Abstract
Small molecules are useful tools for probing the biological function and therapeutic potential of individual proteins, but achieving selectivity is challenging when the target protein shares structural domains with other proteins. The Bromo and Extra-Terminal (BET) proteins have attracted interest because of their roles in transcriptional regulation, epigenetics, and cancer. The BET bromodomains (protein interaction modules that bind acetyl-lysine) have been targeted by potent small-molecule inhibitors, but these inhibitors lack selectivity for individual family members. We developed an ethyl derivative of an existing small-molecule inhibitor, I-BET/JQ1, and showed that it binds leucine/alanine mutant bromodomains with nanomolar affinity and achieves up to 540-fold selectivity relative to wild-type bromodomains. Cell culture studies showed that blockade of the first bromodomain alone is sufficient to displace a specific BET protein, Brd4, from chromatin. Expansion of this approach could help identify the individual roles of single BET proteins in human physiology and disease.
The Bromo and Extra-Terminal (BET) proteins Brd2, Brd3, Brd4, and Brdt play key roles in transcriptional regulation by controlling networks of genes involved in cellular proliferation and cell-cycle regulation as part of multiprotein complexes. Misregulation of BET protein activity has been linked to disease states, notably in NUT-midline carcinoma and other cancers (1). Key to the activity of BET proteins are paired, highly homologous bromodomains present in their amino-terminal regions (Fig. 1A) that direct recruitment to nucleosomes by specifically binding to acetylated lysines within histone tails. Elucidation of the complex biological processes controlled by BET proteins would benefit greatly from chemical probes that allow perturbation of individual bromodomains with high selectivity.
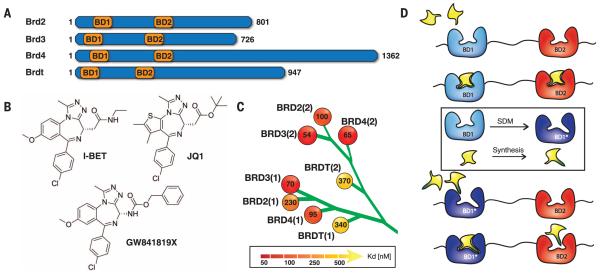
Fig. 1. BET bromodomains, pan-selective inhibitors, and bump-and-hole approach.
(A) Domain organization of BET proteins. The name and length of the proteins are shown together with the position of their first and second bromodomains. (B) Chemical structures of BET bromodomain inhibitors I-BET, JQ1, and GW841819X that share a common triazolodiazepine scaffold. (C) Dissociation constants (Kd, in nanomolar) determined by means of ITC are shown for I-BET binding to the eight individual BET bromodomains distributed as subfamily branch in a human bromodomain phylogenetic tree. (D) Rationale for a bump-and-hole approach to engineer selectivity of BET bromodomain inhibitors.
Potent cell-active small molecules based on a triazolodiazepine scaffold including I-BET (2), JQ1 (3), and GW841819X (4) (Fig. 1B) were recently discovered that bind to the acetyl-lysine (KAc) binding pocket of BET bromodomains [dissociation constant (Kd) 50 to 370 nM for I-BET] (Fig. 1C, table S1, and fig. S1). These molecules display activity in vivo (5) against NUT-midline carcinoma (6), multiple myeloma (7), mixed-lineage leukemia (8), and acute myeloid leukemia (9, 10). Several compounds, including I-BET, are now in clinical trials (11). These and other inhibitors developed to date are pan-selective for the BET members relative to other bromodomains (3) but show poor selectivity within the subfamily (Fig. 1C). Lack of selectivity confounds association of the pharmacology of BET bromodomain inhibitors to a particular target, which has fueled interest in finding more selective inhibitors. However, it is not clear which BET bromodomains should be the target of such an effort, and the high structural conservation and sequence identity of their KAc-binding sites hamper achievement of single-target selectivity.
To address these problems, we hypothesized that a chemical genetic approach could be devised based on engineered shape complementarity between the bromodomain and a small-molecule inhibitor, allowing systematic generation of an orthogonal high-affinity protein-ligand variant pair. In a “bump-and-hole” strategy conceptually related to that developed to selectively target the adenosine 5′-triphosphate–binding site of protein kinases (12, 13), a conserved hydrophobic residue on the bromodomain would be mutated to a smaller residue to generate a “hole” on the protein. The mutant would then be specifically targeted with analogs of a known ligand bearing a sterically bulky “bump” that can be accommodated by the pocket engineered on the mutant protein. Wild-type (WT) bromodomains would then bind the bulky analog more weakly as a result of a steric clash between the bump and the naturally occurring residue (Fig. 1D).
We first inspected sequence alignments of the BET bromodomains (Fig. 2A) and their crystal structures with bound I-BET, JQ1, and GW841819X (Fig. 2B and fig. S2) to identify a suitable position for mutagenesis. These analyses revealed a leucine residue [L94 in Brd4(1)] from the ZA loop that is strictly conserved throughout the BET subfamily and forms side-chain hydrophobic contacts with the inhibitors, thus providing a general strategy. To introduce a hole that could accommodate bumped ligands, we mutated leucine to an alanine (L/A). All L/A mutants displayed melting temperatures (Tm) above 40°C and thermal shifts (ΔTm) within +1° and −3°C relative to wild type in differential scanning fluorimetry (DSF) (table S2) and showed overall similar binding profiles toward multi-acetylated histone H4 peptides compared with that of the respective wild type, by means of biolayer interferometry (BLI) (fig. S3). Representative L/A mutants of both bromodomains of Brd2 and Brd4 as well as singly and doubly mutated tandem constructs retained the ability to bind to a tetra-acetylated H4 peptide in isothermal titration calorimetry (ITC), showing Kd within five- to ninefold (first bromodomain) and 0.3- to 1.6-fold (second bromodomain) relative to that of the wild type (table S3). Furthermore, singly and doubly mutated full-length Brd4 constructs conditionally expressed with tetracycline could restore c-Myc mRNA levels in a background of small interfering RNA knockdown of endogenous Brd4 in U2OS osteosarcoma cells (fig. S4). Together, these results demonstrate that L/A mutants of BET bromodomains are stable, retain KAc-specific histone binding, and can substitute for WT functionality.
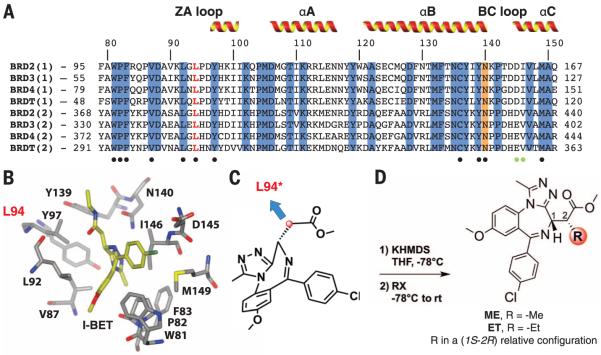
Fig. 2. Identification of L94 as mutational position and bumped ligand design.
(A) Sequence alignment of the eight BET bromodomains. Conserved residues (blue) and a conserved asparagine (orange) that directly hydrogen bonds to acetyl-lysine are highlighted, and positions of α-helices are shown. Conserved and nonconserved residues making contacts with I-BET are highlighted with black and green dots, respectively.The targeted leucine residue is highlighted in red.The sequence numbering shown on top of the alignment is for Brd4(1). (B) Stick representation of I-BET (yellow carbons) and binding site residues of Brd4(1) [PDB 3P5O (2)]. L94 is highlighted in red. (C) Chemical structure of a methylester derivative of I-BET and position selected for derivatization to target a hole introduced by mutation at L94. (D) Synthetic route to introduce bumps at position 2 of the methylester side chain. R, alkyl; X, I; KHDMS = potassium bis(trimethylsilyl)amide.
Analyses of crystal structures suggested that functionalization of the side chain methylene of I-BET (Fig. 2C, red carbon) could provide a desired bump to fill the hole introduced by the L/A mutation. Docking studies suggested optimal substitutions to be in a (R)-configuration (Fig. 2D and fig. S5). To minimize alteration in electrostatics and physicochemical properties of the ligand scaffold, a methylester analog of I-BET was functionalized with alkyl groups at the R position (Fig. 2D). Compounds ME (R = methyl) and ET (R = ethyl) were synthesized and tested by means of DSF and ITC to determine whether they are more selective for L/A than for WT proteins (Fig. 3, A and B). Ligand ME bound to WT Brd2(1) and Brd2(2) with Kd of 1.5 and 0.3 μM, respectively, which are a seven- and threefold weaker binding than I-BET as a result of introducing the bump. ME displayed Kd of 17 and 22 nM (ΔTm of 7.9° and 9.3°C, respectively) against the corresponding L/A mutants, which are a 90- and 14-fold stronger binding relative to wild type. This promising selectivity profile was improved upon introducing a bulkier ethyl group. Compound ET showed Kds of 74 and 86 nM (ΔTm of 7.6° and 8.1°C) against Brd2(1)L110A and Brd2(2)L383A, respectively, which are 120- and 200-fold increases in affinity compared with wild type (Kd, 9 and 17 μM) (Fig. 3, A and B).
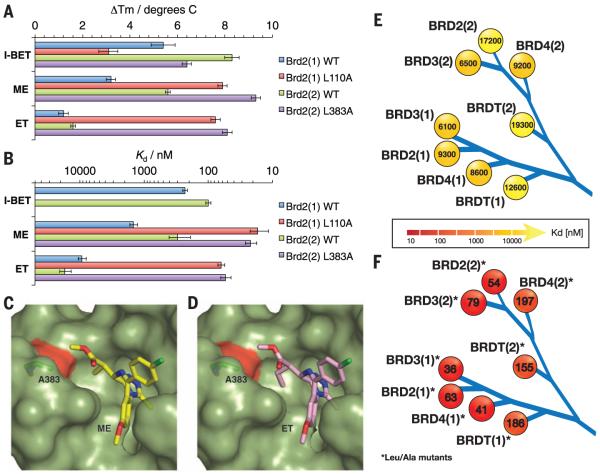
Fig. 3. Bumped ligand ET binds to engineered L/A bromodomains with high selectivity.
(A) Thermal stabilization (ΔTm, in degrees Celsius) of WT Brd2 bromodomains and their L/A mutants by I-BET, ME, and ET measured by means of DSF. The data shown are mean ± SD of three measurements. (B) Dissociation constants (Kd, in nanomolar) determined by means of ITC for I-BET binding to WT and for ME and ET binding to WT and L/A mutant Brd2 bromodomains. The error bars reflect the quality of the fit between the nonlinear least-squares curve and the experimental points. (C and D) Cocrystal structures of Brd2(2)L383A (green, surface representation) in complex with ME [(C) stick, yellow carbons] and ET [(D) stick, pink carbons].The L/A mutation is shown in red, and A383 is shown as stick, green carbons. (E and F) Affinity profile (Kd, in nanomolar) of ET binding to (E) wild-type and (F) L/A mutant BET bromodomains measured by means of ITC.
To validate our design strategy, we solved high-resolution structures of Brd2(2)L383A apoprotein (apo) and in complex with ME and ET using x-ray crystallography (table S4 and figs. S6 to S8). The L/A mutant retained the same overall fold as the wild type (root mean square deviation of backbone atoms = 0.22 Å) (fig. S6). As predicted by docking, bumped ligands adopt the same binding mode as that of I-BET and JQ1 (fig. S8, A and B), positioning the respective methyl and ethyl substituents toward the hole introduced by the L/A mutation (Fig. 3, C and D, and fig. S8C). In particular, the ethyl group of ET achieves optimal shape complementarity with the engineered pocket (Fig. 3D) and desired steric clash against the Leu side chain present in the wild type (fig. S8D).
We next profiled selectivity across the entire BET bromodomain subfamily by characterizing the binding of ET to all WT and L/A mutant proteins by means of DSF (table S5) and ITC (Fig. 3, E and F, table S5, and fig. S9). ET induced high thermal stabilization of all L/A mutants, with ΔTm ranging from 5.4° to 13.4°C, while stabilizing all WT proteins by less than 3°C (table S5). Pleasingly, ET maintained high binding affinity toward all L/A mutants, with Kd ranging from 36 to 200 nM (Fig. 3F) and large favorable binding enthalpies ΔH from −8 to −22 kcal/mol (table S5). Crucially, ET was notably less potent against WT bromodomains, with Kd ranging from 6 to 19 μM (Fig. 3E). A complete profile of ET binding affinity for L/A versus the wild type shows that binding selectivity of up to 540-fold and no less than 30-fold (average of 160-fold) is afforded across the subfamily (table S6). Taken together, these results demonstrate that our orthogonal ET and L/A mutant pairs achieve tailored high selectivity at the level of individual bromodomains.
To further demonstrate the feasibility of targeting the L/A mutation selectively, we characterized binding affinities and stoichiometries of ET within the context of a tandem bromodomain construct of Brd2 using ITC (Fig. 4A). In contrast to I-BET (Kd = 360 nM and expected stoichiometry of 2:1), no binding of ET to wild type was observed (Kd > 10 μM). The inactivity of ET against WT was further evidenced by its inability to induce upregulation of p21 mRNA levels, as reporter of downstream c-Myc activity (14), when compared with I-BET treatment in U2OS cells (fig. S10). However, ET exhibited Kd of 140 to 150 nM for the two single L/A mutants and 24 nM for the double mutant, with the expected 1:1 and 2:1 stoichiometries, respectively, confirming potent and selective targeting of mutant versus WT bromodomain (Fig. 4A).
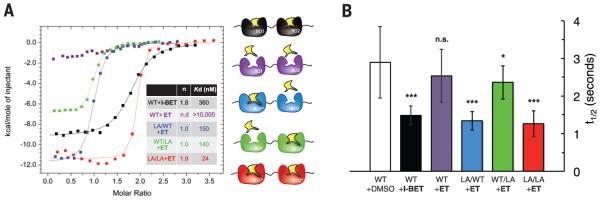
Fig. 4. ET is highly selective for L/A BET bromodomains in vitro and in cells.
(A) ITC titrations of I-BET against WT tandem bromodomain construct of Brd2 (black) and of ET against the wild type (purple) and the same construct containing the L/A mutation in the first only (blue), second only (green), and both first and second bromodomains (red) at 30°C. Stoichiometry n and dissociation constants Kd are given in the inset table. (B) Evaluation of the selectivity of ET in human osteosarcoma cells (U2OS) cells by using FRAP. Quantitative comparison of half-time of fluorescence recovery are shown for cells transfected with full-length human green fluorescent protein (GFP)–Brd4 and treated with dimethyl sulfoxide (white, vehicle control) or 1 μM I-BET (black), and for cells expressing wild type (purple), L/A-WT (blue), WT-L/A (green), or double L/A (red) GFP-Brd4 and treated with 1 μM ET.The data shown represent the mean ± SEM (n = 16 to 21 biological replicates). Statistical significance was determined with one-tailed t tests: *P < 0.05; **P < 0.01; ***P < 0.001; n.s. not significant.
To assess probe selectivity inside cells, we developed fluorescence recovery after photobleaching (FRAP) assays in U2OS cells transfected with full-length human Brd4. Control treatment with 1 μM I-BET accelerated the fluorescence recovery of the photobleached nuclear region of cells transfected with wild type (Fig. 4B, black, and fig. S11) relative to vehicle (Fig. 4B, white), indicating displacement of Brd4 from chromatin, as reported with JQ1 (3). As expected, exposure with 1 μM ET against wild type showed no significant reduction of recovery times relative to vehicle-treated cells (Fig. 4B, purple). Crucially, exposure of 1 μM ET against a double L(94,387)/A mutant showed recovery times comparable with the I-BET control in FRAP assays (Fig. 4B, red), and similarly fast recoveries were seen when the first domain only was mutated (Fig. 4B, blue) but not the second (Fig. 4B, green). Together, our data show that ET retains selectivity in cells and suggest that blockade of the first domain alone is sufficient to displace Brd4 from chromatin.
We describe a bump-and-hole approach to engineer controlled selectivity onto small-molecule modulation of BET bromodomains. We demonstrate that mutation of a conserved leucine residue within the bromodomain can be targeted by an ethyl derivative of I-BET with high potency and BET-subfamily selectivity in vitro and in cells. We also show proof of concept of applying orthogonal bromodomain:ligand pairs to dissect the role of individual bromodomains of Brd4 in chromatin binding. Future application of this approach could help identify which BET bromodomain target would be the most relevant therapeutic target in a given disease condition. To this end, recent advances in site-specific nuclease technologies for targeted genome engineering by use of clustered regulatory interspaced short palindromic repeat (CRISPR)/Cas9-based RNA-guided DNA endonucleases, among others (15, 16), have opened up the possibility of systematically generating knock-in mutants in cells and living rodents (17). If a desired selectivity cannot be achieved at the KAc-binding site of WT bromodomains, it could be achieved instead by targeting allosteric sites or by modulating other specific protein-protein interactions of BET multiprotein complexes. Last, our approach could be extended to engineer selective chemical control within other subfamilies of the human bromodomain phylogenetic tree.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Knapp, O. Fedorov, and their team for constructs, assistance with BLI, and discussions; S. Swift for assistance with the Light Microscopy Facility; C. Conte, E. Griffis, V. Cowling, and M. Peggie for materials and discussions; and D. Chirgadze for assistance with the Crystallographic X-ray Facility. This work was supported by awards to A.C. from the UK Biotechnology and Biological Sciences Research Council (BBSRC, grant BB/J001201/1 and David Phillips Fellowship BB/G023123/1). E.L.S. and A.P. were supported by European Commission Erasmus work placement grants. Microscopy and biophysics were supported by Wellcome Trust strategic awards to the University of Dundee (097945/Z/11/Z and 100476/Z/12/Z, respectively). The University of Dundee and the authors have filed patent applications (GB1320994.5 and GB1401001.1) related to the use of the bump-and-hole BET bromodomain chemical probes and mutant pairs for examining the biological function of BET bromodomain proteins. Coordinates and structure factors have been deposited with the Protein Data Bank (PDB) under accession code 4QEU [Brd2(2)L383A apo], 4QEV (in complex with ME), and 4QEW (in complex with ET).
Footnotes
www.sciencemag.org/content/346/6209/638/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S11
Tables S1 to S6
References (18–36)
REFERENCES AND NOTES
- 1.Belkina AC, Denis GV. Nat. Rev. Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicodeme E, et al. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, et al. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung CW, et al. J. Med. Chem. 2011;54:3827–3838. doi: 10.1021/jm200108t. [DOI] [PubMed] [Google Scholar]
- 5.Prinjha RK, Witherington J, Lee K. Trends Pharmacol. Sci. 2012;33:146–153. doi: 10.1016/j.tips.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 6.GlaxoSmithKline A study to investigate the safety, pharmacokinetics, pharmacodynamics, and clinical activity of GSK525762 in subjects with NUT midline carcinoma (NMC) ClinicalTrials.gov identifier NCT01587703; available at www.clinicaltrials.gov/show/NCT01587703.
- 7.Delmore JE, et al. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson MA, et al. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J, et al. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mertz JA, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, Knapp S. Nat. Rev. Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 12.Shah K, Liu Y, Deirmengian C, Shokat KM. Proc. Natl. Acad. Sci. U.S.A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishop AC, et al. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 14.Lamoureux F, et al. Nat. Commun. 2014;5:3511. doi: 10.1038/ncomms4511. [DOI] [PubMed] [Google Scholar]
- 15.Cong L, et al. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, et al. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaj T, Gersbach CA, Barbas CF., 3rd Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.