Abstract
Identification of the specific HLA locus and allele presenting an epitope for recognition by specific T cell receptors (HLA restriction) is necessary to fully characterize the immune response to antigens. Experimental determination of HLA restriction is complex and technically challenging. As an alternative, the restricting HLA locus and allele can be inferred by genetic association, utilizing response data in an HLA typed population. However, simple odds ratio calculations can be problematic when dealing with large numbers of subjects and antigens and because the same epitope can be presented by multiple alleles (epitope promiscuity). Here, we develop a tool, denominated Restrictor Analysis Tool for Epitopes (RATE), to extract inferred restriction from HLA class II -typed epitope responses. This automated method infers HLA class II restriction from large datasets of T cell responses in HLA class II typed subjects by calculating Odds Ratios and relative frequencies from simple data tables. The program is validated by 1. Analyzing data of previously determined HLA restrictions. 2. Experimentally determining in selected individuals new HLA restrictions using HLA transfected cell lines 3. Predicting HLA restriction of particular peptides, and showing that corresponding HLA class II tetramers efficiently bind to epitope specific T cells. We further design a specific iterative algorithm to account for promiscuous recognition by calculation of Odds Ratio values for combinations of different HLA molecules while incorporating predicted HLA binding affinity. The RATE program streamlines the prediction of HLA class II restriction across multiple T cell epitopes and HLA types.
Keywords: HLA restriction, MHC, epitope, T cell, tetramer
Introduction
Determination of the HLA restriction of human T cell responses is becoming increasingly necessary, as new approaches to immunophenotyping such as CyTOF, Fluidigm, and RNA profiling, rely on tetramer staining as a way to gate or isolate antigen specific T cells (1-3). In molecular terms, HLA restriction reflects the formation of a trimolecular complex, encompassing the antigen specific T cell receptor (TCR) and a bimolecular complex formed by a specific epitope, and a specific HLA molecule. TCR binding occurs when the epitope/HLA complex “fits” the antigen-binding site of the specific TCR. Hence, recognition of that epitope by the TCR is “restricted “ by that particular HLA type. In cellular terms, a T cell response to a given epitope being restricted by a given HLA refers to the fact that T cells recognition will only occur when that epitope is presented by an APC or a target expressing the specific HLA molecule that was involved in the original priming and elicitation of the T cell response.
The production of tetramer staining reagents relies on the exact identification not only of the epitope recognized by the specific T cells, but also the specific HLA molecule(s) that bind and present the specific epitope to T cell scrutiny (4). Since HLA molecules are polygenic (encoded by multiple loci) and polymorphic (each gene can be encoded by different allelic variants) this task is not trivial, also due to the extreme HLA diversity present in human populations (5). HLA class II molecules are heterodimers consisting of alpha (less polymorphic) and beta (highly polymorphic) chains, and HLA class I molecules are encoded at multiple loci (-A, B, C as the main ones). As of January 2015, the three major HLA class I alpha chain loci are comprised of 9,308 alleles (HLA-A (2,995), HLA-B (3,760) and HLA-C (2,553)), which all bind the invariant beta2-microglobulin chain. The class II loci consist of an alpha (less polymorphic) and a beta (highly polymorphic) chain, and the four major class II loci include 97 alpha and 2,963 beta alleles (HLA-DPA (38 alleles) and -DPB (489), HLA-DQA (52) and -DQB (734) and HLA-DRA (7), B1 and B3/4/5 (1,740)) (5).
The exact HLA type of human subjects in a population under study can be readily determined by a variety of methods, increasingly relying on second generation sequencing methods (1-3). The gene and allelic variant restricting specific T cell responses is determined by additional experimentation relying on classical immunobiological approaches, such as inhibition by HLA locus specific antibodies, and use of matched/mismatched or single HLA molecule transfected cell lines (6).
An alternative is to use classic genetic tools based on calculations of Odds Ratios (OR) (7). The OR method has been used extensively to estimate the likelihood that a certain genetic trait is associated with a certain biological condition or outcome (8-10). The method is based on comparing the frequency with which a certain outcome is observed in individuals carrying a given gene or allelic variant to the frequency of outcome in individuals not expressing the given gene or allelic variant. For example, OR have been extensively used to pinpoint and quantify the relative contribution of various genes to autoimmune diseases (11, 12).
The OR method can be utilized to determine likely restrictions, by considering the presence or absence of a response to a given epitope as the biological outcome, and calculating the OR and associated statistical significance for each HLA molecule expressed in human subjects in which a response, or lack thereof, is observed. The method is simple and powerful. However, by definition, statistical significance is reached only when a suitably large number of subjects are assayed. As a result, calculations of OR and statistical significance may become cumbersome, especially when a relatively large number of epitopes is simultaneously analyzed in a cohort represented by a large number of allelic polymorphisms.
Additional complexity may arise due to “HLA linkage” and/or “epitope promiscuity”. Certain HLA class I and II alleles are sometimes in very strong linkage disequilibrium, thus a positive OR can be obtained for an HLA molecule that is not the real restricting element of the epitope but is in linkage disequilibrium with the real restricting allele. Further, an epitope can be restricted by multiple HLA molecules (a phenomenon called epitope promiscuity). Therefore obtaining a positive OR may be problematic, because restriction by multiple alleles may proportionally increase the noise (“rider” alleles) without increasing the signal, thereby masking relevant restricting alleles. This can further complicate identification of restricting allele for an epitope.
Here we report the development and initial validation of the Restrictor Analysis Tool for Epitopes (RATE), a computational application which allows the user to obtain reports describing HLA class II restrictions inferred from response patterns in an HLA typed human population based on a standardized process flow and statistical evaluation.
Materials & Methods
Programming
The RATE tool is a Python 2.6.5+ CGI script. The web interface is implemented using HTML and python-CGI with the statistical analysis done using RPy (http://rpy.sourceforge.net/).
Statistical analysis
OR assess the strength of association of one property to another in a sample population (7). In the current application, OR are used to quantify the strength of associations between expression of a specific allele and detection of positive immune response. An OR > 1 indicates a positive association between the two properties in question (i.e., expressing the specific allele increases the “odds” of having positive immune response). OR are calculated according to the formula:
Where
A+ = Number of donors expressing a specific allele
A− = Number of donors not expressing the specific allele
R+ = Number of donors with a positive immune response to the specific peptide
R− = Number of donors that do not have a positive immune response to the specific peptide
Thus, for example, A+R+ indicates number of donors expressing the specific allele AND having a positive response against a specific peptide.
The OR becomes infinity when none of the donors that do not express the allele have a positive response, i.e. A-R+ = 0. While this cannot be avoided, a “Relative frequency” (RF) can be used to estimate the enrichment of responders expressing a given allele relative to the whole population. Since this value will never be zero, the RF measure will never be “infinity” due to division by zero, even in instances where the OR measure is “infinity” due to division by zero. Accordingly we also calculate RF, which is expressed as the ratio of the response in donors expressing the specific allele to the response in all donors, and is calculated as follows:
The Fisher's exact test is applied here to calculate the statistical significance of the difference in immune response between the donors who express a specific allele and those who do not, thus highlighting the restricting allele for each peptide. A p-value < 0.05 was considered statistically significant.
No adjustments are made on the p-value to correct for multiple statistical tests. Accordingly, the p-values cannot be taken as actual probabilities of a given restriction to be true, but rather serve as a relative ranking which restriction has most statistical support. The purpose of these rankings is to guide further experiments that are necessary to fully confirm restrictions, and that allow the experimenter to focus on prioritized candidate HLA alleles.
Iterative algorithm for detection of promiscuous binding alleles
A specific algorithm was designed to address epitope promiscuity. The algorithm first identifies the HLA alleles expressed in each of the donors that gave a positive response to an epitope. The binding affinity of the epitope for each of the alleles identified is then predicted using the IEDB's MHC binding prediction tools using RESTful web services (http://tools.immuneepitope.org/main/html/tools_api.html) (13). The binding prediction is done using the consensus method, which uses a combination of NN-align, SMM-align and CombLib/Sturniolo. If the specified allele is not available under the consensus method, the NetMHCIIpan method is chosen by default. More details on prediction methods is available in Paul et al (14). All alleles predicted to bind with the peptide (IEDB consensus percentile ≤15.0) are selected for further screening. For general binding predictions for individual alleles, the recommended threshold for considering a peptide to be binder is IEDB consensus percentile 10.0 whereas for predicting promiscuous binding, the recommended threshold is 20.0 (14, 15). The cutoff utilized here (15.0) was chosen as a midway between these two thresholds, and based on the reasoning that too stringent of a cutoff might be overlooking potential promiscuous restrictions.
After the algorithm calculates the OR, RF and p-value for each individual allele, it then combines response data for all possible allele pairs, and evaluates if an improved p-value is obtained. Subsequent iterations combine various allele groups and p-values are calculated with each iteration. Iteration cycles continue until the p-value cannot be improved further. The algorithm reports the allele combinations associated with the best p-value for each epitope. For example, consider alleles A, B, C and D with the following values for an epitope:
| Allele | A+R+ | A−R+ | A+R− | A−R− | RF | OR | p-value |
|---|---|---|---|---|---|---|---|
| A | 8 | 3 | 27 | 45 | 1.7 | 4.4 | 0.046 |
| B | 4 | 7 | 10 | 62 | 2.2 | 3.5 | 0.084 |
| C | 3 | 8 | 8 | 64 | 2.1 | 2.9 | 0.157 |
| D | 4 | 7 | 9 | 63 | 2.3 | 3.9 | 0.065 |
| B+C+D | 10 | 1 | 20 | 52 | 2.5 | 17.4 | <0.001 |
While only allele A is associated with p-value < 0.05 and no other allele showed significant p-value, iterative analysis by combining the data for the other alleles may reveal that the combination of the three alleles (B, C & D) gives a more significant p-value. The algorithm reports this combination of alleles as potential promiscuous restricting alleles.
Availability
The tool is available online at http://iedb-rate.liai.org/.
Immune Response datasets
Immune response datasets were generated as described previously (16) and by McKinney et al (manuscript in preparation). Briefly, immune responses to various Mycobacterium tuberculosis (MTB) epitopes in PBMCs from individuals with latent MTB infection (LTBI) were measured by IFN-γ-specific ELISPOT as representative of TH1 responses and reported as spot-forming cells (SFC) per million cells.
Additionally, CD4+ T cell immune responses to 15-mer peptides from the acellular Bordetella pertussis vaccine (Dillon et al, manuscript in preparation) were measured as previously described (17, 18). Briefly, PBMCs isolated from whole blood were stimulated with isolated B. pertussis vaccine proteins for 14 days, with fresh human recombinant IL-2 added every three days. Subsequently, the cells were restimulated with peptides and lymphokine production was measured with a dual IFN-γ and IL-5 ELISPOT assay, representative of TH1 and TH2 CD4+ subsets, respectively (18).
The criteria for positivity for the ELISPOT assay we utilized was as follows; responses were considered positive if the stimulus had >20 SFC per 106 PBMCs, p<0.05 by Student's t-test, and a stimulation index > 2.0. These criteria are the ones we have consistently used for over 10 years, and have been maintained for consistency's sake herein as well. (15, 17, 19-22).
HLA typing
HLA class II typing was performed using next generation sequencing methods (McKinney et al, submitted). Specifically, amplicons were generated from the appropriate Class II locus for exons 2 through 4 by PCR amplification. Sequencing libraries were generated (Illumina Nextera XT) from these amplicons, and sequenced with MiSeq Reagent Kit v3 as per manufacturer instructions (Illumina, San Diego, CA). Sequence reads were matched to HLA alleles and donor genotyping assigned (McKinney et al, submitted).
Tetramer staining experiments
B.pertussis biotinylated HLA class II tetramers conjugated to streptavidin-PE were provided by the Tetramer Core Laboratory at Benaroya Research Institute. For B.pertussis ex vivo tetramer staining experiments, CD4+ T cells were isolated from cryopreserved PBMCs using CD4+ T cell Isolation Kit (Miltenyi) according to manufacturer's instructions. For B.pertussis in vitro tetramer staining, isolated PBMCs were stimulated as above with the tetramer-specific peptide for 14 days and subsequently harvested for analysis. Purified CD4+ cells or expanded cells were incubated with a 1:50 dilution of tetramer-PE for 2 hours at room temperature and stained for 30 min at room temperature in FACS buffer (PBS with 2% FBS) with antibodies to following surface markers: CD3-AF700 (BD Bioscience), CD4-APCef780 (eBioscience), CD8-V500 (BD Bioscience), CD45RA-ef450 (eBioscience), and CCR7-PerCPCy5.5 (Biolegend). After washing, cells were resuspended in PBS and read on a BD LSRII and analyzed with FlowJo. MTB biotinylated HLA class II tetramers were generously provided by the NIH tetramer core facility. For MTB ex vivo tetramer staining, PBMCs were stained with tetramer-PE, CD4-FITC, CD8a-PECy5, CD19-PECy5, CD11b-PECy5, CD56-PECy5, and Live-Dead Aqua (Invitrogen). Tetramer stained cells were enriched with anti-PE magnetic beads (Miltenyi) and analyzed.
Experimental restriction determination by single HLA transfected cells
To verify if HLA/epitope predicted restrictions were correct, antigen presentation assays with single HLA transfectants were performed using cell lines as described (6). Briefly, PBMCs isolated from whole blood were incubated ex vivo with peptide-pulsed EBV-transformed cell lines expressing selected HLA molecules in an IFN-γ ELISPOT assay, as described above. The cell lines used for transfection were DAP.3 (for DRB1*03:01, DRB1*04:01, DRB1*07:01, DRB1*11:01, DRB1*13:01, DRB1*15:01, DRB4*01:01, and DRB5*01:01) and RM3 (for DRB3*02:02, DPA1*02:01/DPB1*01:01, DPA1*01:01/DPB1*04:01, DQA1*05:01/B1*02:01, and DQA1*01:02/DQB1*06:02) (6). All DR lines used DRA1*01:01 as the alpha chain. Allele restriction was determined by comparing responses of peptide-pulsed cell lines with media only-pulsed cell lines. The level of statistical significance was determined with a Student's t-test using the mean of triplicate values of the response against peptide pulsed cell lines versus the response against the media pulsed cell line control.
Results
Standardized import format generation for subjects and epitope-donor response
To afford analysis of congruent datasets in a standardized fashion, RATE was designed to allow importing test results from a number of different epitopes in a given biological assay, and for responses in a cohort of donors/test subjects. A tab delimited plain text file format was chosen as it is typically available as a data export option from most instrumentation, or can easily be generated from commonly utilized graphing and spreadsheet software. Supplemental Table I shows sample response data in a spreadsheet.
The main focus of our efforts was to design and validate a method to facilitate determination of restrictions for HLA class II molecules, and accordingly we mostly utilized data obtained in our laboratory where HLA class II responses were measured in a number of different settings, utilizing 15-mer peptides and ELISPOT or ICS assays, in a sufficient number of donors. To measure immune responses we routinely use ELISPOT. However, data from any assay may be utilized, as long as operative criteria for positivity can be defined. While the two statistical measures employed (OR and RF, as described below) depend on a binary outcome (positive or negative), the absolute values determined by the assay can be entered directly, and a threshold for positivity can be chosen prior to calculation of the metrics. Positivity thresholds for ELISPOT measures have been described previously by our group (6, 16). The tool does not require that all epitopes be tested in all donors.
Similar to the response data, RATE uploads HLA typing data provided as a tab delimited plain text file (Supplemental Table II). While different typing methods determine HLA type to differing levels of resolution (e.g., allele group, protein, etc.) (23), the OR and RF methods are independent from the HLA methods used for typing. Indeed, the MHC type analyzed may be processed using any user preferred nomenclature system as long as the typing categories are mutually exclusive. In our laboratory, we routinely HLA type to the protein level (four digit typing, e.g. HLA-DRB*01:01), but serological (HLA-DRB1), allele group (HLA-DRB*01), and even typing formats down to the level demarcating synonymous DNA substitutions (HLA-DRB*01:01:01) are also compatible with the approach. Furthermore, complete HLA class II typing is not required, and data for a given locus or even a partial list of alleles can be used, with the caveat that the tool will output the most likely statistical association given the data at hand, while a better candidate could have been identified with a more complete dataset.
Two alternative outputs of calculated ORs, and RFs
For each epitope/HLA combination, RATE generates a matrix tabulating the number of positive versus negative responders amongst the subjects expressing that particular HLA type, as well as the number of positive and negative responders among the subjects NOT expressing that HLA type. Next, for each epitope RATE calculates the ORs and RFs corresponding to each of the HLA molecules following classical formulas, as described in the Materials and Methods. The addition of RF allows the user to calculate a numerical value for epitope/HLA pairs for which an OR would be incalculable due to lack of response among individuals not expressing that particular HLA allele. The Fisher's exact test is used to calculate the significance (p-value) associated with each epitope/HLA combination.
For each peptide, the OR, RF, and p-values for each of the HLA types expressed in the responder subjects are ranked and tabulated from high to low OR. In this reporting format, the number of responding individuals positive in the assay (R+) and expressing each particular HLA (A+) are also presented (A+R+). The number of similarly defined A+R−, A−R+ and A−R− individuals is also reported. A partial example of this type of report is shown in Table I.
Table I.
RATE HLA restriction complete results.
| Peptide# | Peptide id | Peptide seq | Allele# | Allele | A+ R+ | A− R+ | A+ R− | A− R− | No. of Donors | Response n/a | RF | OR | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 1 | DPA1*01:03 | 13 | 9 | 48 | 11 | 81 | 6 | 0.78 | 0.34 | 0.048 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 1 | DPA1*01:03 | 7 | 1 | 54 | 19 | 81 | 6 | 1.16 | 2.44 | 0.672 |
| 3 | 3531.0511 | GEEYLILSARDVLAV | 1 | DPA1*01:03 | 6 | 0 | 59 | 22 | 87 | 0 | 1.34 | inf | 0.330 |
| 4 | 3550.0065 | STHEANTMAMMARDT | 1 | DPA1*01:03 | 3 | 1 | 34 | 9 | 47 | 40 | 0.95 | 0.80 | 1.000 |
| 5 | 3550.0063 | DLVRAYHSMSSTHEA | 1 | DPA1*01:03 | 5 | 0 | 32 | 10 | 47 | 40 | 1.27 | inf | 0.569 |
| 6 | 3550.0061 | DLVRAYHAMSSTHEA | 1 | DPA1*01:03 | 4 | 0 | 33 | 10 | 47 | 40 | 1.27 | inf | 0.564 |
| 7 | 3550.0060 | AMEDLVRAYHAMSST | 1 | DPA1*01:03 | 2 | 0 | 35 | 10 | 47 | 40 | 1.27 | inf | 1.000 |
| 8 | 3550.0059 | IMYNYPTMLGHAGDM | 1 | DPA1*01:03 | 4 | 1 | 33 | 9 | 47 | 40 | 1.02 | 1.09 | 1.000 |
| 9 | 3550.0058 | MSQIMYNYPTMLGHA | 1 | DPA1*01:03 | 4 | 1 | 33 | 9 | 47 | 40 | 1.02 | 1.09 | 1.000 |
| 10 | 3550.0057 | IMYNYPAMLGHAGDM | 1 | DPA1*01:03 | 7 | 1 | 30 | 9 | 47 | 40 | 1.11 | 2.07 | 0.667 |
| 11 | 3550.0056 | MSQIMYNYPAMLGHA | 1 | DPA1*01:03 | 7 | 2 | 30 | 8 | 47 | 40 | 0.99 | 0.93 | 1.000 |
| 12 | 3550.0055 | TEIRRSNAPRLVDLV | 1 | DPA1*01:03 | 1 | 0 | 36 | 10 | 47 | 40 | 1.27 | inf | 1.000 |
| 13 | 3550.0052 | GTEIRRSDAPRLVDL | 1 | DPA1*01:03 | 2 | 0 | 35 | 10 | 47 | 40 | 1.27 | inf | 1.000 |
| 14 | 3550.0051 | SNIKIIRIDEFRRCG | 1 | DPA1*01:03 | 1 | 0 | 36 | 10 | 47 | 40 | 1.27 | inf | 1.000 |
| 15 | 3550.0046 | HSNIKIIRIDEFRRY | 1 | DPA1*01:03 | 1 | 0 | 36 | 10 | 47 | 40 | 1.27 | inf | 1.000 |
| 16 | 3550.0028 | PYVIELDGQFCGQLT | 1 | DPA1*01:03 | 1 | 0 | 42 | 14 | 57 | 30 | 1.33 | inf | 1.000 |
| 17 | 3550.0026 | EWTVRHTVAAWPAVC | 1 | DPA1*01:03 | 1 | 0 | 42 | 14 | 57 | 30 | 1.33 | inf | 1.000 |
| 18 | 3550.0024 | GTEIRRSNAPRLVDLV | 1 | DPA1*01:03 | 1 | 0 | 38 | 12 | 51 | 36 | 1.31 | inf | 1.000 |
| 19 | 3550.0020 | HSNIKIIRIDEFRRYG | 1 | DPA1*01:03 | 1 | 0 | 38 | 12 | 51 | 36 | 1.31 | inf | 1.000 |
| 20 | 3550.0006 | AAVLRFQEAANKQKQ | 1 | DPA1*01:03 | 3 | 0 | 38 | 12 | 53 | 34 | 1.29 | inf | 1.000 |
| 21 | 3536.0170 | THSWEYWGAQLNAMK | 1 | DPA1*01:03 | 1 | 0 | 41 | 11 | 53 | 34 | 1.26 | inf | 1.000 |
| 22 | 3536.0147 | AGSLSALLDPSQGMG | 1 | DPA1*01:03 | 2 | 1 | 41 | 12 | 56 | 31 | 0.87 | 0.59 | 0.555 |
| 23 | 3536.0144 | SAMILAAYHPQQFIY | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 24 | 3536.0139 | PQWLSANRAVKPTGS | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 25 | 3536.0138 | LTSELPQWLSANRAV | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 26 | 3536.0136 | GCQTYKWETFLTSEL | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 27 | 3536.0133 | SSFYSDWYSPACGKA | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 28 | 3536.0132 | PVGGQSSFYSDWYSP | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 29 | 3536.0131 | LSIVMPVGGQSSFYS | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 30 | 3536.0121 | PSMGRDIKVQFQSGG | 1 | DPA1*01:03 | 1 | 0 | 42 | 13 | 56 | 31 | 1.30 | inf | 1.000 |
| 31 | 3536.0109 | ALGATPNTGPAPQGA | 1 | DPA1*01:03 | 1 | 0 | 40 | 12 | 53 | 34 | 1.29 | inf | 1.000 |
| 32 | 3536.0102 | GGHNGVFDFPDSGTH | 1 | DPA1*01:03 | 1 | 0 | 43 | 14 | 58 | 29 | 1.32 | inf | 1.000 |
| 33 | 3536.0071 | QTYKWETFLTSELPG | 1 | DPA1*01:03 | 1 | 0 | 42 | 14 | 57 | 30 | 1.33 | inf | 1.000 |
The table shows a part of the “complete results” obtained for a sample data set.
This complete report is usually too large and cumbersome to evaluate. For example, even considering only the 25 most common variants of the four polymorphic HLA class II molecules yields 100 different HLA molecules, and in the case of a set of 200 peptides this generates an output with 20,000 entries. For this reason the tool also generates a concise report (Table II) which lists, for each peptide, only RF values greater than 2.0. This threshold is utilized since the determination of negative HLA association is not within the scope of the present application, and stronger RF (or OR) values are more likely to reflect HLA restrictions. HLA molecules that are associated with significant ORs and RFs AND predicted to bind the corresponding epitope with high affinity (IEDB consensus prediction score less than the 15th percentile) (13) are considered as potential restrictions.
Table II.
RATE HLA restriction concise results.
| Peptide# | Peptide id | Peptide seq | Allele# | Allele | A+ R+ | A− R+ | A+ R− | A− R− | No. of Donors | Response n/a | RF | OR | p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 66 | DQB1*06:01 | 4 | 18 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.004 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 101 | DRB1*15:02 | 4 | 18 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.004 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 111 | DRB5*01:02 | 2 | 20 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.071 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 3 | DPA1*01:05 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 18 | DPB1*104:01 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 31 | DPB1*27:02 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 33 | DPB1*40:01 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 43 | DQA1*03 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 58 | DQB1*03:08 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 76 | DRB1*04:02 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 81 | DRB1*04:10 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 87 | DRB1*08:06 | 1 | 21 | 0 | 59 | 81 | 6 | 3.68 | inf | 0.272 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 100 | DRB1*15:01 | 11 | 11 | 1 | 58 | 81 | 6 | 3.38 | 53.97 | 0.000 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 110 | DRB5*01:01 | 14 | 8 | 5 | 54 | 81 | 6 | 2.71 | 17.84 | 0.000 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 25 | DPB1*14:01 | 2 | 20 | 1 | 58 | 81 | 6 | 2.45 | 5.65 | 0.178 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 62 | DQB1*05:02 | 2 | 20 | 1 | 58 | 81 | 6 | 2.45 | 5.65 | 0.178 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 84 | DRB1*08:02 | 2 | 20 | 1 | 58 | 81 | 6 | 2.45 | 5.65 | 0.178 |
| 1 | 3531.0375 | MSQIMYNYPAMMAHA | 39 | DQA1*01:03 | 5 | 17 | 4 | 55 | 81 | 6 | 2.05 | 3.96 | 0.056 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 43 | DQA1*03 | 1 | 7 | 0 | 73 | 81 | 6 | 10.13 | inf | 0.099 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 76 | DRB1*04:02 | 1 | 7 | 0 | 73 | 81 | 6 | 10.13 | inf | 0.099 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 84 | DRB1*08:02 | 2 | 6 | 1 | 72 | 81 | 6 | 6.75 | 21.84 | 0.025 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 111 | DRB5*01:02 | 1 | 7 | 1 | 72 | 81 | 6 | 5.06 | 9.69 | 0.189 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 62 | DQB1*05:02 | 1 | 7 | 2 | 71 | 81 | 6 | 3.38 | 4.90 | 0.271 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 66 | DQB1*06:01 | 1 | 7 | 2 | 71 | 81 | 6 | 3.38 | 4.90 | 0.271 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 86 | DRB1*08:04 | 1 | 7 | 2 | 71 | 81 | 6 | 3.38 | 4.90 | 0.271 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 101 | DRB1*15:02 | 1 | 7 | 2 | 71 | 81 | 6 | 3.38 | 4.90 | 0.271 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 71 | DRB1*01:01 | 3 | 5 | 7 | 66 | 81 | 6 | 3.04 | 5.47 | 0.055 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 60 | DQB1*04:02 | 2 | 6 | 5 | 68 | 81 | 6 | 2.89 | 4.40 | 0.140 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 47 | DQA1*04:01 | 3 | 5 | 8 | 65 | 81 | 6 | 2.76 | 4.73 | 0.072 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 13 | DPB1*04:01 | 6 | 2 | 17 | 56 | 81 | 6 | 2.64 | 9.54 | 0.006 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 68 | DQB1*06:03 | 1 | 7 | 3 | 70 | 81 | 6 | 2.53 | 3.26 | 0.346 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 14 | DPB1*04:02 | 3 | 5 | 10 | 63 | 81 | 6 | 2.34 | 3.70 | 0.113 |
| 2 | 3531.0461 | AGCQTYKWETFLTSE | 37 | DQA1*01:01 | 2 | 6 | 7 | 66 | 81 | 6 | 2.25 | 3.08 | 0.216 |
The table shows a part of the “concise results” obtained for a sample data set.
RATE predicts previously validated restrictions
To validate the approach, we examined whether RATE would successfully re-identify HLA class II restrictions experimentally determined in previous studies (16). The immune response and HLA allele-typing data from 3 previously validated epitope-HLA restrictions were analyzed using the program (16). As seen in Table III, donors expressing HLA DRB1*15:01 accounted for 11 of 22 responders for MTB Rv0288/Rv3019c epitope MSQIMYNYPAMMAHA, with an OR of 54.0 and an RF of 3.4, (p < 0.001). The MTB Rv3804c epitope AGCQTYKWETFLTSE and Rv3418c epitope GEEYLILSARDVLAV, were predicted to be restricted by DPB1*04:01 (p = 0.0056), and DRB1*01:01 (p = 0.0249), respectively (Table III). All three restrictions had been previously validated by peptide-tetramer staining of PBMC from HLA-matched donors (16). Thus, RATE correctly re-identified known HLA-epitope restrictions.
Table III.
Epitope/RATE Predictions Match Validated Tetramer Data.
| Sequence | Allele | A+a R+c | A−b R+ | A+ R−d | A− R− | RF | OR | p-valuee |
|---|---|---|---|---|---|---|---|---|
| MSQIMYNYPAMMAHA | DRB1*15:01 | 11 | 11 | 1 | 58 | 3.4 | 54.0 | <0.001 |
| AGCQTYKWETFLTSE | DPB1*04:01 | 6 | 2 | 17 | 56 | 2.6 | 9.5 | 0.006 |
| GEEYLILSARDVLAV | DRB1*01:01 | 3 | 3 | 8 | 73 | 4.0 | 8.7 | 0.025 |
The response data and HLA typing from three previously validated tetramers (16) were matched using the RATE Program.
A+: genotyped HLA allele positive
A−: genotyped HLA allele negative
R+: epitope response positive
R−: epitope response negative.
p-values calculated by Fisher's exact test.
Discovery of novel HLA-peptide restrictions
To further validate the use of RATE to predict novel restrictions, we considered a dataset generated by testing various MTB derived epitopes as described previously (16) and by McKinney et al (manuscript in preparation). Briefly, immune responses to various MTB epitopes in PBMCs from individuals with LTBI were measured by IFN-γ-specific ELISPOT as representative of TH1 responses and reported as spot-forming cells (SFC) per million cells. These data were utilized to generate RATE predicted restrictions.
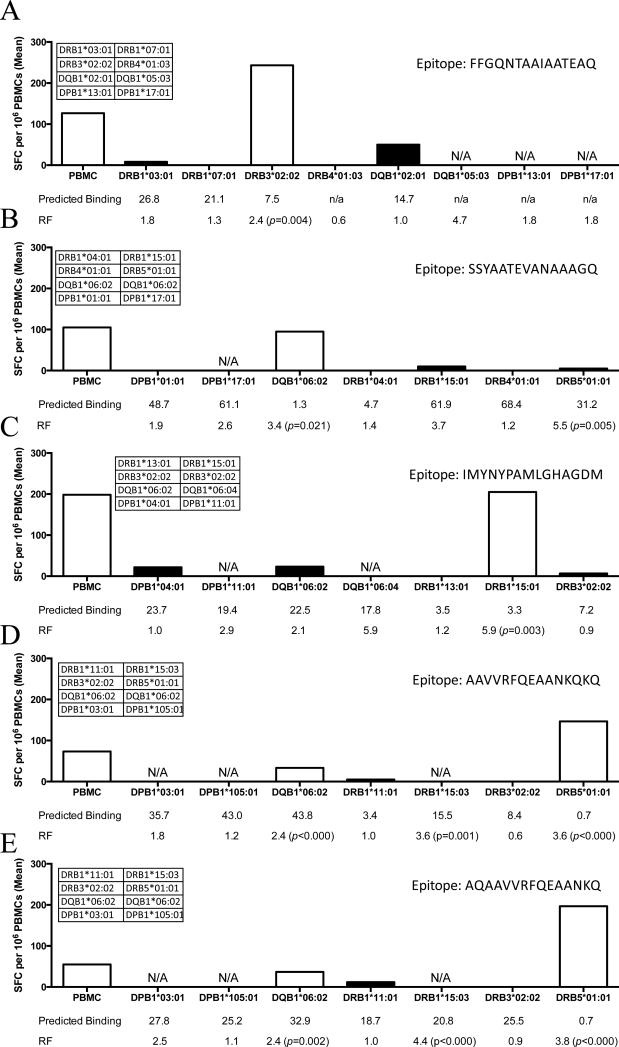
In parallel, the HLA class II restrictions of five selected MTB derived epitopes (Rv1705c; sequence FFGQNTAAIAATEAQ, Rv1195 epitope SSYAATEVANAAAGQ, Rv0288 epitope IMYNYPAMLGHAGDM, and Rv3874 epitopes AAVVRFQEAANKQKQ and AQAAVVRFQEAANKQ) were determined by single HLA transfected cell lines (6). PBMCs from LTBI were incubated together with a panel of cell lines presenting the specific epitopes and expressing HLA molecules matching those expressed in the donors (Figure 1). Responses were evaluated in a standard IFN-γ ELISPOT assay to measure TH1 responses. In the case of LTBI donors, the strong CD4+ T cell responses allow detection of IFN-gamma responses directly ex vivo (16).
Figure 1. Novel HLA-Epitope Restrictions Predicted by RATE.
PBMCs were incubated with single HLA-transfected cells pulsed with (A) Rv1705c epitope FFGQNTAAIAATEAQ, (B) Rv1195 epitope SSYAATEVANAAAGQ, (C) Rv0288 epitope IMYNYPAMLGHAGDM, or (D) Rv3874 epitopes AAVVRFQEAANKQKQ and (E) AQAAVVRFQEAANKQ for 24 hours. IFN-γ release was measured by ELISPOT. The white bars show significant responses (p-values < 0.05), while the black bars represent non-significant responses. HLA alleles expressed by donor are presented in the table insert. HLA-transfected cell lines not available are labeled N/A. Predicted binding from IEDB is listed below each allele tested. RFs and Fisher's exact test p-values (when significant) are listed for predicted restrictions.
As can be seen in Figure 1a, a significant response to the peptide was observed in the case of DRB3*02:02 transfected cells, but not for any of the other lines transfected with HLA molecules expressed by the donor. These results match those obtained with the RATE program, where DRB3*02:02 was associated with a RF of 2.4 (p-value = 0.004) for the FFGQNTAAIAATEAQ epitope. Similar patterns of restrictions and RATE predictions were observed in the four additional epitopes tested (Figures 1b-e). Thus, the RATE approach correctly predicted five new HLA restrictions.
Use of RATE to guide generation of HLA tetramers
As a next step towards validation, we tested whether RATE could predict HLA-peptide restrictions de novo as a means to guide generation of specific tetrameric staining reagents. Tetramer staining was utilized as an alternative method of validating restrictions to the transfection assay technique above to show the versatility of the RATE predictions. In an initial set of experiments, we used the immune response dataset from a study of 31 HLA class II -typed donors vaccinated with the acellular B. pertussis vaccine (Dillon et al manuscript in preparation). In this cohort, little to no reactivity was detected ex vivo, but good T cell reactivity was detected after expansion in vitro with the vaccine component proteins. Following expansion, epitopes were defined using a set of 785 overlapping peptides (16-mers overlapping by 8) completely spanning the component proteins of which 154 non-redundant epitopes induced positive TH1 and/or TH2 CD4+ T cell responses. Of those, RATE indicated potential restrictions for 35 of them.
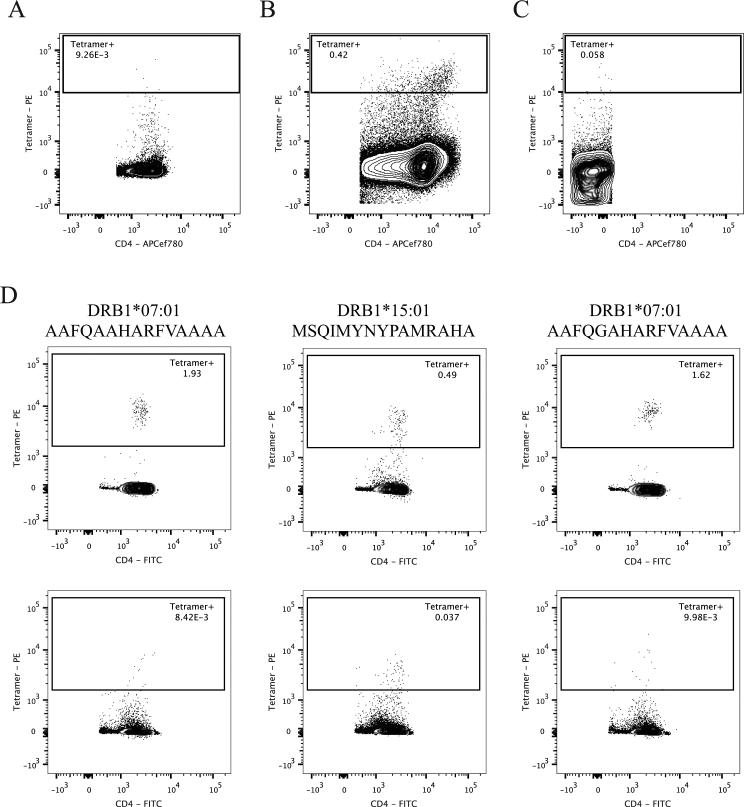
More specifically, when the combined IFNγ and IL-5 CD4+ T cell immune response reactivity was analyzed by RATE, epitope YYSNVTATRLLSSTNS from PTxB129-144 was found to be associated with significant OR values (p < 0.05; Table IV for DRB1*07:01. Accordingly, the corresponding PE-conjugated tetramer was generated and staining measured on PBMCs (Figure 2). As expected, little staining was detected directly ex vivo, with positive staining for YYSNVTATRLLSSTNS of less than 0.01 % (Figure 2a). However, after 14 days of stimulation with the corresponding peptide, the tetramer displayed significant binding to CD3+CD4+CD8− cells, representing an increase of more than 20-fold compared to ex vivo staining (0.42 %)(Figure 2b). The staining was specific since, as expected, no significant staining was detected on CD3+CD4−CD8+ cells from the same expanded PBMCs cultures (Figure 2c).
Table IV.
Selection of Significant Predicted Restrictions to Guide Tetramer Generation.
| Sequence | Allele | A+a R+c | A−b R+ | A+ R−d | A− R− | RF | p-valuee | Predicted bindingf (percentile) |
|---|---|---|---|---|---|---|---|---|
| YYSNVTATRLLSSTNS | DRB1*07:01 | 3 | 1 | 5 | 22 | 2.9 | 0.043 | 0.76 |
| AAFQAAHARFVAAAA | DRB1*07:01 | 7 | 12 | 4 | 58 | 2.7 | 0.003 | 0.03 |
| MSQIMYNYPAMRAHA | DRB1*15:01 | 11 | 11 | 1 | 58 | 3.4 | 0.000 | 0.90 |
| AAFQGAHARFVAAAA | DRB1*07:01 | 8 | 14 | 3 | 56 | 2.7 | 0.001 | 0.13 |
The response data and HLA typing from donors recently vaccinated with acellular B.pertussis or with LTBI were matched using the RATE Program.
A+: genotyped HLA allele positive
A−: genotyped HLA allele negative
R+: epitope response positive
R−: epitope response negative.
p-values calculated by Fisher's exact test.
Predicted binding values were obtained from IEDB (13).
Figure 2. Novel HLA-Epitope Restrictions Validated by Tetramer Binding.
A) Purified CD4+CD3+CD8− cells stained with YYSNVTARTLLSSTNS tetramer-PE. B) CD3+CD4+CD8− PBMCs stained with YYSNVTARTLLSSTNS tetramer-PE after 14 days of stimulation with peptide. C) CD3+CD4−CD8+ PBMCs stained with YYSNVTARTLLSSTNS tetramer-PE after 14 days of stimulation with peptide. D) Top: CD3+CD8−CD19−CD11b−CD56− PBMCs stained with the indicated tetramer-PE combinations after anti-PE magnetic bead enrichment. Bottom: Flow-through from magnetic bead enrichment. Numbers shown are percentages of tetramer+ CD4+ or CD8+ cells.
RATE predictions to generate MTB-specific tetramers
To expand the results obtained with the B.pertussis epitope, we examined additional instances of RATE predicted restrictions, in the context of the MTB response rates in LTBI donors (as described above in the case of Figure 3). In the case of LTBI and MTB epitopes, as stated above the strong CD4+ T cell responses allow detection of IFN-gamma responses directly ex vivo (16).
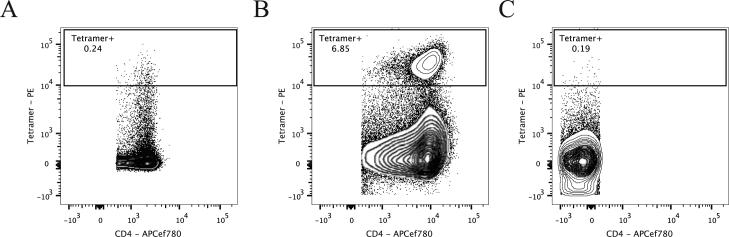
Figure 3. Predicted Promiscuous Binding Epitope Validated by Tetramer Binding.
A) Purified CD4+CD3+CD8− cells stained with VKAQNITNKRAALIEA tetramer-PE. B) CD3+CD4+CD8− PBMCs stained with VKAQNITNKRAALIEA tetramer-PE after 14 days of stimulation with peptide. C) CD3+CD4−CD8+ PBMCs stained with VKAQNITNKRAALIEA tetramer-PE after 14 days of stimulation with peptide. Numbers shown are percentages of tetramer+ CD4+ or CD8+ cells.
Three different instances of restrictions predicted from the RATE approach were selected for further investigation, as shown in Table IV. More specifically, Rv0287 epitope AAFQAAHARFVAAAA and Rv3020c epitope AAFQGAHARFVAAAA were predicted to be restricted to DRB1*07:01 (p = 0.003 and 0.001, respectively), and Rv3019c epitope MSQIMYNYPAMRAHA to DRB1*15:01 (p = 0.000). Accordingly, PBMCs from epitope responsive LTBI subjects were stained ex vivo with the respective tetramer and enriched with anti-PE magnetic beads. In all three epitope/allele combinations, tetramer staining was detected at 13- to 160-fold higher percentages than those detected in the negative control flow-through from the magnetic bead enrichment (Figure 2d).
These results further validate the use of the RATE approach to predict HLA restrictions for the purpose of generating functional tetrameric staining reagents. This method thus allows to rapidly transition from HLA typing and response data to tetramers, essentially skipping the usual HLA association determination steps.
Combined RATE calculations to identify promiscuous restrictions
Many HLA allelic variants are functionally similar (24-26). As a result, a given epitope may be restricted by multiple HLA molecules encoded by a particular locus (especially if they are close variants), or even molecules from different loci (promiscuous restriction). In these cases, the fact that multiple HLA molecules may restrict the response to a single epitope will (paradoxically) lower the statistical significance of each individual HLA restriction. To overcome this issue, we have developed an algorithm that calculates for a given peptide OR and RF values of all possible combination of alleles for which predicted binding is within the 15th percentile, and tabulates particular combinations of alleles associated with the best p-value.
To validate the approach, we selected the epitope EEWEPLTKKGNVWEV from Phlp341-55 which was previously (15) determined to be promiscuously restricted by the two HLA alleles, DRB1*08:01 and DRB1*11:01, as determined by single HLA-transfected cell lines. Indeed, when the reactivity of EEWEPLTKKGNVWEV in the cohort of allergic individuals (15) was analyzed by RATE, of 14 HLA alleles (including DRB1*08:01 and DRB1*11:01) expressed by donors responsive to the epitope, 8 combinations had RF values greater than 1.5, but none were associated with a significant p value (p > 0.05).
When the results were analyzed by the promiscuous restrictions algorithm, it was found that, the allele combination of DRB1*08:01 and DRB1*11:01 was associated with a significant p-value (p < 0.05 and RF = 3.1; Table V). We thus concluded that RATE was able to correctly predict promiscuous restriction of this previously described example of promiscuous restriction.
Table V.
Combined RF for Responsive Alleles Predicts Restriction for EEWEPLTKKGNVWEV.
| Allele(s) | A+a R+c | A−b R+ | A+ R−d | A− R− | RF | p-valuee | Predicted binding (percentile)f |
|---|---|---|---|---|---|---|---|
| DRB1*08:01 | 1 | 2 | 0 | 22 | 8.3 | 0.120 | 2.58 |
| DRB1*11:01 | 2 | 1 | 5 | 17 | 2.4 | 0.180 | 3.99 |
| DRB1*08:01 + DRB1*11:01 | 3 | 0 | 5 | 17 | 3.1 | 0.024 | n/a |
The response data and HLA typing from Timothy Grass allergic donors (19) were matched using the RATE Program promiscuity algorithm.
A+: genotyped HLA allele positive
A−: genotyped HLA allele negative
R+: epitope response positive
R−: epitope response negative. The combined predictions are shown in the bottom four rows.
p-values calculated by Fisher's exact test.
Predicted binding values were obtained from IEDB (13).
Use of RATE to de novo identify promiscuous HLA restrictions
To further validate that the RATE algorithm could also predict novel promiscuous restrictions, we selected the epitope VKAQNITNKRAALIEA from FHA1753-1768. When the reactivity of this epitope in the cohort of individuals vaccinated with the acellular B. pertussis vaccine (Dillon et al manuscript in preparation) was analyzed by RATE, no alleles were predicted as potential restrictions (p > 0.05) (Table VI). However when analyzed by the promiscuous restriction algorithm, the allele combination of DQB1*06:02 and DRB1*14:04 was associated with a significant p-value (p < 0.05 and RF ≥ 2.6; Table VI), as the predicted binding of the epitope for DQA1*01:02/DQB1*06:02 is 9.98 percentile and for DRB1*14:04 is 9.92 percentile.
Table VI.
Combined RF for Responsive Alleles Predicts Restriction for VKAQNITNKRAALIEA.
| Allele(s) | A+a R+c | A−b R+ | A+ R−d | A− R− | RF | p-valuee | Predicted bindingf (percentile) |
|---|---|---|---|---|---|---|---|
| DQB1*06:02 | 2 | 1 | 5 | 23 | 3.0 | 0.120 | 9.98 |
| DRB1*14:04 | 1 | 2 | 0 | 28 | 10.3 | 0.097 | 9.92 |
| DQB1*06:02 + DRB1*14:04 | 3 | 0 | 5 | 23 | 3.9 | 0.012 | - |
The response data and HLA typing from donors recently vaccinated with acellular B.pertussis were matched using the RATE Program promiscuity algorithm.
A+: genotyped HLA allele positive
A−: genotyped HLA allele negative
R+: epitope response positive
R−: epitope response negative. The combined predictions are shown in the bottom row.
p-values calculated by Fisher's exact test.
Predicted binding values were obtained from IEDB (13).
To validate this predicted restriction, we developed a DQB1*06:02/KAQNITNKRAALIEA tetramer. In order to match the most common alpha/beta combination in the population, DQA1*01:02 was selected for the alpha chain. The staining of the PE-conjugated tetramer of VKAQNITNKRAALIEA with DQB1*06:02 measured on PBMCs from a responsive donor (Figure 3). As above with YYSNVTATRLLSSTNS and DRB1*07:01, limited staining was detected ex vivo, (Figure 3a). In contrast, 14 days of stimulation with the corresponding peptide dramatically increased the tetramer binding to CD3+CD4+CD8− cells, changing the ex vivo percentage of 0.24 % to 6.85 % in the stimulated cells (Figure 3b). Specificity of the stain was confirmed by low staining in CD3+CD4−CD8+ cells of 0.19 % (Figure 3c). The ideal validation for promiscuous restriction of this epitope would also include the corresponding DRB1*14:04 tetramer stain. Unfortunately, DRB1*14:04 is not available as a tetramer at this time and a binding assay for this allele has not been developed as yet. However, the promiscuous algorithm of RATE does take into account the in silico predicted binding to this allele. In conclusion, the promiscuous RATE algorithm allowed identification of additional restrictions that could not be detected statistically when considered alone, which was experimentally verified where production of tetramer reagents was technically feasible.
Validation using class I allele and response data
While the RATE tool was designed with a focus on class II alleles, we speculate that it might be also applicable to the determination of HLA class I restrictions. Datasets related to epitopes of known restriction tested in groups of HLA typed donors is not generally available in our laboratory, since we and most other groups routinely infer HLA class I restriction on the basis of presence of specific motifs and HLA binding, and only test HLA matched donors for reactivity.
However, analysis of a data set on class I alleles obtained from the HIV Molecular Immunology Database by the Los Alamos National Laboratory (27) was able to address the applicability to HLA class I restrictions. The data set (Study-4 in section “HLA typing and epitope mapping”)(28) - the largest among the available studies - contained class I HLA typing data for 631 HIV patients and the reaction data (SFC values) for 409 HIV-1 peptides in each of the respective positive patient (patient that gave a positive immune response to the peptide). The HIV database provides “A list HIV CTL epitopes” which is defined as best-defined CTL/CD8+ epitopes and lists the restricting class I alleles for each epitope.
There were 118 A list epitopes that were embedded in 108 peptides in the data set and the data from these 108 peptides and the HLA typing of the 631 patients were used to validate the RATE tool. The 118 epitopes (embedded in 108 peptides) were restricted by 67 class I alleles and 45 of them were expressed by the patients in the study. These 45 alleles restricted 86 peptides (out of 108) in the data set. The results from the RATE tool showed 33 peptide-allele combinations where the allele was relatively frequent among the patients (present in ≥ 10% of patients) and expressed in at least one positive donor (A+R+ ≥ 1) (Table VII). 27 peptide-allele restrictions out of the above 33 combinations were significant hits (p-value ≤ 0.05) i.e. the RATE tool could confirm 82% of the relevant peptide-allele combinations as restrictions.
Table VII.
Validation using class I data from Los Alamos HIV Molecular Immunology database.
| Peptide sequence | Allele | A+R+ | A−R+ | A+R− | A−R− | RF | OR | p-value | Allele frequency | Embedded epitopes from “A list epitopes” |
|---|---|---|---|---|---|---|---|---|---|---|
| NDIQKLVGKLNWASQIY | A*30:02 | 9 | 3 | 58 | 561 | 7.1 | 29.0 | 0.000 | 10.62% | KLNWASQIY |
| TKELQKQIIKIQNFRVYY | A*30:02 | 18 | 8 | 49 | 556 | 6.5 | 25.5 | 0.000 | 10.62% | KIQNFRVYY |
| SKLNWASQIYPGIKVRQL | A*30:02 | 6 | 28 | 61 | 536 | 1.7 | 1.9 | 0.160 | 10.62% | KLNWASQIY |
| IKIQNFRVYYRDSRDPIW | A*30:02 | 24 | 16 | 43 | 548 | 5.7 | 19.1 | 0.000 | 10.62% | KIQNFRVYY |
| TGTEELRSLYNTVATLY | A*30:02 | 30 | 66 | 37 | 498 | 2.9 | 6.1 | 0.000 | 10.62% | RSLYNTVATLY |
| GIWQLDCTHLEGKIILVA | B*15:10 | 22 | 5 | 77 | 527 | 5.2 | 30.1 | 0.000 | 15.69% | THLEGKIIL |
| GGHQAAMQMLKDTINEEA | B*15:10 | 11 | 17 | 88 | 515 | 2.5 | 3.8 | 0.002 | 15.69% | GHQAAMQML |
| LQTGERDWHLGHGVSIEW | B*15:10 | 51 | 15 | 48 | 517 | 4.9 | 36.6 | 0.000 | 15.69% | WHLGHGVSI |
| NTMLNTVGGHQAAMQMLK | B*15:10 | 7 | 7 | 92 | 525 | 3.2 | 5.7 | 0.003 | 15.69% | GHQAAMQML |
| QMVHQAISPRTLNAWVKV | B*15:10 | 20 | 14 | 79 | 518 | 3.7 | 9.4 | 0.000 | 15.69% | HQAISPRTL |
| QGYFPDWQNYTPGPGVRY | A*29 | 9 | 45 | 95 | 482 | 1.0 | 1.0 | 1.000 | 16.48% | YFPDWQNYT |
| QITLWQRPLVSIKVGGQI | A*68:02 | 1 | 12 | 109 | 509 | 0.4 | 0.4 | 0.709 | 17.43% | ITLWQRPLV |
| QLEKEPIAGAETFYVDGA | A*68:02 | 25 | 17 | 85 | 504 | 3.4 | 8.7 | 0.000 | 17.43% | GAETFYVDGA |
| GLGQYIYETYGDTWTGV | A*68:02 | 40 | 13 | 70 | 508 | 4.3 | 22.3 | 0.000 | 17.43% | ETYGDTWTGV |
| ETYGDTWTGVEALIRIL | A*68:02 | 35 | 11 | 75 | 510 | 4.4 | 21.6 | 0.000 | 17.43% | ETYGDTWTGV |
| GAETFYVDGAANRETKI | A*68:02 | 65 | 61 | 45 | 460 | 3.0 | 10.9 | 0.000 | 17.43% | GAETFYVDGA |
| GIQQEFGIPYNPQSQGVV | B*15:03 | 7 | 12 | 106 | 506 | 2.1 | 2.8 | 0.060 | 17.91% | IQQEFGIPY |
| YHCLVCFQTKGLGISYGR* | B*15:03 | 11 | 7 | 102 | 511 | 3.4 | 7.9 | 0.000 | 17.91% | FQTKGLGISY |
| VKAACWWAGIQQEFGIPY* | B*15:03 | 38 | 11 | 75 | 507 | 4.3 | 23.4 | 0.000 | 17.91% | IQQEFGIPY |
| PRTLNAWVKVIEEKAF* | B*15:03 | 58 | 15 | 55 | 503 | 4.4 | 35.4 | 0.000 | 17.91% | VKVIEEKAF |
| AVFIHNFKRKGGIGGYSA* | B*15:03 | 80 | 12 | 33 | 506 | 4.9 | 102.2 | 0.000 | 17.91% | FKRKGGIGGY |
| YVDRFFKTLRAEQATQDV | B*15:03 | 18 | 132 | 95 | 386 | 0.7 | 0.6 | 0.038 | 17.91% | YVDRFFKTL |
| GPKEPFRDYVDRFFKTLR | B*15:03 | 24 | 105 | 89 | 413 | 1.0 | 1.1 | 0.798 | 17.91% | YVDRFFKTL |
| GKKAIGTVLVGPTPVNII | B*15:03 | 22 | 19 | 91 | 499 | 3.0 | 6.3 | 0.000 | 17.91% | GKKAIGTVL |
| EVNIVTDSQYALGII | B*15:03 | 1 | 21 | 112 | 497 | 0.3 | 0.2 | 0.152 | 17.91% | VTDSQYALGI |
| WVKVIEEKAFSPEVIPMF* | B*15:03 | 2 | 39 | 111 | 479 | 0.3 | 0.2 | 0.020 | 17.91% | VKVIEEKAF |
| IYPGIKVRQLCKLLRGAK | B*42:01 | 21 | 12 | 99 | 499 | 3.3 | 8.8 | 0.000 | 19.02% | YPGIKVRQL |
| NYTPGPGVRYPLTFGWCF* | B*42:01 | 64 | 136 | 56 | 375 | 1.7 | 3.2 | 0.000 | 19.02% | TPGPGVRYPL |
| SKLNWASQIYPGIKVRQL | B*42:01 | 16 | 18 | 104 | 493 | 2.5 | 4.2 | 0.000 | 19.02% | YPGIKVRQL |
| EVGFPVRPQVPLRPMTFK | B*42:01 | 67 | 150 | 53 | 361 | 1.6 | 3.0 | 0.000 | 19.02% | RPQVPLRPM |
| MASEFNLPPIVAKEIVA* | B*42:01 | 47 | 23 | 73 | 488 | 3.5 | 13.7 | 0.000 | 19.02% | LPPIVAKEI |
| GATPQDLNTMLNTVGGH | B*42:01 | 90 | 90 | 30 | 421 | 2.6 | 14.0 | 0.000 | 19.02% | TPQDLNTML |
| GIKQLQTRVLAIERYLK | B*58:02 | 36 | 3 | 96 | 496 | 4.4 | 62.0 | 0.000 | 20.92% | QTRVLAIERYL |
Epitope allele restrictions confirmed by Larsen et al. (28)
The analysis of the data resulted in 33 peptide-allele combinations where the allele was relatively frequent among the patients (present in ≥ 10% of patients) and expressed in at least one positive donor (A+R+ ≥ 1). Out of these 33 combinations, 27 peptide-allele restrictions were significant hits (p-value ≤ 0.05).
We also analyzed the peptide-allele restrictions confirmed by Larsen et al. (2010) (29). Out of the 18 peptides and respective restricting alleles, 7 peptide-allele combinations were present in RATE results for Los Alamos HIV database data set with the allele being expressed in at least one positive donor. All of these peptide-allele restrictions were significant hits as per the RATE results (p-value ≤ 0.05) (Table VII).”
Discussion
Herein we describe a novel approach to determine restriction of CD4+ T cell epitopes, at the population level using genetic association methods. The main focus of our efforts was to design and validate a method to facilitate determination of restrictions for HLA class II molecules, and accordingly we mostly utilized data obtained in our laboratory where HLA class II responses were measured in a number of different settings, utilizing 15-mer peptides and ELISPOT or ICS assays, in a sufficient number of donors. We anticipate that this methodology will greatly facilitate the rapid generation of tetrameric staining reagents for investigations of immune reactivity in human populations.
Identification of the specific HLA locus and allele responsible for presenting an epitope for recognition by specific T cell receptors (HLA restriction) is necessary to fully characterize the immune response to an antigen. In this context, it is helpful to distinguish determination of the HLA molecule restricting responses to a given epitope in a particular donor (i.e. individual restriction of that peptide in that donor) from the HLA molecule(s) restricting the response in a population (i.e. general restriction of that peptide in that population).
Individual and general restrictions are not necessarily, and in reality are not frequently, the same. This is because not all donors expressing a given allele will generate T cells recognizing an epitope presented by that allele, due to immunodominance at the epitope and HLA levels (15, 18, 30, 31). Thus, population restriction does not always predict individual restriction. Conversely, because the same epitope can be presented by multiple alleles (epitope promiscuity) (30, 32-36), the fact that a given epitope is presented in a particular individual by a specific allele does not fully predict its general pattern of restriction in a population.
Experimental determination of HLA class II restriction is complex and technically challenging. Since HLA molecules are polygenic (genes are encoded by multiple loci) and polymorphic (genes are encoded by different allelic variants), this task is complicated due to the extreme HLA class II diversity present in human populations (5). To determine which gene and allelic variant act as restriction element, classic approaches such as inhibition by HLA locus specific antibodies and/or the use of matched/mismatched or single HLA transfected cell lines are employed.
Inhibition with anti-HLA class II antibodies is a good way to determine the locus (but not the allele). A primary limitation is that there are no available antibodies that can distinguish the DRB1 and DRB3/4/5 loci. Additionally, questions remain in the field as to whether pan reactive antibodies for the DQ locus truly detect all DQ alpha and beta combinations. Finally, anti-HLA antibodies that inhibit T cell recognition may act by simply killing or inhibiting the APC, and hence need to be titrated to find a “sweet spot”. The use of HLA class II transfected cell lines as described by McKinney (6) is technically the most accurate, but using transfected cell lines is cumbersome and transfected cell lines are available for only the most frequent HLA alleles.
HLA binding and motif predictions could be utilized, as they are widely available. However, HLA binding in itself is only a necessary, but not sufficient, requisite for T cell recognition. Because of the considerations listed above, it is of interest to develop alternative strategies. There have been similar works in this direction, mostly focusing on class I alleles. For example, Kiepiela et al. has applied a statistical approach similar to ours to identify the HLA Class-I restriction of HIV peptides (37). Another statistical method designed by Listgarten et al. can identify restricting HLA alleles for a specific epitope from ELISpot data from a set of patients and their respective allele type (38). The HLArestrictor tool developed by Larsen et al. is based on the class I binding prediction method NetMHCpan and can predict the patient-specific epitopes restricted by alleles based on the patient HLA allele type (29). However, these methods are focused on class I alleles and are applicable to only one peptide or one patient data at a time, while the RATE tool that we describe in this study was developed to address class II restrictions and relies on datasets describing response of multiple donors, based on genetic inference. Accordingly the output of our method is based on odds ratios, relative frequency and p-value from Fisher's exact test for each peptide-allele combination to assess the strength of association between the peptide and allele.
The RATE tool described herein represents a novel approach for determining HLA class II allele restriction of epitopes. In particular, it does not depend on experimental work, and is most suited to analyze and extract immunological information from complex datasets encompassing large numbers of peptides and donors (as long as HLA typing data for each donor is available) as generated in clinical studies and vaccine trials. As such the method is also likely to be of value in system biology studies where large amounts of data are generated. Furthermore, since the program is agnostic to the MHC nomenclature used, it can be expanded to other species and we have in fact utilized it to analyze class II immunogenicity data obtained in the rhesus macaque system (Mothé et al, submitted).
While the experimental examples and data analyses provided here are focused on HLA class II, we speculate that the program can also be applied to class I, or any set of responses associated with large polymorphisms. The limited validation we performed to date utilizing the data from the Los Alamos HIV Molecular Immunology database suggest that this is likely to be the case.
However, certain caveats should be kept in mind when interpreting results derived from the approach. Despite the various options provided herein it is likely that there are instances of ambiguous results, especially for peptides weakly or infrequently recognized. This is most commonly observed when too few subjects have been tested, or in the case of alleles that are either very rare or very frequent. As a rule of thumb, strong associations can be detected with as few as 10-15 subjects, but about 30 seems to be required in most cases, with the power of the analysis increasing dramatically as more subjects are included. However, the additional calculation of RF as described here increases the likelihood of detection for strong association even with the use of a limited number of subjects. These instances are usually relatively few, and the ambiguity can be resolved with additional testing using transfected cell lines (39) or direct test of tetrameric staining reagents.
Second, it is possible that HLA molecules encoded at different loci might be associated with statistically significant OR values for the same epitope. While in some cases this may indeed be due to the promiscuity of the epitope, in others it may reflect the fact that the different HLA loci are physically close to one another in their chromosomal location, and thus are in strong linkage disequilibrium with each other (40, 41). For this reason, if alleles for more than one HLA locus are associated with significant OR values for a specific epitope, further analysis is warranted. We recommend that the locus with the best p-value be considered first. If the combination of the data from this locus with any other locus does not lead to a better p-value for the combined data, the association is likely due to linkage disequilibrium and should be discarded. In addition, instances where an allele encoded by a particular locus is not predicted to bind the epitope under consideration likely reflect an association due to linkage disequilibrium and should be considered with caution or discarded.
Third, while one of the advantages of RATE is to be able to globally analyze a dataset generated over multiple experiments (since it is in our experience impossible to determine restrictions in a single experiment by cellular methods for many donors and many peptides), the issue of reproducibility from experiment to experiment needs to be carefully considered. If significant experiment-to-experiment variability is present in a given dataset, this would correspondingly affect the conclusions. Therefore, the application of appropriate positive and negative controls within each experiment is necessary. In our experience, we always include a negative control and a positive PHA control, and ensure that each falls within acceptable ranges, based on our routine QC of experimental assays.
Finally, we acknowledge that the experimental validation of the RATE approach is still somewhat limited. In total, RATE correctly predicted ten novel restrictions (eight MTB and two B.pertussis) and five previously validated restrictions (three MTB and two Timothy Grass). Evaluation of additional epitopes mapped by other investigators is limited by the fact that immunogenicity data needs to be described on a donor-by-donor basis, and HLA typing must be available for each donor. Still, clearly more experimental work will more firmly establish the success rate of the approach.
In conclusion, we have developed an automated method to infer HLA restriction from large datasets of T cell responses in HLA-typed subjects. The web-accessible program calculates OR and relative frequencies from simple data tables, incorporates prediction of HLA binding capacity, and accounts for linkage disequilibrium and promiscuous recognition by iterative calculation of OR values for different combinations of HLA molecules. We consider the current algorithm and software implementation a proof of principle that it is possible to derive HLA restrictions based on genetic associations. To the best of our knowledge, the program presented here is the first that allows determination of restriction at the population level, and estimates response rate and immunodominance, as well as promiscuous restrictions. Accordingly, we believe is important that the current prototype is made available to the scientific community. The tool is indeed available online at http://iedb-rate.liai.org/, and we look forward to receiving user feedback, for its improvement and optimization. We expect that future refinements of the approach will lead to improved results, for example by more precisely modeling the statistical underpinnings of HLA linkage, promiscuous binding, and incorporating the predicted binding affinities as statistical priors rather than binary cutoffs.
Supplementary Material
Acknowledgments
Funding provided by NIH contracts HHSN272201200010C, HHSN272200900042C, HHSN27220140045C, and HHSB27220900044C, and Bill and Melinda Gates Foundation grant OPP1066265
Abbreviations
- FHA
Filamentous hemagluttinin
- HLA
Human leukocyte antigen
- ICS
Intracellular cytokine staining
- LTBI
Latent MTB infection
- MTB
Mycobacterium tuberculosis
- OR
Odds Ratio
- PBMCs
Peripheral blood mononuclear cells
- PTxB
Pertussis toxin subunit B
- RF
Relative frequency
- RATE
Restrictor analysis tool for epitopes
- SFC
Spot-forming cell
References
- 1.Lange V, Böhme I, Hofmann J, Lang K, Sauter J, Schöne B, Paul P, Albrecht V, Andreas JM, Baier DM, Nething J, Ehninger U, Schwarzelt C, Pingel J, Ehninger G, Schmidt AH. Cost-efficient high-throughput HLA typing by MiSeq amplicon sequencing. BMC Genomics. 2014;15:63. doi: 10.1186/1471-2164-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boegel S, Löwer M, Schäfer M, Bukur T, de Graaf J, Boisguérin V, Türeci O, Diken M, Castle JC, Sahin U. HLA typing from RNA-Seq sequence reads. Genome Med. 2013;4:102. doi: 10.1186/gm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moonsamy PV, Williams T, Bonella P, Holcomb CL, Höglund BN, Hillman G, Goodridge D, Turenchalk GS, Blake LA, Daigle DA, Simen BB, Hamilton A, May AP, Erlich HA. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array™ System for simplified amplicon library preparation. Tissue Antigens. 2013;81:141–149. doi: 10.1111/tan.12071. [DOI] [PubMed] [Google Scholar]
- 4.Nepom GT. MHC Class II Tetramers. The Journal of Immunology. 2012;188:2477–2482. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA database. Nucleic Acids Research. 2011;39:D1171–6. doi: 10.1093/nar/gkq998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKinney DM, Southwood S, Hinz D, Oseroff C, Arlehamn CSL, Schulten V, Taplitz R, Broide D, Hanekom WA, Scriba TJ, Wood R, Alam R, Peters B, Sidney J, Sette A. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics. 2013;65:357–370. doi: 10.1007/s00251-013-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris JA, Gardner MJ. Calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. Br Med J (Clin Res Ed) 1988;296:1313–1316. doi: 10.1136/bmj.296.6632.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmed I, Tamouza R, Delord M, Krishnamoorthy R, Tzourio C, Mulot C, Nacfer M, Lambert J-C, Beaune P, Laurent-Puig P, Loriot M-A, Charron D, Elbaz A. Association between Parkinson's disease and the HLA-DRB1 locus. Mov Disord. 2012;27:1104–1110. doi: 10.1002/mds.25035. [DOI] [PubMed] [Google Scholar]
- 9.Klein NP, Bartlett J, Fireman B, Rowhani-Rahbar A, Baxter R. Comparative Effectiveness of Acellular Versus Whole-Cell Pertussis Vaccines in Teenagers. PEDIATRICS. 2013;131:e1716–e1722. doi: 10.1542/peds.2012-3836. [DOI] [PubMed] [Google Scholar]
- 10.Yucesoy B, Talzhanov Y, Johnson VJ, Wilson NW, Biaginie RE, Wang W, Frye B, Weissman DN, Germolec DR, Luster MI, Barmada MM. Genetic variants within the MHC region are associated with immune responsiveness to childhood vaccinations. Vaccine. 2013:1–11. doi: 10.1016/j.vaccine.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin H, Arase N, Hirayasu K, Kohyama M, Suenaga T, Saito F, Tanimura K, Matsuoka S, Ebina K, Shi K, Toyama-Sorimachi N, Yasuda S, Horita T, Hiwa R, Takasugi K, Ohmura K, Yoshikawa H, Saito T, Atsumi T, Sasazuki T, Katayama I, Lanier LL, Arase H. Autoantibodies to IgG/HLA class II complexes are associated with rheumatoid arthritis susceptibility. Proc. Natl. Acad. Sci. U.S.A. 2014 doi: 10.1073/pnas.1401105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu L, Guo S, Yang D, Ma Y, Ji H, Chen Y, Zhang J, Wang Y, Jin L, Wang J, Liu J. Copy number variations of HLA-DRB5 is associated with systemic lupus erythematosus risk in Chinese Han population. Acta Biochim Biophys Sinica. 2014;46:155–160. doi: 10.1093/abbs/gmt137. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Ponomarenko J, Zhu Z, Tamang D, Wang P, Greenbaum J, Lundegaard C, Sette A, Lund O, Bourne PE, Nielsen M, Peters B. Immune epitope database analysis resource. Nucleic Acids Research. 2012;40:W525–30. doi: 10.1093/nar/gks438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul S, Kolla RV, Sidney J, Weiskopf D, Fleri W, Kim Y, Peters B, Sette A. Evaluating the immunogenicity of protein drugs by applying in vitro MHC binding data and the immune epitope database and analysis resource. Clinical and Developmental Immunology. 2013;2013:467852. doi: 10.1155/2013/467852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oseroff C, Sidney J, Kotturi MF, Kolla R, Alam R, Broide DH, Wasserman SI, Weiskopf D, McKinney DM, Chung JL, Petersen A, Grey H, Peters B, Sette A. Molecular Determinants of T Cell Epitope Recognition to the Common Timothy Grass Allergen. The Journal of Immunology. 2010;185:943–955. doi: 10.4049/jimmunol.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindestam Arlehamn CS, Gerasimova A, Mele F, Henderson R, Swann J, Greenbaum JA, Kim Y, Sidney J, James EA, Taplitz R, McKinney DM, Kwok WW, Grey H, Sallusto F, Peters B, Sette A. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 2013;9:e1003130. doi: 10.1371/journal.ppat.1003130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulten V, Greenbaum JA, Hauser M, McKinney DM, Sidney J, Kolla R, Lindestam Arlehamn CS, Oseroff C, Alam R, Broide DH, Ferreira F, Ferreira-Briza F, Grey HM, Sette A, Peters B. Previously undescribed grass pollen antigens are the major inducers of T helper 2 cytokine-producing T cells in allergic individuals. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3459–3464. doi: 10.1073/pnas.1300512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oseroff C, Sidney J, Tripple V, Grey H, Wood R, Broide DH, Greenbaum J, Kolla R, Peters B, Pomés A, Sette A. Analysis of T cell responses to the major allergens from German cockroach: epitope specificity and relationship to IgE production. The Journal of Immunology. 2012;189:679–688. doi: 10.4049/jimmunol.1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulten V, Tripple V, Sidney J, Greenbaum J, Frazier A, Alam R, Broide D, Peters B, Sette A. Association between specific timothy grass antigens and changes in TH1- and TH2-cell responses following specific immunotherapy. J. Allergy Clin. Immunol. 2014;134:1076–1083. doi: 10.1016/j.jaci.2014.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oseroff C, Sidney J, Vita R, Tripple V, McKinney DM, Southwood S, Brodie TM, Sallusto F, Grey H, Alam R, Broide D, Greenbaum JA, Kolla R, Peters B, Sette A. T Cell Responses to Known Allergen Proteins Are Differently Polarized and Account for a Variable Fraction of Total Response to Allergen Extracts. The Journal of Immunology. 2012;189:1800–1811. doi: 10.4049/jimmunol.1200850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindestam Arlehamn CS, Sidney J, Henderson R, Greenbaum JA, James EA, Moutaftsi M, Coler R, McKinney DM, Park D, Taplitz R, Kwok WW, Grey H, Peters B, Sette A. Dissecting Mechanisms of Immunodominance to the Common Tuberculosis Antigens ESAT-6, CFP10, Rv2031c (hspX), Rv2654c (TB7.7), and Rv1038c (EsxJ). The Journal of Immunology. 2012;188:5020–5031. doi: 10.4049/jimmunol.1103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moutaftsi M, Bui H-H, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 23.Lefranc M-P. IMGT, the International ImMunoGeneTics Information System. Cold Spring Harbor Protocols. 2011;2011:595–603. doi: 10.1101/pdb.top115. [DOI] [PubMed] [Google Scholar]
- 24.Doolan DL, Southwood S, Chesnut R, Appella E, Gomez E, Richards A, Higashimoto YI, Maewal A, Sidney J, Gramzinski RA, Mason C, Koech D, Hoffman SL, Sette A. HLA-DR-promiscuous T cell epitopes from Plasmodium falciparum pre-erythrocytic-stage antigens restricted by multiple HLA class II alleles. J. Immunol. 2000;165:1123–1137. doi: 10.4049/jimmunol.165.2.1123. [DOI] [PubMed] [Google Scholar]
- 25.Schulze zur Wiesch J, Lauer GM, Day CL, Kim AY, Ouchi K, Duncan JE, Wurcel AG, Timm J, Jones AM, Mothe B, Allen TM, McGovern B, Lewis-Ximenez L, Sidney J, Sette A, Chung RT, Walker BD. Broad repertoire of the CD4+ Th cell response in spontaneously controlled hepatitis C virus infection includes dominant and highly promiscuous epitopes. J. Immunol. 2005;175:3603–3613. doi: 10.4049/jimmunol.175.6.3603. [DOI] [PubMed] [Google Scholar]
- 26.Cecconi V, Moro M, Del Mare S, Sidney J, Bachi A, Longhi R, Sette A, Protti MP, Dellabona P, Casorati G. The CD4+ T-cell epitope-binding register is a critical parameter when generating functional HLA-DR tetramers with promiscuous peptides. Eur. J. Immunol. 2010;40:1603–1616. doi: 10.1002/eji.200940123. [DOI] [PubMed] [Google Scholar]
- 27.Yusim K, Korber BTM, Brander C, Haynes BF, Koup R, Moore JP, Walker BD, Watkins DI. HIV Molecular Immunology 2009. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, New Mexico: 2009. LA-UR 09-05941. [Google Scholar]
- 28.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nature Medicine. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 29.Erup Larsen M, Kloverpris H, Stryhn A, Koofhethile CK, Sims S, Ndung'u T, Goulder P, Buus S, Nielsen M. HLArestrictor--a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics. 2011;63:43–55. doi: 10.1007/s00251-010-0493-5. [DOI] [PubMed] [Google Scholar]
- 30.Assarsson E, Bui H-H, Sidney J, Zhang Q, Glenn J, Oseroff C, Mbawuike IN, Alexander J, Newman MJ, Grey H, Sette A. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. Journal of Virology. 2008;82:12241–12251. doi: 10.1128/JVI.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larché M. Determining MHC restriction of T-cell responses. Methods Mol. Med. 2008;138:57–72. doi: 10.1007/978-1-59745-366-0_6. [DOI] [PubMed] [Google Scholar]
- 32.Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzeciak A, Gillessen D, Pink JR. A malaria T-cell epitope recognized in association with most mouse and human MHC class II molecules. Nature. 1988;336:778–780. doi: 10.1038/336778a0. [DOI] [PubMed] [Google Scholar]
- 33.Panina-Bordignon P, Demotz S, Corradin G, Lanzavecchia A. Study on the immunogenicity of human class-II-restricted T-cell epitopes: processing constraints, degenerate binding, and promiscuous recognition. Cold Spring Harb. Symp. Quant. Biol. 54 Pt. 1989;1:445–451. doi: 10.1101/sqb.1989.054.01.053. [DOI] [PubMed] [Google Scholar]
- 34.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur. J. Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 35.Krieger JI, Karr RW, Grey HM, Yu WY, O'Sullivan D, Batovsky L, Zheng ZL, Colón SM, Gaeta FC, Sidney J. Single amino acid changes in DR and antigen define residues critical for peptide-MHC binding and T cell recognition. J. Immunol. 1991;146:2331–2340. [PubMed] [Google Scholar]
- 36.Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J. Immunol. 1990;144:1849–1856. [PubMed] [Google Scholar]
- 37.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S, Rathnavalu P, Moore C, Pfafferott KJ, Hilton L, Zimbwa P, Moore S, Allen T, Brander C, Addo MM, Altfeld M, James I, Mallal S, Bunce M, Barber LD, Szinger J, Day C, Klenerman P, Mullins J, Korber B, Coovadia HM, Walker BD, Goulder PJR. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 38.Listgarten J, Frahm N, Kadie C, Brander C, Heckerman D. A statistical framework for modeling HLA-dependent T cell response data. PLoS Comput. Biol. 2007;3:1879–1886. doi: 10.1371/journal.pcbi.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mothé BR, Southwood S, Sidney J, English AM, Wriston A, Hoof I, Shabanowitz J, Hunt DF, Sette A. Peptide-binding motifs associated with MHC molecules common in Chinese rhesus macaques are analogous to those of human HLA supertypes and include HLA-B27-like alleles. Immunogenetics. 2013;65:371–386. doi: 10.1007/s00251-013-0686-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, Ling KL, Barnardo M, Goldthorpe S, Walton R, Bunce M, Jewell DP, Welsh KI. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Human Molecular Genetics. 2003;12:647–656. [PubMed] [Google Scholar]
- 41.Hviid TVF, Christiansen OB. Linkage Disequilibrium Between Human Leukocyte Antigen (HLA) Class II and HLA-G—Possible Implications for Human Reproduction and Autoimmune Disease. Hum. Immunol. 2005;66:688–699. doi: 10.1016/j.humimm.2005.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.