Abstract
Small interfering ribonucleic acid (siRNA), 20 – 25 base pairs in length, can interfere with the expression of specific genes. Recently, many groups reported the therapeutic intervention of siRNA in various cancer cells. In this study, dendrimer type bio-reducible polymer (PAM-ABP) which was synthesized using arginine grafted bio-reducible poly(cystaminebisacrylamide-diaminohexane) (ABP) and polyamidoamine (PAMAM) was used to deliver anti-VEGF siRNA into cancer cell lines including human hepatocarcinoma (Huh-7), human lung adenocarcinoma (A549), and human fibrosarcoma (HT1080) cells and access their potential as a siRNA delivery carrier for cancer therapy. PAM-ABP and siRNA formed polyplexes with average diameter of 116 nm and charge of around +24.6 mV. The siRNA in the PAM-ABP/siRNA polyplex released by 5 mM DTT and heparin. VEGF gene silencing efficiency of PAM-ABP/siRNA polyplexes was shown to be more effective than PEI/siRNA polyplexes in three cell lines with the following order HT1080>A549>Huh-7.
Keywords: Dendrimer, bio-reducible polymer, siRNA, VEGF
1. Introduction
In the gene delivery field, the study of small interfering RNA (siRNA) has increased due to their potential as a therapeutic macromolecule for the treatment of cancer therapy. siRNA can inhibit production of oncogenic regulators associated with tumor growth, transformation, and metastasis by a sequence-specific gene silencing effect [1-4]. The introduced siRNA interacts with the RNA-induced silencing complex (RICS) in the cell cytoplasm and promotes cleavage of messenger RNA (mRNA) which consists of complementary sequences [5-7]. Therefore, siRNA has a very powerful mechanism of silencing specific genes that has been reported both in vitro and in vivo with various cell types and diseases including solid tumors, respiratory syncytial virus infection, inherited skin disorder, and age-related macular degeneration [8-12]. However, siRNA still has limited to clinical applications due to the lack of efficiency and the low stability in the human body. Therefore, siRNA has not been approved by the Food and Drug Administration (FDA or USFDA).
To overcome the drawbacks of siRNA, various non-viral vectors, such as polycationic polymer, polyanionic polymer, liposomes, and micelles have been studied. These non-viral vectors shown lower transfection efficiency than viral vectors but have several advantages including lower cytotoxicity, non-immunogenicity, stability, and large-scale production. Hence, the studies using non-viral vector in the field of gene therapy has increased and various strategies have been developed to enhance transfection efficiency. These include 1) nano-sized polyplexes that easily accumulate at the tumor site through the enhanced permeability and retention (EPR) effect [13-15], 2) cell penetrating peptide (CPP) modified non-viral vectors to increase cellular uptake [16, 17], 3) increased numbers of amine groups in non-viral vectors to enhance the gene condensing ability and increase escape property from endosome to cytoplasm [14, 18-20], 4) bio-reducible non-viral vectors for easily release of genetic materials from polyplex in reductive environments [16, 17, 21, 22], and 5) targeting ligand conjugated non-viral vectors for specific delivery to the target site [23, 24]. The results of these researches showed the enhanced transfection efficiency.
In our previous work, dendrimer type bio-reducible polymer (PAM-ABP) was synthesized using arginine grafted bio-reducible poly(cystaminebisacrylamide-diaminohexane) (ABP) and polyamidoamine (PAMAM) and evaluated for application as a pDNA delivery carrier [22, 25]. PAM-ABP consist of dendrimers, arginine, and di-sulfide bonds. These components form a polyplex with genetic material at a low complex ratio (weight ratio or N/P ratio) to increase cellular uptake of polyplex into the cells and to easily release genetic material from polyplex. In this study, we evaluate the potential of PAM-ABP as a siRNA delivery carrier for cancer therapy in human hepatocarcinoma (Huh-7), human lung adenocarcinoma (A549), and human fibrosarcoma (HT1080) cells using anti-vascular endothelial growth factor (VEGF) siRNAto silence the vasculogenesis and angiogenesis of cancer cells.
2. Materials and Methods
2.1. Materials
ABP was synthesized as in our previous work.[16] Poly(amido amine) (PAMAM) dendrimer (G0), poly(ethylenimine) (PEI; branched form, Mw = 25 kDa), dimethylformamide (DMF), dithiothreitol (DTT), 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), and trypan blue solution (0.4%) were purchased from Sigma-Aldrich (St. Louis, MO). Traut's reagent and N-succinimidyl-3-(2-pyridyldithio) propionate (SPDP) were purchased from pierce (Rockford, IL). Spectrapor dialysis membranes (molecular weight cut off, MWCO = 500 Da, 1,000 Da, and 3,500 Da) were purchased from Spectrum Laboratories, Inc. (Rancho Dominguez, CA). YOYO-1 iodide (1 mM solution in DMSO) and all cell culture products such as fetal bovine serum (FBS), SYBR safe DNA gel stain, Dulbecco's phosphate buffered saline (DPBS), Dulbecco's modified Eagle's medium (DMEM), and trypsin-like enzyme (TrypLE™ Express) were purchased from Invitrogen (Carlsbad, CA). A siRNA sequence targeting the human VEGF gene was purchased from Bioneer Co. (Alameda, CA). The sense sequence was 5′-GGAGUACCCUGAUGAGAUCdTdT-3′ and the anti-sense sequence was 5′-GAUCUCAUC AGGGUACUCCdTdT-3′.
2.2. Synthesis of PAM-ABP
PAM-ABP was synthesized and characterized as previously detailed.[22] In brief, ABP was reacted in 0.1 M phosphate buffered saline (pH 7.2, 0.15 M NaCl) with SPDP (eq. 1.2, in DMF) at room temperature (RT) for 1 h. After 1 h, the product dialyzed using a dialysis membrane (MWCO = 1,000 Da) against ultrapure water and then lyophilized. To introduce thiol groups to the PAMAM back bone, PAMAM was reacted in 0.1 M phosphate buffered saline (pH 8.0, 0.15 M NaCl, 2.0 mM EDTA) with Traut's reagent (eq. 2, per primary amine group of PAMAM) at RT for 2 h and then dialyzed using a dialysis membrane (MWCO = 500 Da), followed by lyophilization. The purified PAMAM-SH was reacted in 50 mM phosphate buffered saline (pH 7.2, 0.15 M NaCl, 10 mM EDTA), with ABP-SPDP (eq. 4.4) at RT for 4 h. To detect the presence of released pyridine-2-thione, mixture solution was monitored by UV spectroscopy at 343 nm. The final product was purified by dialysis using a dialysis membrane (MWCO = 3,500 Da) and then lyophilization.
2.3. Agarose gel retardation assay
2.3.1. siRNA condensation ability
PAM-ABP/siRNA polyplexes were formed by simple vortexing at various weight ratios ranging from 1:0.2 to 1:5 based on siRNA (300 ng) in Hepes buffered saline (10 mM HEPES, 1 mM NaCl, pH 7.4), followed by incubation for 30 min. PEI/siRNA polyplexes were prepared at the same condition. Then, the polyplexes containing siRNA were electrophoresed onto a 1% agarose gel plate containing a SYBR safe gel staining solution. The electrophoresis was run in TAE (10 mM Tris/HCl, 1% acetic acid (v/v), 1 mM EDTA) buffer at 100 V for 30 min.
2.3.2. siRNA release assay
To confirm the siRNA release from PAM-ABP/siRNA polyplexes by degradation of the disulfide bond of PMA-ABP, each polyplex was incubated in the presence of 5 mM DTT for 30 min in 37°C incubator. PEI was used as a control group at the same condition. In addition, the release of siRNA from PAM-ABP/siRNA polyplexes was carried out by treatment of heparin. Briefly, PAM-ABP/siRNA polyplex (1:5, weight ratio, based on siRNA (300 ng)) was incubated with different amounts of heparin ranging from 0 to 5 units for 30 min in a 37°C incubator. After incubation, the samples were electrophoresed as described above. The nucleic acids bands were visualized on an UV illuminator (gel Documentation System, Bio-Rad, Hercules, CA).
2.4. Particle size and zeta-potential measurement
The particle sizes and zeta potentials of PAM-ABP/siRNA polyplexes were characterized using a Nano ZS (ZEN3600, Malvern Instruments) with a He-Ne ion laser (633 nm). The polyplexes were formed in HEPES buffer with various weight ratios from 0.2 to 10 based on siRNA (4 μg). After incubation, the each of the polyplex solutions were diluted using distilled water to a final volume of 600 μL before measurement. In addition, to confirm the dissociated polyplex by DTT, the particle size and zeta potential of PAM-ABP/siRNA polyplex (1:5, weight ratio, based on siRNA (μg)) was measured with both conditions (with 5 mM DTT and without DTT). PEI was used as a control group at the same experimental condition.
2.5. Cell culture
Human hepatocarcinoma (Huh-7) cells, human lung adenocarcinoma (A549) cells, and human fibrosarcoma (HT1080) cells were selected to investigate cell cytotoxicity, cellular uptake, and VEGF silencing activity. The cells were cultured in full medium (high glucose DMEM, 10% FBS, without antibiotic agent) at 37°C in a humidified atmosphere containing 5% CO2. When the confluence reached 70 ∼ 80%, the cells were harvested by trypsinization (TrypLE™ Express) and centrifugation. Then, the cell suspensions were used to measure for the following cell tests.
2.6. MTT assay
The cytotoxicity assay of the PAM-ABP was confirmed by the MTT assay in Huh-7, A549, and HT1080 cells. The cells were seeded in a 24-well plate at a density of 3 × 104 cells/well in 500 L DMEM medium containing 10% FBS and then incubated at 37°C in a humidified atmosphere containing 5% CO2. When the cell confluence reached 70 ∼ 80%, the cells were treated the various amounts of PAM-ABP ranging from 5 to 10 μg in 500 μL fresh pure DMEM for another 48 h. PEI was also treated to the cells with the various amounts ranging from 1 to 2 μg. The amounts of PAM-ABP and PEI are same the used amounts to prepare polyplexes with siRNA. Then 10 μL of stock solution of MTT (5 mg/mL in PBS) was added to each well. After an additional 2 h, the medium and unreacted MTT were removed by aspiration and 300 μL DMSO was procedurally added to each well.
The Tecan Infinite M200 Pro was used to measure the optical density at 570 nm and the relative cell viability was calculated by the following equation: Relative cell viability (%) = (OD sample – OD blank)/(OD control – OD blank) × 100.
2.7. Cellular uptake assay
To investigate the cellular uptake, siRNA was labeled by YOYO-1 with the method recommended by manufacturer. The cellular uptake behavior was assessed using a BD FACScan analyzer at a minimum of 5 × 103 cells gated per sample. Huh-7, A549, and HT1080 cells (6 × 104 cells/well) were seeded into 12-well plate in DMEM medium containing 10% FBS and incubated for 24 h. PAM-ABP was formed the polyplex with YOYO-1 labeled siRNA at weight ratio 1:5 based on siRNA (2 μg) for 30 min. When the confluence reached 70 ∼ 80%, the media in each well was replaced with serum-free DMEM, and then cells were treated with polyplex. After 4 h, the cells washed with PBS and trypsinized. The cells were collected by centrifugation for 3 min at 3,000 rpm. The collected cells were re-suspended in 300 μL of PBS and measured to assess cellular uptake. Analysis of cellular uptake was performed by using De NoVo FCS Express 4 Plus software. The quantitative cellular uptake of the polyplexes was calculated as a percentage of cell counts in the M gate region.
2.8. In vitro gene silencing
Huh-7, A549, and HT1080 cells (6 × 104 cells/well) were seeded using DMEM medium containing 10% FBS in a 12 well plate and incubated for 24 h. PAM-ABP/siRNA polyplexes were prepared with 2 μg of anti-VEGF siRNA at weight ratio of 1:5 based on siRNA in 50 μL. PEI/siRNA polyplexes were also prepared at weight ratio of 1:1 as a control group. After 24 h, the media in each well was replaced by 950 μL serum-free DMEM and then cells were treated with polyplexes. After 4 h, the medium was exchanged with 1 mL of fresh medium containing 10% FBS. Cells were incubated further for 48 h and then the levels of secreted VEGF in culture supernatants were obtained by using a human VEGF ELISA kit (Pierce, Rockford, IL) by the method of recommended by the manufacturer. Briefly, 50 μL of assay diluent and 200 μL of culture supernatants were added to each VEGF microplate well which was coated with a monoclonal antibody against VEGF and then incubated for 2 h at room temperature. The wells were aspirated and washed three times with wash buffer. The wash buffer clearly removed then 200 μL of VEGF conjugate added to each microplate well. After additional incubation for 2 h, the wells were aspirated and washed three times and then 200 μL of substrate solution added to each microplate well in the dark. After 20 min, the reaction was stopped by adding 50 μL stop solution to each well. The absorbance was investigated at 450 nm using the Tecan Infinite M200 Pro.
3. Results and Discussion
3.1. Formation and characterization of PAM-ABP/siRNA polyplex
In our previous work, we assessed the optimal weight ratio to form the polyplex with DNA by gel retardation assay [22]. However, siRNA consists of smaller sequences than DNA. Therefore, the siRNA condensing ability of PAM-ABP was measured by an agarose gel retardation assay at various weight ratios ranging from 0.2 to 5. As shown Figure 1A, free siRNA showed distinct bands, whereas polyplexes formed with PAM-ABP were retarded at weight ratio above 1:0.5 based on siRNA. PAM-ABP/siRNA displayed no siRNA migration at weight ratio above 1:0.5, indicating that PAM-ABP completely formed polyplexes with siRNA. These results were similar to the positive control PEI group. In the case of DNA, PAM-ABP formed polyplexes at weight ratio above 1:2 [22]. These results suggests that the genetic material condensing ability of PAM-ABP is dependent on sequence of genetic materials.
Fig. 1.
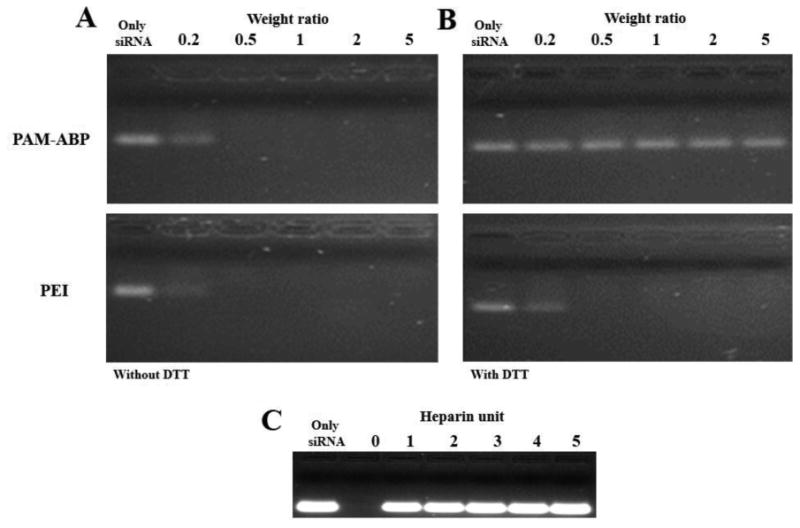
Agarose gel electrophoresis retardation assay of PAM-ABP/siRNA and PEI/siRNA polyplexes at weight ratios ranging from 0.2 to 5 based on siRNA (300 μg). (A) siRNA condensation ability, (B) siRNA releasing behavior from polyplexes by 5 mM DTT, and (C) siRNA releasing behavior from PAM-ABP/siRNA polyplex (weight ratio 1:5, siRNA (300 μg)) by increasing unit of heparin.
To confirm the siRNA release ability from the polyplex through the cleaved disulfide bond of PAM-ABP, DTT was used to simulate an intracellular reducing agent like glutathione (GSH). After cellular uptake, 0.5–10 mM of an intracellular reducing agent can easily cleave the disulfide bond of bio-reducible polymer in the cytoplasm [21, 26]. Figure 1B showed that the band of PAM-ABP/siRNA polyplexes was located in same position with free siRNA at all weight ratios after incubation with 5 mM DTT. Meanwhile, PEI was not influenced by 5 mM DTT. This result means that the disulfide bond of PAM-ABP can be cleaved by reducing agents in the cytoplasm and siRNA can easily release from polyplex. We also confirmed the release of siRNA after incubation with the anionic polysaccharide, heparin as an electrostatic competition agent, in Figure 1C. The released amount of siRNA from polyplex was increased with increasing heparin ranging from 0 to 5 units. This result predicts an easy release of Particle size and zeta-potential values of PAM-ABP/siRNA polyplexes were measured at various weight ratios ranging from 0.2 to 10 to confirm the details of polyplex formation (Figure 2A). Zeta-potential values of PAM-ABP/siRNA polyplexes were increased with increasing weight ratios from 0.2 to 2, and at weight ratio above 2, PAM-ABP/siRNA polyplexes had similar zeta-potential values. At weight ratios 0.2 and 0.5, particle sizes were below 100 nm. However, at the same weight ratios, the zeta-potentials were negatively charged. This result means that PAM-ABP was not surrounded by siRNA and polyplexes were not formed. When the weight ratio was increased 0.5 to 1, the particle size was increased sharply. This phenomenon means that the PAM-ABP/siRNA were formed polyplexes at a weight ratio 1 with positive surface charge. However, PAM-ABP did not completely and tightly bind to siRNA. Therefore, the particle size is bigger than other weight ratios such as 2, 5, and 10. At the weight ratio 2 to 10, the particle sizes of PAM-ABP/siRNA were 154 ∼ 130 nm. The particle size was not dramatically changed when increased weight ratios 2 to 10. Our previous work, PAM-ABP/pDNA polyplexes showed similar transfection efficiency both weight ratio 5 and 10. However, at a weight ratio 10, PAM-ABP/pDNA polyplexes showed slight cytotoxicity [22]. Therefore, we optimized the weight ratio to form the polyplex with siRNA at 5. Particle size and zeta-potential values of PAM-ABP/siRNA polyplexes were determined to be 116 nm and + 24.6 mV, respectively. In addition, after incubation with 5 mM DTT, particle size and zeta-potential values were measured at a weight ratio 5 (Figure 2B). Particle size was increased and zeta-potential was decreased after incubation with 5 mM DTT, while PEI/siRNA polyplexes were not influenced by 5 mM DTT. When the di-sulfide bond of bio-reducible carrier cleaved by DTT, the particle size of polyplexes was increased due to the decreased condensing ability of bio-reducible carrier [27]. This results indicate that the disulfide bond of PAM-ABP is cleaved by DTT and PAM-ABP/siRNA polyplex can more easily release the siRNA from polyplexes in reductive environments by dissociation than PEI/siRNA polyplexes.
Fig. 2.
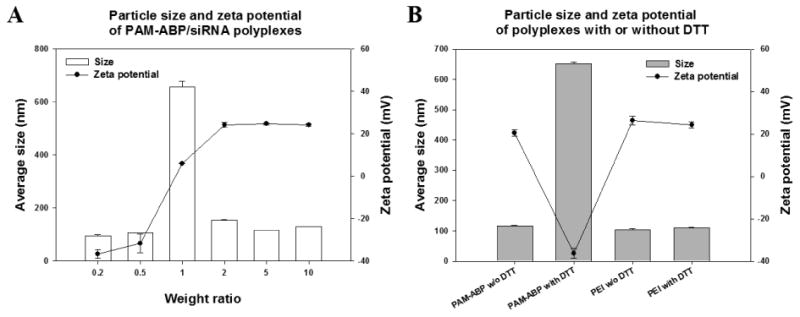
(A) Particle size and zeta potential of PAM-ABP/siRNA polyplexes at weight ratios ranging from 0.2 to 10 based on siRNA (4 μg). (B) Particle size and zeta potential of PAM-ABP/siRNA polyplex (weight ratio 1:5, siRNA(4 μg)) and PEI/siRNA polyplex (weight ratio 1:1, siRNA (4 μg)) with 5 mM DTT or without DTT. The results are presented as mean ± SD (n=3).
3.2. MTT assay to investigate cytotoxicity of PAM-ABP
The cytotoxicity of PAM-ABP at the transfection condition was investigated in Huh-7, A549, and HT1080. The amounts of PAM-ABPs were 5, 7.5, and 10 μg/mL. PEI were also tested as a control group. The amounts of PEIs were 1, 1.5, and 2 μg/mL. As shown in Figure 3, PAM-ABP has approximately 100% of the relative cell viability at all concentrations in all of the cell lines. This result suggests that PAM-ABP is a stable polymer when used as a siRNA delivery carrier at the transfection condition of this study. The cytotoxicity of PAM-ABP was reported to be low, retaining approximately 80% of the relative cell viability at 60 μg/mL of polymer concentration in both HeLa human epithelial carcinoma (HeLa) and C2C12 (mouse myoblast) cells [22]. In the case of the PEI control group, the relative cell viability was decreased with increasing concentration from 1 to 2 μg/mL in all of the cell lines.
Fig. 3.
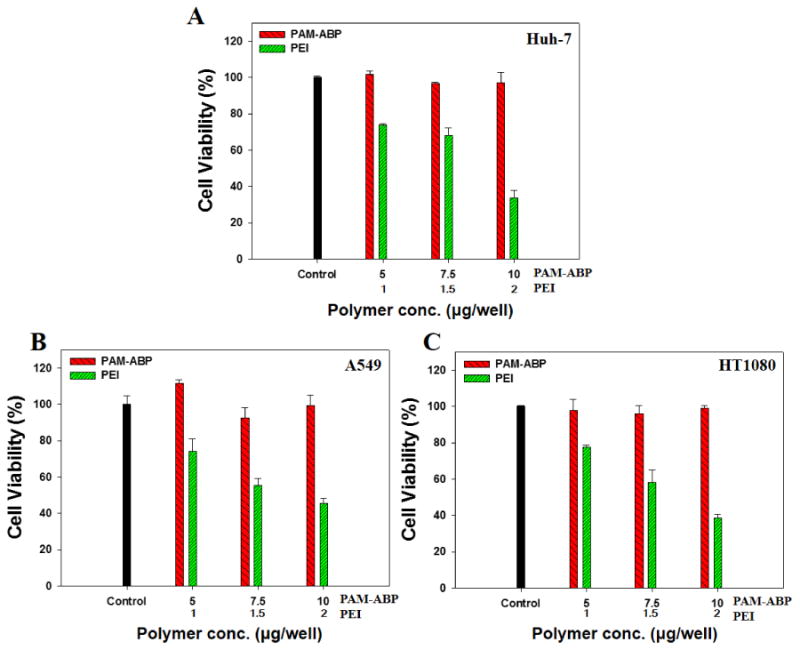
Cytotoxicity of PAM-ABP at various amounts ranging from 5 to 10 μg in (A) Huh-7, (B) A549, and (C) HT1080 cells (3 × 104 cell/well) without anti-VEGF siRNA. PEI was used the compared group at various amounts ranging from 1 to 2 μg. The results are presented as mean ± SD (n=3).
3.3. Cellular uptake of PAM-ABP/siRNA polyplexes
Cellular uptake of PAM-ABP/siRNA polyplexes were evaluated by using flow cytometry in various cell lines. siRNA was labeled by YOYO-1 to verify the cellular uptake ability of the polyplexes after treating the cells. PEI/siRNA polyplexes were also used as a positive control group. Figure 4 showed the cellular uptake efficiency of PAM-ABP/siRNA polyplexes at weight ratio 1:5 and PEI/siRNA polyplexes at weight ratio 1:1 based on YOYO-1 labeled siRNA (2 μg) in Huh-7, A549, and HT1080 cells. Both PAM-ABP and PEI polyplexes with siRNA showed similar cellular uptake and quantified cellular uptake (%) of 39.9, 96.7, and 90.4 (PAM-ABP) and 46.9, 94.1, and 96.7 (PEI) in Huh-7, A549, and HT1080 cells, respectively. As mentioned above, PAM-ABP/siRNA formed nano-sized polyplex and has positive surface charge. The small size and positive surface charge of polyplexes are important to delivering genetic material into the cell. Several studies reported that small sized and positive charged carriers can facilitate better uptake by the cell in comparison with both large sized and negatively charged carriers [14, 16, 28-30]. As mentioned above, nano-sized polyplexes can preferentially accumulate in the tumor site via the EPR effect and tend to circulate for longer times when injected via intravenous injection [13-15, 30]. Our previous work demonstrated that the ability of PAM-ABP to form compact polyplexes showed the enhanced cellular uptake about 5 folds more than ABP at a weight ratio 5 [22]. In addition, arginine residues, a constituent of the cell penetrating peptides (CPP), in PAM-ABP can increase cellular uptake [16, 22]. Generally, arginine residues is well known that assist in enhancing membrane permeability, facilitating nuclear localization, and adsorptive endocytosis [31, 32]. Zhang and co-works reported that the cellular uptake of arginine conjugated chitosan nanoparticles is significantly higher in comparison with arginine unconjugated chitosan nanoparticles [33].
Fig. 4.
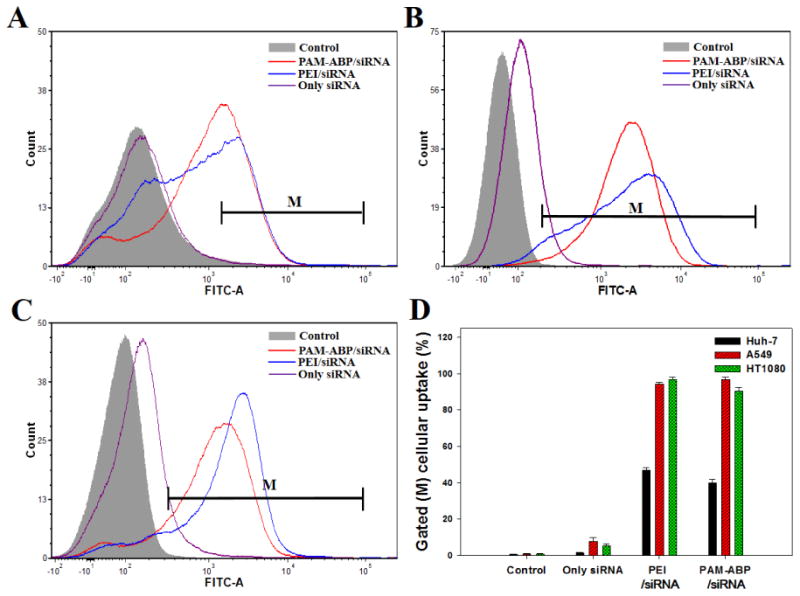
Cellular uptake by FACs in (A) Huh-7, (B) A549, and (C) HT1080 cells (6 × 104 cell/well). PAM-ABP/YOYO-1 labeled siRNA poloyplex was formed at weight ratio 1:5 and PEI/YOYO-l labeled siRNA polyplex was formed at weight ratio 1:1 based on YOYO-1 labeled siRNA (2 μg). (D) Bar graph representing the mean percentages of cellular uptake by M region gating. The results are presented as mean ± SD (n=3).
3.4. Down-regulation of gene expression in vitro
In order to estimate the gene silencing efficiency of PAM-ABP polyplex with anti-VEGF siRNA, the VEGF expression was determined by ELISA protein assay in three cell lines (Huh-7, A549, and HT1080). The gene silencing efficiency was measured at the fixed weight ratio 1:5 (PAM-ABP) and 1:1 (PEI), respectively. An anti-VEGF siRNA only treated group was used a negative control. As shown Figure 5, the negative control (only siRNA) did not show gene silencing effects in the three cell lines. Meanwhile, PAM-ABP and PEI polyplexes caused decreased VEGF expression compared with non-treated cells and the siRNA only group. PAM-ABP/siRNA polyplexes showed the lowest VEGF expression in all of the cell lines. Figure 5D shows the relative VEGF expression (%), after treatment with PAM-ABP/siRNA polyplexes, VEGF expressions were decreased to 36.7%, 33.9%, and 21.6% in Huh-7, A549, and HT1080 cells, respectively. Gene inhibition efficiency of PAM-ABP in cell lines followed order HT1080>A549>Huh-7. In the case of PEI/siRNA polyplex, VEGF expressions were 65.9%, 78.0%, and 78.2% in Huh-7, A549, and HT1080 cells, respectively. The cellular uptake of PEI/siRNA polyplexes was similar to or above that of PAM-ABP/siRNA polyplexes (Figure 4). However, the gene down-regulation effect was actually lower than PAM-ABP. This phenomenon suggests that siRNA was not completely release from PEI/siRNA polyplexes because PEI is not degraded by intracellular reducing agents in reductive environments. While, PAM-ABP which has disulfide bonds, can be cleaved by reducing agents, leading to easy release of siRNA from PAM-ABP/siRNA polyplexes. This hypothesis was supported by the result of the gel retardation assay (Figure 1). As mentioned above, several researches reported that the gene delivery carriers which are consisted disulfide bond, showed the increased transfection efficiency [16, 17, 21, 22]. In addition, Hahn and co-works demonstrated that the reducible polyethyleneimine-hyaluronic acid ((PEI-ss)-b-HA) formed polyplexes with VEGF siRNA and showed more significant VEGF gene silencing than PEI/siRNA polyplexes [34]. Kim and co-works also reported that the reducible poly(oligo-d-arginine) (rPOA) rapidly dissociated in the cytoplasm and showed enhanced bioactivity of siRNA against VEGF [35]. These reports supported that the di-sulfide bond of bio-reducible polymer can increase transfection efficiency, leading to high gene silencing activity.
Fig. 5.
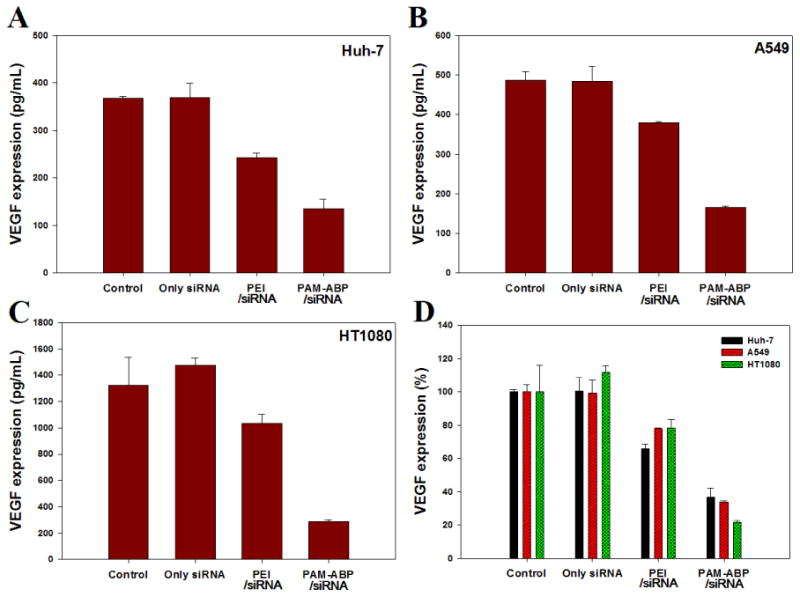
Expression of VEGF determined using ELISA kit in (A) Huh-7, (B) A549, and (C) HT1080 cells (6 × 104 cell/well). PAM-ABP/siRNA poloyplex was formed at weight ratio 1:5 and PEI/siRNA polyplex was formed at weight ratio 1:1 based on siRNA (2 μg). (D) Quantification of VEGF expression is shown by relative VEGF expression %. Relative VEGF expression (%) = Amount of VEGF (Treated)/Amonut of VEGF (Control) × 100. The results are presented as mean ± SD (n=3).
4. Conclusion
In this study, we accessed the potential of PAM-ABP as a siRNA delivery carrier for applications in cancer therapy. PAM-ABP formed nano-sized (116 nm) and positively charged (+24.6 mV) polyplexes with siRNA, and the siRNA in the PAM-ABP/siRNA polyplex released by 5 mM DTT and heparin. In addition, VEGF gene silencing efficiency of PAM-ABP/siRNA polyplexes was shown to be more effective than PEI/siRNA in three cell lines with the following order HT1080>A549>Huh-7. PAM-ABP/siRNA polyplexes showed no cytotoxicity in all cell lines. Our results demonstrated that PAM-ABP consisting of arginine groups, disulfide bonds, and dendrimers is an effective siRNA carrier for applications in cancer therapy.
Acknowledgments
This work was financially supported by the NIH Grant CA177932. The authors would like to thank Joshua P. Jones for help with preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chemistry & biology. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nature reviews. Genetics. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Miele E, Spinelli GR, Miele E, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. International journal of nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Y, Qiao H, Su Z, Chen M, Ping Q, Sun M. PEGylated carboxymethyl chitosan/calcium phosphate hybrid anionic nanoparticles mediated hTERT siRNA delivery for anticancer therapy. Biomaterials. 2014;35:7978–7991. doi: 10.1016/j.biomaterials.2014.05.068. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 6.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 8.McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nature reviews. Genetics. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 9.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seth S, Matsui Y, Fosnaugh K, Liu Y, Vaish N, Adami R, Harvie P, Johns R, Severson G, Brown T, Takagi A, Bell S, Chen Y, Chen F, Zhu T, Fam R, Maciagiewicz I, Kwang E, McCutcheon M, Farber K, Charmley P, Houston ME, Jr, So A, Templin MV, Polisky B. RNAi-based therapeutics targeting survivin and PLK1 for treatment of bladder cancer. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:928–935. doi: 10.1038/mt.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser PK, Symons RC, Shah SM, Quinlan EJ, Tabandeh H, Do DV, Reisen G, Lockridge JA, Short B, Guerciolini R, Nguyen QD. I. Sirna-027 Study, RNAi-based treatment for neovascular age-related macular degeneration by Sirna-027. American journal of ophthalmology. 2010;150:33–39. e32. doi: 10.1016/j.ajo.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.DeVincenzo J, Cehelsky JE, Alvarez R, Elbashir S, Harborth J, Toudjarska I, Nechev L, Murugaiah V, Van Vliet A, Vaishnaw AK, Meyers R. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral research. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Liu X. Chitosan-siRNA complex nanoparticles for gene silencing. Sheng wu yi xue gong cheng xue za zhi = Journal of biomedical engineering = Shengwu yixue gongchengxue zazhi. 2010;27:97–101. [PubMed] [Google Scholar]
- 14.Park SC, Nam JP, Kim YM, Kim JH, Nah JW, Jang MK. Branched polyethylenimine-grafted-carboxymethyl chitosan copolymer enhances the delivery of pDNA or siRNA in vitro and in vivo. International journal of nanomedicine. 2013;8:3663–3677. doi: 10.2147/IJN.S50911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malhotra M, Tomaro-Duchesneau C, Saha S, Prakash S. Systemic siRNA Delivery via Peptide-Tagged Polymeric Nanoparticles, Targeting PLK1 Gene in a Mouse Xenograft Model of Colorectal Cancer. International journal of biomaterials. 2013;2013:252531. doi: 10.1155/2013/252531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TI, Rothmund T, Kissel T, Kim SW. Bioreducible polymers with cell penetrating and endosome buffering functionality for gene delivery systems. Journal of controlled release : official journal of the Controlled Release Society. 2011;152:110–119. doi: 10.1016/j.jconrel.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjugate chemistry. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 19.Bello Roufai M, Midoux P. Histidylated polylysine as DNA vector: elevation of the imidazole protonation and reduced cellular uptake without change in the polyfection efficiency of serum stabilized negative polyplexes. Bioconjugate chemistry. 2001;12:92–99. doi: 10.1021/bc0000738. [DOI] [PubMed] [Google Scholar]
- 20.Yue ZG, Wei W, Lv PP, Yue H, Wang LY, Su ZG, Ma GH. Surface charge affects cellular uptake and intracellular trafficking of chitosan-based nanoparticles. Biomacromolecules. 2011;12:2440–2446. doi: 10.1021/bm101482r. [DOI] [PubMed] [Google Scholar]
- 21.Breunig M, Hozsa C, Lungwitz U, Watanabe K, Umeda I, Kato H, Goepferich A. Mechanistic investigation of poly(ethylene imine)-based siRNA delivery: disulfide bonds boost intracellular release of the cargo. Journal of controlled release : official journal of the Controlled Release Society. 2008;130:57–63. doi: 10.1016/j.jconrel.2008.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Nam HY, Nam K, Lee M, Kim SW, Bull DA. Dendrimer type bio-reducible polymer for efficient gene delivery. Journal of controlled release : official journal of the Controlled Release Society. 2012;160:592–600. doi: 10.1016/j.jconrel.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Yhee JY, Lee SJ, Lee S, Song S, Min HS, Kang SW, Son S, Jeong SY, Kwon IC, Kim SH, Kim K. Turnor-Targeting Transferrin Nanoparticles for Systemic Polymerized siRNA Delivery in Tumor-Bearing Mice. Bioconjugate chemistry. 2013 doi: 10.1021/bc400226b. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Liu P, Du J, Sun Y, Li F, Duan Y. Targeted siRNA delivery by anti-HER2 antibody-modified nanoparticles of mPEG-chitosan diblock copolymer. Journal of biomaterials science. Polymer edition. 2013;24:1219–1232. doi: 10.1080/09205063.2012.745716. [DOI] [PubMed] [Google Scholar]
- 25.Won YW, Lee M, Kim HA, Nam K, Bull DA, Kim SW. Synergistically combined gene delivery for enhanced VEGF secretion and antiapoptosis. Molecular pharmaceutics. 2013;10:3676–3683. doi: 10.1021/mp400178m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. The Journal of nutrition. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 27.Yu H, Russ V, Wagner E. Influence of the molecular weight of bioreducible oligoethylenimine conjugates on the polyplex transfection properties. The AAPS journal. 2009;11:445–455. doi: 10.1208/s12248-009-9122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, McCrate JM, Lee JC, Li H. The role of surface charge on the uptake and biocompatibility of hydroxyapatite nanoparticles with osteoblast cells. Nanotechnology. 2011;22:105708. doi: 10.1088/0957-4484/22/10/105708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. International journal of nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi H, Watanabe R, Choyke PL. Improving conventional enhanced permeability and retention (EPR) effects; what is the appropriate target? Theranostics. 2013;4:81–89. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris VB, Sharma CP. Enhanced in-vitro transfection and biocompatibility of L- arginine modified oligo (-alkylaminosiloxanes)-graft-polyethylenimine. Biomaterials. 2010;31:8759–8769. doi: 10.1016/j.biomaterials.2010.07.073. [DOI] [PubMed] [Google Scholar]
- 32.Lu S, Morris VB, Labhasetwar V. Codelivery of DNA and siRNA via arginine-rich PEI-based polyplexes. Molecular pharmaceutics. 2015;12:621–629. doi: 10.1021/mp5006883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Zhu D, Song L, Liu L, Dong X, Liu Z, Leng X. Arginine conjugation affects the endocytic pathways of chitosan/DNA nanoparticles. Journal of biomedical materials research. Part A. 2011;98:296–302. doi: 10.1002/jbm.a.33115. [DOI] [PubMed] [Google Scholar]
- 34.Park K, Lee MY, Kim KS, Hahn SK. Target specific tumor treatment by VEGF siRNA complexed with reducible polyethyleneimine-hyaluronic acid conjugate. Biomaterials. 2010;31:5258–5265. doi: 10.1016/j.biomaterials.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 35.Won YW, Yoon SM, Lee KM, Kim YH. Poly(oligo-D-arginine) with internal disulfide linkages as a cytoplasm-sensitive carrier for siRNA delivery. Molecular therapy : the journal of the American Society of Gene Therapy. 2011;19:372–380. doi: 10.1038/mt.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]


