Abstract
Lack of genetic tools in the Lemnaceae (duckweed) has impeded full implementation of this organism as model for biological research, despite its rapid doubling time, simple architecture and unusual metabolic characteristics. Here we present technologies to facilitate high-throughput genetic studies in duckweed. We developed a fast and efficient method for producing Lemna minor stable transgenic fronds via agrobacterium-mediated transformation and regeneration from tissue culture. Additionally, we engineered an artificial microRNA (amiRNA) gene silencing system. We identified a Lemna gibba endogenous miR166 precursor and used it as a backbone to produce amiRNAs. As a proof of concept we induced the silencing of CH42, a Magnesium Chelatase subunit, using our amiRNA platform. Expression of CH42 in transgenic Lemna minor fronds was significantly reduced, which resulted in reduction of chlorophyll pigmentation. The techniques presented here will enable tackling future challenges in the biology and biotechnology of Lemnaceae.
Keywords: RNA silencing, amiRNA, stable transformation, Lemnaceae, CH42, microRNA
INTRODUCTION
Lemnaceae are the smallest aquatic flowering plants, commonly known as duckweeds, and have seen a revival in research interest during the past few years as an attractive alternative to algae biofuel feedstock, due to their robust growth in marginal environments and their particular metabolic traits, such as high starch accumulation and low lignin content (Xu et al., 2011; Xu et al., 2012; Yan et al., 2013). To increase its value in this respect, it is necessary to investigate the complexity of its metabolic pathways using genetic assays. Genetic engineering of duckweed has already been used to produce recombinant proteins and human monoclonal antibodies (Cox et al., 2006; Nguyen et al., 2012). Still, the lack of an efficient system for genetic manipulation, along with the extremely low efficiency of existing transformation methods, have held back efforts to perform major genetic studies. Therefore, most genetic tools are either unavailable in Lemna and related species, or else require major improvement in order to be able to perform high-throughput genetic manipulation.
Polyploidy and difficulty in obtaining Lemnaceae seeds from sexual crosses, make chemical mutagenesis and sequence-indexed insertion mutants challenging for high-throughput genetics, especially for homozygous loss-of-function alleles (Urbanska-Worytkiewicz, 1975). Therefore, a dominant silencing approach is needed to down-regulate genes. RNA interference (RNAi) regulates the expression of protein-coding genes by a mechanism known as posttranscriptional gene silencing (PTGS), and is ideally suited to this purpose (Fire et al., 1998; Hamilton and Baulcombe, 1999). Small RNAs target specific sequences via Argonaute proteins in the RNA-induced silencing complex (RISC), and, by recognizing complementary motifs in nucleic acids, they can protect against viral infection, prevent transposon mobilization and regulate endogenous genes. microRNAs (miRNAs) are derived from imperfect hairpin RNA precursors that typically give rise to one predominant 21 nucleotide (nt) miRNA, along with an imperfectly complementary antisense 21nt miRNA* (Axtell, 2013). Artificial microRNAs (amiRNAs) can be designed to down-regulate genes of interest in plants and animals. The amiRNA precursor (pre-amiRNA) is typically processed such that one stable small RNA is preferentially generated, which facilitates prediction of the spectrum of amiRNA targets, thus avoiding off-target effects. This technology has been successfully developed and adapted in several plant model organisms, and has proven to be an efficient and reproducible tool for highly specific gene silencing (Molnar et al., 2009; Schwab et al., 2006; Warthmann et al., 2008). The use of modified versions of endogenous miRNA overcomes the self-silencing and ‘off-target’ silencing associated with long dsRNAs (Jackson et al., 2003).
Here we describe an artificial microRNA system based on an endogenous Lemna gibba miR166a precursor. We demonstrate how this system can be used as a gene silencing platform in Lemna minor. Additionally, we introduce a new stable transformation protocol for obtaining Lemna minor transgenic lines. We propose that both techniques will facilitate efficient high-throughput genetic manipulation in Lemnaceae.
MATERIALS AND METHODS
Biological material
The clones used in this study were Lemna gibba G3 DWC131 and Lemna minor 8627 (both originally obtained from the Rutgers Duckweed Stock cooperative of Rutgers University). Bacterial strains include Escherichia coli TOP10 (Invitrogen) for cloning purposes, and Agrobacterium tumefaciens CV3101 with pSoup for callus transformation.
Plant and tissue culture conditions
Fronds of both species were cultivated for 2–3 weeks in 50 mL Schenk and Hildebrandt (SH) medium with 10 g/L sucrose at pH 5.6 (Schenk and Hildebrandt, 1972). Fronds and calli were maintained at 23 °C under a 16 hour photoperiod of approximately 30 μmol/m2/s per second of white florescent light.
Tissue cultures were induced from L. minor fronds using a modification of existing protocols (Moon and Stomp, 1997). Fronds were incubated on Induction Medium containing 4.4 g/L Murashige and Skoog (MS) basal salts, 30 g/L sucrose, 5 μM 2,4-Dichlorophenoxyacetic (2,4-D) and 0.5 μM Thidiazuron (TDZ) and 5 g/L of bacteriological agar at pH 5.6. After three to four weeks, light green masses of unorganized cells were selected and transferred to solid Propagation Medium containing 4.4 g/L Murashige and Skoog (MS) basal salts, 30 g/L sucrose, 1 μM 2,4-Dichlorophenoxyacetic (2,4-D) and 2 μM 6-Benzylaminopurine (BAP) and 5 g/L of Agar at pH 5.6. After 7 to 10 days, the fastest growing calli were propagated in fresh media and used in transformation assays.
Transformation and regeneration of L. minor
A. tumefaciens CV3101 carrying the vector of interest was cultured in LB with selective antibiotics and 100 μM acetosyringone at 28 °C to an OD600 of 1.0. Cells were resuspended in 10 mM magnesium sulfate, 10 g/L sucrose and 200 μM acetosyringone and incubated at room temperature for 1 hour. Calli of approximately 3 mm in diameter were submerged in the bacterial suspension for 5 min. Calli were then placed on Propagation Medium with 100 μM acetosyringone and co-cultivated for two days. Transformed calli were transferred to solid Regeneration Medium containing 10 g/L sucrose, 200 mg/L carbenicillin, 500 mg/L cefotaxamin, 10 mg/L of phosphinothricin (PPT) and 5 g/L of Agar at pH 5.6 for two more days.
All the transformations performed in this study use eGFP as a reporter gene. Calli with the greatest number of GFP expressing cells were transferred individually to 75 cm2 cell culture flask with vented cap containing 50 mL of liquid Regeneration Medium and cultured in a shaking incubator at 100 rpm. After 4 weeks, refreshing the media weekly, calli were placed back on solid Regeneration media. Regenerated fronds arising from the callus were transferred to standard growth media and screened for fluorescence.
Lemna miRNA precursor prediction
L. gibba small RNA sequences were mapped to genomic scaffolds from the L. gibba v0.1 assembly (http://www.lemna.org), and filtered to remove t/r/sn/snoRNA, redundant and high copy reads. The sRNAs that passed this filter were tested for their precursor structure using miREAP. Precursors were defined by extending from the potential miRNA on either side, up to 200 nt. After this, two additional filters (strand-bias and top1+top2 ratio) were applied to distinguish miRNA from siRNA loci. Strand-bias is the sum of sRNA abundance on sense strand divided by the total abundance on both strands. Top1+top2 is the proportion of the abundance of top two abundant tags, also referred as the “distribution filter”. The cut-off was picked based on known miRNAs from Arabidopsis and rice. During each step, the remaining number of known Ath-miRNA (miRBase v17) was tracked as an indicator of the efficiency of the filtering.
Artificial microRNA construct design
To automate the amiRNA design process, we installed the WMD3 (http://wmd3.weigelworld.org) webservice on our lemna.org server for use with L. gibba de novo transcriptome assemblies. After identifying transcripts of target genes by homology with A. thaliana, we used WMD3 to generate candidate 21nt mature amiRNA sequences that resemble natural miRNAs while minimizing possible off-target effects to other transcripts (Ossowski et al., 2008).
The 21nt candidates were introduced into a L. gibba pre-miR166a backbone as described (Schwab et al., 2006) and then cloned into a pB7WG2D vector (Karimi et al., 2002) using the Gateway system (Invitrogen).
DNA and RNA isolation
Genomic DNA from L. gibba and L. minor was extracted as previously described (Murray and Thompson, 1980). For the extraction of total RNA from L. minor, the Direct-zol RNA MiniPrep kit (ZYMO RESEARCH Corp.) was used following the instructions of the manufacturer.
Quantitative RT-PCR analysis
Gene expression was analyzed by quantitative PCR using the iQ SYBR-Green (Bio Rad). An L. minor polyubiquitin gene was used as a standard control. The threshold cycle (Ct) values of PCR reactions were obtained from three independent biological replicates with three technical replicates each. The relative quantification of expression levels was performed using the comparative Ct method (Livak and Schmittgen, 2001).
RT-PCR detection of microRNAs
RT-PCR analyses of miRNAs and amiRNAs was carried out as described (Varkonyi-Gasic et al., 2007).
Thermal Asymmetric Interlaced PCR (TAIL-PCR)
To identify the insertion sites of the T-DNA, TAIL-PCR was performed as described (Liu et al., 1995), except that Ex Taq Polymerase (TaKaRa) was used in the first amplification to increase sensitivity.
Chlorophyll quantification
Chlorophyll was extracted in ethanol and quantified by spectrophotometry (Ritchie, 2006) using three biological replicates.
5′-RACE PCR
The RACE PCR was performed using a standard protocol for non-capped RNA. 2–5 μg of total RNA were used for the 5′RACE adapter ligation reaction (T4 RNA ligase 5 U/μl from Ambion). After the ligation an RT PCR was performed using SuperScript® III First-Strand Synthesis SuperMix (Invitrogen™) following the manufacturer indications. Two rounds of nested PCRs were performed using Taq DNA polymerase (NEB). The bands obtained were gel extracted (QIAquick Gel Extraction Kit – QIAGEN), cloned using TOPO® TA Cloning Kit (Invitrogen™) and transformed into One Shot ® TOP10 Competent Cells (Invitrogen™) following the manufacturer indications. White colonies were selected for amplification in liquid medium and plasmid was extracted using the QIAprep Spin Miniprep Kit (QIAGEN). Plasmid DNA was sequenced using a gene specific primer.
RESULTS AND DISCUSSION
Design of the artificial microRNA
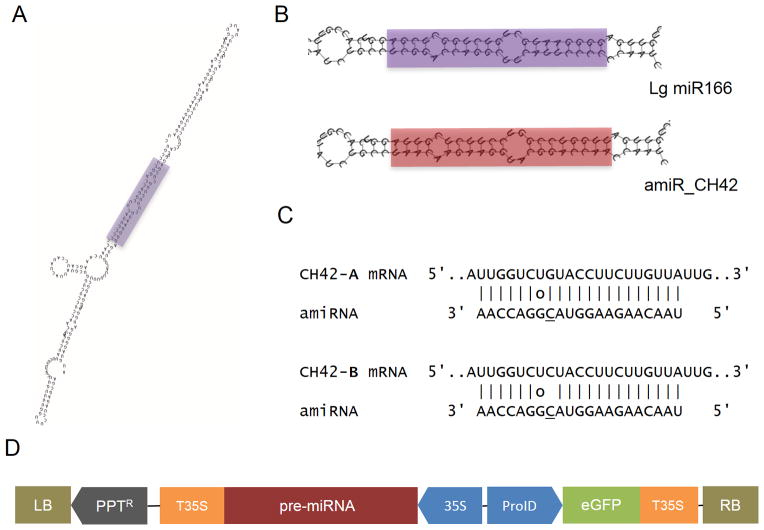
We identified 152 miRNA precursors in small RNA libraries based on hairpin secondary structure (Methods). Among these candidates, 9 miRNAs matched known miRNAs in A. thaliana, while 105 novel miRNAs were identified based on the general criteria for annotating plant miRNA (Meyers et al., 2008). Among the conserved miRNAs, we selected miR166 to be used as backbone. MiR166 is one of the few miRNAs present in both seed plants and mosses, therefore, its fidelity of processing should be more robustly conserved, which is a determining consideration when it comes to designing miRNA/amiRNA substitutions. The sequence of the predicted precursor produced a short hairpin of 138 nt with imperfect complementarity of the passenger sequence (*miR166) in 3 different positions (Figure 1A).
Figure 1. L. gibba pre-miR166a as backbone for amiRNA expression.
Secondary structure of the miRNA precursor Lg-miR166, predicted by RNAfold (ViennaRNA package 2). The sequence corresponding to miRNA/*miRNA duplex is colored in purple (A). Comparison between secondary structures of the endogenous and the modified precursor (B). Predicted binding site of amiRNA to the CH42 mRNA. Circle indicates G:U wobble pairing (C). Schematic diagram of the construct designed to express the amiRNA precursor. The construct contains a selectable marker that confers resistance to PPT as well as an independent GFP expression cassette to track regeneration of the callus (D). Transcription of the amiRNA precursor is driven by the CaMV 35S promoter.
To evaluate the usefulness of this precursor as amiRNA backbone, we designed a proof of concept assay targeting an endogenous gene, the homolog of A. thaliana CHLORINA 42 (CH42). CH42 encodes a magnesium chelatase subunit (CHLI), which is required for chlorophyll biosynthesis (Apchelimov et al., 2007). Its inactivation in Arabidopsis causes yellow - pale green tissue (Koncz et al., 1990). amiRNA knockdown approaches in Arabidopsis have been evaluated by targeting this gene due to this easily recognizable bleaching phenotype (Felippes et al., 2011; Felippes and Weigel, 2009; Werner et al., 2010). For Lemna, we used WMD3 with our L. gibba transcript assembly database to select candidate amiRNAs that would potentially lead to PTGS of CH42. Both the miRNA and the *miRNA from the miR166 precursor were substituted with the amiRNA/*amiRNA. For the *amiRNA we took into account the structural and energetic features of the endogenous pairing, considered to be important for correct processing (Figure 1B). The resulting amiR_CH42 targets the 3′ UTR of the L. minor CH42. However, L. minor has two copies of CH42 with several single nucleotide polymorphisms (SNPs). One of these is located specifically in the binding region, opposite of position 13 of the small RNA (Figure 1C). We used overlapping PCR to generate the amiRNA-containing precursor, and then introduced it into pB7WG2D using Gateway®-assisted cloning (Figure 1D).
Generation of L. minor transgenic lines through callus transformation
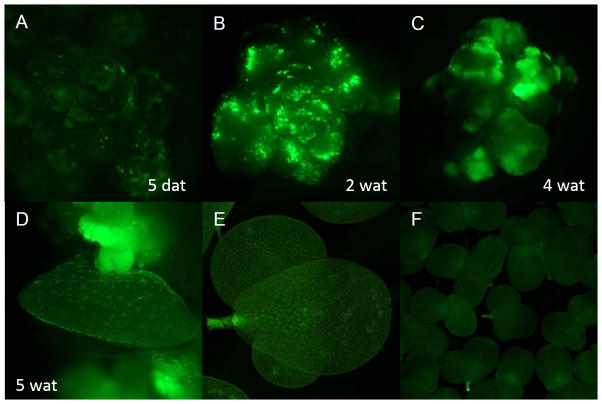
A stable transformation protocol for L. minor using Agrobacterium-mediated gene transfer has previously been described (Stomp and Rajbhandari, 2011; Yamamoto et al., 2001), but low efficiency and long cultivation times limits the utility of this approach, especially for high throughput genetic manipulation. Therefore, we made a number of modifications to the protocol, including the use of a more virulent Agrobacterium strain (CV3101) in a determined infection medium and the use of PPT as the selective agent. We also reduced overall selection and regeneration time by simultaneous execution in liquid media. This method for shortening selection time was first reported in cucumber (Tabei et al., 1994) and may promote uptake of selective agents. In order to visually assist the selection process, we included eGFP, an in vivo reporter gene. Transgenic calli presenting the greatest number of cells expressing GFP after co-cultivation with A. tumefaciens were transferred to selective liquid media (Figure 2A). Progressive growth of eGFP positive cells was observed (Figure 2A–C). After five weeks in selection media, transgenic fronds began to regenerate from transformed calli (Figure 2D).
Figure 2. Stable transformation and regeneration of L. minor callus with a GFP expression construct.
Green fluorescence signal detected at various stages of selection in liquid media (A, B, C). 5 weeks after transformation transgenic fronds began to regenerate from callus (D). Transformed fronds expressing GFP were regenerated with no phenotypic abnormalities (E), and the transgenes were stably maintained through generations (F). day = days after transformation; wat = weeks after transformation.
Individual fronds regenerated from callus were transferred to standard growth media, and normal morphological characteristics were observed after the first frond generation (Figure 2E). Transgenic lines had ubiquitous expression of GFP that was stably maintained for generation after generation (Figure 2F).
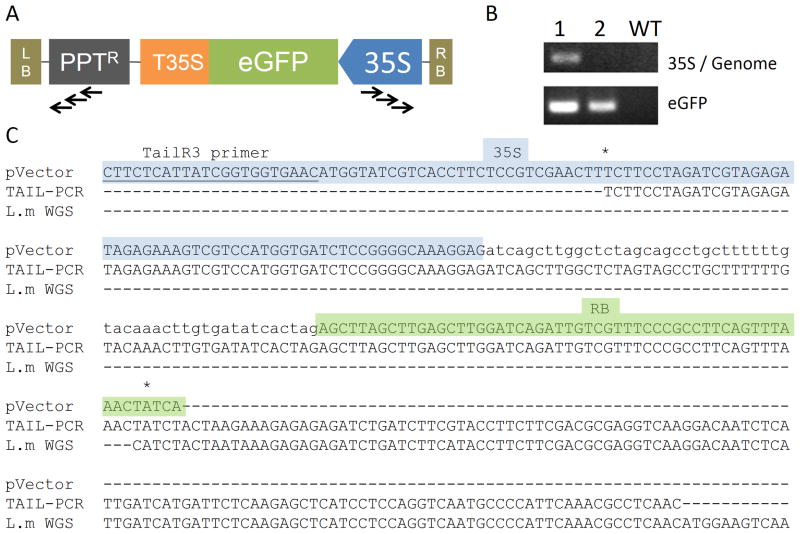
In order to confirm successful integration of the T-DNA into the L. minor genome we performed TAIL-PCR. Using random degenerate and T-DNA specific primers, we screened individual transformation events (Figure 3A). Several different border sequences were detected and then validated using a primer from the T-DNA and another from the flanking genome region. We used internal cassette primers as a control for unique T-DNA insertions (Figure 3B). With this technique we were able recover several single insertion sites that were efficiently mapped to our preliminary sequence contigs of the L. minor genome (Figure 3C).
Figure 3. Verification of T-DNA integration by TAIL-PCR.
Design of primers (arrows) for mapping genomic sequences flanking T-DNA insertions (A). PCR verification of the integration in two different lines and a L. minor wild type (B). Alignments of TAIL-PCR right junction sequences of transgenic L. minor with de novo assembled whole-genome shotgun contigs (C).
Testing the functionality of the amiRNA
To assess the silencing potency of our artificial precursor, we generated transgenic lines containing the amiRNA construct described above for targeting CH42 (Figure 4C-D). Expression of the amiRNA resulted in reduced frond pigmentation (Figure 4C), resembling that observed in A. thaliana knockdown lines (Felippes et al., 2011). To confirm the bleaching phenotype was due to a decrease in chlorophyll content, we extracted chlorophyll from wild type and a transformed regenerant using 3 biological replicates (Methods). A significant decrease of approximately 40% in chlorophyll content was observed (Figure 4E), consistent with previously reported observations in CH42-defective Arabidopsis mutants (Apchelimov et al., 2007; Soldatova et al., 2005).
Figure 4. Phenotype of L. minor expressing amiRNA targeting CH42.
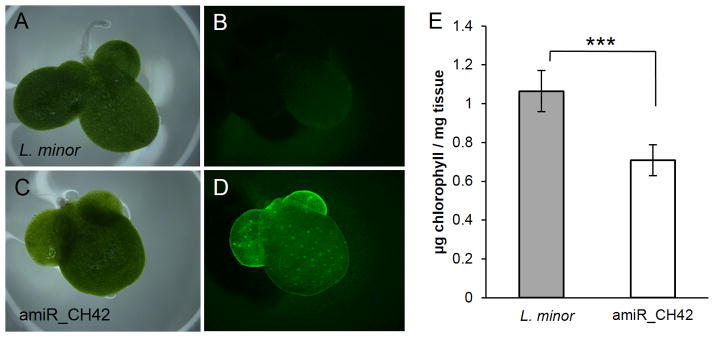
Wild type L. minor (A, B) and transformed amiR_CH42 (C,D) fronds observed under bright field (A, C) and fluorescence (B, D) conditions. Quantification of the chlorophyll content in L. minor and amiR_CH42 (E). The s.d. values are shown, n=3. Stars indicate significance in two-tailed z-test, ***P<0.0001
To estimate the degree of gene silencing, we quantified the accumulation levels of the amiRNA and the expression changes of CH42. Using stem-loop end-point RT-PCR amplification, we showed that our transgenic lines accumulated amiRNA at significant levels (Figure 5A). Once the presence of the amiRNA was confirmed, we further characterized the extent of gene silencing using quantitative reverse-transcription PCR (qRT-PCR) analysis. We observed a significant decrease of approximately 60% in transcript abundance (Figure 5B). Expression level of CH42, relative to α-Tubulin mRNA, was reduced by more than half; which was consistent with the phenotypic observations.
Figure 5. Expression of the amiR_CH42 and down-regulation of CH42.
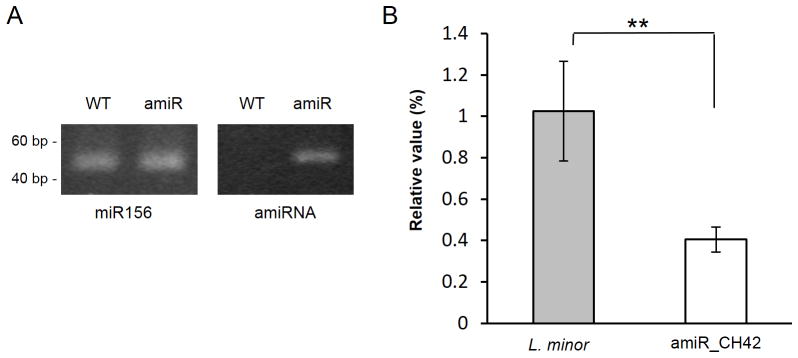
Stem-loop end-point RT-PCR amplification of miRNAs, using endogenous miRNA (miR156) as an internal control (A). Changes in relative expression levels of CH42 mRNA were measured by quantitative RT–PCR and normalized to Tubulin α mRNA (B). The s.d. values for the qRT-PCR are shown, n=9. Stars indicate statistical significance in two-tailed Student’s t-test, **P<0.002
Numerous factors impact the degree of amiRNA-mediated silencing (Alvarez et al., 2006; Schwab et al., 2006) including mismatches at the binding site of the amiRNA, which could reduce or abolish endonucleolytic cleavage of mRNA targets by Argonaute-miRNA endoribonucleases (Molnar et al., 2009). To ensure that amiRNA expression resulted in gene down-regulation, we performed 5′-rapid amplification of cDNA ends (RACE) analysis to detect cleaved mRNA. The RACE products mapped to only one of the CH42 gene copies (CH42A), very close to the predicted cleavage site (Figure 6). All 20 cleavage products mapped downstream of nucleotide position 10/11 of the amiRNA, where Argonaute typically cleaves its targets, which could indicate secondary degradation of cleaved RNAs.
Figure 6. 5′-RACE analysis of the amiR-guided mRNA cleavage.

Alignment of the two CH42 sequences present in L. minor genome with the sequences obtained from the cloning of 3 different bands. 10 out of 10 clones from each band present the sequence depicted here. Sequences corresponding to the amiRCH42 target (green) and the stop codon (purple) are highlighted. Stars indicate polymorphisms between the two different gene copies.
The mismatch at position 14 of the target site in the second copy CH42B seems to have a strong negative impact on the amiRNA-mediated silencing as no CH42B cleavage products were detected by RACE-PCR (Figure 6). Similar mismatches have been shown to reduce silencing efficiency in rice (Warthmann et al., 2008). These data confirm the high specificity of our amiRNA silencing platform, as our amiRNA could specifically direct cleavage of only one of the two copies of the gene.
CONCLUSION
The ease with which duckweed can be axenically cultured, its fast clonal growth rate, and small size are strong arguments for using L. minor as a model system for genetic studies. Lemna has historically been an important model plant, but the development of sophisticated genetic tools for manipulation of Arabidopsis and rice has gradually displaced duckweed research. To enable researchers to take advantage of duckweed, we have initiated studies to develop robust protocols for genetic manipulation. The first step in this process was to develop a facile reproducible transformation protocol. In order to make possible reverse genetic studies we developed a transcript silencing protocol initiated by amiRNA similar to that developed for other model plants. Because it is preferable to use an orthologous miRNA as a backbone (Ossowski et al., 2008) with the endogenous amiRNA system for gene silencing, we developed a system based on a modified miR166 precursor. This provides a powerful platform for reverse genetic studies in the Lemnaceae. Together with an efficient transformation system, it will facilitate genetic studies of duckweed that may lead to improvements in its agronomic performance and economic value.
Supplementary Material
Acknowledgments
We thank Vitaly Citovsky for advice on Agrobacterium strains and infection media, and Leo Guthart for his support. This work was supported by a grant from US Department Of Energy (DE-EE0003298) to R.A.M. and J.S.
References
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. The Plant Cell Online. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apchelimov A, Soldatova O, Ezhova T, Grimm B, Shestakov S. The analysis of the ChlI 1 and ChlI 2 genes using acifluorfen-resistant mutant of Arabidopsis thaliana. Planta. 2007;225:935–943. doi: 10.1007/s00425-006-0390-1. [DOI] [PubMed] [Google Scholar]
- Axtell MJ. Classification and Comparison of Small RNAs from Plants. Annual Review of Plant Biology. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- Cox KM, Sterling JD, Regan JT, Gasdaska JR, Frantz KK, Peele CG, Black A, Passmore D, Moldovan-Loomis C, Srinivasan M. Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nature Biotechnology. 2006;24:1591–1597. doi: 10.1038/nbt1260. [DOI] [PubMed] [Google Scholar]
- Felippes FF, Ott F, Weigel D. Comparative analysis of non-autonomous effects of tasiRNAs and miRNAs in Arabidopsis thaliana. Nucleic acids research. 2011;39:2880–2889. doi: 10.1093/nar/gkq1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO reports. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science (New York, NY) 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nature Biotechnology. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Koncz CS, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. The EMBO journal. 1990;9:1337. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant Journal. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2^(−ΔΔCT). Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe DC, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. Criteria for annotation of plant MicroRNAs. The Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe DC. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. The Plant Journal. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Moon HK, Stomp AM. Effects of medium components and light on callus induction, growth, and frond regeneration in Lemna gibba (Duckweed) In Vitro CellDevBiol-Plant. 1997;33:20–25. [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LV, Cox KM, Ke JS, Peele CG, Dickey LF. Genetic engineering of a Lemna isoleucine auxotroph. Transgenic Res. 2012;21:1071–1083. doi: 10.1007/s11248-012-9594-2. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. The Plant Journal. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Ritchie RJ. Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynthesis Research. 2006;89:27–41. doi: 10.1007/s11120-006-9065-9. [DOI] [PubMed] [Google Scholar]
- Schenk RU, Hildebrandt AC. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Canadian Journal of Botany. 1972;50:199–204. [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatova O, Apchelimov A, Radukina N, Ezhova T, Shestakov S, Ziemann V, Hedtke B, Grimm B. An Arabidopsis mutant that is resistant to the protoporphyrinogen oxidase inhibitor acifluorfen shows regulatory changes in tetrapyrrole biosynthesis. Molecular Genetics and Genomics. 2005;273:311–318. doi: 10.1007/s00438-005-1129-6. [DOI] [PubMed] [Google Scholar]
- Stomp AM, Rajbhandari N. Genetically engineered duckweed (EP Patent. 2011;1:037–523. [Google Scholar]
- Tabei Y, Nishio T, Kurihara K, Kanno T. Selection of Transformed Callus in a Li uid Medium and Regeneration of Transgenic Plants in Cucumber (C ucumls sa lvus L.) Breeding Science. 1994;44:47–51. [Google Scholar]
- Urbanska-Worytkiewicz K. Cytological variation within Lemna L. Aquatic Botany. 1975;1:377–394. [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Hervé P. Highly specific gene silencing by artificial miRNAs in rice. PLoS One. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Wollmann H, Schneeberger K, Weigel D. Structure Determinants for Accurate Processing of miR172a in Arabidopsis thaliana. Current Biology. 2010;20:42–48. doi: 10.1016/j.cub.2009.10.073. [DOI] [PubMed] [Google Scholar]
- Xu J, Cui W, Cheng J, Stomp AM. Production of high-starch duckweed and its conversion to bioethanol. Biosystems Engineering. 2011;110:67–72. [Google Scholar]
- Xu J, Zhao H, Stomp AM, Cheng JJ. The production of duckweed as a source of biofuels. Biofuels. 2012;3:589–601. [Google Scholar]
- Yamamoto YT, Rajbhandari N, Lin X, Bergmann BA, Nishimura Y, Stomp AM. Genetic transformation of duckweed Lemna gibba and Lemna minor. In Vitro CellDevBiol-Plant. 2001;37:349–353. [Google Scholar]
- Yan Y, Candreva J, Shi H, Ernst E, Martienssen RA, Schwender J, Shanklin J. Survey of the total fatty acid and triacylglycerol composition and content of 30 duckweed species and cloning of a Delta6-desaturase responsible for the production of gamma-linolenic and stearidonic acids in Lemna gibba. BMC plant biology. 2013;13:201. doi: 10.1186/1471-2229-13-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.