Abstract
BACKGROUND:
Triggering receptor expressed on myeloid cells-1 (TREM-1) is a cell surface receptor expressed on neutrophils and monocytes. TREM-1 acts to amplify inflammation and serves as a critical mediator of inflammatory response in the context of sepsis. To date, the predisposition of TREM-1 gene polymorphisms to septic shock has not been reported. This study was designed to investigate whether TREM-1 genomic variations are associated with the development of septic shock.
METHODS:
We genotyped two TREM-1 single nucleotide polymorphisms (SNPs, rs2234237 and rs2234246) and evaluated the relationships between these SNPs and septic shock on susceptibility and prognosis.
RESULTS:
TREM-1 rs2234246 A allele in the promoter region was significantly associated with the susceptibility of septic shock in recessive model (AA, OR=3.10, 95%CI 1.15 to 8.32, P=0.02), and in codominant model (AG, OR=0.72, 95%CI 0.43–1.19, P=0.02; AA, OR=2.71, 95%CI 1.00–7.42; P=0.03). However, in three inherited models (dominant model, recessive model, and codominant model), none of the assayed loci was significantly associated with the prognosis of septic shock. The non-survivor group demonstrated higher plasma IL-6 levels (99.7±34.7 pg/mL vs. 61.2±26.5 pg/mL, P<0.01) than the survivor group. Plasma concentrations of IL-6 among the three genotypes of rs2234246 were AA 99.4±48.9 pg/mL, AG 85.4±43 pg/mL, and GG 65.3±30.7 pg/mL (P<0.01). The plasma concentrations of IL-6 in patients with AA genotypes were significantly higher than those in patients with GG genotypes (P<0.01).
CONCLUSION:
TREM-1 genetic polymorphisms rs2234246 may be significantly correlated only with susceptibility to septic shock in the Chinese Han population.
KEY WORDS: Triggering receptor expressed on myeloid cells-1, Single nucleotide polymorphisms, Septic shock, Association study
INTRODUCTION
Sepsis is one of the most frequent complications in surgical patients and one of the leading causes of mortality in intensive care units. It is an infection-initiated systemic inflammatory reaction syndrome resulting from the interactions between environmental and genetic factors.[1–3] Septic shock, the most severe complication of sepsis, is a deadly disease. Many mechanisms are involved in the pathophysiology of septic shock, including the release of cytokines, the activation of neutrophils, monocytes, and microvascular endothelial cells, as well as the activation of neuroendocrine reflex and plasma protein cascade systems (complement system, the intrinsic and extrinsic pathways of coagulation, and the fibrinolytic system).[4] Innate immune responses to invading pathogens provide a first line of host defense that occurs rapidly and involves the coordinate action of diverse cells and proteins. Neutrophils and monocytes are key effector cells during sepsis by virtue of their capability to recognize pathogen-associated molecular patterns, which in turn results in initiation of the inflammatory cascade reaction after infection.[5–7]
Triggering receptor expressed on myeloid cells-1 (TREM-1) is a recently identified cell surface receptor expressed on neutrophils and a subset of monocytes.[8] This receptor belongs to the immunoglobulin superfamily and activates downstream signaling pathways by interaction with an adaptor protein DAP12.[9–11] Engagement of TREM-1 has been shown to induce the production of proinflammatory chemokines such as interleukin (IL)-8 and cytokines such as tumor necrosis factor (TNF)-α and interleukin (IL)-1β.[9,12] Meanwhile, TREM-1 acts synergistically with Toll-like receptors (TLRs) and Nod-like receptors to enhance the inflammatory reaction.[9] Furthermore, in an animal model of septic shock, blocking of TREM-1 clearly attenuated systemic inflammatory response and protected mice against lethality.[13] In addition, elevated concentrations of a soluble form of TREM-1 (sTREM-1) were detectable in fluids from patients with infections, whereas a progressive decline in plasma sTREM-1 concentrations indicates a favorable clinical evolution of septic shock, although the mechanism of generation of sTREM-1 remains unclear.[9,14,15] Therefore, TREM-1 functions as an amplifier as well as a critical mediator of inflammatory response in the context of septic shock. However, the association between genomic variations within the TREM-1 gene and septic shock remains unknown.
Studies[16–20] confirmed that single nucleotide polymorphisms (SNPs) in some particular genes, such as interleukin-6 gene polymorphisms G-174>C and G-572>C, bactericidal permeability increasing protein (BPI) (G545>C) Taq and BPI (A645>G) 216, β-defensin 1 variant haplotype -20G/-44G/-52G, IRAK-1 variant haplotype, protein C-1641 AA genotype, etc, play a critical role in severe sepsis or septic shock.
We therefore determined the genotype and allelic frequencies of two common variants (minor allelic frequency >10%) within the TREM-1 gene in patients with septic shock and ethnically matched healthy control subjects. Haplotype distributions were also estimated in the related groups. We evaluated their associations with both susceptibility to and prognosis of septic shock in a Chinese Han cohort.
METHODS
Subjects
After the study was approved by the local Ethics Committee, informed written consent was obtained from all subjects or a close kin of the patient. A total of 130 patients with septic shock treated from January 2011 to August 2012 and 157 healthy control subjects were included in the study; all of them were Han Chinese. The patients with septic shock were enrolled from the Department of Intensive Care Unit, Affiliated Hospital of Guangdong Medical College. The patients were followed up for 28 days. Survival or non-survival was noted at the end of the observation period. The diagnosis of sepsis met the criteria recommended by the American College of Chest Physicians and Society of Critical Care Medicine Consensus Conference 1992 and 2001.[21,22] Septic shock was considered as any value of systolic pressure below 90 mmHg requiring the administration of vasopressors.[22] The patients were bundle treated referring to “the adult serious infection and septic shock hemodynamic monitoring and support guide” (2006). The exclusion criteria included: patients with acquired immune deficiency syndrome; patients receiving chemotherapy in the past 8 weeks; patients receiving immunosuppressive treatment after organ transplantation; and patients receiving corticosteroids in the past 4 weeks. The control subjects were ethnically matched, healthy individuals comprising 86 men and 53 women, with a mean age of 57.2±17.7 years.
Data collection
Demographic and disease data of the patients included age, gender, chief complaint for admission, vital signs, routine blood test results, liver and kidney functions, and coagulation indicators. The Acute Physiology and Chronic Health Evaluation II (APACHE II) score was obtained within 24 hours of admission. The SOFA score was calculated daily; the maximum SOFA score (SOFAmax) was defined as the highest SOFA score during the stay in the intensive care unit and was used to express the worst status of organ dysfunction during the stay. Plasma was collected at the same time points and IL-6 levels were determined. Records were also kept for the 28-day survival patients.
Extraction of genomic DNA
Five mL of peripheral whole blood was drawn from each subject. Genomic DNA was extracted with the selective binding properties of a silica-based membrane with the speed of microspin technology (Blood/Liquid Sample Total DNA Rapid Extraction Kit, Aidlab Biotechnologies) according to the manufacturer’s instructions. The DNA was qualified in a spectrophotometer and stored at –20 °C until the next analysis.
ELISA assays
Blood samples from the patients were drawn from venous line for culture and measurement of IL-6. After centrifugation, plasma was kept at −80 °C until the start of assay. IL-6 was determined using a double antibody sandwich enzymeimmunoassay (Diaclone, Paris, France). ELISA was performed in duplicate and in strict accordance with the manufacturer’s instructions. Lowest limits of detection were 6.25 pg/mL for IL-6.
Genotyping of single nucleotide polymorphisms
Single nucleotide polymorphisms (SNPs), rs2234246, which are located within the 3'flanking region of the reference TREM-1 gene and may influence the transcriptional activity, and one nonsynonymous variation rs2234237 (Ser25Thr), which is in the second exon of the TREM-1 gene and may also influence the biologic function of TREM-1, were genotyped using the PCR/ligase detection reaction assay.[23] Primers were synthesized from Shanghai BioWing Applied Biotechnology Company. Each set of ligase detection reaction probes comprised one common probe and two discriminating probes for the two types.
The target DNA sequences were amplified using a multiplex PCR method. Multiplex PCR primers are shown in Table 1. PCRs for each subject were carried out in a final volume of 10 μL containing 1×PCR buffer, 3.0 mmol/L MgCl2, 2.0 mmol/L deoxynucleotide triphosphates, 0.4 μL primers, 0.2 μL Qiagen HotStarTaq polymerase (QIAGEN, Shenzhen, China), 4 μL of 1× Q-solution, and 10–20 ng genomic DNA. Thermal cycling was performed for rs2234237 and rs2234246 in Gene Amp PCR system 9600 (PerkinElmer, Waltham, MA) with an initial denaturation of 15 minutes at 95 °C, followed by 35 cycles of denaturation at 94 °C for 30 seconds, annealing at 57 °C for 1 minute and 30 seconds, and extension at 72 °C for 1 minute, followed by a final extension at 72 °C for 10 minutes.
Table 1.
Multiple PCR primers

The ligation reaction for each subject was carried out in a final volume of 10 μL containing 1×NEB Taq DNA ligase buffer, 12.5 pmol of each probe mix, 0.05 μL Taq DNA ligase [NEB Biotechnology (Beijing, China)], and 1 μL of multi-PCR product. Ligase detection reaction was performed using at 2 minutes at 95 °C, followed by 30 cycles of denaturation at 94 °C for 30 seconds, and 50 °C for 25 seconds. The fluorescent products of ligase detection reaction were differentiated by an ABI sequencer 377 (Applied Biosystems, Foster City, CA). Ligase detection reaction probes are shown in Table 2.
Table 2.
Ligase detection reaction probes

Statistical analysis
Quantitative data with normal distributions including age, APACHE II scores, and body temperature were presented as mean±standard deviation. Student’s t test was used to compare the mean between the two groups. All SNP data were evaluated for Hardy-Weinberg equilibrium using the Chi-square test and the goodness-of-fit test. Allele frequency and genotype frequency between patients with septic shock and control subjects, or between survivors and nonsurvivors among patients with septic shock, were compared using Fisher’s exact test or the Chi-square test. Analysis of variance and Student-Neuman-Keuls procedure were used to describe the changes in IL-6 levels between the three genotypes of rs2234246. Multiple logistic regression was used to model the effect of the related genetic variations on the incidence of septic shock, and Cox regression was used to evaluate the influence of the polymorphisms on the prognosis of septic shock. Different models of inheritance were tested using SNPStats software.[24] Linkage disequilibrium among genotyped SNPs was obtained and haplotypes were estimated using the Haploview 4.2 program.[25] Haplotypes with frequencies <3% were not included in the analysis. Haplotype distribution was analyzed using the Chi-square test. Power estimates were calculated using the PS program.[26] Statistical analysis used SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). A P value below 0.05 after adjustment for multiple comparisons was considered statistically significant.
RESULTS
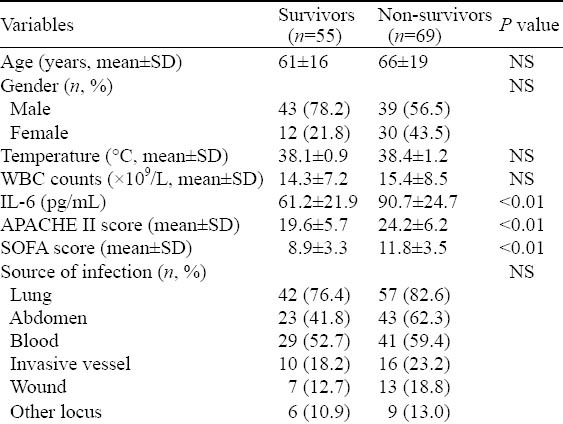
We enrolled 287 subjects into this study, including 152 in the control group (five samples failed sequencing) and 130 in the septic shock group. The patients with septic shock eventually comprised 124 Han Chinese people (six sample failed sequencing) who came from Guangdong province. The characteristics of patients with septic shock (divided into survivors and non-survivors) at admission to ICU are listed in Table 3. The non-survivor group demonstrated higher serum IL-6 levels (P<0.01). APACHE II and SOFA scores were significantly different between survivors and non-survivors (P<0.01). In terms of age, gender, temperature, WBC counts, and underlying diseases, a cross-group comparison upon admission was devoid of statistical significance. The lung was the most frequent initial source of infection, followed by abdomen, blood, invasive vessel, wound and other locus.
Table 3.
Characteristics of patients with septic shock
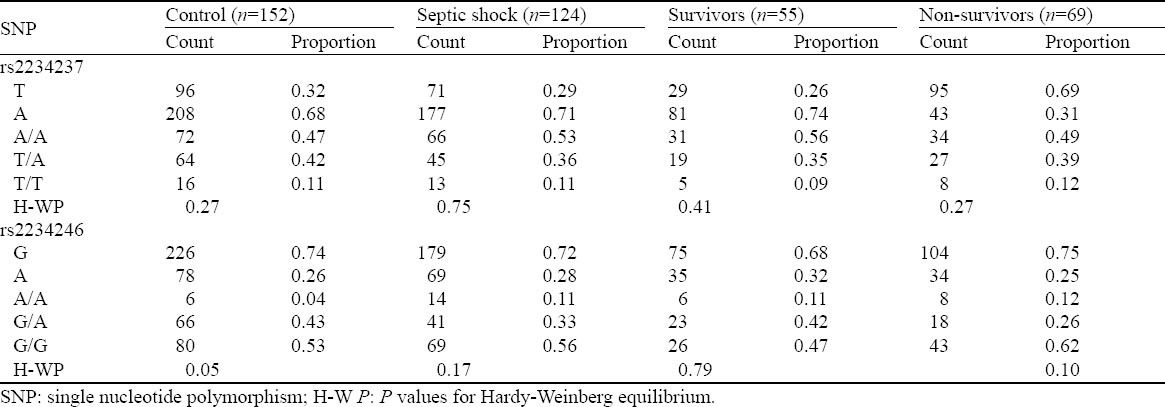
The genotype distribution for the assayed loci followed the Hardy–Weinberg equilibrium in patients with septic shock and controls (Table 4). All pairwise r2 values for linkage disequilibrium among SNPs rs2234237 and rs2234246 in both groups were analyzed. Control group D′ was 1.0, and r2 was 0.158; septic shock group D′ was 0.93, and r2 was 0.139. Based on high LD values defined as r2>1/3 as suggested by Ardlie et al[27] or D′>0.7 as suggested by Gabriel et al,[28] there was a weak linkage disequilibrium between rs2234237 and rs2234246.
Table 4.
Evaluation of genotype distribution for Hardy–Weinberg equilibrium in the assayed loci in the survivors and non-survivors
Different models of inheritance were tested for the impact of the two polymorphisms on septic shock. We found that TREM-1 genetic polymorphisms rs2234246 was significantly correlated only with susceptibility to septic shock. TREM-1 rs2234246 A allele in the promoter region was significantly associated with the occurrence of septic shock in recessive model (AA, OR=3.10, 95%CI 1.15 to 8.32, P=0.02), and in codominant model (AG, OR=0.72, 95%CI 0.43–1.19, P=0.02; AA, OR=2.71, 95%CI 1.00–7.42; P=0.03). However, in three inherited models (dominant model, recessive model, and codominant model), none of the assayed loci was significantly associated with the outcome of septic shock (Tables 5 and 6).
Table 5.
Comparison of different models of inheritance for rs2234237 and rs2234246 in the sepsis and control groups
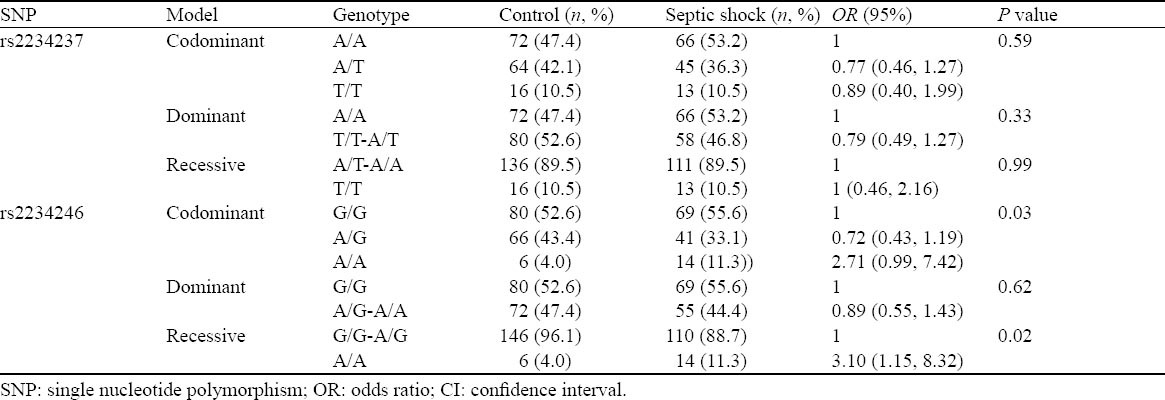
Table 6.
Comparison of different models of inheritance for rs2234237 and rs2234246 in the survivors and non-survivors
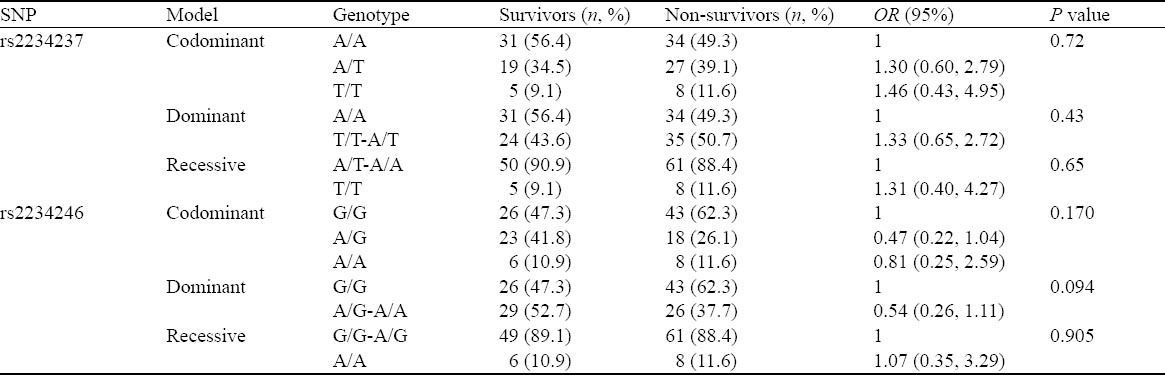
SNP, odds ratio, CI
The assayed locus of rs2234246 was significantly associated with susceptibility of septic shock in the recessive and codominant inheritance models. We speculated whether different genotypes could affect IL-6 expression. Therefore, all patients with septic shock (124 cases) were divided into three groups according to rs2234246 genotype. Patients were divided into either a surviving group (>28 days survival) or a non-surviving group (<28 days survival) based on 28-day survivals. The plasma IL-6 levels of patients in the non-surviving group were significantly higher than those in the surviving group (Figure 1). Based on analysis of variance, patients with the AA genotype had increased susceptibility compared with patients with the AG or GG genotypes (P<0.01) (Figure 2). Based on the Student-Neuman-Keuls procedure, the concentrations of IL-6 in patients with AA genotypes were significantly higher than those in patients with GG genotypes (P<0.01).
Figure 1.
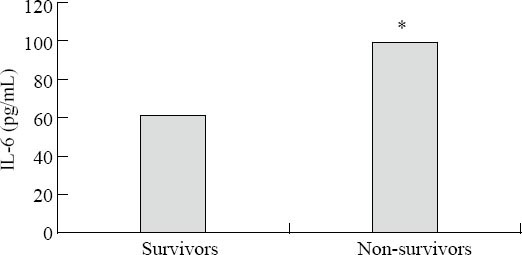
Serum concentrations of IL-6 among the survivors and non-survivors from septic shock. Survivors vs. non-survivors, *P<0.01.
Figure 2.
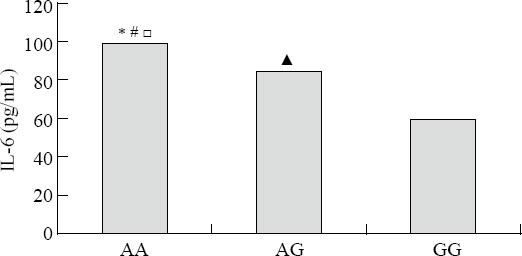
Serum concentrations of IL-6 among the three genotypes (AA, AG, GG rs2234246). Comparisons between the three genotypes, *P<0.01; AA genotypes vs. GG genotypes, #P<0.01; AA genotypes vs. AG genotypes, □P<0.01; AG genotypes vs. GG genotypes, ▲P<0.01.
The frequencies of the three major patterns of haplotypes were comparable in the defined groups (Table 7). Multiple logistic regression analysis or Cox regression analysis also showed that none of the genetic variations within the TREM-1 gene contributed to the incidence or fatal outcome of septic shock, respectively (all P>0.05).
Table 7.
Haplotype analysis of rs2234237and rs2234246 in patients with septic shock and control subjects and in the survivors and non-survivors

DISCUSSION
With recent advances in molecular biology and modern genetics, studies have shown that sepsis is a complex disease resulting from the interactions between environmental factors and genetic factors. In particular, SNPs in the genes participating in the pathophysiologic process of sepsis may contribute to the susceptibility to and outcome of sepsis.[19,20] Recent studies have identified that IL-6 -174G/C and -572G/C, bactericidal permeability increasing (BPI) protein gene Taq polymorphism, human β-defensin 1 gene (DEFB1) -44G/C, and protein C-1641AA genotype as well as IL-1 receptor–associated kinase (IRAK-1) haplotype were associated with the clinical course of severe sepsis.[16–20] It is anticipated that these findings will lead to the use of new diagnostic markers, prognostic indicators, and therapeutic strategies for sepsis.[29]
In this study, two common SNPs in the TREM-1 gene were evaluated with regard to the susceptibility and prognosis of septic shock. The case-control association study showed that TREM-1 genetic polymorphisms rs2234246 was significantly correlated only with susceptibility to septic shock. TREM-1 rs2234246 A allele in the promoter region was significantly associated with the occurrence of septic shock in the recessive model (AA, OR=3.10, 95%CI 1.15 to 8.32, P=0.02), and in the codominant model (AG, OR=0.72, 95%CI 0.43–1.19, P=0.02; AA, OR=2.71, 95%CI 1.00–7.42; P=0.03). But, there was no significant association with the susceptibility of septic shock in the dominant model. The reason may be due to the sample size of this study, which only included fourteen patients with genotype AA in the septic shock group and six patients in the control group. The SNP (rs2234246) located within the 3′ flanking region of the reference TREM-1 gene may influence the transcriptional activity. Therefore, we conclude that G allele may be a protective factor for septic shock susceptibility, and A allele may be a risk factor. The susceptibility analysis showed that patients with the AA genotype had an increased susceptibility compared with those with the GG or AG genotype. TREM-1 genetic polymorphisms show a significant association with the development of septic shock. These findings suggest that TREM-1 genetic polymorphisms have a strong effect on septic shock susceptibility. None of the assayed loci was significantly associated with prognosis of septic shock in the three inheritance models. However, further studies are needed to rule out the possibility that this finding may be attributable to the small sample size, or to exclude other unknown SNPs.
Our results are inconsistent with those reported by Chen et al,[30] who reported that no association was found between the variations rs7768162, rs9471535 and rs2234237 within the TREM-1 gene and either the susceptibility to or the fatal outcome of severe sepsis. However, a study, which investigated an association between TREM-1 gene polymorphisms and severe sepsisconducted in a Chinese cohort, came to another conclusion.[31] They found that there was no association between the variations rs2234246 and rs2234237 within the TREM-1 gene and susceptibility to sepsis. However, the TREM-1 rs2234237 polymorphisms associated with increased 28-day mortality in patients with sepsis. Different conclusions in similar studies may be due to differences in the study population. Patients involved in our study mainly came from Guangdong province rather than other regions of China. Jung et al[32] recently proved that TREM-1 SNPs (rs7768162, rs9471535, and rs2234237) may play a significant role in the development of intestinal Behçet ’s disease and may have modest effects on disease severity. Therefore, TREM -1 gene polymorphism is still a problem and is worthy of investigation in further studies.
According to r2 and D′ values, linkage disequilibrium analysis showed that rs2234237 and rs2234246 were in weak linkage although the physical distance of these two polymorphisms was very close, whereas rs2234237 and rs2234246 were in perfect linkage (D′>0.7). These findings might be explained by the variable recombination rate, natural selection, or other unknown genetic reasons. According to the data from a publicly available database (dbSNP, http://www.ncbi.nlm.nih.gov/SNP), the allelic frequencies of the assayed variations in individuals of different ethnicities were not identical (for example, rs7768162 G-allele: 0.302 in European, 0.47 in Han Chinese, 0.43 in Japanese, and 0.034 in African), which may result in diverse genetic roles of these polymorphisms in different populations in the context of certain diseases. Therefore, the current findings would only apply to Han Chinese rather than to other ethnic populations.
The assayed locus of rs2234246 was significantly associated with susceptibility of sepsis in the recessive and codominant inheritance models. Then we speculated whether different genotypes could affect IL-6 expression. The concentrations of IL-6 were significantly higher in patients with AA genotypes than in those with GG genotypes (Figure 2), i.e., the expression of IL-6 was highly up-regulated in patients with A allele. The engagement of TREM-1 has been shown to induce the production of proinflammatory chemokines such as interleukin (IL)-8 and cytokines such as tumor necrosis factor (TNF)-α and interleukin (IL)-1β. We conclude that patients with AA genotypes are significantly associated with susceptibility of septic shock. Possibly, TREM-1 rs2234246 A allele increases the expression of inflammatory cytokines such as IL-6, TNF-α, etc.
When the association of one or several SNPs in certain diseases is studied, a negative result could not be considered as evidence of lack of implication of the gene in predisposition to the disease, or as evidence to rule out a contribution of this molecule to the pathophysiology of the disease. The results here suggest that the two variations rs2234246 and rs2234237 do not play a major role in the fatal outcome of septic shock in a Chinese Han population. Thus, no major functional differences result from these polymorphisms, or alternatively, possible functional alterations are not relevant to the etiology of septic shock.
Our study has several limitations. First, our study was limited by its small sample size. Because of the small sample size, we were unable to demonstrate strong associations between TREM-1 genetic polymorphisms in the dominant model. Hence, a larger Chinese cohort study is necessary to validate our data. Second, our study investigated only 2 SNPs of TREM-1. Supplementary research is required to uncover new SNPs that might be further related to septic shock susceptibility and prognosis through the full sequencing of the TREM-1 gene and haplotype-based association study. In addition, it would be necessary to search new genes that have similar action on TREM-1, and then analyze the gene-to-gene interaction and functional relevance of these genes. Finally, our results were obtained from a sample in Guangdong Province and should be validated in other ethnic groups.
In conclusion, in a Chinese Han population, TREM-1 rs2234246 A allele in the promoter region was significantly associated with the occurrence of septic shock in the recessive and codominant models. However, in the three inherited models (dominant model, recessive model, and codominant model), none of the assayed loci was significantly associated with the outcome of septic shock. Further studies are needed to clarify its association with other similar molecules involved in the inflammatory response to septic shock.
ACKNOWLEDGEMENT
We wish to thank all of the doctors and nurses in the Department of Intensive Care Unit, Affiliated Hospital of Guangdong Medical College.
Footnotes
Funding: This study was supported by Science & Technology Pillar Program of Guangdong Province (2009BAI86B03).
Ethical approval: The study was approved by the local ethical committee.
Conflicts of interest: There is no conflict of interest in this study.
Contributors: Peng LS proposed and wrote the study. All authors contributed to the design and interpretation of the study, and approved the final manuscript.
REFERENCES
- 1.Sands KE, Bates DW, Lanken PN, Graman PS, Hibberd PL, Kahn KL, et al. Epidemiology of sepsis syndrome in 8 academic medical centers. JAMA. 1997;278:234–240. [PubMed] [Google Scholar]
- 2.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–150. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 3.Dunn DL. Gram-negative bacterial sepsis and sepsis syndrome. Surg Clin North Am. 1994;74:621–635. [PubMed] [Google Scholar]
- 4.Tsiotou AG, Sakorafas GH, Anagnostopoulos G, Bramis J. Septic shock: current pathogenetic concepts from a clinical perspective. Med Sci Monit. 2005;11:RA76–85. [PubMed] [Google Scholar]
- 5.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365:63. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 6.Russell JA. Management of sepsis. N Engl J Med. 2006;355:1699. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 7.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 8.Bouchon A, Dietrich J, Colonna M. Cutting edge: Inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- 9.Klesney-Tait J, Turnbull IR, Colonna M. The TREM receptor family and signal integration. Nat Immunol. 2006;7:1266. doi: 10.1038/ni1411. [DOI] [PubMed] [Google Scholar]
- 10.Colonna M, Facchetti F. TREM-1 (triggering receptor expressed on myeloid cells): a new player in acute inflammatory responses. J Infect Dis. 2003;187(Suppl 2):S397–401. doi: 10.1086/374754. [DOI] [PubMed] [Google Scholar]
- 11.Radaev S, Kattah M, Rostro B, Colonna M, Sun PD. Crystal structure of the human myeloid cell activating receptor TREM-1. Structure (Camb) 2003;11:1527–1535. doi: 10.1016/j.str.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Gu Q, Zheng YS. Expression of triggering receptor-1 in myeloid cells of mice with acute lung injury. World J Emerg Med. 2010;1:144–148. [PMC free article] [PubMed] [Google Scholar]
- 13.Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 14.Chang RM, Wen LQ, Chang JX, Fu YR, Jiang ZP, Chen S. Repair of damaged intestinal mucosa in a mouse model of sepsis. World J Emerg Med. 2013;4:223–228. doi: 10.5847/wjem.j.issn.1920-8642.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibot S, Cravoisy A, Kolopp-Sarda MN, Béné MC, Faure G, Bollaert PE, et al. Time-course of sTREM (soluble triggering receptor expressed on myeloid cells)– 1, procalcitonin, and C-reactive protein plasma concentrations during sepsis. Crit Care Med. 2005;33:792. doi: 10.1097/01.ccm.0000159089.16462.4a. [DOI] [PubMed] [Google Scholar]
- 16.Michalek J, Svetlikova P, Fedora M, Klimovic M, Klapacova L, Bartosova D, et al. Interleukin-6 gene variants and the risk of sepsis development in children. Hum Immunol. 2007;68:756–760. doi: 10.1016/j.humimm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Michalek J, Svetlikova P, Fedora M, Klimovic M, Klapacova L, Bartosova D, et al. Bactericidal permeability increasing protein gene variants in children with sepsis. Intensive Care Med. 2007;33:2158–2164. doi: 10.1007/s00134-007-0860-3. [DOI] [PubMed] [Google Scholar]
- 18.Chen QX, Lv C, Huang LX, Cheng BL, Xie GH, Wu SJ, et al. Genomic variations within DEFB1 are associated with the susceptibility to and the fatal outcome of severe sepsis in Chinese Han population. Genes Immun. 2007;8:439–443. doi: 10.1038/sj.gene.6364401. [DOI] [PubMed] [Google Scholar]
- 19.Arcaroli J, Silva E, Maloney JP, He Q, Svetkauskaite D, Murphy JR, et al. Variant IRAK-1 haplotype is associated with increased nuclear factor-kappa B activation and worse outcomes in sepsis. Am J Respir Crit Care Med. 2006;173:1335–1341. doi: 10.1164/rccm.200603-341OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walley KR, Russell JA. Protein C -1641 AA is associated with decreased survival and more organ dysfunction in severe sepsis. Crit Care Med. 2007;35:12–17. doi: 10.1097/01.CCM.0000249823.44726.4E. [DOI] [PubMed] [Google Scholar]
- 21.Members of the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit Care Med. 1992;20:864. [PubMed] [Google Scholar]
- 22.Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, et al. SCCM/ESICM/ACCP/ATS/SIS, 2001SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit Care Med. 2003;31:1250. doi: 10.1097/01.CCM.0000050454.01978.3B. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Z, Xiao J, Jiang Y, Zhang S, Yu M, Zhao J, et al. A novel method based on ligase detection reaction for low abundant YIDD mutants detection in hepatitis B virus. Hepatol Res. 2006;34:150–155. doi: 10.1016/j.hepres.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: A web tool for the analysis of association studies. Bioinformatics. 2006;22:1928. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. Epub 2004 Aug 5. [DOI] [PubMed] [Google Scholar]
- 26.Dupont WD, Plummer WD. PS power and sample size program available for free on the Internet. Control Clin Trials. 1998;19:589. doi: 10.1016/s0197-2456(98)00037-3. [DOI] [PubMed] [Google Scholar]
- 27.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Cobb JP, O'Keefe GE. Injury research in the genomic era. Lancet. 2004;363:2076. doi: 10.1016/S0140-6736(04)16460-X. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Zhou H, Wu S, Wang H, Lv C, Cheng B, et al. Lack of as sociation between TREM- 1 gene polymorphisms and severe sepsis in a Chinese Han population. Hum Immunol. 2008;69:220–226. doi: 10.1016/j.humimm.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Su L, Liu C, Li C, Jiang Z, Xiao K, Zhang X, et al. Dynamic changes in serum soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) and its gene polymorphisms are associated with sepsis prognosis. Inflammation. 2012;35:1833–1843. doi: 10.1007/s10753-012-9504-z. [DOI] [PubMed] [Google Scholar]
- 32.Jung ES, Kim SW, Moon CM, Shin DJ, Son NH, Kim ES, et al. Relationships between genetic polymorphisms of triggering receptor expressed on myeloid cells-1 and inflammatory bowel diseases in the Korean population. Life Sci. 2011;89:289–294. doi: 10.1016/j.lfs.2011.06.018. [DOI] [PubMed] [Google Scholar]


