Abstract
Biomarkers for predicting chemotherapy response are important to treatment of colorectal cancer (CRC) patients. Cryptochrome 2 (CRY2) is a circadian clock protein involved in cell cycle, but the biological consequences of this activity in cancer are poorly understood. We set up biochemical and cell biology analyses to analyze CRY2 expression and chemoresistance. Here we report that CRY2 is overexpressed in chemoresistant CRC samples, and CRY2 overexpression is correlated with poor patient survival. Knockdown CRY2 increased colorectal cancer sensitivity to oxaliplatin in colorectal cancer cell. We also identify FBXW7 as a novel E3 ubiquitin ligase for targeting CRY2 through proteasomal degradation. Mechanistic studies show that CRY2 is regulated by FBXW7, in which FBXW7 binds directly to phosphorylated Thr300 of CRY2. Furthermore, FBXW7 expression leads to degradation of CRY2 through enhancing CRY2 ubiquitination and accelerating CRY2’s turnover rate. High expressed FBXW7 downregulates CRY2 and increases colorectal cancer cells sensitivity to chemotherapy. Low FBXW7 expression is correlated with high CRY2 expression in CRC patient samples. Also, low FBXW7 expression is correlated with poor patient survival. Taken together, our findings indicate that the upregulation of CRY2 caused by downregulation of FBXW7 may be a novel prognostic biomarker and may represent a new therapeutic target in colorectal cancer.
Keywords: Colorectal Cancer, FBXW7, Cryptochrome 2, Chemotherapy, circadian clock, Ubiquitination
INTRODUCTION
Colorectal cancer (CRC) is one of the leading causes of cancer mortality. 5-fluorouracil (5-Fu) based chemotherapy is routinely employed to treat those patients at high risk of developing recurrence or those with advanced or metastatic disease. As a consequence of early diagnosis, prevention, and adjuvant chemotherapy, death rate of CRC has fallen by 40% during the past four decades (1). However, a significant proportion of patients receiving chemotherapy become chemoresistant (2). Therefore, understanding the mechanisms underlying chemoresistance can help us to identify a subgroup of patients who may benefit from chemotherapy and avoid over-treatment. Studies have shown that multiple cellular processes including DNA repair, cell apoptosis and proliferation may play important role in chemoresistance (3–6). Several clinical studies have performed in an attempt to find biomarkers predicting the benefit from chemotherapy. For example, BRAFV660E mutations confer poor prognosis, but limited data suggest lack of antitumor activity from anti-EGFR monoclonal antibodies in the presence of a BRAF mutation status (7) (8, 9). However, except KRAS /NRAS activating mutations on exon2 and other RAS mutations (10) (11, 12), few of the studied markers have impact on the clinical management of colorectal cancer so far. There is an urgent need for discovering better markers that can enhance the prognostic strength.
Circadian clock is an endogenous biochemical mechanism shared by most organisms, which can influence nearly all aspects of physiology and behavior. Deregulation of circadian rhythm have been found to accelerate malignant growth and increase the risk of certain kinds of cancer such as breast cancer, prostate cancer and colorectal cancer, implying the involvement of circadian clock in cancer development and tumor progression (13–19). Circadian genes are also related to the clinical outcome of cancer patients (14, 18). Several studies have shown that anticancer drugs were more efficacious at a certain circadian time, indicating chronotherapy, a circadian-based chemotherapy, may be considered with patient treatment (20–23). It has been suggested prevention of chemotherapy-induced circadian disruption might reduce toxicity and improve efficacy in cancer patients (22). Cryptochrome 2 (CRY2), one of the identified circadian clock proteins, has been shown to be involved in DNA damage checkpoint control and regulating important cell cycle progression genes (24, 25). In addition, CRY mutation can increase the cell sensitivity to apoptosis induced by genotoxic agents and also protect p53 mutant mice from early onset of cancer (26). Also, breast cancer cells with reduced CRY2 have accumulated greater mutagen-induced DNA damage (15, 26, 27). These studies indicated that CRY2 might correlate with DNA damage and chemotherapeutic outcome of patients. Understanding these molecular alterations can help improving clinical care.
FBXW7 (F-box and WD repeat domain-containing 7 or FBW7) is a component of conserved SCF (complex of SKP1, CUL1 and F-box protein)-type ubiquitin ligase (28, 29). SCFFBXW7 is known to degrade several proto-oncogenes that function in cellular growth and division pathways, including cyclin E (30), Aurora B (31), Aurora A(32), MYC(33) (34), JUN(35) (36) and Notch (37). Loss-of-function of tumor suppressor FBXW7 increased pro-survival protein MCL1 levels and conferred resistance to Taxol-induced cell death, which indicated that FBXW7 is also involved in drug resistance (38). Interestingly, the ubiquitin-mediated protein degradation pathway plays an essential role in maintaining normal circadian clock function (39, 40). It is not clear whether FBXW7 and CRY2 have a link involved in chemoresistance.
In this study, we found that CRY2 was overexpressed in chemoresistant CRC patient samples. We showed that FBXW7 negatively regulated CRY2 via the ubiquitination pathway and sensitized colorectal cancer cells to chemotherapy. Low FBWX7 expression was correlated with CRY2 overexpression in CRC patient samples. Our studies provide important insight into the signal deregulation of the FBXW7-CRY2 axis in the chemosensitivity of colon cancer and help elucidate CRY2’s role as a therapeutic intervention target in colorectal cancer treatment.
MATERIALS AND METHODS
Human tissue
First, sixteen patients with stage III primary rectal cancer patients who consecutively underwent neoadjuvant chemotherapy (fluorouracil-leucovorin-oxaliplatin) were randomly selected from the Biobank of the Department of Colorectal Surgery, the Sixth Affiliated Hospital of Sun Yat-Sen University. Tumor specimens were obtained by colonoscopy prior to neoadjuvant therapy. The effect of chemotherapy on the tumor was assessed as the three-dimensional volume reduction rate or tumor response rate. The tumor response was evaluated by the Response Evaluation Criteria in Solid Tumors (RECIST), which is defined as the following: complete response (CR; disappearance of the disease), partial response (PR; reduction of ≥30%), stable disease (SD; reduction <30% or enlargement ≤20%), or progressive disease (PD; enlargement ≥20%). Among them, eight patients were defined as CR/PR and others were defined as SD/PD. Paraffin-embedded samples of primary colorectal adenocarcinomas were included from 289 patients from the First Affiliated Hospital of Sun Yat-Sen University. Our study protocol was approved by the ethics committee. Overall survival was the end-point of this study. Survival time was calculated from the date of surgery to the date of death or the last follow-up time. A written informed consent from all patients regarding tissue sampling had been obtained.
TMA construction and immunohistochemistry
Tissue microarrays were constructed; in brief, paraffin-embedded tissue blocks were overlain with corresponding slides of hematoxylin and eosin–stained tissue sections for tissue microarray sampling. Duplicate 0.6-mm-diameter cylinders were punched from representative tumor areas of an individual donor tissue block and re-embedded into a recipient paraffin block at predefined positions using a tissue-arraying instrument (Beecher Instruments, Silver Spring, MD). Immunohistochemical staining was performed on 5-μm tissue sections. The sections were placed on polylysine-coated slides, deparaffinized in xylene, and then rehydrated using graded ethanol. The slides were then placed 3% hydrogen peroxide to quench for endogenous peroxidase and then processed for antigen retrieval by microwave heating for 10 min in 10 mM citrate buffer (pH 6.0). The primary antibody against CRY2 (1:1000, Abcam, USA) or FBXW7 (1:1000, Invitrogen, USA) was diluted in phosphate-buffered saline (PBS) containing 1% bovine serum albumin and incubated at 4°C overnight. The next day, immunostaining was performed using the Invitrogen SuperPicture 3rd Gen IHC Detection Kit, which resulted in a brown precipitate at the antigen site. Then, the slides were counterstained with hematoxylin (Zymed Laboratories), mounted in nonaqueous mounting medium, and coverslipped. The original immunohistochemistry slides were scanned with the Advanced CCD Imaging Spectrometer that captured digital images of the immunostained slides. The ACIS calculates a score for regions selected by the pathologist. The receiver operating characteristic curve was used to define the cut-off point.
Data mining
The CRY2 gene mRNA expressions in colon tissue were obtain by Gene Expression Omibus. Cohort: GSE-14333 consisting of 223 colon cancer patients.
DNA constructs and reagents
PCR-amplified human CRY2 was cloned into PCMV5 or PDEST15. CRY2 T300A mutant was made by the Herculase II Fusion Enzyme Kit (Agilent Technologies). siRNA oligos were designed and manufactured by Ruibo (Guangzhou, China), targeting CRY2, sense: 5′-GAACGAAUGAAGCAGAUUU. Flag-FBXW7 was previously described (31). Cycloheximide and MG132 were obtained from Sigma. Ni-NTA Agarose was obtained from Invitrogen (#R901-15). Antibodies: Flag (M2 monoclonal antibody, Sigma, F3165), CRY2 (Abcam), myc (Santa cruz, sc-40).
Cell culture and transfection
The colon cancer cell lines DLD-1 and SW480 and were purchased from ATCC (American Type Culture Collection, Manassas, VA; authenticated by short-tandem repeat analysis). Cells were maintained in DME/F12 (Gibco), supplemented with 10% (v/v) fetal calve serum (FCS). HCT116 FBXW7+/+ and HCT116 FBXW7−/− (a kind gift from Dr. Bert Vogelstein, authenticated with Western blot routinely) were cultured in McCoy’s 5A media (Hyclone) supplemented with 10% (v/v) fetal calve serum. All cells were incubated in a humidified atmosphere of 5% CO2 at 37°C. Plasmids and CRY2 siRNA were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. Cells were harvested 48 hours after transfection to assay for transfection efficiency by qPCR.
RNA isolation and quantitation
RNA samples were isolated from harvested cells using Trizol® reagent (Invitrogen) according to the manufacturer’s instructions. A total of 1μg RNA from each sample was reversed transcribed using oligo-dT primers and ReverAid First Strand cDNA Synthesis Kit (Fermentas, EU). Quantitative real-time PCR was performed using the Thunderbird SYBR qPCR Mix (TOYOBO, Japan) in the Applied Biosystems 7500 Real-Time PCR system (ABI, USA). The primers used for CRY2 amplification were: (Forward: GTCCTGCAGTGCTTTCTTCC, Reverse: CCACACAGGAAGGGACAGAT). mRNA quantity was normalized using GAPDH content, and fold change of expression was calculated according to the ΔΔCT method.
Cell proliferation assay
Cell proliferation was determined using a MTT assay as previously described(41). Briefly, 5×103 cells per well were plated into 96-well plates, incubated at 37°C, 5% CO2 overnight. After transfection, cells were treated with different concentrations of fluorouracil or oxaliplatin (Sigma, USA) for 72 hours. Cells were incubated in medium containing 0.1 mg/ml MTT (Sigma, USA) for an additional 4 hours. The cells were then lysed in 150 μl DMSO (Sigma, USA). Cell viability was determined by measuring the absorbance at 550 nm using a 680 BioRad Microplate Absorbance Reader (BIO-RAD, UK).
Annexin V/PI staining
The cell was seeded on 96-well plates at a density of 105cells per well and cultured overnight. Cells were transfected with 100 nM negative control or CRY2 siRNAs. After 48 hours treatment with oxaliplatin, cells were collected and washed twice with PBS by spinning at 1000 rpm for 10 min. Cell pellets were resuspended in a FITC-labeled annexin V and propidium iodide (PI) staining solution (BD Bioscience, USA) and incubated for 15 min at room temperature. The samples were then analyzed on a FACSCalibur (FACSCanton II, BD, USA).
TUNEL assay
Apoptosis was determined by TUNEL method using the TMR Red kit (Roche, Mississauga, ON) according to the manufacturer’s suggested protocol. Briefly, cells were rinsed with PBS, fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Cells were then incubated in TUNEL reaction mixture at 37°C for 60 minutes, protected from light. After three rinses with PBS, the samples were analyzed under a fluorescence microscope (Leica DMI4000B, German) using an excitation wavelength of 540 nm and emission wavelength of 580nm. The number of cells was determined by DAPI staining. At least 200 cells were counted for each experiment.
Immunoprecipitation and Immunoblotting
Total cell lysates were processed as previously decribed (42) (43, 44). Cell lysates for western blot or immunoprecipitation were collected from tissue culture dishes after two rinses with cold PBS. Cells were centrifuged at low speed for 10min and supernatants discarded. Pellets were then lysed with 200μl 1x lysis buffer [0.5L batch:7.5g 1M Tris (Fisher), 15ml 5M NaCl (Fisher), 0.5ml NP-40(USB Corp.), 0.5ml Triton X-100(Sigma) and 1ml 0.5M EDTA(Fisher)] for 20min at 4o C. Lysis buffer also contained a cocktail of protease/phosphatase inhibitors: 5 mM NaV, 1mM NaF, 1μM DTT, 0.1mg/ml Pepstatin A, 1mM PMSF and 1,000x Complete Cocktail Protease Inhibitor (Roche). Lysates were immunoblotted with indicated antibodies. For immunoprecipitations, cell lysates were prepared and standardized as before, and 1mg protein was immunoprecipitated with appropriately diluted antibody in lysis buffer overnight. Antibody was pulled down with 50μl of either Protein A or G beads (Santa Cruz Biotechnology) for 1h. Beads were centrifuged at low speed for 10min and supernatant discarded. Dried beads were mixed with 2x loading dye and boiled for 5min. Lysate samples were loaded onto gels and SDS-PAGE was performed as before.
In vitro binding assay
Flag-FBXW7 and myc-CRY2 were prepared by in vitro transcription and translation using the TNT coupled system (Promega, San Luis Obispo, CA, USA). TNT products were mixed and immunoprecipitated followed by immunoblotting as described in figure legends.
Molecular docking of CRY2-FBXW7 Complex Structure
The information driven docking program HADDOCK (version 2.0)(45) was used to generate the Cry2-FBXW7 complex model. The starting structures for docking were an X-ray crystal structure of FBXW7 (30) (PDBID: 2ovp) and the homology model of Cry2. The Cry2 model was constructed using crystal structure of Cryptochrome-2 from Mus musculus (PDBID: 4i6g) as the template model, and the modeled residues were from residue 22 to 511. Based on the Skp1-FBXW7-Cyclin E peptide complex structure (PDB: 2ovp), specific degron-motif (TPXXS) binding to narrow face of WD40 domain of FBXW7 was used to generate this complex model. The WD40 domain of FBXW7 structure contained sets that included the active residues W425, T439, S462, T463, R465, R479, Y519, Y545, S585 and the neighbors of these active residues were selected as passive residues. The Cry2 model contained sets that included the active residues from R275 to S304, and the neighbors of these active residues were selected as passive residues. The active residues were chosen on the basis of the Skp1-FBXW7-Cyclin E peptide complexes structure (PDB: 2ovp). During the rigid body energy minimization, 10,000 structures were calculated, and the 162 best solutions based on the intermolecular energy were used for the semi-flexible, simulated annealing. The best 162 docked models were clustered a cutoff of 3.5 Å with a minimum of 4 structures in each clusters, which yielded 7 clusters. In terms of HADDOCK score and the basis of FBXW7-CycE peptides complex binding motif, the best structure was selected as a final model of Cry2-FBXW7 complex structure.
Turnover assay
The cells were transfected with indicated plasmids and incubated in the 37°C with 5%(v/v) CO2 for 24h. Turnover rate was analyzed as previously described (46) (47, 48). Then cycloheximide was added into the media at 60ug/ml of final concentration. The cells were harvested at the indicated time points after CHX treatment. The protein levels were analyzed by immunoblotting.
Ubiquitination assay
Ubiquitination was processed as previously described (34, 43). DLD-1 cells cotransfected with indicated plasmids. At 24h post-transfection, cells were treated with 50μg/ml of MG132 for 6h. The ubiquitinated proteins were pulled down by the Ni-NTA. The protein complexes were then resolved by SDS-polyacrylamide gel and probed with indicated antibody to observe the ubiquitianted level.
Statistical analysis
Statistical analyses were performed using SPSS (standard version 16.0; SPSS, Chicago, IL). The correlation between Cry2 expression and clinicalpathologic features of CRC patients was analyzed by χ2-test or Fisher’s exact test. For univariate survival analysis, survival curves were obtained by the Kaplan-Meier method and differences between survival curves were assessed with the log-rank test. The Cox proportional hazards regression model was used to identify the independent prognostic factors. All in vitro experiments were performed in triplicate. The results are described as mean±SD. Statistical analysis was performed by one-way analysis of variance (ANOVA), and comparisons among groups were performed by the independent-sample t-test. Results were considered significant at a p-value less than 0.05.
RESULTS
CRY2 expression is correlated with chemoresistance and poor outcome in CRC patients
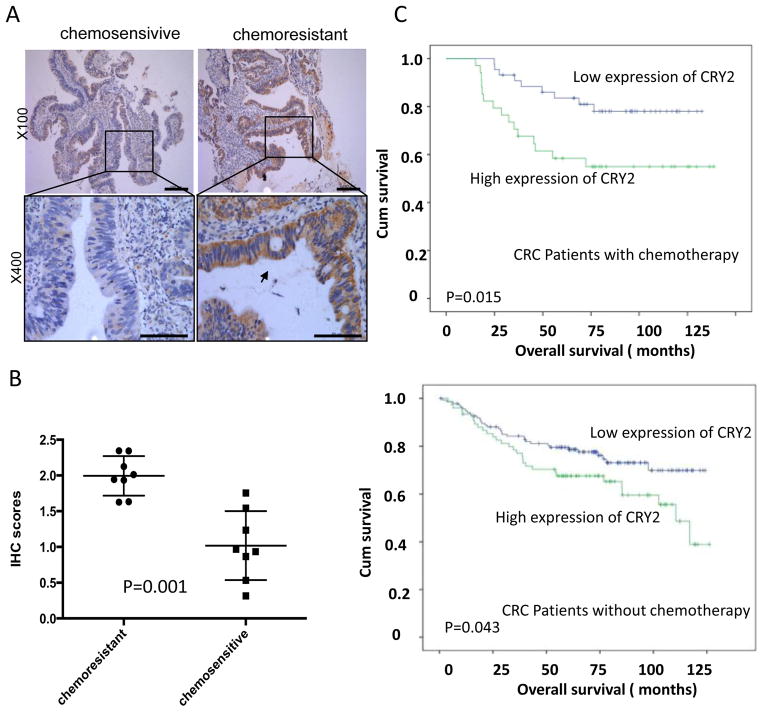
To investigate whether CRY2 expression relates to chemosensitivity in CRC patients, we first compared the expression of CRY2 levels in CRC colonoscopy samples from 16 rectal cancer patients who subsequently underwent neoadjuvant chemotherapy. IHC revealed that the level of CRY2 expression was higher in cancer tissue samples from stable/progressive patients than in samples from complete/partial response patients (Figure 1A). Statistic analysis revealed IHC score between two groups were significantly different (Figure 1B, P=0.001).
Figure 1. Chemoresistant colorectal cancer samples express high level of CRY2.
(A) Representative immunohistochemical staining for CRY2 in chemosensitive and chemoresistant colorectal cancer samples from colonoscopy. In chemoresistant patient sections, strong CRY2 signals were detected. In chemosensitive patient sections, weak staining was detected. Top panel: ×100; bottom panel: ×400. Scale bars: 100μm (top panel), 50 μm (bottom panel).
(B) Statistic analysis revealed IHC score between two groups were significantly different (P=0.001).
(C) Kaplan-Meier survival curves of overall survival duration based on CRY2 expression in the CRC tissues. The receiver operating characteristic curve was used to define the cutoff, and log-rank analysis was used to test for significance.
To determine the clinical relevance, we subjected a tissue microarray containing 289 human CRC specimens to immunohistochemical staining for CRY2. The clinicopathological characteristics of the CRC patients are summarized in Table 1. Among the CRC patients, 9 of 30 metastatic CRC patients (30%) received the oxaliplatin based chemotherapy, and 69 of 259 patients (26.7%) without metastasis received the chemotherapy. To assess statistical significance, ROC curve was plotted to determine cutoff scores for CRY2 expression. We divided the CRC patients into high and low CRY2 expression group according to cutoff scores. In the cohort, high expression of CRY2 was found in 124 of 289 (42.9%) of CRC patients. There was no significant association between CRY2 expression and clinicopathological features, such as patient gender, age, T classification, N classification, distant metastasis, and clinical stage (Table 1). The expressions of CRY2 as well as other clinicopathologic factors were further examined by multivariate Cox regression analysis. Results indicated that histological grade, N stage, M stage, and CRY2 overexpression were independent prognostic factors for CRC patients (Supplemental Table 1). Importantly, Kaplan-Meier analysis showed that high expression of CRY2 was correlated with poor survival, especially in CRC patients who received adjuvant chemotherapy (Figure 1C). Therefore, CRY2 can be identified as a biomarker that can predict outcomes of CRC patients.
Table 1.
Correlation between expression of CRY2 and clinicopathological features in 289 case of colorectal cancer
| All cases | CRY2
|
|||
|---|---|---|---|---|
| Low expression | Overexpression | P Value | ||
| Gender | 0.8131 | |||
| Male | 147(50.9) | 85(51.5) | 62(50.0) | |
| Female | 142(49.1) | 80(48.5) | 62(50.0) | |
| Age2 | 0.0731 | |||
| <59 | 130(45.0) | 82(49.7) | 48(38.7) | |
| ≥59 | 159(55.0) | 83(50.3) | 76(61.3) | |
| Histological grade | 0.2331 | |||
| G1 | 20(6.9) | 12(7.3) | 8(6.5) | |
| G2 | 230(79.6) | 126(76.4) | 104(83.9) | |
| G3 | 39(13.5) | 27(16.4) | 12(9.7) | |
| pT status | 0.1113 | |||
| T1 | 7(2.4) | 7(4.2) | 0(0.0) | |
| T2 | 39(13.5) | 24(14.5) | 15(12.1) | |
| T3 | 237(82.0) | 131(79.4) | 106(85.5) | |
| T4 | 6(2.1) | 3(1.8) | 3(2.4) | |
| pN status | 0.2261 | |||
| N0 | 173(59.9) | 104(63.0) | 69(55.6) | |
| N1 | 116(40.1) | 61(37.0) | 55(44.4) | |
| pM status | 0.2461 | |||
| M0 | 259(89.6) | 151(91.5) | 108(87.1) | |
| M1 | 30(10.4) | 14(8.5) | 16(12.9) | |
| Clinical stage | 0.4301 | |||
| I | 30(10.4) | 20(12.1) | 10(8.1) | |
| II | 124(42.9) | 73(44.2) | 51(41.1) | |
| III | 105(36.3) | 58(35.2) | 47(37.9) | |
| IV | 30(10.4) | 14(8.5) | 16(12.9) | |
| Chemotherapy | 0.2211 | |||
| No | 211(73.0) | 136(75.6) | 75(68.8) | |
| Yes | 78(27.0) | 44(24.4) | 34(31.2) | |
CRY2 knockdown leads to chemosensitivity of CRC cell lines
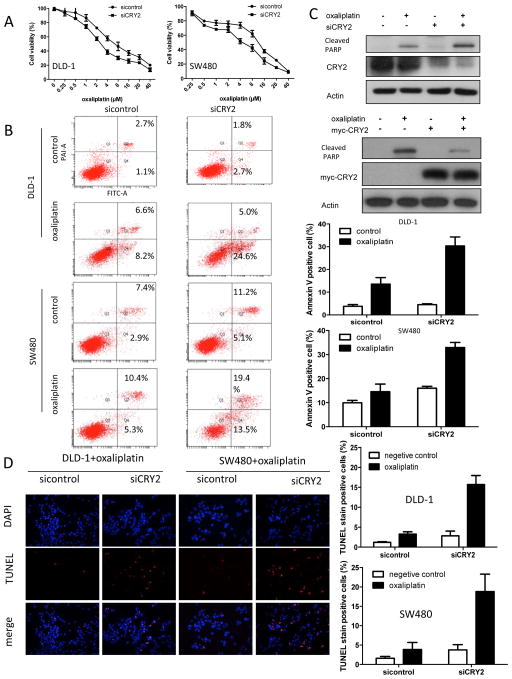
To examine further whether CRY2 expression has an impact on cheomoresistance of colon cancer cells in vitro, CRY2 was downregulated by specific siRNA in two CRC cell lines DLD-1, SW480. We found that DLD-1 and SW480 cells with reduced CRY2 were more sensitive to oxaliplatin compared with control cells (Figure 2A). Because CRY2 was involved in DNA damage-specific control of cell death, we then examined the contribution of CRY2 to oxaliplatin-induced apoptosis in DLD-1 and SW480 cells. In the presence of 4uM oxaliplatin, CRY2 depletion significantly increased number of apoptotic cells (P<0.001, Figure 2B), as evident by Annexin V staining. Cells were also analyzed for the presence of the cleaved form of poly-(ADP-ribose) polymerase cleavage (PARP), an apoptosis marker. CRY2 knockdown cells showed more PARP cleavage compared with the control cells (Figure 2C). Conversely, overexpression of CRY2 prevents oxaliplatin-induced PARP cleavage (Figure 2C). In addition, CRY2 depletion by siRNA resulted in six times more apoptotic cells than the cells transfected with scramble siRNA, as evident by TUNEL staining (Figure 2D). Taken together, these results indicate that CRY2 expression leads to chemoresistance.
Figure 2. CRY2 regulates colon cancer chemoresistance.
(A) Knockdown of CRY2 accelerates apoptosis induced by oxaliplatin. DLD-1 and SW480 cells were treated with increasing concentrations of oxaliplatin from 0.25 to 80μM after transfected with siCRY2 or control. The proportion of viable cells upon transduction with siCRY2 dropped more rapidly than in the control cells.
(B) Down-regulation of CRY2 expression enhances the pro-apoptotic effects of oxaliplatin on DLD-1 and SW480 cell line. Binding of Annexin V and uptake of propidium iodide were analyzed by flow cytometry. The mean of three data sets was taken and calculated from the corresponding quadrants (bar graphs, right panel). Error bars represent 95% confidence intervals. Treatment of 4μM oxaliplatin dramatically increased the portion of Annexin V+PI+/Annexin V+PI− in DLD-1 cells and SW480 transduced with siCRY2.
(C) CRY2 knockdown leads to increased poly-(ADP-ribose)polymerase (PARP) cleavage. DLD-1 cells transfected with siCRY2 or control siRNA were treated with or without 4μM oxaliplatin for 48 hours. Cell lysates were immunoblotted with indicated antibodies. CRY2 overexpression reduced cleavage of PRAP. DLD-1 cells transfected with myc-CRY2 or control vector were treated with or without 4μM oxaliplatin for 48 hours. Cell lysates were immunoblotted with indicated antibodies.
(D). TUNEL assay of DLD-1 and SW480 cell line. Cells transfected with control, or SiCRY2 were assayed for TUNEL signals. TUNEL positive cells were measured and presented as bar graphs. (**, P<0.01, n=3 experiments)
FBXW7 negatively regulates CRY2
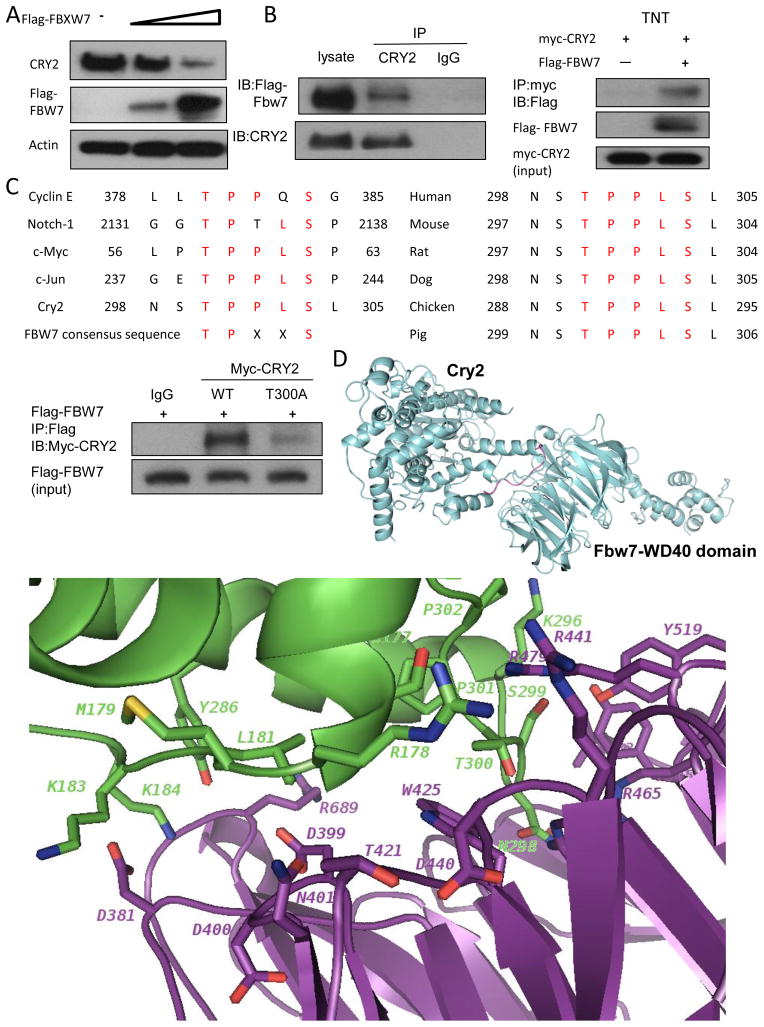
Because FBXW7 can regulate the chemosensitivity of cancer cells (38), we explored whether the FBXW7 is involved in CRY2-mediated chemosensitivity. CRY2 levels were decreased when cells were transfected with FBXW7 in a dose-dependent manner (Figure 3A). Given that FBXW7 is involved in the destabilization of CRY2, it is possible that there is an interaction between FBXW7 and CRY2. To investigate this potential interaction, we analyzed cell lysates cotransfected with FBXW7 and CRY2. Indeed, exogenous expressed FBXW7 was able to associate with endogenous CRY2 as assayed by Co-IP (Figure 3B). In-vitro binding assay confirmed the interaction between FBXW7 and CRY2 (Figure 3B). FBXW7 preferentially binds to target proteins with the degron TPXXS motifs(36). We found that CRY2 contains a consensus sequence (298NSTPPLSL305) that is highly similar to the FBXW7 binding sites in c-Myc, cyclin E, Notch1 and c-Jun (Figure 3C). FBXW7 binding site on Cry2 is conserved in different species (Figure 3C). To determine whether the consensus sequence is critical in mediating the binding between FBXW7 and CRY2, we constructed T300A site mutant of CRY2 and assessed the binding by immunoprecipatiation and Immunoblotting. The result showed that CRY2 T300A mutant reduced its binding to FBXW7 (Figure 3C), suggesting that T300 of CRY2 is important site for mediating FBXW7 binding.
Figure 3. FBXW7 interacts with CRY2.
(A) FBXW7 negatively regulates the steady-state expression of CRY2. DLD-1 cells were transfected with indicated plasmids and increasing amounts of FBXW7. Equal amounts of cell lysates were immunoblotted with the indicated antibodies.
(B) Interaction of FBXW7 with CRY2. Equal amounts of DLD-1 cell lysates transfected with Flag-FBXW7 were immunoprecipitated with anti-CRY2 and immunoblotted with anti-Flag. FBXW7 interacts with CRY2 in vitro. Flag-FBXW7 and Myc-CRY2 cDNAs were transcribed and translated in vitro (TNT). FBXW7 and CRY2 proteins were incubated overnight and immunoprecipitated with anti-Myc followed by immunoblotting with anti-Flag.
(C) The consensus FBXW7 binding motif is highlighted. Sequences of CRY2 and other known FBXW7 substrates are shown for comparison (left panel). Sequence alignment of CRY2 containing FBXW7 binding motifs from different species is shown (right panel). Equal amounts of cell lysates from DLD-1 cotransfected with WT myc-CRY2 or myc-CRY2-T300A and Flag-FBXW7 plasmids were immunoprecipitated with anti-Flag and immunoblotted with indicated antibodies.
(D) Ribbon presentation of CRY2-FBXW7 complex model. The loop of CRY2 degron-motif (residues T300-S304) interacts with the WD40 domain of FBXW7. Close-up view of the interface in CRY2-FBXW7 complex. The interaction residues involving in the interface of CRY2-FBXW7 molecules were shown in stick.
To gain insight into the structural basis of CRY2 recognition by FBXW7, we generated a model of the CRY2-FBXW7 complex using HADDOCK docking protocol (45). Briefly, the crystal structures of WD40 domain of FBXW7 and CRY2 were used for modeling the protein–protein complex. We defined structure-based degron motif (ϕ-X-ϕ-ϕ-ϕ-pT/S-P-P-X-pS/T, with ϕ representing a hydrophobic residue and X any amino acid) as active bases in CRY2. Residues 298-305 (comprising degron motif) are acid positions shared by degron motif in cyclin E, Notch-1, c-Myc, and c-Jun (Figure 3C). Structures of the FBXW7-CycE peptide complexes shows that the CycE peptide binds to the narrow face of the WD40 domain of FBXW7 extending from blades six and seven at one side of the surface across the central channel. After the HADDOCK docking protocol similar to FBXW7-CycE peptide complexes, we showed the best structure interactions between CRY2 and WD40 domain of FBXW7 (Figure 3D). The degron-motif of CRY2 residues 298-305 is docked to the narrow face of WD40 domain of FBXW7 (Figure 3D). We also plotted the inter-molecule H-bonds and hydrophobic interactions observed in the binding interface (Figure 3D). The degron-motif of CRY2 binds to the narrow face WD40 domain of FBXW7. Residues T300 and P301 (in the TPXXS motif) of CRY2 make hydrogen bonds with R465 and R479 of FBXW7 (Figure 3D). Besides the interactions involving in the TPXXS-motif, some additional interactions are also observed from this CRY2-FBXW7 complex model. Some electrostatic interactions are made between the charged residues R178, K183, K184, and K296 of CRY2 and the charged residues D381, D399, D440, and R689 of FBXW7 (Figure 3D). This complex structural model reveals the interactions between CRY2 and WD40 domain of FBXW7.
FBXW7 regulates CRY2 via enhancing ubiquitin-mediated degradation
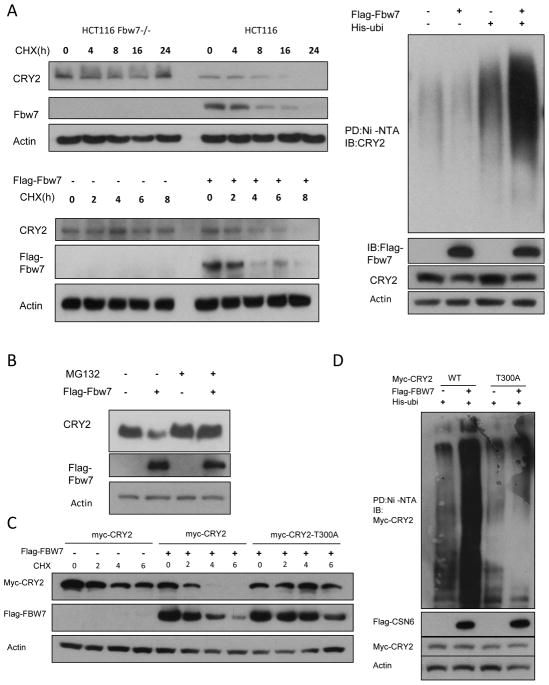
To investigate the impact of FBXW7 on CRY2 stability, we examined the turnover rate of CRY2 in the presence of de novo protein synthesis inhibitor cycloheximide (CHX) in HCT116 and HCT116 FBXW7−/− cells, and found that HCT116 FBXW7−/− cells had a decelerated turnover rate of endogenous CRY2 (Figure 4A). On the other hand, we transfected FBXW7 and performed the CHX experiments. The data showed that CRY2 had a faster turnover rate in the presence of FBXW7 (Figure 4A). FBXW7-mediated CRY2 downregulation was suppressed by MG132, a proteasome inhibitor, suggesting the involvement of the 26S proteasome (Figure 4B). Indeed, we found that FBXW7 increased the ubiquitination level of CRY2 (Figure 4A). We then identified that CRY2 T300A mutant had a slower turnover rate than WT CRY2 even in the presence of FBXW7 (Figure 4C). Accordingly, CRY2 T300A mutant is resistant to FBXW7-mediated ubiquitination (Figure 4D). These results suggest that FBXW7 activity negatively regulates CRY2 stability via enhancing the levels of CRY2 ubiquitination, thereby increasing CRY2 turnover rate.
Figure 4. FBXW7 regulates CRY2 stability via ubiquitination.
(A) CRY2 turnover rate and polyubiquitination are regulated by FBXW7. HCT116 and FBXW7−/− HCT116 cells were treated with CHX (100μg/ml) for the indicated times. After transfected with the FBXW7 plasmid, DLD-1 cells were treated with cycloheximide (CHX) (100μg/ml) for the indicated times. Equal amounts of cell lysates were immunoblotted with the antibodies. DLD-1 cells were cotransfected with FBXW7 and His-Ubi. MG132 was added 6h before harvesting cells with guanidine-HCl containing buffer. The cell lysates were pulled down with nickel beads and immunoblotted by anti-CRY2 antibody.
(B) DLD-1 cells were transfected with the FBXW7 plasmids. Cells were treated with MG 132 for 6h before harvesting. Equal amounts of cell lysates were immunoblotted with the antibodies.
(C) DLD-1 cells were transfected with myc-CRY2 or myc-CRY2 (T300A). After treated with CHX for indicated times, cells were harvested. Equal amounts of cell lysates were immunoblotted with the antibodies.
(D) DLD-1 cell were transfected with myc-CRY2 or myc-CRY2 (T300A). MG132 was added 6h before harvesting cells with guanidine-HCl containing buffer. The cell lysates were pulled down with nickel beads and immunoblotted by anti-myc antibody.
FBXW7 enhances chemosensitivity of CRC cell line
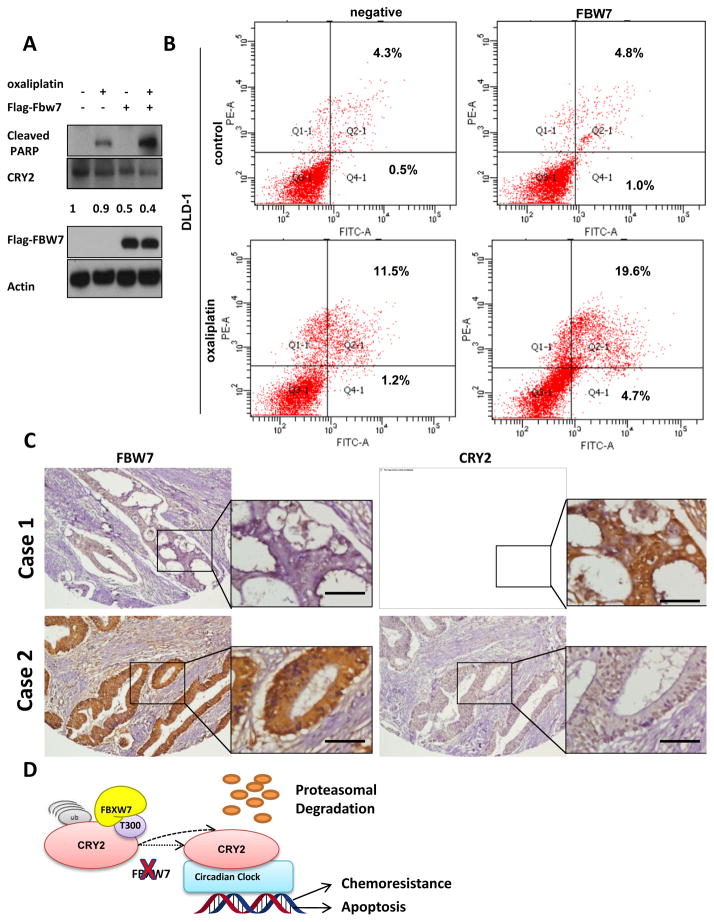
We examined whether FBXW7 expression had an impact on cheomoresistance of colon cancer cells and found that DLD-1 cells with overexpressed FBXW7 were more sensitive to oxaliplatin compared to control cells (Supplemental Figure 1A). Accordingly, overexpressed FBXW7 resulted in more oxaliplatin-induced PARP cleavage than the control cells (Figure 5A). As expected, FBXW7 overexpression led to downregulation of CRY2 during this process (Figure 5A). Another line of evidence showed that in the presence of 4uM oxaliplatin, overexpressed FBXW7 significantly potentiated the drug-mediated apoptosis, as evident by annexin V staining (Figure 5B & Supplemental Figure 1B). These data suggest that FBXW7-CRY2 axis is critical for chemosensitivity.
Figure 5. FBXW7 regulates CRY2 expression and colon cancer chemoresistance.
(A) Overexpression of FBXW7 expression leads to PARP cleavage. DLD-1 cells transfected with FBXW7 or control vector were treated with or without 4μM oxaliplatin for 48 hours. Cell lysates were immunoblotted with indicated antibodies. Ratio of CRY2 expression was shown.
(B) Overexpression of FBXW7 enhances the pro-apoptotic effects of oxaliplatin. DLD-1 cells transfected with FBXW7 or control vector were treated with or without 4μM oxaliplatin for 48 hours. Binding of Annexin V and uptake of propidium iodide were analyzed by flow cytometry.
(C) Representative IHC staining for FBXW7 and CRY2 in serial sections from colon cancer patient samples. Case 1 represents CRY2 high expressing patient. Case 2 represents CRY2 low expressing patient. The sample used was derived from colon cancer cases. Scale bars represent 50μm.
(D) Model of FBXW7 modulating CRY2 stability and chemosensitivity. Model of FBXW7 in modulating CRY2 ubiquitination and chemosensitivity. FBXW7 binds to phosphorylated T300 of CRY2 to ubiquitinate CRY2 and causes subsequent degradation, which reduces CRY2’s activity.
Low FBXW7 expression correlates with high CRY2 expression in CRC and manifests poor survival
To determine the clinical relevance, we subjected a tissue microarray containing 289 human CRC specimens to immunohistochemical staining for CRY2 and FBXW7. The CRC samples had high CRY2 expression, which correlated with low FBXW7 expression (representative case 1). Accordingly, CRC samples with low CRY2 expression had high FBXW7 expression (representative case 2) (Figure 5C). The CRY2 and FBXW7 expression levels were reversely correlated with each other (Table 2). Importantly, Kaplan-Meier analysis showed that high expression of CRY2 and low expression of FBXW7 were correlated with poor survival (Supplemental Figure 1C). These results strongly suggest that the FBXW7-CRY2 axis is deregulated during the development of human CRC.
Table 2.
Correlation between CRY2 and FBXW7 expression in CRC patient samples (P<0.001)
| Low CRY2 | High CRY2 | Total | ||
|---|---|---|---|---|
| Low | FBXW7 | 48 (17%) | 86 (30%) | 134 (46%) |
| High | FBXW7 | 117 (40%) | 38 (13%) | 155 (54%) |
|
| ||||
| Total | 165 (57%) | 124 (43%) | 289 (100%) | |
Fisher’s exact test.
Discussion
Adjuvant chemotherapy with 5FU and oxaliplatin currently remains a standard treatment for patients with advanced CRC. However, chemotherapy resistance leading to treatment failure and local recurrence is still a critical problem. One of the biggest challenges is to identify the subpopulation of patients who are most likely to respond to a specific therapy. If one or more biomarkers could predict patient response to chemotherapy, one could spare the non-responders from ineffective treatment and direct them to alternative treatment strategies that could be more effective. There was some evidence showed that circadian clock can modulate drug metabolism and efficacy of anti-tumor therapy. Patient response to chemotherapy was closely correlated to the functional status of the circadian genes. However, chronotherapeutic approach has not become routine in clinical practice, perhaps in part, because of the lack of a clear mechanistic basis (49). Noticeably, we found that circadian clock protein CRY2 is overexpressed in chemoresistant CRC and that FBXW7 has unprecedented biological activity in downregulating CRY2 to mediate chemosenstivity; this demonstrates cicadian clock proteins may play an important role in CRC chemosensitivity.
There has been evidence that circadian clock could modulate multidrug resistance genes, thereby playing important roles in chemoresistance (50), as mdr1a gene can be affected by the circadian clockwork. Also, overexpression of circadian clock gene Bmal1 can increase sensitivity to oxaliplatin in colorectal cancer (51). These findings suggest that dysregulation of circadian clock may be associated with drug sensitivity. Paradoxically, we showed that high expression of CRY2 was significantly associated with poor chemotherapeutic outcomes in CRC patients who received standard adjuvant chemotherapy. It is possible that CRY2 expression is involved in DNA damage response, thereby affecting the chemosensitivity. It was reported that mutation of Cry could protect p53 mutant mice from early onset of cancer and extend their survival time after ionizing radiation (26), suggesting that Cry may be involved in p53-mediated DNA damage response. However, there was no study regarding the correlation between CRY2 expression and p53 in patients in terms of clinical outcome. Interestingly, our in vitro study did show that, repression of CRY2 in DLD-1 and SW480 cells (both are p53 mutant) leads to decreased cell viability and increased apoptosis in the presence of oxaliplatin, thereby resulting in an increased sensitivity to chemotherapy drugs. Thus, previous findings and current study indicate that CRY2 expression level and p53 status might be factors in predicting chemoresistance in CRC patients.
Ubiquitination-mediated degradation of clock proteins has been highlighted as a key regulatory process in clockwork. The FBXL3 and FBXL21 are critical for generating the CRY expression dynamics and behavior rhythms (39, 40, 52). Both CRY1 and CRY2 repressor proteins are regulated by ubiquitin-mediated degradation for temporally orchestrating the transcription of clock genes. Our studies have shown that FBXW7 can bind to and destabilize CRY2 through enhancing ubiquitin-mediated degradation of CRY2. FBXW7 is tumor suppressor (53) protein through recognizing and degrading several oncoproteins such as cyclinE, c-JUN, Aurora B, and MYC (33, 35, 54). Mutant FBXW7 or downregulation of FBXW7 causes tumorigenesis as it fails to decrease target oncoproteins (37, 55). Although FBXW7 has a critical function in cancer regulation, very few circadian clock proteins are the known targets of FBXW7. Giving that three F-Box proteins are all involved in CRY2 ubiquitination, the relationship between FBXW7 and FBXL3 and FBXL21 in terms of regulating CRY2 stability certainly warrants further investigation. Nonetheless, it makes sense that a tumor suppressor protein FBXW7 is involved in degrading CRY2 when CRY2’s overexpression leads to poor clinical outcome. Indeed, we found that CRY2 overexpression is quite common in chemoresistant colorectal cancer. CRY2 overexpression was negatively correlated with FBXW7 protein expression in human colon cancer samples. These findings demonstrate that FBXW7 downregulation can at least partially account for CRY2 overexpression in colon cancer and provide important insights into the mechanisms underlying CRY2 overexpression in chemoresistant colorectal cancer. The inverse relationship between FBXW7 and CRY2 is demonstrated for the first time in cancer.
In summary, our mechanistic studies explain the regulatory relationship between CRY2, FBXW7, and chemosensitivity (Figure 5D). That CRY2 overexpressed in chemoresistant CRC raises the possibility that inhibiting the CRY2 is an efficient therapeutic approach in overcoming chemoresistance.
Supplementary Material
Acknowledgments
Financial Information: This work was supported by the National Institutes of Health through grant R01CA089266 (M.H. Lee), by the Fidelity Foundation (M.H. Lee), by the Susan G. Komen Breast Cancer Foundation grant KG081048 (M.H. Lee), and by the NIH CA16672 (MDACC Core grant). This work was also supported by the Program of Introducing Talents of Discipline to Universities B12003 (J. Wang) and the International S&T Cooperation Program of China 2011DFA32570 (J. Wang).
We thank Joseph Munch of the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for reading.
Footnotes
Potential conflicts of interest: The authors declare that there is no conflict of interest.
References
- 1.DeVita VT, Jr, Rosenberg SA. Two hundred years of cancer research. The New England journal of medicine. 2012;366:2207–14. doi: 10.1056/NEJMra1204479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biagi JJ, Raphael MJ, Mackillop WJ, Kong W, King WD, Booth CM. Association between time to initiation of adjuvant chemotherapy and survival in colorectal cancer: a systematic review and meta-analysis. Jama. 2011;305:2335–42. doi: 10.1001/jama.2011.749. [DOI] [PubMed] [Google Scholar]
- 3.Ebert MP, Tanzer M, Balluff B, Burgermeister E, Kretzschmar AK, Hughes DJ, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. The New England journal of medicine. 2012;366:44–53. doi: 10.1056/NEJMoa1009473. [DOI] [PubMed] [Google Scholar]
- 4.Brahimi-Horn MC, Ben-Hail D, Ilie M, Gounon P, Rouleau M, Hofman V, et al. Expression of a truncated active form of VDAC1 in lung cancer associates with hypoxic cell survival and correlates with progression to chemotherapy resistance. Cancer research. 2012;72:2140–50. doi: 10.1158/0008-5472.CAN-11-3940. [DOI] [PubMed] [Google Scholar]
- 5.Tamada M, Nagano O, Tateyama S, Ohmura M, Yae T, Ishimoto T, et al. Modulation of glucose metabolism by CD44 contributes to antioxidant status and drug resistance in cancer cells. Cancer research. 2012;72:1438–48. doi: 10.1158/0008-5472.CAN-11-3024. [DOI] [PubMed] [Google Scholar]
- 6.Fang L, Li H, Wang L, Hu J, Jin T, Wang J, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–87. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Cutsem E, Kohne CH, Lang I, Folprecht G, Nowacki MP, Cascinu S, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2011–9. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 8.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 9.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. European journal of cancer. 2015;51:587–94. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 10.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:1254–61. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 11.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 12.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. The New England journal of medicine. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 13.Mostafaie N, Kallay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Molecular carcinogenesis. 2009;48:642–7. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 14.Climent J, Perez-Losada J, Quigley DA, Kim IJ, Delrosario R, Jen KY, et al. Deletion of the PER3 gene on chromosome 1p36 in recurrent ER-positive breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3770–8. doi: 10.1200/JCO.2009.27.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman AE, Zheng T, Ba Y, Stevens RG, Yi CH, Leaderer D, et al. Phenotypic effects of the circadian gene Cryptochrome 2 on cancer-related pathways. BMC cancer. 2010;10:110. doi: 10.1186/1471-2407-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu J, et al. A role for the clock gene per1 in prostate cancer. Cancer research. 2009;69:7619–25. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Dauchy RT, Tirrell PC, Wu SS, Lynch DT, Jitawatanarat P, et al. Light at night activates IGF-1R/PDK1 signaling and accelerates tumor growth in human breast cancer xenografts. Cancer research. 2011;71:2622–31. doi: 10.1158/0008-5472.CAN-10-3837. [DOI] [PubMed] [Google Scholar]
- 18.Alhopuro P, Bjorklund M, Sammalkorpi H, Turunen M, Tuupanen S, Bistrom M, et al. Mutations in the circadian gene CLOCK in colorectal cancer. Molecular cancer research : MCR. 2010;8:952–60. doi: 10.1158/1541-7786.MCR-10-0086. [DOI] [PubMed] [Google Scholar]
- 19.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 20.Levi F, Focan C, Karaboue A, de la Valette V, Focan-Henrard D, Baron B, et al. Implications of circadian clocks for the rhythmic delivery of cancer therapeutics. Advanced drug delivery reviews. 2007;59:1015–35. doi: 10.1016/j.addr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nature reviews Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 22.Innominato PF, Levi FA, Bjarnason GA. Chronotherapy and the molecular clock: Clinical implications in oncology. Advanced drug delivery reviews. 2010;62:979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiology international. 2002;19:237–51. doi: 10.1081/cbi-120002600. [DOI] [PubMed] [Google Scholar]
- 24.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell cycle. 2010;9:1097–103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell cycle. 2009;8:1665–7. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 26.Ozturk N, Lee JH, Gaddameedhi S, Sancar A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2841–6. doi: 10.1073/pnas.0813028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JH, Sancar A. Regulation of apoptosis by the circadian clock through NF-kappaB signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12036–41. doi: 10.1073/pnas.1108125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skaar JR, Pagan JK, Pagano M. SCF ubiquitin ligase-targeted therapies. Nature reviews Drug discovery. 2014;13:889–903. doi: 10.1038/nrd4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system - implications for cell cycle control and the targeted treatment of cancer. Biochimica et biophysica acta. 2014;1843:150–62. doi: 10.1016/j.bbamcr.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Molecular cell. 2007;26:131–43. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Teng CL, Hsieh YC, Phan L, Shin J, Gully C, Velazquez-Torres G, et al. FBXW7 is involved in Aurora B degradation. Cell cycle. 2012;11:4059–68. doi: 10.4161/cc.22381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu CC, Yang TY, Yu CT, Phan L, Ivan C, Sood AK, et al. p53 negatively regulates Aurora A via both transcriptional and posttranslational regulation. Cell cycle. 2012;11:3433–42. doi: 10.4161/cc.21732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welcker M, Orian A, Grim JE, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14:1852–7. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Shin JH, Zhao R, Phan L, Wang H, Xue Y, et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat Commun. 2014;5:5384. doi: 10.1038/ncomms6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nateri AS, Riera-Sans L, Da Costa C, Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–8. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- 36.Wei W, Jin J, Schlisio S, Harper JW, Kaelin WG., Jr The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nature reviews Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 38.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–4. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 39.Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, et al. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–18. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- 40.Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi HH, Gully C, Su CH, Velazquez-Torres G, Chou PC, Tseng C, et al. COP9 signalosome subunit 6 stabilizes COP1, which functions as an E3 ubiquitin ligase for 14–3–3sigma. Oncogene. 2011;30:4791–801. doi: 10.1038/onc.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao R, Yeung SC, Chen J, Iwakuma T, Su CH, Chen B, et al. Subunit 6 of the COP9 signalosome promotes tumorigenesis in mice through stabilization of MDM2 and is upregulated in human cancers. J Clin Invest. 2011;121:851–65. doi: 10.1172/JCI44111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao R, Phan L, Chen B, Yang HY, Chen J, Che TF, et al. Ubiquitination-mediated p57Kip2 Degradation by CSN5 Confers Cancer Cell Proliferation. Cancer Hallmarks. 2013;1:133–44. [Google Scholar]
- 44.Fuentes-Mattei E, Velazquez-Torres G, Phan L, Zhang F, Chou PC, Shin JH, et al. Effects of obesity on transcriptomic changes and cancer hallmarks in estrogen receptor-positive breast cancer. Journal of the National Cancer Institute. 2014;106 doi: 10.1093/jnci/dju158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Vries SJ, Bonvin AM. CPORT: a consensus interface predictor and its performance in prediction-driven docking with HADDOCK. PloS one. 2011;6:e17695. doi: 10.1371/journal.pone.0017695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gully CP, Velazquez-Torres G, Shin JH, Fuentes-Mattei E, Wang E, Carlock C, et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1513–22. doi: 10.1073/pnas.1110287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen YY, Chou PC, Pham L, Su CH, Chen J, Hsieh YC, et al. DNA damage-mediated c-Myc degradation requires 14–3–3 sigma. Cancer Hallmarks. 2013;1:3–17. [Google Scholar]
- 48.Choi HH, Su CH, Fang LK, Zhang J, Yeung SC, Lee MH. CSN6 deregulation impairs genome integrity in a COP1-dependent pathway. Oncotarget. 2015 doi: 10.18632/oncotarget.3151. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nature reviews Cancer. 2003;3:350–61. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 50.Murakami Y, Higashi Y, Matsunaga N, Koyanagi S, Ohdo S. Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology. 2008;135:1636–44 e3. doi: 10.1053/j.gastro.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 51.Zeng ZL, Luo HY, Yang J, Wu WJ, Chen DL, Huang P, et al. Overexpression of the circadian clock gene Bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:1042–52. doi: 10.1158/1078-0432.CCR-13-0171. [DOI] [PubMed] [Google Scholar]
- 52.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–4. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 53.Fuchs SY. Tumor suppressor activities of the Fbw7 E3 ubiquitin ligase receptor. Cancer Biol Ther. 2005;4:506–8. doi: 10.4161/cbt.4.5.1703. [DOI] [PubMed] [Google Scholar]
- 54.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–9. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crusio KM, King B, Reavie LB, Aifantis I. The ubiquitous nature of cancer: the role of the SCF(Fbw7) complex in development and transformation. Oncogene. 2010;29:4865–73. doi: 10.1038/onc.2010.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.