Abstract
Acute myeloid leukemia (AML) is the most common form of leukemia in adults. Unfortunately, the standard therapeutic agents used for this disease have high toxicities and poor efficacy. The one exception to these poor outcomes is the use of the retinoid, all-trans retinoic acid (ATRA), for a rare subtype of AML (APL). The use of the differentiation agent, ATRA, in combination with low-dose chemotherapy leads to the long-term survival and presumed cure of 75–85% of patients. Unfortunately ATRA has not been clinically useful for other subtypes of AML. Though many non-APL leukemic cells respond to ATRA, they require significantly higher concentrations of ATRA for effective differentiation. Here we show that the combination of ATRA with glycogen synthase kinase 3 (GSK3) inhibition significantly enhances ATRA-mediated AML differentiation and growth inhibition. These studies have revealed that ATRA's receptor, the retinoic acid receptor (RAR), is a novel target of GSK3 phosphorylation and that GSK3 can impact the expression and transcriptional activity of the RAR. Overall, our studies suggest the clinical potential of ATRA and GSK3 inhibition for AML and provide a mechanistic framework to explain the promising activity of this combination regimen.
Keywords: AML, differentiation, retinoids
INTRODUCTION
Acute myeloid leukemia (AML) is a broad range of disorders that all have the defining feature of leukemic cells with a maturation arrest. The current AML therapeutics which target rapidly dividing cells and not the maturation arrest have poor efficacy and high toxicities. For example, only 20% of patients over age 56 survive 2 years.1 Despite the poor therapeutics, AML is the most common form of leukemia in adults leading to an enormous unmet need for novel agents to improve the morbidity and mortality of these patients. Unfortunately even with these challenges, there has not been any change in the standard AML clinical agents in over 30 years.
By targeting the maturation arrest instead of rapidly dividing cells, a more efficacious and less-toxic therapeutic regimen may be developed. AML differentiation can result in cells that are irreversibly growth arrested and eventually die off without the necessity for overt cytotoxicity. In fact, all-trans retinoic acid (ATRA) is an AML differentiation agent that has revolutionized the treatment of a rare subtype of AML (APL, 5–10% of AML) leading to the long-term survival and presumed cure of 75–85% of patients.2 Unfortunately ATRA is not clinically useful for the other subtypes of AML. Though most AML cells respond to ATRA, non-APL cells require significantly higher concentrations for significant differentiation and growth inhibitory effects.
In efforts to identify novel AML differentiation agents which exhibit activity in non-APL cells, we have performed several screens of small molecule collections for novel AML differentiation-inducing agents.3,4 From these ongoing screening efforts, we recently identified that a glycogen synthase kinase 3 (GSK3) inhibitor can lead to evidence of modest AML differentiation. This finding is consistent with previous studies that have found that GSK3 inhibition can inhibit the growth of AML cells.5–7
Though GSK3 inhibition alone leads to modest AML differentiation and growth inhibition, here we report that it significantly enhances ATRA-mediated differentiation even in non-APL cells because of the ability of GSK3 to directly modulate ATRA's receptor. GSK3 is a constitutively active serine/threonine kinase that is important in signaling pathways involved in the regulation of cell fate, protein synthesis, glycogen metabolism, cell mobility, proliferation and survival.8–10 There are two isoforms of GSK3 (α and β), however, all known inhibitors block both forms. This kinase has been widely studied as a target for the treatment of diabetes, inflammation, and multiple neurological diseases, including Alzheimer's, stroke and bipolar disorders.8,11 Though lithium (a relatively non-specific GSK3 inhibitor) has been widely used clinically for biopolar disease, specific GSK3 inhibitors are just starting to enter clinical trials primarily for neurological diseases.12 Recently, the therapeutic potential of GSK3 inhibitors in cancer has become an area of interest, mostly for solid tumors.13–15 While GSK3 is a currently under active investigation as a target for many malignancies, our preclinical studies provide significant support and mechanistic insights into the combination of ATRA and GSK3 inhibition for AML, including non-APL leukemia.
MATERIALS AND METHODS
Reagents and cells
SB415286 (SB), TWS119, Bio, Adapalene and AM80 were obtained from Tocris Biosciences (Minneapolis, MN, USA). Lithium, MTT and NBT and noble agar were from Sigma (St Louis, MO, USA). Methylcellulose with cytokines was from Stem Cell Technologies (Vancouver, BC, Canada). CHIR99021 was from Cayman Chemical (Ann Harbor, MI, USA). Anti GSK3β and β-catenin, α-tubulin, histone 3 antibodies were obtained from Cell Signaling Technology Inc. (Beverly, MA, USA). p21, cyclin A and retinoic acid receptor (RAR)α antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). OCI-AML3-AML3, WEHI-3 and M1 cells were obtained from DSMZ (Braunschweig, Germany) and 293T, HL-60, NB4, THP-1, U937 and Hela cells were obtained from ATCC (Manassas, VA, USA). Primary normal and AML human bone marrow cells were obtained from the CWRU Hematopoietic Stem cell core facility (Cleveland, OH, USA). The AML samples that had fewer than 80% leukemic cells were purified by flow sorting (FACSAria) using CD34+ antibody (BD Biosciences, Franklin Lakes, NJ, USA). RAR–GFP cloned into pcr3.1 was kindly provided by Noa Noy (CWRU). The serine to alanine mutant RAR constructs were made using the site directed mutagenesis kit (Stratagene, Santa Clara, CA, USA). The GSK3S9A construct was from Addgene (Cambridge, MA, USA).
Cell culture
Cells were cultured in RPMI 1640 media (Hyclone, Waltham, MA, USA) except for 293T cells, which were cultured in DMEM media (Hyclone). All media were supplemented with 10% FBS and 1% penicillin and streptomycin. Cells were cultured in a humidified atmosphere of 95% (v/v) air and 5% (v/v) CO2 at 37 °C.
Differentiation
NBT reduction assay was performed in the same manner as described earlier (Wald et al.3). Immunophenotyping was performed by staining cells with CD11b-PE (Becton Dickinson, Franklin Lakes, NJ, USA). The stained samples were run on a Beckman Coulter Cytomics FC 500 cytometer (Becton Coulter, Brea, CA, USA).
Transfections and lentivirus infections
293T cells were co-transfected with either shGSK3β, shβcatenin or empty vector pLK (Sigma) and the packaging plasmids pCMVÄR8.74 and pMD.G using lipofectamine (Invitrogen, Carlsbad, CA, USA). AML cells were infected with the virus containing supernatant concentrated overnight in PEG in the presence of 6 μg/ml of polybrene and stable cell lines were generated by selection with puromycin (1 μg/ml). Hela cells were also transfected with lipofectamine (Invitrogen).
Western blot analysis/immunoprecipitations
Cells treated as indicated were washed with PBS, centrifuged and lysed with a triton containing lysis buffer for whole cell extracts. Immunoprecipitations were performed overnight with the indicated antibodies and Protein A beads (Roche, Penzberg, Upper Bavaria, Germany) at 4 °C followed by washing four times. Protein lysates (50 μg/lane) were resolved on appropriate SDS-PAGE gel and transferred to PVDF membrane (Millipore, Billerica, MA, USA) using Bio-Rad transfer apparatus (Bio-Rad, Hercules, CA, USA). The membranes were blocked, incubated with the indicated primary antibodies at 4 °C overnight, and then the appropriate horseradish peroxidase-conjugated secondary antibodies. For the immunoprecipitations, light-chain-specific secondary antibodies were used (Jackson Laboratories, Bar Harbor, ME, USA). Immunoreactive protein bands were detected by enhanced chemiluminescence (Pierce, Waltham, MA, USA) using XAR-5 film.
Luciferase assay
Hela cells were transfected with RARE-luciferase (kindly provided by Noa Noy, CWRU) and treated as indicated. After treatment the luminescence was measured from an equal number of cells using a Spectramax L luminometer (Molecular Devices, Sunnyvale, CA, USA) with the Bright-glo reagent (Promega, Madison, WI, USA).
Mouse xenograft study
Six-week-old female nude mice were injected bilaterally s.c. with 10 × 106 HL-60 cells. Drug treatment was started 10 days after tumor cell injection. Palpable tumors were present for the established tumor model before initiating drug treatment. SB (10 mg/kg), ATRA (15 mg/kg) or vehicle (30 μl of DMSO and 70 μl of water) were injected i.p. 2× a day for 1 week, followed by once a day for 1 week and every other day for a third week. Lithium chow (0.4% lithium carobanate from Teklad, Indianapolis, IN, USA) was fed to mice for the entire 3 weeks that the mice received the other drug treatments. Mouse tumor measurements were made at the indicated times after treatment was started. The Case Western Reserve University Animal Research Committee approved all of the animal protocols used in this study.
Real-time PCR
Total RNA was isolated from cells treated with SB for 72 h using TRIzol reagent (Invitrogen). RNA was transcribed into cDNA using the Enhanced Avian RT First Strand Synthesis Kit (Sigma). Relative quantitative PCR was performed in triplicate using the FastStart SYBR Green Master (Roche Diagnostics) on an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA).
Mass spectrometry based identification of GSK3-mediated RAR phosphorylation sites
Constitutively active recombinant GSK3β (NEB) was incubated with recombinant RARα (ProteinOne, Rockville, MD, USA) in the NEB supplied buffer for 2 h at 37 °C. Then sample containing phosphorylated RARá was reduced with dithiothreitol, alkylated with iodoacetamide and digested overnight with a modified trypsin at an enzyme to protein ratio of 1:40 (wt/wt) at 37 °C The peptide digest was loaded onto a TiO2 column (GL Sciences, Inc., Torrance, CA, USA) for enrichment of phosphorylated peptides. Phosphopeptides were eluted with 3% ammonium hydroxide, then concentrated in a vacuum centrifuge to 2–3 μl and adjusted to a final volume of 5 μl with 1% formic acid. Liquid chromatography was performed on a U 3000 nano-HPLC system (Dionex, Co., Waltham, MA, USA). The resulting peptides were loaded onto a 300-μm inner diameter ×5-mm C18 PepMap nano-reverse phase trapping column (Dionex, Co.) to pre-concentrate and wash away excess salts. Peptides separations were accomplished using buffer A (100% water and 0.1% formic acid) and buffer B (20% water, 80% acetonitrile and 0.1% formic acid). Mixture of phosphorylated peptides eluted from the PepMap100, 75 μm × 15 mm, 5 μm particle size and 100° column (Dionex Co.) with the gradient of acetonitrile of 1% per min were introduced into the LTQ FT (ThermoFisher, Waltham, MA, USA) mass analyzer equipped with a nanospray ion source (2.4 kV). The phospho-enriched peptides were analyzed using the data-dependent multitask capability of the LTQ FT acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino-acid sequence in successive instrument scans. The Mascot search engine (Matrix Science, Boston, MA, USA) was used to search a single protein data base (RARα protein) considering Ser, Thr or Tyr phosphorylation. All spectra were manually validated and the following acceptance criteria were applied: all phospho-Ser and phospho-Thr peptides were required to show a pronounced neutral loss-dependent MS3 scan and extensive coverage of β- and/or γ-type series ions. Besides, in vitro analysis, the RAR phosphorylation at Ser445 was confirmed in cells. Briefly, RAR–GFP was transfected into Hela cells and after 24 h the cells were treated with vehicle or SB (30 μm) for 6 h. RAR was immunoprecipitated and the amount of phosphorylation at Ser445 was quantified by mass spectrometry.
RESULTS
GSK3 inhibition alone induces moderate AML differentiation
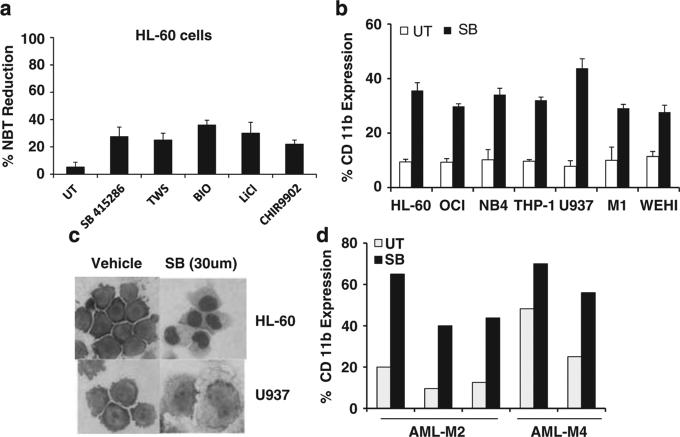
Through screening a collection of kinase inhibitors for AML differentiation activity, we found that GSK3 inhibition can induce AML differentiation through identifying a GSK3 inhibitor, SB415286 (SB), as a hit using a compound library screen to uncover novel AML differentiation agents. As no compounds are entirely specific, we confirmed GSK3 inhibition induces differentiation with five structurally distinct GSK3 inhibitors using the NBT reduction assay in HL-60 cells (Figure 1a). The NBT assay is a highly specific and commonly used method to quantitate myeloid differentiation. It measures the functional differentiation by detecting the respiratory burst capacity, a process that only occurs in differentiated cells.16–20 We further confirmed the ability of GSK3 inhibition to induce differentiation in HL-60 cells and six other AML cell lines by measuring the upregulation of CD11b surface expression, a commonly used marker of AML differentiation (Figure 1b). Of note, only one of these seven cell lines (NB4) tested falls into the APL subtype for which ATRA is clinically efficacious with current regimens. Morphological assessment of several cell types demonstrated monocytic differentiation as can be seen from increased cytoplasm, vacuoles and altered nuclear morphology (Figure 1c). In addition to AML cell lines, GSK3 inhibition is also able to lead to evidence of differentiation of primary AML cells (Figure 1d).
Figure 1.
GSK3 inhibitors induce monocytic differentiation. (a) GSK3 inhibitors induce NBT reduction activity consistent with myelomonocytic differentiation. HL-60 cells were treated with SB415286 (30 μm), TWS116 (5 μm), Bio (1 μm), LiCl (10 mm) or CHIR9902 (10 μm) for 4 days and the NBT reduction assay was performed to assess functional evidence of differentiation. (b) GSK3 inhibitors induce immunophenotypic changes consistent with differentiation. After treatment for 4 days with SB (30 μm), cells were stained with CD11b-PE and flow analysis was performed. (c) GSK3 inhibition induces morphological changes consistent with monocytic differentiation. After treatment for 4 days with SB (30 μm), cytospin preparations were prepared and the cells were stained with Wright-Giemsa. (d) GSK3 inhibition induces differentiation in primary non-M3 AML cells. Leukemic cells (>80% pure) derived from five AML patients from AML-M2 and AML-M4 subtypes were treated with SB (30 μm) for 5 days and differentiation was assessed by CD11b staining.
GSK3 inhibition dramatically inhibits the growth of AML cells
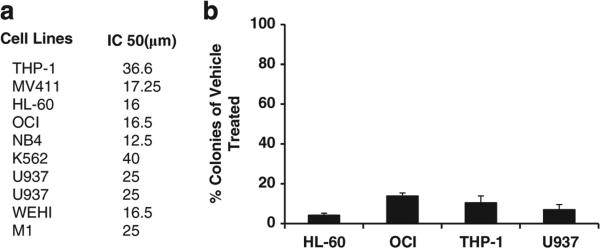
Besides differentiation, GSK3 inhibition leads to significant growth inhibition of AML cells as has also been recently reported by others.5,7 For example, utilizing a panel of nine different AML cell lines, the IC50 of SB ranged from 12.5 to 40 μm at 72 h after treatment using the MTT assay (Figure 2a). As the primary goal of AML differentiation therapy is to permanently prevent the growth of AML cells, colony assays were performed to test for irreversible growth arrest after limited treatment with GSK3 inhibitors. For this assay, AML cells are exposed to drug for 3 days, drug is washed off and an equal number of viable cells are plated in soft agar. At optimal doses for differentiation and GSK3 inhibition, dramatic inhibition of colony growth was observed in several AML cell lines after GSK3 inhibition (Figure 2b).
Figure 2.
GSK3 inhibition inhibits the growth of AML cells. (a) GSK3 inhibition inhibits the growth of a panel of diverse AML cell lines. The indicated cell lines were treated with increasing doses of SB and the MTT assay was performed after 72 h. (b) GSK3 inhibition dramatically inhibits AML colony formation. The indicated cell lines were treated with SB (30 μm) for 72 h, the drug was washed off and an equal number of viable cells were tested for colony formation in soft agar after 7–10 days.
GSK3 inhibitors dramatically enhance ATRA's activity both in vitro and in vivo
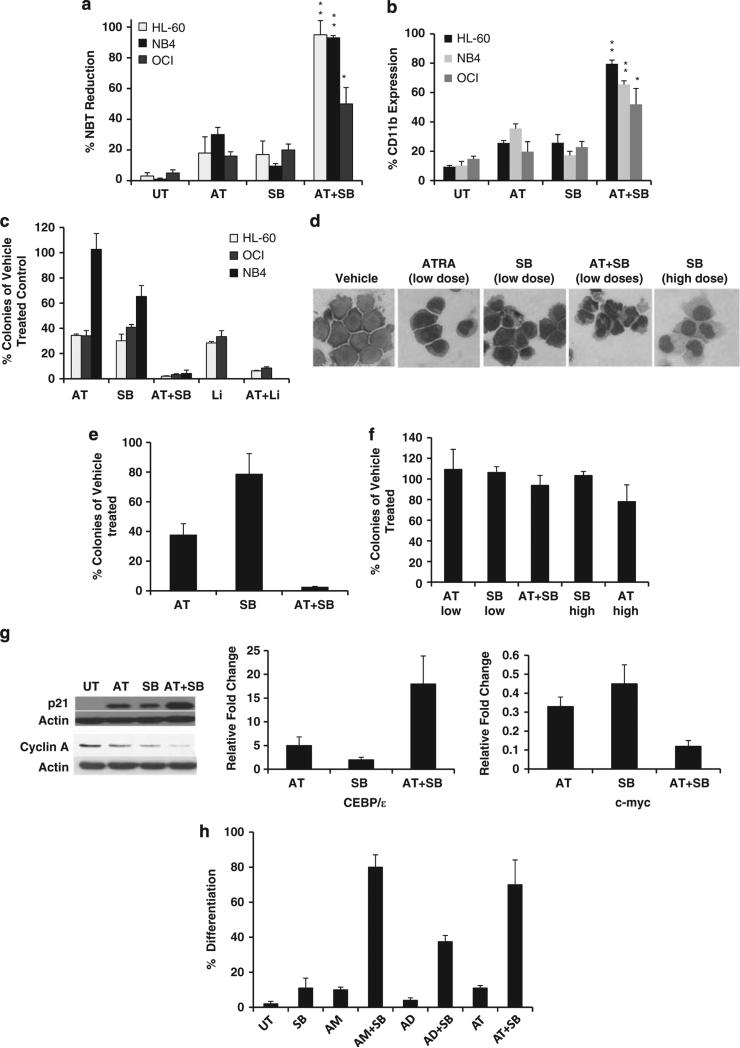
ATRA has seen enormous clinical success, but only for ~5% of AML patients. This is thought to be because of the requirement for high doses of ATRA for efficacy in non-APL leukemia as virtually all AML cell lines and patient samples express RAR and many can differentiate in vitro when exposed to significantly higher concentrations of ATRA than are required for APL cells. For example, NB4 cells (APL-M3 subtype) undergo significant differentiation when exposed to nanomolar concentrations of ATRA (~100 nm) whereas other AML cells such as HL-60 (M2 subtype) or OCI-AML3 (M4 subtype) require micromolar concentrations. Because of ATRA's success for APL, significant efforts have been made to enhance its activity for the remaining ~95% of patients with non-APL leukemia. We found that low doses of GSK3 inhibitors that alone lead to little differentiation induce significant differentiation and irreversible growth arrest in several AML cell lines and primary AML patient samples in combination with low doses of ATRA (Figure 3). For example, ATRA (50 nm) leads to 18% differentiation and SB (15 μm) leads to 17% differentiation whereas the combination induces 95% differentiation as measured by NBT reduction in HL-60 cells (Figure 3a). Utilizing colony assays to test for irreversible growth arrest, we observe a dramatic reduction of colonies with the combined treatment as compared with low-dose ATRA (10 or 50 nm) and SB (15 μm) treatments (Figure 3c). The ability of these pathways to interact to induce differentiation can also be seen by morphological assessment as low doses of ATRA (50 nm) and SB (15 μm) have only minor effects on the morphology of HL-60 cells whereas the combination of these agents causes morphological evidence of differentiation of nearly all cells into neutrophils. This finding is in contrast to GSK3 inhibition alone that differentiates HL-60 cells into monocytic cells (Figure 3d). In addition to cell lines, non-APL leukemic patient samples also exhibited significant differentiation and growth inhibition with the combined treatment of ATRA and GSK3 inhibition (Figure 3e).
Figure 3.
GSK3 inhibition significantly enhances ATRA's activity. (a, b) GSK3 inhibitors enhance ATRA-mediated differentiation. Cells were treated with SB (15 μm), AT (50 nm for HL-60 and OCI-AML3 and 10 nm for NB4) or a combination for 4 days and differentiation was assessed by NBT reduction and CD11b staining. *P < 0.05, **P < 0.001 when comparing single treatments (AT or SB) to the combination treatment. In all cases, SB and AT demonstrated synergistic effects in inducing NBT reduction and CD11b expression as measured by the excess over Bliss independence method. (c) GSK3 inhibition and ATRA dramatically inhibit colony formation in AML cell lines. Cells were treated with vehicle, ATRA (10 or 50 nm), SB (15 μm), Lithium (10 mm) or GSK3 inhibitor and ATRA for 72 h and a colony assay was performed as described in Figure 2. (d) The combination of GSK3 inhibition and ATRA can differentiate HL-60 cells into neutrophils. HL-60 cells were treated as indicated and stained as described in Figure 1c to assess morphology. (e) GSK3 inhibition and ATRA can inhibit colony growth in an AML patient sample. An AML patient sample (AML-M2) was treated as described in Figure 3c and tested for colony growth. (f) GSK3 inhibition does not significantly impair colony growth from normal bone marrow. Normal human bone marrow cells were treated as described in Figure 3c and plated in methylcellulose with cytokines and the resulting erythroid and myeloid colony types were counted after 14 days. (g) The combination of GSK3 inhibition and ATRA lead to enhanced changes in the expression of growth and differentiation related genes. OCI-AML3 cells were treated with ATRA (0.1 μm), SB (15 μm) or a combination for 72 h and the expression of p21 and cyclin A was measured by western and c-myc and CEBP/ε by real-time PCR. (h) GSK3 inhibition can enhance differentiation mediated by retinoids besides ATRA. HL-60 cells were treated for 4 days with AM80 (10 μm), Adapalene (1 μm), ATRA (0.1 μm), SB (15 μm) or a combination and the NBT reduction assay was performed.
Though lithium has been widely used clinically and is known to increase and not decrease white blood cell counts in patients, we tested the effect of GSK3 inhibition on normal bone marrow cells as a major problem with AML therapeutics is toxicity on normal bone marrow.21 Importantly, GSK3 inhibition at the doses used did not significantly change the number of colonies formed after standard colony assays performed with normal human bone marrow (Figure 3f). Interestingly, the same regimens of ATRA and GSK3 inhibition that dramatically inhibit leukemic cell growth do not have significant effects on the growth of normal bone marrow progenitor cells (Figure 3f).
Consistent with our biological findings, we identified downstream AML differentiation and growth inhibition pathways can be significantly modulated by ATRA and GSK3 inhibition. In particular, the cell proliferation related factors cyclin A, c-myc and p21 and the differentiation-related gene CEBP/ε are all affected by the combined treatment of ATRA and GSK3 inhibition (Figure 3g).
GSK3 inhibitors enhance the ability of other retinoids besides ATRA to induce differentiation
As other retinoids besides ATRA have seen clinical use, we tested whether or not other RAR ligands could also enhance the differentiation activity of GSK3 inhibition. Similarly to our results with ATRA, we observed that differentiation induced by other retinoids such as Adapalene and AM80 is significantly enhanced by GSK3 inhibition. This result demonstrates that the ability of GSK3 inhibition to enhance retinoid activity is not simply limited to ATRA (Figure 3h).
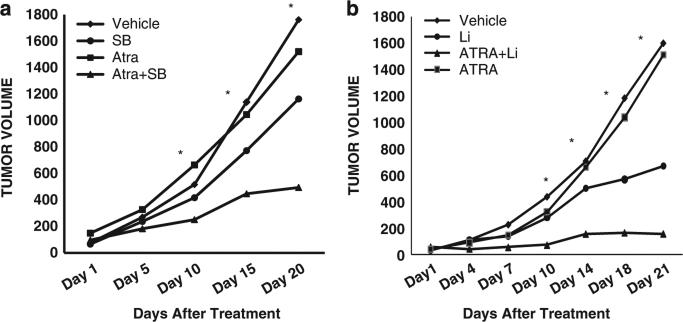
Besides cell based studies, GSK3 inhibitors and ATRA also show a significant enhancement in impairing tumor growth in a mouse xenograft model system using a non-APL leukemia cell line (AML-M2). Although neither ATRA nor GSK3 inhibition alone (with lithium or SB) had dramatic effects on tumor growth, the combination led to significant reductions in tumor growth (Figure 4). Of note our mouse studies using lithium demonstrated efficacy with circulating lithium levels (~0.7 mm),22 which are well within the normal therapeutic range of lithium for bipolar disease (0.6–1.2 mm).23 This work suggests that GSK3 inhibition is a promising strategy to achieve a long-held goal in AML therapy of enhancing the clinical activity of ATRA in non-APL leukemia.
Figure 4.
The combination of GSK3 inhibition and ATRA significantly impairs the growth of AML tumors. Nude mice (five per group) were injected with HL-60 cells bilaterally into the flank. Seven days after tumor cell inoculation, the mice were started on treatment regimens including vehicle, ATRA, SB (a), Lithium (b), or a combination of GSK3 inhibition and ATRA. The average volume of tumors (mm3) measured on indicated days after treatment initiation is shown. *Indicates P < 0.05 when comparing vehicle to combination treated mice.
Genetic evidence of GSK3 in AML differentiation
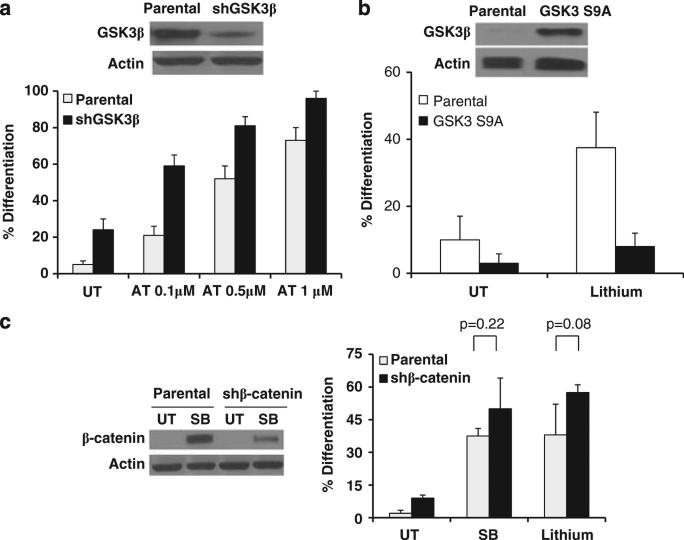
Though we utilized well-established and specific GSK3 inhibitors, no inhibitors are entirely specific. Therefore, we performed genetic experiments to validate the role of GSK3 in AML differentiation. After knocking down GSK3β in a variety of AML cell lines (HL-60, OCI-AML3 and U937), we observed an increase in the basal level of differentiation as well as an enhanced sensitivity to ATRA-mediated differentiation (Figure 5a and data not shown). In an opposite approach, we found that GSK3 inhibitor-mediated differentiation is impaired in AML cells that highly overexpress a mutant form of GSK3β (Ser9 to Ala) that renders the kinase resistant to inactivation (Figure 5b).24 Lithium was used for this study as lithium, unlike SB, has been clearly shown to inhibit GSK3 through Ser9 phosphorylation.25 In addition, as β-catenin induction is a major consequence of GSK3 inhibition, we knocked down the expression of β-catenin. Utilizing these cells, we found that GSK3-inhibition mediated differentiation is completely independent of α-catenin (Figure 5c).
Figure 5.
Genetic evidence of the role of GSK3 in AML differentiation. (a) Knockdown of GSK3β induces partial AML differentiation. OCI-AML3 cells transduced with shGSK3β or empty vector were treated with ATRA for 4 days and the NBT reduction assay was performed. The knockdown of GSK3β was confirmed by western blot. (b) Overexpression of GSK3 S9A impairs GSK3 inhibitor-mediated differentiation. Parental OCI-AML3 cells or cells overexpressing GSK3 S9A were treated with lithium (20 mm) for 4 days and the NBT reduction assay was performed. The overexpression of GSK3S9A was confirmed by western blot. (c) β-catenin knockdown does not impact GSK3 inhibitor-mediated differentiation. OCI-AML3 cells were treated with SB (30 μm) or lithium (20 mm) for 4 days and the NBT reduction assay was performed. The knockdown of β-catenin was confirmed by western blot. Cells were treated with SB to visualize β-catenin as the expression is low in untreated cells.
GSK3 modulates RAR expression and activity
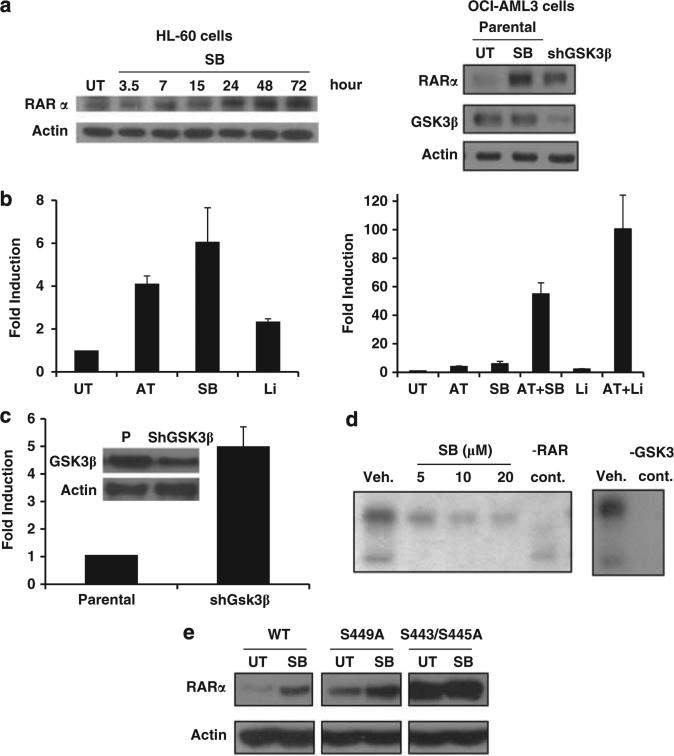
As GSK3 is a kinase that is known to target a wide variety of substrates, to explain the ability of GSK3 inhibition to regulate ATRA-mediated differentiation, we assessed if GSK3 affects ATRA's receptor, the RAR. Initially we assessed the ability of GSK3 inhibition to modulate the expression of RAR. GSK3 inhibition using small molecules or GSK3 knockdown was found to lead to RAR protein induction in both AML cells as well as Hela cells (Figures 6a and e). The induction of RAR occurs in the presence of cycloheximide suggesting that GSK3 inhibition stabilizes RAR posttranslationally (data not shown).
Figure 6.
Mechanisms through which GSK3 modulates RAR signaling. (a) GSK3 inactivaton induces RARα expression. First panel: HL-60 cells were treated for the indicated times with SB (30 μm) and western analysis was performed. Second panel: OCI-AML3 cells in which GSK3β was stably knocked down demonstrate an increase in RARα expression as compared with empty vector infected parental cells. The parental cells also exhibit an induction in RARα expression after 72 h of SB (30 μm) treatment. (b) GSK3 inhibition enhances RAR transcriptional activity. Hela cells transfected with RARE-luc (RAR response element-luciferase) were treated for 16 h with ATRA (1 μm), SB (30 μm), lithium (20 mm) or a combination and the luciferase signal was measured. The single treatments are shown in a separate graph to demonstrate the induction of RARE with GSK3 inhibition alone. (c) GSK3β knockdown induces RAR transcriptional activity. Parental and GSK3β knockdown Hela cells were transfected with RARE-luc and assessed for luciferase activity after 72 h. (d) GSK3β phosphorylates RARα. Recombinant GSK3β phosphorylates recombinant RARα (as well as itself) using a P32 labelled in vitro kinase assay. GSK3 autophosphorylation and RAR phosphorylation are inhibited by SB treatment. (e) Mutant RAR shows increased expression and a loss of GSK3-mediated RAR induction. Hela cells were transfected with wild-type RAR, a S449A RAR mutant or a S443A/S445A RAR mutant and assessed for RAR protein expression with and without SB treatment (30 μm) for 24 h. Transfection efficiency was found to be equal with these constructs. The cells were all co-transfected with prL-CMV (Promega) and the Renilla luciferase signal was measured to control for transfection efficiency (data not shown).
Besides RAR induction, GSK3 inactivation also enhances the transcriptional activity of RAR (even in the absence of retinoids) (Figure 6b). For this work charcoal treated serum was used to eliminate any effects of low levels of endogenous retinoids present in the serum. Although GSK3 inhibition alone induced RAR activity (approximately fivefold), consistent with our synergistic biological effects, the combination of ATRA and GSK3 inhibition led to a remarkable increase in RAR transcriptional activation again using multiple GSK3 inhibitors in Hela cells (Figure 6b second panel). This dramatic enhancement in transcriptional activity suggests that GSK3 modulates the transcriptional activity of RAR in addition to simply increasing RAR levels. Besides measuring RAR transcriptional activity using GSK3 chemical inhibition, we also found that GSK3β knockdown leads to a similar induction of RAR transcriptional activity as observed with SB treatment suggesting that GSK3 inhibition can lead to an increase in RAR activity in the absence of ligand (Figure 6c). Consistent with our findings in Hela cells, Collins and coworkers26 has recently reported that GSK3 inhibition can enhance RAR transcriptional activity in AML cells.
To further investigate how GSK3 modulates RAR, we found GSK3β directly phosphorylates RARα using an in vitro kinase assay with recombinant GSK3β and RARα. This phosphorylation is blocked by GSK3 inhibition in a dose-dependent fashion (Figure 6d). Next, we mapped the phosphorylation sites by mass spectrometry and identified that GSK3β can phosphorylate RARα on at least three serine residues (Ser443, 445 and 449) located in the F-domain. Although the function and regulation of this domain is poorly understood, consistent with our model, deletion of the F-domain has been found to lead to constitutive RAR transcriptional activity in the absence of ligand.27 After mutation of these serine residues, we observed a significant enhancement of RAR levels using a S443A/S445A RAR mutant, but not a S449A RAR mutant as compared with wild-type RAR (Figure 6e). Importantly, although GSK3 inhibition upregulates wild-type RAR and the S449A RAR mutant, it does not modulate the expression of the S443A/S445A RAR mutant. This finding suggests that GSK3 negatively regulates RAR expression through phosphorylating this portion of RAR. To further confirm that GSK3 can phosphorylate RAR in vivo, the relative amount of Ser445 RAR phosphorylation in cells with or without SB treatment was quantitated by mass spectrometric analysis of immunoprecipitated RAR. We found that Ser445 RAR phosphorylation decreased significantly from 61 to 24% after SB treatment.
GSK3β interacts with RARα
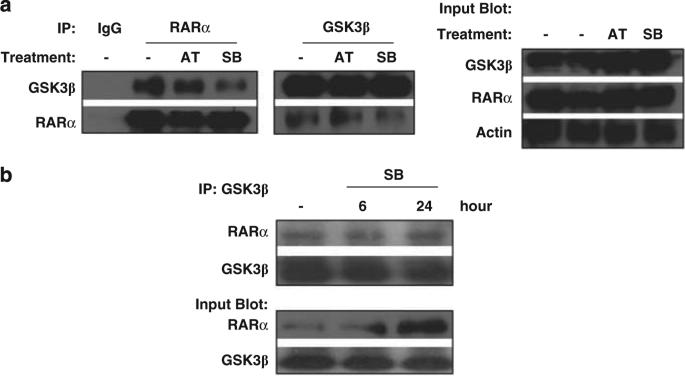
To further explore how nuclear GSK3 modulates RAR, we assessed for the ability of these proteins to bind. Initially utilizing Hela cells overexpressing RARα, we observed that GSK3 and RARα can physically interact by co-immunoprecipitation assays (Figure 7a). In addition utilizing OCI-AML3 cells, we found that endogenous GSK3β and RARα physically interact in AML cells (Figure 7b).
Figure 7.
GSK3β binds RARα. (a) GSK3β can bind RARα. Co-immunoprecipitations were performed as indicated to test for the interaction of GSK3â and RARá in Hela cells treated for 6 h with AT (1 μm) or SB (30 μm). RARα was overexpressed in these Hela cells. (b) Endogenous GSK3β and RARα interact in AML cells. The interaction of GSK3β and RARα was assessed by co-immunoprecipiation in OCI-AML3 cells with or without treatment with SB (30 μm).
DISCUSSION
Retinoids are compounds related to vitamin A that regulate a large range of biological processes including development, differentiation, proliferation and apoptosis. Because of these functions, synthetic and naturally occurring retinoids have been tested clinically as therapeutics for numerous cancers such as leukemia, liver, lung, colon and breast cancers. In addition, retinoids are used for a wide variety of skin diseases.28
One disease in which the retinoid therapy has made an enormous impact is AML where the RAR-specific ligand ATRA is efficacious for a small subset of patients with the APL subtype. In this case ATRA has revolutionized the treatment of APL and leads to the long-term survival and presumed cure of 75–85% of patients in combination with low-dose chemotherapy.2 By inducing differentiation, leukemic cells can be forced to lose their ability to proliferate and eventually die off without the necessity for high doses of cytotoxic chemotherapeutics. Unfortunately ATRA is not efficacious for the majority of AML patients that fall into the other AML subtypes. As AML is the most common form of leukemia in adults, there is an enormous unmet need for novel agents to improve the morbidity and mortality of these patients.
We identified a novel therapeutic approach for AML that involves combining RAR ligands with GSK3 inhibition. This study has elucidated that RAR is a novel and clinically relevant target of GSK3 particularly for AML. We have found that GSK3 can impact RAR through a region of the F-domain, a poorly understood domain on RAR. Importantly, we have identified in cells that GSK3 inhibition dramatically reduces phosphorylation of Ser445.
Interestingly, during our preparation of this manuscript, it was reported that GSK3 inhibition could enhance the activity of ATRA in NB4 cells, an APL cell line.26 Although our work is in agreement with this finding, here we demonstrate that besides APL, which already has an excellent prognosis and is highly sensitive to ATRA, GSK3 inhibition significantly enhances ATRA's activity in non-APL leukemia, which is a major therapeutic challenge.
Our studies as well as work from others have demonstrated that GSK3 is an attractive therapeutic target for AML.5–7,26 One major advantage of targeting GSK3 is that GSK3 inhibition preferentially inhibits the growth of AML, but not normal hematopoietic progenitor cells. This activity is in stark contrast to the standard AML chemotherapeutics that often lead to significant bone marrow toxicities.
Although further investigations are necessary to understand the preferential activity of GSK3 inhibition on AML cells, this differential activity has high-clinical importance. Though these studies have focused on RARα, it is feasible that GSK3 also phosphorylates or interacts with the other RAR isotypes (RARβ and RARγ), which are known to also be clinically significant in many contexts. In particular, RARβ is known to lead to strong growth inhibition in a variety of cancer types (reviewed in Soprano et al.29). The development of a further understanding of how GSK3 regulates the family of RAR proteins will help further determine the utility of using GSK3 inhibition to enhance retinoid signaling in a variety of other diseases besides AML. Although this study focused primarily on AML, the use of cervical cancer cells (Hela) in several mechanistic studies suggest that the findings are not limited to AML alone.
Overall, this work has identified a novel and clinically significant interaction between GSK3 and RARα. As retinoids are already in clinical use for a variety of disease and specific GSK3 inhibitors are actively being tested in clinical trials, it may be possible to significantly enhance the efficacy of AML therapy by combining these two approaches. Because of the importance of modulating the RARs in a wide variety of diseases, it is also feasible that this therapeutic approach may be significantly expanded in the future for other diseases where enhanced RAR activity is desirable.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Estey EH. General approach to, and perspectives on clinical research in, older patients with newly diagnosed acute myeloid leukemia. Semin Hematol. 2006;43:89–95. doi: 10.1053/j.seminhematol.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Tallmann MS. Curative therapeutic approaches to APL. Ann Hematol. 2004;83(Suppl 1):S81–S82. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- 3.Wald DN, Vermaat HM, Zang S, Lavik A, Kang Z, Peleg G, et al. Identification of 6-benzylthioinosine as a myeloid leukemia differentiation-inducing compound. Cancer Res. 2008;68:4369–4376. doi: 10.1158/0008-5472.CAN-07-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta K, Chakrabarti A, Rana S, Ramdeo R, Roth B, Agarwal M, et al. Securinine, a myeloid differentiation agent with therapeutic potential for AML. PLoS One. 2011;6:e21203. doi: 10.1371/journal.pone.0021203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455:1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes T, O'Brien TA, Knight R, Lindeman R, Shen S, Song E, et al. Glycogen synthase kinase-3beta inhibition preserves hematopoietic stem cell activity and inhibits leukemic cell growth. Stem Cells. 2008;26:1288–1297. doi: 10.1634/stemcells.2007-0600. [DOI] [PubMed] [Google Scholar]
- 7.Song EY, Palladinetti P, Klamer G, Ko KH, Lindeman R, O'Brien TA, et al. Glycogen synthase kinase--3beta inhibitors suppress leukemia cell growth. Exp Hematol. 2010;38:908–921 e901. doi: 10.1016/j.exphem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Rayasam GV, Tulasi VK, Sodhi R, Davis JA, Ray A. Glycogen synthase kinase 3: more than a namesake. Br J Pharmacol. 2009;156:885–898. doi: 10.1111/j.1476-5381.2008.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116(Part 7):1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimes CA, Jope RS. The multifaceted roles of glycogen synthase kinase 3beta in cellular signaling. Prog Neurobiol. 2001;65:391–426. doi: 10.1016/s0301-0082(01)00011-9. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez F, Nido JD, Avila J, Villanueva N. GSK3 inhibitors and disease. Mini Rev Med Chem. 2009;9:1024–1029. doi: 10.2174/138955709788922647. [DOI] [PubMed] [Google Scholar]
- 12.Medina M, Castro A. Glycogen synthase kinase-3 (GSK-3) inhibitors reach the clinic. Curr Opin Drug Discov Devel. 2008;11:533–543. [PubMed] [Google Scholar]
- 13.Guzman ML, Li X, Corbett CA, Rossi RM, Bushnell T, Liesveld JL, et al. Rapid and selective death of leukemia stem and progenitor cells induced by the compound 4-benzyl, 2-methyl, 1,2,4-thiadiazolidine, 3,5 dione (TDZD-8). Blood. 2007;110:4436–4444. doi: 10.1182/blood-2007-05-088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y, Uddin S, Zimmerman T, Kang JA, Ulaszek J, Wickrema A. Growth control of multiple myeloma cells through inhibition of glycogen synthase kinase-3. Leuk Lymphoma. 2008;49:1945–1953. doi: 10.1080/10428190802304966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez A. Preclinical efficacy on GSK-3 inhibitors: towards a future generation of powerful drugs. Med Res Rev. 2008;28:773–796. doi: 10.1002/med.20119. [DOI] [PubMed] [Google Scholar]
- 16.Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J Exp Med. 1979;149:969–974. doi: 10.1084/jem.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newburger PE, Chovaniec ME, Greenberger JS, Cohen HJ. Functional changes in human leukemic cell line HL-60. A model for myeloid differentiation. J Cell Biol. 1979;82:315–322. doi: 10.1083/jcb.82.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins SJ, Bodner A, Ting R, Gallo RC. Induction of morphological and functional differentiation of human promyelocytic leukemia cells (HL-60) by componuds which induce differentiation of murine leukemia cells. Int J Cancer. 1980;25:213–218. doi: 10.1002/ijc.2910250208. [DOI] [PubMed] [Google Scholar]
- 19.Guglielmo P, Pagano MC, Giustolisi R. [The nitroblue tetrazolium activated test in the study of granulocytic function. Proposed methodology]. Boll Soc Ital Biol Sper. 1980;56:183–187. [PubMed] [Google Scholar]
- 20.Newburger PE, Speier C, Borregaard N, Walsh CE, Whitin JC, Simons ER. Development of the superoxide-generating system during differentiation of the HL-60 human promyelocytic leukemia cell line. J Biol Chem. 1984;259:3771–3776. [PubMed] [Google Scholar]
- 21.Prakash R. A review of the hematologic side effects of lithium. Hosp Community Psychiatry. 1985;36:127–128. doi: 10.1176/ps.36.2.127. [DOI] [PubMed] [Google Scholar]
- 22.Dixon JF, Hokin LE. Lithium acutely inhibits and chronically up-regulates and stabilizes glutamate uptake by presynaptic nerve endings in mouse cerebral cortex. Proc Natl Acad Sci USA. 1998;95:8363–8368. doi: 10.1073/pnas.95.14.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grandjean EM, Aubry JM. Lithium: updated human knowledge using an evidence-based approach. Part II: clinical pharmacology and therapeutic monitoring. CNS Drugs. 2009;23:331–349. doi: 10.2165/00023210-200923040-00005. [DOI] [PubMed] [Google Scholar]
- 24.MacAulay K, Blair AS, Hajduch E, Terashima T, Baba O, Sutherland C, et al. Constitutive activation of GSK3 down-regulates glycogen synthase abundance and glycogen deposition in rat skeletal muscle cells. J Biol Chem. 2005;280:9509–9518. doi: 10.1074/jbc.M411648200. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Friedman AB, Zhu W, Wang L, Boswell S, May RS, et al. Lithium regulates glycogen synthase kinase-3beta in human peripheral blood mononuclear cells: implication in the treatment of bipolar disorder. Biol Psychiatry. 2007;61:216–222. doi: 10.1016/j.biopsych.2006.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si J, Mueller L, Collins SJ. GSK3 inhibitors enhance retinoic acid receptor activity and induce the differentiation of retinoic acid-sensitive myeloid leukemia cells. Leukemia. 2011;25:1914–1918. doi: 10.1038/leu.2011.171. [DOI] [PubMed] [Google Scholar]
- 27.Farboud B, Privalsky ML. Retinoic acid receptor-alpha is stabilized in a repressive state by its C-terminal, isotype-specific F domain. Mol Endocrinol. 2004;18:2839–2853. doi: 10.1210/me.2004-0236. [DOI] [PubMed] [Google Scholar]
- 28.Bushue N, Wan YJ. Retinoid pathway and cancer therapeutics. Adv Drug Deliv Rev. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr. 2004;24:201–221. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]