Abstract
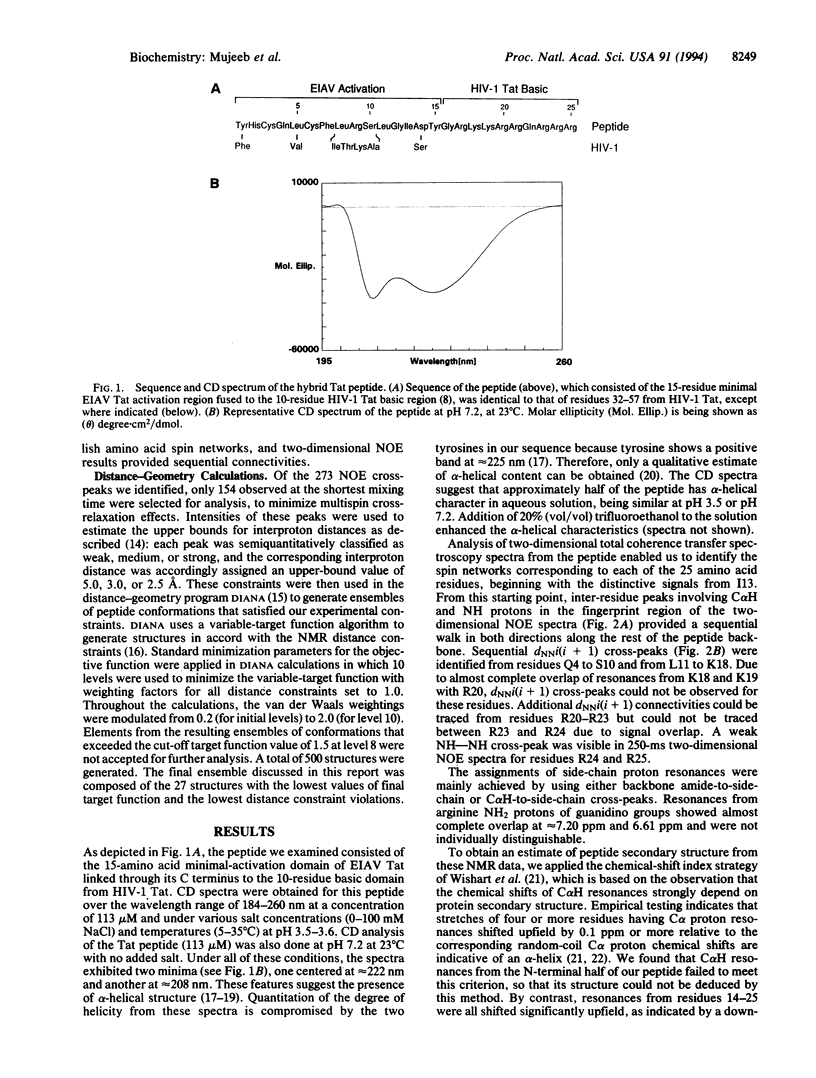
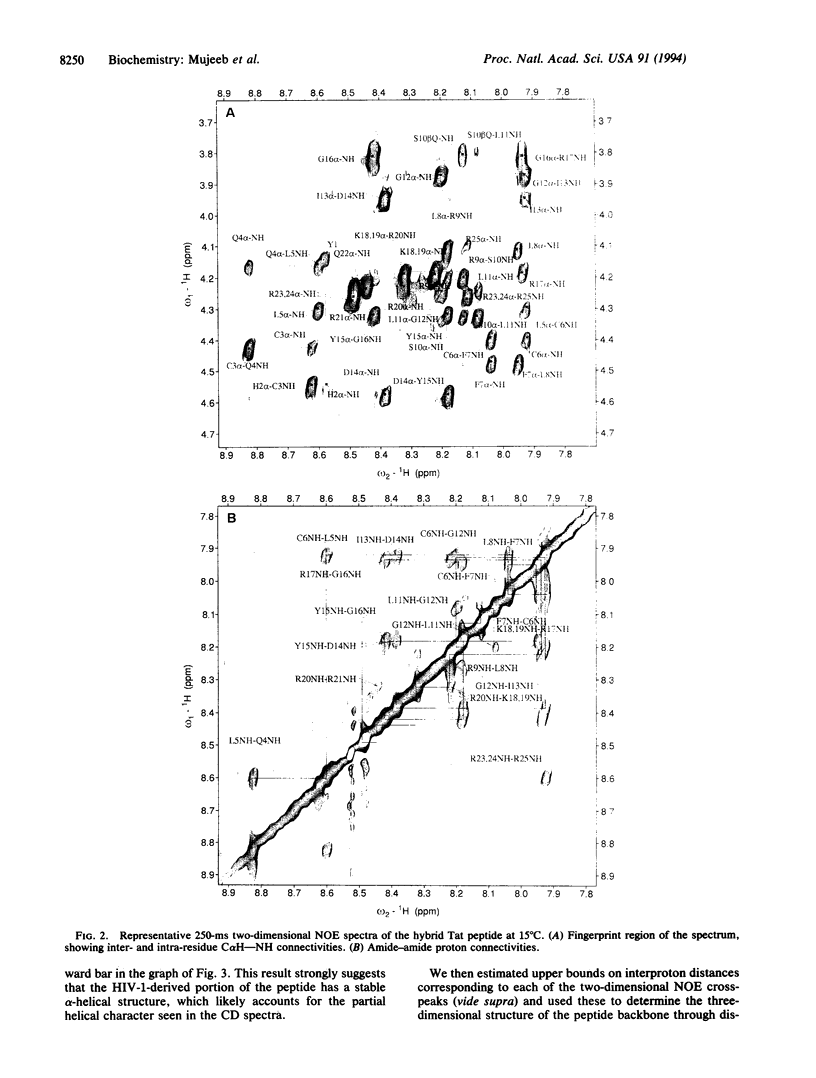
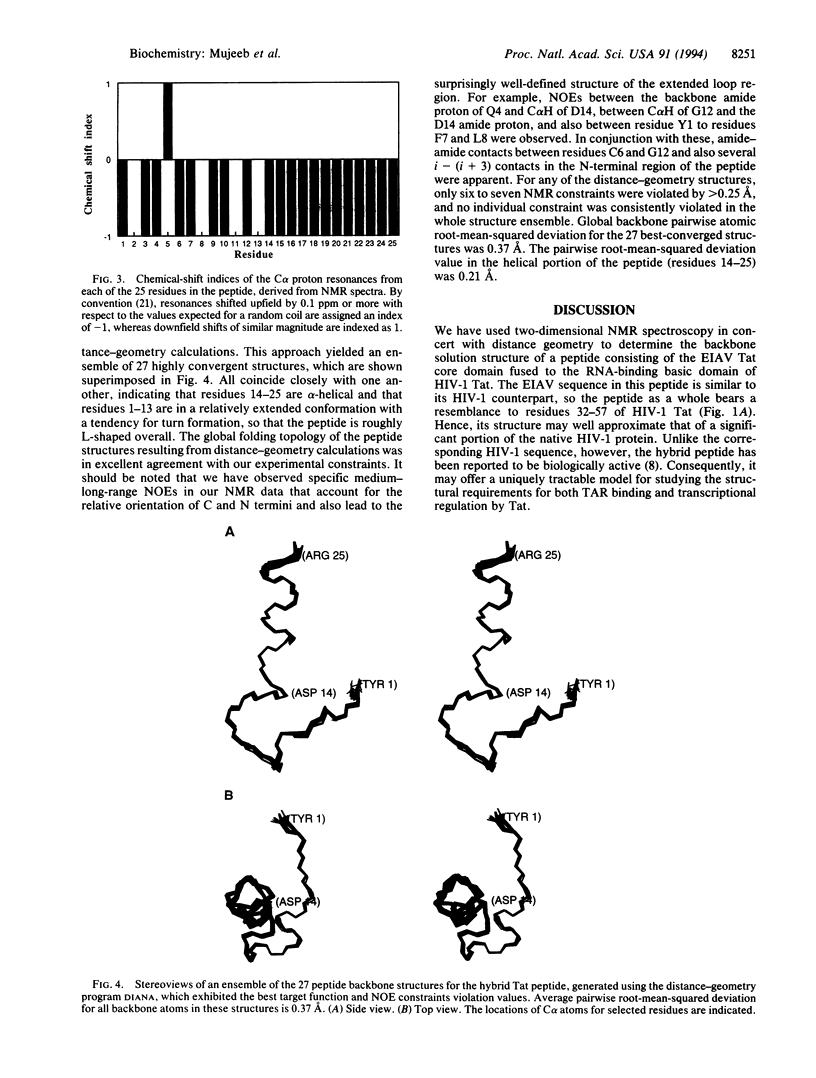
The Tat protein of human immunodeficiency virus type 1 enhances transcription by binding to a specific RNA element on nascent viral transcripts. Binding is mediated by a 10-amino acid basic domain that is rich in arginines and lysines. Here we report the three-dimensional peptide backbone structure of a biologically active 25-mer peptide that contains the human immunodeficiency virus type 1 Tat basic domain linked to the core regulatory domain of another lentiviral Tat--i.e., that from equine infectious anemia virus. Circular dichroism and two-dimensional proton NMR studies of this hybrid peptide indicate that the Tat basic domain forms a stable alpha-helix, whereas the adjacent regulatory sequence is mostly in extended form. These findings suggest that the tendency to form stable alpha-helices may be a common property of arginine- and lysine-rich RNA-binding domains.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler A. J., Greenfield N. J., Fasman G. D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- Auer M., Gremlich H. U., Seifert J. M., Daly T. J., Parslow T. G., Casari G., Gstach H. Helix-loop-helix motif in HIV-1 Rev. Biochemistry. 1994 Mar 15;33(10):2988–2996. doi: 10.1021/bi00176a031. [DOI] [PubMed] [Google Scholar]
- Bradley E. K., Thomason J. F., Cohen F. E., Kosen P. A., Kuntz I. D. Studies of synthetic helical peptides using circular dichroism and nuclear magnetic resonance. J Mol Biol. 1990 Oct 20;215(4):607–622. doi: 10.1016/S0022-2836(05)80172-X. [DOI] [PubMed] [Google Scholar]
- Braun W., Go N. Calculation of protein conformations by proton-proton distance constraints. A new efficient algorithm. J Mol Biol. 1985 Dec 5;186(3):611–626. doi: 10.1016/0022-2836(85)90134-2. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Derse D., Carvalho M., Carroll R., Peterlin B. M. A minimal lentivirus Tat. J Virol. 1991 Dec;65(12):7012–7015. doi: 10.1128/jvi.65.12.7012-7015.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Güntert P., Braun W., Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991 Feb 5;217(3):517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kashanchi F., Piras G., Radonovich M. F., Duvall J. F., Fattaey A., Chiang C. M., Roeder R. G., Brady J. N. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994 Jan 20;367(6460):295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989 Oct 20;59(2):283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Loret E. P., Vives E., Ho P. S., Rochat H., Van Rietschoten J., Johnson W. C., Jr Activating region of HIV-1 Tat protein: vacuum UV circular dichroism and energy minimization. Biochemistry. 1991 Jun 18;30(24):6013–6023. doi: 10.1021/bi00238a027. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Sharp P. A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991 Dec;10(13):4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Peterlin B. M. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990 Aug 24;62(4):769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- Slice L. W., Codner E., Antelman D., Holly M., Wegrzynski B., Wang J., Toome V., Hsu M. C., Nalin C. M. Characterization of recombinant HIV-1 Tat and its interaction with TAR RNA. Biochemistry. 1992 Dec 8;31(48):12062–12068. doi: 10.1021/bi00163a014. [DOI] [PubMed] [Google Scholar]
- Southgate C., Zapp M. L., Green M. R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990 Jun 14;345(6276):640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- Spolar R. S., Record M. T., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994 Feb 11;263(5148):777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Sticht H., Willbold D., Bayer P., Ejchart A., Herrmann F., Rosin-Arbesfeld R., Gazit A., Yaniv A., Frank R., Rösch P. Equine infectious anemia virus Tat is a predominantly helical protein. Eur J Biochem. 1993 Dec 15;218(3):973–976. doi: 10.1111/j.1432-1033.1993.tb18455.x. [DOI] [PubMed] [Google Scholar]
- Tan R., Chen L., Buettner J. A., Hudson D., Frankel A. D. RNA recognition by an isolated alpha helix. Cell. 1993 Jun 4;73(5):1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- Vaishnav Y. N., Wong-Staal F. The biochemistry of AIDS. Annu Rev Biochem. 1991;60:577–630. doi: 10.1146/annurev.bi.60.070191.003045. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Willbold D., Krüger U., Frank R., Rosin-Arbesfeld R., Gazit A., Yaniv A., Rösch P. Sequence-specific resonance assignments of the 1H-NMR spectra of a synthetic, biologically active EIAV Tat protein. Biochemistry. 1993 Aug 24;32(33):8439–8445. doi: 10.1021/bi00084a008. [DOI] [PubMed] [Google Scholar]
- Williamson M. P., Havel T. F., Wüthrich K. Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol. 1985 Mar 20;182(2):295–315. doi: 10.1016/0022-2836(85)90347-x. [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Sykes B. D., Richards F. M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992 Feb 18;31(6):1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]




