Abstract
X-ray Repair Cross Complementing protein 1 (XRCC1) plays an important role in base excision DNA repair (BER) as a scaffolding protein for BER enzymes. BER is one of the basic DNA repair pathways repairing greater than 20,000 endogenous lesions per cell per day. Proper functioning of XRCC1, one of the most important players in BER, was suggested to be indispensable for effective DNA repair. Despite accumulating evidence of an important role that XRCC1 plays in maintaining genomic stability, the relationship between one of its most predominant variants, R280H (rs25489), and cancer prevalence remains ambiguous. In the current study we functionally characterized the effect of the R280H variant expression on immortal non-transformed mouse mammary epithelial C127 and human breast epithelial MCF10A cells. We found that expression of R280H results in increased focus formation in mouse C127 cells and induces cellular transformation in human MCF10A cells. Cells expressing R280H showed significantly increased levels of chromosomal aberrations and accumulate double strand breaks in the G1 cell cycle phase. Our results confirm a possible link between R280H and genomic instability and suggest that individuals carrying this mutation may be at increased risk of cancer development.
1. Introduction
Endogenous DNA damage, as a result of the presence of reactive oxygen and nitrogen species (RONs), induces DNA damage at a rate of 20,000-30,000 lesions per cell per day (1). Much of this damage is repaired by base excision repair (BER). BER is initiated by lesion-specific DNA glycosylases that recognize and remove damaged bases (for a review see (2)). After removal of the damaged base, bifunctional glycosylases catalyze removal of the resulting abasic site (AP) by either ß or ßδ elimination. If the base is excised by a monofunctional DNA glycosylase, apyrimidinic endonuclease 1 processes the AP site. AP site removal is followed by end remodeling, if necessary, and filling in of the single nucleotide gap by DNA polymerase beta (Pol ß). The X-Ray Cross Complementing 1 (XRCC1)-Ligase 3α (Lig3α) complex seals the nick. Due to its repair of 20,000-30,000 lesions per cell per day, the BER pathway plays a major role in maintaining genomic stability.
XRCC1 interacts with a number of different proteins that function in BER and single-strand break repair (SSBR) including UNG2, NEIL2, OGG1, MPG, NTH1 (3-5), Pol ß, PARP1, APE1, LIG3a, polynucleotide kinase, and PCNA (6-10). XRCC1 functions during BER and (SSBR) by acting as a scaffold to bring proteins into proximity to one another in order to catalyze DNA repair and ensure efficient handoff of intermediate substrates (for reviews see (11,12)). XRCC1 deficient cells exhibit sensitivity to a number of DNA damaging agents including methylmethane sulfonate (MMS), ethylmethane sulfonate (EMS), hydrogen peroxide and camptothecin (13-18) as a result of defective or inefficient joining of single-strand breaks. Cells deficient in XRCC1 also exhibit genomic instability (13,19-23).
The 1000 Genomes Project reports a total of 6,469 variations in the XRCC1 gene, including missense mutations, indels -, and variation in 3′ and 5′ UTRs. The rs25489 single nucleotide polymorphism (SNP) is present with a minor allele frequency of 6% and is predominantly found in individuals of Asian ancestry. This SNP encodes the R280H XRCC1 variant, which has been a topic of epidemiological studies focused predominantly on the relationship between the presence of this variant to the development of cancer. R280H has been found to be associated with a number of cancers including bladder, gastric, hepatocellular, breast, and with cancer in general (24-27). The R280H protein was found to exhibit reduced focal localization in response to micro-irradiation (28). This variant was also found to dissociate more rapidly than WT XRCC1 from sites of DNA damage induced by micro-irradiation (29).
The goal of this study was to determine if the R280H germline variant possesses a functional phenotype related to cancer. We found that expression of R280H in both mouse and human non-transformed cells induces genomic instability and cellular transformation. Taken together with epidemiological studies, our results imply that subsets of individuals who harbor this variant could be at increased risk for the development of cancer.
2. Material and Methods
2.1 Plasmids and Cloning
To generate the pRVYTet-hXRCC1 constructs the human XRCC1 sequence was PCR amplified using a downstream primer containing the hemagglutinin (HA) tag and then cloned into the pRVYTet retroviral vector as described (30,31). The single base mutation resulting in the R280H variant was introduced into the human WT XRCC1 cDNA sequence using site-directed mutagenesis (Stratagene) following the manufacturer’s protocol.
2.2 Cell Lines and Cell Culture
All cell lines used in the present study were grown at 37°C in a 5% CO2 humidified incubator.
C127 cells are a non-transformed epithelial cell line derived from a mammary tumor of an RIII mouse (American Type Culture Collection, ATCC). The cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS, Gemini Bio Products) and 1% penicillin-streptomycin (Invitrogen).
MCF10A cells are diploid immortalized, non-transformed mammary epithelial cells derived from a female with fibrocystic disease (ATCC). These cells were maintained in DMEM/F12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 1% penicillin-streptomycin (Invitrogen), epidermal growth factor (20 ng/ml)(Gemini Bio Products), hydrocortisone (0.5 μg/ml)(Sigma-Aldrich), cholera toxin (100 ng/ml) (Sigma-Aldrich), insulin (10 μg/ml) (Sigma-Aldrich).
The GP2-293 virus packaging cell line (Clontech) was used for retrovirus preparation. These cells were maintained in Dulbecco modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Gemini Bio Products), 1% L-glutamine (Invitrogen), 1% penicillin-streptomycin (Invitrogen), and 1mM HEPES (Invitrogen).
2.3 Transfection, Infection and Expression Analysis
To generate retrovirus containing the WT or R280H XRCC1 construct, the GP2-293 packaging cell line was co-transfected with the pRVYTet construct and pVSVG plasmid using the method as described in (32) GP2-293 cells were incubated for 72h before virus was harvested. MCF10A cells were grown to 30% confluence and infected with retrovirus in the presence of 4 μg/ml polybrene (American Bioanalytical) (30). For selection of pools, cells were split 1:3 the day of infection and integrants were selected with Hygromycin B (15 μg/ml). Single cell clones were selected using cloning rings.
Expression of exogenous HA-tagged hXRCC1 in resulting cell lines was confirmed by Western blot. WT and R280H expressing cells were passaged in parallel and harvested at 80-90% confluence by scraping with hot SDS Loading Buffer (50mM Tris pH 6.8, 10 mM DTT, 2% SDS, 10% glycerol). Lysates were boiled for 5 min, resolved on a 10% SDS-polyacrylamide gel, and transferred to the nitrocellulose membrane. Exogenous XRCC1 was detected using mouse monoclonal anti-HA antibody (Covance, MMS101P), and mouse monoclonal anti-β-actin antibody (Sigma-Aldrich, A54441) was used as a loading control. Blots were imaged using BioRad ChemiDoc instrument, and quantified by Image Lab software. Cell lines (either pools or single cell clones) expressing equal levels of WT and R280H hXRCC1 were selected for subsequent studies.
2.4 Genomic Instability Analysis
MCF10A pools expressing either WT or R280H hXRCC1 were plated at 106 per 10cm dish and grown overnight. The next day fresh media with colcemid (0.1 μg/ml)(Invitrogen) was added to arrest cells in mitosis. Three hours later cells were trypsinized, collected by centrifugation, washed with PBS, and resuspended dropwise in 0.8% sodium citrate. Following lysis, cells were incubated at 37°C for 30 minutes before fixing in Carnoy’s Fixative (75% methanol, 25% acetic acid). Cells were dropped onto microscope slides, dried, and stained with 5% KaryoMax Giemsa stain (Invitrogen). Well spread metaphases were identified under ×100 objective (Zeiss) and images were taken using Spot Camera software (Diagnostic Instruments). Metaphase spreads were scored by eye for chromosomal fusions, breaks, and fragments as described (33).
2.5 Cellular Transformation
The anchorage independent growth assay was conducted as described in (32). Briefly, MCF10A cells expressing either WT or R280H hXRCC1 were mixed with media containing 0.7% noble agar (USB) and poured onto a layer of media containing 1% noble agar in a well of a 6-well dish. Cells were fed twice weekly for 4 weeks. The number of colonies present in each of five microscope fields per well from a total of 6 wells per experiment were counted after 4 weeks of growth.
2.6 Cellular Proliferation Assay
To determine the effect of R280H or WT hXRCC1 expression on cellular proliferation, MCF10A single cell clones were plated into 10 cm dishes at a density of 50,000 cells/dish and counted using automated cell counter (Nexelcom) every day for 5 consecutive days as described (33). Experiments were performed in triplicate and the data were plotted as change in cell number per day.
2.7 Flow Cytometry
MCF10A cells expressing either WT or R280H hXRCC1 were plated at a density of 106 cells per 10 cm dish and allowed to attach overnight. For H2O2 treatment, plates were incubated with 30 mM H2O2 in serum free media for 30 minutes on ice, then replaced with fresh complete media and allowed to recover for 1, 2, and 6 hours as described (34). For MMS treatment, cells were incubated with 2 mM MMS for 2h and then allowed to recover for 1, 2, and 4 hours (33). Cells were then trypsinized and rinsed with PBS, pelleted and resuspended by adding 70% ice cold ethanol dropwise while vortexing. Cells were fixed overnight at −20°C. To detect double strand breaks, cells were first incubated with primary phospho-γH2AX antibody (Millipore 05-636) 1:500 overnight at 4°C, washed twice with PBS, then incubated with anti-mouse secondary antibody conjugated to FITC 1:500 for 1h at room temperature in dark. Finally, cells were washed twice with PBS and resuspended in 200μl PI/RNase staining buffer (BD Pharmingen). Fluorescence was analyzed by flow cytometry using the BD FACSCalibur and analyzed using FlowJo 8.8.6 software.
2.8 Immunofluorescence microscopy
For γH2AX/53BP1 immunofluorescence analysis, cells were plated in 8-well chamber slides (BD Falcon) at 105cells/ml and cultured overnight. Then cells were treated with 2mM MMS for 1h and allowed to recover for 2 hours. Cells were fixed for 15 minutes at room temperature (1% paraformaldehyde/2% sucrose in PBS), permeabilized in pre-chilled 100% methanol, and stained with corresponding antibodies. First, cells were incubated with a primary mouse anti-phospho-H2AX antibody (Millipore, 05-636) for 2 hours at room temperature, then with a primary rabbit anti-53BP1 antibody (Santa Cruz Biotech, sc-22760) at 4C overnight. The next day cells were labeled with an Alexa Fluor-488 conjugated goat anti-rabbit and Alexa Fluor-546 conjugated goat anti-mouse antibodies for 2 hours at room temperature. Resulting slides were mounted using DAPI containing SlowFade Gold anti-fade mounting solution (Invitrogen, S36938) for cell nuclei visualization. Analysis was performed on an inverted fluorescence microscope (Axiovert 200, Carl Zeiss) using the Plan-Apochromat 63× objective with immersion oil. Images were processed using ImageJ software (NIH).
2.9 Statistics
All statistics were performed using GraphPad Prism version 5 (GraphPad Software, San Diego, CA). Data were presented as mean ± SEM. Two-tailed t test, One-way or Two-way analysis of variance (ANOVA) were applied as appropriate and Tukey’s, Sidak’s or Bonferroni’s multiple comparison tests were used to determine significant differences between the means of each group.
3. Results
3.1 Expression of R280H XRCC1 in mouse C127 cells results in increased focus formation
Several studies have suggested that the R280H XRCC1 germline variant is associated with human cancer. We recently developed methods to determine if expression of germline variants of genes in the BER pathway confer cancer-associated phenotypes to cells in which they are expressed (33,34). Our strategy is to express either the germline variant or the WT protein using a Tet off system and the pRVY-Tet vector (32) in non-transformed but immortalized epithelial cells and to subsequently characterize the levels of genomic instability and cellular transformation.
First we infected immortalized but non-transformed C127 mouse mammary epithelial cells with retroviruses carrying either WT or the R280H XRCC1 variant in the pRVY-Tet vector. In the retroviral vectors, the tetO/CMV promoter drives expression of XRCC1 protein in a Tet-repressible manner and an internal SV40 early promoter drives expression of Hygromycin (Hyg) resistance gene (32). We selected cell pools resistant to Hyg and then selected single colonies of C127 cells expressing either WT or R280H XRCC1 and expanded them into stable cell lines. The HA-tagged XRCC1 constructs (either WT or R280H) were expressed and cell extracts from early passage clonal cell lines were subjected to Western blotting using monoclonal anti-HA antibody to reveal exogenous XRCC1. Figure 1A shows that the mouse clones express either the WT or R280H XRCC1 variant at levels similar to each other.
Figure 1. Expression of R280H hXRCC1 results in increased focus formation in mouse C127 epithelial cells.
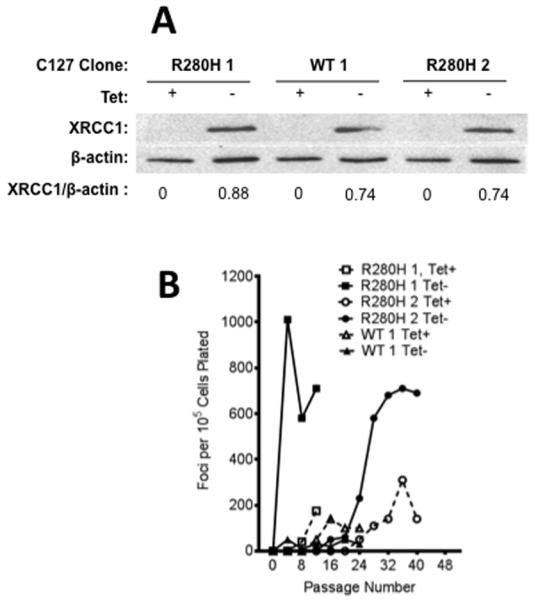
A. Western blotting of XRCC1 expression in C127 clones #1 and #2 of R280H and clone #1 of WT XRCC1 was performed on cellular extracts and antibodies raised against monoclonal XRCC1 were used to reveal expression of exogenous XRCC1. Lanes 1, 3, and 5 show expression of XRCC1 in the presence of Tet (not detected) while lanes 2, 4, and 6 show expression in absence of Tet. Quantification of the bands showed similar levels of XRCC1 expression in all clones. B. Focus formation assay with two clonal C127 cell lines either inducing (Tet−, filled symbols, solid line) or not inducing (Tet+, open symbols, dashed line) expression of exogenous WT or R280H XRCC1. Both individual clones carrying R280H show a significant increase in focus formation under inducing (Tet−) conditions while expression of exogenous WT XRCC1 resulted in very low levels of focus formation.
We monitored cellular transformation by characterizing the ability of the mouse C127 cells expressing either WT or R280H XRCC1 to form foci. Three different clones of C127 cells carrying either WT (WT#1) or R280H (clone R280H#1 and clone R280H#2) XRCC1 were serially passaged in media with or without Tet, to repress or induce expression, respectively, of exogenous XRCC1. The level of focus formation was assessed every four passages. Only clones expressing the R280H variant showed significant focus formation whereas expression of exogenous WT XRCC1 did not result in a significant increase in the number of foci even at late passages (Figure 1B). Focus formation is dependent upon expression of the protein, as cells that were not induced to express R280H XRCC1 by growth in medium containing Tet exhibited a lower level of focus formation versus cells induced to express R280H.
3.2 Expression of R280H XRCC1 induces cellular transformation in human MCF10A cells
Next, we assessed cellular transformation in the human immortalized, non-transformed mammary epithelial MCF10A cells. Similarly to the C127 mouse cell line, MCF10A cells were infected with retrovirus carrying the pRVYTet vector with HA-tagged WT or R280H XRCC1. We selected and expanded stable pools expressing either WT or the R280H variant at similar levels as shown in Figure 2A. We also selected single colonies of MCF10A cells expressing either WT or R280H XRCC1 and expanded them into stable cell lines all expressing similar amounts of exogenous XRCC1 as shown in Figure 2B.
Figure 2. Expression of R280H hXRCC1 induces cellular transformation in human MCF10A cells.
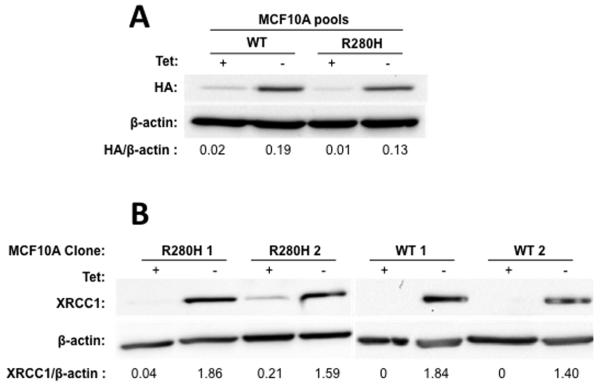
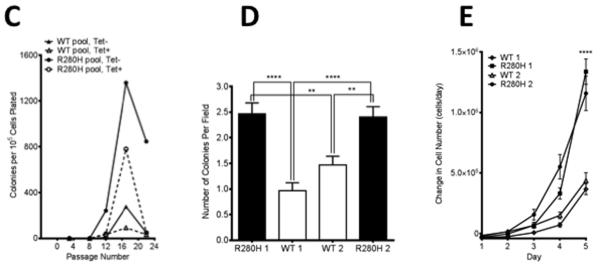
Western blotting was performed on cellular extracts and antibodies raised against the HA-tag or/and monoclonal anti-XRCC1 were used to reveal expression of exogenous XRCC1. A. WT and R280H pools. Lanes 1 and 3 show WT and R280H pools, respectively, grown in the presence of Tet. Lanes 2 and 4 are WT and R280H pools, respectively, grown in the absence of Tet. Quantification of the bands showed that the corresponding pools expressed similar levels of WT and R280H exogenous XRCC1. B. WT and R280H clones. Expression of WT and R280H XRCC1 is shown in the presence (lanes 1, 3, 5, 7) or absence (lanes 2, 4, 6, 8) of Tet. Quantification of the bands revealed similar levels of XRCC1 expression in all clones. C. Anchorage independent growth assay with MCF10A pools with either induced (WT Tet−, R280H Tet−) or not induced (WT Tet+, R280H Tet+) exogenous XRCC1 expression. D. Anchorage independent growth assay with four MCF10A clonal cell lines expressing either WT (Clones 1 and 2, white columns) or R280H (Clones 1 and 2, black columns) at passage 12. The mean number of colonies per field ± S.E is plotted on the Y-axis. E. Proliferation rate assay with four MCF10A clonal cell lines expressing either WT or R280H grown in the absence of tet. ** and **** denote p < 0.01 and 0.0001, respectively.
The ability of the constructs expressing either WT or R280H XRCC1 to induce cellular transformation was assessed by characterizing the formation of colonies in soft agar. First, the numbers of colonies formed by WT- or R280H -expressing MCF10A pools were assessed every four passages. As shown on Figure 2C, the R280H-expressing pool started to form colonies in soft agar as early as passage 8 and reached a value of approximately 1,500 colonies per 105 cells plated by passage 16. Significantly fewer colonies were observed in the WT expressing pool relative to the R280H expressing pool. A similar result was obtained with MCF10A clonal cell lines (Figure 2D). At passage 12, both Clones 1 and 2 of cells expressing R280H XRCC1 exhibit significantly increased anchorage independent growth when compared to Clones 1 and 2 of WT XRCC1, as shown in Figure 2D. We also demonstrate that R280H Clones 1 and 2 exhibit significantly increased rates of cellular proliferation, another hallmark of transformed cells, as shown in Figure 2E. Taken together, these results show that expression of R280H XRCC1 in immortal but non-transformed mouse or human epithelial cells confers cancer-associated phenotypes.
3.3 Expression of R280H hXRCC1 in MCF10A cells results in genomic instability
To determine the mechanism of cellular transformation, we characterized the ability of either WT or R280H XRCC1 expression in human MCF10A cells to induce chromosomal aberrations. Chromosomal aberrations can arise from aberrant BER through the accumulation of BER intermediates, including DNA breaks. Therefore we wished to determine if expression of the exogenous R280H variant results in any changes in chromosomal appearance compared to the WT XRCC1. We counted different types of aberrations, including chromosomal breaks, fusions, and fragments in metaphase spreads prepared from early passage cells expressing either WT or R280H XRCC1 (examples in Figure 3A and B). A total of at least 50 metaphase spreads were scored for each cell line. Cells expressing R280H have significantly increased levels of breaks and fragments, as shown in Figure 3C. These results demonstrate the R280H XRCC1 induces genomic instability when expressed in the human MCF10A cells, providing a possible mechanism leading to cellular transformation.
Figure 3. Expression of R280H hXRCC1 in MCF10A cells results in an increased level of chromosomal aberrations.
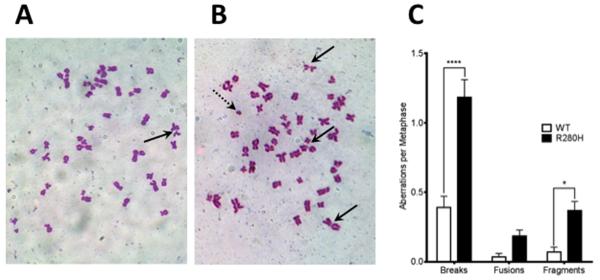
Representative metaphase spreads of MCF10A expressing WT (A.) or R280H hXRCC1 (B.). Chromosomal breaks are shown with the solid black arrow and fragments are shown with the dotted arrow. C. Numbers of aberrations per metaphase. A total of at least 50 metaphases were scored for each cell line. Data are plotted as mean ± S.E. * and **** denote p < 0.05 and 0.0001, respectively.
3.4 MCF10A cells expressing R280H hXRCC1 accumulate double strand breaks in the G1 cell cycle phase
The increased levels of chromosomal aberrations we observe in cells expressing R280H XRCC1 indicates that double-strand breaks (DSBs) may be present at increased levels in these cells. To determine if this was the case, we treated cells with either H2O2 or methylmethane sulfonate (MMS) to initiate BER, and monitored the levels of γH2AX, a marker that is reflective of the presence of DSBs, as a function of cell cycle. The levels of γH2AX were assessed just after treatment and as a function of recovery from treatment. Strikingly, the levels of γH2AX are significantly increased during G1 in cells expressing R280H XRCC1 compared to WT-expressing cells, for both treatments, as shown in as shown in Figure 4A (H2O2) and 4B (MMS).. DSB continue to be present in R280H expressing cells at significantly higher levels even 2-4 hours after recovery.
Figure 4. MCF10A cells expressing R280H hXRCC1 accumulate double strand breaks in the G1 cell cycle phase.
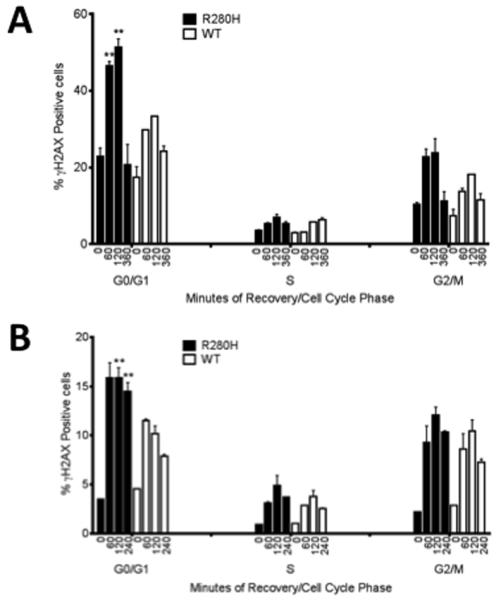
MCF10A pools expressing either WT or R280H hXRCC1 were treated with (A) 30 mM H2O2 for 30 min on ice and allowed to recover for 1, 2, and 6 hours or (B) 2mM MMS for 2 hours at 37°C and allowed to recover for 1, 2, and 4 hours. Cells were stained with FITC-conjugated γH2AX antibody and propidium iodide to assess the levels of double-strand breaks and the cell cycle phase, respectively, and analyzed by flow cytometry. Data are plotted as mean ± S.E. ** denotes p < 0.01.
To confirm that the increased levels of γH2AX observed by flow cytometry indeed correspond to increased levels of double strand breaks, immunofluorescence staining of MMS treated MCF10A cells was performed for the two major DSB markers, γH2AX and 53BP1, as described in the Materials and Methods section. γH2AX/53BP1 co-localizing foci (yellow staining in Figure 5) were counted and data were compared between WT and R280H hXRCC1 expressing cells. A statistically significant increase in the number of DSBs was observed in the case of cells expressing the R280H variant.
Figure 5. Expression of R280H hXRCC1 in MCF10A cells results in an increased level of double strand breaks.
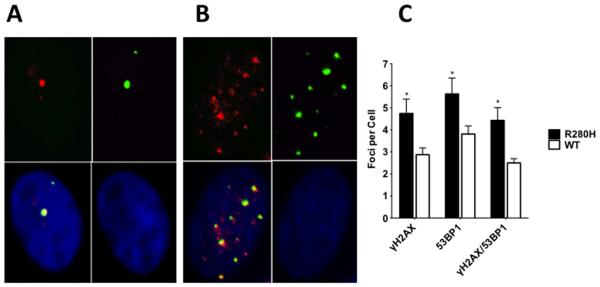
Representative γH2AX/53BP1 immunostaining images of MCF10A expressing WT (A.) or R280H hXRCC1 (B.). Left upper panel represents γH2AX staining (red), right upper - 53BP1 staining (green), right bottom - DAPI staining (blue), and left bottom is a composite picture of all three panels (yellow for co-localizing foci). C. Only co-localizing foci (yellow on composite picture) were counted in WT or R280H hXRCC1 expressing cells. A total of at least 30 cells were scored for each cell line. Data are plotted as mean ± S.E. ** denotes p < 0.01.
These results show that DSBs accumulate during the G1 phase of the cell cycle in the MCF10A cells expressing R280H XRCC1 indicating that damage induced by either reactive oxygen species or alkylating agents is repaired in an aberrant manner. Accumulation of damage could lead to genomic instability and, as a result, to cellular transformation.
4. Discussion
We found that expression of the R280H germline variant of XRCC1 induces genomic instability and cellular transformation in mouse and human epithelial cells. Expression of WT XRCC1 in the cells did not lead to significantly increased levels of cellular transformation and genomic instability. The genomic instability we observe In the R280H variant likely arises as a result of the accumulation of DSBs, as there are significantly higher levels of DSBs in cells expressing R280H compared to cells expressing WT XRCC1. Our results suggest that individuals harboring the R280H germline variant of XRCC1 may be at increased risk for cancer.
It is not presently clear how exactly XRCC1 is recruited to the site of DNA damage. XRCC1 recruitment appears to be mediated by PARylation, either directly through interaction with PAR chains (35) or indirectly as a response to chromatin reorganization induced by PARylation (36). Recruitment of XRCC1 to near-UVA micro-irradiated sites of the nuclei is strongly influenced by the region encompassing amino acids 166-310 (28). Moreover, the R280H variant exhibits a decreased retention time, compared to WT XRCC1, at near-UV micro-irradiation induced DNA damage sites and decreased affinity for binding to DNA gaps (29). Altogether this suggests that the compromised genomic stability that we observed is primarily caused by changes in the recruitment of R280H XRCC1 to the site of DNA damage.
XRCC1 is a heavily phosphorylated protein and the most well verified phosphorylations cluster in two domains, amino acids 183-315 and 403-538, respectively (37). Particularly, checkpoint kinase 2 (CHK2) phosphorylates Thr284, which is in close proximity to Arg280. Phosphorylation of Thr284 of XRCC1 leads to recruitment of BER proteins (37) and is suggested to promote BER. It is possible that alteration of Arg280 to His results in an XRCC1 protein variant that cannot be phosphorylated or that is inappropriately phosphorylated, resulting in defective recruitment of BER proteins. However, Berquist et al. showed that R280H interacts normally with proteins that function in BER (29).
The unstable recruitment of the R280H variant to DNA damage (28,29) suggests that scaffolding of the BER machinery is less than optimum in cells expressing this variant, leading to less efficient completion of BER and the accumulation of DSBs. Interestingly, cells expressing R280H XRCC1 have higher levels of DSBs during the G1 phase of the cell cycle only, although equivalent levels of DSBs are observed in S and G2/M in cells expressing either WT or R280H XRCC1. We suggest that the DSBs that are observed in the S and G2/M phases of the cell cycle arise during collision of the replication fork with single nucleotide gaps, which are a BER intermediate. If this is indeed the case, it would suggest that scaffolding of the BER reaction by R280H has an efficiency similar to that of WT XRCC1 and that in some cases incomplete BER in cells expressing either WT or R280H XRCC1 leads to DSB formation during DNA replication. Alternatively, the DSBs we observe during S and G2/M could be frank DSBs induced by MMS and H2O2, although we have not observed this to be the case in a previous study (34).
It is possible that the accumulation of DSBs during G1 in cells treated with H2O2 or MMS could result from defective scaffolding of canonical BER at oxidized or alkylated bases by R280H. Defective scaffolding of canonical BER by R280H could lead to clusters of BER intermediates in the form of SSBs or nicks in the DNA eventually resulting in the formation of DSBs during the G1 phase of the cell cycle. However, as stated above, BER scaffolding appears to be similar for WT and R280H, given the relatively equivalent accumulation of DSBs during S and G2/M in cells expressing these proteins. In addition to BER, XRCC1 has also been reported to play a role in non-homologous end joining (NHEJ) (38), and specifically in the process of alternative end-joining (39,40), although this is controversial (41). In this pathway of end joining, MRE11 interacts with XRCC1/Lig3α to promote the joining of incompatible ends. Because NHEJ occurs during the G1 phase of the cell cycle, we suggest that the DSBs that accumulate during G1 in cells expressing R280H XRCC1 arise from deficient end joining. Our results indicate that the retention of XRCC1 at sites of damage during G1 is more critical for end joining than for scaffolding of the BER reaction or that XRCC1 has distinct roles in these different repair processes.
At the present moment the relationship between the R280H variant and increased risk of various types of cancer remains ambiguous. This can be partly explained by significant differences in XRCC1 genomic single polymorphism (SNP) distribution between populations. R280H substitution is most prevalent in Asia (1.0% homozygous, 16.8% heterozygous), less common in Europe and America (0.5% and 0.6% homozygous, 11.6% and 12.2% heterozygous, respectively) while very rare in Africa (no data for homozygous, 2.8% heterozygous) (36). Also most of the studies published were performed on relatively small populations. Our results suggest that further major epidemiological studies may reveal a more substantial connection between R280H (rs25489) carriers and increased risk of decreased latency of cancer development or perhaps faster progression of the disease.
Highlights.
-
-
Expression of the R280H variant of XRCC1 results in genomic instability and cellular transformation.
-
-
Expression of R280H XRCC1 leads to accumulation of double-strand breaks during the G1 phase of the cell cycle.
-
-
Individuals with the R280H XRCC1 variant may be at increased risk for various types of cancer.
Acknowledgements
This research was supported by RO1 CA116753 from the National Cancer Institute.
References
- 1.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annual review of genetics. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 2.Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer letters. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akbari M, Solvang-Garten K, Hanssen-Bauer A, Lieske NV, Pettersen HS, Pettersen GK, Wilson DM, 3rd, Krokan HE, Otterlei M. Direct interaction between XRCC1 and UNG2 facilitates rapid repair of uracil in DNA by XRCC1 complexes. DNA repair. 2010;9:785–795. doi: 10.1016/j.dnarep.2010.04.002. 2901844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campalans A, Marsin S, Nakabeppu Y, O’Connor T,R, Boiteux S, Radicella JP. XRCC1 interactions with multiple DNA glycosylases: a model for its recruitment to base excision repair. DNA repair. 2005;4:826–835. doi: 10.1016/j.dnarep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Marsin S, Vidal AE, Sossou M, Menissier-de Murcia J, Le Page F, Boiteux S, de Murcia G, Radicella JP. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. The Journal of biological chemistry. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 6.Caldecott KW, Aoufouchi S, Johnson P, Shall S. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. 146288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 8.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM., 3rd XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res. 2004;32:2193–2201. doi: 10.1093/nar/gkh556. 407833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Molecular and cellular biology. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. 108937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldecott KW. XRCC1 and DNA strand break repair. DNA repair. 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 12.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7:176–178. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 13.Thompson LH, Brookman KW, Dillehay LE, Carrano AV, Mazrimas JA, Mooney CL, Minkler JL. A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutation research. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 14.Cantoni O, Murray D, Meyn RE. Induction and repair of DNA single-strand breaks in EM9 mutant CHO cells treated with hydrogen peroxide. Chemico-biological interactions. 1987;63:29–38. doi: 10.1016/0009-2797(87)90102-5. [DOI] [PubMed] [Google Scholar]
- 15.Christie NT, Cantoni O, Evans RM, Meyn RE, Costa M. Use of mammalian DNA repair-deficient mutants to assess the effects of toxic metal compounds on DNA. Biochemical pharmacology. 1984;33:1661–1670. doi: 10.1016/0006-2952(84)90289-2. [DOI] [PubMed] [Google Scholar]
- 16.Churchill ME, Peak JG, Peak MJ. Correlation between cell survival and DNA single-strand break repair proficiency in the Chinese hamster ovary cell lines AA8 and EM9 irradiated with 365-nm ultraviolet-A radiation. Photochemistry and photobiology. 1991;53:229–236. doi: 10.1111/j.1751-1097.1991.tb03927.x. [DOI] [PubMed] [Google Scholar]
- 17.Churchill ME, Peak JG, Peak MJ. Repair of near-visible- and blue-light-induced DNA single-strand breaks by the CHO cell lines AA8 and EM9. Photochemistry and photobiology. 1991;54:639–644. doi: 10.1111/j.1751-1097.1991.tb02068.x. [DOI] [PubMed] [Google Scholar]
- 18.Thompson LH, West MG. XRCC1 keeps DNA from getting stranded. Mutation research. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 19.Zdzienicka MZ, van der Schans GP, Natarajan AT, Thompson LH, Neuteboom I, Simons JW. A Chinese hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis. 1992;7:265–269. doi: 10.1093/mutage/7.4.265. [DOI] [PubMed] [Google Scholar]
- 20.Carrano AV, Minkler JL, Dillehay LE, Thompson LH. Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutation research. 1986;162:233–239. doi: 10.1016/0027-5107(86)90090-4. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez I, Daza P, Natarajan AT, Cortes F. A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutation research. 1998;398:67–73. doi: 10.1016/s0027-5107(97)00241-8. [DOI] [PubMed] [Google Scholar]
- 22.Op het Veld CW, Jansen J, Zdzienicka MZ, Vrieling H, van Zeeland AA. Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CHO9 and its xrcc1-deficient derivative EM-C11. Mutation research. 1998;398:83–92. doi: 10.1016/s0027-5107(97)00243-1. [DOI] [PubMed] [Google Scholar]
- 23.Op het Veld CW, Zdzienicka MZ, Vrieling H, Lohman PH, van Zeeland AA. Molecular analysis of ethyl methanesulfonate-induced mutations at the hprt gene in the ethyl methanesulfonate-sensitive Chinese hamster cell line EM-C11 and its parental line CHO9. Cancer Res. 1994;54:3001–3006. [PubMed] [Google Scholar]
- 24.Liu C, Yin Q, Li L, Jiao G, Wang M, Wang Y. XRCC1 Arg194Trp and Arg280His polymorphisms in bladder cancer susceptibility: a meta-analysis. Critical reviews in eukaryotic gene expression. 2013;23:339–354. doi: 10.1615/critreveukaryotgeneexpr.2013007781. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Wang M, Gu D, Wu D, Zhang X, Gong W, Tan Y, Zhou J, Wu X, Tang C, Zhang Z, Chen J. Association of XRCC1 gene polymorphisms with the survival and clinicopathological characteristics of gastric cancer. DNA and cell biology. 2013;32:111–118. doi: 10.1089/dna.2012.1840. [DOI] [PubMed] [Google Scholar]
- 26.Zhang K, Zhou B, Wang Y, Rao L, Zhang L. The XRCC1 Arg280His polymorphism contributes to cancer susceptibility: an update by meta-analysis of 53 individual studies. Gene. 2012;510:93–101. doi: 10.1016/j.gene.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Ha TC, Tai BC. XRCC1 gene polymorphisms and breast cancer risk in different populations: a meta-analysis. Breast. 2009;18:183–191. doi: 10.1016/j.breast.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Hanssen-Bauer A, Solvang-Garten K, Gilljam KM, Torseth K, Wilson DM, 3rd, Akbari M, Otterlei M. The region of XRCC1 which harbours the three most common nonsynonymous polymorphic variants, is essential for the scaffolding function of XRCC1. DNA repair. 2012;11:357–366. doi: 10.1016/j.dnarep.2012.01.001. 3319167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berquist BR, Singh DK, Fan J, Kim D, Gillenwater E, Kulkarni A, Bohr VA, Ackerman EJ, Tomkinson AE, Wilson DM., 3rd Functional capacity of XRCC1 protein variants identified in DNA repair-deficient Chinese hamster ovary cell lines and the human population. Nucleic Acids Res. 2010;38:5023–5035. doi: 10.1093/nar/gkq193. 2926592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosa JL, Sweasy JB. The E249K mutator mutant of DNA polymerase beta extends mispaired termini. The Journal of biological chemistry. 1999;274:35866–35872. doi: 10.1074/jbc.274.50.35866. [DOI] [PubMed] [Google Scholar]
- 31.Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB. The E295K DNA polymerase beta gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Molecular and cellular biology. 2007;27:5587–5596. doi: 10.1128/MCB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweasy JB, Dalal S, Sun KW, Lai C-C, DiMaio D, Lang T. Expression of DNA Polymerase Beta Mutants in Mouse Cells Results in Cellular Transformation. Proc Natl Acad Sci U S A. 2005;102:14350–14355. doi: 10.1073/pnas.0505166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamtich J, Nemec AA, Keh A, Sweasy JB. A germline polymorphism of DNA polymerase Beta induces genomic instability and cellular transformation. PLoS genetics. 2012;8:e1003052. doi: 10.1371/journal.pgen.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galick HA, Kathe S, Liu M, Robey-Bond S, Kidane D, Wallace SS, Sweasy JB. Germ-line variant of human NTH1 DNA glycosylase induces genomic instability and cellular transformation. Proc Natl Acad Sci U S A. 2013;110:14314–14319. doi: 10.1073/pnas.1306752110. 3761600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Khamisy SF MM, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanssen-Bauer A, S GK, Akbari M, Otterlei M. X-ray repair cross complementing protein 1 in base excision repair. International journal of molecular sciences. 2012;13:17210–17229. doi: 10.3390/ijms131217210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou WC, W H, Wong FH, Ding SL, Wu PE, Shieh SY, Shen CY. Chk2-dependent phosphorylation of XRCC1 in the DNA damage response promotes base excision repair. EMBO J. 2008;27:3140–3150. doi: 10.1038/emboj.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Audebert M, S B, Calsou P. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun. 2008;369:982–988. doi: 10.1016/j.bbrc.2007.11.132. [DOI] [PubMed] [Google Scholar]
- 39.Della-Maria J, Zhou Y, Tsai MS, Kuhnlein J, Carney JP, Paull TT, Tomkinson AE. Human Mre11/human Rad50/Nbs1 and DNA ligase IIIalpha/XRCC1 protein complexes act together in an alternative nonhomologous end joining pathway. The Journal of biological chemistry. 2011;286:33845–33853. doi: 10.1074/jbc.M111.274159. 3190819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saribasak H, Maul RW, Cao Z, McClure RL, Yang W, McNeill DR, Wilson DM, 3rd, Gearhart PJ. XRCC1 suppresses somatic hypermutation and promotes alternative nonhomologous end joining in Igh genes. The Journal of experimental medicine. 2011;208:2209–2216. doi: 10.1084/jem.20111135. 3201205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]


