Abstract
The process of peptide bond synthesis by ribosomes is conserved between species, but the initiation step differs greatly between the three kingdoms of life. This is illustrated by the evolution of roughly an order of magnitude more initiation factor mass found in humans compared with bacteria. Eukaryotic initiation of translation is comprised of a number of sub-steps: (i) recruitment of an mRNA and initiator methionyl-tRNA to the 40S ribosomal subunit; (ii) migration of the 40S subunit along the 5′ UTR to locate the initiation codon; and (iii) recruitment of the 60S subunit to form the 80S initiation complex. Although the mechanism and regulation of initiation has been studied for decades, many aspects of the pathway remain unclear. In this review, I will focus discussion on what is known about the mechanism of mRNA selection and its recruitment to the 40S subunit. I will summarize how the 43S preinitiation complex (PIC) is formed and stabilized by interactions between its components. I will discuss what is known about the mechanism of mRNA selection by the eukaryotic initiation factor 4F (eIF4F) complex and how the selected mRNA is recruited to the 43S PIC. The regulation of this process by secondary structure located in the 5′ UTR of an mRNA will also be discussed. Finally, I present a possible kinetic model with which to explain the process of mRNA selection and recruitment to the eukaryotic ribosome.
1. Overview of translation initiation in eukaryotes
It has long been recognized that initiation serves as the rate-limiting step of the translation pathway on the majority of cellular mRNAs. However, rare codons located in open reading frames (ORFs) have been shown to control protein abundance, implying that elongation can serve as the rate-limiting step on some abundant mRNAs [1–6]. To directly address which step limits translation in yeast, a recent study tested if the abundance or body sequence of the rare AGG tRNA is able to control translation efficiency [7]. Using the recently developed ribosome profiling technique to monitor ribosome pauses, the experiments clearly revealed that translation efficiency is unchanged even when rare tRNA levels are dramatically altered [7]. This reaffirms that initiation likely serves as the rate-limiting step on the majority of mRNAs, even when rare codons are found in ORFs. The apparent codon bias observed in mRNAs may therefore exist in part to ensure the efficient use of the translational machinery in highly translated mRNAs. Ultimately, the overall rate of protein production in the cell depends primarily on the availability of free ribosomes to enter a translation cycle. To this end, the rate of ribosome recycling will likely play a significant role in controlling translational efficiency during low ribosomal availability [8]. As discussed later, the competition between mRNAs for this limiting pool of free ribosomes will likely determine the translation efficiency of individual mRNAs. Interestingly, a recent computational model generated from available data for translation rates in yeast has predicted that initiation events on mRNAs can range by two orders of magnitude (from ~4 seconds to ~240 seconds; [9]). This clearly provides a cell with a substantial capacity with which to fine tune protein synthesis by regulating initiation efficiency. In eukaryotes, translation initiation requires the coordinated action of a large number of initiation factors and two ribosomal subunits. The initiation phase essentially proceeds through three main steps (Figure 1). In the first step, the mRNA and initiation factors are recruited to the 40S subunit to form the 43S–mRNA–preinitiation complex (43S–mRNA–PIC). In step two, this complex is converted into the 43S–mRNA–initiation complex (43S–mRNA–IC) when the anticodon of the initiator tRNA interacts productively with the initiation codon of the mRNA. In the third step, the 60S subunit binds to the 40S subunit, forming the 80S initiation complex (80S–mRNA–IC). Each step is promoted by interactions between different initiation factors and the two ribosomal subunits. The entire process must occur with high fidelity so that the correct initiation codon is selected to ensure accurate translation. Although this simplified pathway is shown that includes three main steps, it is important to note that a number of key sub-steps are likely important in mRNA selection and recruitment, as will be discussed later. In this review, I will discuss our current understanding of the mechanism by which capped mRNAs are recruited to the 40S subunit. In particular, I will discuss how thermodynamic and kinetic frameworks are beginning to reveal how 40S subunits are prepared for mRNA recruitment, and how different mRNAs can be selected for translation. For an extensive review of the mechanism of eukaryotic initiation, I encourage the reader to refer to a number of excellent recent reviews [10–13]. More specific reviews discussing initiation codon selection [14–16], ribosome recycling and reinitiation [17–19] are also available. In addition, tremendous advances in our understanding of the structures of eukaryotic initiation complexes have been made and are summarized in earlier reviews [20–24].
Figure 1.
Pathway of eukaryotic translation initiation. In the first step of initiation (step 1), a mRNA is recruited to the 40S subunit. The eIF4F complex (eIF4E, eIF4A, and eIF4G) specifically binds to the m7G cap structure located on the 5′ end of the mRNA. Although not shown, an interaction between PABP, the poly(A) tail and eIF4F can also occur. The eIF4F bound mRNA is recruited to the 43S PIC by virtue of numerous interactions between initiation components described in the text. In the second step of initiation (step 2), the 40S subunit scans along the 5′ UTR in a 5′ to 3′ direction. Codons are continuously sampled by the initiator tRNA anticodon until one is selected (shown here as an AUG). In the final step of initiation (step 3), two GTP hydrolysis steps occur, resulting in the recruitment of the 60S subunit and initiation factor release. The newly formed 80S ribosome then enters the elongation cycle.
1.1. Initiation components
The core set of components needed to recruit an mRNA to the eukaryotic ribosome and initiate protein synthesis was first identified in the 1970s. Using biochemical techniques, the Staehelin, Hershey, Anderson, and Merrick laboratories successfully reconstituted the initiation process using purified components. The activities of individual factors were determined by omitting one factor at a time, providing the first pathway models for assembly of initiation complexes [25–29]. Due to the relatively poor separation qualities of the ion exchange columns of the time, some initiation factor preparations were in fact “contaminated” with more than one initiation factor. This resulted in some initiation factors possessing several activities that were not established until later; the presence of eIF4G in early eIF3 preparations is a good example [30]. Nevertheless, these early experiments provided a general pathway for eukaryotic initiation that has essentially stood the test of time. However, the parts list of the initiation machinery has continued to grow, with activities of new initiation factors such as DHX29 and DDX3 still being discovered (see below).
2. The 43S pre-initiation complex
Prior to the recruitment of an mRNA to the 40S subunit, initiation factors must bind to the 40S subunit to form what is generally named the 43S pre-initiation complex (PIC). These factors include eIF1, eIF1A, eIF2-GTP-Met-tRNAi, eIF3 and eIF5, which through a complex set of interactions stabilize each other on the surface of the 40S subunit. These initiation factors function in part by altering the conformation of the 40S subunit mRNA decoding site to promote: (i) mRNA recruitment; (ii) scanning; and (iii) initiation codon selection. A considerable amount of biochemical, biophysical, genetic, and structural data has converged to generate models for the eukaryotic 43S PIC. Here, I will briefly summarize what is known regarding the binding sites of each 43S PIC factor on the 40S subunit and how they alter the 40S subunit conformation. I will then discuss what is known about the maturation pathway of the 43S PIC.
2.1. Initiation factor binding sites on the 40S subunit
The mRNA decoding site of the 40S subunit extends across the width of the 40S subunit interface between the head and body domains (Figure 2; 40S subunit interface view). The mRNA entry and exit channels are located at each end of the decoding site, and as the names suggest, constitute the entry and exit sites of the mRNA. The binding sites for the 43S PIC initiation factors have been established using different methods, including crosslinking, footprinting, electron microscopy and crystallography (Figure 2; reviewed in [22, 31]). Biochemical and structural models position eIF1 and eIF1A close to the peptidyl (P) and aminoacyl (A) sites respectively, where they are able to play an important role in controlling mRNA binding and the fidelity of initiation codon selection (Figure 2; 40S–eIF1–eIF1A). These proteins are structurally and functionally related to bacterial IF1 (eIF1A) and IF3 (eIF1), which bind to analogous regions of the 30S subunit [32–37]. Two recent cryo-EM models have revealed how the eIF2-GTP-Met-tRNAi ternary complex (TC) is positioned in the P-site of the 40S subunit in the absence and presence of mRNA (Figure 2; 40S–eIF1–eIF1A–TC) [36, 38]. Interestingly, the TC appears to accommodate more deeply into the P-site of the 40S subunit upon mRNA binding and codon recognition [36]. The majority of the large eIF3 complex binds to the solvent exposed surface of the 40S subunit (Figure 2; 43S–mRNA) [35, 38]. Consistent with its known interactions with eIF1, eIF2 and eIF5, it is expected that a number of its subunits extend into the 40S subunit interface and decoding site. To date, only the eIF3j subunit has been shown to bind to the mRNA entry channel and A-site of the 40S subunit [35, 39]. The position of the GTPase activating protein (GAP) eIF5 on the 40S subunit is less clear. Presumably, it would likely bind to the 40S subunit interface close to eIF1, eIF1A, and eIF2 since it is known to make direct and/or indirect interactions with these proteins on the 40S subunit [40]. Consistent with this, a recent cryo-EM reconstruction of a yeast 43S PIC intermediate complex (minus eIF3) indicates that eIF5 may in fact bind in between eIF2 and eIF1 on the surface of the 40S subunit [36].
Figure 2.
40S subunit structure and substrate binding sites. The atomic model of the yeast 40S subunit is shown from the interface view according to a recent 40S–mRNA–eIF1–eIF1A–TC cryo-EM model (PDB: 3J81 [36]). Landmarks for the 40S subunit are labeled: A, A-site; P, P-site; E, E-site; bk, beak; b, body; pt, platform; and h, head. The latch of the 40S is formed between helix 18 of the body and helix 34 of the head. The mRNA entry and exit channels either side of the neck are labeled accordingly. Binding sites of mRNA (40S–mRNA), eIF1 and eIF1A (40S–eIF1–eIF1A), TC (40S–eIF1–eIF1A–TC) are shown individually for clarity by removal of individual substrates from the original atomic model (PDB: 3J81). The general binding site for the bulk of the eIF3 complex is shown on the solvent exposed surface of the 40S subunit according to a recent low-resolution cryo-EM model of the mammalian 43S complex [38].
The 40S subunit is a dynamic machine that can adopt a number of conformations [31]. In the absence of initiation factors, the 40S subunit is likely to rapidly move between these conformations. Initiation factor binding stabilizes specific conformations, helping to prepare the 40S subunit for mRNA binding and subsequent steps in the initiation pathway. Footprinting, hydroxyl radical probing, fluorescent assays, electron microscopy and crystallography can observe changes in 40S subunit conformation at different degrees of resolution and time (reviewed in [31]). A well-characterized conformational rearrangement of the 40S subunit is one that occurs upon the binding of eIF1 and eIF1A to the subunit interface. These small initiation factors stabilize a conformation that “opens” the mRNA binding channel [34, 41]. This conformation is consistent with the function of these small initiation factors in promoting mRNA recruitment into the 40S subunit decoding site and subsequent scanning [42]. Interestingly, a similar opening of the mRNA binding channel can be observed upon binding eIF3. This was revealed using a hydroxyl radical probing assay that monitored the position of eIF3j relative to the 18S rRNA in the absence and presence of eIF1 and eIF1A, or eIF3 [43]. Interestingly, the binding of the HCV-IRES also promotes a similar conformational rearrangement of the mRNA binding channel in the 40S structure, implying that there are multiple ways to achieve a similar change in 40S subunit conformation [43, 44]. Recent advances in cryo-EM are beginning to generate exciting new high-resolution structural models for ribosomal subunits and intermediates in the initiation pathway [45, 46]. It is anticipated that new high-resolution structural models will help to reveal how the dynamic nature of the 40S subunit is fine-tuned by the different components of the 43S PIC in order to prepare it for mRNA recruitment.
2.2. Assembly of the 43S PIC
Genetic, biochemical, and biophysical approaches have begun to reveal how a complex network of direct and indirect interactions exists between the components of the 43S PIC. Non-equilibrium methods such as sucrose gradients and size exclusion chromatography have provided some information about the relative affinity of each component for the 40S subunit in vitro and in vivo. However, a detailed thermodynamic framework of the 43S PIC requires binding experiments that are able to measure equilibrium dissociation constants. The Lorsch laboratory was first to address this by measuring a number of interactions between 43S PIC components in yeast using equilibrium binding assays. The elucidated thermodynamic frameworks have generated profound mechanistic insight into how interactions between initiation factors stabilize the 43S PIC. Importantly, they have also rigorously tested their in vitro models using in vivo experiments in collaboration with the Hinnebusch laboratory (reviewed in [10]). This extensive body of work has quantitatively shown how eIF1, eIF1A, eIF5 and TC all stabilize one another on the yeast 40S subunit in the absence of mRNA [41, 47–51]. One particularly important interaction within this framework is the thermodynamic coupling between eIF1 and eIF1A on the surface of the 40S subunit [50, 52, 53]. As mentioned earlier, these small initiation factors regulate the opening and closing of the mRNA binding channel, which ensures mRNA recruitment, scanning and the fidelity of initiation codon selection [41, 42, 49]. Although these small initiation factors independently possess high affinity to the 40S subunit (15–50 nM), thermodynamic coupling between them increases their affinity for the 40S subunit roughly five-fold [47, 50, 52]. In addition, this coupling provides a central mechanism by which the 43S PIC stabilizes the TC and eIF5 on the 40S subunit prior to mRNA binding [48, 51].
One limitation of the binding experiments using yeast components is that technical difficulties prevented the inclusion of the large eIF3 complex when measuring equilibrium binding affinities. Since non-equilibrium binding assays indicate that eIF3 increases the relative affinity of eIF1, eIF1A and TC for the 40S subunit, it would be expected to appreciably increase the affinity of these initiation factors in the 43S PIC. It has now been possible to monitor how human eIF3 changes the affinity of eIF1, eIF1A and eIF3j in the 43S PIC [52]. Interestingly, eIF3j binds to the 40S subunit A-site and mRNA entry channel and reduces the affinity of eIF1, eIF1A and TC for the 40S subunit [39, 52]. To overcome this negative cooperativity, the binding of the eIF3 complex to the 40S subunit ensures that all of these components possess high affinity (≤10 nM). One limitation of this study is that it did not address a possible contribution of eIF5 on the stability of the 43S PIC [52]. Nevertheless, the frameworks developed for the yeast and human systems are starting to generate important quantitative information regarding the connections between components of the 43S PIC that ultimately stabilizes it. It is important to note that the cellular concentration of the 43S PIC initiation factors is likely to be around 0.5–1 μM [54]. This means that the 40S subunit would likely be saturated with the 43S PIC components at equilibrium, even without the observed thermodynamic coupling between them. However, the positive cooperativity between initiation factors indicates that their lifetime on the surface of the 40S subunit could be many times longer when all factors are present. Maintaining the integrity of the 43S PIC for long enough to productively engage an mRNA is therefore likely to rely on these stabilizing interactions. An important goal for the future will therefore include determining the rates of binding and release of individual 43S PIC components. It should be noted that the concentration of initiation components and ribosomes might vary considerably between cell types and species [55–58]. Although in vitro assays are able to determine how the concentration of initiation components can regulate the formation of different initiation intermediates, it will be important to relate these models to in vivo concentrations of initiation components.
The pathway of 43S PIC formation is expected to depend on whether the 40S subunit is selected from free ribosomal subunits or 40S subunits generated during ribosome recycling after the termination of mRNA translation [17]. When generated from free ribosomal subunits, the independent binding of each component could generate the 43S PIC. This may be accomplished by a random order of binding, or potentially by kinetically favoured routes dictated by the arrival times of individual components. This has recently been shown to be the case in bacteria, where sophisticated kinetic experiments have revealed different pathways of 30S PIC assembly that partly depend on initiation factor concentrations [59, 60]. We currently know very little about the kinetically favoured routes of 43S PIC assembly on free ribosomal subunits because association rates for individual components have not been rigorously determined. It has been shown that a multifactor complex (MFC) containing eIF1, eIF2, eIF3 and eIF5 can form independently of the 40S subunit in yeast and mammals [61, 62]. It is therefore entirely possible that formation of this complex prior to 40S subunit binding may provide a favourable kinetic pathway of 43S PIC formation. Even though kinetic and equilibrium binding constants can help reveal how the 43S PIC forms on a free 40S subunit, it is currently unclear how a free 40S subunit can be derived from an 80S ribosome that is not actively translating a message. The eIF3 complex possesses anti-association activity by binding to the 40S subunit following 80S dissociation, but it is not clear if eIF3 can accelerate the dissociation of a 60S ribosomal subunit from the 40S subunit. Ultimately, if different 43S PIC assembly pathways do in fact exist in eukaryotic cells, it will be important to determine if these different pathways affect the recruitment of specific mRNAs to the 40S subunit.
An alternative pathway of 43S PIC formation exists when ribosomal subunits are derived from terminating ribosomes (Figure 3). Generation of the 43S PIC by ribosome recycling appears to follow a better-defined pathway than from free ribosomal subunits [17, 63-65]. During stop codon recognition, eRF1 and eRF3 bind into the available A-site and promotes termination of protein synthesis. Following eRF3 release, an ATPase named ABCE1 is recruited to the 80S complex and functions to dissociate eRF1 and split the 40S-mRNA-tRNA complex from the 60S ribosomal subunit upon ATP hydrolysis. The deacylated tRNA remaining in the P-site of the 40S is then ejected during the binding of eIF1, eIF1A and eIF3. However, the order of binding of these three initiation factors is not known and will need kinetic experiments to precisely define them. Importantly, the binding of these initiation factors to known bridge contacts likely ensures that re-association of the ribosomal subunits is prevented. Finally, eIF3j associates with the A-site of the 40S subunit to aid mRNA dissociation from the 40S subunit. Whether mRNA dissociates spontaneously from the 40S subunit (as found for mRNA release from 30S subunits during recycling [66, 67]) or if eIF3j somehow accelerates mRNA dissociation is currently unknown. Finally, the binding of the TC and eIF5 completes the formation of the 43S PIC, which likely allows it to enter into another round of initiation. At present, the timing of eIF5 recruitment to complete the 43S PIC is poorly understood. Interestingly, since the MFC is able to form independently of the 40S subunit, it is also possible that the TC will be recruited to the 40S subunit via this complex.
Figure 3.
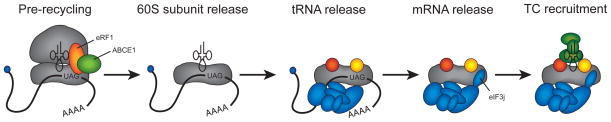
Ribosome recycling pathway in eukaryotes. One possible pathway of 43S PIC generation is shown according to recent models [63-65]. A termination/prerecycling complex containing eRF1 and ABCE1 is shown as the first complex in the pathway, although it should be noted that additional steps in the termination pathway exist prior to this step [17]. The ATPase activity of ABCE1 is responsible for the dissociation of the 60S subunit and eRF1 in the first step of the pathway. The deacylated tRNA in the P-site is then released, most likely upon recruitment of eIF1, eIF1A and eIF3. The mRNA is then released from the complex upon binding of eIF3j into the mRNA entry channel and A-site of the 40S subunit. Finally, the TC is recruited to the 40S subunit to form the 43S PIC. In addition to the labelling of eRF1 and ABCE1, individual initiation components are shown according to the key provided in Figure 1.
3. Selection and Recruitment of mRNA to the 43S PIC
All mRNAs in the cell must compete with one another for the translation machinery through their efficiency in recruitment to the cap-binding complex (eIF4F) and the 43S PIC. It is 40 years since Harvey Lodish first proposed a kinetic rate equation to model eukaryotic mRNA translation [68]. His model postulated that an increase in the rate of initiation at the mRNA recruitment step would result in a preferential increase in the translation of poorly competing mRNAs. A subsequent model from the Thach laboratory further separated initiation into substeps and introduced the concept of a “discriminatory factor” binding to the mRNA prior to its recruitment to the 40S subunit [69]. A mechanism to explain how mRNAs can compete for such a discriminatory factor was then provided by a correlation between m7G cap accessibility and translation efficiency [70, 71]. Nevertheless, the molecular details for how different classes of mRNAs actually compete for recruitment to eIF4F and the ribosome in cells is still poorly defined. Moreover, possible mechanisms to regulate the recruitment of different subsets of mRNAs in response to environmental changes are not clear.
A large amount of data indicates that an increase in the amount and stability of secondary structure in the 5′ UTR of an mRNA leads to a decrease in the rate of initiation (reviewed in [72]). This is thought to be due to the fact that the thermodynamic barrier of any secondary structure in the 5′ UTR must be overcome to enable efficient recruitment to eIF4F and the 43S PIC. Consistent with this, mRNAs containing large amounts of secondary structure in their 5′ UTRs are more dependent on the unwinding activity of eIF4F [73]. Cellular mRNAs that appear to fall into this category include VEGF, ODC and cyclin D1, all of which possess relatively long G–C rich 5′ UTRs that are predicted to possess extensive secondary structure [72]. In contrast, highly translated mRNAs such as GAPDH are not predicted to possess significant secondary structure and are translated much more efficiently [72]. While it is likely that many different classes of mRNAs possessing various amounts of secondary structure in their 5′ UTR exist in cells, experimentally determined mRNA 5′ UTR secondary structure is still limited. It is hoped that high-throughput chemical probing techniques will help to address this in the future [74–78].
3.1. Assembly of the eIF4F complex
Eukaryotic mRNAs are modified with a 7-methyl-guanosine cap (m7G cap) on the 5′ end and a polyadenylated (poly(A)) tail on the 3′ end. These modifications promote nuclear export, stabilize the mRNA, but also act in a cooperative manner to stimulate the rate of initiation [79, 80]. A key initiation factor complex that interacts directly and indirectly with both of these mRNA features is eIF4F, which was first identified in mammals three decades ago [81, 82]. This complex consists of three subunits: the m7G cap binding protein, eIF4E; the DEAD-box helicase, eIF4A; and the molecular scaffold protein, eIF4G. These proteins consist of several conserved domains that have been shown to directly interact with different initiation components (Figure 4A). Some structural models for these proteins and their interaction with other components are available (Figure 4B). However, we still lack structures for the entire eIF4F complex from any species. It is important to note that while mammalian, plant, and yeast eIF4F are likely to function similarly in mRNA recruitment, their structure and regulation appears to be somewhat different. Although I will focus discussion primarily on the mammalian eIF4F complex, some aspects of yeast and plant eIF4F function will be discussed in the following sections.
Figure 4.
High-resolution structures of eIF4F components. (A) Cartoon representations of human eIF4GI and eIF4AI proteins. Colored boxes represent the general positions of conserved domains in each protein and are labelled accordingly. The binding sites of other initiation components with each conserved domain in eIF4G are indicated. (B) A selection of high-resolution structures for domains of eIF4G together with eIF4E and eIF4A are shown. Each structural model is colored according to the cartoons shown in panel A. The HEAT-1 domain of yeast eIF4G bound to eIF4AI is shown with eIF4AI in an “open” conformation (bound to AMP), which is not compatible with RNA binding (PDB: 2VSO [178]). As a comparison, the “closed” conformation of eIF4AIII from the exon junction complex is presented (bound to ADPNP), which is bound to RNA (PDB: 2HYI [179]). Other components of the exon-junction complex have been omitted for clarity. A solution structure of the yeast eIF4E bound to m7GDP (cap) and its eIF4G-binding domain is shown with eIF4E colored orange (PDB: 1RF8 [180]). A structure of the C-terminal HEAT-2 and HEAT-3 domains of human eIF4GI is shown in the absence of its eIF4AI and MNK1 binding partners (PDB: 1UG3 [181]). The N-terminal PAM domain of eIF4G is shown bound to RRM1 and RRM2 of PABP together with a short stretch of poly(A) mRNA (PDB: 4F02 [182]).
The availability of eIF4F for binding to an mRNA is an important regulated step early in the initiation pathway. Accordingly, the eIF4E component can be sequestered from the complex by a family of eIF4E binding proteins (4E-BPs). These proteins bind to eIF4E and act as competitive inhibitors to prevent its binding to eIF4G [83]. This mechanism takes advantage of the fact that eIF4E is generally believed to be the lowest abundant initiation factor in many cells, although this fact has been questioned [54, 84]. There are three known mammalian isoforms named 4E-BP1, 4E-BP2, and 4E-BP3. Together with eIF4G, these proteins share a YXXXXLΦ motif that binds to eIF4E [85, 86]. Non-phosphorylated 4E-BPs interact tightly with eIF4E, preventing it from interacting with eIF4G. A hierarchical phosphorylation pathway converts 4E-BPs to proteins that possess a roughly 4000-fold reduced affinity to eIF4E [87, 88]. Interestingly, recent structural information has revealed how an extended surface in 4E-BPs beyond the YXXXXLΦ motif also interacts with eIF4E [89, 90]. It has been proposed that this surface may play a key role in promoting the dissociation of eIF4E from eIF4G [89]. Although this is an interesting possibility, it will be important to test this hypothesis using equilibrium and kinetic assays. As discussed later, an important goal of future work is to fully elucidate the thermodynamic and kinetic frameworks for the interaction between eIF4E and all of its binding partners.
Despite its importance in mRNA recruitment to the 43S PIC, the molecular details of how eIF4F selects mRNAs from the cellular pool is poorly defined. To this end, a number of laboratories have worked to elucidate the thermodynamic and kinetic frameworks for eIF4F complex formation in the absence and presence of mRNA. Recent work with purified yeast factors has revealed a strong thermodynamic coupling between eIF4E and eIF4A binding to eIF4G [91]. Interestingly, this coupling increases the affinity of eIF4A for eIF4G roughly 10-fold from 200 nM to ≤30 nM in the presence of eIF4E. However, eIF4A is one of the most abundant initiation factors in all cells, so it is possible that this change in affinity doesn’t appreciably change the saturation of eIF4A in the eIF4F complex at equilibrium in the absence of mRNA [58]. Although not directly measured, this coupling would also be expected to increase the affinity of eIF4E for eIF4G when eIF4A is bound to the complex. Again, since eIF4E binds eIF4G with a high affinity in the absence of eIF4A (≤30 nM), this increase in affinity may not be important for controlling eIF4E saturation in the eIF4F complex. Nevertheless, the cooperative binding between eIF4A and eIF4E with eIF4G could have an important role in increasing the lifetime of the eIF4F complex in the absence of mRNA. To date, no thermodynamic framework for the formation of the human eIF4F complex has been determined using full-length eIF4G. The affinities of key interactions between eIF4E and eIF4A with domains of eIF4G have been determined, but whether these binding affinities are relevant in the context of the complete eIF4F complex is unknown. Interestingly, it does appear that the affinity of eIF4A for human eIF4G is likely to be higher than that of the yeast complex. This is inferred from the fact that human eIF4A is readily purified as a complex with eIF4G, while this tends not to be the case in yeast [92]. Whether this is due to the fact that human eIF4G possesses an additional eIF4A-binding domain in its C-terminal extension remains to be determined. While the thermodynamic framework for eIF4F complex formation is beginning to take shape, more work is clearly needed to precisely define it and its kinetic framework. As mentioned above, high-resolution structural models are available for some eIF4F components and their interacting domains (Figure 4B), but the field eagerly awaits high-resolution structural models of the complete eIF4F complex to help us better understand how this complex functions in the initiation pathway.
3.2. mRNA selection by eIF4F
Cellular mRNAs exist as messenger ribonucleoprotein particles (mRNPs) [93]. These mRNPs can be found in actively translating polysomes, repressed in granules such as P-bodies, or awaiting selection by eIF4F. The composition and amount of mRNAs in each of these pools change over time, depending on variables such as the growth state of the cell. Importantly, de novo mRNPs newly exported from the nucleus likely differ in their protein content from previously translated mRNPs, although precisely what proteins are associated with different mRNPs is poorly defined [93]. The pioneer round of translation is promoted by the cap-binding heterodimer CBP80-CBP20 and functions to remove the exon-junction complex to ensure that the mRNA is suitable for translation [94]. Additional remodelling of mRNPs likely occurs during translation, further altering the composition of mRNPs and perhaps the mRNA structure. These rearrangements may provide an important mechanism in controlling translation efficiency of subsequent translation cycles.
How eIF4F selects an mRNA from a complex mixture in the available pool is poorly defined. One possibility is that mRNAs might possess different affinities for this complex, resulting in preferential selection. While this is an attractive model, it is not clear how the concentration of eIF4F relates to the concentration of mRNAs awaiting recruitment. Should the concentration of eIF4F be appreciably lower than that of the mRNA pool, selection likely becomes more important than if the concentration of eIF4F approaches that of the mRNA pool. Appropriately, regulating the amount of available eIF4E through sequestration by 4E-BPs will clearly have an important role to play in controlling the concentration of eIF4F [83]. Once again, the specific mRNP pool that the mRNA is derived from may also significantly alter the selection process. For example, the recruitment efficiency of mRNA to eIF4F may be greater when an mRNA is actively being translated compared to one that is awaiting selection. This would be consistent with the finding that initiation rates on actively translating mRNAs appear to be higher than de novo initiation events [95]. This could be due to preferential selection of these mRNAs by eIF4F as the m7G cap becomes available during, or following the previous initiation event.
A crosslinking approach first identified the eIF4E subunit of eIF4F as the m7G cap-binding protein [96]. High-resolution structures later revealed how this protein uses two conserved tryptophan amino acids to sandwich the modified guanosine of the m7G cap with a third tryptophan interacting with the N7-methyl group [97, 98] (Figure 4). The interaction of this protein with the m7G cap structure of an mRNA has been extensively studied using different quantitative approaches including fluorescence (bulk and single molecule), surface plasmon resonance (SPR), and isothermal titration calorimetry (ITC) [99–104]. These approaches have all revealed that eIF4E has a reasonably high affinity to the m7G cap structure (~100 nM) and any change in m7G cap proximal secondary structure can change the affinity of eIF4E for the m7G cap by up to 5-fold [102, 105]. Therefore, it is possible that a change in affinity by secondary structure might explain how different mRNAs could compete for eIF4E binding. However, one limitation of these experiments is that they typically employ short (~50 nt) oligonucleotides in place of a functional mRNA. It will therefore be important to determine the rate of eIF4E binding to different functional mRNAs that possess well-characterized secondary structures located at different distances from the m7G cap structure.
In addition to the binding of eIF4E to the m7G cap, eIF4G and eIF4A both possess RNA binding domains that could combine to increase the affinity of eIF4F for an mRNA (Figure 4). This has been recently investigated using yeast eIF4F in an elegant single molecule FRET assay. Using this approach, the affinity of eIF4E for the m7G cap structure was found to be increased about 5-fold from ~100 nM to ~20 nM upon binding full-length yeast eIF4G [102]. Although not tested, it is likely that the multiple RNA binding domains in eIF4G play a critical role in this coupling. This would be consistent with the fact that these RNA binding domains promote mRNA recruitment and subsequent stages of initiation in vivo [106]. Because of the positive coupling between the components of eIF4F in the absence of mRNA, the recruitment of mRNA to the complex would be expected to substantially increase the affinity of eIF4E and eIF4A to eIF4G in order to stabilize the eIF4F complex on the mRNA. Presently, it is not clear what the role of the RNA binding property of the eIF4A helicase is on the affinity of the eIF4F complex for an mRNA. It has also been shown that ATP increases the affinity of eIF4A for the HEAT-1 repeat of human eIF4G, but it has not been determined if this would be true for full-length eIF4G [107].
Although eIF4F is a stable complex that can be purified in the absence of other proteins, accessory proteins that can associate with it include the eIF4E kinase (MNK1) and poly(A) binding protein (PABP) (Figure 4). It has been shown that PABP binds to a conserved domain in the N-terminus of eIF4G [108–110]. This interaction possesses a very high affinity in plants (~40 nM; [111]), while the human factors have been reported to interact with a much weaker affinity (27 μM; [112]). However, the affinity of the human components has only been tested using a minimal binding domain of eIF4G and it remains to be determined if full-length eIF4G will possess an affinity closer to that shown for plant eIF4G. In yeast, the relative affinity between eIF4G and PABP appears to be dramatically increased by the poly(A) tail, although no quantitative data are yet available [108]. Importantly, the interaction of PABP with the eIF4F complex has been proposed to explain why addition of a m7G cap and poly(A) tail synergistically stimulate translation in vivo [79, 108, 113, 114]. However, there may be some amount of redundancy in this interaction since a mutation in yeast eIF4G that lowers its affinity to PABP still shows a synergistic activation of translation by the m7G cap and poly(A) tail [80]. An alternative model whereby PABP stimulates 60S subunit joining has been proposed to explain these data [80]. Consistent with PABP playing an important role in mRNA recruitment to the ribosome, the binding of PABP to eIF4F has been shown to increase the affinity of eIF4E for a m7G cap analogue (minus mRNA) by 40-fold [115]. As expected, this coupling also increases the affinity of eIF4F to PABP by the same degree. While these changes in affinity are striking, it will be important to verify the extent to which this coupling changes these affinities using a functional mRNA. A small, but appreciable 2-fold increase in PABP binding to the poly(A) tail has been shown in the presence of eIF4F [111, 116]. However, the importance of stabilizing this interaction is not clear since the affinity of PABP for the poly(A) tail on its own is very high (7 nM; [117]). Taken together, the interaction between eIF4G and PABP stabilizes the eIF4F complex on the mRNA m7G cap. This may substantially increase the lifetime of the complex so that it can efficiently recruit the selected mRNA to the 43S PIC. A widely adopted model is that an mRNA can be circularized as a result of this interaction. While this can be shown using in vitro components [113], whether this is important or even occurs in vivo is still unclear [118, 119]. Therefore, despite a considerable amount of work being carried out on the interaction between eIF4F and PABP, it remains to be determined mechanistically how this contributes to mRNA selection and the rate of mRNA recruitment to the 43S PIC.
3.3. Duplex unwinding by eIF4F
It is likely that one of the main determinants of successful eIF4F docking onto an mRNA is the amount of secondary structure located in the m7G cap proximal region. As mentioned above, relatively stable hairpin structures can lower the affinity of eIF4F for the m7G cap structure when tested using short oligonucleotides [102]. This is presumably due to eIF4F possessing a reduced affinity to double stranded RNA regions, although this has not been rigorously tested. To overcome the inhibitory effect of secondary structure close to the m7G cap structure, eIF4F can melt this structure by the duplex unwinding activity of the eIF4A component. It is not clear whether there is a minimal amount of single stranded RNA that is needed to enable eIF4G to productively bind to the m7G cap proximal region of mRNA. It has been shown that the rate of duplex unwinding by the eIF4F complex on an uncapped substrate is strongly stimulated by the presence of a single-stranded RNA region in vitro [120]. Presumably, this is due to a greater affinity of eIF4G/eIF4F to single-stranded regions of mRNA when they are available, although this has not been rigorously tested using a m7G capped mRNA. Therefore, spontaneous unwinding of any m7G cap-proximal secondary structure would likely enable eIF4F to be stabilized on the mRNA by virtue of an increase in available single stranded RNA. The unwinding potential of eIF4F in vitro is limited to melting short hairpins with modest stability (~ΔG –30 kcal/mol; [121]). This may be due to the fact that eIF4A, like all DEAD box proteins, is not thought to be a processive helicase [121]. However, it is entirely possible that it becomes processive in the presence of eIF4G and/or eIF4B [120]. Importantly, the in vitro helicase activity of eIF4A is synergistically activated by its binding to eIF4G and the accessory factors eIF4B and eIF4H [120, 122–124]. These accessory proteins have been shown to accelerate the cycling of eIF4A between open and closed conformations, which is needed to destabilize RNA duplex regions [124–126] (Figure 4B; compare open and closed eIF4A conformations). This activity of eIF4B is consistent with the finding that mRNAs containing large amounts of predicted secondary structure in their 5′ UTRs are translated less efficiently when eIF4B protein is reduced using RNA interference (RNAi) [127]. Importantly, the binding of eIF4E to eIF4G is also required to counteract an autoinhibitory domain within human eIF4G that otherwise inhibits eIF4A unwinding [128]. Therefore, the rate of duplex unwinding by eIF4A will be greatest when bound to the eIF4F complex and in the presence of eIF4B/eIF4H. Interestingly, eIF4B also interacts directly with PABP in plants and mammals, which may play an additional role in stabilizing this accessory protein on the eIF4F complex [111, 129]. Taken together, any increase or decrease in the rate of unwinding by eIF4A could be an important factor in determining whether eIF4F will fully accommodate on a selected mRNA. While this is an attractive model to explain the pathway of mRNA selection by the eIF4F complex, a significant amount of quantitative data that will be needed to fully support it is still missing. It is noteworthy to mention that the helicase activity of eIF4F may also function to disrupt RNA–protein interactions in addition to its function in melting duplex regions. This would be consistent with the finding that other RNA helicases can lead to the dissociation of proteins from RNA substrates independently of their duplex unwinding activity [130].
3.4. Recruitment of mRNA to the 43S PIC
The mRNA binding channel on the 40S subunit can only accommodate single stranded RNA. Therefore, any secondary structure located in the m7G cap proximal region of the mRNA must be melted in order to bind the 43S PIC. Consistent with this, changing the position of hairpin structures within the first 10 nt of a reporter mRNA can modulate translation by more than 50-fold [131]. Whether such hairpin structures inhibit recruitment of mRNA to the eIF4F complex, or subsequent binding of this complex to the 43S PIC is not known. The stable binding of the eIF4F complex to the m7G cap proximal region of an mRNA serves as a docking site for the 43S PIC. Using a non-equilibrium gel shift assay, eIF4F has been shown to possess the highest affinity to ~60 nt of RNA, although the precise footprint of the complex has not yet been determined [132]. Footprinting and structural models indicate that the 40S subunit accommodates ~30 nt of mRNA, while the eIF4F–43S PIC protects ~60 nt of mRNA [133, 134]. The additional 30 nt of protected mRNA beyond that protected by the 40S subunit appears to be a 5′ extension on the solvent side of the 40S subunit where eIF3 is located and can crosslink to the mRNA [134]. Thus, the minimum 5′ UTR length that would likely enable full protection would be ~45 nt long, since the 40S subunit protects ~15 nt of RNA downstream of the initiation codon when it is located in the P-site. Accordingly, the average length of 5′ UTRs across species is ~100 to ~200 nucleotides, which would indicate that full accommodation of the eIF4F-43S PIC would be possible on the majority of 5′ UTRs [135]. A well-characterized hairpin structure that can specifically inhibit 43S PIC binding is the iron responsive element (IRE; [136]). Generally found in mRNAs that regulate iron metabolism, these mRNAs possess a ~30 nt hairpin structure within ~50 nts from the m7G cap structure. In the absence of iron, the IRE-binding protein binds to the hairpin with high affinity and prevents 43S PIC binding. This inhibition occurs despite the fact that eIF4F can still bind to the m7G cap with a very high affinity (9 nM; [136, 137]). Although not directly tested, the limited unwinding potential of the eIF4F complex is likely not able to melt the IRE hairpin when it is stabilized by IRE-binding protein. Upon binding of iron to the IRE-binding protein, the protein dissociates from the hairpin, which enables unwinding of the IRE hairpin and the recruitment of the 43S PIC. While this is an extreme case of secondary structure regulating mRNA recruitment to the 43S PIC, it is likely that many m7G cap proximal hairpin structures could also regulate 43S PIC recruitment to some extent.
In mammals, productive recruitment of the mRNA–eIF4F complex to the 43S PIC requires a direct interaction between eIF4G and eIF3 [138, 139]. This interaction likely stabilizes the 43S PIC on the mRNA for long enough to enable it to engage the unstructured mRNA in its decoding site, although this has not been directly measured. Immunoprecipitation assays have indicated that the relative affinity of eIF4G binding to eIF3 is increased following mTORC1 activation [140, 141]. Prolonging the lifetime of this interaction in this way could significantly increase the likelihood of mRNA recruitment to the 43S PIC. Surprisingly, a direct interaction between eIF4G and eIF3 does not appear to exist in yeast [91, 142]. The reason for this is not clear, but it is possible that other interactions between initiation components could accomplish the same goal. Such stabilizing interactions may include the eIF4G–eIF5, eIF4B–eIF3, and even the eIF4B–40S interaction [91, 142, 143].
Equilibrium and non-equilibrium binding assays have indicated that the large eIF3 complex stabilizes the 43S PIC prior to mRNA binding [28, 52, 61, 144–147]. In addition, eIF3 also plays a critical role in promoting mRNA recruitment to the 43S PIC in vitro and in vivo (reviewed in [10]). Using a gel-shift assay to monitor mRNA recruitment to the 43S PIC, the Lorsch laboratory has recently shown that yeast eIF3 increases the affinity and the rate of m7G capped mRNA binding to the 43S PIC [91]. Yeast eIF3 increases the apparent affinity of mRNA for the 43S PIC 12-fold when the mRNA possesses a 37 nt 5′ UTR [91]. Interestingly, this increase in apparent affinity is only 2-fold when the mRNA possess a 4 nt 5′ UTR, indicating that eIF3 stimulates mRNA recruitment to the 43S PIC when the mRNA extends beyond the mRNA exit channel. Since eIF3a and eIF3d both crosslink to an mRNA region on the 5′ side of the 40S subunit binding channel, this may explain how eIF3 increases the affinity of mRNA to the 43S PIC [134]. Nevertheless, recent cryo-EM models also indicate that eIF3 may make additional contacts with the mRNA close to the mRNA entry channel [35, 38]. Both of these interaction sites are consistent with the fact that mutations in different eIF3 subunits reduce the apparent affinity of mRNA for the 43S PIC in vivo [148, 149]. However, since eIF3 also changes the conformation of the mRNA binding channel, it is also possible that this complex plays an additional role beyond direct mRNA binding in stabilizing mRNA in the 40S subunit decoding site [43]. Although the eIF3 complex increases the affinity and rate of mRNA recruitment to the 43S PIC, the eIF3j subunit actually decreases the affinity of mRNA for the 40S subunit [39, 91]. This apparent paradox may be explained by the finding that the TC overcomes the eIF3j-mediated inhibition, implying that eIF3j may function in part to ensure TC is recruited to the 43S PIC prior to mRNA. However, the role of TC in overcoming eIF3j mediated inhibition of mRNA recruitment has only been quantitatively demonstrated in the absence of the eIF3 complex [39]. Although not investigated using equilibrium binding assays, the HCV IRES promotes the release of eIF3j from the mRNA binding channel in a manner that is dependent on the TC and domain II of the IRES [43]. Since this IRES domain induces a conformational change in the 40S subunit that opens the mRNA binding channel [44], it is possible that other canonical initiation factors will play a similar role in releasing eIF3j during the mRNA binding step.
As mentioned earlier, eIF1 and eIF1A play an important role in stabilizing a 40S subunit conformation that promotes mRNA recruitment [41, 42]. In addition, equilibrium and non-equilibrium assays have revealed that eIF1 and eIF1A enhance the affinity of TC binding to the 40S subunit [41, 47, 52, 144, 145, 150]. Increasing the affinity and rate of TC binding to the 40S subunit is likely to be important since initiation codons could be bypassed if scanning were initiated prior to TC loading. Although eIF3j may help to minimize this potential problem, there is evidence that mRNA recruitment and scanning can occur, at least to some extent, in the absence of TC. In particular, this appears to be true for GCN4 regulation, which is dependent on the fact that following termination after a uORF, a rescanning 40S subunit doesn’t require the presence of the TC [151]. For de novo initiation events, a 5′ m7G cap proximal AUG codon is bypassed more readily when TC availability is reduced by the mechanism of eIF2 phosphorylation in vivo [152]. Bypassing a m7G cap proximal AUG codon has also been demonstrated in vitro when the concentration of mRNA is increased [153]. Importantly, preferential selection of the first AUG codon was recovered when the eIF2 concentration was increased in the lysate. While these data are consistent with mRNA recruitment to the 43S PIC prior to TC binding, it is not yet clear to what extent these events may occur naturally for de novo initiation events. Since aberrant initiation codon selection could occur if mRNA recruitment and scanning occurs in the absence of TC, it is likely that more than one mechanism could be in place to prevent this.
It is worth noting that the eIF4A accessory protein, eIF4B, appears to play multiple roles in mRNA recruitment to the 43S PIC [10]. As mentioned above, this protein plays a key role in stimulating the ATPase and helicase activity of eIF4A in eukaryotic cells [122, 154, 155]. In yeast, eIF4B plays an additional role in increasing the affinity between eIF4A and eIF4G [143, 156]. In spite of this data, the precise mechanism by which eIF4B promotes duplex unwinding is still not clear. A Brownian Ratchet model has been proposed whereby eIF4B functions as the pawl of the ratchet [157]. This activity would enable eIF4B to restrict the backward diffusional sliding of the unwinding complex, while enabling stochastic forward movement. Interestingly, yeast eIF4B also binds directly to the 40S subunit, which appears to be critical for increasing the affinity and rate of mRNA recruitment to the 43S PIC [143, 158]. It remains to be determined if mammalian eIF4B will play an equally important role in mRNA recruitment to the 43S PIC, or if it primarily functions to stimulate the helicase activity of eIF4A within the eIF4F complex. Interestingly, plant eIF4B binds to both PABP and the eIF3g subunit of eIF3 [159]. This additional interaction may be important in promoting mRNA recruitment to the 43S PIC. In contrast, it has been shown that a truncated form of mammalian eIF4B binds eIF3a [160]. However, full-length eIF4B appears to bind eIF3g, which would be consistent with plant eIF4B (John Hershey, personal communication). Interestingly, mammalian eIF4B is phosphorylated on S422 by S6K in response to serum and insulin [161-163]. Immunoprecipitation has indicated that phosphorylation increases the affinity of eIF4B to eIF3 and the 43S PIC [162]. It is possible that phosphorylation may promote duplex unwinding by eIF4A, but this has not yet been directly tested. Curiously, phosphomimetic amino acid substitutions S422D and S422E activate eIF4B in cells, but these mutations do not alter translation rates in cell-free translation systems [161, 164]. It is hoped that obtaining high-resolution structures of unwinding intermediates will help reveal how this protein functions in promoting duplex unwinding and mRNA recruitment to the 43S PIC. In addition, further single molecule assays should also help provide additional biophysical information regarding the mechanism by which the eIF4F complex together with eIF4B is able to step through hairpin structures. Lastly, it is still unclear exactly what function eIF4H plays in promoting eIF4A helicase activity. This protein has been shown to function in a very similar way to eIF4B in its ability to stimulate eIF4A activity, although it appears to have reduced activity in terms of duplex unwinding kinetics [120, 122, 165]. It is curious to note that eIF4H exhibits some amount of differential expression across tissues, which may suggest that it plays a specific role in the translation of certain mRNAs [166].
4. Scanning and initiation codon selection
Following accommodation of the mRNA into the decoding site of the 40S subunit, the 43S PIC migrates along the 5′ UTR in a 5′ to 3′ direction until the anticodon of the Met-tRNAi base-pairs with the initiation codon [10]. I will only comment briefly on the mechanism of scanning and initiation codon selection here since it takes place after the main focus of this review, which is mRNA recruitment to the 43S PIC. For a more detailed discussion about these steps in the initiation pathway I recommend some excellent recent reviews [10, 11, 13, 157, 167]. The scanning mechanism was originally proposed by Kozak and Shatkin over three decades ago, and has remained the primary mechanism to explain the process of initiation codon selection on the majority of eukaryotic mRNAs [168]. The migration of the 43S PIC along the 5′ UTR requires ATP energy, which is presumably used to promote the unwinding of secondary structure and/or backtracking [157]. Consistent with this, the Pestova laboratory has elegantly shown that ATP energy is not absolutely required for migration if the 5′ UTR is devoid of secondary structure [169]. While it was generally accepted for many years that eIF4A constituted the only helicase protein in the initiation pathway, recent genetic and biochemical work has revealed that at least two other helicases may also play an important role in scanning [167]. The DEAD-box protein DDX3/ded1 has been shown to associate with eIF4G and to increase the rate of initiation on mRNAs that contain extensive secondary structure in their 5′ UTR [170, 171]. Interestingly, DDX3 can stimulate initiation on mRNAs with secondary structure positioned close to the m7G cap structure [170]. Since this protein interacts with eIF4G, it is possible that it plays a role in mRNA recruitment as well as scanning. It will be important in future to quantitatively determine if this protein accelerates the rate of mRNA recruitment to the 43S PIC. The second helicase that has been shown to stimulate initiation is the DExH-box protein named DHX29. This protein was recently identified to selectively stimulate initiation on mRNAs containing secondary structure in their 5′ UTRs in vitro and in vivo [172, 173]. This protein binds close to the mRNA entry channel of the 40S subunit and could conceivably unwind mRNA as it enters the decoding site [38].
The scanning 43S PIC will pause when the initiation codon enters into the P-site of the 40S subunit. The Kozak consensus sequence, GCC(A/G)CCAUGG, regulates the fidelity of initiation codon selection, at least in mammals. In the sequence shown here, the A of the AUG (underlined) is defined as the +1 position. The more critical bases for recognition at the −3 and +4 positions are indicated in bold. Any deviation of this sequence can cause the scanning 40S subunit to bypass the initiation codon, a process generally called leaky scanning. Although AUG is the most common initiation codon for the primary ORF, CUG and GUG can be used as initiation codons, especially for short upstream open reading frames (uORFs) [174]. These alternative initiation codons are not as efficiently recognized by the scanning 40S subunit, but they play an important role in regulating the number of ribosomes that initiate at the main ORF. Presumably, any secondary structure in the region around the initiation codon that alters the rate of scanning around the initiation codon could change the likelihood of initiation codon selection, either in a positive or negative way.
Kinetic experiments have revealed that mRNA recruitment to the 43S PIC results in hydrolysis of the eIF2 bound GTP [175]. Consistent with other GTPase reactions, it is actually the release of inorganic phosphate (Pi) that ultimately results in the committed step in the pathway. To this end, it is the base-pairing between the anti-codon of the initiator tRNA and the codon in the P-site that results in Pi release [175]. This is brought about by coordinated movements between eIF1, eIF1A and eIF5 on the surface of the 40S subunit (reviewed in [10, 14]). Similar to the role of IF3 in bacteria, eIF1 plays a key role in ensuring the fidelity of initiation codon selection in eukaryotes. It has been proposed that the release of eIF1 from the 40S subunit ultimately helps to commit the scanning 43S PIC to a particular initiation codon.
5. Towards a kinetic model for mRNA recruitment to the 40S subunit
Tremendous advances have been made in recent years to elucidate the kinetic framework of mRNA recruitment in bacteria [176]. In contrast, the additional complexity of the eukaryotic initiation mechanism has hampered efforts to elucidate the kinetic framework of the eukaryotic pathway. Nevertheless, a possible model for mRNA recruitment to the 40S subunit can be proposed with likely kinetic partitions in the pathway. This pathway is depicted in Figure 5 with appropriate forward and reverse rates indicated for each step.
Figure 5.

A possible kinetic pathway of eukaryotic mRNA recruitment. In the step 1 of the pathway, initial binding of the eIF4F complex to the m7G cap structure of the mRNA occurs through the eIF4E subunit of eIF4F. In step 2, any m7G cap proximal secondary structure must be melted to enable eIF4F accommodation to occur. This likely involves the interaction of eIF4G RNA binding domains with single stranded mRNA. In step 3, the eIF4F–mRNA complex is recruited to the 43S PIC. This step involves the positioning of single stranded mRNA into the decoding site of the 40S subunit. In step 4, additional mRNA secondary structure downstream of the m7G cap structure is melted as the 43S PIC migrates along the 5′ UTR in search of the initiation codon. While dissociation of the m7G cap structure from eIF4E is depicted during this step, the precise timing of eIF4E detachment from the m7G cap and/or eIF4G is not known. For each step in the pathway, forward and reverse rate constants are shown, as described in the text. Individual initiation components are shown according to the key provided in Figure 1. Although not shown, it should be noted that ATP hydrolysis is likely to accelerate each forward step.
The rate of initial binding of a m7G cap structure by the eIF4F complex will depend on the abundance of the mRNA and the concentration of eIF4F that is available to bind it (k1; Figure 5). As mentioned earlier, the concentration of eIF4F is directly controlled by the family of 4E-BPs [83]. This is important since less abundant mRNAs are expected to compete more effectively for the eIF4F complex when the amount of eIF4F is increased [68]. In addition, the interaction of PABP with the poly(A) tail and eIF4F may play a significant role in promoting mRNA recruitment to the eIF4F complex. This could occur by increasing the association rate (k1; Figure 5) or decreasing the dissociation rate (k−1; Figure 5) of the m7G cap structure with eIF4F. Although it has not been rigorously tested on physiologically relevant mRNAs, any occlusion of the m7G cap structure by secondary structure would be expected to reduce the association rate (k1; Figure 5) and/or increase the dissociation rate (k–1; Figure 5) of the mRNA to the eIF4F complex. Following initial m7G cap binding by the eIF4E subunit of eIF4F, complete accommodation of eIF4F onto the 5′ end of the mRNA will likely be regulated by the availability of single stranded mRNA for eIF4G to bind to. Therefore, the fate of an mRNA at this stage in the pathway will depend of the rate of dissociation (k–1; Figure 5) and accommodation (k2; Figure 5) of eIF4F on the mRNA. The latter rate will likely be reduced if secondary structure prevents the RNA binding domains of eIF4G binding to the m7G cap proximal region of the mRNA. As a result, combining the kinetic rates of steps 1 and 2 will determine how the eIF4F complex selects an mRNA from the pool.
The rate of 43S PIC recruitment to the eIF4F-mRNA complex will in part depend on the concentration of the 43S PIC. The presence of TC on the 43S PIC will likely be important in promoting mRNA recruitment (high k3; Figure 5). Thus, reducing the availability of TC through eIF2α phosphorylation will decrease the rate of 43S PIC recruitment and/or possibly increase the dissociation rate (low k3 and high k–3; Figure 5). Importantly, stable secondary structure located within ~50 nucleotides of the m7G cap structure can reduce the rate of 43S PIC recruitment (k3; Figure 5). This has been particularly well demonstrated by IRE containing mRNAs, which block 43S PIC [136]. However, understanding the direct effect of secondary structure on the kinetics of mRNA recruitment to the 43S PIC is still needed. In addition, the possible kinetic roles of additional helicase proteins, such as DDX3/ded1 and DHX29, have not been quantitatively determined. These proteins may increase the rate of 43S PIC recruitment to the mRNA (k3; Figure 5), but sophisticated kinetic assays will be needed to test this possibility. It is also possible that eIF3 plays an important role in stabilizing the mRNA on the 43S PIC. This is apparent from mutations in eIF3 that inhibit 43S-mRNA-PIC stability (reviewed in [10]). Interestingly, the over-expression of specific eIF3 subunits in cells can selectively increase the translation of growth promoting “structured” mRNAs [177]. Whether the binding of these eIF3 subunits to the eIF3 complex plays a role in promoting mRNA recruitment to the 43S PIC remains to be determined.
The rate of scanning to the initiation codon is expected to depend on the amount of secondary structure in the 5′ UTR of an mRNA. Highly structured 5′ UTRs will likely delay the movement of the 43S PIC along the mRNA, resulting in a low k4 value (Figure 5). The role of secondary structure in controlling the transition towards initiation codon selection (step 4) will therefore dictate the translation efficiency of an mRNA. Increasing the rate of helicase activity associated with the scanning 43S PIC would likely increase the rate of scanning (high k4; Figure 5). This could be achieved by increasing the rate of eIF4A helicase activity, or by recruiting additional helicase proteins, such as DHX29 [172, 173]. Unfortunately, the actual rate of scanning (k4; Figure 5) is difficult to determine using current assays, which are generally not kinetic in nature. It should be noted that although a single step (4) is shown to constitute the scanning mechanism and initiation codon selection, multiple sub-steps likely exist to explain the mechanism of initiation codon selection [10, 11, 14].
6. Perspectives
Despite many years of research, mechanistic insight into mRNA selection by the eukaryotic machinery is still poorly defined. Although the role of 5′ UTR secondary structure in controlling the rate of translation in vivo is apparent, how it precisely controls different stages of the initiation pathway is still not clear. Without this knowledge, we will only possess a superficial understanding of how initiation rates can be controlled in cells. In addition, we will not appreciate how mRNA selection and initiation can be dysregulated in disease states. Our lack of kinetic understanding of the initiation pathway is in part due to the fact that it has been very difficult to generate kinetic assays to monitor pathway intermediates. While gel shift assays can be used to some degree, this approach is not able to monitor rapid association rates. A powerful assay often used to capture the mRNA-eIF4F–43S PIC complex is that of primer extension, or toeprinting. This assay is able to monitor the position of the 40S subunit on an mRNA. However, this assay is limited by the fact that it only monitors relative stability of complexes that reach the initiation codon and is not able to detect changes in kinetics. Therefore, a challenge to overcome in the future will be to generate new assays in vitro and in vivo to monitor the kinetic checkpoints of mRNA recruitment to the 43S PIC (Figure 5). It will also be important to include natural full-length mRNAs in in vitro assays to avoid the limitations that short oligonucleotides can introduce. Interestingly, it has also become apparent that the translation machinery can be extensively modified by phosphorylation. Understanding how these modifications modulate mRNA recruitment and initiation codon selection will be essential for determining how cells change the proteome in response to environmental signals.
Highlights.
The process of mRNA selection by the eukaryotic ribosome is poorly defined.
Cap proximal secondary structure likely regulates mRNA selection by the eIF4F complex.
Secondary structure in the 5′ UTR regulates mRNA binding into the 40S subunit decoding site and subsequent scanning.
Acknowledgments
I wish to thank John Hershey for so many enjoyable thought-provoking conversations about eukaryotic initiation. I am also grateful for his insightful comments and critical reading of the manuscript. I also wish to thank Richard Jackson for his many insightful comments about the mechanism of mRNA recruitment. I am also indebted to the Fraser laboratory for stimulating discussion and comments on the manuscript. I gratefully acknowledge support for this work from NIH grant RO1 GM092927.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. Translation elongation can control translation initiation on eukaryotic mRNAs. Embo J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nature reviews Genetics. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciandrini L, Stansfield I, Romano MC. Ribosome traffic on mRNAs maps to gene ontology: genome-wide quantification of translation initiation rates and polysome size regulation. PLoS computational biology. 2013;9:e1002866. doi: 10.1371/journal.pcbi.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kemp AJ, Betney R, Ciandrini L, Schwenger AC, Romano MC, Stansfield I. A yeast tRNA mutant that causes pseudohyphal growth exhibits reduced rates of CAG codon translation. Molecular microbiology. 2013;87:284–300. doi: 10.1111/mmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M, Guo J, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495:111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 7.Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Molecular systems biology. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall E, Stansfield I, Romano MC. Ribosome recycling induces optimal translation rate at low ribosomal availability, Journal of the Royal Society. Interface/the Royal Society. 2014;11:20140589. doi: 10.1098/rsif.2014.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annual review of biochemistry. 2014;83:779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 11.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nature structural & molecular biology. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 12.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and molecular biology reviews : MMBR. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. first page of table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Algire MA, Lorsch JR. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr Opin Chem Biol. 2006;10:480–486. doi: 10.1016/j.cbpa.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Advances in protein chemistry and structural biology. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 18.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harbor perspectives in biology. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nurenberg E, Tampe R. Tying up loose ends: ribosome recycling in eukaryotes and archaea. Trends Biochem Sci. 2013;38:64–74. doi: 10.1016/j.tibs.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Franckenberg S, Becker T, Beckmann R. Structural view on recycling of archaeal and eukaryotic ribosomes after canonical termination and ribosome rescue. Current opinion in structural biology. 2012;22:786–796. doi: 10.1016/j.sbi.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Voigts-Hoffmann F, Klinge S, Ban N. Structural insights into eukaryotic ribosomes and the initiation of translation. Current opinion in structural biology. 2012;22:768–777. doi: 10.1016/j.sbi.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Jenner L, Melnikov S, Garreau de Loubresse N, Ben-Shem A, Iskakova M, Urzhumtsev A, Meskauskas A, Dinman J, Yusupova G, Yusupov M. Crystal structure of the 80S yeast ribosome. Current opinion in structural biology. 2012;22:759–767. doi: 10.1016/j.sbi.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harbor perspectives in biology. 2012;4 doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreier MH, Erni B, Staehelin T. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. Journal of molecular biology. 1977;116:727–753. doi: 10.1016/0022-2836(77)90268-6. [DOI] [PubMed] [Google Scholar]
- 26.Schreier MH, Staehelin T. Translation of rabbit hemoglobin meessenger RNA in vitro with purified and partially purified components from brain or liver of different species. Proc Natl Acad Sci U S A. 1973;70:462–465. doi: 10.1073/pnas.70.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safer B, Adams SL, Kemper WM, Berry KW, Lloyd M, Merrick WC. Purification and characterization of two initiation factors required for maximal activity of a highly fractionated globin mRNA translation system. Proc Natl Acad Sci U S A. 1976;73:2584–2588. doi: 10.1073/pnas.73.8.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benne R, Hershey JW. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J Biol Chem. 1978;253:3078–3087. [PubMed] [Google Scholar]
- 29.Adams SL, Safer B, Anderson WF, Merrick WC. Eukaryotic initiation complex formation. Evidence for two distinct pathways. J Biol Chem. 1975;250:9083–9089. [PubMed] [Google Scholar]
- 30.Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 31.Fraser CS, Doudna JA. Quantitative studies of ribosome conformational dynamics. Q Rev Biophys. 2007;40:163–189. doi: 10.1017/S0033583507004647. [DOI] [PubMed] [Google Scholar]
- 32.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & development. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, Ban N. Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hussain T, Llácer Jose L, Fernández Israel S, Munoz A, Martin-Marcos P, Savva Christos G, Lorsch Jon R, Hinnebusch Alan G, Ramakrishnan V. Structural Changes Enable Start Codon Recognition by the Eukaryotic Translation Initiation Complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nature structural & molecular biology. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- 38.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell. 2013;153:1108–1119. doi: 10.1016/j.cell.2013.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Molecular cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Nanda JS, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J Biol Chem. 2013;288:5316–5329. doi: 10.1074/jbc.M112.440693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Molecular cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 43.Fraser CS, Hershey JW, Doudna JA. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nature structural & molecular biology. 2009;16:397–404. doi: 10.1038/nsmb.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SH. Molecular architecture of a eukaryotic translational initiation complex. Science. 2013;342:1240585. doi: 10.1126/science.1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. Embo J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. Journal of molecular biology. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 49.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Molecular cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 50.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. Journal of molecular biology. 2003;330:917–924. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- 51.Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. Journal of molecular biology. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sokabe M, Fraser CS. Human Eukaryotic Initiation Factor 2 (eIF2)-GTP-Met-tRNAi Ternary Complex and eIF3 Stabilize the 43S Preinitiation Complex. J Biol Chem. 2014;289:31827–31836. doi: 10.1074/jbc.M114.602870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Molecular cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 54.Duncan R, Hershey JW. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem. 1983;258:7228–7235. [PubMed] [Google Scholar]
- 55.Singh CR, Udagawa T, Lee B, Wassink S, He H, Yamamoto Y, Anderson JT, Pavitt GD, Asano K. Change in nutritional status modulates the abundance of critical preinitiation intermediate complexes during translation initiation in vivo. Journal of molecular biology. 2007;370:315–330. doi: 10.1016/j.jmb.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 57.von der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Molecular microbiology. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 58.Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. An in vivo control map for the eukaryotic mRNA translation machinery. Molecular systems biology. 2013;9:635. doi: 10.1038/msb.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. Real-time assembly landscape of bacterial 30S translation initiation complex. Nature structural & molecular biology. 2012;19:609–615. doi: 10.1038/nsmb.2285. [DOI] [PubMed] [Google Scholar]
- 60.Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–393. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes & development. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokabe M, Fraser CS, Hershey JW. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res. 2012;40:905–913. doi: 10.1093/nar/gkr772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci U S A. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Molecular cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011;108:E1392–1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Molecular cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 67.Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Molecular cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 68.Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature. 1974;251:385–388. doi: 10.1038/251385a0. [DOI] [PubMed] [Google Scholar]
- 69.Godefroy-Colburn T, Thach RE. The role of mRNA competition in regulating translation. IV. Kinetic model. J Biol Chem. 1981;256:11762–11773. [PubMed] [Google Scholar]
- 70.Godefroy-Colburn T, Ravelonandro M, Pinck L. Cap accessibility correlates with the initiation efficiency of alfalfa mosaic virus RNAs. European journal of biochemistry/FEBS. 1985;147:549–552. doi: 10.1111/j.0014-2956.1985.00549.x. [DOI] [PubMed] [Google Scholar]
- 71.Godefroy-Colburn T, Thivent C, Pinck L. Translational discrimination between the four RNAs of alfalfa mosaic virus. A quantitative evaluation. European journal of biochemistry/FEBS. 1985;147:541–548. doi: 10.1111/j.0014-2956.1985.00541.x. [DOI] [PubMed] [Google Scholar]
- 72.Livingstone M, Atas E, Meller A, Sonenberg N. Mechanisms governing the control of mRNA translation. Physical biology. 2010;7:021001. doi: 10.1088/1478-3975/7/2/021001. [DOI] [PubMed] [Google Scholar]
- 73.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. Rna. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A. 2013;110:5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aviran S, Trapnell C, Lucks JB, Mortimer SA, Luo S, Schroth GP, Doudna JA, Arkin AP, Pachter L. Modeling and automation of sequencing-based characterization of RNA structure. Proc Natl Acad Sci U S A. 2011;108:11069–11074. doi: 10.1073/pnas.1106541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci U S A. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome–wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes & development. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 80.Searfoss A, Dever TE, Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Molecular and cellular biology. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tahara SM, Morgan MA, Shatkin AJ. Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1981;256:7691–7694. [PubMed] [Google Scholar]
- 82.Grifo JA, Tahara SM, Morgan MA, Shatkin AJ, Merrick WC. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983;258:5804–5810. [PubMed] [Google Scholar]
- 83.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 84.Rau M, Ohlmann T, Morley SJ, Pain VM. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 85.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Molecular and cellular biology. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Molecular cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 87.Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2014 doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 88.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes & development. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Igreja C, Peter D, Weiler C, Izaurralde E. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nature communications. 2014;5:4790. doi: 10.1038/ncomms5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lukhele S, Bah A, Lin H, Sonenberg N, Forman-Kay JD. Interaction of the eukaryotic initiation factor 4E with 4E-BP2 at a dynamic bipartite interface. Structure. 2013;21:2186–2196. doi: 10.1016/j.str.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 91.Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Molecular cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lanker S, Muller PP, Altmann M, Goyer C, Sonenberg N, Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992;267:21167–21171. [PubMed] [Google Scholar]
- 93.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Molecular cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 94.Maquat LE, Hwang J, Sato H, Tang Y. CBP80-promoted mRNP rearrangements during the pioneer round of translation, nonsense-mediated mRNA decay, and thereafter. Cold Spring Harbor symposia on quantitative biology. 2010;75:127–134. doi: 10.1101/sqb.2010.75.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nelson EM, Winkler MM. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- 96.Sonenberg N, Morgan MA, Merrick WC, Shatkin AJ. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc Natl Acad Sci U S A. 1978;75:4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 98.Matsuo H, Li H, McGuire AM, Fletcher CM, Gingras AC, Sonenberg N, Wagner G. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nature structural biology. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 99.Niedzwiecka A, Marcotrigiano J, Stepinski J, Jankowska-Anyszka M, Wyslouch-Cieszynska A, Dadlez M, Gingras AC, Mak P, Darzynkiewicz E, Sonenberg N, Burley SK, Stolarski R. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. Journal of molecular biology. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 100.Slepenkov SV, Darzynkiewicz E, Rhoads RE. Stopped-flow kinetic analysis of eIF4E and phosphorylated eIF4E binding to cap analogs and capped oligoribonucleotides: evidence for a one-step binding mechanism. J Biol Chem. 2006;281:14927–14938. doi: 10.1074/jbc.M601653200. [DOI] [PubMed] [Google Scholar]
- 101.Slepenkov SV, Korneeva NL, Rhoads RE. Kinetic mechanism for assembly of the m7GpppG.eIF4E.eIF4G complex. J Biol Chem. 2008;283:25227–25237. doi: 10.1074/jbc.M801786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.O’Leary SE, Petrov A, Chen J, Puglisi JD. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure. 2013;21:2197–2207. doi: 10.1016/j.str.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carberry SE, Darzynkiewicz E, Goss DJ. A comparison of the binding of methylated cap analogues to wheat germ protein synthesis initiation factors 4F and (iso)4F. Biochemistry. 1991;30:1624–1627. doi: 10.1021/bi00220a026. [DOI] [PubMed] [Google Scholar]
- 104.Friedland DE, Wooten WN, LaVoy JE, Hagedorn CH, Goss DJ. A mutant of eukaryotic protein synthesis initiation factor eIF4E(K119A) has an increased binding affinity for both m7G cap analogues and eIF4G peptides. Biochemistry. 2005;44:4546–4550. doi: 10.1021/bi047645m. [DOI] [PubMed] [Google Scholar]
- 105.Carberry SE, Friedland DE, Rhoads RE, Goss DJ. Binding of protein synthesis initiation factor 4E to oligoribonucleotides: effects of cap accessibility and secondary structure. Biochemistry. 1992;31:1427–1432. doi: 10.1021/bi00120a020. [DOI] [PubMed] [Google Scholar]
- 106.Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. Embo J. 2011;30:302–316. doi: 10.1038/emboj.2010.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. Embo J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 109.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. Embo J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. Translation initiation factors eIF-iso4G and eIF-4B interact with the poly(A)-binding protein and increase its RNA binding activity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 112.Groft CM, Burley SK. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Molecular cell. 2002;9:1273–1283. doi: 10.1016/s1097-2765(02)00555-5. [DOI] [PubMed] [Google Scholar]
- 113.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Molecular cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 114.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-Poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J Biol Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 115.Wei CC, Balasta ML, Ren J, Goss DJ. Wheat germ poly(A) binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry. 1998;37:1910–1916. doi: 10.1021/bi9724570. [DOI] [PubMed] [Google Scholar]
- 116.Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- 117.Gorlach M, Burd CG, Dreyfuss G. The determinants of RNA-binding specificity of the heterogeneous nuclear ribonucleoprotein C proteins. J Biol Chem. 1994;269:23074–23078. [PubMed] [Google Scholar]
- 118.Amrani N, Ghosh S, Mangus DA, Jacobson A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J Biol Chem. 2012;287:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 121.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 122.Ozes AR, Feoktistova K, Avanzino BC, Fraser CS. Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. Journal of molecular biology. 2011;412:674–687. doi: 10.1016/j.jmb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 2011;39:2678–2689. doi: 10.1093/nar/gkq1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harms U, Andreou AZ, Gubaev A, Klostermeier D. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 2014;42:7911–7922. doi: 10.1093/nar/gku440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Y, Atas E, Lindqvist LM, Sonenberg N, Pelletier J, Meller A. Single-molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure. 2014;22:941–948. doi: 10.1016/j.str.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 126.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39:2260–2270. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shahbazian D, Parsyan A, Petroulakis E, Hershey J, Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell cycle. 2010;9:4106–4109. doi: 10.4161/cc.9.20.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A. 2013;110:13339–13344. doi: 10.1073/pnas.1303781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ, Clemens MJ. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J Biol Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- 130.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 131.Babendure JR, Babendure JL, Ding JH, Tsien RY. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12:851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaye NM, Emmett KJ, Merrick WC, Jankowsky E. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J Biol Chem. 2009;284:17742–17750. doi: 10.1074/jbc.M109.009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kozak M, Shatkin AJ. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J Biol Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- 134.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. Embo J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- 136.Muckenthaler M, Gray NK, Hentze MW. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Molecular cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 137.Ma J, Haldar S, Khan MA, Sharma SD, Merrick WC, Theil EC, Goss DJ. Fe2+ binds iron responsive element-RNA, selectively changing protein-binding affinities and regulating mRNA repression and activation. Proc Natl Acad Sci U S A. 2012;109:8417–8422. doi: 10.1073/pnas.1120045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Villa N, Do A, Hershey JW, Fraser CS. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem. 2013;288:32932–32940. doi: 10.1074/jbc.M113.517011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hinton TM, Coldwell MJ, Carpenter GA, Morley SJ, Pain VM. Functional analysis of individual binding activities of the scaffold protein eIF4G. J Biol Chem. 2007;282:1695–1708. doi: 10.1074/jbc.M602780200. [DOI] [PubMed] [Google Scholar]
- 140.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Harris TE, Chi A, Shabanowitz J, Hunt DF, Rhoads RE, Lawrence JC., Jr mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. Embo J. 2006;25:1659–1668. doi: 10.1038/sj.emboj.7601047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valasek L, Donahue TF, Hinnebusch AG. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. Embo J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Walker SE, Zhou F, Mitchell SF, Larson VS, Valasek L, Hinnebusch AG, Lorsch JR. Yeast eIF4B binds to the head of the 40S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA. 2013;19:191–207. doi: 10.1261/rna.035881.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Majumdar R, Bandyopadhyay A, Maitra U. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40 S preinitiation complex. J Biol Chem. 2003;278:6580–6587. doi: 10.1074/jbc.M210357200. [DOI] [PubMed] [Google Scholar]
- 145.Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40 S ribosomal preinitiation complex. J Biol Chem. 1999;274:17975–17980. doi: 10.1074/jbc.274.25.17975. [DOI] [PubMed] [Google Scholar]
- 146.Trachsel H, Erni B, Schreier MH, Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. Journal of molecular biology. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- 147.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. Rna. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chiu WL, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Molecular and cellular biology. 2010;30:4415–4434. doi: 10.1128/MCB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jivotovskaya AV, Valasek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Molecular and cellular biology. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Jr, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, Hinnebusch AG, Lorsch JR. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 152.Palam LR, Baird TD, Wek RC. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J Biol Chem. 2011;286:10939–10949. doi: 10.1074/jbc.M110.216093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dasso MC, Milburn SC, Hershey JW, Jackson RJ. Selection of the 5′-proximal translation initiation site is influenced by mRNA and eIF-2 concentrations. European journal of biochemistry/FEBS. 1990;187:361–371. doi: 10.1111/j.1432-1033.1990.tb15313.x. [DOI] [PubMed] [Google Scholar]
- 154.Andreou AZ, Klostermeier D. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. Journal of molecular biology. 2014;426:51–61. doi: 10.1016/j.jmb.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 155.Bi X, Ren J, Goss DJ. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry. 2000;39:5758–5765. doi: 10.1021/bi992322p. [DOI] [PubMed] [Google Scholar]
- 156.Park EH, Walker SE, Zhou F, Lee JM, Rajagopal V, Lorsch JR, Hinnebusch AG. Yeast eukaryotic initiation factor 4B (eIF4B) enhances complex assembly between eIF4A and eIF4G in vivo. J Biol Chem. 2013;288:2340–2354. doi: 10.1074/jbc.M112.398537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Spirin AS. How does a scanning ribosomal particle move along the 5′-untranslated region of eukaryotic mRNA? Brownian Ratchet model. Biochemistry. 2009;48:10688–10692. [Google Scholar]
- 158.Zhou F, Walker SE, Mitchell SF, Lorsch JR, Hinnebusch AG. Identification and characterization of functionally critical, conserved motifs in the internal repeats and N-terminal domain of yeast translation initiation factor 4B (yeIF4B) J Biol Chem. 2014;289:1704–1722. doi: 10.1074/jbc.M113.529370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park HS, Browning KS, Hohn T, Ryabova LA. Eucaryotic initiation factor 4B controls eIF3-mediated ribosomal entry of viral reinitiation factor. Embo J. 2004;23:1381–1391. doi: 10.1038/sj.emboj.7600140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Methot N, Song MS, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Molecular and cellular biology. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. Embo J. 2004;23:1761–1769. doi: 10.1038/sj.emboj.7600193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 163.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, Hershey JW, Blenis J, Pende M, Sonenberg N. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. Embo J. 2006;25:2781–2791. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shagam LI, Terenin IM, Andreev DE, Dunaevsky JE, Dmitriev SE. In vitro activity of human translation initiation factor eIF4B is not affected by phosphomimetic amino acid substitutions S422D and S422E. Biochimie. 2012;94:2484–2490. doi: 10.1016/j.biochi.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 165.Richter-Cook NJ, Dever TE, Hensold JO, Merrick WC. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- 166.Richter NJ, Rogers GW, Jr, Hensold JO, Merrick WC. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- 167.Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- 168.Kozak M. Evaluation of the “scanning model” for initiation of protein synthesis in eucaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- 169.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & development. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Soto-Rifo R, Rubilar PS, Limousin T, de Breyne S, Decimo D, Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. Embo J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Molecular cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen CU, Pestova TV, Svitkin YV, Sonenberg N. The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci U S A. 2009;106:22217–22222. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Molecular cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 176.Milon P, Rodnina MV. Kinetic control of translation initiation in bacteria. Critical reviews in biochemistry and molecular biology. 2012;47:334–348. doi: 10.3109/10409238.2012.678284. [DOI] [PubMed] [Google Scholar]
- 177.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 178.Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 180.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- 181.Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two Structurally Atypical HEAT Domains in the C-Terminal Portion of Human eIF4G Support Binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 182.Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Molecular cell. 2012;48:375–386. doi: 10.1016/j.molcel.2012.09.001. [DOI] [PubMed] [Google Scholar]





