Abstract
Allergic asthma is a chronic inflammatory disorder that affects approximately 20% of the population worldwide. Microarray analyses of nasal epithelial cells from acute asthmatic patients detected a 50% decrease in expression of Stard7, an intracellular phosphatidylcholine (PC) transport protein. To determine if loss of Stard7 expression promotes allergic responses, mice were generated in which one allele of the Stard7 locus was globally disrupted (Stard7+/− mice). Ovalbumin (OVA) sensitization and challenge of Stard7+/− mice resulted in a significant increase in pulmonary inflammation, mucous cell metaplasia, airway hyperreactivity (AHR), and OVA-specific IgE compared to OVA sensitized/challenged WT mice. This exacerbation was largely Th2-mediated with a significant increase in CD4+IL-13+ T cells and IL-4, IL-5 and IL-13 cytokines. The loss of Stard7 was also associated with increased lung epithelial permeability and activation of proinflammatory DCs in sensitized and/or challenged Stard7+/− mice. Notably, OVA-pulsed DCs from Stard7+/− mice were sufficient to confer an exaggerated allergic response in OVA-challenged WT mice, although AHR was greater in Stard7+/− recipients compared to WT recipients. Enhanced allergic responses in the lung were accompanied by age-dependent development of spontaneous atopic dermatitis. Overall, these data suggest that Stard7 is an important component of a novel protective pathway in tissues exposed to the extracellular environment.
Introduction
Asthma frequently begins in childhood and is a leading cause of morbidity in this population. There is a strong genetic component to the disease and distinct sets of genes are differentially regulated in the respiratory epithelium of children experiencing an asthma exacerbation compared to age-matched patients with stable disease(1). One cluster of genes included a PC transfer protein, Stard7, whose expression was decreased by 50% during acute exacerbations, raising the possibility that loss of Stard7 expression contributes to the development of asthma.
Stard7 was first identified as a transcript that was upregulated in a choriocarcinoma cell line(2, 3) and belongs to the Stard2 subfamily of START (steroidogenic acute regulatory protein-related lipid transfer) domain proteins that transfers phospholipids among intracellular membranes(4, 5). The Stard2 subfamily includes Stard2/phosphatidylcholine transfer protein (PC-TP, hereafter referred to as Stard2) and Stard10, all of which have high specificity for PC binding/transfer. Deficiency of Stard2 or Stard10 resulted in dysregulation of glucose and lipid metabolism(6) or bile acid metabolism(7), respectively. Very little is known about the function of Stard7 apart from the observation that transient overexpression in HEPA-1 cells promoted association of Stard7 with the mitochondrial membrane and uptake of PC into this organelle(8). Although it remains unclear if Stard7-mediated transport of PC is important for mitochondrial homeostasis, there is increasing evidence that asthma pathogenesis is linked to mitochondrial dysfunction(9–12). Taken together, these data support a model in which members of the Stard2 subfamily modulate metabolic and allergic responses.
Given that Stard7 is linked to acute asthma, as well as a pathway associated with asthma pathogenesis (i.e. mitochondrial dysfunction), we hypothesized that mice deficient in Stard7 would have an exaggerated response to allergen challenge. Disruption of the Stard7 locus in Stard7−/− mice resulted in embryonic lethality; however, haploinsufficiency resulted in increased IgE and mucous production, eosinophilia and AHR in response to OVA sensitization and challenge. Consistent with increased susceptibility to allergy, heterozygous Stard7 mice developed spontaneous dermatitis with age. Overall, these results suggest that Stard7 plays a protective role in mucosal tissues by preventing pathogenic immune responses to environmental antigens.
MATERIALS AND METHODS
Animals
ES cells carrying a “knockout first” Stard7 allele were generated by the International Knockout Mouse Consortium (Davis, CA). Two correctly targeted ES cell clones were expanded and used for the production of chimeras. Injection of ES cells into C57BL/6J blastocytes was carried out by the ES Core at the University of Cincinnati College of Medicine (Cincinnati, OH). Chimeric mice were crossed with C57BL/6J mice to identify offspring carrying the targeted allele in germ cells (Stard7+/− mice). Animals were maintained according to protocols approved by the Institutional Animal Care and Use Committee at the Cincinnati Children’s Research Foundation (Cincinnati, OH). Mice were housed in a pathogen-free barrier facility in humidity- and temperature-controlled rooms on a 12:12 h light/dark cycle and were allowed food and water ad libitum.
OVA sensitization and challenge
Mice were sensitized by intraperitoneal (i.p.) injection of 50 μg OVA (Sigma, St. Louis, MO) emulsified with 2 mg alum (Sigma) in 100 μL PBS on days 0 and 7, and then challenged daily by intratracheal instillation of 10 μg OVA dissolved in 50 μl PBS from days 18 to 21. Control mice were injected with alum at days 0 and 7 followed by intratracheal challenge with PBS from days 18 to 21 (Fig. 2A). Other control groups included mice sensitized with OVA (alum) and challenged with PBS, or mice sensitized with PBS (alum) and challenged with OVA. All analyses were performed 24 hours after the last challenge.
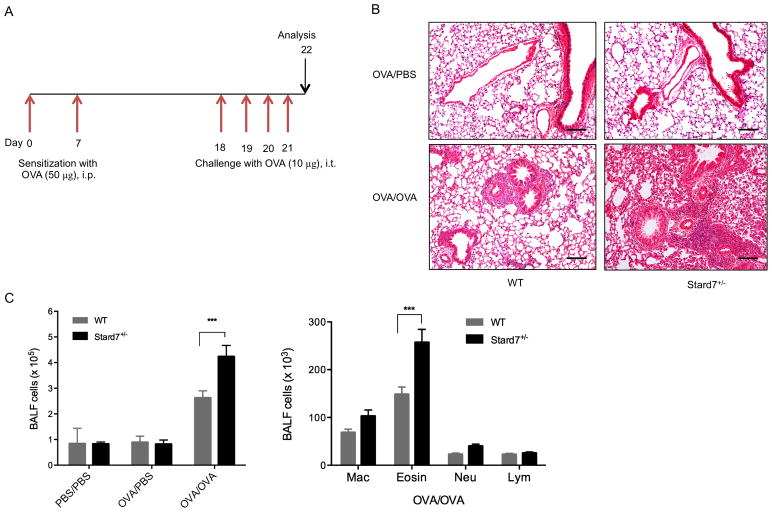
Fig. 2. Deficiency of Stard7 is associated with exaggerated allergic disease.
(A) Schematic of allergen sensitization and challenge in WT and Stard7+/− littermates in the C57BL/6J background. Mice were intraperitoneally (i.p.) sensitized with 50 μg of OVA (alum) on days 0 and 7, followed by intratracheal (i.t.) challenge with 10 μg of OVA on days 18 through 21. Control mice were sensitized with PBS (alum) or OVA (alum) and challenged with PBS. Mice were analyzed on day 22. (B) H&E staining of lung tissue following sensitization and challenge. n = 6–8 mice/group. Scale bars = 50 μm. (C) Total (left panel) and differential (right panel) BALF cell counts in mice following sensitization and challenge. n = 6–8 mice/group. Mac, macrophages; Eosin, eosinophils; Neu, neutrophils; Lym, lymphocytes.
Airway Hyperresponsiveness (AHR)
AHR was assessed in anesthetized mice, as described previously(13). Briefly, a tracheostomy was performed and the mouse was connected to a flexiVent system (SCIREQ, Montreal, QC, Canada). Airway resistance was measured after nebulization of PBS (baseline) followed by increasing doses of methacholine (acetyl-β-methylcholine chloride, Sigma).
Preparation of tissues for histological, immunohistochemical and immunofluorescence analyses
Mice were sacrificed by intraperitoneal overdose of pentobarbital, exsanguinated, and intratracheally cannulated with an 18 gauge luer stub adaptor. Lungs were instilled with 4% paraformaldehyde (PFA) at a pressure of 25 cmH2O, harvested, and fixed at 4°C overnight in 4% PFA. Fixed lungs were sub-dissected into 4–8 mm pieces prior to PBS wash, alcohol dehydration, and paraffin embedding. For skin tissue, 1–2 mm thick sections were harvested from the back of the mouse, hair and fat removed, and processed as described above. Tissue sections (5 μm) were deparaffinized in xylene, rehydrated and processed for H&E, Alcian blue, toluidine blue and Luna staining. For immunohistochemistry, antigen retrieval was performed by boiling sections in citrate buffer (pH 6.0) for 21 minutes. Sections were incubated with guinea pig anti-mouse Stard7 antibody (1:2500; generated in this laboratory, see below), rabbit anti-mouse TSLP antibody (1:400; LSBio, Seattle, WA) or rabbit anti-mouse CLCA3 antibody (1:12,500; Abcam, Cambridge, MA) overnight at 4°C, followed by a goat anti-guinea pig (1:200; Life Technologies) or goat anti-rabbit IgG secondary Ab (1:200; Life Technologies). Antibody staining was revealed by developing in DAB substrate solution: 0.05% DAB/0.015% H2O2. Microscopic images were acquired using a Zeiss Axio A2 microscope equipped with an AxioCam MRc5 camera (Carl Zeiss). For immunofluorescence, blocking was performed in 4% normal donkey serum in PBST (0.2% Triton-X100 in PBS). Sections were incubated at 4°C overnight with mouse antibodies against keratin 14 (1:1000; NeoMarkers, Fremont, CA), CCSP (1:2000, Seven Hills Bioreagents) and/or Stard7 (1:250). After washing in PBST, sections were incubated with secondary antibody (Life Technologies) conjugated to Alexa-Fluor 488, Alexa-Fluor 555, or Alexa-Fluor 647 at a 1:200 dilution for one hour at room temperature. Sections were then washed in PBST and mounted with ProLong Gold (Life Technologies). Confocal images of labeled tissue sections were acquired using a Nikon C1si confocal microscope (Nikon Instruments) at a scanning resolution of 1024×1024 pixels.
Generation of Stard7 antibody
In order to generate Stard7 antigen, the START domain (encoding amino acids 136–327 of mouse Stard7) was amplified from mouse type II epithelial cell cDNA using upstream primer 5′ - CGG GAT CCA AGT CAA AAG ACA AG - 3′ and downstream primer 5′ - ATA AGA ATG CGG CCG CGG CTT TCA GAG TGG C - 3′; restriction sites for BamH I and Not I were encoded in the upstream and downstream primers, respectively. The amplified 579-bp fragment was cloned into the BamH I/Not I sites of pGEX-6p-1 vector (GE Healthcare Bio-Sciences, Piscataway, NJ). Stard7 LBD was expressed in Escherichia coli BL21 (DE3). Transformed bacteria were grown in Luria-Bertani medium, supplemented with 50 mg/ml carbenicillin, to an OD600 of 0.6; protein expression was induced by the addition of 0.1 mM isopropyl b-d-thiogalactoside (IPTG) for 3 h at 37°C. Ten to 20% tricine-SDS-PAGE of bacterial lysates expressing Stard7 LBD detected a band, Mr ≈ 49 kDa, following IPTG induction. The broth was centrifuged, and the isolated bacterial pellet was lysed by sonication in 20 mM Tris buffer (pH 7.4) at 4°C. Inclusion bodies were recovered by centrifugation, washed in Tris buffer, and solubilized in the BugBuster buffer (EMD Chemicals, Philadelphia, PA) with 0.3% N-lauroylsarcosine (Sigma) and 1 mM DTT. Denatured, solubilized inclusion body protein was diluted (1:10) in 20 mM Tris, 0.1 mM DTT (pH 8.5), and dialyzed twice against 10 volumes of the same buffer for 3 h and then dialyzed twice against 10 volumes of 20 mM Tris (pH 8.5) for 3 h, followed by dialysis twice in PBS. After centrifugation, the supernatant was applied to prepacked GSTrap columns (GI Healthcare Bio-Sciences). The column was washed and eluted according to the manufacturers’ protocol. Eluted protein was dialyzed against sodium phosphate buffer (pH 7.4), and stored in aliquots at −80°C.
Purified, recombinant mouse Stard7 START domain was injected into guinea pigs to generate polyclonal antibodies. Stard7 antibodies were initially characterized by immunohistochemistry of lung sections from 8 week-old WT and surviving Stard7−/− littermates (Fig. 1).
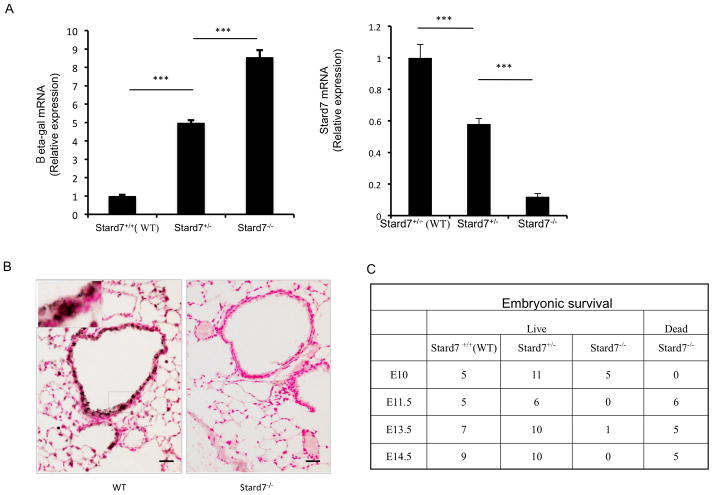
Fig. 1. Generation and initial characterization of Stard7+/− and Stard7−/− mice.
(A) qRT-PCR of β-gal (left panel) and Stard7 (right panel) mRNAs in lung tissue from 8 week-old mice. β-actin was used as an endogenous control. n = 3 samples/group. (B) Immunohistochemical staining of Stard7 in lung tissue of 8 week-old mice. Scale bars = 25 μm. (C) Table depicting the number of surviving/dead embryos from Stard7+/− intercrosses.
Cytokine and quantitative RT-PCR
Total lung RNA was purified using the RNeasy Mini Kit (Qiagen, Valencia, CA) and reverse transcribed into cDNA using the iScript™ cDNA synthesis kit (BIO-RAD, Hercules, CA). qRT-PCR was performed using TaqMan primer/probe sets (Table I, Life Technologies, Grand Island, NY) and data was normalized to mouse β-actin using SDS software (Life Technologies).
Table 1.
TaqMan primer/probe sets
| Primer | Product number |
|---|---|
| IL-4 | Mm00445259_m1 |
| IL-5 | Mm99999063_m1 |
| IL-6 | Mm99999064_m1 |
| IL-13 | Mm99999190_m1 |
| IL-25 | Mm00499822_m1 |
| IL-33 | Mm01195784_m1 |
| Ccl-17 | Mm01244826_g1 |
| CLCA 3 | Mm01320697_m1 |
| TSLP | Mm01157588_m1 |
| TSLPR | Mm 00497362_m1 |
| Actin | Mm00607939_m1 |
Quantitation of IgE
Serum total and OVA-specific IgE were quantitated by ELISA. Briefly, plates were coated at 4°C overnight with 2 μg/ml IgE (Biolegend, San Diego, CA) antibody. For OVA-specific IgE, plates were coated at 4°C overnight with 100 μg/ml OVA. The following day, plates were blocked with 1% BSA in PBS for 1 h and then coated with serum samples. Biotin-conjugated IgE (2.0 μg/ml) and streptavidin-HRP (1:1000; Biolegend) were sequentially added, and the reaction was developed using tetramethylbenzidine as a substrate (Biolegend). Serum total IgE concentration was determined from a standard curve generated from a known concentration of mouse IgE provided by the manufacturer. OVA-specific IgE levels were expressed at an optical density of 450 nm.
Inflammatory cells
BALF was collected by lavaging lungs with 1 ml PBS containing BSA (1%) and EDTA (2 mM). The BALF was centrifuged (5000 rpm for 10 minutes) and the cell pellet was resuspended in RBC lysis buffer (Sigma). The remaining cells were counted using a hemocytometer, spun onto slides, and stained with Diff-Quick (Shandon Lipshaw, Pittsburgh, PA). Two hundred cells were counted per sample to determine the percentage of eosinophils, macrophages, neutrophils, and lymphocytes.
Flow cytometry
Single lung or LN cell suspensions (1×106 cells/100 μl) were incubated in FACS buffer (PBS, 1% FBS, 0.05% NaN3) containing CD16/32 Fc-blocking antibody (2.4G2, BD Biosciences, San Jose, CA) for 30 minutes at 4°C and then stained with fluorescently-labeled antibodies for 30 minutes at 4°C. Cells were washed repeatedly in FACS buffer, fixed in 2% PFA for 20 minutes at room temperature, washed and resuspended in FACS buffer for flow cytometry analysis. Data was acquired using the LSR II flow cytometer (BD Biosciences) and analyzed using FACSDiVa (BD Biosciences) and FlowJo software (TreeStar, Inc. Ashland, OR). The following mouse-specific monoclonal antibodies were purchased from eBioscience with the clone indicated in parentheses: CD11c-Alexa Fluor 647 (HL3), CD11b-PE-Cy7 (M1/70), Gr1-APC-eFluor780 (RB6-8C5), CD317-Alexa Fluor 488 (eBio927), CD80-PE (16-10A1), CD86-PE (GL1), PD-L1-PE (MIH5), B7-DC-PE (TY25), CD4-PE-Cy7 (RM4-5), IL-13-PE (eBio13A) and IL-17A-Alexa Fluor 647 (eBio17b7). Myeloid DCs were defined as CD11c+CD11b+Gr1−CD317−.
Isolation and adoptive transfer of BMDCs
Bone marrow cells (3×105 cells/ml) were isolated from the femurs of mice and cultured in complete RPMI supplemented with GM-CSF (10ng/ml, Peprotech, Rocky Hill, NJ). Medium was changed on day 3. On day 6, BMDCs were pulsed overnight with PBS or 100 μg/ml OVA (Sigma). The next day, PBS- or OVA-pulsed BMDCs (2.5×105/50μl PBS) were administered intratracheally to mice. Recipients were intratracheally challenged with 50 μl of PBS or OVA (10 μg) from days 10 to 13. AHR was assessed on day 14 and mice were subsequently sacrificed for analyses of lung structure, inflammation and mucous cell hyperplasia.
Permeability Studies
In vivo baseline permeability studies were performed as previously described by Davidovich and colleagues(14). Each mouse was injected with 0.3 ml of FITC-conjugated albumin (12 mg/ml; Sigma) in the lateral tail vein after sedation with isoflurane. After three hours, BALF was harvested as described above, and whole blood was collected via cardiac puncture and allowed to clot. Albumin fluorescence in BALF and serum was determined using Synergy2 Multimode Microplate Reader (BioTek, Winooski, VT) with absorption/emission wavelengths of 480/520 nm. Alveolar epithelial permeability was defined as the ratio of BALF to serum fluorescence.
Statistics
Data were analyzed using Prism 5 software (GraphPad Software, San Diego, CA) by unpaired Student’s t test, one-way ANOVA or two-way ANOVA with a Bonferroni post hoc test. Data were presented as mean ± SEM with *p≤0.05, **p≤0.01 and ***p≤0.001 significance.
RESULTS
Global deletion of the Stard7 gene
Guajardo et al identified 253 genes that were either activated or repressed by at least 3-fold in children experiencing an acute exacerbation compared to those with stable disease (1). Of interest, Stard7, a member of the START domain protein family involved in intracellular lipid transport, was decreased by approximately 50% in acute asthmatic children compared to either stable asthmatics or healthy controls (Fig. S1A). To identify whether Stard7 deficiency influences asthma susceptibility, embryonic stem (ES) cells carrying a targeted Stard7 allele were generated by the International Knockout Mouse Consortium using a promoterless targeting cassette designed to produce a “knockout first” allele (Fig. S1B). Targeted ES cells were injected into blastocysts and the resulting chimeric mice were crossed with C57BL/6J mice to identify offspring carrying the targeted allele in germ cells (Stard7+/− mice) (Fig. S1C). Expression of the targeted Stard7 allele resulted in splicing of exon 1 with the targeting cassette leading to transcription of the β-galactosidase reporter gene and termination of the Stard7 transcript (Fig. 1A). As expected, expression of β-galactosidase mRNA was inversely correlated with expression of Stard7 mRNA; similar results were observed in both lung (Fig. 1A) and skin tissue (not shown). Immunohistochemical staining of lung tissue detected Stard7 in epithelial cells of airways, peripheral bronchioles and alveoli of adult WT mice but not in surviving Stard7−/− mice, validating specificity of the Stard7 antibody (Fig. 1B). There were no obvious changes in lung structure or function through one year of age (not shown).
Intercrosses of Stard7+/− mice resulted in the expected number of WT and heterozygous mice at 3 weeks of age (Fig. S1D); however, the number of Stard7−/− offspring was greatly reduced (Fig. S1D). Analyses of developing embryos indicated that the viability of Stard7−/− embryos was dramatically decreased after E10 (Fig. 1C). Surviving male Stard7−/− mice were infertile but both male and female Stard7−/− mice had a normal life expectancy. Because of the low numbers of surviving Stard7−/− mice, all subsequent experiments were performed with Stard7+/− mice, a model that mimics the expression of Stard7 in humans with acute asthma exacerbations(1).
Haploinsufficiency for Stard7 is associated with exaggerated allergic disease
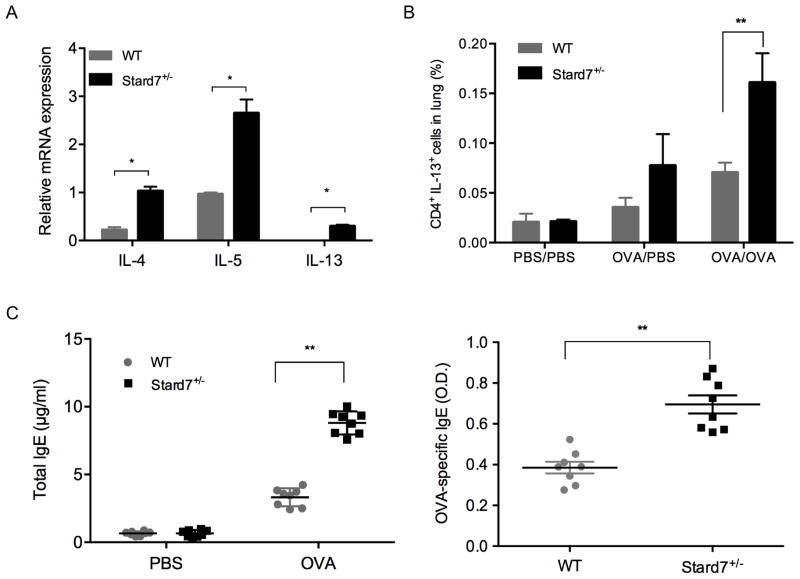
To determine whether the deficiency of Stard7 was accompanied by an exaggerated allergic response, mice in the C57BL/6J genetic background were sensitized by intraperitoneal injection of OVA on days 0 and 7 followed by intratracheal challenge with OVA on days 18 through 21 (Fig. 2A). Histological evaluation of lung sections 24 hours after the last challenge detected peribronchial and perivascular inflammatory cell infiltration that was markedly increased in OVA-challenged Stard7+/− mice compared to controls (Fig. 2B). Consistent with this finding, the total number of cells in BALF was significantly elevated in Stard7+/− mice (Fig. 2C, left panel) with eosinophils being the predominant cell type (Fig. 2C, right panel). Th2 cytokines, including IL-4, IL-5, and IL-13, were increased to a greater extent in Stard7+/− mice (Fig. 3A); further, the subset of CD4+ cells expressing IL-13 was significantly increased in Stard7+/− mice (Fig. 3B – gating strategy shown in Fig. S2A, B) whereas the frequency of T cells expressing IL-17 was similar to WT littermates (Fig. S2C). Cytokines associated with Th2-biased epithelial cell signaling to dendritic cells, including GM-CSF, TSLP, TSLP receptor and CCL17/TARC, were also selectively increased in lung homogenates of OVA-challenged Stard7+/− mice (Fig. S3A); in contrast, two other epithelial cell-derived cytokines, IL-25 and IL-33, were not different between groups (not shown). TSLP protein localized to airway epithelial cells and infiltrating inflammatory cells and was increased in OVA-challenged Stard7+/− mice compared to WT littermates (Fig. S3B). This Th2-dominated phenotype in Stard7+/− mice was accompanied by elevated serum total IgE, as well as OVA-specific IgE (Fig. 3C), consistent with an enhanced allergic response.
Fig. 3. Deficiency of Stard7 is associated with enhanced Th2 inflammation.
(A) Quantitation of Th2 cytokines by qRT-PCR in lung tissue on day 22 after OVA sensitization and challenge. n = 4 mice/group. (B) Lung cell suspensions from sensitized and challenged mice were analyzed by flow cytometry using antibodies directed against CD4 and IL-13. n = 4–8 mice/group. (C) Total (left panel) and OVA-specific (right panel) IgE in the serum of PBS or OVA challenged mice. n = 6 mice/group.
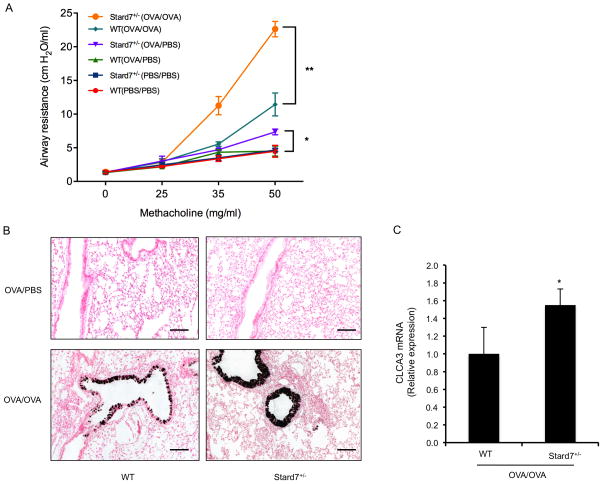
In addition to a prominent Th2-mediated inflammatory response, clinical asthma is characterized by mucous cell metaplasia and increased AHR. AHR was significantly increased in OVA-sensitized and challenged Stard7+/− mice compared to WT littermates (Fig. 4A). Baseline airway resistance was similar in unchallenged WT and Stard7+/− mice; however, OVA sensitization in the absence of OVA challenge produced a modest but significant increase in AHR in Stard7+/− animals compared to similarly treated WT littermates (Fig. 4A). Mucous cell metaplasia was detected by immunostaining and qRT-PCR for CLCA3 (Fig. 4B, C), and was confirmed by alcian blue staining (Fig. S3C). In each case, mucous cell metaplasia was more pronounced in airways of OVA-challenged Stard7+/− mice compared to WT littermates. Overall, haploinsufficiency for Stard7 was associated with enhanced pulmonary inflammation, Th2 cytokines, total and OVA-specific IgE, mucous cell metaplasia and AHR.
Fig. 4. Deficiency of Stard7 is associated with increased AHR and mucous cell metaplasia following OVA sensitization and challenge.
(A) AHR was evaluated in PBS and/or OVA-challenged mice. n = 8 mice/group; *p value, Stard7+/− (OVA/PBS) vs. WT (OVA/PBS); **p value, Stard7+/− (OVA/OVA) vs. WT (OVA/OVA). (B) CLCA3 immunohistochemistry of lung tissue from OVA-sensitized and OVA or PBS challenged mice. Scale bars = 50 μm. (C) qRT-PCR of CLCA3 mRNA in lung tissue of OVA-sensitized and challenged mice. n = 4 mice/group.
Haploinsufficiency for Stard7 increases epithelial barrier permeability and activation of pro-asthmatic DCs
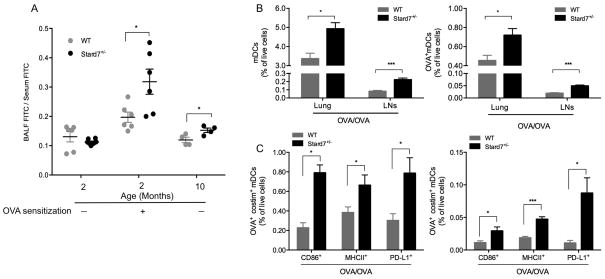
Because Stard7 is prominently expressed in the respiratory epithelium, we tested the hypothesis that decreased expression of Stard7 was associated with increased permeability of the epithelial barrier. Lung epithelial barrier permeability was assessed in non-sensitized or sensitized WT and Stard7+/− mice following intravenous administration of FITC-conjugated albumin. Accumulation of FITC-albumin in BALF was similar in nonsensitized 2 month-old WT and Stard7+/− littermates; however, epithelial permeability was significantly increased in sensitized 2 month-old or nonsensitized 10 month-old Stard7+/− mice (Fig. 5A). Consistent with increased epithelial barrier permeability, we observed an increase in the frequency of both total myeloid DCs (mDCs, CD11c+CD11b+Grl−CD317−), as well as OVA-bearing mDCs in the lungs and lung-draining lymph nodes (LNs) of OVA-challenged Stard7+/− mice (Fig. 5B). Further, a greater frequency of lung and LN mDCs were positive for CD86, MHC II or PD-L1 (Fig. 5C). Taken together, the development of an enhanced airway response in Stard7+/− mice was associated with increased epithelial barrier permeability and activation of mDCs.
Fig. 5. Deficiency of Stard7 is associated with increased lung epithelial permeability and activation of pro-asthmatic mDCs.
(A) Epithelial barrier permeability was assessed by tail vein injection of FITC-albumin followed by quantitation of FITC in BALF and serum of non-sensitized or OVA-sensitized mice. Results expressed as BALF FITC/Serum FITC. n = 4–6 mice/group. (B) The frequency of total mDCs (left panel) and OVA+ mDCs (right panel) in the lung or LNs of OVA-sensitized and challenged mice. (C) The frequency of mDCs in the lung (left panel) or LNs (right panel) expressing OVA and either CD86, MHC-II or PD-L1 in OVA-sensitized and challenged mice. n = 4–8 mice/group.
Adoptive transfer of OVA-pulsed DCs from Stard7+/− mice confers an exaggerated allergic response
Since deficiency of Stard7 was associated with an increased activation of pro-asthmatic DCs, we next determined if DCs from Stard7+/− mice were sufficient to drive an allergic response in WT mice. Bone marrow cells from WT and Stard7+/− mice were cultured in the presence of GM-CSF for 6 days, to drive development of DCs, with the addition of OVA on the last day of culture (Fig. 6A). Stard7 mRNA in bone marrow-derived DCs (BMDCs) from Stard7+/− mice was less than the expected 50% (Fig. S4A). OVA-pulsed BMDCs were subsequently transferred to the airways of WT/Stard7+/− mice (day 0) followed by intratracheal challenge with OVA on days 10 through 13 (Fig 6A). Histological evaluation 24 hours after the last OVA challenge revealed extensive peribronchial cellular infiltration (Fig. 6B) in association with a significant increase in the total number of BALF cells (Fig. S4B, left panel), eosinophilia (Fig. S4B, right panel) and elevated serum IgE (Fig. S4C) in mice that received OVA-pulsed BMDCs from Stard7+/− mice. Mucous cell metaplasia was also significantly increased in WT recipient mice that received OVA-pulsed BMDCs from Stard7+/− mice (Fig. S4D). Consistent with these findings, Stard7+/− BMDCs conferred a significant increase in airway resistance when transferred to WT recipient mice (Figs. 6C and S4E, blue line); however, the magnitude of the response was significantly increased in Stard7+/− recipient mice compared to WT control mice (Fig. 6C, green vs. blue line). In contrast, WT BMDCs were not sufficient to induce AHR when transferred to WT recipient mice (Fig. 6C, S4E, orange line), but caused a significant increase in airway resistance when transferred to Stard7+/− recipient mice (Fig. 6C, purple line). AHR remained at baseline in the PBS-challenged WT recipient mice regardless of whether the OVA-pulsed BMDCs were from WT or Stard7+/− mice (Fig. S4E). Overall, BMDCs from Stard7+/− mice were sufficient to confer an exaggerated allergic response, but the magnitude of the response was modulated by the pulmonary environment, with increased allergeneity in Stard7+/− recipients.
Fig. 6. Adoptive transfer of BMDCs from Stard7+/− mice is sufficient to confer an exaggerated allergic phenotype in WT mice.<.
br>(A) OVA-pulsed BMDCs were injected into the airways of WT or Stard7+/− littermates (Day 0) followed by OVA challenge (Days 10–13) and analysis on day 14. (B) H&E staining of lung tissue from WT recipient mice challenged with OVA. Scale bars = 50 μm. (C) AHR was evaluated in OVA-challenged recipient mice. n = 8 mice/group.
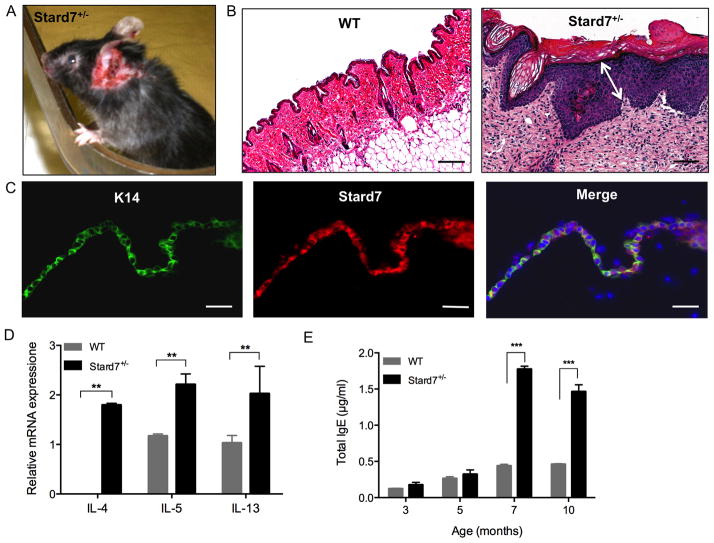
Deficiency of Stard7 results in spontaneous dermatitis
Although lung structure and function remained normal in unchallenged Stard7+/− mice (not shown), approximately 30% of Stard7+/− mice developed skin lesions, predominately on the ears and back beginning at 6 to 7 months of age (Fig. 7A). Lesions were accompanied by vigorous scratching and worsened over time. Histological analyses demonstrated significant thickening of the epidermis, including the cornified layer (Fig. 7B). Immunofluoresence microscopy detected prominent staining for Stard7 in skin tissue of WT mice that colocalized with Keratin 14 (K14, Fig. 7C), indicating that Stard7 is normally expressed in the basal cell layer of the epidermis. Th2 cytokine mRNAs, including IL-4, IL-5, and IL-13, were significantly increased in lesion skin of Stard7+/− mice compared to non-lesion skin of WT littermates (Fig. 7D). Serum IgE was normal in 3 and 5 month-old Stard7+/− mice, but was significantly increased beginning at 7 months of age (Fig. 7E), correlating with the appearance of skin lesions. This Th2-mediated inflammatory response occurred in the absence of any detectable microorganisms in cultures from lesion skin of Stard7+/− mice (not shown). Taken together, these data indicate that loss of Stard7 in the epidermis is associated with spontaneous atopic dermatitis.
Fig. 7. Stard7+/− mice develop spontaneous dermatitis.
(A) A skin lesion on the ear of a 7 month-old Stard7+/− mouse. (B) H&E staining of skin tissue from 7 month-old WT and Stard7+/− mice. A section through the skin lesion shows expansion of basal, spinous, granular and cornified layers of the epidermis (white arrow) and cellular infiltration of the dermis in Stard7+/− mice. Scale bars = 50 μm. (C) Immunofluorescence of the epithelial cell marker K14 and Stard7 in skin tissue from 8 week-old WT mice. Scale bars = 25 μm. (D) qRT-PCR of Th2 cytokine mRNAs in skin tissue of 7 month-old WT and Stard7+/− littermates. β-actin was used as an endogenous control. n = 3 mice/group. (E) Total IgE in the serum of mice over time. n = 4 mice/group.
DISCUSSION
In the current study, haploinsufficiency for Stard7 was associated with exaggerated allergic lung disease and atopic dermatitis in a mouse strain (C57BL/6J) that is widely considered to be resistant to allergic disease. OVA sensitization followed by intratracheal allergen challenge of Stard7+/− mice resulted in significantly increased pulmonary inflammation, mucous cell metaplasia, AHR, and OVA-specific IgE compared to similarly treated WT littermates. Furthermore, approximately one-third of Stard7+/− mice developed pruritic skin lesions associated with marked Th2 inflammation, and elevated IgE. Taken together, these data suggest that Stard7 is an important component of a novel protective pathway that dampens allergic responses at these two mucosal-environmental interfaces.
The current study provides evidence that deficiency of Stard7 expression in both DCs and epithelial cells contributes to pathogenesis in Stard7+/− mice. Stard7 was expressed in BMDCs, and transfer of OVA-sensitized BMDCs from Stard7+/− mice was sufficient to confer an exaggerated allergic response in WT mice. However, the magnitude of the response in WT recipient mice was significantly lower than that in Stard7+/− mice. Given that the major site of pulmonary Stard7 expression in the lung is in respiratory epithelial cells, it is likely that loss of Stard7 in epithelial cells also contributed to an exaggerated allergic response. Consistent with this hypothesis, lung permeability was significantly increased in aged unchallenged Stard7+/− mice; further, OVA sensitization alone was sufficient to induce AHR, suggesting that Stard7+/− mice are predisposed to allergic disease. Epithelial barrier integrity plays a critical role in limiting access of environmental allergens, microorganisms and toxicants to tissues(15, 16). Overall, decreased epithelial barrier function likely leads to local sensitization and DC recruitment/activation. Generation of mice in which Stard7 is specifically deleted in lung epithelial cells is currently underway to directly test this hypothesis.
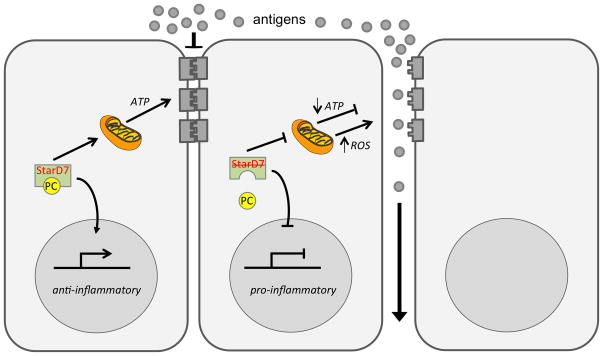
Stard7 is a member of the subfamily of START domain proteins that transfers phospholipids (Stard2, Stard7, and Stard10) or ceramide (Stard11) among intracellular membranes(4, 5). Two Stard7 isoforms, with predicted masses of 43 kDa (Stard7-I) and 35 kDa (Stard7-II) have been identified and both isoforms bind/transfer PC(2, 8). The Stard7-I isoform contains a 75 amino acid NH2-terminal extension that serves as a mitochondrial targeting signal and overexpression of this isoform facilitates uptake of PC by mitochondria. PC is the major phospholipid in mitochondria, comprising 44% of total phospholipid(17) and, since mitochondria lack the capacity to synthesize PC, import of this phospholipid is likely critical for mitochondrial homeostasis. Mitochondrial dysfunction in airway epithelial cells has previously been linked to oxidant stress and asthma pathogenesis(9–12). Thus, one possible explanation for exaggerated allergic disease in Stard7+/− mice is oxidant damage and/or diminished generation of ATP, secondary to mitochondrial injury, that leads to reduced epithelial barrier function, increased allergen penetration and recruitment/activation of dendritic cells (Fig. 8). Consistent with this model, reactive oxygen species arising from mitochondrial respiration were previously shown to promote differentiation of dendritic cells(18). Thus, mitochondrial dysfunction could contribute to the increased frequency of pro-asthmatic DCs, as well as altered barrier function in Stard7+/− mice. Although the precise mechanisms are unclear, our study suggests that loss of Stard7 promotes recruitment and/or activation of mDCs resulting in an exaggerated Th2 inflammatory response to allergens.
Fig. 8. Potential mechanisms for Stard7 action.
Stard7 facilitates uptake of PC by mitochondria, promoting mitochondrial homeostasis and the energy required for maintenance of epithelial barrier integrity. Stard7 may also suppress inflammatory responses, leading to decreased DC activation and Th2 inflammation. Haploinsufficiency for Stard7 could lead to decreased ATP production, oxidative stress (generation of reactive oxygen species, ROS) and pro-inflammatory responses that promote epithelial barrier permeability and DC maturation, as discussed in the text.
Stard7-II lacks the mitochondrial targeting sequence present in Stard7-I and may arise from translation initiation at a downstream, in-frame start codon in the Stard7 mRNA and/or processing of Stard7-I(3). The relative abundance and subcellular localization of Stard7-II to the cytosol and plasma membrane in trophoblast cells (19) and nucleus of testis cells (20) strongly suggest that this isoform performs a non-mitochondrial function(s) that could also impact the phenotype of Stard7+/− mice (Fig. 8). Consistent with a nuclear function, transcription factors that contain both a START domain and a homeobox domain are relatively common in plants(21, 22). Nuclear localization of Stard7 would require association with a partner protein containing a nuclear localization sequence. In this regard, Stard2, a closely related PC transfer protein, was shown to bind to the homeodomain transcription factor Pax3 and colocalize to the nucleus of transfected cells(23). Moreover, Stard10 was recently shown to modulate activity of the transcription factor PPARα and its downstream target genes(7). Both PPARα and -γ antagonize several transcription factors involved in the effector phase of allergic inflammation, including nuclear factor-κB (NF-κB) and activator protein-1 (AP-1)(24–27). Humans with polymorphisms in PPARγ are at an increased risk for developing asthma(28, 29). Consistent with these findings, mice deficient in PPARα presented with increased eosinophilia, IgE production and AHR following allergen challenge(30), while mice treated with PPARγ agonists were protected against disease(30–34). Importantly, PC was identified as an endogenous ligand of PPARα and LRHI/NR5A2(35–37). Although the mechanism by which PC is delivered to PPAR remains an important knowledge gap, it is conceivable that Stard7 and/or Stard2 and Stard10 serve as PC chaperones for the nuclear PPAR/RXR complex. Notably, PPAR signaling negatively regulates DC immunogenicity(38, 39); thus, it is plausible that the loss of Stard7 prevented PPAR-mediated suppression of DC activation, resulting in the ability of OVA-pulsed Stard7+/− DCs to confer an exaggerated allergic response in WT recipient mice. This model may also explain the remarkable phenotypic similarities among Stard7+/− mice, RXRα/β−/− mice and PPARα−/− mice, including an increased susceptibility to allergic lung disease and/or atopic dermatitis(30, 40). Overall, a potential nuclear function for Stard7 merits further investigation.
Global deletion of Stard7 resulted in embryonic lethality between E10 and E11, most likely related to disruption of cardiovascular development, although this hypothesis has yet to be tested. In contrast to Stard7−/− mice, Stard2/Pctp−/−(41) and Stard10−/−(7) mice survived and exhibited relatively subtle and distinct metabolic phenotypes in adulthood. These results suggest that, despite significant structural homology among these PC transfer proteins, overlap of biologic function may be limited. It is important to note that a role for Stard2 or Stard10 in allergic disease has not been directly tested and this remains an important knowledge gap. The observation that haploinsufficiency for Stard7 was sufficient for predisposition to allergic disease in mice may have important implications for human disease. Indeed, expression of Stard7 mRNA was decreased by approximately 50% in nasal mucosal brushings from human patients experiencing an asthma exacerbation compared to stable asthmatics and healthy controls (1). Importantly, respiratory viruses are a major cause of asthma exacerbations and may contribute to suppression of Stard7 expression. Overall these findings underscore the need for studies to determine if loss of Stard7 expression is causally linked to asthma susceptibility in humans.
Supplementary Material
Footnotes
This work is supported by NIH, HL122130 (TEW)
References
- 1.Guajardo JR, Schleifer KW, Daines MO, Ruddy RM, Aronow BJ, Wills-Karp M, Hershey GK. Altered gene expression profiles in nasal respiratory epithelium reflect stable versus acute childhood asthma. The Journal of allergy and clinical immunology. 2005;115:243–251. doi: 10.1016/j.jaci.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 2.Durand S, Angeletti S, Genti-Raimondi S. GTT1/StarD7, a novel phosphatidylcholine transfer protein-like highly expressed in gestational trophoblastic tumour: cloning and characterization. Placenta. 2004;25:37–44. doi: 10.1016/S0143-4004(03)00214-5. [DOI] [PubMed] [Google Scholar]
- 3.Flores-Martin J, Rena V, Angeletti S, Panzetta-Dutari GM, Genti-Raimondi S. The Lipid Transfer Protein StarD7: Structure, Function, and Regulation. International journal of molecular sciences. 2013;14:6170–6186. doi: 10.3390/ijms14036170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark BJ. The mammalian START domain protein family in lipid transport in health and disease. The Journal of endocrinology. 2012;212:257–275. doi: 10.1530/JOE-11-0313. [DOI] [PubMed] [Google Scholar]
- 5.Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 6.Kang HW, Wei J, Cohen DE. PC-TP/StARD2: Of membranes and metabolism. Trends in endocrinology and metabolism: TEM. 2010;21:449–456. doi: 10.1016/j.tem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito M, Yamanashi Y, Toyoda Y, Izumi-Nakaseko H, Oda S, Sugiyama A, Kuroda M, Suzuki H, Takada T, Adachi-Akahane S. Disruption of Stard10 gene alters the PPARalpha-mediated bile acid homeostasis. Biochimica et biophysica acta. 2013;1831:459–468. doi: 10.1016/j.bbalip.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Horibata Y, Sugimoto H. StarD7 mediates the intracellular trafficking of phosphatidylcholine to mitochondria. The Journal of biological chemistry. 2010;285:7358–7365. doi: 10.1074/jbc.M109.056960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabalirajan U, Dinda AK, Kumar S, Roshan R, Gupta P, Sharma SK, Ghosh B. Mitochondrial structural changes and dysfunction are associated with experimental allergic asthma. Journal of immunology. 2008;181:3540–3548. doi: 10.4049/jimmunol.181.5.3540. [DOI] [PubMed] [Google Scholar]
- 10.Mabalirajan U, Rehman R, Ahmad T, Kumar S, Singh S, Leishangthem GD, Aich J, Kumar M, Khanna K, Singh VP, Dinda AK, Biswal S, Agrawal A, Ghosh B. Linoleic acid metabolite drives severe asthma by causing airway epithelial injury. Scientific reports. 2013;3:1349–1361. doi: 10.1038/srep01349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas B, Rutman A, Hirst RA, Haldar P, Wardlaw AJ, Bankart J, Brightling CE, O’Callaghan C. Ciliary dysfunction and ultrastructural abnormalities are features of severe asthma. The Journal of allergy and clinical immunology. 2010;126:722–729. doi: 10.1016/j.jaci.2010.05.046. [DOI] [PubMed] [Google Scholar]
- 12.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. Journal of immunology. 2009;183:5379–5387. doi: 10.4049/jimmunol.0900228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer EL, Mushaben EM, Pastura PA, Acciani TH, Deutsch GH, Khurana Hershey GK, Korfhagen TR, Hardie WD, Whitsett JA, Le Cras TD. Early growth response-1 suppresses epidermal growth factor receptor-mediated airway hyperresponsiveness and lung remodeling in mice. American journal of respiratory cell and molecular biology. 2009;41:415–425. doi: 10.1165/rcmb.2008-0470OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidovich N, DiPaolo BC, Lawrence GG, Chhour P, Yehya N, Margulies SS. Cyclic stretch-induced oxidative stress increases pulmonary alveolar epithelial permeability. American journal of respiratory cell and molecular biology. 2013;49:156–164. doi: 10.1165/rcmb.2012-0252OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holgate ST, Roberts G, Arshad HS, Howarth PH, Davies DE. The role of the airway epithelium and its interaction with environmental factors in asthma pathogenesis. Proceedings of the American Thoracic Society. 2009;6:655–659. doi: 10.1513/pats.200907-072DP. [DOI] [PubMed] [Google Scholar]
- 16.Holgate ST. The sentinel role of the airway epithelium in asthma pathogenesis. Immunological reviews. 2011;242:205–219. doi: 10.1111/j.1600-065X.2011.01030.x. [DOI] [PubMed] [Google Scholar]
- 17.Horvath SE, Daum G. Lipids of mitochondria. Progress in lipid research. 2013;52:590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Del Prete A, Zaccagnino P, Di Paola M, Saltarella M, Oliveros Celis C, Nico B, Santoro G, Lorusso M. Role of mitochondria and reactive oxygen species in dendritic cell differentiation and functions. Free radical biology & medicine. 2008;44:1443–1451. doi: 10.1016/j.freeradbiomed.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 19.Angeletti S, Rena V, Nores R, Fretes R, Panzetta-Dutari GM, Genti-Raimondi S. Expression and localization of StarD7 in trophoblast cells. Placenta. 2008;29:396–404. doi: 10.1016/j.placenta.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Leman ES, Magheli A, Yong KM, Netto G, Hinz S, Getzenberg RH. Identification of nuclear structural protein alterations associated with seminomas. Journal of cellular biochemistry. 2009;108:1274–1279. doi: 10.1002/jcb.22357. [DOI] [PubMed] [Google Scholar]
- 21.Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome biology. 2004;5:R41.1–16. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schrick K, Bruno M, Khosla A, Cox PN, Marlatt SA, Roque RA, Nguyen HC, He C, Snyder MP, Singh D, Yadav G. Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity. BMC biology. 2014;12:70–89. doi: 10.1186/s12915-014-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. The Journal of biological chemistry. 2007;282:30728–30736. doi: 10.1074/jbc.M703745200. [DOI] [PubMed] [Google Scholar]
- 24.Yang XY. Activation of Human T Lymphocytes Is Inhibited by Peroxisome Proliferator-activated Receptor gamma (PPARgamma) Agonists. PPARgamma co-association with transcription factor NFAT. Journal of Biological Chemistry. 2000;275:4541–4544. doi: 10.1074/jbc.275.7.4541. [DOI] [PubMed] [Google Scholar]
- 25.Delerive PBK, Besnard S, Berghe WV, Peters J, Gonzalez F, Fruchart J-C, Tedgui A, Haegeman G, Staels B. Peroxisome Proliferator-activated Receptorα Negatively Regulates the Vascular Inflammatory Gene Response by Negative Cross-talk with Transcription Factors NF-κ B and AP-1. J Biol Chem. 1999;274:32048–54. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- 26.Desmet CGP, Pajak B, Cataldo D, Bentires-Alj M, Lekeux P, Bureau F. Selective Blockade of NF-κB Activity in Airway Immune Cells Inhibits the Effector Phase of Experimental Asthma. J Immunol. 2004;173:5766–75. doi: 10.4049/jimmunol.173.9.5766. [DOI] [PubMed] [Google Scholar]
- 27.Desmet C, Gosset P, Henry E, Garze V, Faisca P, Vos N, Jaspar F, Melotte D, Lambrecht B, Desmecht D, Pajak B, Moser M, Lekeux P, Bureau F. Treatment of experimental asthma by decoy-mediated local inhibition of activator protein-1. American journal of respiratory and critical care medicine. 2005;172:671–678. doi: 10.1164/rccm.200410-1431OC. [DOI] [PubMed] [Google Scholar]
- 28.Oh SH, Park SM, Lee YH, Cha JY, Lee JY, Shin EK, Park JS, Park BL, Shin HD, Park CS. Association of peroxisome proliferator-activated receptor-gamma gene polymorphisms with the development of asthma. Respiratory medicine. 2009;103:1020–1024. doi: 10.1016/j.rmed.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Benayoun L, Letuve S, Druilhe A, Boczkowski J, Dombret MC, Mechighel P, Megret J, Leseche G, Aubier M, Pretolani M. Regulation of peroxisome proliferator-activated receptor gamma expression in human asthmatic airways: relationship with proliferation, apoptosis, and airway remodeling. American journal of respiratory and critical care medicine. 2001;164:1487–1494. doi: 10.1164/ajrccm.164.8.2101070. [DOI] [PubMed] [Google Scholar]
- 30.Woerly G, Honda K, Loyens M, Papin JP, Auwerx J, Staels B, Capron M, Dombrowicz D. Peroxisome proliferator-activated receptors alpha and gamma down-regulate allergic inflammation and eosinophil activation. The Journal of experimental medicine. 2003;198:411–421. doi: 10.1084/jem.20021384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda K, Marquillies P, Capron M, Dombrowicz D. Peroxisome proliferator-activated receptor gamma is expressed in airways and inhibits features of airway remodeling in a mouse asthma model. The Journal of allergy and clinical immunology. 2004;113:882–888. doi: 10.1016/j.jaci.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 32.Mueller C, Weaver V, Vanden Heuvel JP, August A, Cantorna MT. Peroxisome proliferator-activated receptor γ ligands attenuate immunological symptoms of experimental allergic asthma. Archives of Biochemistry and Biophysics. 2003;418:186–196. doi: 10.1016/j.abb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Trifilieff A, Bench A, Hanley M, Bayley D, Campbell E, Whittaker P. PPAR-alpha and -gamma but not -delta agonists inhibit airway inflammation in a murine model of asthma: in vitro evidence for an NF-kappaB-independent effect. British journal of pharmacology. 2003;139:163–171. doi: 10.1038/sj.bjp.0705232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward JE, Fernandes DJ, Taylor CC, Bonacci JV, Quan L, Stewart AG. The PPARgamma ligand, rosiglitazone, reduces airways hyperresponsiveness in a murine model of allergen-induced inflammation. Pulmonary pharmacology & therapeutics. 2006;19:39–46. doi: 10.1016/j.pupt.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JM, Lee YK, Mamrosh JL, Busby SA, Griffin PR, Pathak MC, Ortlund EA, Moore DD. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musille PM, Pathak MC, Lauer JL, Hudson WH, Griffin PR, Ortlund EA. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nature structural & molecular biology. 2012;19:532–537. S531–532. doi: 10.1038/nsmb.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nencioni A, Grunebach F, Zobywlaski A, Denzlinger C, Brugger W, Brossart P. Dendritic cell immunogenicity is regulated by peroxisome proliferator-activated receptor gamma. Journal of immunology. 2002;169:1228–1235. doi: 10.4049/jimmunol.169.3.1228. [DOI] [PubMed] [Google Scholar]
- 39.Klotz L, Dani I, Edenhofer F, Nolden L, Evert B, Paul B, Kolanus W, Klockgether T, Knolle P, Diehl L. Peroxisome proliferator-activated receptor gamma control of dendritic cell function contributes to development of CD4+ T cell anergy. Journal of immunology. 2007;178:2122–2131. doi: 10.4049/jimmunol.178.4.2122. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Messaddeq N, Teletin M, Pasquali JL, Metzger D, Chambon P. Retinoid X receptor ablation in adult mouse keratinocytes generates an atopic dermatitis triggered by thymic stromal lymphopoietin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14795–14800. doi: 10.1073/pnas.0507385102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Helvoort A, de Brouwer A, Ottenhoff R, Brouwers JF, Wijnholds J, Beijnen JH, Rijneveld A, van der Poll T, van der Valk MA, Majoor D, Voorhout W, Wirtz KW, Elferink RP, Borst P. Mice without phosphatidylcholine transfer protein have no defects in the secretion of phosphatidylcholine into bile or into lung airspaces. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:11501–11506. doi: 10.1073/pnas.96.20.11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.