Abstract
Macrophage polarization plays a critical role in tissue homeostasis, disease pathogenesis, and inflammation and its resolution. IL-4-induced macrophage polarization involves induction of STAT6 and KLF4 that induce each other and promote M2 polarization. However, how these transcription factors implement M2 polarization is not understood. We report that in murine macrophages MCPIP, induced by KLF4, inhibits M1 polarization by inhibiting NF-κB activation and implements M2 polarization using both its deubiquitinase and RNase activities that cause sequential induction of reactive oxygen species (ROS), endoplasmic reticulum (ER) stress and autophagy required for M2 polarization. MCPIP also induces C/EBPβ and PPARγ that promote M2 polarization. Macrophages from mice with myeloid-targeted overexpression of MCPIP show elevated expression of M2 markers and reduced response to LPS, whereas macrophages from mice with myeloid-specific deletion of MCPIP manifest elevated M1 polarization with enhanced phagocytic activity. Thus, both in vivo and in vitro experiments demonstrate that the transcription factors STAT6 and KLF4 implement IL-4-induced M2 polarization via the dual catalytic activities of MCPIP.
Keywords: Macrophage polarization, MCPIP, M2 polarization by IL-4 mediated via MCPIP, STAT6/KLF4 induce MCPIP, induction of polarization using both catalytic activities of MCPIP
Introduction
Macrophages are central players in the initiation and resolution of inflammation. They show remarkable plasticity that helps them to modulate their phenotype in response to environmental signals. Such signals include exogenous ones such as LPS from invading bacteria or cytokines produced by injury or inflammation. Macrophages, under the influence of such signals, undergo classical activation and differentiate into M1 macrophages, characterized by their production of inflammatory cytokines such as TNF-α and IL-6, and reactive oxygen and nitrogen species (1–6). Their cytotoxic activities enable the M1 macrophage to eliminate pathogens and initiate inflammation. Even though this activity protects the host, sustained inflammation can be very detrimental to the host. To protect the host against such damage, macrophages can undergo alternate activation to generate M2 macrophages (1–6). M2 macrophages produce anti-inflammatory cytokines such as IL-10 and have increased expression of arginase-1(Arg1), which competes with eNOS for a common limiting substrate L-arginine (1–4). IL-4 and IL-13, produced most highly by eosinophils, induce M2 polarization. M2 macrophages are primarily involved in atopic disease, parasite response, phagocytosis of apoptotic cells, resolution of inflammation, angiogenesis, tissue repair, and remodeling (3,6).
Several signaling molecules, transcription factors, and post-transcriptional regulators are known to regulate M1/M2 macrophage polarization (4, 7). GM-CSF promotes M1 and M-CSF promotes M2 polarization (6, 8). Transcription factors NF-κB, AP-1, C/EBPα, PK1, IRFS, and STAT1 activation promote M1, whereas STAT3/6, IRF4, PPARγ, and C/EBPβ activation promote M2 macrophage polarization (4, 6, 7). IL-4 induces M2 polarization via STAT6 and KLF4 that induce each other and cooperatively induce M2 polarization (9). Even though these transcription factors are known to eventually cause induction of M2 markers such as Arg1, the proteins that connect KLF4 to the biological processes involved in M2 polarization remain to be identified.
IL-4-induced M2 polarization, mediated via KLF4, involves inhibition of M1 polarization and promotion of M2 polarization (9). NF-κB is a key player that promotes M1 polarization (4, 6, 9). MCPIP, first identified as a protein induced in human peripheral blood monocytes upon MCP-1 treatment (10), was also found to be induced by other inflammatory agents (11, 12). MCPIP is known to inhibit NF-κB activation (11, 13). Thus, MCPIP might contribute to IL-4-induced M2 polarization by inhibiting M1 polarization. MCPIP was also found to have RNase activity including anti-Dicer activity and thus, has been also called Regnase (12, 14, 15). The RNase activity causes suppression of production of inflammatory cytokines and their receptors (16), thus could inhibit M1 polarization. In addition, MCPIP is known to induce reactive oxygen species (ROS) production, endoplasmic reticulum (ER) stress, and autophagy in some cellular contexts (17–19). All of these three biological processes have been reported to be required for IL-4-induced M2 polarization (20–22). Furthermore, the promoter of MCPIP shows multiple KLF4 sites. Therefore we postulated that IL-4 induced M2 polarization, mediated via KLF4, would involve KLF4 induction of MCPIP that would induce ROS production, ER stress, and autophagy required for M2 polarization. Here, we present experimental evidence to support this hypothesis.
We demonstrate that treatment of murine peritoneal macrophages with IL-4 induces MCPIP via induction of KLF4 and that the M2 polarization induced by the IL-4 is mediated via MCPIP. We also demonstrate that macrophages from mice with specific deletion of MCPIP are incapable of IL-4-induced M2 polarization. We show that macrophages from transgenic animals with macrophage-specific over expression of MCPIP, inhibited expression of M1 markers and stimulated expression of M2 markers. Thus, both in vitro and in vivo experiments show that MCPIP plays a critical role in M2 polarization. MCPIP is known to have deubiquitinase and RNase activities including anti-Dicer activity. With MCPIP mutants that have only one of the two catatylic activities we demonstrate that both of these catalytic powers of MCPIP implement the IL-4 induction of differentiation mediated via transcription factors STAT6 and KLF4, and thus establish MCPIP as the catalyst that connects the transcription factors, STAT6 and KLF4, to the biological processes they regulate.
Materials and Methods
Preparation and characterization of deubiquitinase mutant of MCPIP that retains RNase activity
Deletion mutants for the four potential ubiquitin interacting domains were prepared, the mutant proteins were expressed in HEK cells and purified and assayed for deubiquitinase activity with a model substrate Ub-AFC and with high molecular weight K63-linked polyubiquitin (Boston Biochem) as described (23). One of the four mutants that showed loss of deubiquitinase activity is designated Dub-mutant. This mutant was also assayed for RNase activity as per manufacturer’s instructions (Applied Biosystem). Anti-Dicer RNAse activity of MCPIP and Dub mutant was measured using a synthetic pre miRNA-135a tagged with a fluorophore in the loop and a quencher in the stem (5’-rCrArG rCrCrC rUrArU rGrUrG rArUrU rGrC/i6-FAMK rGrUrC rCrCrA rArArC rUrCrA rUrGrU rArGrG /iBHQ-1 /rGrCrA −3’) (IDT). Purified MCPIP (5µg) was incubated with 50 pmole of pre miRNA-135a in buffer containing 30 mM HEPES pH 7.5, 100 mM potassium acetate, 10mM magnesium acetate, 10 mM DTT and 10% glycerol in a final volume of 200 µl. Dicer activity was measured by the increase in fluorescence caused by release of the fluorophore from the loop. The Dub mutant retained full RNase and anti-Dicer activities. Experiments were performed in triplicates.
Generation of animals with myeloid specific MCPIP knockout mice
A bacterial artificial chromosome clone containing 223,095 bp of mouse chromosome 4, including the entire MCPIP gene, was used to subclone the full length MCPIP gene into a minimal vector containing an origin of replication and an Ampicillin resistance gene. The Gene Bridges’ BAC subcloning kit by RED/ET recombination was used to subclone a 9kb segment of MCPIP gene according to the manufacturer’s protocol. The subcloned 9kb containing exon 2 through 6 along with the intervening introns was used to introduce loxP sites at intron 2 and intron 4 of the MPCIP gene using Gene Bridges’ Quick and Easy Conditional Knockout Kit (LoxP/Cre) by Red/ET recombination according to the manufacturer’s protocol. Plasmid DNA from the final clone was purified and sequence confirmed prior to producing a linear fragment of the construct by EcoRV digestion. The linearized DNA segment containing the MCPIP-LoxP construct was electroporated into C57/BL6/7 ES cells and selection was made with neomycin. PCR based screening and southern blot analysis were used to confirm homozygous recombination. ES cells containing the MCPIP-LoxP construct were injected into blastocysts from coisogenic strain C57BL6 Ty(c)2J and homozygous line for MCPIP-loxP allele was produced by breeding and genotyping with PCR. The macrophage-specific MCPIP knock out mice (myelo-KO) were generate by crossing MCPIP-LoxP +/+ mice with LysM-Cre mice (Jackson Laboratory) and LoxP +/+, Cre+ (myelo-KO) mice were identified by PCR genotyping.
Generation of mice with myeloid targeted overexpression of MCPIP
Murine LysM promoter (5532bp) from mouse chromosome 10 position 116724852 to 116719328 was fused to murine MCPIP-FLAG in a pBluescript vector. A7332bp NotI-XhoI fragment containing the LysM promoter fused to MCPIP was purified by gel electrophoresis and microinjected into fertilized C57BL/6J mouse ova at the MD Anderson Cencer Center, Houston Texas. Genotying was carried out using PCR with specific primers in the LysM promoter region and the transgenic coding region. The transgene containing founders were bred with C57BL/6J mice to generate F1 transgenic mice; homozygous myelo-MCPIP mice were produced by interbreeding.
Murine peritoneal macrophage isolation and culture
Thioglycollate-elicited macrophages were plated in 6-well plates at a density of 1 × 106 cells per well in DMEM medium containing 10% fetal calf serum and 1% penicillin-streptomycin and 1% glutamine. After 4 hr incubation, nonadherent cells were removed with PBS, and new culture medium was added to the wells and cells were subjected to treatment after 48 hr culture. The following strains of mice were used for these experiments: MCPIP mice homozygous for targeting myeloid-specific expression of MCPIP or myeloid-specific deletion of MCPIP on the C57/BL background. C57/BL and MCPIP-Loxp homozygous mice were used as wild-type (WT) controls.
Plasmid construction for generating deletion mutants of MCPIP
Constructs with in frame deletions of the four putative ubiquitin interacting motifs identified using bioinformatics were created within MCPIP. The deletions encompassed the following sequences; U1-nucleotide 1 to 63, U2- nucleotide 379 to 417, U3- nucleotide 681 to 717 and U4- nucleotide 1112 to 1156. The wild type MCPIP subcloned into the pCMV-MAT-FLAG vector was used as PCR template and PfuUltra II Fusion HS DNA Polymerase (Agilent) was used for the PCR reactions generating the deletion mutants. The deletions for the internal motifs were achieved using a PCR technique known as gene splicing by overlap extension (24).The final PCR product was cloned into the pCR- Blunt II-TOPO vector; after sequencing to confirm the integrity of the sequence and the in frame deletion, it was subcloned into pCMV-MAT-FLAG vector digested with HindIII and XbaI. The N-terminal deletion mutant MCPIP U1 was directly amplified using primer pair U1A for and UDrev. To be able to monitor the transfection efficiency the mutated MCPIP constructs were subcloned into the EcoRI- BamH1 sites of pEGFP-N1 containing an enhanced green fluorescent protein (GenBank:U55762.1).
Cell treatment, transfection and siRNA knockdown
Macrophages were seeded at 1 X 106 cells per well in six-well plates and cultured in the complete DMEM medium for 2 days. After removal of the nonadherent cells, the attached cells were treated with LPS (100ng/ml; Sigma) or IL-4 (20ng/ml; Cell Signaling) for the reported time. For cell transfection, cells were transfected with either 1 µg of pEGFP/N1 vector or 1 µg of pEGFP-MCPIP expression plasmid, or expression vectors for D141N mutant or DUB mutant for 48 hr as previously described (10). For siRNA knockdown studies, the attached cells were preincubated with Opti-MEM I medium containing lipofectamine (in vitrogen)/siRNA mixture (final concentration 100 nM siRNA) for 24 hr before any treatments. siRNA for MCPIP, KLF4 and STAT6 was purchased from Life Technologies Ambion. For inhibitor intervention experiments, the attached cells were preincubated with 1 µM CeO2 nanoparticles, 100 µM of tauroursodeoxycholate (TUDC; Sigma-Aldrich) or 50 µM LY294002 (Sigma) in the complete DMEM medium for 6 hr before IL-4 treatment. Experiments were repeated at least three times.
Determination of intracellular ROS
The presence of free radicals in the macrophages after different stimulation was determined using dihydrorhodamine (DHR) 123 (In vitrogen), as described previously (18). Fluorescence images were obtained with a Nikon fluorescence microscope and NIS elements software (Nikon). In all cases at least six different fields covering at least 200 cells were examined for quantifying the data.
Dual luciferase reporter assay
Macrophages were seeded at a concentration of (1 X 106 cells per well) in six-well plates and cotransfected with 2 µg of KLF4 expression vector (Addgene) and vector containing 1.5 Kb MCPIP promoter fused to a luciferase gene in a 1:1 ratio using the Lipofectamine Transfection Reagent (In vitrogen) in FBS-free cell culture media. After 24 hr exposure to the transfection mixture, the media was replaced with complete medium for 24 hr, and then the cells were washed with DPBS, lysed with 1× passive lysis buffer from Promega, and luciferase activity was measured by using the Dual Luciferase Reporter assay kit (Promega) according to the manufacturer's protocol. NF-κB activity was determined using the NF-κB Reporter kit (BPS Bioscience). Macrophages were seeded at 1 × 106 cells per well in six-well plates and cultured in the complete DMEM medium for 2 days. After removal of the non-adherent cells, the attached cells were transfected with NF-κB reporter and negative control reporter, for 24 hr following the kit protocol. The cells were then incubated with Opti-MEM I medium containing Lipofectamine (in vitrogen)/MCPIP siRNA mixture (final concentration 100 nM siRNA) for 24 hr. After which, the cells were treated with LPS (100ng/ml) for 6 hr. After LPS treatment the cells were lysed according to the manufacturer's instructions, and dual luciferase assay was performed using Dual-Glo luciferase Assay System (Promega). NF-κB activity is reported as a ratio of firefly luminescence to Renilla luminescence. Experiments were performed in triplicates.
Quantitative real-time RT-PCR
Total RNA was extracted from cell cultures using the Exiqon Micurry RNA isolation kit (Exiqon) according to the manufacturer's instructions. cDNA was generated using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Real-time RT-PCR was performed with 7500 Fast real-time PCR detection system (Applied Biosystems) using the SYBR Green master mix (Life Technologies). mRNA levels for M1 markers (TNF-α, IL-1β, IL-6, and iNOS), M2 markers (Arg1, MRC1, and FIZZ1), ER stress markers (GRP78, IRE-1) and autophagy marker (Beclin-1) were determined relative to the housekeeping gene β-actin, and fold differences within each group were calculated. Primers for gene expression assays were synthesized by Integrated DNA technology (Table 1). The same cDNA was used to measure the levels of miR-155, −125, −223, and −146. Primers for miRs were purchased from Exiqon.
Table 1.
List of primers used for qRT-PCR
| FIZZ1 | Forward | 5' CCAATCCAGCTAACTATCCCTCC 3' |
| Reverse | 5' ACCCAGTAGCAGTCATCCCA 3' | |
| YM1 | Forward | 5'AGAAGGGAGTTTCAAACCTGGT3' |
| Reverse | 5'CTCTTGCTGATGTGTGTAAGTGA 3' | |
| MRC1 | Forward | 5' CTCTGTTCAGCTATTGGACGC 3' |
| Reverse | 5' CGGAATTTCTGGGATTCAGCTTC 3' | |
| Arg1 | Forward | 5' TGGCTTGCGAGACGTAGAC 3' |
| Reverse | 5' GCTCAGGTGAATCGGCCTTTT 3' | |
| GAPDH | Forward | 5' GAAGGTGAAGGTCGGAGTCAAC 3' |
| Reverse | 5' CAGAGTTAAAAGCAGCCTTGG 3' | |
| Beclin-1 | Forward | 5'ATGGAGGGGTCTAAGGCGTC 3' |
| Reverse | 5' AGACACCATCCTGGCGAGTTTCAA 3' | |
| GRP78 | Forward | 5' GCCCAAGTGGCTGTTTACTGCTTT 3' |
| Reverse | 5' ATCCAAGGTGAACACACACCCTGA 3' | |
| IRE-1 | Forward | 5' TATGCCTCTCCCTCAATGGTGCAT 3' |
| Reverse | 5' TCAAACTTGAGGTCTGTGCTGGGA 3' | |
| MCPIP | Forward | 5'-TGAGCCATGGGAAGAAGGAAGTCT 3' |
| Reverse | 5' TGTGCTGGTCTGTGATAGGCACAT 3' | |
| TNF-α | Forward | 5'GTGGAACTGGCAGAAGAGGC 3' |
| Reverse | 5' AGACAGAAGAGCGTGGTGGC 3' | |
| IL-1β | Forward | 5' GGAAGATTCTGAAGAAGAGACGG 3' |
| Reverse | 5' TGAGATTTTTAGAGTAACAGG 3' | |
| IL-6 | Forward | 5' ACAAGTCGGAGGCTTAATTACACAT 3' |
| Reverse | 5'AATCAGAATTGCCATTGCACAA 3' | |
| β-actin | Forward | 5' ATGTTTGAGACCTTCAACA 3' |
| Reverse | 5' CACGTCAGACTTCATGATGG 3' |
Immunoblot analysis
Macrophages were lysed with Cell Lytic Buffer (Sigma) and the cell lysate was collected. For p47 phox estimation the cell lysate was centrifuged at 600g for 10 min at 4°C to remove unbroken cells and nuclei. The supernatant was then ultracentrifuged at 100,000g for 1hr at 4°C to isolate the membrane fraction. Protein was estimated by Bradford’s reagent and equal amounts of protein sample from each experimental condition was subjected to immunoblot analysis using the following primary antibodies: goat poly anti-MCPIP (1:500), rabbit anti-IRE-1 (1:500), rabbit anti-LC3 II (1:500), rabbit anti-C/EBPβ (1:100), rabbit anti-PPARγ (1:100), rabbit anti-FIZZ1 (1:500), monoclonal anti–p47phox (1:200), and rabbit anti-Fas (1:1000; Santa Cruz Biotechnology); mouse polyclonal anti-GAPDH (1:1000); rabbit anti-GRP78 (1:500); rabbit anti-Beclin 1(1:1000), goat anti-Arg1 (1:2000; Cell signaling). The immune complexes were detected autoradiographically using appropriate peroxidase-labeled secondary antibodies (Santa Cruz Biotechnology) and enhanced chemiluminescence detection reagent ECL (GE Healthcare). Anti-β-actin and anti-GAPDH antibodies served as loading controls. Specific bands were quantified by densitometry using analytic software (Image J) and expressed as a ratio over loading controls.
Autophagasome measurement
The presence of autophagic vacuoles was determined using cyto-ID autophagic detection kit (Enzo Life Sciences) using the manufacturers’ protocol. Fluorescence images were obtained with a Nikon fluorescence microscope and fluorescence intensity was measured by using NIS elements software (Nikon).In all cases at least six different fields covering at least 200 cells were examined for quantifying the data.
Zymosan phagocytosis assay
Zymosan phagocytosis assay was done using pHrodo™ Red Zymosan Bioparticles (Life Technologies) according to the manufacturer’s instructions. Macrophages were plated at a density of 1 × 106 cells per well in 24-well plates in a final volume of 1 mL. After 24 hours, fluorescently labeled (red) Zymosan A (Life technologies) was added at an approximate ratio of 1:5 (macrophage/zymosan). At the reported time points, cells were washed three times with PBS to remove free particles and the amount of fluorescent intensity was analyzed by a fluorescence microscope, with three random fields per well (n = 3 wells per sample).
Animal protocol
The experiments with mice were approved by the animal care and use committee of the University of Central Florida and were in accordance with the Guide for the care and use of laboratory animals from the National Institutes of Health.
Statistical analysis
All values are presented as means ± standard deviation. Significant differences were determined by two-way analysis of variance for multiple comparisons. A p value of < 0.05 is considered significant.
Results
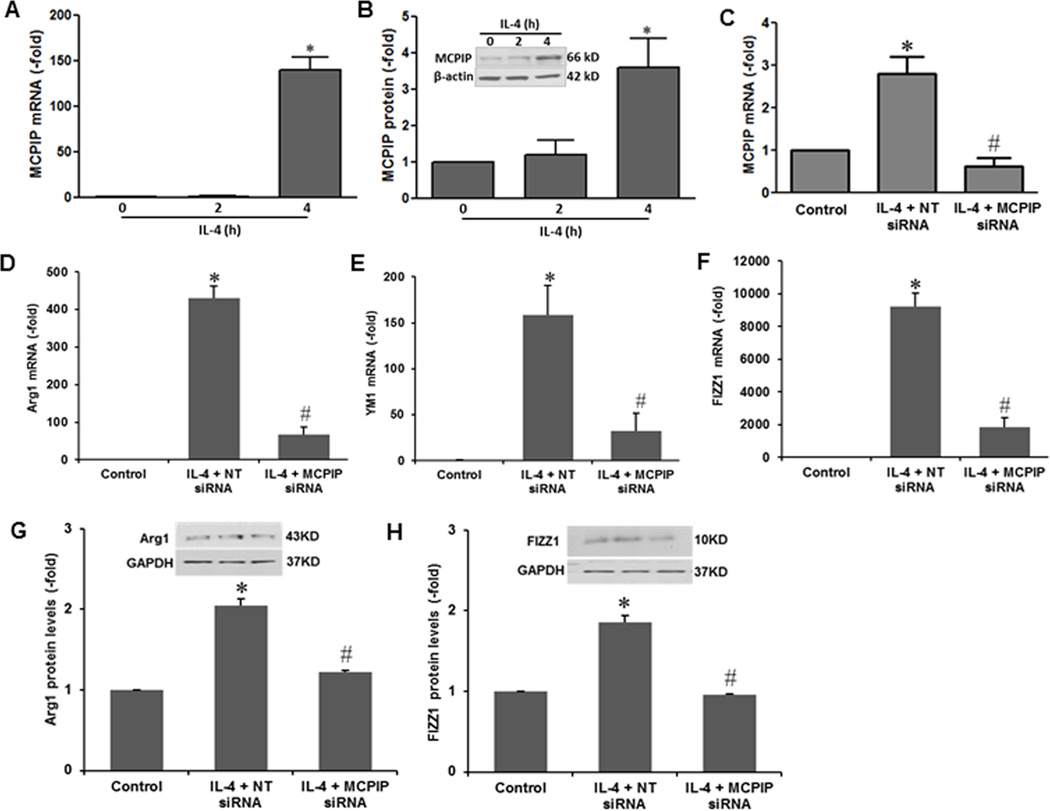
To test whether IL-4 induced M2 polarization is mediated via MCPIP, we tested whether IL-4 treatment of murine macrophages induces MCPIP. The results showed that IL-4 treatment caused induction of MCPIP at both transcript and protein levels (Fig 1A, B). If IL-4-induced expression of the M2 markers is mediated via MCPIP, knockdown of MCPIP should inhibit the IL-4-induced M2 marker expression. In fact, MCPIP specific siRNA, that knocked down MCPIP expression (Fig. 1C), inhibited the IL-4-induced expression of M2 markers Arg1, YM1/chitinase-like lectin, and FIZZ1/Relm-α (Fig. 1D, E, F, G, H), whereas nontarget siRNA did not affect the induction of these markers at both transcript and protein levels. If IL-4-induced M2 polarization is mediated via induction of MCPIP, forced expression of MCPIP should induce M2 polarization of murine macrophages without IL-4 treatment. To test for this possibility we transfected murine macrophages with MCPIP expression vector without IL-4 addition. The results showed that MCPIP expression alone induced expression of M2 markers (Arg1, FIZZ1) at both transcript and protein levels (Fig.2A–D).
FIGURE 1. MCPIP is required for IL-4-induced M2 macrophage polarization.
The peritoneal macrophages isolated from C57BL/6 mice were treated with IL-4 (20ng/ml) at the indicated time points. Expression of MCPIP was assayed by qRT-PCR and immunoblots (A, B). *P < 0.05 vs untreated cells. Mouse macrophages were transfected with non-targeted siRNA or siRNA against MCPIP for 24 hr, and then treated with 20ng/ml IL-4 for 4 hr. After 4 hr, RNA and protein were isolated. Knockdown of MCPIP by treatment with siRNA against MCPIP inhibits IL-4-induced expression of MCPIP, assayed by qRT-PCR (C). Knockdown of MCPIP by siRNA against MCPIP inhibits IL-4-induced expression of M2 markers (Arg1, YM1, FIZZ1), assayed by qRT-PCR (D, E, F) and immunoblot (G, H).*P < 0.05 vs controls; #P <0.05 vs nontarget (NT) siRNA. Each experiment was repeated three times.
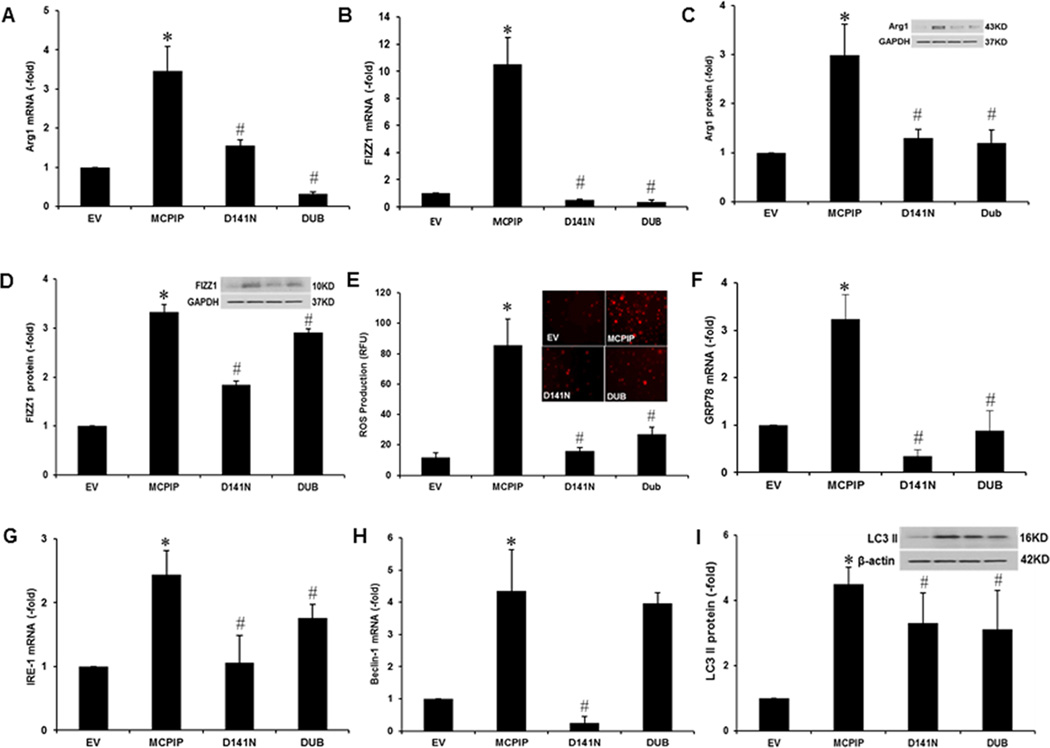
FIGURE 2. Both RNase and deubiquitinase activities of MCPIP are required for M2 polarization.
The peritoneal macrophages isolated from C57BL/6 wild-type mice were transfected with either control vector, or MCPIP expression plasmid, or expression vectors for D141N mutant, or Dub mutant for 48 hr. Forced expression of MCPIP induced expression of M2 markers, Arg1 and FIZZ1 (A, B, C, D). Expression of M2 markers (Arg1, FIZZ1), assayed by qRT-PCR (A, B) and immunoblot (C, D), were inhibited by loss of deubiquitinase or RNase activity of MCPIP. ROS production, assayed by DHR 123 (E), ER stress markers GRP78 and IRE-1, assayed by qRT-PCR (F, G) and autophagy as measured by expression of Beclin-1 and LC3II assayed by qRT-PCR, and immunoblot (H, I), were inhibited by loss of either catalytic activity of MCPIP. *P < 0.05 vs EV (empty vector); #P <0.05 vs MCPIP. Experiments were repeated three times. Six different fields containing at least 200 cells were analyzed for ROS production.
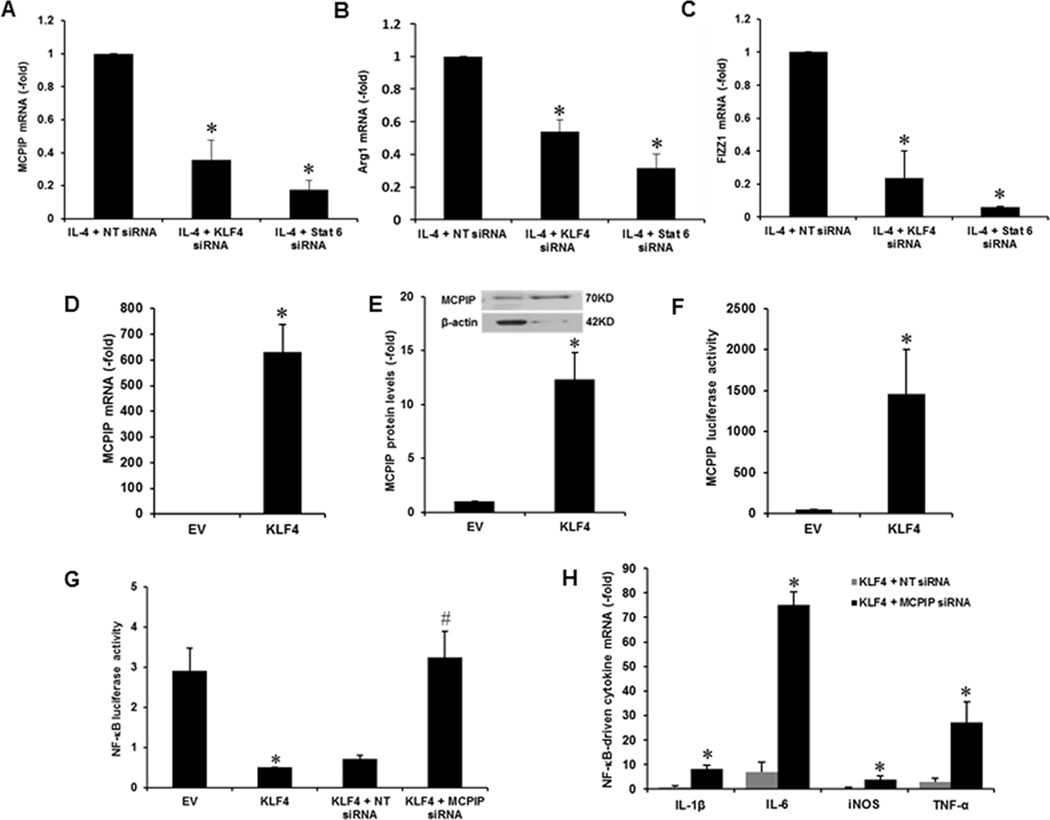
Since IL-4 is known to function via induction of STAT6 that is known to induce KLF4 to mediate M2 polarization (9), we tested whether IL-4 induction of MCPIP, whose promoter contains multiple KLF4 sites, is mediated via KLF4. Knockdown of either STAT6 or KLF4 with specific siRNA inhibited IL-4-induced MCPIP induction, whereas nontargeted siRNA had no effect (Fig. 3A). Knockdown of either STAT6 or KLF4 also inhibited the induction of M2 marker genes, Arg1 and FIZZ1, by IL-4 treatment (Fig. 3B, C). Thus, STAT6 and KLF4 cooperatively induce MCPIP. To directly test for KLF4 induction of MCPIP, we tested whether forced expression of KLF4 could induce MCPIP. Transfection of murine macrophages with KLF4 expression vector induced MCPIP at both transcript and protein levels (Fig. 3D, E). To further test whether the multiple KLF4 binding sites in MCPIP promoter is responsible for KLF4 induction of MCPIP, we fused a 1.0kb 5’-flanking region of MCPIP gene, containing the multiple KLF4 sites, to luciferase gene and tested whether KLF4 expression would drive luciferase expression. Murine macrophages transfected with the luciferase fusion construct showed luciferase expression only upon co-transfection with KLF4 expression vector (Fig. 3F). These results clearly showed that IL-4 induced MCPIP expression was mediated via KLF4. IL-4 induction of M2 polarization of macrophages was reported to be mediated by inhibition of M1 polarization mediated via NF-κB activation and KLF4 was reported to mediate this inhibition (9). Since MCPIP is known to suppress NF-κB activation (12, 13), we tested whether the KLF4-mediated NF-κB inhibition is implemented via MPCIP. LPS induced expression of NF-κB target genes, iNOS, IL-1β, TNFα and IL-6 was suppressed by forced expression of KLF4 and this suppression was reversed by knocking down MCPIP with specific siRNA (Fig 3H). To further test whether KLF4-mediated suppression of M1 polarization is mediated through suppression of NF-κB activation via MCPIP, we used luciferase reporter assay. LPS induction of luciferase driven by NF-κB was suppressed by forced expression of KLF4 (Fig. 3G). If this suppression is mediated via MCPIP, knockdown of MCPIP with specific siRNA should reverse the suppression by KLF4 expression. In fact, luciferase activity was recovered by treatment with siRNA specific for MCPIP but not with nontargeted siRNA (Fig. 3G). These results provide direct evidence for the involvement of MCPIP in the KLF4-induced suppression of NF-κB activation.
FIGURE 3.
IL-4-induced MCPIP induction is via STAT6/KLF4, and KLF4 induced NF-κB inhibition is mediated via MCPIP. The peritoneal macrophages isolated from C57BL/6 mice were pretreated with siRNA against KLF4 and STAT6 for 24 hr, and then treated with IL-4 (20ng/ml) for 4 hr. Expression of MCPIP and M2 markers (Arg1, FIZZ1) was determined by qRT-PCR (A, B, C). Transfection with KLF4 expression plasmid induced MCPIP expression in murine macrophages as determined by qRT-PCR (D) and western blot (E). Murine macrophages were transfected with MCPIP promoter-luciferase construct with or without co-transfection with KLF-4 expression vector for 24hr and luciferase activity was measured in the lysate. Expression of KLF4 enhanced MCPIP-luciferase reporter activity (F). Mouse macrophages were pretreated with siRNA against MCPIP for 24 hr then treated with 100ng/ml of LPS for 6 hr. KLF4 suppression of LPS-induced NF-κB activity was prevented by siRNA knockdown of MCPIP as measured by the NF-κB reporter kit (G). KLF4 suppression of LPS-induced NF-κB target genes (iNOS, IL-1β, TNF-α, and IL-6), assayed by qRT-PCR, were prevented by siRNA knockdown of MCPIP (H). *P <0.05 vs untreated Control cells, #P <0.05 vs non-targeted (NT) siRNA. Experiments were repeated three times.
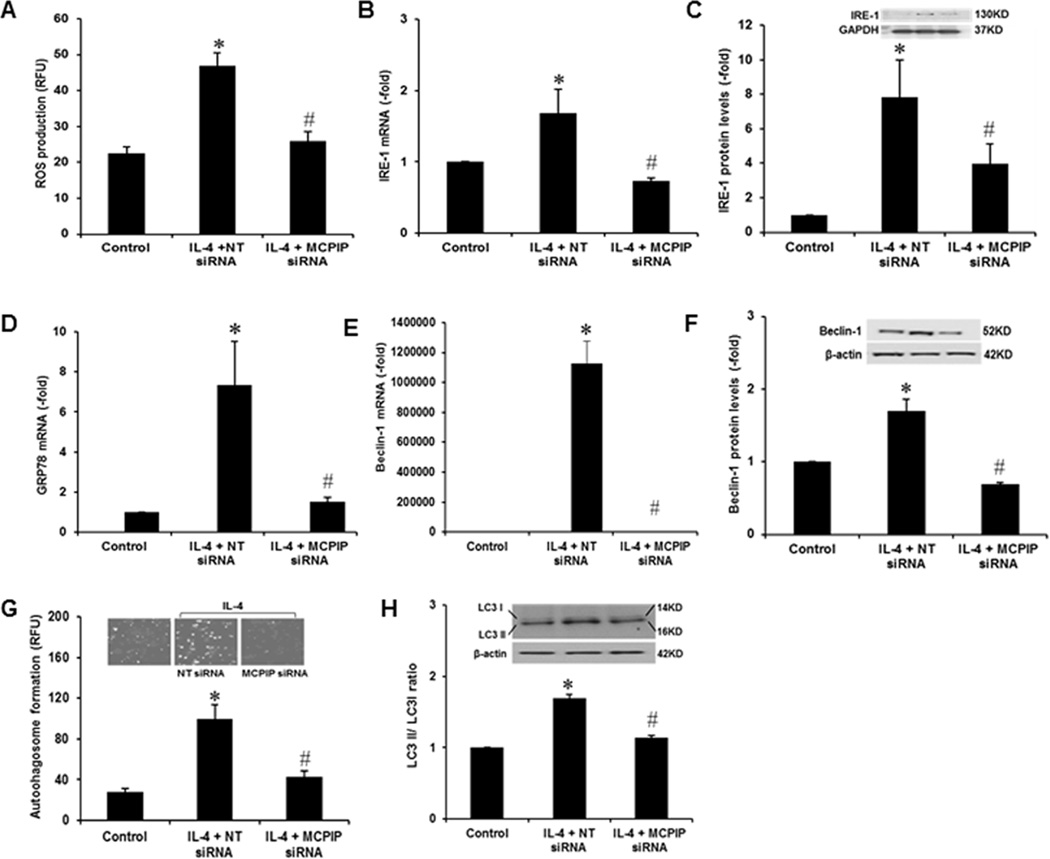
ROS production, ER stress, and autophagy have been reported to be involved in IL-4-induced M2 polarization (20–22). MCPIP is known to induce oxidative stress, ER stress and autophagy in several cell types (17–19). We tested whether MCPIP mediates the IL-4-induced generation of ROS, ER stress, and autophagy in murine macrophages. Knockdown of MCPIP with specific siRNA inhibited IL-4-induced ROS production as measured with DHR123 (Fig. 4A). IL-4 induction of ER stress, as measured by induction of GRP78 and IRE-1, was inhibited by knock down of MCPIP with specific siRNA in murine macrophages (Fig 4B, C, D). IL-4 induction of autophagy, as measured by Beclin-1 expression, LC3 II:LC3 I ratio, and by autophagosome staining was inhibited by knockdown of MCPIP with specific siRNA (Fig 4E, F, G, H). These results show that IL-4 induction of oxidative and ER stress and autophagy is mediated via MCPIP.
FIGURE 4. Knockdown of MCPIP inhibited IL-4-induced ROS, ER stress and autophagy.
The peritoneal macrophages isolated from C57BL/6 mice were transfected with non-targeted siRNA (NT) or siRNA against MCPIP for 24 hr, and then treated with 20ng/ml of IL-4 for 4 hr. IL-4-induced production of ROS was measured with DHR123 (A), ER stress was measured by expression of IRE-1 and GRP78 (B, C, D) and autophagy was measured by expression of Beclin-1 transcript (E) and protein (F), autophagosome staining (G) and LC3II:LC3I ratio (H). ROS production, ER stress and autophagy were inhibited by siRNA against MCPIP, but not by nontargeted siRNA. *P < 0.05 vs Control, #P <0.05 vs NT siRNA. Experiments were repeated three times. Six different fields containing at least 200 cells were analyzed for ROS production and autophagosome staining.
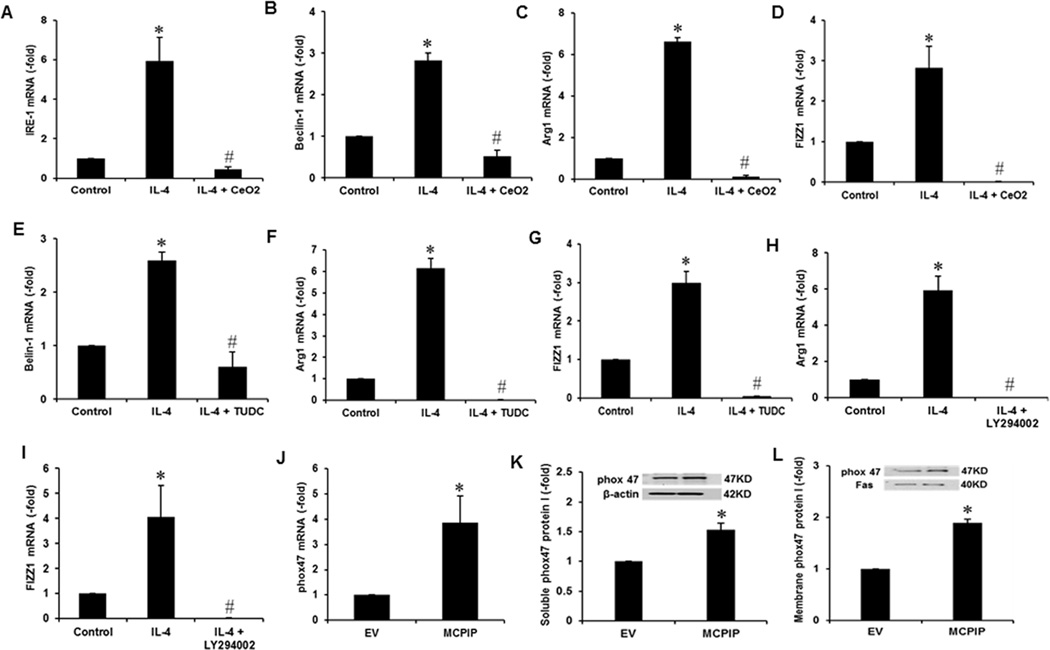
Even though ROS production, ER stress, and autophagy induced by IL-4 treatment of murine macrophages were reported to be involved in M2 polarization (20–22), the relationships among these processes have not been investigated. We used selective inhibitors of these processes to test the sequence of MCPIP mediated processes involved in M2 polarization. Inhibition of ROS production with CeO2 nanoparticles inhibited ER stress formation as measured by GRP78 and IRE-1 expression levels (Fig. 5A, S1A, S1B). This inhibition of ROS production also blocked autophagy as measured by Beclin-1 level and autophagosome staining (Fig. 5B, S1C, S1D) and induction of M2 markers, Arg1 and FIZZ1, at both transcript and protein levels (Fig. 5C, D, S1E, S1F).
FIGURE 5. Inhibition of ROS, ER stress or autophagy inhibits IL-4-induced M2 polarization and MCPIP induces expression of p47phox.
The peritoneal macrophages isolated from C57BL/6 mice were pre-incubated with 1µM of CeO2 nanoparticles for 6 hr, then treated with 20ng/ml of IL-4 for 4 hr. Blockage of IL-4-induced ROS production by antioxidant CeO2 nanoparticles resulted in suppression of IL-4-mediated ER stress marker, IRE-1 expression (A) autophagy marker, Beclin-1 expression (B) and expression of M2 markers Arg1 and FIZZ1 (C, D). Mouse macrophages were pre-incubated with 100µM of TUDC for 6 hr, then treated with 20ng/ml of IL-4 for 4 hr. Inhibition of IL-4-induced ER stress by TUDC resulted in inhibition of IL-4-mediated autophagy marker, (Beclin-1) expression (E) and expression of M2 markers, Arg1 and FIZZ1 (F, G). Mouse macrophages were pre-incubated with 20µM of LY294002 for 6 hr, then treated with 20ng/ml of IL-4 for 4 hr. Inhibition of autophagy by LY294002 resulted in inhibition of IL-4-induced expression of M2 markers, Arg1 and FIZZ1 (H, I), assayed by qRT-PCR. Macrophage transfection with MCPIP expression vector for 48 hr resulted in induction of p47phox mRNA assayed by qRT-PCR (J) and p47phox protein in soluble (K) and membrane (L) fractions assayed by Western blots*P < 0.005 vs Control. #P <0.05 vs cells treated with IL-4. Experiments were repeated three times.
Inhibition of ER stress with TUDC blocked autophagy as measured by Beclin-1 expression, LC3II:LC3I ratio and autophagosome staining (Fig. 5E, S2A, S2B, S2C) and induction of M2 markers, Arg1 and FIZZ1, at both transcript and protein levels, caused by IL-4 treatment (Fig. 5F, G, S2D, S2E). Inhibition of autophagy with LY294002 inhibited induction of expression of M2 markers, caused by IL-4 treatment (Fig. 5H, I, S2F, S2G). How MCPIP might initiate the sequential process was examined by testing whether MCPIP induces, p47phox, a critical component of NADPH oxidase responsible for ROS production. Transfection of murine macrophages with MCPIP expression plasmid induced p47phox (Fig. 5J) suggesting that induction of ROS production by MCPIP expression is mediated via p47phox. The level of p47phox protein also showed statistically significant elevation in both soluble and membrane fractions of the cell lysate (Fig. 5K, L).
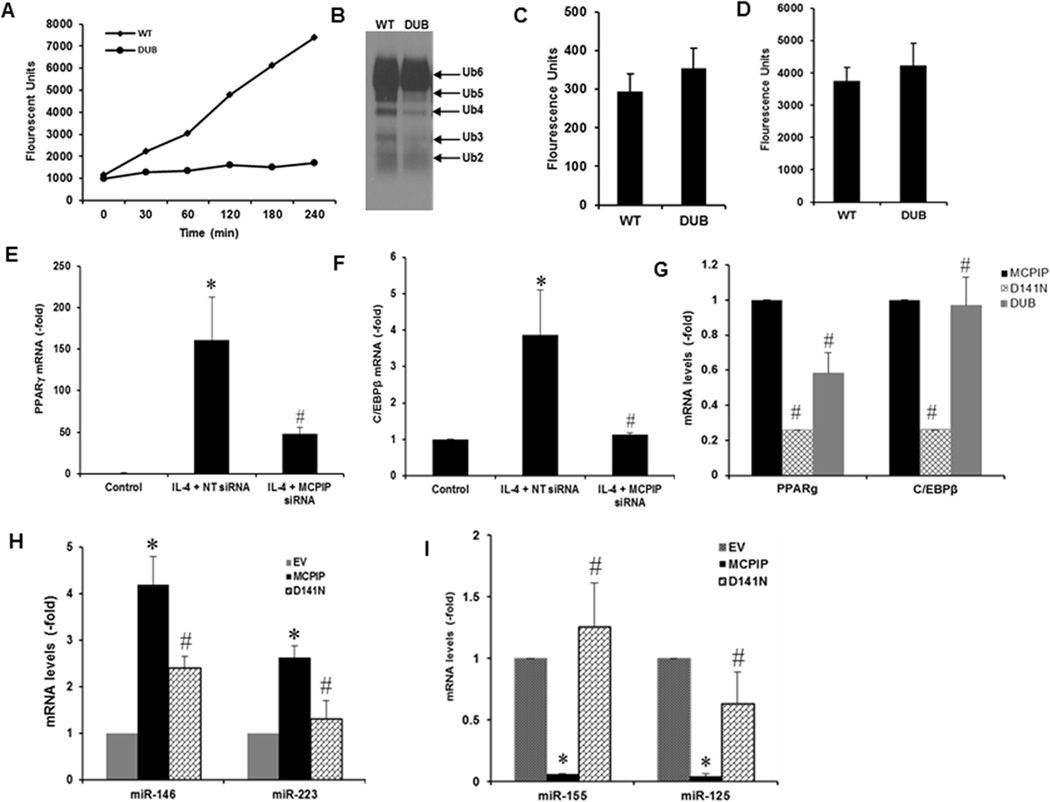
MCPIP has deubiquitinase (13) and RNase activities including anti-Dicer activity (12, 14, 15). The anti-Dicer RNase activity cleaves the loop from the pre-miR and thus depletes the substrate for Dicer causing inhibition of miR synthesis (25). To determine whether either or both of these activities of MCPIP are involved in M2 polariazation we needed MCPIP mutants with only one of these activities. It has been shown that D141N mutant of MCPIP has lost RNase and anti-Dicer activities (14, 24) but retains its deubiquitinase activity against model substrate, high molecular weight polyubiquitin and in vitro ubiquitinated HIF1α (23). Since no mutant was available that has lost only deubiquitinase activity with retention of RNase activity, we attempted to generate such a mutant. We deleted the four potential ubiquitin interacting domains in MCPIP and all four mutant proteins were assayed for deubiquitinase activity. Only one of them was found to have lost deubiquitinase activity against both a model substrate (Fig. 6A) and high molecular weight polyubiquitin (Fig 6B). This mutant showed full retention of RNase activity as measured with a commercially available general RNase activity kit (Fig. 6C). Anti-Dicer activity was measured by a novel assay we designed in which we used synthetic pre-miR135a tagged with a fluorophore in the loop and a quencher in the stem. Anti-Dicer activity released the fluorophore from the loop causing increase in fluorescence. With this assay this mutant showed full retention of anti-Dicer activity (Fig. 6D).
FIGURE 6. Generation of Dub-dead mutant and evidence that Il-4-induced expression of PPARγ and C/EBPβ is via the dual catalytic activities of MCPIP.
Ub-AFC assay showed loss of Dub activity by Dub-mutant compared to the wild-type MCPIP (A). K63-linked polyubiquitin was cleaved by MCPIP, but not by its Dub-mutant (B). RNase activity measurement and anti-dicer activity measurement with a novel pre-miR 135a tagged with a fluorophore in the loop and a quencher in the stem showed that the Dub mutant retained full RNase and anti-Dicer activities (C, D). The peritoneal macrophages isolated from C57BL/6 wild-type mice were transfected with non-targeted siRNA (NT) or siRNA against MCPIP for 24 hr, and then treated with 20ng/ml of IL-4 for 4 hr. Knockdown of MCPIP inhibited IL-4-induced expression of PPARγ and C/EBPβ assayed by qRT-PCR (E, F). Mouse macrophages were transfected with MCPIP expression plasmid, or expression vectors for D141N mutant, or DUB mutant for 48 hr. Expression of PPARγ and CEBP β was severely inhibited by RNase mutation, assayed by qRT-PCR (G). miRNAs involved in M2 polarization were affected by RNase activity of MCPIP (H, I). *P < 0.05 vs Control/EV; #P <0.05 vs NT siRNA/MCPIP. Experiments were repeated three times.
We tested the RNase mutant and Dub mutant for their activity to induce M2 polarization by measuring induction of M2 marker gene expression after transfection of murine macrophages with expression vector for wild type or the two mutants of MCPIP. The results showed that both Dub mutant and RNase mutant were very much less effective in inducing the expression of M2 markers, Arg1 and FIZZ1, at both transcript and protein levels than the wild type MCPIP, indicating that both catalytic activities of MCPIP were necessary for the induction of M2 polarization (Fig. 2A, B, C, D).
Since ROS production was reported to be critical for IL-4-induced M2 polarization (20) we tested whether production of ROS was affected by loss of either catalytic activity of MCPIP. When murine macrophages were transfected with expression vectors for wild type MCPIP and the two mutants, it was found that ROS production was inhibited by loss of either of the two activities of MCPIP (Fig. 2E). We tested whether loss of either catalytic activity of MCPIP would affect induction of ER stress or autophagy. Induction of ER stress as indicated by expression of GRP78 was inhibited by loss of either deubiquitiase or RNase activity of MCPIP but IRE-1 expression was inhibited by its mutations only at the transcript level (Fig. 2F, G, S3A, S3B). Induction of autophagy, as indicated by Beclin-1 expression, was severely inhibited by loss of RNase activity but not by loss of deubiquitinase activity (Fig. 2H, S3C, S3D). LC3 II protein levels induced by MCPIP were significantly lower with both mutants (Fig 2I).
PPARγ and C/EBPβ, two transcription factors known to promote M2 polarization (26–28), are known to be induced by IL-4 treatment of murine macrophages (27, 28). Knockdown of MCPIP with specific siRNA, but not nontargeted siRNA, inhibited induction of PPARγ and C/EBPβ by IL-4 treatment of murine macrophages (Fig. 6E, F). We tested whether either catalytic activity of MCPIP is involved in the induction of these M2-associated transcription factors. PPARγ induction was severely inhibited by loss of RNase activity and less severly inhibited by loss of deubiquitinase activity (Fig. 6G). Induction of C/EBPβ was severely inhibited by loss of RNase activity of MCPIP but was not significantly affected by loss of deubiquitinase activity.
Since anti-Dicer RNase activity would be involved in miR synthesis that might be associated with macrophage polarization we tested whether the loss of anti-Dicer RNase activity of MCPIP affects the production of miR known to be involved in macrophage polarization. qRT-PCR measurements of miR levels showed that expression of M2 associated miRs 223 and 146 was enhanced by MCPIP expression but this induction was suppressed by loss of RNase activity of MCPIP (Fig.6H). On the other hand production of M1 associated miRs, 155 and 125, was dramatically suppressed by MCPIP when compared to the RNase mutant of MCPIP (Fig. 6I). These results suggest that anti-Dicer activity of MCPIP participates in the regulation of M2 polarization via control of miR production.
We tested whether MCPIP plays a critical role in M2 polarization in vivo, by examining macrophages from mice with myeloid cell-specific deletion of MCPIP and myeloid cell-targeted over expression of MCPIP. We generated myeloid cell specific MCPIP knockout mice using Cre/Lox system (29). Recombinant ES cells containing MCPIP-LoxP construct with LoxP inserted in intron 2 and intron 4 (Fig. S4A) were used to generate homozygous MCPIP-LoxP+/+ mice. Crossing these mice with LysMCre+ mice generated MCPIP-LoxP+/+, Cre+ mice (myelo-KO) as shown by PCR (Fig. S4B). Myelo-KO mice showed normal growth with some splenomegaly with no other pathology.
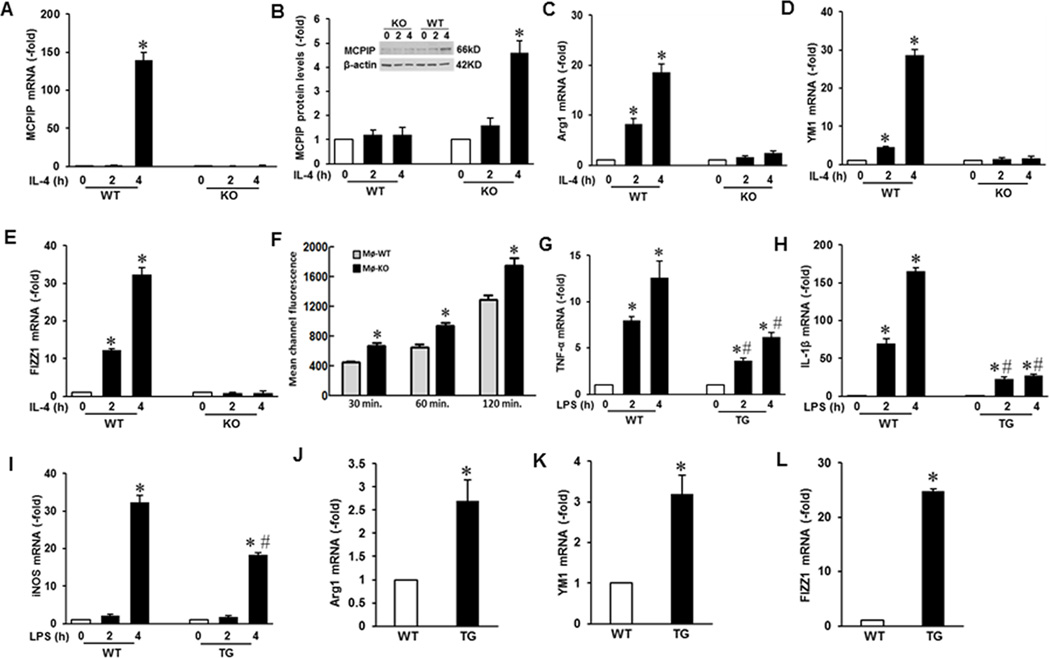
IL-4 treatment of peritoneal macrophages from myelo-KO mice failed to induce MCPIP upon treatment with IL-4 as shown by qRT-PCR measurements of the transcript levels and immunoblot analysis of the MCPIP protein levels (Fig 7A, B). Macrophages from myelo-KO mice showed enhanced expression of M1 markers such an iNOS and TNF-α as a result of LPS treatment (Fig. S4C, D).
FIGURE 7. Macrophages deficient in MCPIP failed to undergo M2 polarization and macrophages expressing MCPIP promoted M2 polarization.
Peritoneal macrophages from wild type and myelo-KO mice were treated with IL-4 for the indicated times and expression of MCPIP transcript was measured by qRT-PCR (A) and protein by immunoblot (B). Expression of M2 markers Arg1, YM1 and FIZZR1 in the IL-4 treated macrophages was measured by qRT-PCR (C, D, E). Phagocytosis by macrophages from myelo-KO and WT mice was measured by Zymosan internalization assay (F). Macrophages from WT and myelo-MCPIP mice were treated with LPS for the indicated periods and expression of M1 markers TNF-α, IL-1β, and iNOS was measured by qRT-PCR (G, H, I). Expression of M2 macrophage markers Arg1, YM1, and FIZZ1 in myelo-MCPIP and WT macrophages was measured by qRT-PCR (J, K, L). P < 0.05 compared with macrophages isolated from wild-type mice (n = 3 per each genotype).
We tested whether the absence of MCPIP affected IL-4 induction of M2 markers in murine macrophages. Macrophages from wild-type (WT) mice showed induction of Arg1, YM1, and FIZZ1 upon IL-4 treatment within 4 hr whereas induction of these M2 markers was drastically reduced in the myelo-KO macrophages when compared to WT controls (Fig. 7C, D, E) clearly showing the inability of IL-4 to induce M2 polarization in vivo in absence of MCPIP.
If myelo-KO promoted M1 polarization of macrophages, the phagocytic capability of macrophages from these mice should reflect this. Phagocytic capability of macrophages from myelo-KO mice, as measured by a zymosanA internalization assay, showed enhanced zymosan uptake than those from wild type mice (Fig. 7F).
To determine the effect of MCPIP in M2 polarization in vivo, we generated mice with myeloid-targeted expression of MCPIP (myelo-MCPIP). LysM promoter was used to drive MCPIP expression specifically in differentiated macrophages (29). The peritoneal macrophages isolated from the myelo-MCPIP mice clearly showed elevated MCPIP expression at both the transcript level and protein level when compared to the very low levels found in the macrophages from the wild type mice (Fig. S4E, S4F).
Peritoneal macrophages from myelo-MCPIP mice showed suppressed expression of M1 marker genes upon treatment with LPS (Fig. 7G). Thus, 2 and 4 hr treatment of the macrophages from the transgenic (TG) mice with 100 ng/ml of LPS showed suppressed expression iNOS, TNF-α, and IL-1 β (Fig. 7G, H, I). On the other hand, peritoneal macrophages from myelo-MCPIP mice displayed significant induction of M2 marker genes, Arg1, YM1, and FIZZ1 (Fig 7J, K, L). These results show that MCPIP plays a critical role in macrophage polarization in vivo.
Discussion
The remarkable plasticity of macrophages allows them to respond to environmental signals. Infection, injury, and inflammatory stimuli cause activation of macrophages to M1 state that allow them to defend the host against invading pathogens or other harmful agents. However sustained inflammatory condition, represented by M1 activated macrophages, can damage the host. To protect the host from such damage the macrophages undergo M2 polarization that is anti-inflammatory. This differentiation is triggered by clues generated by the inflammatory state. MCP-1 and other inflammatory chemokine and cytokine are produced by macrophages via NF-κB activation that occurs during macrophage M1 activation. MCP-1 interaction with its receptor CCR2 causes signal transduction events that results in the induction of a zinc-finger protein called MCPIP (10). MCPIP is also induced by other inflammatory signals such as IL-Iβ, TNF-α and IL-6 (11, 12). Inflammatory signals also trigger formation of agents that lead to the eventual production of anti-inflammatory components that mediate resolution of inflammation to protect the host from damage from sustained/excessive inflammation. Our results strongly suggest that MCPIP is a critical inflammation-induced agent that promotes the polarization of macrophages into anti-inflammatory M2 state using its multiple catalytic activities.
It was reported that IL-4-induced M2 polarization involved STAT6 and KLF4 that induce each other and function cooperatively to induce M2 polarization (9). It was also pointed out that KLF4 mediated induction of PPARγ probably contributes to the inhibition of M1 polarization. IL-4 binding to its receptor causes signal transduction that results in tyrosine phosphorylation in STAT6 leading to dimerization and nuclear entry to cause transcriptional activation of genes including PPARγ (27). PPARγ regulates fatty acid metabolism promoting the aerobic respiration that occurs in M2 macrophages. Our results indicate that IL-4 induced PPARγ induction is mediated by MCPIP.
The role of MCPIP in implementing the functions of the transcription factors, STAT6 and KLF4, is mediated via its deubiquitinase activity and RNase activity. Results presented here indicate that both catalytic activities are involved in mediating M2 polarization. IL-4 induction of M2 polarization mediated by KLF4 inhibits M1 polarization and promotes M2 polarization. Deubiquitinase activity of MCPIP can prevent NF-κB activation via removal of ubiquitins from the components like TRAF6 and high molecular weight polyubiquitins involved in IKK activation required for NF-κB activation (13). Since ubiquitination state regulates the stability and function of many proteins it is yet to be determined whether other components involved in inhibition of M1 and promotion of M2 polarization are regulated by the deubiquitinase activity of MCPIP. Recently, immunopurifications and mass spectrometric analysis revealed that more than 20 proteins may be directly or indirectly associated with ubiquitin in IFN-γ/TLR stimulated RAW cells implicating the role of ubiquitin modification involved in immune responses (30).
The role of RNase activity of MCPIP includes previously reported inhibition of synthesis of inflammatory cytokines such as IL-1β and IL-6 by causing degradation of their mRNA (12, 14,15). The anti-Dicer activity would inhibit miR synthesis by its cleavage of the loop from pre-miR. The role of miR in macrophage polarization is only beginning to be elucidated. Using microarrays, changes in miR levels associated with macrophage polarization in human (31) and murine (32) macrophages were examined. The number of miRs that showed significant changes ranged from 109 in murine to 249 in human macrophages with a dozen or so showing major changes in expression. Obviously, the regulation of processes involved in macrophage polarization by miR remains poorly understood. However, there are examples that point to possible ways by which the anti-Dicer activity of MCPIP would regulate macrophage polarization. Suppression of production of miRs that negatively regulate M2 polarization may promote M2 polarization. miR155 is upregulated in M1 macrophages where it promotes production of inflammatory MCP-1 and TNF-α (33, 34). It also reduces expression of some M2 markers such as Arg1. We provide evidence that the suppression of synthesis of miR155 and miR125 by MCPIP is probably involved in the induction of M2 polarization. The MCPIP mutant, that has lost anti-Dicer activity, does not suppress the production of these miRs that are known to inhibit M2 polarization and thus lost the ability to induce M2 polarization in murine macrophages. Expression of M2-associated miRs (miR223 and miR146) was enhanced by MCPIP expression but was down regulated by the RNase mutant. These results suggest that the anti-Dicer activity of MCPIP is involved in M2 polarization. How the miRs regulate macrophage polarization is yet to be fully elucidated.
CREB/CEBP cascade causes transcriptional activation of M2 macrophage markers Arg1, IL-10 and MRC1 (28). Arg1 gene promoter has STAT6 and C/EBP sites that are involved in M2 polarization. C/EBP synergizes STAT factors on anti-inflammatory promoters. The M2 program was reported to be specifically sensitive to C/EBP levels (28). An E3 ligase, Nrdp1, was reported to K-63 ubiquitinate C/EBPβ and activate it to enhance transcription of Arg1 in Il-4-polarized macrophages (35). In the macrophages of transgenic mice expressing Nrdp1, not only Arg 1 but also other M2 markers such as YM1, FIZZ1, and IL-10 were upregulated. Nrdp1 expression promoted M2 polarization also by inhibiting LPS induction of iNOS, TNF-α, IL-1β and IL-6 in murine macrophages. Results presented here show that IL-4 induces C/EBPβ via MCPIP.
IL-4-induced M2 polarization was reported to require ROS production (20–22). Tumor associated macrophages that are mostly M2 type, are involved in tumorigenesis owing to their proangiogenic functions. Antioxidants that prevented the formation of ROS were found to inhibit M2 polarization and markedly suppressed tumorigenesis (36). We have previously reported that MCPIP induces oxidative stress in other cellular contexts, at least in the part, via induction of p47phox (17, 18). Here we demonstrate that ROS production involved in IL-4-induced M2 polarization is mediated via MCPIP, as indicated by the finding that knockdown of MCPIP inhibited IL-4-induced ROS production.
It has been reported that ER stress controls M2 macrophage differentiation (22, 37). Alternative activation of macrophages from diabetic human patients, induced by IL-4, manifested unfolded protein response (37). Inhibition of ER stress with chemical chaperone, phenylbutyric acid, prevented the M2 gene expression pattern and showed low IL-10 production. Furthermore, induction of ER stress by thapsigargin treatment of human macrophages induced M2 marker expression (37). These results indicated that inhibition of ER stress inhibited M2 marker expression. MCPIP expression has been reported to induce ER stress in some cell types (17–19). In the present-case, IL-4-induced ER stress, that is required for M2 polarization, is shown to be mediated via MCPIP, as indicated by the finding that knockdown of MCPIP with specific siRNA inhibited IL-4-induced ER stress and M2 polarization.
Recently it was reported that cathepsin S-mediated autophagic flux is involved in promoting M2 polarization in tumor associated macrophages (36). Cathepsin S deficient macrophages failed to manifest autophagic flux and M2 polarization in a tumor environment (36). Treatment with an inhibitor of autophagy, chloroquine, also inhibited M2 polarization (21). The present results that IL-4-induced M2 polarization involves autophagy are consistent with previous reports. Since knockdown of MCPIP with specific siRNA inhibited IL-4-induced autophagy and M2 polarization, it is highly likely that IL-4-induced M2 polarization in murine macrophages involves MCPIP-mediated autophagy. Autophagy would allow degradation of proteins that represent M1 state to provide amino acids for the synthesis of proteins involved in M2 state. The role of autophagy in differentiation is to provide amino acids for the synthesis of proteins needed for the differentiated state.
Even though ROS, ER stress, and autophagy were reported to be involved in IL-4-induced M2 polarization, if and how such biological processes cooperatively function in M2 polarization have not been examined. The present results indicate that IL-4-induced M2 polarization involves ROS production that causes ER stress that leads to autophagy involved in the differentiation process involved in M2 polarization. Thus, inhibition of each step leads to inhibition of subsequent steps in the sequential process. CeO2 nanoparticles that inhibit oxidative stress inhibit ER stress and autophagy and M2 marker expression. Inhibition of ER stress with a chemical chaperon (TUDC), that is known to selectively inhibit ER stress (38), inhibits autophagy and M2 marker expression. Inhibition of autophagy with a selective chemical inhibitor, LY294002, inhibits M2 marker expression. These results are consistent with previous reports about the involvement of sequential induction of oxidative stress, ER stress, and autophagy in adipogenesis, angiogenesis, and osteoclastogenesis induced by MCPIP (17–19).
Results presented here demonstrate that mediation of M2 polarization by MCPIP involves both its deubiquitinase and RNase activities. Thus, MCPIP mutants that lost either one of the two catalytic activities failed to induce ROS production, ER stress, autophagy and expression of M2 markers. Results presented here demonstrate that IL-4-induced M2 polarization is mediated via KLF4-mediated induction of MCPIP that implements M2 polarization via exerting its post transcriptional regulation by its RNase activity and its post translational level regulation by its deubiquitinase activities. It is demonstrated that MCPIP mediates ROS production that cause ER stress that leads to autophagy involved in M2 polarization. The central role played by MCPIP in M2 polarization is summarized in Fig 8. Transcription factors implement their biological function via the catalytic property of proteins that function as a link between the transcription factors and the biological processes they regulate. The results presented here show that the dual catalytic activities of MCPIP implement a function of the two transcription factors STAT6 and KLF4. Even though RNase activity of MCPIP has been well documented by several laboratories (14, 15), whether MCPIP has deubiquitinase activity has been questioned (39). With a new MCPIP mutant that lost deubiquitinase activity but retains full RNase activity, we demonstrate that the deubiquitinase activity of MCPIP plays an important biological function. The substrates for the two catalytic activities that play critical roles in the biological processes involved in M2 polarization remain to be identified.
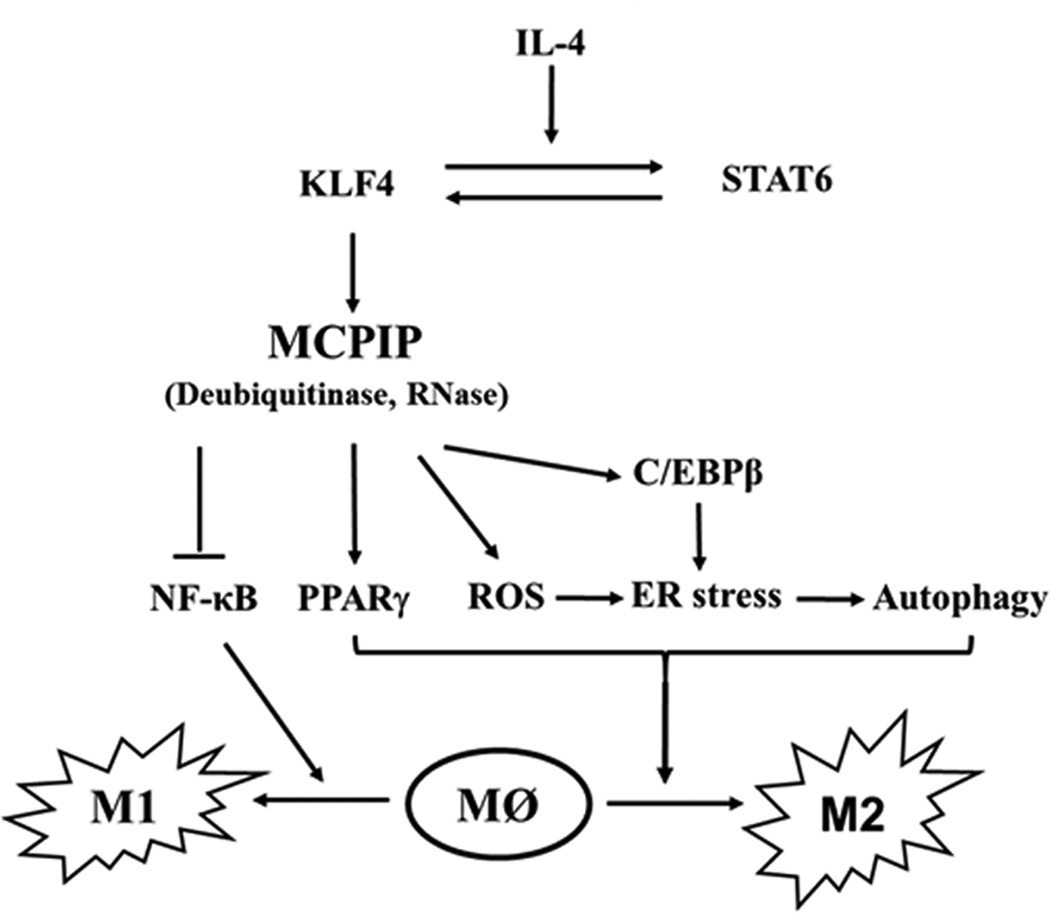
FIGURE 8. Model for the regulation of macrophage polarization by MCPIP.
Scheme showing how KLF4 and STAT6 co-operately mediate IL-4-induced M2 polarization via the KLF4 induction of MCPIP that mediates inhibition of M1 polarization via inhibition of NF-κB activation and promotes M2 polarization by sequentially inducing ROS production, ER stress and autophagy required for M2 polarization.
Macrophage polarization plays important roles in some major diseases. For example, obesity and type 2 diabetes, that are major health problems in the world, involve macrophages that infiltrate the adipose tissue (26). KLF4, that mediates MCPIP induction, is expressed at a reduced level in obese people (9). Furthermore, this inducer of MCPIP is expressed at a lower level in visceral versus subcutaneous fat in obese subjects. Visceral fat is associated with insulin resistance. This difference in KLF4 levels probably would results in lower MCPIP levels and consequently macrophages associated with visceral fat would be more M1 than M2. M1 macrophages are well known to be associated with insulin resistance (26, 40). Macrophage polarization is also relevant to cardiovascular diseases. For example, M1/M2 plays a role in atherosclerosis. M1 phenotype is more prevalent in rupture prone places in the plaques, such as plaque shoulder (8, 41). M1 macrophages are exclusively found in plaques of symptomatic patients and at higher levels in unstable plaques, whereas M2 macrophages are found to be higher in stable plaques in both symptomatic and asymptomatic patients. Thus, M1/M2 ratio and their location within the atherosclerotic lesion can be critical for plaque stability.
The tumor-associated macrophages tend to be of M2 phenotype (42,43). These macrophages promote angiogenesis and thus promote tumor growth. M2 macrophages also promote wound healing presumably by promoting angiogenesis involved in effective wound healing, cardiovascular disease and cancer (6, 44). Absence of KLF4 in macrophages caused increased phagocytic activity, as absence of KLF4 favored M1 polarization (9). This effect of KLF4 is probably mediated via its ability to induce MCPIP demonstrated here. The present results show that absence of MCPIP in macrophages favored M1 phenotype with higher bactericidal activity. Thus the critical role of MCPIP in M2 polarization is relevant to major health issues such as obesity and insulin resistance in type 2 diabetes, control of bacterial infection, angiogenesis involved in wound healing, and cancer. Thus, MCPIP could be a suitable target for intervention in such human health problems.
Supplementary Material
Acknowledgements
We thank Kelly Brussel for assistance with the manuscript preparation and Dr. Janet Parker-Thornburg of MD Anderson Cancer Center, Houston, TX for help in generating nice with myeloid targeted expression of MCPIP.
This work was supported by HL69458 from the National Institutes of Health.
Abbreviations used in this article
- MCPIP
MCP-1-induced protein
- Arg1
arginase 1
- KLF4
Krüppel-like factor 4
- qRT-PCR
quantitative real-time PCR
- ROS
reactive oxygen species
- YM1
chitinase-like lectin
- FIZZ1
Relm-α
- LPS
lipopolysaccharides
- ER
endoplasmic reticulum
- DHR123
dihydrorhodamine 123
- TUDC
tauroursodeoxycholate
References
- 1.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epelman S, Lavine KJ, Randolph GJ. Origin and Functions of Tissue Macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu. Rev. Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tugal D, Liao X, Jain MK. Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 2013;33:1135–44. doi: 10.1161/ATVBAHA.113.301453. [DOI] [PubMed] [Google Scholar]
- 5.Yang Z, Ming X. Functions of Arginase Isoforms in Macrophage Inflammatory Responses: Impact on Cardiovascular Diseases and Metabolic Disorders. Front. Immunol. 2014;5:533. doi: 10.3389/fimmu.2014.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Zhou D, Huang C, Lin Z, Zhan S, Kong L, Fang C, Li J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 8.De Paoli F, Staels B, Chinetti-Gbaguidi G. Macrophage phenotypes and their modulation in atherosclerosis. Circ. J. 2014;78:1775–1781. doi: 10.1253/circj.cj-14-0621. [DOI] [PubMed] [Google Scholar]
- 9.Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. Krüppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Azfer A, Niu J, Graham S, Choudhury M, Adamski FM, Younce C, Binkley PF, Kolattukudy PE. Monocyte chemoattractant protein-1 induces a novel transcription factor that causes cardiac myocyte apoptosis and ventricular dysfunction. Circ. Res. 2006;98:1177–1185. doi: 10.1161/01.RES.0000220106.64661.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, Wang J, Azfer A, Song W, Tromp G, Kolattukudy PE, Fu M. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 2008;283:6337–6346. doi: 10.1074/jbc.M707861200. [DOI] [PubMed] [Google Scholar]
- 12.Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-kappaB and MCP-1-induced protein 1 RNase. FEBS. J. 2009;276:5892–5905. doi: 10.1111/j.1742-4658.2009.07273.x. [DOI] [PubMed] [Google Scholar]
- 13.Liang J, Saad Y, Lei T, Wang J, Qi D, Yang Q, Kolattukudy PE, Fu M. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 15.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B, Jura J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS. J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 16.Jeltsch KM, Hu D, Brenner S, Zöller J, Heinz GA, Nagel D, Vogel KU, Rehage N, Warth SC, Edelmann SL, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote TH17 differentiation. Nat. Immunol. 2014;15:1079–1089. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 17.Younce CW, Kolattukudy PE. MCP-1 causes cardiomyoblast death via autophagy resulting from ER stress caused by oxidative stress generated by inducing a novel zinc-finger protein, MCPIP. Biochem. J. 2010;426:43–53. doi: 10.1042/BJ20090976. [DOI] [PubMed] [Google Scholar]
- 18.Wang K, Niu J, Kim H, Kolattukudy PE. Osteoclast precursor differentiation by MCPIP via oxidative stress, endoplasmic reticulum stress, and autophagy. J. Mol. Cell Biol. 2011;3:360–368. doi: 10.1093/jmcb/mjr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy A, Kolattukudy PE. Monocyte chemotactic protein-induced protein (MCPIP) promotes inflammatory angiogenesis via sequential induction of oxidative stress, endoplasmic reticulum stress and autophagy. Cell Signal. 2012;24:2123–2131. doi: 10.1016/j.cellsig.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Choksi S, Chen K, Pobezinskaya Y, Linnoila I, Liu ZG. ROS play a critical role in the differentiation of alternatively activated macrophages and the occurrence of tumor-associated macrophages. Cell Res. 2013;23:898–914. doi: 10.1038/cr.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Morgan MJ, Chen K, Choksi S, Liu ZG. Induction of autophagy is essential for monocyte-macrophage differentiation. Blood. 2012;119:2895–2905. doi: 10.1182/blood-2011-08-372383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh J, Riek AE, Weng S, Petty M, Kim D, Colonna M, Cella M, Bernal-Mizrachi C. Endoplasmic reticulum stress controls M2 macrophage differentiation and foam cell formation. J. Biol. Chem. 2012;287:11629–11641. doi: 10.1074/jbc.M111.338673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy A, Zhang M, Saad Y, Kolattukudy PE. Antidicer RNAse activity of monocyte chemotactic protein-induced protein-1 is critical for inducing angiogenesis. Am. J. Physiol. Cell Physiol. 2013;305:C1021–C1032. doi: 10.1152/ajpcell.00203.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton RM, Cai Z, Hom SM, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 2013;54:129–133. doi: 10.2144/000114017. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki HI, Arase M, Matsuyama H, Choi YL, Ueno T, Mano H, Sugimoto K, Miyazono K. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2010;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Charo IF. Macrophage polarization and insulin resistance: PPARgamma in control. Cell Metab. 2007;6:96–98. doi: 10.1016/j.cmet.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Chawla A. Control of macrophage activation and function by PPARs. Circ. Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 30.Kim JY, Anderson ED, Huynh W, Dey A, Ozato K. Proteomic survey of ubiquitin-linked nuclear proteins in interferon-stimulated macrophages. J. Interferon Cytokine Res. 2011;31:619–628. doi: 10.1089/jir.2011.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 2012;287:21816–21825. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhang M, Zhong M, Suo Q, Lv K. Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 2013;31:797–802. doi: 10.3892/ijmm.2013.1260. [DOI] [PubMed] [Google Scholar]
- 33.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He M, Xu Z, Ding T, Kuang DM, Zheng L. MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta. Cell. Mol. Immunol. 2009;6:343–352. doi: 10.1038/cmi.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye S, Xu H, Jin J, Yang M, Wang C, Yu Y, Cao X. The E3 ubiquitin ligase neuregulin receptor degradation protein 1 (Nrdp1) promotes M2 macrophage polarization by ubiquitinating and activating transcription factor CCAAT/enhancer-binding Protein β (C/EBPβ) J. Biol. Chem. 2012;287:26740–26748. doi: 10.1074/jbc.M112.383265. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Riek AE, Oh J, Bernal-Mizrachi C. 1,25(OH)2 vitamin D suppresses macrophage migration and reverses atherogenic cholesterol metabolism in type 2 diabetic patients. J. Steroid Biochem. Mol. Biol. 2013;136:309–312. doi: 10.1016/j.jsbmb.2012.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang M, Liu J, Shao J, Qin Y, Ji Q, Zhang X, Du J1. Cathepsin S-mediated autophagic flux in tumor-associated macrophages accelerate tumor development by promoting M2 polarization. Mol. Cancer. 2014;13:43–57. doi: 10.1186/1476-4598-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 39.Uehata T, Akira S. mRNA degradation by the endoribonuclease Regnase-1/ZC3H12a/MCPIP-1. Biochim Biophys Acta. 2013;1829:708–713. doi: 10.1016/j.bbagrm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Lee J. Adipose tissue macrophages in the development of obesity-induced inflammation, insulin resistance and type 2 diabetes. Arch. Pharm. Res. 2013;36:208–222. doi: 10.1007/s12272-013-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stöger JL, Gijbels MJ, van der Velden S, Manca M, van der Loos CM, Biessen EA, Daemen MJ, Lutgens E, de Winther MP. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis. 2012;225:461–468. doi: 10.1016/j.atherosclerosis.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Schmieder A, Michel J, Schönhaar K, Goerdt S, Schledzewski K. Differentiation and gene expression profile of tumor-associated macrophages. Semin Cancer Biol. 2012;22:289–297. doi: 10.1016/j.semcancer.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Sica A, Schioppa T, Mantovani A, Allavena P. Tumour-associated macrophages are a distinct M2 polarised population promoting tumour progression: potential targets of anti-cancer therapy. Eur. J. Cancer. 2006;42:717–727. doi: 10.1016/j.ejca.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Chambers SE, O'Neill CL, O'Doherty TM, Medina RJ, Stitt AW. The role of immune-related myeloid cells in angiogenesis. Immunobiology. 2013;218:1370–1375. doi: 10.1016/j.imbio.2013.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.