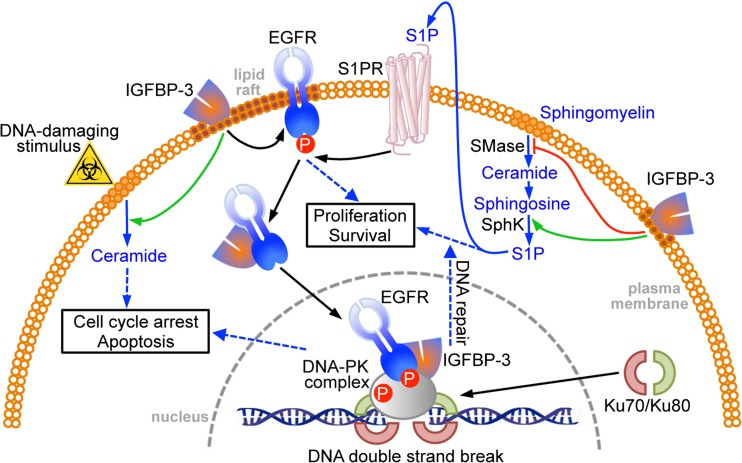
The idea that IGFBP-3 may promote survival rather than apoptosis under some conditions has strong experimental support. Although it appears to function as a tumor suppressor in some cancers, high IGFBP-3 expression is a poor prognostic feature in some other tumor types. IGFBP-3 has also been shown to have growth-promoting activity in a number of cell culture studies (Baxter 2014). These disparate observations raise the possibility that IGFBP-3 may stimulate cell proliferation and/or survival pathways under some conditions. The observation of enhanced nuclear entry by IGFBP-3 in response to phosphorylation by DNA-PKcs suggests that nuclear IGFBP-3 might be involved in DNA-PK-dependent DNA damage repair (Fig. 1). This has been investigated in triple-negative breast cancer cell lines that have high IGFBP-3 expression. In MDA-MB-468 cells, that have particularly high IGFBP-3 production, the formation of nuclear complexes between DNA-PKcs and EGFR in response to etoposide treatment was found to be inhibited by the EGFR kinase inhibitor, gefitinib, suggesting that they were dependent on EGFR autophosphorylation at Tyr1068 (Lin et al. 2014). These nuclear complexes also appear to include IGFBP-3, since gefitinib also inhibited the formation of nuclear DNA-PKcs-IGFBP-3 and EGFR-IGFBP-3 complexes, as demonstrated both by co-immunoprecipitation studies and by direct visualization using the proximity ligation assay.
Fig. 1.
Involvement of IGFBP-3 in the DNA damage response. IGFBP-3 interacts with EGFR in lipid rafts and, in response to a DNA-damaging stimulus, translocates to the nucleus where both proteins complex with DNA-PK to facilitate non-homologous end-joining. This effect is blocked by EGFR kinase inhibition. IGFBP-3 promotes survival by inhibiting the production of pro-apoptotic ceramide, and increasing sphingosine-1-phosphate, but in response to genotoxic stress, it can increase apoptosis by activating ceramide synthesis. SMase, sphingomyelinase; SphK, sphingosine kinase; S1P, sphingosine-1-phosphate
However, whereas complexes between DNA-PKcs and IGFBP-3 were found almost exclusively within the nucleus by confocal microscopy, those between EGFR and IGFBP-3 had a significant extranuclear presence (Lin et al. 2014). The complexes initially appeared associated with lipid rafts, but this co-localization decreased 2–4 h after etoposide treatment, consistent with their translocation to the nucleus (Fig. 1). IGFBP-3 silencing by either of two siRNAs decreased the DNA-PKcs autophosphorylation response to etoposide, the formation of nuclear EGFR-DNA-PKcs complexes, and the repair of DNA by NHEJ, as measured by a direct assay using nuclear extracts (Lin et al. 2014). These findings suggest an integral role for IGFBP-3 in the events following exposure of breast cancer cells to DNA double-strand breaks, eventually leading to DNA repair.
Although direct binding between IGFBP-3 and DNA-PKcs has been observed, no interaction between IGFBP-3 and the Ku70/80 subunits of the DNA-PK complex has been reported. In contrast, both Ku70 and Ku80 have been purified by affinity with IGFBP-6, and Ku80 was shown to have a high-affinity interaction with IGFBP-6 (Iosef et al. 2010). IGFBP-6 overexpression decreased the association of Ku70/80 with DNA ends, suggesting that it might be inhibitory to DNA repair by NHEJ, in contrast to IGFBP-3 which appears to participate in this process.
Concluding comments
The regulation of DNA damage repair is of considerable interest clinically because it is an important mechanism by which cancer cells resist the actions of chemotherapy and radiotherapy, but it is also of much broader interest considering that DNA damage occurs in all cells every day. The observations that all of the IGFBP genes have p53 binding sites, and that several of the IGFBPs are functionally inducible by p53 activation, have provided important clues that this family of proteins might play integral roles in cell responsiveness to genotoxic stress. It is perhaps surprising that, to date, there have been few explicit demonstrations of IGFBPs fulfilling these roles, despite an increasing body of literature suggesting the possibility of direct or indirect involvement. The evidence is certainly strongest for IGFBP-3, the IGFBP that is most abundant in adult humans and also the most extensively studied for its diverse IGF-dependent and –independent actions.
How might the involvement of IGFBPs in DNA repair mechanisms be exploited for therapeutic advantage? It seems unlikely that the IGFBPs themselves would be ideal targets, as it may be difficult to distinguish therapeutically between their dual endocrine and autocrine roles. A more focused approach may be to discover and target key intracellular interactions between IGFBPs and other effector proteins; in the case of IGFBP-3 or IGFBP-6 this might be their interaction with components of the DNA-PK complex. Since mutations in IGFBP genes are relatively rare, biochemical discovery tools (e.g. proteomics) are likely to be more effective at uncovering novel interactions involved in DNA repair than the genetic approaches that have revealed the involvement of many other components of DNA repair complexes. Regardless of the discovery technique, increased knowledge of the involvement of IGFBPs in the cancer cell response to DNA damage will not only improve understanding of the ways in which cells maintain genomic integrity, but may also yield unexpected benefits in the form of new cancer therapy.
Acknowledgments
This work was supported in part by grants from the Australian Research Council (DP140100137) and the Cancer Council NSW (RG 11-09).
Abbreviations
- IGFBP
Insulin-like growth factor binding protein
- DSB
Double-strand break
- NHEJ
Non-homologous end-joining
- EGFR
Epidermal growth factor receptor
- DNA-PK
DNA-dependent protein kinase
- IGF1R
Type 1 IGF receptor
- ATM
Ataxia-telangiectasia mutated
- ATR
Ataxia-telangiectasia and Rad3-related
- PIKK
PI3 kinase-related kinases
- HR
Homologous recombination
- MRN
Mre11 Rad50 and Nbs1
- ERK
Extracellular signal-regulated kinase
- S1P
Sphingosine-1-phosphate
References
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Abraham RT. PI 3-kinase related kinases: ‘Big’ players in stress-induced signaling pathways. DNA Repair. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Achary MP, Jaggernauth W, Gross E, Alfieri A, Klinger HP, Vikram B. Cell lines from the same cervical carcinoma but with different radiosensitivities exhibit different cDNA microarray patterns of gene expression. Cytogenet Cell Genet. 2000;91:39–43. doi: 10.1159/000056815. [DOI] [PubMed] [Google Scholar]
- Aureli M, Murdica V, Loberto N, Samarani M, Prinetti A, Bassi R, Sonnino S. Exploring the link between ceramide and ionizing radiation. Glycoconj J. 2014;31:449–459. doi: 10.1007/s10719-014-9541-y. [DOI] [PubMed] [Google Scholar]
- Barbieri CE, Perez CA, Johnson KN, Ely KA, Billheimer D, Pietenpol JA. IGFBP-3 is a direct target of transcriptional regulation by DeltaNp63alpha in squamous epithelium. Cancer Res. 2005;65:2314–2320. doi: 10.1158/0008-5472.CAN-04-3449. [DOI] [PubMed] [Google Scholar]
- Baxter RC. Insulin-like growth factor (IGF)-binding proteins: interactions with IGFs and intrinsic bioactivities. Am J Physiol Endocrinol Metab. 2000;278:E967–E976. doi: 10.1152/ajpendo.2000.278.6.E967. [DOI] [PubMed] [Google Scholar]
- Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer. 2014;14:329–341. doi: 10.1038/nrc3720. [DOI] [PubMed] [Google Scholar]
- Beattie J, McIntosh L, van der Walle CF. Cross-talk between the insulin-like growth factor (IGF) axis and membrane integrins to regulate cell physiology. J Cell Physiol. 2010;224:605–611. doi: 10.1002/jcp.22183. [DOI] [PubMed] [Google Scholar]
- Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, Glover JNM. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, Polynucleotide Kinase. Nucleic Acids Res. 2009;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkarev VA, Komarova EA, Siebenhaar F, Botchkareva NV, Komarov PG, Maurer M, Gilchrest BA, Gudkov AV. p53 is essential for chemotherapy-induced hair loss. Cancer Res. 2000;60:5002–5006. [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBα/Akt1 Acts Downstream of DNA-PK in the DNA Double-Strand Break Response and Promotes Survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Firth SM, King MA, Baxter RC. Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J Biol Chem. 2000;275:39174–39181. doi: 10.1074/jbc.M908888199. [DOI] [PubMed] [Google Scholar]
- Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003;278:29676–29685. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- Chen DJ, Nirodi CS. The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- Chuang ST, Patton KT, Schafernak KT, Papavero V, Lin F, Baxter RC, Teh BT, Yang XJ. Over expression of insulin-like growth factor binding protein 3 in clear cell renal cell carcinoma. J Urol. 2008;179:445–449. doi: 10.1016/j.juro.2007.09.106. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Liu B, Lee KW, Cohen P. Phosphorylation by DNA-dependent protein kinase is critical for apoptosis induction by insulin-like growth factor binding protein-3. Cancer Res. 2006;66:10878–10884. doi: 10.1158/0008-5472.CAN-06-0585. [DOI] [PubMed] [Google Scholar]
- Cobb LJ, Mehta H, Cohen P. Enhancing the apoptotic potential of insulin-like growth factor-binding protein-3 in prostate cancer by modulation of CK2 phosphorylation. Mol Endocrinol. 2009;23:1624–1633. doi: 10.1210/me.2008-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel J, Frit P, Bombarde O, Salles B, Négrel A, Bernard S, Jeggo PA, Lieber MR, Modesti M, Calsou P. A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol. 2013;200:173–186. doi: 10.1083/jcb.201203128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst) 2014;17:21–29. doi: 10.1016/j.dnarep.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced Epidermal Growth Factor Receptor Nuclear Import Is Linked to Activation of DNA-dependent Protein Kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair (Amst) 2010;9:1307–1314. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drivdahl RH, Sprenger C, Trimm K, Plymate SR. Inhibition of Growth and Increased Expression of Insulin-Like Growth Factor-Binding Protein-3 (IGFBP-3) and -6 in Prostate Cancer Cells Stably Transfected with Antisense IGFBP-4 Complementary Deoxyribonucleic Acid. Endocrinology. 2001;142:1990–1998. doi: 10.1210/endo.142.5.8158. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Forbes BE, McCarthy P, Norton RS. Insulin-like growth factor binding proteins: a structural perspective. Front Endocrinol (Lausanne) 2012;3:38. doi: 10.3389/fendo.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CA, Perks CM, Newcomb PV, Savage PB, Farndon JR, Holly JMP. Insulin-like growth factor binding protein-3 (IGFBP-3) potentiates paclitaxel-induced apoptosis in human breast cancer cells. Int J Cancer. 2000;88:448–453. doi: 10.1002/1097-0215(20001101)88:3<448::AID-IJC18>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Gill ZP, Perks CM, Newcomb PV, Holly JMP. Insulin-like Growth Factor-binding Protein (IGFBP-3) Predisposes Breast Cancer Cells to Programmed Cell Death in a Non-IGF-dependent Manner. J Biol Chem. 1997;272:25602–25607. doi: 10.1074/jbc.272.41.25602. [DOI] [PubMed] [Google Scholar]
- Golding SE, Rosenberg E, Neill S, Dent P, Povirk LF, Valerie K. Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res. 2007;67:1046–1053. doi: 10.1158/0008-5472.CAN-06-2371. [DOI] [PubMed] [Google Scholar]
- Golding SE, Morgan RN, Adams BR, Hawkins AJ, Povirk LF, Valerie K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol Ther. 2009;8:730–738. doi: 10.4161/cbt.8.8.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata R, De Petrini M, Trovato L, Ponti R, Pons N, Ghe C, Graziani A, Ferry RJ, Jr, Muccioli G, Ghigo E. Insulin-like growth factor binding protein-3 mediates serum starvation- and doxorubicin-induced apoptosis in H9c2 cardiac cells. J Endocrinol Invest. 2003;26:1231–1241. doi: 10.1007/BF03349163. [DOI] [PubMed] [Google Scholar]
- Granata R, Trovato L, Garbarino G, Taliano M, Ponti R, Sala G, Ghidoni R, Ghigo E. Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: involvement of the sphingolipid signaling pathways. FASEB J. 2004;18:1456–1458. doi: 10.1096/fj.04-1618fje. [DOI] [PubMed] [Google Scholar]
- Grimberg A, Coleman CM, Burns TF, Himelstein BP, Koch CJ, Cohen P, El-Deiry WS. p53-Dependent and p53-independent induction of insulin-like growth factor binding protein-3 by deoxyribonucleic acid damage and hypoxia. J Clin Endocrinol Metab. 2005;90:3568–3574. doi: 10.1210/jc.2004-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg A, Coleman CM, Shi Z, Burns TF, MacLachlan TK, Wang W, El-Deiry WS. Insulin-like growth factor factor binding protein-2 is a novel mediator of p53 inhibition of insulin-like growth factor signaling. Cancer Biol Ther. 2006;5:1408–1414. doi: 10.4161/cbt.5.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy GJ, Moulding HA, Caldecott KW, Rulten SL. One ring to bring them all–the role of Ku in mammalian non-homologous end joining. DNA Repair (Amst) 2014;17:30–38. doi: 10.1016/j.dnarep.2014.02.019. [DOI] [PubMed] [Google Scholar]
- Han S, Li Z, Master LM, Master ZW, Wu A. Exogenous IGFBP-2 promotes proliferation, invasion, and chemoresistance to temozolomide in glioma cells via the integrin beta1-ERK pathway. Br J Cancer. 2014;111:1400–1409. doi: 10.1038/bjc.2014.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa T, Shinji T, Shiraha H, Nouso K, Iwasaki Y, Yumoto E, Ono T, Koide N. Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation. BMC Cancer. 2005;5:9. doi: 10.1186/1471-2407-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms KL, Chen X. The C terminus of p53 family proteins is a cell fate determinant. Mol Cell Biol. 2005;25:2014–2030. doi: 10.1128/MCB.25.5.2014-2030.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollowood AD, Lai T, Perks CM, Newcomb PV, Alderson D, Holly JMP. IGFBP-3 prolongs the p53 response and enhances apoptosis following UV irradiation. Int J Cancer. 2000;88:336–341. doi: 10.1002/1097-0215(20001101)88:3<336::AID-IJC3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iosef C, Vilk G, Gkourasas T, Lee KJ, Chen BP, Fu P, Bach LA, Lajoie G, Gupta MB, Li SS, et al. Insulin-like growth factor binding protein-6 (IGFBP-6) interacts with DNA-end binding protein Ku80 to regulate cell fate. Cell Signal. 2010;22:1033–1043. doi: 10.1016/j.cellsig.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and human disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javvadi P, Makino H, Das AK, Lin YF, Chen DJ, Chen BP, Nirodi CS. Threonine 2609 phosphorylation of the DNA-dependent protein kinase is a critical prerequisite for epidermal growth factor receptor-mediated radiation resistance. Mol Cancer Res. 2012;10:1359–1368. doi: 10.1158/1541-7786.MCR-12-0482-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A, Jeggo PA. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br J Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan E, Heidinger J, Levy M, Lisanti M, Ravid T, Goldkorn T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J Biol Chem. 2006;281:14486–14493. doi: 10.1074/jbc.M509332200. [DOI] [PubMed] [Google Scholar]
- Kim KS, Seu YB, Baek SH, Kim MJ, Kim KJ, Kim JH, Kim JR. Induction of cellular senescence by insulin-like growth factor binding protein-5 through a p53-dependent mechanism. Mol Biol Cell. 2007;18:4543–4552. doi: 10.1091/mbc.E07-03-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs M, Kasten-Pisula U, Rieckmann T, Holst K, Saker J, Dahm-Daphi J, Dikomey E. The epidermal growth factor receptor modulates DNA double-strand break repair by regulating non-homologous end-joining. DNA Repair (Amst) 2010;9:889–897. doi: 10.1016/j.dnarep.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Krietsch J, Caron MC, Gagne JP, Ethier C, Vignard J, Vincent M, Rouleau M, Hendzel MJ, Poirier GG, Masson JY. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JI, George DL. Hepatic IGFBP1 is a prosurvival factor that binds to BAK, protects the liver from apoptosis, and antagonizes the proapoptotic actions of p53 at mitochondria. Genes Dev. 2007;21:3095–3109. doi: 10.1101/gad.1567107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, Marzec KA, Martin JL, Baxter RC. The role of insulin-like growth factor binding protein-3 in the breast cancer cell response to DNA-damaging agents. Oncogene. 2014;33:85–96. doi: 10.1038/onc.2012.538. [DOI] [PubMed] [Google Scholar]
- Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 2014;42:7666–7680. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, Lin MZ, McGowan EM, Baxter RC. Potentiation of growth factor signaling by insulin-like growth factor-binding protein-3 in breast epithelial cells requires sphingosine kinase activity. J Biol Chem. 2009;284:25542–25552. doi: 10.1074/jbc.M109.007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JL, de Silva HC, Lin MZ, Scott CD, Baxter RC. Inhibition of insulin-like growth factor-binding protein-3 signaling through sphingosine kinase-1 sensitizes triple-negative breast cancer cells to EGF receptor blockade. Mol Cancer Ther. 2014;13:316–328. doi: 10.1158/1535-7163.MCT-13-0367. [DOI] [PubMed] [Google Scholar]
- Meek K, Gupta S, Ramsden DA, Lees-Miller SP. The DNA-dependent protein kinase: the director at the end. Immunol Rev. 2004;200:132–141. doi: 10.1111/j.0105-2896.2004.00162.x. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku Recruits the XRCC4-Ligase IV Complex to DNA Ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/MCB.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen BB, Issinger OG, Guerra B. Regulation of DNA-dependent protein kinase by protein kinase CK2 in human glioblastoma cells. Oncogene. 2010;29:6016–6026. doi: 10.1038/onc.2010.337. [DOI] [PubMed] [Google Scholar]
- Pawelczak KS, Bennett SM, Turchi JJ. Coordination of DNA-PK activation and nuclease processing of DNA termini in NHEJ. Antioxid Redox Signal. 2011;14:2531–2543. doi: 10.1089/ars.2010.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchejetski D, Doumerc N, Golzio M, Naymark M, Teissie J, Kohama T, Waxman J, Malavaud B, Cuvillier O. Chemosensitizing effects of sphingosine kinase-1 inhibition in prostate cancer cell and animal models. Mol Cancer Ther. 2008;7:1836–1845. doi: 10.1158/1535-7163.MCT-07-2322. [DOI] [PubMed] [Google Scholar]
- Perks CM, Bowen S, Gill ZP, Newcomb PV, Holly JM. Differential IGF-independent effects of insulin-like growth factor binding proteins (1-6) on apoptosis of breast epithelial cells. J Cell Biochem. 1999;75:652–664. doi: 10.1002/(SICI)1097-4644(19991215)75:4<652::AID-JCB11>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Perks CM, McCaig C, Clarke JB, Clemmons DR, Holly JM. A non-IGF binding mutant of IGFBP-3 modulates cell function in breast epithelial cells. Biochem Biophys Res Commun. 2002;294:988–994. doi: 10.1016/S0006-291X(02)00569-7. [DOI] [PubMed] [Google Scholar]
- Pintens S, Neven P, Drijkoningen M, Van Belle V, Moerman P, Christiaens M-R, Smeets A, Wildiers H, Bempt IV. Triple negative breast cancer: a study from the point of view of basal CK5/6 and HER-1. J Clin Pathol. 2009;62:624–628. doi: 10.1136/jcp.2008.061358. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Witherspoon S, Fox T. The infant mouse as a in vivo model for the detection and study of DNA damage-induced changes in the liver. Mol Carcinog. 2004;40:62–72. doi: 10.1002/mc.20017. [DOI] [PubMed] [Google Scholar]
- Riballo E, Kühne M, Rief N, Doherty A, Smith GCM, Recio M-J, Reis C, Dahm K, Fricke A, Krempler A, et al. A Pathway of Double-Strand Break Rejoining Dependent upon ATM, Artemis, and Proteins Locating to γ-H2AX Foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of Eukaryotic Homologous Recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Santana P, Pena LA, Haimovitz-Friedman A, Martin S, Green D, McLoughlin M, Cordon-Cardo C, Schuchman EH, Fuks Z, Kolesnick R. Acid sphingomyelinase-deficient human lymphoblasts and mice are defective in radiation-induced apoptosis. Cell. 1996;86:189–199. doi: 10.1016/S0092-8674(00)80091-4. [DOI] [PubMed] [Google Scholar]
- Sbisa E, Catalano D, Grillo G, Licciulli F, Turi A, Liuni S, Pesole G, De Grassi A, Caratozzolo MF, D’Erchia AM, et al. p53FamTaG: a database resource of human p53, p63 and p73 direct target genes combining in silico prediction and microarray data. BMC Bioinforma. 2007;8(Suppl 1):S20. doi: 10.1186/1471-2105-8-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedlich LJ, Nilsen T, John AP, Jans DA, Baxter RC. Phosphorylation of insulin-like growth factor binding protein-3 by deoxyribonucleic acid-dependent protein kinase reduces ligand binding and enhances nuclear accumulation. Endocrinology. 2003;144:1984–1993. doi: 10.1210/en.2002-220798. [DOI] [PubMed] [Google Scholar]
- Shao ZM, Sheikh MS, Ordonez JV, Feng P, Kute T, Chen JC, Aisner S, Schnaper L, LeRoith D, Roberts CT, Jr, et al. IGFBP-3 gene expression and estrogen receptor status in human breast carcinoma. Cancer Res. 1992;52:5100–5103. [PubMed] [Google Scholar]
- Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, Kakarougkas A, Meek K, Taucher‐Scholz G, Löbrich M, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30:1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikazono N, Noguchi M, Fujii K, Urushibara A, Yokoya A. The Yield, Processing, and Biological Consequences of Clustered DNA Damage Induced by Ionizing Radiation. J Radiat Res. 2009;50:27–36. doi: 10.1269/jrr.08086. [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Cortez D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol. 2013;5:a012724. doi: 10.1101/cshperspect.a012724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitar T, Popowicz GM, Siwanowicz I, Huber R, Holak TA. Structural basis for the inhibition of insulin-like growth factors by insulin-like growth factor-binding proteins. Proc Natl Acad Sci U S A. 2006;103:13028–13033. doi: 10.1073/pnas.0605652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nat Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Toulany M, Kehlbach R, Florczak U, Sak A, Wang S, Chen J, Lobrich M, Rodemann HP. Targeting of AKT1 enhances radiation toxicity of human tumor cells by inhibiting DNA-PKcs-dependent DNA double-strand break repair. Mol Cancer Ther. 2008;7:1772–1781. doi: 10.1158/1535-7163.MCT-07-2200. [DOI] [PubMed] [Google Scholar]
- Truman JP, Garcia-Barros M, Obeid LM, Hannun YA. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim Biophys Acta. 2014;1841:1174–1188. doi: 10.1016/j.bbalip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, Van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, Veronesi P, Intra M, Torrisi R, Cardillo A, et al. Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat. 2009;116:317–328. doi: 10.1007/s10549-008-0206-z. [DOI] [PubMed] [Google Scholar]
- Vikhanskaya F, Lee MK, Mazzoletti M, Broggini M, Sabapathy K. Cancer-derived p53 mutants suppress p53-target gene expression–potential mechanism for gain of function of mutant p53. Nucleic Acids Res. 2007;35:2093–2104. doi: 10.1093/nar/gkm099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Williams AC, Collard TJ, Perks CM, Newcomb P, Moorghen M, Holly JM, Paraskeva C. Increased p53-dependent apoptosis by the insulin-like growth factor binding protein IGFBP-3 in human colonic adenoma-derived cells. Cancer Res. 2000;60:22–27. [PubMed] [Google Scholar]
- Woods D, Turchi JJ. Chemotherapy induced DNA damage response: convergence of drugs and pathways. Cancer Biol Ther. 2013;14:379–389. doi: 10.4161/cbt.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao CY, Hubner S, Jans DA. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J Biol Chem. 1997;272:22191–22198. doi: 10.1074/jbc.272.35.22191. [DOI] [PubMed] [Google Scholar]
- Xie L, Tsaprailis G, Chen QM. Proteomic identification of insulin-like growth factor-binding protein-6 induced by sublethal H2O2 stress from human diploid fibroblasts. Mol Cell Proteomics. 2005;4:1273–1283. doi: 10.1074/mcp.M500032-MCP200. [DOI] [PubMed] [Google Scholar]
- Xue A, Scarlett CJ, Jackson CJ, Allen BJ, Smith RC. Prognostic significance of growth factors and the urokinase-type plasminogen activator system in pancreatic ductal adenocarcinoma. Pancreas. 2008;36:160–167. doi: 10.1097/MPA.0b013e31815750f0. [DOI] [PubMed] [Google Scholar]
- Yamada PM, Lee KW. Perspectives in mammalian IGFBP-3 biology: local vs. systemic action. Am J Physiol Cell Physiol. 2009;296:C954–C976. doi: 10.1152/ajpcell.00598.2008. [DOI] [PubMed] [Google Scholar]
- Yoshino K, Motoyama S, Koyota S, Shibuya K, Usami S, Maruyama K, Saito H, Minamiya Y, Sugiyama T, Ogawa J. IGFBP3 and BAG1 enhance radiation-induced apoptosis in squamous esophageal cancer cells. Biochem Biophys Res Commun. 2011;404:1070–1075. doi: 10.1016/j.bbrc.2010.12.115. [DOI] [PubMed] [Google Scholar]
- Zhao L, Li QQ, Zhang R, Xi M, Liao YJ, Qian D, He LR, Zeng YX, Xie D, Liu MZ. The overexpression of IGFBP-3 is involved in the chemosensitivity of esophageal squamous cell carcinoma cells to nimotuzumab combined with cisplatin. Tumour Biol. 2012;33:1115–1123. doi: 10.1007/s13277-012-0352-0. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]