Abstract
N-linked glycans are required to maintain appropriate biological functions on proteins. Underglycosylation leads to many diseases in plants and animals; therefore, characterizing the extent of glycosylation on proteins is an important step in understanding, diagnosing, and treating diseases. To determine the glycosylation site occupancy, protein N-glycosidase F (PNGase F) is typically used to detach the glycan from the protein, during which the formerly glycosylated asparagine undergoes deamidation to become an aspartic acid. By comparing the abundance of the resulting peptide containing aspartic acid against the one containing non-glycosylated asparagine, the glycosylation site occupancy can be evaluated. However, this approach can give inaccurate results when spontaneous chemical deamidation of the non-glycosylated asparagine occurs. To overcome this limitation, we developed a new method to measure the glycosylation site occupancy that does not rely on converting glycosylated peptides to their deglycosylated forms. Specifically, the overall protein concentration and the non-glycosylated portion of the protein are quantified simultaneously by using heavy isotope-labeled internal standards coupled with LC-MS analysis, and the extent of site occupancy is accurately determined. The efficacy of the method was demonstrated by quantifying the occupancy of a glycosylation site on bovine fetuin. The developed method is the first work that measures the glycosylation site occupancy without using PNGase F, and it can be done in parallel with glycopeptide analysis because the glycan remains intact throughout the workflow.
Keywords: N-linked glycosylation, Site occupancy, PNGase F, Liquid chromatography/mass spectrometry, Chemical deamidation
Introduction
N-glycosylation is a common post-translational modification that is closely related to various biological events, including cancer metastasis, viral infection of cells, and antibody-antigen interactions [1-2]. This modification occurs on an asparagine (N) that is within a consensus sequence of N-X-T/S/C (X could be any amino acid except proline). However, the glycosylation site occupancy depends on the enzymes that catalyze glycan biosynthesis, and the extent of glycosylation can change even for the same glycoprotein that is produced from different cell lines [3-4]. The variability in glycosylation site occupancy is a key indicator of cellular activities, as demonstrated by the correlation between a reduction in glycosylation site occupancy of serum proteins and the severity of congenital disorders of glycosylation (CDGs) [5]. Therefore, it is significant to determine the site occupancy accurately in order to fully understand the impact of protein glycosylation on human health [6].
In measuring the N-glycosylation site occupancy, the most frequently adopted procedure uses PNGase F to detach the glycan from the protein [7-8]. As a result, the glycosylated asparagine (N) is converted to aspartic acid (D) through the PNGase F reaction, inducing an increase in mass of 0.984 Da. This N to D conversion is measured by mass spectrometry (MS), and a larger mass discrimination is achieved by 18O-labeling of the resulting aspartic acid to facilitate the assignment [7, 9]. The ratio of the formerly glycosylated asparagine over the non-glycosylated asparagine (and thus the site occupancy) is calculated by comparing the signal of the deglycosylated peptide against the non-glycosylated peptide. This PNGase F method is widely used either to measure the occupancy of partially glycosylated asparagine or to identify novel glycosylation sites [8, 10].
Nevertheless, it has been found that chemical deamidation of asparagine could occur spontaneously during sample preparation [11-12]. Consequently, in the typical approach that uses the PNGase F reaction, the non-glycsoylated asparagine that undergoes chemical deamidation would be incorrectly assigned as the product of the formerly glycosylated asparagine, which leads to inaccurate quantitation in measuring the site occupancy [12]. Moreover, to quantify the occupancy level by the existing method, the MS signal of the deglycosylated peptide (that contains the aspartic acid) is compared to the signal of the non-glycosylated peptide (that contains the asparagine), and the underlying assumption is that the response factors of these two peptides are the same. However, a recent study indicates the deglycosylated peptide showed reduced signal intensity of up to 50% compared with the non-glycosylated counterpart of equal molar concentration for certain peptide sequences [13]. Clearly, the currently implemented method of using PNGase F in the quantitative analysis of the N-glycosylation site occupancy has limitations that should be addressed.
Herein we have developed an improved approach for determining the glycosylation occupancy that avoids the disadvantages described above. The key innovation in our strategy is to quantify the natively non-glycosylated form of the glycopeptide, using an isotopically labeled internal standard. No glycosidase is added to the sample so that the N-glycan stays intact. Instead, two sets of heavy isotope labeled peptide standards are spiked into the sample before proteolysis, and the digested sample is analyzed by LC-MS. One set of peptide standards is employed to determine the total glycoprotein concentration, while the other standard monitors the non-glycosylated part of the glycoprotein. In this way, the abundance of the glycosylated portion of the protein is calculated by subtracting the non-glycosylated protein abundance from the overall protein concentration, and the site occupancy is then determined. To demonstrate the effectiveness of the PNGase F-free approach we developed, the method was applied to characterize fetuin, which has one partially-occupied N-glycosylation site at Asn-158.
Experimental
Materials and Reagents
Four purified synthetic peptides labeled with 13C and 15N on terminal lysine or arginine (denoted as *P1-4, sequences contained in Supplementary Table 1) were obtained from JPT Peptide Technologies (Berlin, Germany). Bovine fetuin was purchased from Sigma Aldrich (St. Louis, MO) and sequencing grade trypsin was acquired from Promega (Madison, WI). All reagents were of analytical purity or better.
Sample Preparation
A glycoprotein solution of 10 μg/μL was prepared in 100 mM Tris buffer (pH 8.0) containing 6 M urea. The sample was treated with 5 mM tris(2-carboxyethyl)-phosphine (TCEP) and 20 mM iodoacetamide (IAM) in the dark for 1 h at room temperature to reduce and alkylate the disulfide bonds, and 40 mM dithiothreitol (DTT) was added to neutralize excess IAM. Subsequently, the sample was subjected to centrifugal filtration to remove excess urea and DTT using a 10 kDa molecular weight cut-off filter (Millipore, Billerica, MA). The purified sample with a volume of 30 μL was collected and serial diluted by Tris buffer to 0.03, 0.15, 0.6 and 1.5 μg/μL. Each solution, containing 3.75 nmol to 75 pmol of protein, was spiked with 50 pmol of the four heavy isotope labeled peptide standards (*P1-4). Trypsin was then added at a 1:30 enzyme-to-glycoprotein ratio, followed by 18 h incubation of the sample at 37 °C. Additional trypsin was added at a 1:100 enzyme/glycoprotein ratio to ensure complete digestion for an additional 4 h at 37 °C. The digestion was stopped by adding 1 μL acetic acid, and samples were stored at -20 °C until analyzed.
N-Deglycosylation
The glycoprotein, 300 μg, was suspended in 30 μL of 100 mM Tris buffer (pH 8.0), and the solution was thermally denatured at 90 °C for 10 min. After the sample was cooled to room temperature, 6 μL PNGase F solution (5000 units/mL, New England Biolabs, MA) was added to the sample, and the mixture was incubated at 37 °C overnight. The deglycosylated sample was subjected to trypsin digestion under the same condition described above except that no isotopically labeled standards were spiked into the sample. The prepared solution was kept at -20 °C prior to the analysis.
LC-MS Analysis
Each sample was analyzed by LC-MS in triplicate. HPLC was conducted on a Waters Acquity UPLC system (Milford, MA), and mass spectrometry was performed on an Orbitrap Velos Pro hybrid ion trap-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA). Samples (5 μL) were separated using an Aquasil C18 capillary column (320 μm i.d. × 15 cm, 300 Å, Thermo Scientific). Mobile phases included eluent A (99.9% H2O+ 0.1% formic acid) and eluent B (99.9% CH3CN+ 0.1% formic acid). The following gradient was used: 5% eluent B for 5 min, followed by a linear increase to 40% B in 50 min, and a ramp to 95% B in 10 min. The column was held at 95% B for another 10 min before re-equilibration [14-15]. The mass spectrometer was operated at an ESI spray voltage of 3.0 kV with the capillary temperature of 250 °C, and full scan mass spectra (m/z 400-2000) were collected at a resolution of 30,000 at m/z 400. A separate LC-MS experiment was also performed to acquire MS/MS data on two analytes of interest, in which the precursor ions of m/z 1006.20 (eluting at 51.0-51.5 min) and 1006.53 (eluting at 49.5-50.0 min) were selected for collision-induced dissociation (CID) at 35% normalized collision energy, with an isolation width of 3 m/z units.
Results and Discussion
The workflow for quantitative glycosylation site occupancy analysis is illustrated in Scheme 1. Isotopically labeled internal standards are spiked into the glycoprotein sample prior to trypsin digestion, and the digested mixture is analyzed by LC-MS. For a specific N-glycosylation site, the site occupancy is determined by equation 1:
| (1) |
Furthermore, the entire protein population could be divided into two categories: one with the occupied glycosylation site and the other with the unoccupied glycosylation site:
| (2) |
By combining equation 1 and 2, the site occupancy is calculated by equation 3:
| (3) |
Accordingly, the total protein concentration is determined by spiking isotopically labeled peptide standards into the protein sample, while the site-unoccupied protein concentration is quantified by using the labeled peptide standard that contains the unoccupied glycosylation site. Therefore, the site occupancy is readily determined without any glycosidase reaction. It should be noted that in order for these equations to be valid, 100% of the partially glycosylated peptide must be accounted for. In other words, if an additional modification were present on the glycosylated peptide, such as a phosphorylation site, this could impact the accuracy of the above-described method. However, these situations rarely occur, and one can verify in advance whether or not other PTMs are present on the peptide containing the glycosylation site to be quantified.
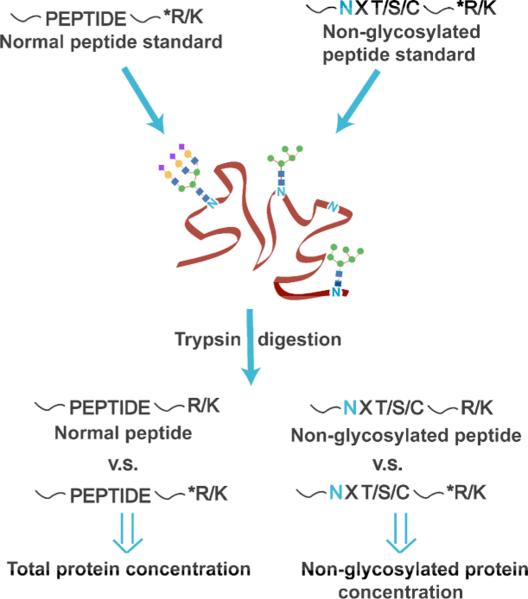
Scheme 1.
Peptide standards with heavy isotope labeled tryptic termini (denoted as *R/K) are spiked to quantify the total protein concentration and the non-glycosylated protein concentration.
As a demonstration of the method, the partially occupied glycosylation site of bovine fetuin at Asn-158 was studied [16-17]. As a first step, we verified that no additional PTMs were present on this peptide. Then, three fetuin peptide standards containing heavy isotopes at the C-terminal ends (denoted as *P1-3, sequences listed in Supplementary Table 1) were spiked into the fetuin sample, followed by trypsin digestion. LC-MS data was used to quantify the total concentration of fetuin by comparing the peak areas from extracted ion chromatograms of the fetuin peptides (P1-3) against those of the corresponding standards. A fourth isotopically labeled peptide standard, *P4, was also included in the experiment. This standard was used to quantify the partially non-glycosylated Asn-158 (contained in fetuin peptide P4, VVHAVEVALATFNAESNGSYLQLVEISR, where N is the potential glycosylation site), by absolute quantitation of P4 in the same way. Extracted ion chromatograms of the fetuin peptides and spiked peptide standards are shown in Supplementary Figure 1; the standards co-eluted with the tryptic peptides of fetuin, as expected.
Supplementary Table 2 summarizes the glycosylation site occupancy values determined by using the quantitation results of the four fetuin peptides (P1-4), based on different concentrations (0.03-1.5 μg/μL) of fetuin spiked with the isotopically labeled internal standards. These data indicate that the glycosylation site occupancy can be measured precisely under different protein concentrations.
The method described in Scheme 1 requires an effective protease digestion because the first three peptide standards are used to determine the concentration of the protein that would be quantified by the fourth standard, if the protein were 100% unglycosylated at the site being studied. In other words, a peptide concentration measured at one part of the protein must be equal to a peptide concentration measured at a different part of the protein. The simplest way to monitor whether or not the peptide concentration is being measured consistently throughout the protein is to use three isotopically labeled peptide standards from different parts of the protein. If each of the three peptide standards produces internally similar results for quantifying the protein concentration, then one can be reasonably assured that the quantitation results of the first three peptide standards are accurately answering the question: How much protein is quantified if the glycosylation site is 100% unoccupied? The fourth standard (P4, which measures the peptide containing the glycosylation site), then, measures the actual (lower) peptide concentration when the glycosylation site is partially occupied.
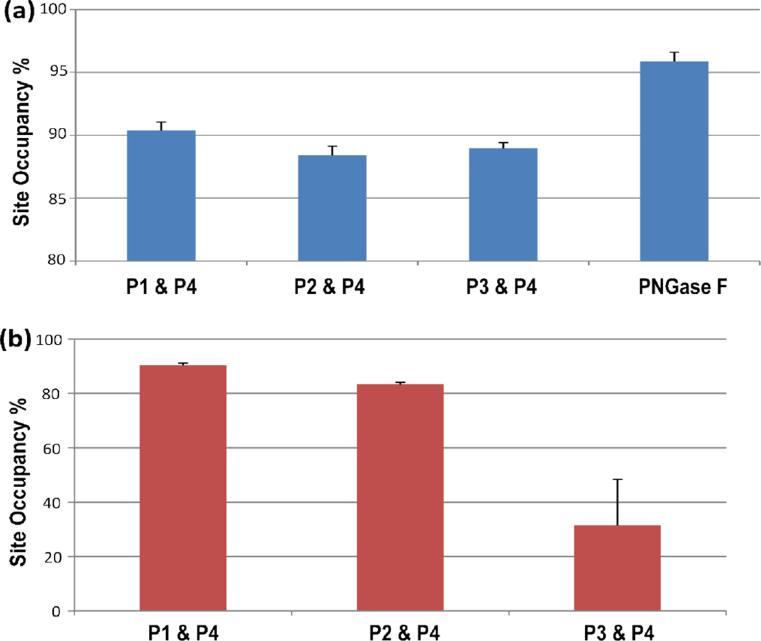
In order to demonstrate that the first three standards (*P1-3) are effective for determining the concentration expected if 100% of the protein is unglycosylated, each of the distinct peptides (P1-3) are quantified, and their concentrations are compared to the concentration of P4, respectively, to give the individual glycosylation site occupancy values. When the variance of the glycosylation site occupancy is low, as calculated by quantifying different combinations of peptides (i.e. P1&P4, P2&P4, P3&P4), this implies that the protein is consistently digested at the four isotopically labeled sites and the quantitative result is reliable. As exemplified in Figure 1a, the glycosylation site occupancy values measured by using the three peptide combinations in a fetuin solution of 0.6 μg/μL are internally consistent (ranging from 88.4-90.3%), indicating that the experiment was successful.
Figure 1.
(a) Mean values of percent site occupancy (n=3) calculated by using the measured concentration of three different peptides (P1-3) and of the non-glycosylated peptide (P4) in a fetuin solution of 0.6 μg/μL; the rightmost bar indicates the site occupancy (n=5) determined by using PNGase to deglycosylate the protein. (b) The percent site occupancy shown was measured in the same way as (a), but herein the analyzed fetuin samples were incompletely digested.
To demonstrate what the data would look like when an imprecise, inaccurate result is obtained, we prepared fetuin solutions with reduced trypsin incubation time of 8 h and analyzed the incompletely digested samples under the same workflow. Under these conditions, the four peptides are not expected to be released from the protein in a consistent manner. The resulting glycosylation site occupancy values measured by different peptides, as shown in Figure 1b, vary significantly from each other (ranging from 31.5-90.4%). In this case, one would readily know that the experiment is problematic and the quantitation using any of the three (non-glycosylated) peptide standards is inaccurate. If these results were obtained on an unknown protein, additional attention to the digestion conditions would be needed prior to quantifying the glycosylation site.
In addition to assuring complete digestion, one other experimentally important detail associated with this method is that the MS signals for the peptides need to be measured in the linear response range. To demonstrate that the experiments above were conducted in the linear range, we plotted four calibration curves measuring the instrument response of the peptides across the concentration range used in this experiment. These data are shown in Supplemental Figure 2, and each calibration curve has good linearity (R2 at or above 0.99).
To compare our new method to the traditional approach, we determined the site occupancy of fetuin using the standard protocol, adding PNGase F to deglycosylate the protein before trypsin digestion then quantifying the percent site occupancy by comparing the peak area of the deglycosylated peptide (m/z 1006.53) to that of the non-glycosylated peptide (m/z 1006.20). The result is included in Figure 1b. In comparison to the approach using labeled internal standards, the PNGase F method results in a higher calculated site occupancy value of 95.8%. We hypothesized that the discrepancy in the measurements is due to inaccuracies in the PNGase F method: Specifically, spontaneous deamidation of the non-glycosylated peptide (P4) is being incorrectly assigned as deglycosylated peptide generated from the PNGase F reaction. If this hypothesis is correct, one would expect to see the spontaneously deamidated peptide even when PNGase F is not present.
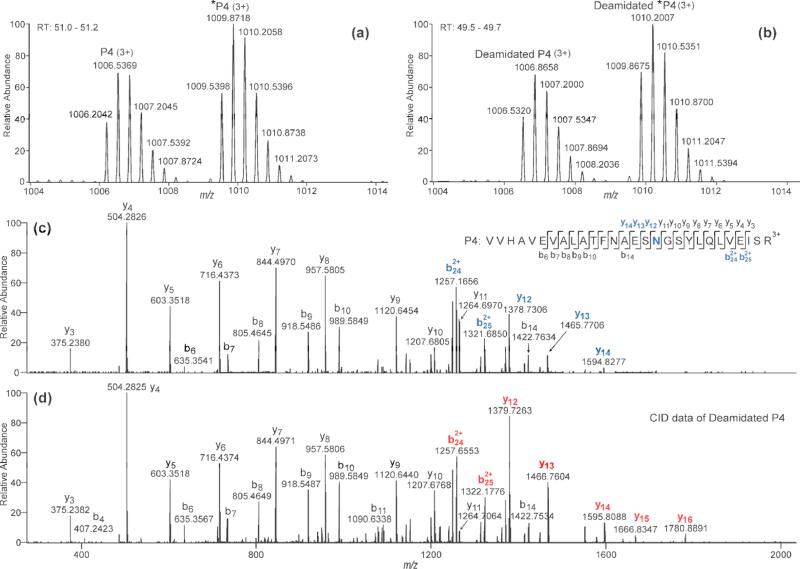
The data in Figure 2 demonstrate that spontaneous deamidation is occurring in the sample with no PNGase F added, thus skewing the quantitation results for the PNGase F approach. Figure 2a shows the high resolution MS data of the native non-glycosylated peptide P4 (monoisotopic m/z 1006.2042), and Figure 2b shows the data of the spontaneously deamidated form of this peptide (monoisotopic m/z 1006.5320), which elutes slightly earlier and is heavier in mass by 0.983 Da. Deamidation is also found for the isotopically labeled internal standard (*P4), where the deamidated *P4 (monoisotopic peak at m/z 1009.8675) co-eluted with the deamidated, unlabeled P4, as shown in Figure 2b. The deamidation site can be localized to Asn-158 by comparing the CID-MS/MS data of the peptide P4 (Figure 2c) against the CID data of the deamidated P4 (Figure 2d). As shown in Figure 2c, b-ions (b6-b14) and y-ions (y3-y11) that do not contain Asn-158 (labeled in blue) have identical m/z values as their counterpart ions in Figure 2d; by contrast, b- and y-ions (b242+-b252+ and y12-y14, labeled in blue, Figure 2c) that contain Asn-158 are 1 Da less in mass compared to the respective ions (labeled in red, Figure 2d) that carry the Asn-158. Hence we can conclude that chemical deamidation happens on the unoccupied N-glycosylation site, Asn-158. Since the spiked internal standard (*P4) is identical to the non-glycosylated peptide (P4) except for its isotopically labeled C-terminus, it must undergo deamidation to the same extent as the native peptide, assuming the deamidation occurs during sample preparation and not at the protein level. We verified that protein-level deamidation is not occurring by comparing the peak area of the deamidated fetuin peptide (deamidated-P4) to that of the deamidated internal standard (deamidated-*P4). The ratios of these two peak areas were nearly the same as the ratios of the non-deamidated forms of the peptides (P4 and *P4). In summary, the accuracy of our quantitation method is not undermined by chemical deamidation during sample prepartion, which induces incorrect quantitative results in the conventional PNGase F approach.
Figure 2.
(a) and (b) Zoomed-in mass spectra at retention times of 51.0-51.2 min and 49.5-49.7 min, respectively, of the digested fetuin solution containing heavy isotope labeled standards, with no PNGase F added. In (b), the MS peaks represent the chemically deamidated peptides. (c) and (d) CID-MS/MS data of the non-glycosylated peptide (P4) and its chemical deamidation product (deamidated P4), respectively. In (c), the potential glycosylation site Asn-158 and fragment ions that contain the site are labeled in blue; in (d), fragment ions that contain the glycosylation site are labeled in red.
Conclusion
We employed stable isotope labeled internal standards in determining the occupancy of a glycosylation site in a protein. The developed method quantifies the overall protein concentration and the amount of the non-glycosylated portion in order to measure the glycosylation site occupancy. No glycosidase is used throughout the protocol. Consequently, the new approach is free from inaccuracies inherent when quantifying using the PNGase F method: Chemical deamidation does not skew the results of the new approach, and one does not need to assume that the deglycosylated peptide and the non-glycosylated peptide have the same response factor. The presented quantitative method can be easily adopted into typical workflows for glycoprotein quantitation and identification.
Supplementary Material
Acknowledgments
The authors acknowledge funding from the National Institute of Health grant 1R01AI094797.
REFERENCES
- 1.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang S-H, Yang X, Zhang M-Y, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake PM, Cho W, Li BS, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the Pot: Adding Glycosylation to the Biomarker Discovery Equation. Clin. Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones J, Krag SS, Betenbaugh MJ. Controlling N-linked glycan site occupancy. Biochim. Biophys. Acta-Gen. Subj. 2005;1726:121–137. doi: 10.1016/j.bbagen.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Petrescu AJ, Milac AL, Petrescu SM, Dwek RA, Wormald MR. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 5.Hulsmeier AJ, Paesold-Burda P, Hennet T. N-glycosylation site occupancy in serum glycoproteins using multiple reaction monitoring liquid chromatography-mass spectrometry. Mol. Cell. Proteomics. 2007;6:2132–2138. doi: 10.1074/mcp.M700361-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Ivancic MM, Gadgil HS, Halsall HB, Treuheit MJ. LC/MS analysis of complex multiglycosylated human alpha(1)-acid glycoprotein as a model for developing identification and quantitation methods for intact glycopeptide analysis. Anal. Biochem. 2010;400:25–32. doi: 10.1016/j.ab.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 7.Kuster B, Mann M. O-18-labeling of N-glycosylation sites to improve the identification of gel-separated glycoproteins using peptide mass mapping and database searching. Anal. Chem. 1999;71:1431–1440. doi: 10.1021/ac981012u. [DOI] [PubMed] [Google Scholar]
- 8.Segu ZM, Hussein A, Novotny MV, Mechref Y. Assigning N-Glycosylation Sites of Glycoproteins Using LC/MSMS in Conjunction with Endo-M/Exoglycosidase Mixture. J. Proteome Res. 2010;9:3598–3607. doi: 10.1021/pr100129n. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Cao L, He YF, Qiao L, Xu CJ, Lu HJ, Yang PY. Tandem O-18 Stable Isotope Labeling for Quantification of N-Glycoproteome. J. Proteome Res. 2010;9:227–236. doi: 10.1021/pr900528j. [DOI] [PubMed] [Google Scholar]
- 10.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision Mapping of an In Vivo N Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Wright HT. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- 12.Palmisano G, Melo-Braga MN, Engholm-Keller K, Parker BL, Larsen MR. Chemical Deamidation: A Common Pitfall in Large-Scale N-Linked Glycoproteomic Mass Spectrometry-Based Analyses. J. Proteome Res. 2012;11:1949–1957. doi: 10.1021/pr2011268. [DOI] [PubMed] [Google Scholar]
- 13.Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Silva DV, Fuchser J, Kaspar S, Rapp E, Seeberger PH, Kolarich D. Quantitative mapping of glycoprotein micro- heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J. Mass Spectrom. 2013;48:627–639. doi: 10.1002/jms.3189. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Z, Hua D, Clark DF, Go EP, Desaire H. GlycoPep Detector: A tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra. Anal. Chem. 2013;85:5023–5032. doi: 10.1021/ac400287n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Z, Su X, Clark DF, Go EP, Desaire H. Characterizing O-linked glycopeptides by electron transfer dissociation: fragmentation rules and applications in data analysis. Anal. Chem. 2013;85:8403–8411. doi: 10.1021/ac401814h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carr SA, Huddleston MJ, Bean MF. Selective identification and differentiation of N-linked and O-linked oligosaccharides in glycoproteins by liquid chromatography-mass spectrometry. Protein Sci. 1993;2:183–196. doi: 10.1002/pro.5560020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Froehlich JW, Briscoe AC, Lee RS. The GlycoFilter: A simple and comprehensive sample preparation platform for proteomics, N-glycomics and glycosylation site assignment. Mol. Cell. Proteomics. 2013;12:2981–2991. doi: 10.1074/mcp.M113.027953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.