Abstract
Autophagy regulates cell differentiation, proliferation and survival in multiple cell types, including cells of the immune system. Herein we examined the effects of a disruption of autophagy on the differentiation of invariant natural killer T cells (iNKT cells). Using mice with a T lymphocyte specific deletion of Atg5 or Atg7,two members of the macroautophagic pathway, we observed a profound decrease in the iNKT cell population. The deficit is cell-autonomous, and it acts predominantly to reduce the number of mature cells, as well as the function of peripheral iNKT cells. In the absence of autophagy, there is reduced progression of iNKT cells in the thymus through the cell cycle, as well as increased apoptosis of these cells. Importantly, the reduction in Th1-biased iNKT cells is most pronounced, leading to a selective reduction in iNKT cell derived IFNγ. Our findings highlight the unique metabolic and genetic requirements for the differentiation of iNKT cells.
Keywords: Autophagy, T lymphocyte, thymus, differentiation, cytokines
Introduction
Autophagy is an evolutionarily conserved catabolic process in which cells sequester cytoplasmic components and deliver the cargo to lysosomes for degradation and recycling (1, 2). Three types of autophagy have been described: macroautophagy, microautophagy and chaperone mediated autophagy (1, 3). Of these forms, macroautophagy, hereafter referred to as autophagy, is the major pathway, and it is the most extensively investigated. Macroautophagy is characterized by the compartmentalization of cytoplasmic molecules or organelles in double-membrane vesicles, termed autophagosomes (1, 3–5). Over thirty autophagy-related gene products (ATG proteins) have been identified in yeast, with orthologs in mammals. The core autophagic machinery includes two ubiquitin-like conjugation systems that contribute to the elongation of autophagic vacuoles. One consists of ATG7, an E1 ubiquitin ligase-like enzyme that participates in the lipid modification of ATG8/LC3 (yeast protein/mammalian ortholog). The other is formed by ATG7-mediated catalysis of ATG12 conjugation to ATG5. The ATG5-ATG12 complex is further non-covalently associated with ATG16/ATG16L (yeast protein/mammalian ortholog) to form a macromolecular complex with E3 ligase-like activity (1, 2, 6–8).
Autophagy plays a role in recycling cytoplasmic materials and is an alternative source for energy production under conditions of nutrition shortage or energy loss. In addition, autophagy has been verified in different contexts to control cell growth, proliferation and differentiation, cell death and functional responses to various stimuli (4, 5, 9–12). In the immune system, naïve T cells maintain a basal level of autophagy that is further induced upon activation through the T cell antigen receptor (TCR) (13–17). Upon depletion of components of the autophagy pathway, conventional T cells exhibited impaired development and homeostasis, with decreased cell numbers and increased markers of cell death (13, 14, 16, 18–20).
Natural killer T (NKT) cells are a T lymphocyte sub-population that shares the properties of both natural killer (NK) cells and T lymphocytes (21–23). The majority of NKT cells in mice express a semi-invariant TCR, with an invariant Vα14-Jα18 chain coupled with a Vβ repertoire consisting mostly of Vβ8, Vβ7 and Vβ2. This major population of NKT cells is often referred to as type I or invariant NKT cells (iNKT cells) (21, 22, 24, 25). iNKT cells recognize lipid antigens (Ag) presented by CD1d, a MHC class I-like Ag presenting molecule (26–29). iNKT cells have the capacity to rapidly secrete large amount of cytokines after activation, consequently amplifying innate and adaptive immune responses (21, 22, 27). The results from recent studies indicate that a functional subset of iNKT cells is dedicated to producing IL-17, and also that there are subsets more prone to the production of Th1 or Th2 cytokines (30–34).
iNKT cells develop in the thymus, but branch off from the main stream of T cell differentiation at the double positive (DP) stage (21, 22, 24, 35–37). Although many factors that contribute to the regulation of iNKT cell development have been identified, the determinants that direct iNKT cell development are still not fully understood (35, 36). It is notable that iNKT cell differentiation requires positive selection by CD1d expressing DP thymocytes rather than cortical epithelial cells (38). iNKT cells also require a number of cell surface molecules, kinases, adaptors, transcription factors and chromatin modifying proteins that are much less important for the differentiation of MHC class I and class II restricted thymocytes (39).
Considering the requirement for autophagy in cell growth and proliferation, we analyzed the development of iNKT cells in mice with a conditional knockout of Atg5 or Atg7. Our results show that a lack of either of these two autophagy genes causes a severe defect in iNKT cells, with selective effects on the subset that produces IFNγ.
Materials and Methods
Mice and reagents
The generation of Atg5f/f and Atg7f/f has been previously described (40, 41). The Atg5f/f mice were a kind gift from Dr. Noboru Mizushima (Tokyo Medical and Dental University, Japan) and the Atg7f/f mice were kindly provided by Dr. Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science, Japan). CD4-Cre transgenic mice, C57BL/6J (B6) and CD45.1 congenic B6.SJL strains were purchased from The Jackson Laboratory and Lck-Cre transgenic mice were obtained from Taconic Farms. The Atg5f/f mice and Atg7f/f mice were bred to CD4-Cre or Lck-Cre mice and littermates were used for analysis and comparison. B6 mice were crossed with the B6.SJL mice to generate CD45.1+CD45.2+ heterozygotes. Mice were maintained under specific pathogen-free conditions, and the experiments were approved by the Institutional Animal Care and Use Committee of the La Jolla Institute for Allergy & Immunology.
Antibodies and reagents
The following antibodies, with clone designation in parentheses, were from BD PharMingen: CD1d–PE (1B1), CD4-APC (RM4–5), CD8-PerCP-Cy5.5 (53–6.7), CD24-FITC (M1/69), CD45.1-FITC (A20), Fas-FITC (Jo2), anti-BrdU-Alexa Fluor 488 (3D4), IFNγ-PE-Cy7 (XMG1.2), IL-4-Alexa Fluor 647 (11B11), Ki-67-PE (B56), NK1.1-PE-Cy7 (PK136), GATA3-PE-Cy7 (L50-823), phospho-Akt(pS473)-PE (M89-61), phospho-Akt(pT308)-PE (J1–223.371) and purified antibody anti-active caspase 3 (C92-605). CD45.2-APC (104), RORγt-PE (B2D) and TCRβ-APC-eFluor 780 (H57-597) were bought from eBioscience (San Diego, CA). CD44-Alexa Fluor 700 (IM7) and CD69 Alexa Fluor 647 (H1.2F3) were obtained from BioLegend (San Diego, CA). p21cip1-Alexa Fluor 647 (C-19), T-bet-Alexa Fluor 488 (4B10) and PLZF-Alexa Fluor 647 (D-9) were from Santa Cruz Biotechnology (Santa Cruz, CA). CD19-PE-Texas Red (6D5) and second antibody goat anti-rabbit IgG (H+L)-AF488 were from Invitrogen (Carlsbad, CA). Purified antibodies recognizing cleaved caspase 8 (D5B2) or phospho-4E-BP1(pT37/pT46) (236B4) were purchased from Cell Signaling Technology (Danvers, MA). Cytofix/Cytoperm buffer, Perm/Wash buffer and Transcription Factor Buffer Set were all from BD Biosciences. αGalCer was kindly provided by Kyowa Hakko Kirin. Live/Dye (Yellow) was obtained from Invitrogen (Carlsbad, CA), and 5-Bromo-deoxyuridine (BrdU) and Annexin V-APC were from BD PharMingen.
Flow cytometry
Thymus, spleen and liver were collected and single cell suspensions were prepared. For cell surface staining, after blocking in staining buffer (PBS, 2% BSA, 10 mM EDTA, and 0.1% sodium azide) containing anti-FcR antibody (2.4G2) for 30 min at 4°C, cells were stained with fluorophore-conjugated antibodies and fixed with fixation buffer (PBS, 1% paraformaldehyde, and 0.1% sodium azide). To stain intracellular cytokines and transcription factors, after cell surface staining, cells were treated with Cytofix/Cytoperm buffer and Transcription Factor buffer, respectively, followed by staining with corresponding fluorophore-conjugated antibodies in Perm/Wash buffer. The data were acquired on an LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). The iNKT cell population was gated as CD19−, TCRβ+ and CD1d-tetramer+ cells.
Mixed bone marrow chimeras
Bone marrow cells were prepared from tibias and femurs, and Thy1+ cells were removed by MACS microbeads (Miltenyi Biotec, Auburn, CA). 10 × 106 bone marrow cells with a 1:1 ratio of B6.SJL (CD45.1+) and either wild type controls (CD45.2+) or Atg5f/f CD4-Cre or Atg7f/f CD4-Cre (CD45.2+) were injected i.v. to eight-to-ten-week old B6 mice (CD45.1+CD45.2+) that had been subjected to twice to 600 Rads irradiation with an X-Ray Irradiator, with 3h between doses. Mice were analyzed 11–12 weeks post bone marrow transfer.
In vitro culture and apoptosis analysis
Thymocytes were purified and single cell suspensions were prepared. 20 × 106 cells were inoculated in 1 ml RPMI-1640 medium supplemented with 10% fetal bovine serum, 50 µM 2-mercaptoethanol as well as antibiotics, and cultured overnight at 37°C in a humidified atmosphere of 5% CO2. Cells were collected and stained with cell surface markers to gate either DP thymocytes for some experiments or iNKT cells in others, as well as with Annexin V and Live/Dead Dye Yellow (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions.
Semi-quantitative PCR
Single cell suspensions of thymocytes were prepared and stained. DP thymocytes were sorted with the αGalCer CD1d-tetramer positive cells removed. Total mRNA was purified by using TriZOL (Invitrogen) and reverse transcribed to cDNA. PCR was carried out with titrated cDNA templates at concentrations of 200 ng, 60 ng and 20 ng. The primer pairs used were the following: forward primer for Vα14-Jα18: 5’-GTGTCCCTGACAGTCCTGGT-3’ and reverse: 5’-CAAAATGCAGCCTCCCTAAG-3’; forward primer for Cα: 5’-CCTCTGCCTGTTCACCGACTT-3’ and reverse: 5’-TGGCGTTGGTCTCTTTGAAG-3’. The PCR program consisted of an initial denaturation step at 95 °C, 4 min; 38 cycles for Vα14-Jα18 or 28 cycles for Cα at 95 °C for 15 sec, 55 °C for 15 sec, 72 °C for 15 sec; and a final extension for 7 min at 72 °C. The PCR products were analyzed by electrophoresis on a 2% agarose gel.
Autophagy and mitochondrial mass detection
After staining with cell surface markers, thymocytes were washed twice with PBS, resuspended with pre-warmed PBS containing 0.1 µM MitoTracker Green FM (Invitrogen, Carlsbad, CA) or DMSO as a control, and incubated at 37°C for 30 min. The cells were washed with warm PBS twice prior to flow cytometry analysis. Similar procedures were carried out for detection of mitochondrial superoxide, except staining was done with freshly made 5 µM MitoSOX Red (Invitrogen, Carlsbad, CA) in pre-warmed HBSS/Ca2+/Mg2+ medium. Thymocytes were stained with the Cyto-ID® Autophagy detection kit (Enzo Life Sciences, Farmingdale, NY) according to the manufacturer’s instructions.
Proliferation and cell cycle analysis
To measure cell turnover in vivo, six-week-old female mice were injected i.v. with 2 mg BrdU and 5 h after injection thymus tissue was collected and a single cell suspension of thymocytes was prepared. Cells were stained with surface markers, followed by staining for BrdU and Ki-67 according to the manufacturer’s manual. For analysis of cell cycle, 2 mg BrdU was introduced into each mouse intravenously. 2 h later, single cell suspensions of thymocytes were prepared and stained to evaluate BrdU incorporation and DNA content with 7-aminoactinomycin D (7-AAD. Invitrogen, Carlsbad, CA)
Lipid Ag immunization
Eight-week-old mice were injected i.v. with 1 µg αGalCer. 2 h after injection, spleens were collected and single cell suspensions were prepared. Cells were stained with surface markers and then for intracellular cytokines, followed by analysis by flow cytometry.
Statistical analysis
Two-tailed Student’s t test was used for analysis of statistical significance. p values < 0.05 were considered statistically significant.
Results
Deficiency in autophagy genes caused decreased iNKT cells
Disruption of either ATG5 or ATG7 expression effectively eliminates the majority of autophagic processes (42–44). While a germ line deletion causes neonatal lethality, mice with cell type-specific deletions of Atg5 or Atg7 have been used as models to evaluate the role of autophagy in various physiological processes. Herein, Atg5f/f or Atg7f/f mice were crossed with either CD4-Cre or Lck-Cre mice, producing mice with the gene deletions specifically restricted to T lymphocytes. T lymphocytes continuously undergo autophagy, and previous results showed that when autophagy gene deficient mice were crossed to Lck-Cre transgenic mice the amount of LC3-II formed in T cells was greatly decreased, indicating that autophagy was highly impaired (14, 18).
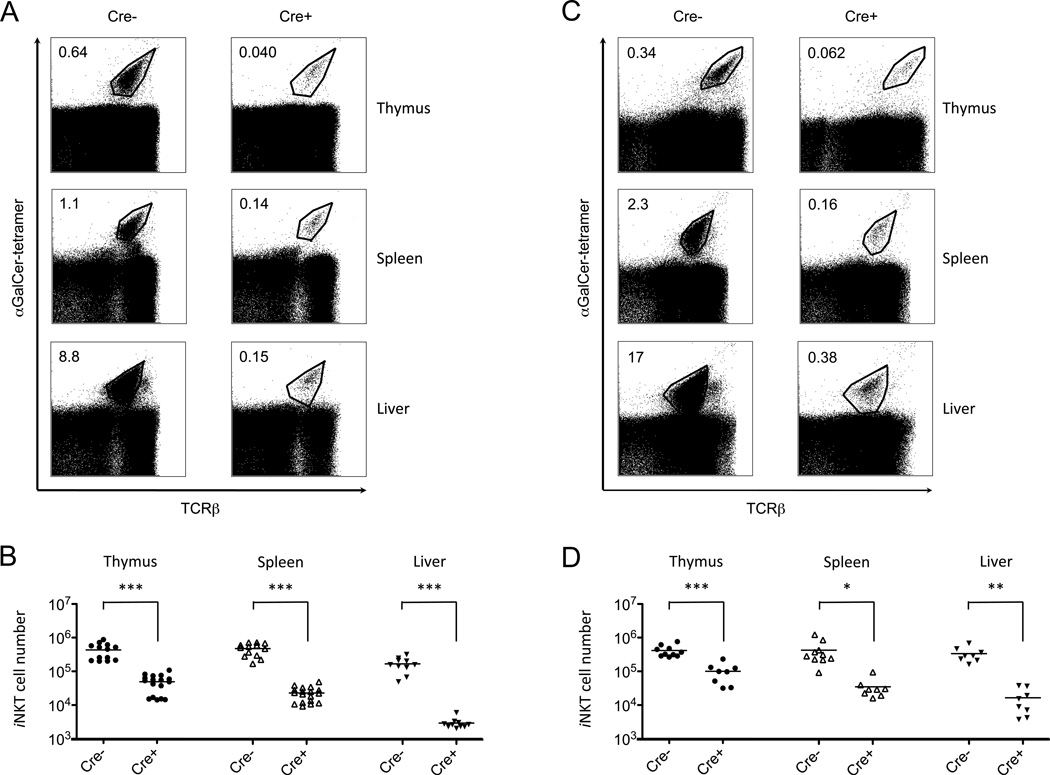
We analyzed iNKT cells from thymus, spleen and liver of eight-to-nine-week old mice. Mice with deletion of either Atg5 or Atg7 had a dramatic reduction in both the percentage and absolute number of iNKT cells compared with controls that do not express Cre recombinase (Fig. 1 and data not shown), although there were subtle differences in the extent of the reduction, depending on the Atg gene deleted and the promoter controlling T cell-specific Cre expression. The effect of Atg5 or Atg7 deficiency on iNKT cells also was somewhat organ-specific, with a relatively smaller decrease in cell frequency in the thymus and the most profound defect occurring in liver (Table 1).
Figure 1.
Mice deficient for autophagy genes had reduced iNKT cells. iNKT cells were analyzed by gating on CD1d tetramer+ cells as described in Experimental Procedures. Depicted are the results from cells from the indicated organs from eight-to-nine-week old mice with a CD4-Cre mediated deletion of Atg5 (A,B) or Atg7 (C,D) and controls. Shown are the percentages (A,C) and absolute cell numbers (B,D) compared with Atg5f/f or Atg7f/f mice without the Cre transgene. Representative flow cytometry analyses are from one of at least three independent experiments. *p<0.01, **p<0.005, ***p<0.0001, n≥8.
Table 1.
Effects on the population size of iNKT cells and conventional T cells in autophagy deficient mice compared with wild type controls.
| Atg5f/f CD4-Cre | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| iNKT cells | DP thymocytes | CD4 T cells | CD8 T cells | |||||||
| Thymus | Spleen | Liver | Thymus | Spleen | Liver | Thymus | Spleen | Liver | ||
| Reduction folds (Cre−/Cre+) | 8.8a | 20a | 56a | 1.1 | 1.2 | 2.9a | 1.7c | 1.1 | 3.1a | 1.5c |
| Atg7f/f CD4-Cre | ||||||||||
| iNKT cells | DP thymocytes | CD4 T cells | CD8 T cells | |||||||
| Thymus | Spleen | Liver | Thymus | Spleen | Liver | Thymus | Spleen | Liver | ||
| Reduction folds (Cre−/Cre+) | 4.1a | 12d | 20c | 1.1 | 1.1 | 1.2 | 1.4 | 1.0 | 1.5e | 0.97 |
| Atg5f/f Lck-Cre | ||||||||||
| iNKT cells | DP thymocytes | CD4 T cells | CD8 T cells | |||||||
| Thymus | Spleen | Liver | Thymus | Spleen | Liver | Thymus | Spleen | Liver | ||
| Reduction folds (Cre−/Cre+) | 15a | 21a | 35a | 2.2d | 2.8c | 3.9b | 3.4b | 2.7c | 3.7b | 2.2c |
| Atg7f/f Lck-Cre | ||||||||||
| iNKT cells | DP thymocytes | CD4 T cells | CD8 T cells | |||||||
| Thymus | Spleen | Liver | Thymus | Spleen | Liver | Thymus | Spleen | Liver | ||
| Reduction folds (Cre−/Cre+) | 13a | 51a | 65c | 3.7b | 3.5b | 3.1b | 2.7e | 3.6b | 7.3b | 3.6c |
Atg5f/f or Atg7f/f crossed with either CD4-Cre or Lck-Cre transgenic mice were analyzed and the average cell numbers of the indicated T cell populations were calculated.
Shown here are the ratios of wild type controls (Cre-) to autophagy deficient iNKT cells and conventional T cells (Cre+). n≥8 in all cases. The differences that reach significance compared to mice wild type for autophagy are indicated
p<0.0001
<0.0005
p<0.005
p<0.01
<0.05
In Atg5f/f CD4-Cre and in Atg7f/f CD4-Cre mice, lack of either of these two autophagy genes did not cause a significant decrease in total thymocyte cellularity (Supplemental Fig. 1). However, consistent with some previous reports (13, 19), there was a reduction in peripheral CD4 and CD8 T cells in Atg5f/f CD4-Cre mice, but this was much smaller in magnitude than the decrease in iNKT cells (Supplemental Fig. 1 and Table 1). In Lck-Cre mice, however, in agreement with earlier reports (18, 20), the total cellularity of the thymus was reduced, which might reflect the earlier expression of the Cre recombinase under the control of this promoter. Similarly, the frequencies of peripheral CD4 and CD8 T cells exhibited more pronounced decreases in Lck-Cre mice. However, in both the CD4-Cre and Lck-Cre strains the reductions in iNKT cells caused by either Atg5 or Atg7 deletion were much more pronounced than the reductions in CD4 and CD8 T cells (Table 1).
Wild type thymocytes were analyzed for autophagy using a dye that detects autophagic vacuoles. By this analysis, iNKT cells exhibited increased autophagy compared to either DP, CD4 single positive or CD8 single positive cells (Supplemental Fig. 2). Therefore, an increased requirement for autophagy correlated with increased autophagy in these cells.
Autophagy related gene deficiency interfered with mature iNKT cell stages
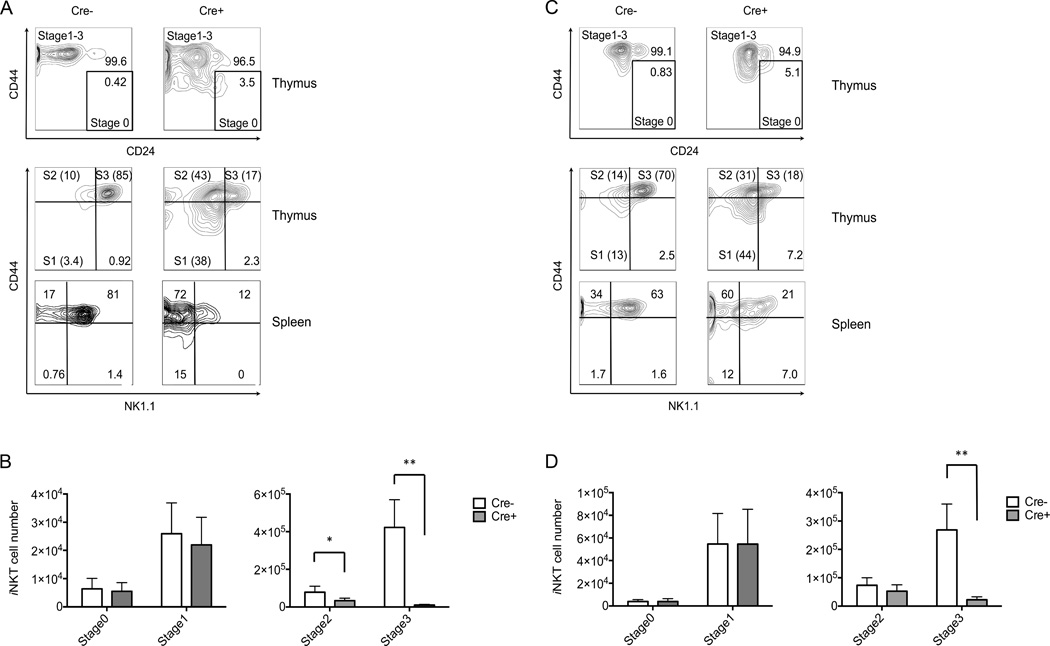
Differentiation of iNKT cells has been divided into four developmental stages, with the most immature or stage 0 cells expressing CD24, but lacking both expression of NK1.1 and a high level of CD44 (45). Stage 1 cells lose CD24 expression (CD24−, CD44low, NK1.1−), while stage 2 cells become CD44high, and finally, stage 3 is characterized by acquisition of NK1.1 expression (CD24−, CD44high, NK1.1+) (21, 24). The final maturation of iNKT cells to stage 3 is not confined to the thymus, however, as cells with a stage 2 phenotype are the majority of iNKT cell recent thymic emigrants, and NK1.1 expression can be acquired in the periphery (46, 47).
In either Atg5 or Atg7 deficient mice thymic and splenic iNKT cells exhibited a dramatic decrease in the number of stage 3 or CD44high, NK1.1+iNKT cells, and a significant but less dramatic decrease for stage 2 cells in Atg5 but not Atg7 deficient mice (Fig. 2). Unless indicated, the data shown in the following figures were derived from CD4-Cre mice, but the results were similar for Lck-Cre strains. While the percentages of iNKT cells in the early stages (stage 0 and 1) were higher in mice deficient for either Atg5 or Atg7, the absolute cell numbers were not significantly different (Fig. 2). These results indicate that ablation of autophagy genes greatly reduced the transition of immature iNKT cells to the more mature stages 2 and 3.
Figure 2.
Deficiency for autophagy genes inhibited iNKT cell maturation. (A,C) Thymus and spleen cells from eight-to-nine-week old mice were analyzed for iNKT cell phenotype. Representative flow cytometry plots from one of four experiments showing gated iNKT cells from Atg5f/f CD4-Cre (A) or Atg7f/f CD4-Cre (C) mice and controls lacking the Cre transgene. Absolute cell numbers of thymic iNKT cells at different stages were shown in (B, Atg5f/f CD4-Cre and D, Atg7f/f CD4-Cre). The analysis of all stages was carried out simultaneously, but because stages 0 and 1 have much fewer cells, for visual clarity their numbers are plotted on a separate graph (left). *p<0.0005, **p<0.0001, n≥10. Error bars are SD.
Autophagy gene mutation caused a cell autonomous defect in iNKT cells
A defect in autophagy genes in T cells could have affected CD1d surface expression or the presentation of positively selecting lipid ligands by thymocytes. However, deletion of either Atg5 or Atg7 did not significantly affect the amount of CD1d on the surface of thymocytes (Supplemental Fig. 3).
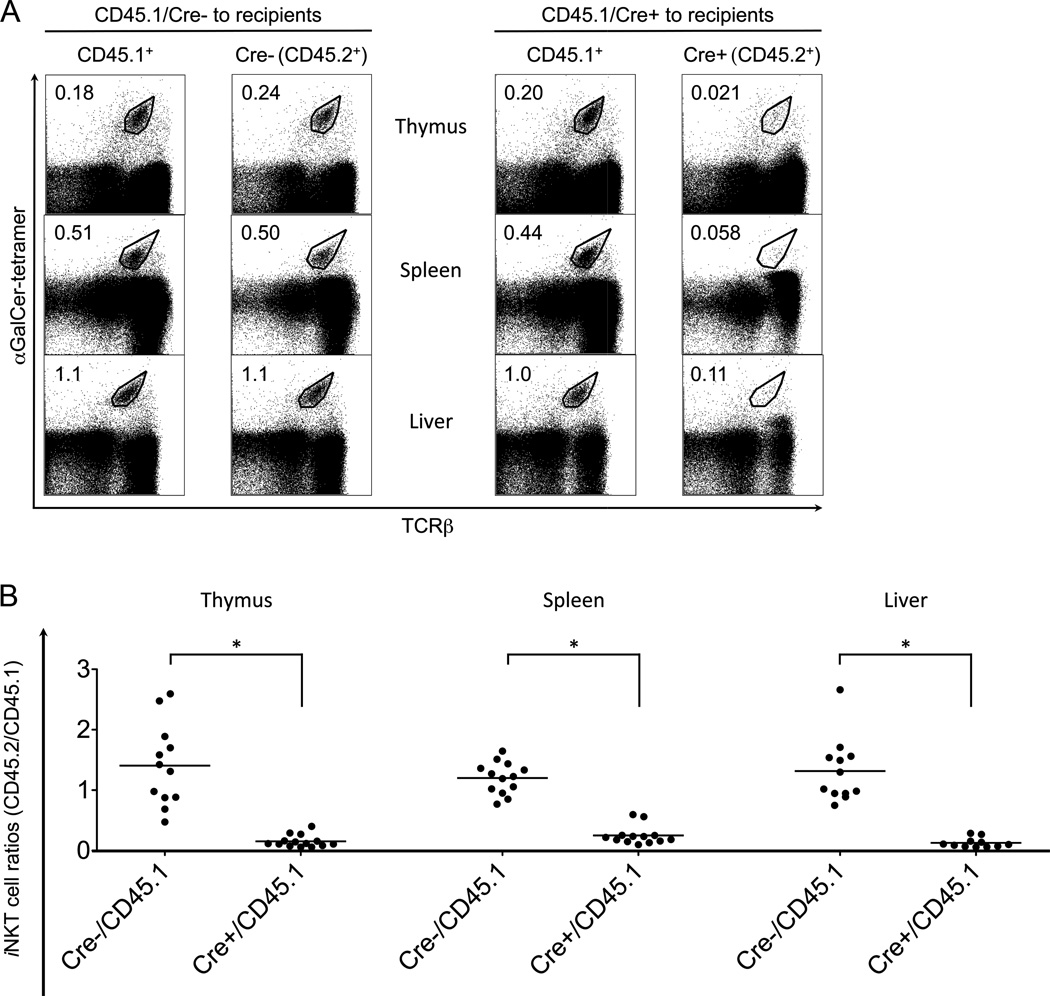
We carried out mixed bone marrow chimera experiments to determine if the decrease in iNKT cells in the autophagy gene deficient mice was cell intrinsic, meaning the problem is located with the iNKT cell progenitor. Alternatively, it could be cell extrinsic, for example due to impaired presentation of a positively selecting ligand, in which case the co-transfer of wild type bone marrow cells should rescue the defect. CD45.2+ marrow cells derived from either autophagy gene deficient mice or controls were transferred together with CD45.1+ wild type bone marrow cells to lethally irradiated recipient mice. After at least eleven weeks, iNKT cells in thymus, spleen and liver were analyzed by flow cytometry. Wild type CD45.1+ and CD45.2+iNKT cells developed equally in the thymus and periphery in the mixed chimeras, while autophagy gene deficient iNKT cells manifested a 4- to 10-fold reduction compared to their wild type counterparts in all three organs tested (Fig. 3 and Atg7 data not shown). These results indicate that the defect in iNKT cells caused by autophagy gene deficiency is cell-intrinsic.
Figure 3.
Cell-intrinsic defects impaired Atg5 deficient iNKT cell development. (A) Bone marrow cells from Atg5f/f CD4-Cre mice or wild type controls (both CD45.2+), together with CD45.1+, wild type bone marrow cells, were co-transferred into recipients. Reconstituted iNKT cells from the indicated organs were analyzed after 11–12 weeks with flow cytometry. Data are representative from one of at least three separate experiments. (B) The ratios of reconstituted iNKT cells (CD45.2+/CD45.1+). *p<0.0001, n>10.
Equivalent viability of DP thymocytes
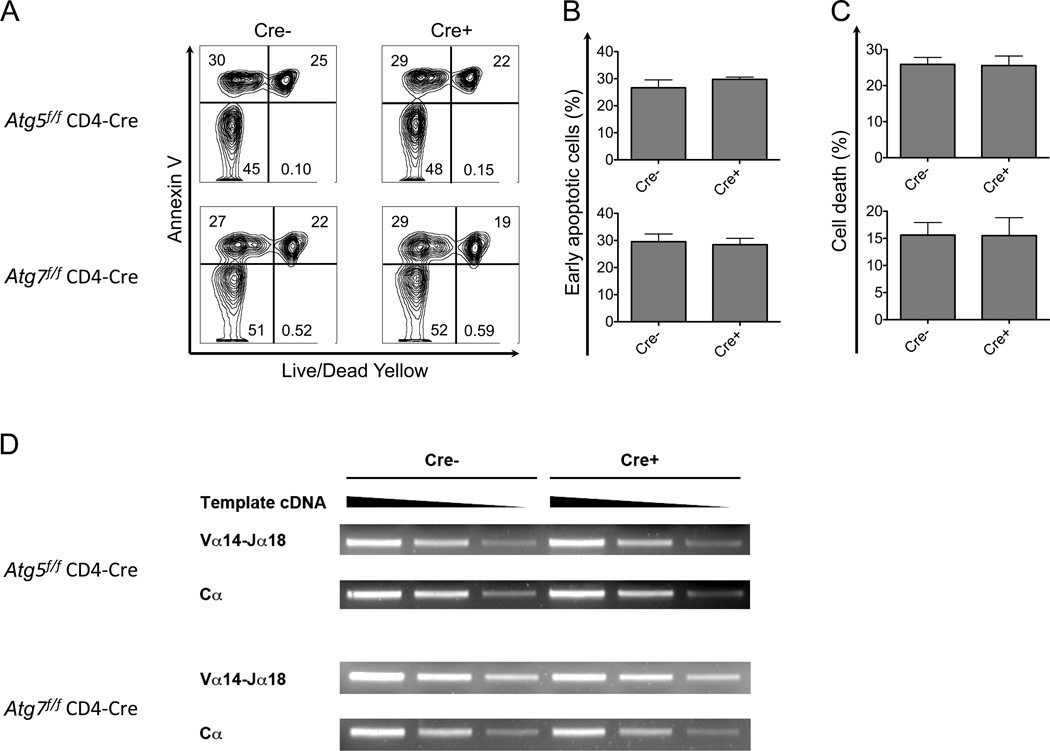
Rearrangements of Jα gene segments distal to the Vα locus, including Jα18, require secondary TCRα rearrangements in DP thymocytes that are undergoing positive selection. The frequency of these secondary rearrangements is dependent on a normal DP thymocyte lifespan (48), and therefore a defect in DP thymocyte survival will cause a cell intrinsic decrease in iNKT cells, due to the negative impact on the frequency of rearrangement of the Jα18 gene segment (48, 49). To determine if an alteration in DP lifespan was the cause of the reduction in iNKT cells, we cultured thymocytes overnight, followed by analysis of cell death by flow cytometry. The data show that the proportions of cells undergoing apoptosis (Annexin V+, Live/Dead Dye not stained) and cells that were already dead, as detected with a vital dye (Live/Dead Dye stained), were very similar for either Atg5 or Atg7-deficient thymocytes and wild type controls (Fig. 4A–C).
Figure 4.
Atg5 or Atg7 deficiency did not cause significant DP thymocyte death and skewed TCR α rearrangements in thymocytes. (A-C) Thymocytes were cultured overnight and cells undergoing apoptosis and dead cells were identified, shown here as flow cytometry analyses (A) and the geometric MFI results indicating cells undergoing apoptosis (B) and dead cells (C). Data in (A) are representative of at least four independent experiments. (B and C) n≥5. Error bars are SD. (D). Equivalent TCR Vα14-Jα18 rearrangements were observed in Atg5 or Atg7 deficient DP thymocytes. cDNA reverse transcribed from total RNA of DP thymocytes depleted of αGalCer-CD1d tetramer+ cells was used as template and titrated at serial dilutions (200 ng, 60 ng and 20 ng) for semi-quantitative PCR. Representative data are from three separate experiments.
If the DP thymocyte lifespan were decreased in vivo in Atg5 or Atg7 deficient mice, we would expect a decrease in the rearrangement of Vα14 to the distal Jα18 locus in DP thymocytes. Therefore we measured Vα14-Jα18 rearrangements in DP thymocytes, which had not yet undergone positive selection, utilizing semi-quantitative PCR. We found that amplified copies of the Vα14-Jα18 rearrangement were not diminished in autophagy gene deficient DP thymocytes (Fig. 4D). In summary, the in vitro and in vivo experiments are consistent in showing that the deletion of either Atg5 or Atg7 did not alter the number of thymic iNKT cell progenitors by reducing the lifespan of DP progenitor thymocytes.
Arrested cell cycle in Atg5 deficient iNKT cells
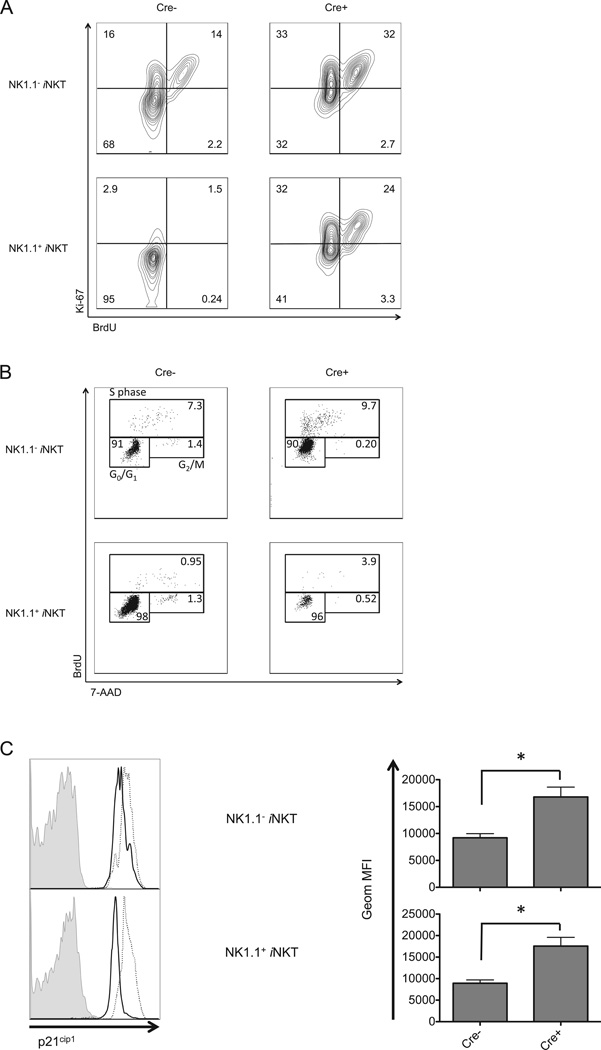
We determined if the decrease in iNKT cells in autophagy gene deficient mice could be attributed to decreased proliferation. Surprisingly, autophagy gene disrupted iNKT cells exhibited a higher rate of incorporation of BrdU and an augmented level of Ki-67, suggesting they are more proliferative (Fig. 5A).
Figure 5.
Loss of Atg5 expression led to cell cycle arrest. (A) Atg5 deficient iNKT cells exhibited enhanced incorporation of BrdU. 5 h after injection, thymic iNKT cells were collected and analyzed for BrdU incorporation and Ki-67 expression. (B) Thymic iNKT cells from Atg5f/f CD4-Cre mice accumulated in S-phase. 2 h post injection of BrdU, thymic iNKT cells were stained with 7-AAD and anti-BrdU antibody to evaluate cell cycle progression. (C) Increased expression of p21cip1 in Atg5 deficient iNKT cells. Shown are the representative flow cytometry analysis (left) and geometric MFI (right) of data pooled from at least three independent experiments gating on either NK1.1 or NK1.1+, CD1d tetramer+ thymocytes. Filled histogram: isotype control; solid line: wild type mice; dotted line: Atg5f/f CD4-Cre mice. * p<0.01, n≥4. Error bars are SD.
An increase in BrdU and Ki-67 staining was observed in NK1.1 negative iNKT cells, but the increase in the absence of Atg5 was even more striking when analyzing the NK1.1+ cells, a population that is normally quiescent in wild type mice (50). This may reflect the fact that this residual NK1.1+ population is not fully differentiated, and the remnant is more similar to the less mature iNKT cells. The results obtained from BrdU and Ki-67 analyses do not exclude an arrest in the cell cycle after chromosome replication, and therefore we analyzed cell cycle progression of Atg5 deficient thymic iNKT cells. Fig. 5B shows an analysis following a short pulse with BrdU and measurement of DNA content with 7-AAD. The results show that Atg5 deficient iNKT cells accumulated in S-phase, with relatively fewer cells accomplishing the S to G2 transition and proceeding to the G2/M phases, indicative of a blockade in the cell cycle in Atg5 knockout iNKT cells. This was especially true for the NK1.1+ subset.
We further determined that a negative regulator of cell cycle progression, p21cip1 (51), was increased in Atg5 deficient, either NK1.1− or NK1.1+iNKT cells (Fig. 5C). Because the limited number of iNKT cells did not permit a biochemical analysis, especially in mice deficient for autophagy genes, in this and subsequent experiments we used flow cytometry to analyze signaling pathways in iNKT cells. Atg5 deficient iNKT cells also had increased expression of p27kip1, another cell cycle inhibitor (51) (data not shown). Activation of cyclin-dependent kinase 1 (CDK1) promotes cell cycle progression (52), and a lack of Atg5 also led to an increase of inhibitory phosphorylation at Tyr15 on CDK1 (data not shown). These results indicate that intact autophagy machinery is required for a normal control of proliferation of iNKT cells and a smooth transition between cell cycle phases for iNKT cells in the thymus.
Autophagy gene deficient iNKT cells exhibited increased mitochondria and superoxide
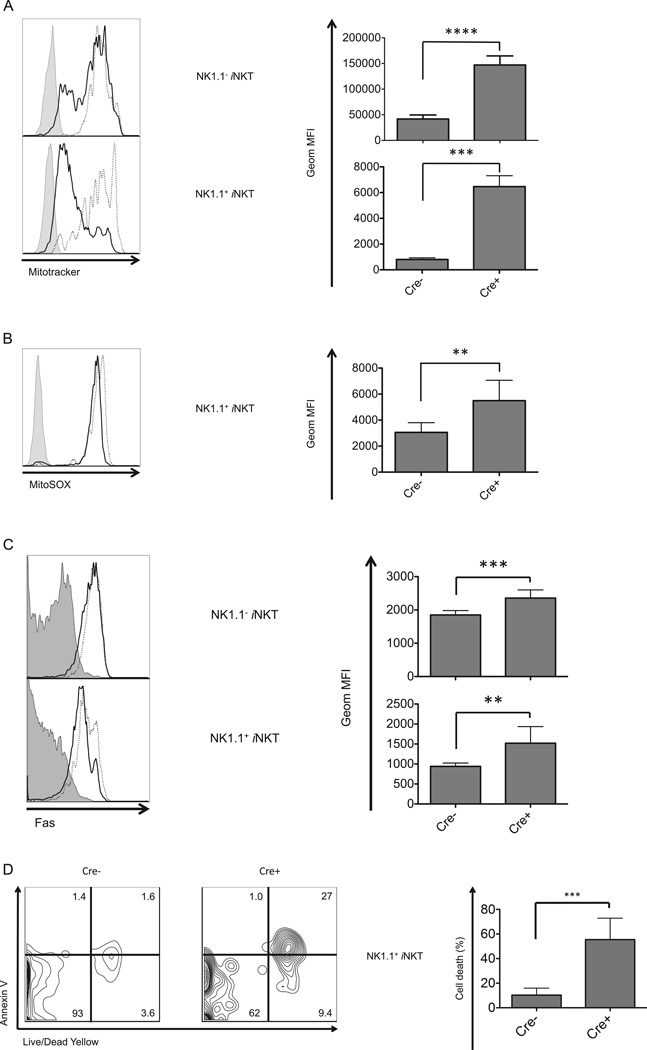
The status of mitochondria, the cellular energy-producing organelle, is closely related to cell cycle and proliferation, although mitochondria also have a central role in apoptotic cell death. Recent studies have shown autophagy involves quality control of mitochondria, removing damaged organelles through mitophagy (53). Therefore, we analyzed mitochondrial mass in Atg5 deficient iNKT cells utilizing MitoTracker Green, a lipophilic thiol-reactive dye that specifically labels mitochondria (54). Disruption of the autophagic machinery led to an increased mitochondrial mass (Fig. 6A) indicating that the organelle accumulated in thymic iNKT cells due to the loss of the autophagic clearance system.
Figure 6.
Atg5 deletion resulted in increased mitochondria, super oxide and cell death. Augmented mitochondrial mass (A) and mitochondrial superoxide production (B) in Atg5 deleted, thymic, NK1.1 and NK1.1+iNKT cells. (C) Higher expression of Fas on Atg5 deficient iNKT cells compared to wild type controls. At least four independent experiments were performed with consistent results, both representative flow cytometry plots (left) and geometric MFI values (right) are shown. Filled histogram: isotype control; solid line: wild type mice; dotted line: Atg5f/f CD4-Cre mice. (D) Dramatically increased apoptosis leading to cell death for NK1.1+ thymic iNKT cells devoid of Atg5. Cells were analyzed after overnight culture and compared with wild type controls. Representative flow cytometry analyses from three separate experiments (left) and geometric MFI (right) of dead cells (Annexin V+, Live/Dead Yellow+) are shown. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, n≥4. Error bars are SD.
Mitochondria are a major site for the production of reactive oxygen species (ROS). ROS positively regulates cellular functions, such as promoting cell growth and proliferation, but excessive ROS can induce apoptosis and cell death. We therefore measured superoxide in iNKT cells. Atg5 deficiency not only led to accumulated mitochondria, but also to one of their products, excessive superoxide, in the NK1.1+ iNKT cells (Fig. 6B).
Autophagy genes are required for optimal survival of iNKT cells
Several recent studies have reported that ablation of ATG proteins caused increased apoptosis in T cells (13, 14, 16, 19, 20). We reasoned that cell death must be increased in iNKT cells from Atg5 or Atg7 deficient mice, given their dramatically decreased cell number, especially in the stage 3 cells. T lymphocytes have the ability to sense external signals by death receptors expressed on their cell surface, such as Fas/CD95 and death receptor 5 (DR5/CD262), which initiate a serial caspase cascade that eventually commits the cells to apoptosis (55). The expression of Fas, as well as DR5 and its ligand Trail (data not shown), were increased on the cell surface of both thymic and splenic iNKT cells from Atg5 or Atg7 deleted mice, more prominently for the NK1.1+ subset (Fig. 6C and Atg7 data not shown).
In order to assess more directly if iNKT cells with either Atg5 or Atg7 deleted were undergoing apoptosis more than wild type controls, we measured the viability of the autophagy gene deficient iNKT cells. Thymocytes derived from either Atg5f/f CD4-Cre, Atg7f/f CD4-Cre, or control strains without Cre recombinase, were cultured at 37°C overnight and then stained with Annexin V and Live/Dead Dye. Unlike the total population of Atg5 or Atg7 deficient DP thymocytes, which did not exhibit decreased survival (Fig. 4A–C), iNKT cells from Atg5 or Atg7 deficient mice exhibited increased proportions of dead cells, especially in the NK1.1+ population (Fig. 6D and Atg7 data not shown).
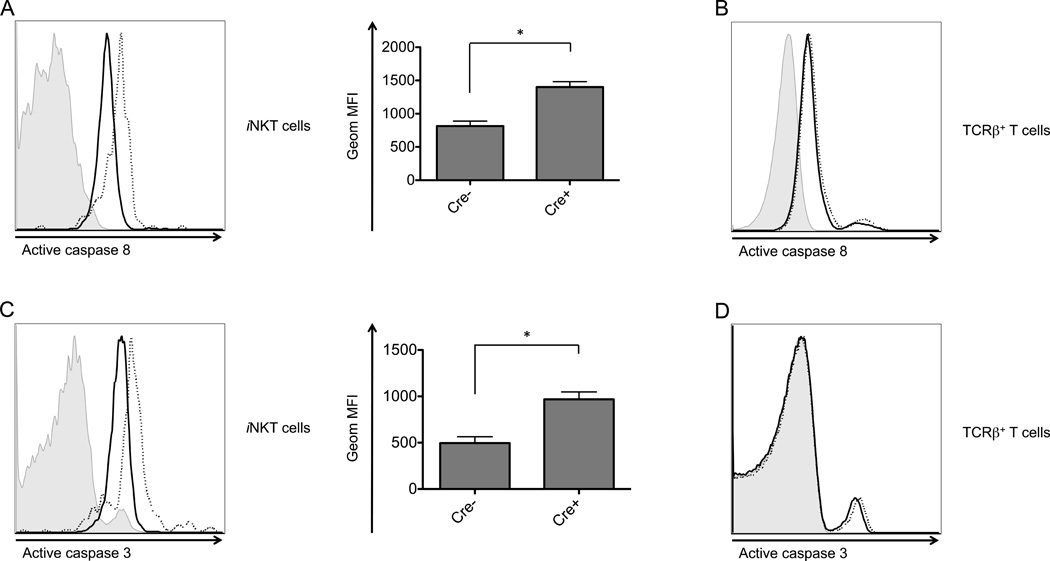
Autophagy gene deficient thymic iNKT cells cultured overnight consistently exhibited a higher level of active caspase 8, an initiator caspase downstream of Fas and DR5 (Fig. 7A). They also had increased active caspase 3, an executioner caspase cleaved and activated by caspase 8 (Fig. 7C). The increases were not observed for the majority population of DP thymocytes (Fig. 7B and D). This is consistent with the result that DP thymocytes derived from the autophagy gene deficient mice did not exhibit reduced viability. Overall, these results demonstrate that the autophagy genes are required specifically for the survival of iNKT cells in the thymus.
Figure 7.
Atg5 deficient thymic iNKT cells exhibited an increase in active caspase 8 and caspase 3. Atg5 deleted thymocytes were cultured overnight, followed by analysis by flow cytometry of active caspase 8 (A and B) and active caspase 3 (C and D) in thymic iNKT cells (A and C) and TCRβ+ thymocytes (B and D). Shown here are both flow cytometry plots (left) and geometric MFI (right). Data are representative of at least two independent experiments. Filled histogram: isotype control; solid line: wild type cells; dotted line: Atg5 deficient. *p<0.05, n≥3. Error bars are SD.
Autophagy gene deficient iNKT cells exhibited an altered cytokine profile
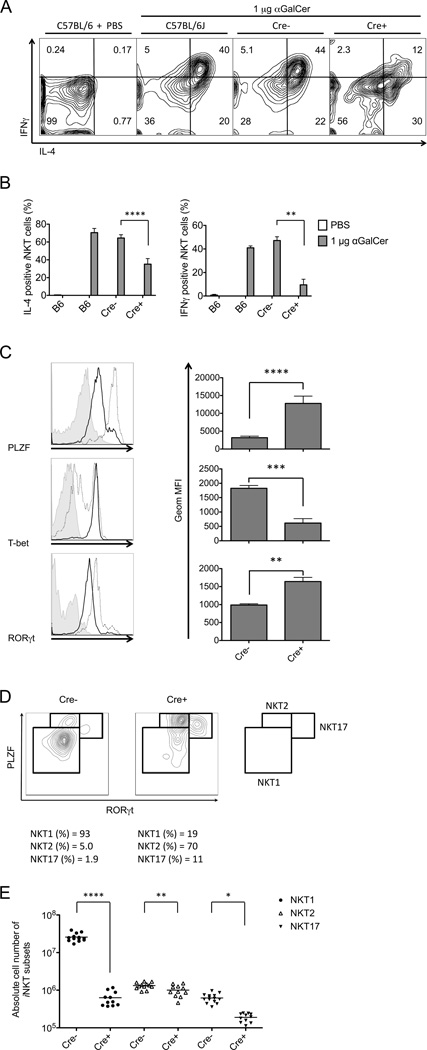
Once activated by strong antigens, the majority of iNKT cells rapidly release large amounts of both Th1 and Th2 cytokines. We determined howAtg5 or Atg7 deficiency impacted the in vivo response of mature iNKT cells to α-galactosylceramide (αGalCer), a potent synthetic glycolipid Ag. 2 h after αGalCer injection, splenic iNKT cells were analyzed directly ex vivo at the single cell level. Compared to wild type controls, the percentage of responding Atg5 deficient iNKT cells was decreased approximately two-fold for IL-4, while the IFNγ response was decreased more than four-fold (Fig. 8A,B). Atg7 deficient mice showed similar results (data not shown). These data suggest that the autophagy machinery is required for optimal responses by mature cells, particularly for antigen-dependent Th1 cytokine secretion by iNKT cells.
Figure 8.
Atg5 deficiency differentially affected iNKT cell functional subsets. (A and B) Impaired iNKT cell activation and polarized cytokine outcome in Atg5f/f CD4-Cre mice after αGalCer immunization. Intracellular cytokine production in splenic iNKT cells was analyzed 2 h after αGalCer injection. Flow cytometry plots (A), geometric MFI of intracellular IL-4 and IFNγ (B) are representative of three independent experiments. (C) Altered expression profile of transcription factors in autophagy deficient thymic iNKT cells. Representative flow cytometry plots from three independent experiments and geometric MFI are indicated. Filled histogram: isotype control; solid line: wild type mice; dotted line: Atg5f/f CD4-Cre mice. (D) iNKT cell subset analysis in autophagy deficient mice compared with wild type controls. Representative flow cytometry plots of three independent experiments are shown. (E) Absolute cell number of thymic iNKT cell functional subsets defined by transcription factor expression levels in Atg5f/f CD4-Cre mice and controls. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, n≥4. Error bars are SD.
Differential requirements of Atg5 by iNKT cell subsets
Recent studies have highlighted functional heterogeneity within mature iNKT cells. During their differentiation in the thymus, immature iNKT cells become committed to cytokine polarized subsets: NKT1 (Th1-biased), NKT2 (Th2-biased) and NKT17 (Th17-biased) (30, 31, 34). These subsets can be distinguished on the basis of their expression of surface proteins and transcription factors. For example, NKT1 cells constitute the majority in C57BL/6 mice, and they are NK1.1+ and express a high level of T-bet (34). By contrast, both NKT2 and NKT17 cells are relatively minor in C57BL/6 mice, but more abundant in BALB/c mice (34). NKT2 and NKT17 cells are NK1.1− and express IL-17RB (30), but NKT2 cells are GATA3high while NKT17 cells express CCR6 along with other surface proteins, and they are characteristically RORγthigh (30, 34). PLZF (promyelocytic leukemia zinc finger) is a transcription factor required for iNKT cell maturation and acquisition of effector function (56, 57). Of the three subsets, NKT2 cells express the highest level of PLZF, followed by NKT17 cells, while PLZF is the lowest in NKT1 cells (34). These functional categories overlap to a degree with the developmental stages, as NKT2 and NKT17 cells have a phenotype similar to stage 2 thymocytes, while stage 3 cells are similar to NKT1 cells. Therefore it is difficult to unambiguously distinguish a maturing, stage 2 iNKT cell from a committed NKT2 cell.
When analyzed by flow cytometry, total tetramer+ iNKT cells from the thymus of Atg5f/f CD4-Cre transgenic mice maintained a higher expression of PLZF, GATA3 and RORγt, but a lower level of T-bet compared to their counterparts from wild type mice (Fig. 8C and data not shown). The differences remain, although to a lesser degree, when NK1.1+ and NK1.1− iNKT cells were analyzed separately (data not shown). This indicated that iNKT cell functional subsets were differently affected by disruption of the autophagic machinery. To confirm this hypothesis, we analyzed individual sub-populations of iNKT cells in Atg5 knockout mice and wild type controls according to their expression level of signature transcription factors. Fig. 8D and E show that Atg5 deficiency caused a severe loss of NKT1 phenotype cells, the majority population in this strain, with an approximate 40-fold reduction in cell number compared with the wild type controls. The absolute numbers of NKT17 and NKT2 phenotype cells were much less affected, with a 3-fold decrease and a marginal 1.3-fold decrease, respectively, although this reached statistical significance. These changes in the pattern of transcription factor expression by thymic iNKT cells are consistent with changes in the expression of surface proteins, including a decreased percentage of iNKT cells expressing NK1.1 (Fig. 2) and an increased percentage expressing IL-17RB (data not shown).
Atg5 deficiency leads to augmented mTOR (mechanistic target of rapamycin) signaling
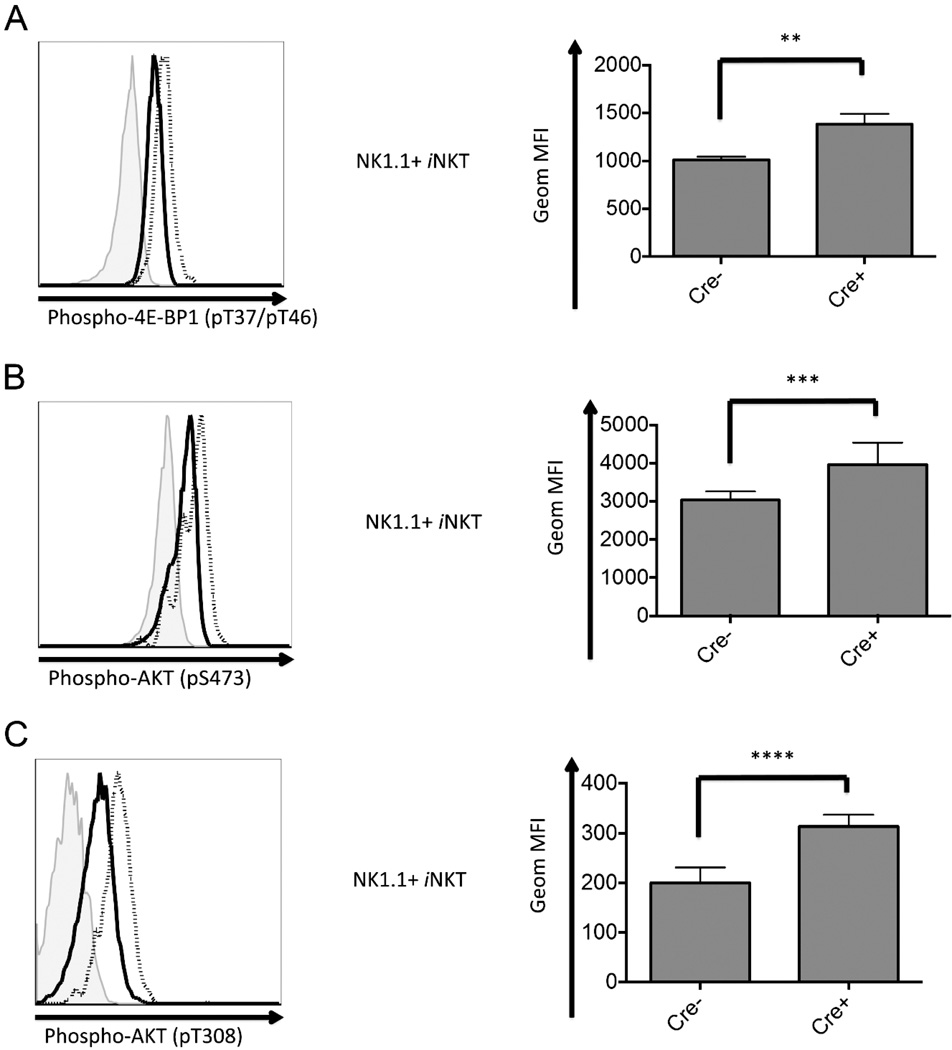
During development and maturation, T lymphocytes experiencing positive and negative selection have an enormous demand of energy and material supply. mTOR, a highly conserved serine/threonine protein of the phosphatidylinositol kinase-related kinase family, functions as a master controller, regulating cellular metabolism to support cell growth and division (58, 59). There are two mTOR complexes both with mTOR as the catalytic subunit, mTORC1 and mTORC2, consisting of different accessory proteins that instruct their substrate specificity (58, 59). Upon activation, mTORC1 phosphorylates 4E-BP1 (eIF4E-binding protein 1) (60), while mTORC2 phosphorylates AKT at Ser473, which primes AKT for subsequent phosphorylation at Thr308 to achieve full activation of the kinase (61).
Fig. 9A shows Atg5f/f CD4-Cre NK1.1+ iNKT cells expressed a higher level of phosphorylated 4E-BP1, indicating an augmented mTORC1 activity in these cells compared with controls. Similarly, phosphorylation of AKT at Ser473 was also elevated in Atg5f/f deficient, NK1.1+ iNKT cells, which is consistent with a hyperactive mTORC2 in these cells (Fig. 9B). Furthermore, combined with the same case for phospho-AKT (pT308) shown in Fig. 9C, the results suggest AKT exhibits a higher activation in Atg5f/f deleted iNKT cells than in the controls.
Figure 9.
Unbalanced proliferative and apoptotic signaling in Atg5 deficient iNKT cells. Thymic iNKT cells were analyzed ex vivo for activation of mTOR signaling pathway. Phosphorylation of mTORC1 substrate, 4E–BP1 (pT37/pT46) (A), and phosphorylation of mTORC2 substrate, AKT(pS473) (B), were analyzed. (C) Phosphorylation of AKT at T308 was measured to evaluate the PI3K/PDK1 signaling pathway. Both Cre- and Cre+ were pooled from at least four samples. Shown here is the representative flow cytometry analysis of three independent experiments. Filled histogram: isotype control; solid line: wild type mice; dotted line: Atg5f/f CD4-Cre mice. *p<0.05, **p<0.01, ***p<0.005, ****p<0.0005, n≥4. Error bars are SD.
Discussion
Our study demonstrates that iNKT cells depend on Atg5 and Atg7 for their differentiation to a far greater extent than mainstream CD4 or CD8 T lymphocytes. Removal of either Atg5 or Atg7 severely reduced both thymic and peripheral iNKT populations. Although some effects of Atg5 deficiency on host defense from infection that do not involve autophagy have been reported (62–64), the results of deletion of either Atg5 or Atg7 were in agreement, supporting the hypothesis that a defect in autophagy was responsible, rather than an effect on a different function of one of these proteins. The mouse strains with deletion mediated either by the CD4 or Lck promoter driven Cre transgenes had similar phenotypes, although a stronger effect in Lck-Cre transgenic mice might be due to the earlier deletion of Atg genes in this strain (65). Peripheral iNKT cells had an even greater reduction than did thymic iNKT cells, suggesting that in addition to effects on thymus differentiation, autophagy also may be required for optimal iNKT cell maintenance in the periphery.
Because iNKT cells are positively selected by DP thymocytes (37), the defect in iNKT cells could have been due to a defect in either the iNKT cell precursor and/or in the positively selecting DP thymocyte population, perhaps related to a problem with presentation of positively selecting lipid ligands. We found, however, that there was no reduction in CD1d expression on DP thymocytes from Atg5 or Atg7 deficient mice. Moreover, the analysis of mixed bone marrow chimeras demonstrated a cell intrinsic defect.
Autophagy is related to cell survival, and even a partial impairment in the survival of DP thymocytes could have caused a cell intrinsic diminution of iNKT cells in the thymus, due to decreased secondary rearrangements in the TCR α gene locus. However, we did not find a reduction in the survival of cultured DP thymocytes in CD4-Cre transgenic mice crossed to mice with floxed Atg5 or Atg7 alleles. In addition, there was no reduction in Vα14-Jα18 rearrangements in DP thymocytes, which would have not occurred if DP survival were impaired, and we observed a normal number of stage 0 iNKT cells in the thymus. Therefore, general effects on DP survival, causing decreases in the secondary TCR α rearrangements needed to form the invariant TCR α chain, do not account for the cell intrinsic iNKT cell deficit in mice deficient for autophagy genes.
Atg5 or Atg7 deficient iNKT cells exhibited a selective defect in their maturation pathway, with the earliest stages 0 and stage 1 iNKT cells maintained, and the later stages depleted due to decreased cell survival. The differentiation of iNKT cells in the thymus along a single developmental pathway has been challenged, however, by recent studies showing there are functional subsets of iNKT cells analogous to T helper cell subsets, such as Th1, Th2 and Th17 cells. It is only the Th1 cytokine-biased NKT1 subset that expresses NK1.1 in C57BL/6 mice and the CD44+, NK1.1low iNKT cell phenotype, defined as less mature stage 2, also includes mature NKT2 and NKT17 cells. Based on the finding of functional subsets, we explored the effects of Atg5 and Atg7 deficiency on iNKT cell function. We found that the production of IFNγ by antigen stimulated autophagy deficient iNKT cells was more severely affected than the production of IL-4. Additionally, based on an analysis of transcription factor expression, the NKT1 subset in the thymus was greatly decreased, and NKT2 and NKT17 cells much less so. There were, however, also defects in the other subsets including a reduced percentage of antigen activated iNKT cells producing IL-4 and decreased expression of key transcription factors, such as RORγt. Additionally, we found reduced expression of CD69 by autophagy deficient iNKT cells, either before or after activation (data not shown). Therefore, while NKT1 cells have by far the most severe survival and functional defects in the absence of either of Atg5 or Atg7, there are deficiencies that affect the entire iNKT cell population.
Given the reduced iNKT cell population in mice with deleted autophagy genes, it would be reasonable to expect reduced proliferation of these cells. However, Atg5 or Atg7 deficient iNKT cells exhibited enhanced incorporation of BrdU and elevated Ki-67 staining. Despite this, there was a blockade in completing the cell cycle, with an arrest at the S-phase. As a consequence, Atg5 deleted thymic iNKT cells accumulated a high DNA content, fewer cells went to the G2/M phases, and they also had decreased survival. Consistent with the decreased survival, Atg5 or Atg7 deficient iNKT cells had increased cell surface expression of several death receptors, and elevated levels of active caspase 8, implicating the cell extrinsic apoptotic pathway. Caspase 8 has been reported to negatively regulate autophagy (66, 67), while the induction of autophagy promotes a reduction in caspase 8 and other pro-apoptotic proteins (19). On the other hand, our data are also consistent with a role for the intrinsic pathway for apoptosis, as evidenced by the increase in mitochondria and ROS in the iNKT cells from Atg5 deficient mice. An increase in mitochondria previously has been observed in autophagy deficient T cells (14, 18, 20). Overexpression of Bcl-XL failed to rescue the defective iNKT cell differentiation in our Atg5 deficient mice, although the frequency of the CD8+ T cells was normalized (Supplemental Fig. 4). This failure to restore the iNKT cell population is not consistent with a major role for the intrinsic apoptosis pathway, but the timing or expression level of the Bcl-XL transgene could have been a mitigating factor preventing restoration. In summary, our data are consistent with the hypothesis that increased apoptosis, which could be induced in the autophagy gene deficient iNKT cells that are in proliferation and in the transition from stage 2 to 3 or in mature stage 3 cells, is responsible for the decrease in the iNKT population, with possible roles for both the cell extrinsic and cell intrinsic pathways.
What drives the increased entry of autophagy deficient iNKT cells into cell cycle? One candidate is mTOR, which is involved in T cell differentiation, including the formation of memory cells (68). There are two mTOR complexes: mTORC1 phosphorylates and activates S6K1 and 4E-BP1, which increase mRNA translation (58, 60, 69). mTORC2, acting in synergy with PDK1 (phosphoinositide-dependent Protein Kinase 1), catalyzes the dual phosphorylations of AKT for its full activity, facilitating cell survival and cell cycle progression (61). We showed that autophagy deficiency led to hyperactivity of mTORC1 and mTORC2 in iNKT cells. The T cell specific deletion of Raptor, an essential component of mTORC1, also impaired the maturation of iNKT cells (70), suggesting the need for a balance, with either too much or too little mTORC1 inhibiting iNKT cell development.
Our findings are consistent with the results from several recent studies indicating a role for proper regulation of metabolism in regulating iNKT cell differentiation. First, mice with a T cell specific deletion of Vps34, a class III PI3 kinase that is an initiator of autophagy, recently were shown to have an even more profound defect in iNKT cells than the one reported here (71), although the mechanism for the effect was not described. Furthermore, while the importance of VPS34 for autophagy in T lymphocytes has been found by several groups (13, 71), this enzyme is also important for IL-7R signaling (72), and therefore some of the deficit in iNKT cells in the absence of VPS34 could have been due to reduced IL-7 signals, or some other effects, including the severe inflammation these mice spontaneously developed. Second, mice with a T cell specific deletion of the gene encoding tuberous sclerosis 1 (TSC1), an inhibitor of mTORC1, also were found to have a decreased number of iNKT cells due to reduced cell survival (73). Because mTOR inhibits autophagy, the increased mTOR activity in the absence of the TSC1 inhibitor in these mice may have led to decreased autophagy in iNKT cells, and therefore a phenotype similar to the one we report here, although this has not been determined. Consistent with this hypothesis are data from a subsequent study finding that the Tsc1 deficient mice retained the NKT17 population, like the Atg5 and Atg7 deficient strains (74). Tsc1 deficiency induced an activated intrinsic apoptotic pathway, however, which was rescued by overexpression of Bcl2. This is different from our findings that Atg5/Atg7 deficiency led to apoptosis through extrinsic pathway as well as possibly intrinsic pathway, which was not rescued by Bcl-XL overexpression (data not shown). Third, an investigation exploring the effect of unbalanced metabolic homeostasis on iNKT cell differentiation showed that folliculin-interacting protein 1 (Fnip1)-null mice had a reduction in mature iNKT cells. Deficiency of Fnip1 led to inability to shut off mTOR mediated energy consuming cellular processes, which could inhibit autophagy. However, inhibition of mTOR was not able to rescue the defect in iNKT cells, indicating Fnip1 is required for iNKT cell development in both an mTOR-dependent and -independent manner (75), and a connection to decreased autophagy was not established in this study. While this manuscript was in revision, a study was published indicating that mice with a Atg7 deficiency driven by CD4 Cre have a reduction in iNKT cells in the thymus and periphery, consistent with the results here (76).
In summary, here we show that the disruption of two autophagy related genes, Atg5 or Atg7, had a particularly severe effect on the later stages of differentiation of iNKT cells, an innate-like T lymphocyte population. The ability of differentiating iNKT cells in the thymus to progress through the cell cycle was decreased and their survival was impaired due to increased apoptosis. The requirement for the autophagy pathway is consistent with the increased autophagy exhibited by thymic iNKT cells compared to other thymocytes populations. It likely reflects the rounds of proliferation that iNKT cells undergo following gene rearrangement and expression of a CD1d autoreactive TCR, which may have similar metabolic requirements to the expansion of antigen-stimulated mainstream T cells in lymphoid organs. In fact, findings from several groups have confirmed that systemic or tissue-specific deletion of genes encoding proteins in the autophagy pathway led to significant reductions in the homeostasis of peripheral T lymphocytes and their ability to respond and produce cytokines (13, 14, 16–20). Alternatively, rather than acting at the effector stage, after expansion, mature iNKT cells are similar to memory cells in their survival requirements (77), and may have similar metabolic requirements that depend on autophagy. Regardless, while all the functional subsets of iNKT cells analyzed were decreased in number and function, the effect was most severe for the dominant NKT1 population. Interestingly, considering B lymphocytes, a more severe defect in innate-like B1a B cells was observed when Atg5 was deleted throughout the B cell lineage (78). This suggests that different types of innate-like lymphocytes, which have a degree of self-reactivity and that undergo rounds of proliferation during their differentiation, may have a selective requirement for autophagy.
Supplementary Material
Acknowledgements
We thank Isaac Engel for helpful discussions, Archana Khurana for preparation of CD1d tetramers, Yiran Wang-Zhu, and the Flow Cytometry Core and the Division of Laboratory Animal Care at the La Jolla Institute for Allergy & Immunology for technical support.
This work was supported by NIH grant R37 AI71922 (M.K.).
Abbreviations
- 4E-BP1
eIF4E-binding protein 1
- 7-AAD
7-aminoactinomycin D
- αGalCer
α-galactosylceramide
- ATG
autophagy-related gene
- AMPK
AMP-activated protein kinase
- CDK
cyclin dependent kinase
- DP
CD4 and CD8 double positive
- DR5
death receptor 5
- Fnip1
folliculin-interacting protein 1
- iNKT cells
invariant natural killer T cells
- MDM2
mouse double minute 2 homolog
- mTORC1 and mTORC2
mechanistic target of rapamycin complex 1 and 2
- PDK1
phosphoinositide-dependent Protein Kinase 1
- PLZF
promyelocytic leukemia zinc finger
- ROS
reactive oxygen species
- S6K
p70 ribosomal S6 kinase 1
- TSC 1 and TSC 2
tuberous sclerosis 1 and 2
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annual review of biochemistry. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Virgin HW, Levine B. Autophagy genes in immunity. Nature immunology. 2009;10:461–470. doi: 10.1038/ni.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Current topics in microbiology and immunology. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature reviews. Molecular cell biology. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 8.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nature cell biology. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 9.Dunkle A, He YW. Apoptosis and autophagy in the regulation of T lymphocyte function. Immunologic research. 2011;49:70–86. doi: 10.1007/s12026-010-8195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu JV, Walsh CM. Programmed necrosis and autophagy in immune function. Immunological reviews. 2012;249:205–217. doi: 10.1111/j.1600-065X.2012.01147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nature cell biology. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunological reviews. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T-cell homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8670–8675. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia W, He YW. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. Journal of immunology. 2011;186:5313–5322. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, Jin S, Lu B. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. Journal of immunology. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 16.Pua HH, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. The Journal of experimental medicine. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolism during effector T cell activation. Journal of immunology. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephenson LM, Miller BC, Ng A, Eisenberg J, Zhao Z, Cadwell K, Graham DB, Mizushima NN, Xavier R, Virgin HW, Swat W. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, Roodman DG, Windle JJ, Zhang X, Lu B. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell death and differentiation. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. Journal of immunology. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 21.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 22.Kronenberg M. Toward an understanding of NKT cell biology: progress and paradoxes. Annual review of immunology. 2005;23:877–900. doi: 10.1146/annurev.immunol.23.021704.115742. [DOI] [PubMed] [Google Scholar]
- 23.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nature immunology. 2013;14:90–99. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Current opinion in immunology. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what’s in a name? Nature reviews Immunology. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 26.Cohen NR, Garg S, Brenner MB. Antigen Presentation by CD1 Lipids, T Cells, and NKT Cells in Microbial Immunity. Advances in immunology. 2009;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 27.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annual review of immunology. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 28.Silk JD, Salio M, Brown J, Jones EY, Cerundolo V. Structural and functional aspects of lipid binding by CD1 molecules. Annual review of cell and developmental biology. 2008;24:369–395. doi: 10.1146/annurev.cellbio.24.110707.175359. [DOI] [PubMed] [Google Scholar]
- 29.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d–restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 30.Watarai H, Sekine-Kondo E, Shigeura T, Motomura Y, Yasuda T, Satoh R, Yoshida H, Kubo M, Kawamoto H, Koseki H, Taniguchi M. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS biology. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Constantinides MG, Bendelac A. Transcriptional regulation of the NKT cell lineage. Current opinion in immunology. 2013;25:161–167. doi: 10.1016/j.coi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coquet JM, Chakravarti S, Kyparissoudis K, McNab FW, Pitt LA, McKenzie BS, Berzins SP, Smyth MJ, Godfrey DI. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1- NKT cell population. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-de-Moraes MC. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. The Journal of experimental medicine. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nature reviews. Immunology. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 36.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nature immunology. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 37.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nature immunology. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 38.Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. The Journal of experimental medicine. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel I, Kronenberg M. Making memory at birth: understanding the differentiation of natural killer T cells. Current opinion in immunology. 2012;24:184–190. doi: 10.1016/j.coi.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 41.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 44.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 45.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. The Journal of experimental medicine. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(−)CD4(+) CD1d–dependent precursor stage. The Journal of experimental medicine. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 48.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nature immunology. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 49.Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 50.Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. Journal of immunology. 2000;164:2412–2418. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 51.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes & development. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 52.Norbury C, Blow J, Nurse P. Regulatory phosphorylation of the p34cdc2 protein kinase in vertebrates. The EMBO journal. 1991;10:3321–3329. doi: 10.1002/j.1460-2075.1991.tb04896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature reviews Molecular cell biology. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Presley AD, Fuller KM, Arriaga EA. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. Journal of chromatography. B, Analytical technologies in the biomedical and life sciences. 2003;793:141–150. doi: 10.1016/s1570-0232(03)00371-4. [DOI] [PubMed] [Google Scholar]
- 55.Pua HH, He YW. Autophagy and lymphocyte homeostasis. Current topics in microbiology and immunology. 2009;335:85–105. doi: 10.1007/978-3-642-00302-8_4. [DOI] [PubMed] [Google Scholar]
- 56.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant’Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews. Molecular cell biology. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung CM, Garcia-Haro L, Sparks CA, Guertin DA. mTOR-dependent cell survival mechanisms. Cold Spring Harbor perspectives in biology. 2012;4:a008771. doi: 10.1101/cshperspect.a008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature reviews Molecular cell biology. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 61.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 62.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell host & microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, Green KY, Lopez-Otin C, Xavier RJ, Thackray LB, Virgin HW. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell host & microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selleck EM, Fentress SJ, Beatty WL, Degrandi D, Pfeffer K, Virgin HWt, Macmicking JD, Sibley LD. Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS pathogens. 2013;9:e1003320. doi: 10.1371/journal.ppat.1003320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolfer A, Wilson A, Nemir M, MacDonald HR, Radtke F. Inactivation of Notch1 impairs VDJbeta rearrangement and allows pre-TCR-independent survival of early alpha beta Lineage Thymocytes. Immunity. 2002;16:869–879. doi: 10.1016/s1074-7613(02)00330-8. [DOI] [PubMed] [Google Scholar]
- 66.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 67.Bell BD, Leverrier S, Weist BM, Newton RH, Arechiga AF, Luhrs KA, Morrissette NS, Walsh CM. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16677–16682. doi: 10.1073/pnas.0808597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng H, Chi H. mTOR and lymphocyte metabolism. Current opinion in immunology. 2013;25:347–355. doi: 10.1016/j.coi.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hung CM, Garcia-Haro L, Sparks CA, Guertin DA. mTOR-dependent cell survival mechanisms. Cold Spring Harbor perspectives in biology. 4:a008771. doi: 10.1101/cshperspect.a008771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L, Tschumi BO, Corgnac S, Ruegg MA, Hall MN, Mach JP, Romero P, Donda A. Mammalian Target of Rapamycin Complex 1 Orchestrates Invariant NKT Cell Differentiation and Effector Function. Journal of immunology. 2014 doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 71.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagomez D, Cover TL, Zong WX, Zhang J, Van Kaer L. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. Journal of immunology. 2013;190:5086–5101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McLeod IX, Zhou X, Li QJ, Wang F, He YW. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. Journal of immunology. 2011;187:5051–5061. doi: 10.4049/jimmunol.1100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature immunology. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu J, Yang J, Yang K, Wang H, Gorentla B, Shin J, Qiu Y, Que LG, Foster WM, Xia Z, Chi H, Zhong XP. iNKT cells require TSC1 for terminal maturation and effector lineage fate decisions. The Journal of clinical investigation. 2014;124:1685–1698. doi: 10.1172/JCI69780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Park H, Tsang M, Iritani BM, Bevan MJ. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7066–7071. doi: 10.1073/pnas.1406473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salio M, Puleston DJ, Mathan TS, Shepherd D, Stranks AJ, Adamopoulou E, Veerapen N, Besra GS, Hollander GA, Simon AK, Cerundolo V. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci U S A. 2014;111:E5678–E5687. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 78.Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, Mizushima NN, Iwasaki A, He YW, Swat W, Virgin HWt. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–314. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.