Abstract
Diabetic complications including diabetic nephropathy, retinopathy, and neuropathy are as major causes of morbidity and mortality in diabetes individuals worldwide and current therapies are still unsatisfactory. One of the reasons for failure to develop effective treatment is the lack of fundamental understanding for underlying mechanisms. Genetic studies are powerful tools to dissect disease mechanism. The heritability (h2) was estimated to be 0.3-0.44 for diabetic nephropathy and 0.25-0.50 for diabetic retinopathy respectively. Previous linkage studies for diabetic nephropathy have identified overlapped linkage regions in 1q43-44, 3q21-23, 3q26, 10p12-15, 18q22-23, 19q13, 22q11-12.3 in multiple ethnic groups. Genome-wide association studies (GWAS) of diabetic nephropathy have been conducted in several populations. However, most of the identified risk loci could not be replicated by independent studies with a few exceptions including those in ELMO1, FRMD3, CARS, MYO16/IRS2, and APOL3-MYH9 genes. Functional studies of these genes revealed the involvement of cytoskeleton reorganization (especially non-muscle type myosin), phagocytosis of apoptotic cells, fibroblast migration, insulin signaling, and epithelial clonal expansion in the pathogenesis of diabetic nephropathy. Linkage analyses of diabetic retinopathy overlapped only in 1q36 region and current results from GWAS for diabetic retinopathy are inconsistent. Conclusive results from genetic studies for diabetic neuropathy are lacking. For now, small sample sizes, confounding by population stratification, different phenotype definitions between studies, ethnic-specific associations, the influence of environmental factors, and the possible contribution of rare variants may explain the inconsistencies between studies.
Keywords: Microvascular complications, Nephropathy, Retionopathy, Neuropathy, Diabetes
Core tip: Most risk genetic loci identified by genome-wide association studies (GWAS) for diabetic nephropathy could not be replicated by independent studies with a few exceptions including those in ELMO1, FRMD3, CARS, MYO16/IRS2, and APOL3-MYH9 genes. These findings highlighted the importance of cytoskeleton reorganization, phagocytosis of apoptotic cells, fibroblast migration, insulin signaling, and epithelial clonal expansion in the pathogenesis of diabetic nephropathy. Conclusive results from GWAS for diabetic retinopathy and diabetic neuropathy are currently lacking.
INTRODUCTION
The prevalence of diabetes mellitus is increasing globally, especially in developing countries[1]. This surge of diabetes mellitus prevalence poses a serious threat to the public and diabetic complications are ranked as major causes of morbidity and mortality worldwide. Several common mechanisms underlying these microvascular complications including the polyol pathway, advanced glycation end products pathway, protein kinase C pathway, the hexosamine pathway, and cytokines such as nuclear factor-κB, tumor growth factor-β, and vascular endothelial growth factor are well described and the unifying mechanism of superoxide production have been proposed[2]. Nevertheless, therapies targeting these pathways have not been very successful[3-5]. One of the reasons is the lack of fundamental understanding for underlying mechanisms.
Genetic studies provide a powerful tool to the understanding of disease mechanism. Previous family linkage analyses have successfully identified mutations responsible for high-penetrating monogenetic disease. Some discoveries, for example, the identification of PCSK9 mutation through linkage analyses in hypercholesterolemic families, have resulted in major breakthroughs in therapy[6,7]. However, family linkage analysis is generally not adequately powered to detect genetic loci of complex disease. Over the last few years, the advent of genome-wide association studies (GWAS) have launched a great leap toward the genetic basis of complex diseases such type 2 diabetes mellitus, cancers, and psychiatric diseases. Intriguingly, many of the identified genetic loci were not previously considered to be related to these diseases and the discoveries indeed illuminated important pathophysiological pathways to these complex diseases. Diabetic microvascular complications are complex traits influenced by both environmental and genetic factors, and compelling evidences indicate that diabetic microvascular complications are heritable[8-12]. Here in this review, we only summarized the progress in the genetics for diabetic microvascular complications.
GENETICS STUDIES OF DIABETIC NEPHROPATHY
Linkage studies of diabetic nephropathy
The heritability (h2) of diabetic nephropathy (DN) defined by reduced glomerular filtration rate (GFR) or albuminuria was estimated to be 0.3-0.44 in multiple Caucasian diabetic populations[8-10]. Previous linkage studies have repeatedly identified linkage region in 1q43-44, 3q21-23, 3q26, 10p12-15, 18q22-23, 19q13, 22q11-12.3 in multiple ethnic groups (Figure 1, Table 1)[13-24]. However, these linkage regions usually spanned over megabases and therefore exact locus or risk gene is unclear. In contrast, the resolution of linkage disequilibrium mapping (also called association mapping) is much higher than linkage studies. The distinction between linkage and association mapping is that family linkage mapping use the small amount of recombination events that occurs in each generation within a pedigree to localize a chromosomal region, which usually contains hundreds of genes; while population-based case-control association mapping uses large amount of recombinations that occurred during the evolutional history of a population to locate the risk loci, which generally did not extend over a few genes. However, population-based case-control association studies are susceptible to the population stratification and independent replication is essential to confirm the result of association studies.
Figure 1.
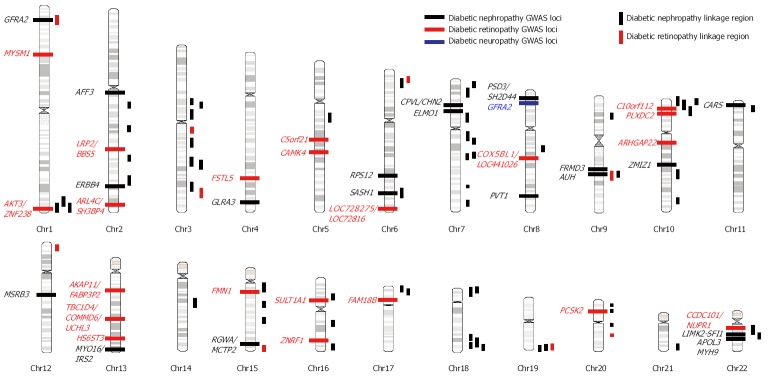
Genetic risk loci for diabetic nephropathy, retinopathy, and neuropathy indentified through genome-wide linkage (vertical bar) and association scans (horizontal bar). GWAS: Genome-wide association study; Chr: Chromosome.
Table 1.
Genome-wide linkage studies for diabetic nephropathy and retinopathy
| Ethnicity and sample size | Type of diabetes | Phenotype definition | Linkage region (LOD score or P-value or MLS) | Ref. |
| Diabetic nephropathy | ||||
| 954 African American, 781 American Indians, 614 European American, 1611 Mexican Americans (FIND) | 1 + 2 | Estimated GFR | 10p12.311 (LOD: 2.16), 1q431 (2.26), 2q31.3 (1.91), 3p12.1 (2.19), 7q11.22 (2.19), 10p141 (2.16), 15q12 (2.84), 20q11.111 (3.34) | [13] |
| 218 African American, 335 American Indians, 119 European American, 469 Mexican Americans (FIND) | 1 + 2 | Urine ACR | 7q21.3 (P = 8.6 x 10-5), 10p15.31 (1.29 x 10-5), 14q23.1 (7.8 x 10-4), 18q22.31 (2.17 x 10-3) | [14] |
| 3972 Americans (African American, American Indians, European American, Mexican Americans) (FIND) | 1 + 2 | DN defined by macroalbuminuria or ESRD, ACR | DN: 1q431 (LOD: 2.00), 6p24.3 (2.84), 7p21.3 (2.81), 10p15.11 (2.10), 11p15.3 (2.28), 15q21.1 (2.04)ACR: 2q22.3 (2.04), 3p13 (2.76), 7q21.2 (2.96), 16q13 (2.31), 22q12.31 (2.29) | [23] |
| 882 American (African American, American Indians, European American, Mexican Americans) (FIND) | 1 + 2 | eGFR | 1q431 (LOD: 1.87), 7q36.1 (4.23), 8q13.3 (2.75), 15q22.3 (2.08), 18q23.3 (1.40) | [24] |
| 100 United States sibling pairs (Joslin Study on Genetics of Diabetic Nephropathy) | 1 | Proteinuria or ESRD | 1q441 (MLS: 1.6), 2q14.1 (2.1), 3p13 (0.6), 5q14.2 (2.7), 10q26.1 (2.4), 17p13.1 (1.9), 19q13.431 (3.1), 20p12.1 (1.8) | [15] |
| 63 extended United States families (Joslin Study on Genetics of Diabetic Nephropathy) | 2 | GFR | 2q33.3 (LOD: 4.1), 10q23.31 (3.1), 18p11.22 (2.2) | [19] |
| 556 Finnish, Danish, and French (FinnDiane) | 1 | Macroalbuminuria or ESRD | 3q21-251 (LOD: 0.76), 6p21 (2.31), 9p21.2, 16p12, 19q131 (1.61), 22q11 (2.78) | [16] |
| 83 Finnish sibling pairs | 1 | Macroalbuminuria or ESRD | 3q21.3-231 (MLS: 2.67) | [21] |
| 18 Turkish family + 101 sibling pairs of Pima India | 2 | Macroalbuminuria | 18q22.3-231 (max LOD:6.14) | [17] |
| 201 Pima India sibling pairs | 2 | Macroalbuminuria or ESRD | 3q26.11 (LOD: 1.48), 7q32.3 (2.04), 20p12.3 (1.83) | [18] |
| 206 African American sibling pairs | 2 | ESRD | 3q13.31 (LOD: 4.55), 7p21.1 (3.59), 18q22.11 (3.72) | [22] |
| 691 West African | 2 | GFR | 7p12.2 (LOD: 1.84), 16q24.1 (3.56), 17p13.2 (2.08) | [20] |
| Diabetic retinopathy | ||||
| 282 Mexican American sibling pairs | 2 | Non-proliferative DR and proliferative DR | 3q12.3 (LOD: 2.41), 12p13.31 (2.47), 20q13.12 (4.47), 6p24.1 (2.28), 15q26.3 (2.53), 19q13.42 (2.21) | [45] |
| 725 Pima Indian sibling pairs | 2 | Worse eye score | 1p36.13 (LOD: 3.1) | [46] |
| 210 Pima Indian sibling pairs | 2 | Hemorrhage, microaneurysm, and proliferative DR | 3q26.31 (LOD: 1.36), 9q22.33 (1.46) | [18] |
Overlapped region. MLS: Maximum LOD score; DN: Diabetic nephropathy; ESRD: End-stage renal disease; GFR: Glomerular filtration rate; ACR: Albumin-to-creatinine ratio; DR: Diabetic retinopathy.
Association studies of DN in type 2 diabetic patients
Several GWAS of DN have been conducted in several ethnic populations (Table 1, Figure 1). ELMO1 (the engulfment and cell motility 1 gene) was first found to be associated with diabetic nephropathy in a GWAS in Japanese 2 diabetic patients (546 DN cases and 334 type 2 diabetic controls)[25]. Replication studies in the GoKinD collection (558 DN cases and 820 type 2 diabetes controls)[26], two African American cohorts [1136 end-stage renal diseae (ESRD) diabetes cases and type 2 diabetic 1160 controls][27], a Chinese population (123 DN cases and 77 type 2 diabetic controls)[28], and a Caucasian GWAS (547 ESRD and 549 type 1 diabetic controls)[29] confirmed this finding although the risk SNPs are not exactly the same with those reported in the original Japanese population (intron 16-20 in original Japanese GWAS, intron 16-20 in GoKinD, intron 13 in African Americans, intron 18 in Chinese). In a large meta-analysis of the GENIE consortium (including UK-ROI, FinnDiane, and GoKinD US) involving 2966 DN cases and 3399 type 1 diabetic controls, an expanded investigation of the ELMO1 locus yielded only nominal associations with DN[30]. Furthermore, another replication studies in Pima Indians of Arizona involving 248 DN cases and 524 diabetic controls found significant association of SNPs in intron 13 but the associations were in the opposite direction from those observed in African Americans[31], and another study in 455 Mexican-American patients with DN and 437 controls failed to replicate the association[32].
A GWAS in Pima Indians comparing 105 diabetic ESRD and 103 controls identified plasmacytoma variant translocation (PVT1), an lncRNA gene, was associated with DN in type 2 diabetic patients[33]. Another large GWAS in an initial set of 965 African American type 2 diabetic patients with ESRD and 1029 controls without type 2 diabetes or kidney disease and further replication studies in 1246 type 2 diabetic patients indentified SASH1 (SH3 Domain Containing 1), RPS12 (ribosomal protein S12), AUH (AU RNA binding protein/enoyl-CoA hydratase), MSRB3 (methionine sulfoxide reductase B3), LIMK2 (LIM domain kinase 2)-SFI1 (Sfi1 homolog, spindle assembly associated), APOL3 (apolipoprotein L, 3), and MYH9 (myosin, heavy chain 9, non-muscle) genes as risk loci[34]. Among them, the association of MYH9 risk variants has been replicated in another study involving 1963 European Americans diabetic patients[35]. Compelling evidence demonstrated that APOL3-MYH9 gene clusters are also associated with non-diabetic nephropathy including focal segmental glomerulosclerosis and hypertensive nephropathy in African American as well as other ethnic populations[36-38].
Association studies of DN in type 1 diabetic patients
A large GWAS in a initial set of 820 DN cases and 885 type 1 diabetic controls in the GoKinD study and a replication set of 1304 participants in the Diabetes Control and Complication Trial/Epidemiology of Diabetes Control and Complication (EDIC) identified FRMD3 (FERM domain containing 3), cysteinyl-tRNA synthase (CARS), carboxypeptidase, vitellogenic-like (CPVL)/chimerin 2, and intergenic region at 13q33.3 between MYO16 and insulin receptor substrate 2 (IRS2) associated with DN[39]. Interestingly, another genome-wide linkage analysis and regional association fine mapping in 1007 general Mongolian also identified SNPs in the FRMD3, glycine amidinotransferase, and spermatogenesis associated 5-like 1 genes associated with estimated glomerular filtration rate[40]. A family-based candidate-gene association study involving 798 type 2 diabetic members in the Joslin Study of Genetics of Nephropathy replicated the association of SNPs in the FRMD3, CARS, and 13q33.3 between MYO16 and IRS2 genes[41]. Another GWAS in 547 Caucasian ESRD cases and 549 type 1 diabetic controls identified ZMIZ1 (zinc finger, MIZ-type containing 1) gene is associated with DN[29]. This study also observed significant association of 13q33 variant near the MYO16/IRS2 genes[29]. However, in a large replication study of 1535 Japanese type 1 and 2 diabetic patients, only variants in 13q33.3 between MYO16/IRS2 gene but not those in FRMD3, CPVL/CHN2, or CARS are significantly associated with DN[42]. Furthermore, a large meta-analysis of the GENIE consortium (UK-ROI, FinnDiane, and GoKinD US) involving 2966 type 1 diabetic cases with DN and 3399 type 1 diabetes controls failed to replicate the association between SNPs in the FRMD3, CARS, and 13q33 loci near MYO16 and IRS2 genes[30].
A recent huge meta-analysis involving 4315 type 1 diabetic nephropathy and ESRD cases and 8568 type 1 diabetic controls of the GENIE consortium and subsequent replication analyses in 9 independent cohorts (1880 cases and 6656 controls) revealed risk SNPs in the AFF3 (AF4/FMR2 family, member 3) and ERBB4 (v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4) genes and an intergenic SNP between RGMA (repulsive guidance molecule family member a)/MCTP2 (multiple C2 domains, transmembrane 2) genes[43]. Another large GWAS for 24-h urine albumin excretion rare in type 1 diabetic patients including an initial set of 1925 patients (FinnDiane) and 3750 additional patients from 7 follow-up studies (Steno Diabetes Center, Italian individuals from the Milano region, Umea Diabetes Study from Sweden, Scania Diabetes Registry, NFS-ORPQ, UK-ROI) identified the strongest signal from the PSD3 (pleckstrin and Sec7 domain containing 3)/SH2D4A (SH2 domain containing 4A) genes[44].
Collectively, current data from GWAS are not very consistent and only genetic loci in the ELMO1, FRMD3, APOL3-MYH9, CARS, and 13q33 between MYO16 and IRS2 genes have been successfully replicated in independent studies.
GENETIC STUDIES OF DIABETIC RETINOPATHY
Linkage studies of diabetic retinopathy
The heritability of diabetic proliferative retinopathy is estimated to be 0.25-0.50 in Caucasian populations[11,12]. Previous results of three family linkage analyses for diabetic retinopathy (DR) are summarized in Table 1 and Figure 1[18,45,46]. However, the only overlapped region is 1q36 between Pima Indians (LOD: 3.1) and Mexican Americans (LOD: 1.24) studies[45,46].
Association studies of DR
Four GWAS of DR have been published till now (Table 2, Figure 1). A large meta-analysis of GWAS in the GoKinD and EDIC cohorts involving 2829 cases of severe diabetic retinopathy defined by proliferative retinopathy and macular edema and 1856 type 1 diabetic controls identified several possible loci including intergenic SNPs between AKT3/ZNF238, LEKR1/CCNL1, KRT18P34/VEPH1 and SNP in the A2BP1 genes with P-value less than 10-6[47]. After excluding cases with concomitant nephropathy to identify DR-specific genes, SNPs in the intergenic region between LOC728275/LOC728316, the CCDC101/NUPR1/SULT1A2/SULT1A1 gene clusters, the FAM18B, AKAP11/FABP3P2/TNFSF11 gene cluster, and intergenic region between COX5BL1/LOC441026, ZNRF1, PCSK2, C10orf112 genes were found to be associated with DR[47]. A GWAS for DR involving 174 Taiwanese type 2 diabetic non-proliferative and proliferative retinopathy cases and 575 controls identified several genetic loci with P-value less than 10-6, including MYSM1, FSTL5, C5orfF21, PLXD2, ARHGAP22, and HS6ST3[48]. Another GWAS in Taiwanese identified three risk loci in TBC1D4-COMMD6-UCHL3, LRP2-BBS5, and ARL4C-SH3BP4 genes in the initial set of 437 cases of proliferative retinopathy and 570 type 2 diabetic controls. However, none of them were replicated in another 585 Hispanic diabetics[49]. A smaller GWAS comparing 103 Mexican-American type 2 diabetics with severe retinopathy and 183 type 2 diabetics identified suggestive signals in the CAMK4 and FMN1 genes[50]. However, the results from these 4 GWAS did not overlap with each other.
Table 2.
Genome-wide association studies for diabetic nephropathy, retinopathy, and neuropathy
| Patients | Ethnic | Case | Control | Gene | Ref. | Replication studies | Non-replication studies |
| Diabetic nephropathy | |||||||
| T2DM | Japanese | 459 DN | 242 | ELMO11 | [25] | 26, 27, 28, 29, 30 | 31, 32 |
| T2DM | European | 105 ESRD | 102 | PVT1 | [33] | ||
| T2DM | African American | 965 ESRD | 1029 | SASH1, RPS12,AUH, MSRB3, LIMK2-SKI1, APOL3-MYH91 | [34] | 35 | |
| T1DM | Caucasian (GoKinD, DCCT/EDIC) | 820 ESRD | 885 | FRMD31, CARS, CPVL/CHN2, 13q3 between MYO16/IRS21 | [39] | 40, 41, 42 | 42, 30 |
| T1DM | Caucasian | 547 ESRD | 549 | ZMIZ1 | [29] | ||
| T1DM | GENIE (UK-ROI, FinnDiane, GoKinUS) + 9 follow-up studies | Stage 1: 4315 ESRDStage 2: 1880 ESRD | Stage 1: 8568Stage 2: 6656 | AFF3, RGMA/MCTP2, ERBB4 | [43] | ||
| T1DM | Caucasian(FinnDiane + 7 follow-up studies) | 5675 T1DMUrine albumin excretion rate | PSD3, SH2D4A | [44] | |||
| Diabetic retinopathy | |||||||
| T1DM | Caucasian (GoKinD and EDIC) | 2829 PDR and macular edema | 1856 | AKT3/ZNF238, LEKR1/CCNL1, KRT18P34/VEPH1, A2BP1 | [47] | ||
| T2DM | Taiwanese | 174 NPDR and PDR | 575 | MYSM1, FSTL5, C5orfF21, PLXD2, ARHGAP22, HS6ST3 | [48] | ||
| T2DM | Taiwanese | 437 PDR | 570 | TBC1D4-COMMD6-UCHL3, LRP2-BBS5, and ARL4C-SH3BP4 | [49] | ||
| T2DM | Mexican-American | 103 severe DR | 183 | CAMK4, FMN1 genes | [50] | ||
| Diabetic neuropathy | |||||||
| United Kingdom | United Kingdom (GoDART) | 572 diabetic neuropathic pain | 2491 | GFRA2 | [51] | ||
Loci that could be replicated in independent studies. T1DM: Type 1 diabetes mellitus; T2DM: Type 2 diabetes mellitus; DN: Diabetic nephropathy; ESRD: End-stage renal disease; GFR: Glomerular filtration rate; DR: Diabetic retinopathy; NPDR: Non-proliferative diabetic retinopathy; PDR: Proliferative diabetic retinopathy.
GENETIC STUDY OF DIABETIC NEUROPATHY
There was no heritability estimation for diabetic neuropathy in human and no family linkage study for diabetic neuropathy. Only GWAS comparing 572 diabetic neuropathic pain cases defined by treatment for diabetic neuropathic pain and positive monofilament test and 2491 diabetic controls in the Genetics of Diabetes Audit and Research Tayside (GoDARTS) identified potential signals from GFRA2 gene[51] (Table 2, Figure 1).
PHYSIOLOGICAL INSIGHT FROM GENETIC STUDIES
The ELMO1 gene encode for a signaling molecule involved in phagocytosis of apoptotic cells[52,53], fibroblast migration[52,54,55], cytoskeleton reorganization[56], and lymphocyte infiltration[57] through interaction with DOCK2 and DOCK180 (Figure 2A). ELMO1 expression was found to be elevated in cells cultured under high glucose conditions and in the kidney of diabetic mice, but was weakly detectable in tubular and glomerular epithelial cells in normal kidney[25].
Figure 2.
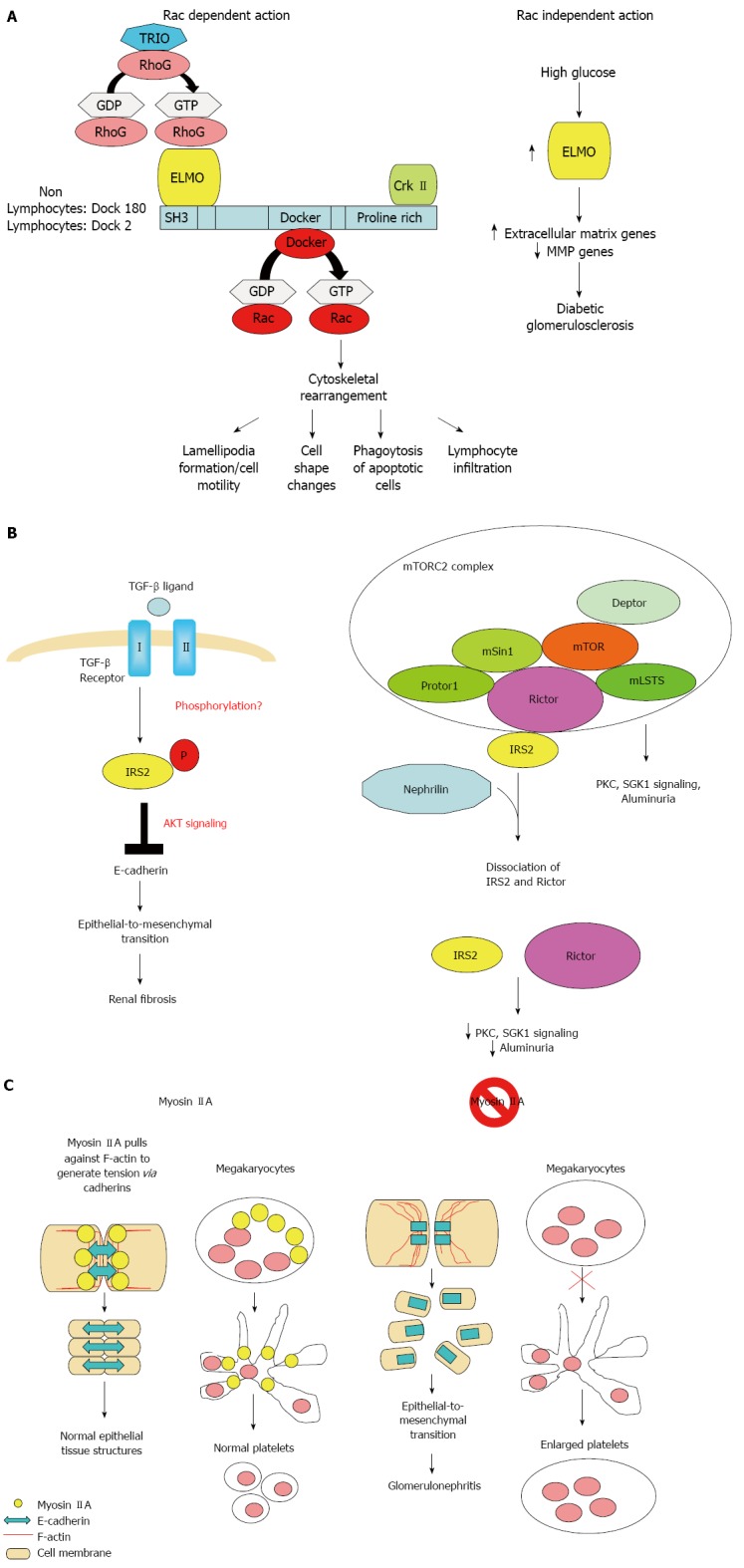
Possible molecular mechanisms. Possible molecular mechanisms by which ELMO1 (A), IRS2 (B), and MYH9 (C) regulate diabetic nephropathy. TRIO: Triple functional domain (PTPRF interacting); RhoG: Ras homolog family member G; GDP: Guanosine diphosphate; GTP: Guanosine triphosphate; MMP: Matrix metalloproteinases; Crk II: V-Crk Avian Sarcoma Virus CT10 Oncogene Homolog II; TGF-β: Transforming growth factor beta; AKT: Protein kinase B; mLSTS: Mammalian lethal with SEC13 protein 8; mTOR: Mammalian target of rapamycin; mTORC2: Mammalian target of rapamycin complex 2; mSin1: Mammalian SAPK interacting protein; PKC: Protein kinase C; SGK1: Serum- and glucocorticoid-induced kinase 1.
The FRMD3 gene encodes for a member of the protein 4.1 superfamily. FRMD3 has been demonstrated to be silenced in lung cancer tissue in genomic screening. FRMD3 overexpression in different epithelial cell lines decreased clonal expansion, indicating FRMD3 as a potential tumor suppressor gene[58]. The CARS encodes for a cysteinyl-tRNA synthetase, which is a frequent gene fusion partner of anaplastic lymphoma kinase found in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor[59,60]. However, the link between FRMD3 or CARS and diabetic nephropathy is currently poorly understood.
The 13q33 risk loci lie between the MYO16 and IRS2 genes. The MYO16 gene encodes a novel unconventional myosin with divergent tails that is presumed to bind to membranous compartments and interact with actin filaments. MYO16 has also been shown to be expressed during brain development and regulate neuronal morphogenesis through interaction with protein phosphatase and modulation of phosphoinositide 3-kinase signaling[61]. A GWAS for autism has identified risk loci within an intergenic region between the MYO16 and IRS2 genes[62]. A genome-wide linkage study and regional fine mapping for schizophrenia[63] and another GWAS of the Framingham Heart Study for pulse pressure[64] have identified MYO16 as risk loci, indicating MYO16 may play pleiotropic functions.
The IRS2 gene encodes for an adaptor protein that interacts directly with the insulin receptors and the insulin-like growth factor I receptor and is a key mediator of insulin signaling. IRS2 was expressed in renal epithelial and tubular cells. Deletion of Irs2 causes reduced kidney size and reduced glomerular number in mice[65]. A study of transcriptome and metabolome profiles of the primary cultured inner medullary collecting duct cells grown in hyperosmolar culture medium identified IRS2 levels to be significantly altered[66]. IRS2 expression in kidney tubules has also been shown to be elevated nine fold in human diabetic nephropathy patients[67]. Transforming growth factor (TGF)-β1 is the primary cytokine shown to induce fibrosis. IRS2 has been shown to mediate TGF-β1 signals in kidney epithelial cells[68]. IRS2 has also been shown to interact with nuclear complex of rictor to regulate albuminuria in diabetic mice[69] (Figure 2B).
Mutations in MYH9 results in a familial autosomal dominant syndrome characterized by a variety of clinical features, including macrothrombocytopenia, deafness, nephritis, and cataract[70]. GWAS also identified common MYH9 polymorphism as risk loci for non-diabetic nephropathy including focal segmental glomerulosclerosis and hypertensive nephropathy[36,27]. MYH9 encodes the non-muscle myosin heavy chain 9, which, with other subunits, forms myosin II. Myosin II is a motor protein that binds actin to regulate cellular motility. MYH9 is expressed in the podocytes, as well as in mesangial cells and arteriolar and peritubular capillaries in kidneys[71]. Classical deletion of Myh9 in mice results in embryonic lethality due to loss of cell-cell adhesion and loss of cell movement during gastrulation. Podocyte-specfic deletion of Myh9 in C57BL/6 mice results in susceptibility to experimental doxorubicin hydrochloride glomerulopathy[71]. Several strains of Myh9 knockin mice showed macrothrombocytopenia, premature cataract formation, kidney abnormalities, including albuminuria, focal segmental glomerulosclerosis and progressive kidney disease, and mild hearing loss[72,73] (Figure 2C).
LIMITATIONS AND PROSPECTIVE
The major limitation of family linkage studies is their low resolution and power to detect variants with small effects, especially for complex genetic diseases. GWAS is a hypothesis-free and unbiased tool with finer resolution and greater power to detect risk loci. However, false positivity often results from population admixture or stratification in GWAS. Therefore, independent replications are essential for genetic association studies. However, current results from GWAS are not consistent since most identified loci are not reproducible except for a few genes such as ELMO1, CARS, FRMD3, MYO16/IRS2, and APOL3/MYH9. Small sample sizes, different phenotype definitions between studies, population-specific associations, and strong influence of environmental factors (medications, co-morbidities) may explain the failure of GWAS for diabetic complications. While GWAS are usually designed for common variants, rare variants with intermediate effects within should also be pursued with next-generation sequencing. The interaction with environmental factors should also be taken into account.
Footnotes
Conflict-of-interest: None.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: September 9, 2014
First decision: December 17, 2014
Article in press: March 18, 2015
P- Reviewer: Das UN, Ido Y, Kaya C, Uehara Y, Verrotti A S- Editor: Gong XM L- Editor: A E- Editor: Zhang DN
References
- 1.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 3.Shamsi HN, Masaud JS, Ghazi NG. Diabetic macular edema: New promising therapies. World J Diabetes. 2013;4:324–338. doi: 10.4239/wjd.v4.i6.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gosmanov AR, Wall BM, Gosmanova EO. Diagnosis and treatment of diabetic kidney disease. Am J Med Sci. 2014;347:406–413. doi: 10.1097/MAJ.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 5.Spallone V, Lacerenza M, Rossi A, Sicuteri R, Marchettini P. Painful diabetic polyneuropathy: approach to diagnosis and management. Clin J Pain. 2012;28:726–743. doi: 10.1097/AJP.0b013e318243075c. [DOI] [PubMed] [Google Scholar]
- 6.Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 7.Stein EA, Mellis S, Yancopoulos GD, Stahl N, Logan D, Smith WB, Lisbon E, Gutierrez M, Webb C, Wu R, et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N Engl J Med. 2012;366:1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 8.Forsblom CM, Kanninen T, Lehtovirta M, Saloranta C, Groop LC. Heritability of albumin excretion rate in families of patients with Type II diabetes. Diabetologia. 1999;42:1359–1366. doi: 10.1007/s001250051450. [DOI] [PubMed] [Google Scholar]
- 9.Fogarty DG, Rich SS, Hanna L, Warram JH, Krolewski AS. Urinary albumin excretion in families with type 2 diabetes is heritable and genetically correlated to blood pressure. Kidney Int. 2000;57:250–257. doi: 10.1046/j.1523-1755.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- 10.Langefeld CD, Beck SR, Bowden DW, Rich SS, Wagenknecht LE, Freedman BI. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis. 2004;43:796–800. doi: 10.1053/j.ajkd.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 11.Arar NH, Freedman BI, Adler SG, Iyengar SK, Chew EY, Davis MD, Satko SG, Bowden DW, Duggirala R, Elston RC, et al. Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest Ophthalmol Vis Sci. 2008;49:3839–3845. doi: 10.1167/iovs.07-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hietala K, Forsblom C, Summanen P, Groop PH. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176–2180. doi: 10.2337/db07-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thameem F, Igo RP, Freedman BI, Langefeld C, Hanson RL, Schelling JR, Elston RC, Duggirala R, Nicholas SB, Goddard KA, et al. A genome-wide search for linkage of estimated glomerular filtration rate (eGFR) in the Family Investigation of Nephropathy and Diabetes (FIND) PLoS One. 2013;8:e81888. doi: 10.1371/journal.pone.0081888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyengar SK, Abboud HE, Goddard KA, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, et al. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the family investigation of nephropathy and diabetes (FIND) Diabetes. 2007;56:1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- 15.Rogus JJ, Poznik GD, Pezzolesi MG, Smiles AM, Dunn J, Walker W, Wanic K, Moczulski D, Canani L, Araki S, et al. High-density single nucleotide polymorphism genome-wide linkage scan for susceptibility genes for diabetic nephropathy in type 1 diabetes: discordant sibpair approach. Diabetes. 2008;57:2519–2526. doi: 10.2337/db07-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wessman M, Forsblom C, Kaunisto MA, Söderlund J, Ilonen J, Sallinen R, Hiekkalinna T, Parkkonen M, Maxwell AP, Tarnow L, et al. Novel susceptibility locus at 22q11 for diabetic nephropathy in type 1 diabetes. PLoS One. 2011;6:e24053. doi: 10.1371/journal.pone.0024053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vardarli I, Baier LJ, Hanson RL, Akkoyun I, Fischer C, Rohmeiss P, Basci A, Bartram CR, Van Der Woude FJ, Janssen B. Gene for susceptibility to diabetic nephropathy in type 2 diabetes maps to 18q22.3-23. Kidney Int. 2002;62:2176–2183. doi: 10.1046/j.1523-1755.2002.00663.x. [DOI] [PubMed] [Google Scholar]
- 18.Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC. Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Pima Diabetes Genes Group. Diabetes. 1998;47:821–830. doi: 10.2337/diabetes.47.5.821. [DOI] [PubMed] [Google Scholar]
- 19.Placha G, Poznik GD, Dunn J, Smiles A, Krolewski B, Glew T, Puppala S, Schneider J, Rogus JJ, Rich SS, et al. A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes. 2006;55:3358–3365. doi: 10.2337/db06-0781. [DOI] [PubMed] [Google Scholar]
- 20.Chen G, Adeyemo AA, Zhou J, Chen Y, Doumatey A, Lashley K, Huang H, Amoah A, Agyenim-Boateng K, Eghan BA, et al. A genome-wide search for linkage to renal function phenotypes in West Africans with type 2 diabetes. Am J Kidney Dis. 2007;49:394–400. doi: 10.1053/j.ajkd.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 21.Osterholm AM, He B, Pitkaniemi J, Albinsson L, Berg T, Sarti C, Tuomilehto J, Tryggvason K. Genome-wide scan for type 1 diabetic nephropathy in the Finnish population reveals suggestive linkage to a single locus on chromosome 3q. Kidney Int. 2007;71:140–145. doi: 10.1038/sj.ki.5001933. [DOI] [PubMed] [Google Scholar]
- 22.Bowden DW, Colicigno CJ, Langefeld CD, Sale MM, Williams A, Anderson PJ, Rich SS, Freedman BI. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 23.Igo RP, Iyengar SK, Nicholas SB, Goddard KA, Langefeld CD, Hanson RL, Duggirala R, Divers J, Abboud H, Adler SG, et al. Genomewide linkage scan for diabetic renal failure and albuminuria: the FIND study. Am J Nephrol. 2011;33:381–389. doi: 10.1159/000326763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schelling JR, Abboud HE, Nicholas SB, Pahl MV, Sedor JR, Adler SG, Arar NH, Bowden DW, Elston RC, Freedman BI, et al. Genome-wide scan for estimated glomerular filtration rate in multi-ethnic diabetic populations: the Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2008;57:235–243. doi: 10.2337/db07-0313. [DOI] [PubMed] [Google Scholar]
- 25.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, Koya D, Babazono T, Tanaka Y, Matsuda M, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 26.Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, Mychaleckyj JC, Rich SS, Warram JH, Krolewski AS. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, Mychaleckyj JC, Rich SS, Kirk JK, Freedman BI, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009;73:152–159. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu HY, Wang Y, Chen M, Zhang X, Wang D, Pan Y, Li L, Liu D, Dai XM. Association of ELMO1 gene polymorphisms with diabetic nephropathy in Chinese population. J Endocrinol Invest. 2013;36:298–302. doi: 10.3275/8525. [DOI] [PubMed] [Google Scholar]
- 29.Craig DW, Millis MP, DiStefano JK. Genome-wide SNP genotyping study using pooled DNA to identify candidate markers mediating susceptibility to end-stage renal disease attributed to Type 1 diabetes. Diabet Med. 2009;26:1090–1098. doi: 10.1111/j.1464-5491.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- 30.Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, Taylor A, Guiducci C, McAteer JB, McKay GJ, Isakova T, et al. Association testing of previously reported variants in a large case-control meta-analysis of diabetic nephropathy. Diabetes. 2012;61:2187–2194. doi: 10.2337/db11-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanson RL, Millis MP, Young NJ, Kobes S, Nelson RG, Knowler WC, DiStefano JK. ELMO1 variants and susceptibility to diabetic nephropathy in American Indians. Mol Genet Metab. 2010;101:383–390. doi: 10.1016/j.ymgme.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim S, Abboud HE, Pahl MV, Tayek J, Snyder S, Tamkin J, Alcorn H, Ipp E, Nast CC, Elston RC, et al. Examination of association with candidate genes for diabetic nephropathy in a Mexican American population. Clin J Am Soc Nephrol. 2010;5:1072–1078. doi: 10.2215/CJN.06550909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson RL, Craig DW, Millis MP, Yeatts KA, Kobes S, Pearson JV, Lee AM, Knowler WC, Nelson RG, Wolford JK. Identification of PVT1 as a candidate gene for end-stage renal disease in type 2 diabetes using a pooling-based genome-wide single nucleotide polymorphism association study. Diabetes. 2007;56:975–983. doi: 10.2337/db06-1072. [DOI] [PubMed] [Google Scholar]
- 34.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, Hester JM, Wing MR, Bostrom MA, Rudock ME, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011;79:563–572. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooke JN, Bostrom MA, Hicks PJ, Ng MC, Hellwege JN, Comeau ME, Divers J, Langefeld CD, Freedman BI, Bowden DW. Polymorphisms in MYH9 are associated with diabetic nephropathy in European Americans. Nephrol Dial Transplant. 2012;27:1505–1511. doi: 10.1093/ndt/gfr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freedman BI, Hicks PJ, Bostrom MA, Cunningham ME, Liu Y, Divers J, Kopp JB, Winkler CA, Nelson GW, Langefeld CD, et al. Polymorphisms in the non-muscle myosin heavy chain 9 gene (MYH9) are strongly associated with end-stage renal disease historically attributed to hypertension in African Americans. Kidney Int. 2009;75:736–745. doi: 10.1038/ki.2008.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park H, Kim HJ, Lee S, Yoo YJ, Ju YS, Lee JE, Cho SI, Sung J, Kim JI, Seo JS. A family-based association study after genome-wide linkage analysis identified two genetic loci for renal function in a Mongolian population. Kidney Int. 2013;83:285–292. doi: 10.1038/ki.2012.389. [DOI] [PubMed] [Google Scholar]
- 41.Pezzolesi MG, Jeong J, Smiles AM, Skupien J, Mychaleckyj JC, Rich SS, Warram JH, Krolewski AS. Family-based association analysis confirms the role of the chromosome 9q21.32 locus in the susceptibility of diabetic nephropathy. PLoS One. 2013;8:e60301. doi: 10.1371/journal.pone.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda S, Araki S, Babazono T, Toyoda M, Umezono T, Kawai K, Imanishi M, Uzu T, Watada H, Suzuki D, et al. Replication study for the association between four Loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes. 2010;59:2075–2079. doi: 10.2337/db10-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandholm N, Salem RM, McKnight AJ, Brennan EP, Forsblom C, Isakova T, McKay GJ, Williams WW, Sadlier DM, Mäkinen VP, et al. New susceptibility loci associated with kidney disease in type 1 diabetes. PLoS Genet. 2012;8:e1002921. doi: 10.1371/journal.pgen.1002921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandholm N, Forsblom C, Mäkinen VP, McKnight AJ, Osterholm AM, He B, Harjutsalo V, Lithovius R, Gordin D, Parkkonen M, et al. Genome-wide association study of urinary albumin excretion rate in patients with type 1 diabetes. Diabetologia. 2014;57:1143–1153. doi: 10.1007/s00125-014-3202-3. [DOI] [PubMed] [Google Scholar]
- 45.Hallman DM, Boerwinkle E, Gonzalez VH, Klein BE, Klein R, Hanis CL. A genome-wide linkage scan for diabetic retinopathy susceptibility genes in Mexican Americans with type 2 diabetes from Starr County, Texas. Diabetes. 2007;56:1167–1173. doi: 10.2337/db06-1373. [DOI] [PubMed] [Google Scholar]
- 46.Looker HC, Nelson RG, Chew E, Klein R, Klein BE, Knowler WC, Hanson RL. Genome-wide linkage analyses to identify Loci for diabetic retinopathy. Diabetes. 2007;56:1160–1166. doi: 10.2337/db06-1299. [DOI] [PubMed] [Google Scholar]
- 47.Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011;20:2472–2481. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang YC, Lin JM, Lin HJ, Chen CC, Chen SY, Tsai CH, Tsai FJ. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011;118:642–648. doi: 10.1016/j.ophtha.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 49.Sheu WH, Kuo JZ, Lee IT, Hung YJ, Lee WJ, Tsai HY, Wang JS, Goodarzi MO, Klein R, Klein BE, et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum Mol Genet. 2013;22:3165–3173. doi: 10.1093/hmg/ddt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu YP, Hallman DM, Gonzalez VH, Klein BE, Klein R, Hayes MG, Cox NJ, Bell GI, Hanis CL. Identification of Diabetic Retinopathy Genes through a Genome-Wide Association Study among Mexican-Americans from Starr County, Texas. J Ophthalmol. 2010;2010 doi: 10.1155/2010/861291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng W, Deshmukh HA, van Zuydam NR, Liu Y, Donnelly LA, Zhou K, Morris AD, Colhoun HM, Palmer CN, Smith BH. A genome-wide association study suggests an association of Chr8p21.3 (GFRA2) with diabetic neuropathic pain. Eur J Pain. 2015;19:392–399. doi: 10.1002/ejp.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 53.deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 54.Grimsley CM, Kinchen JM, Tosello-Trampont AC, Brugnera E, Haney LB, Lu M, Chen Q, Klingele D, Hengartner MO, Ravichandran KS. Dock180 and ELMO1 proteins cooperate to promote evolutionarily conserved Rac-dependent cell migration. J Biol Chem. 2004;279:6087–6097. doi: 10.1074/jbc.M307087200. [DOI] [PubMed] [Google Scholar]
- 55.Yokoyama N, deBakker CD, Zappacosta F, Huddleston MJ, Annan RS, Ravichandran KS, Miller WT. Identification of tyrosine residues on ELMO1 that are phosphorylated by the Src-family kinase Hck. Biochemistry. 2005;44:8841–8849. doi: 10.1021/bi0500832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanui T, Inayoshi A, Noda M, Iwata E, Stein JV, Sasazuki T, Fukui Y. DOCK2 regulates Rac activation and cytoskeletal reorganization through interaction with ELMO1. Blood. 2003;102:2948–2950. doi: 10.1182/blood-2003-01-0173. [DOI] [PubMed] [Google Scholar]
- 57.Janardhan A, Swigut T, Hill B, Myers MP, Skowronski J. HIV-1 Nef binds the DOCK2-ELMO1 complex to activate rac and inhibit lymphocyte chemotaxis. PLoS Biol. 2004;2:E6. doi: 10.1371/journal.pbio.0020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haase D, Meister M, Muley T, Hess J, Teurich S, Schnabel P, Hartenstein B, Angel P. FRMD3, a novel putative tumour suppressor in NSCLC. Oncogene. 2007;26:4464–4468. doi: 10.1038/sj.onc.1210225. [DOI] [PubMed] [Google Scholar]
- 59.Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, De Wolf-Peeters C, Pauwels P, Hagemeijer A, Marynen P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 60.Debelenko LV, Arthur DC, Pack SD, Helman LJ, Schrump DS, Tsokos M. Identification of CARS-ALK fusion in primary and metastatic lesions of an inflammatory myofibroblastic tumor. Lab Invest. 2003;83:1255–1265. doi: 10.1097/01.lab.0000088856.49388.ea. [DOI] [PubMed] [Google Scholar]
- 61.Patel KG, Liu C, Cameron PL, Cameron RS. Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1alpha and 1gamma1. J Neurosci. 2001;21:7954–7968. doi: 10.1523/JNEUROSCI.21-20-07954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang SC, Pauls DL, Lange C, Sasanfar R, Santangelo SL. Sex-specific association of a common variant of the XG gene with autism spectrum disorders. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:742–750. doi: 10.1002/ajmg.b.32165. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-Murillo L, Xu B, Roos JL, Abecasis GR, Gogos JA, Karayiorgou M. Fine mapping on chromosome 13q32-34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropsychopharmacology. 2014;39:934–943. doi: 10.1038/npp.2013.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Basson J, Sung YJ, Schwander K, Kume R, Simino J, de las Fuentes L, Rao D. Gene-education interactions identify novel blood pressure loci in the Framingham Heart Study. Am J Hypertens. 2014;27:431–444. doi: 10.1093/ajh/hpt283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carew RM, Sadagurski M, Goldschmeding R, Martin F, White MF, Brazil DP. Deletion of Irs2 causes reduced kidney size in mice: role for inhibition of GSK3beta? BMC Dev Biol. 2010;10:73. doi: 10.1186/1471-213X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi HJ, Yoon YJ, Kwon YK, Lee YJ, Chae S, Hwang D, Hwang GS, Kwon TH. Patterns of gene and metabolite define the effects of extracellular osmolality on kidney collecting duct. J Proteome Res. 2012;11:3816–3828. doi: 10.1021/pr300309d. [DOI] [PubMed] [Google Scholar]
- 67.Hookham MB, O’Donovan HC, Church RH, Mercier-Zuber A, Luzi L, Curran SP, Carew RM, Droguett A, Mezzano S, Schubert M, et al. Insulin receptor substrate-2 is expressed in kidney epithelium and up-regulated in diabetic nephropathy. FEBS J. 2013;280:3232–3243. doi: 10.1111/febs.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carew RM, Browne MB, Hickey FB, Brazil DP. Insulin receptor substrate 2 and FoxO3a signalling are involved in E-cadherin expression and transforming growth factor-β1-induced repression in kidney epithelial cells. FEBS J. 2011;278:3370–3380. doi: 10.1111/j.1742-4658.2011.08261.x. [DOI] [PubMed] [Google Scholar]
- 69.Singh BK, Singh A, Mascarenhas DD. A nuclear complex of rictor and insulin receptor substrate-2 is associated with albuminuria in diabetic mice. Metab Syndr Relat Disord. 2010;8:355–363. doi: 10.1089/met.2010.0011. [DOI] [PubMed] [Google Scholar]
- 70.Bostrom MA, Freedman BI. The spectrum of MYH9-associated nephropathy. Clin J Am Soc Nephrol. 2010;5:1107–1113. doi: 10.2215/CJN.08721209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnstone DB, Zhang J, George B, Léon C, Gachet C, Wong H, Parekh R, Holzman LB. Podocyte-specific deletion of Myh9 encoding nonmuscle myosin heavy chain 2A predisposes mice to glomerulopathy. Mol Cell Biol. 2011;31:2162–2170. doi: 10.1128/MCB.05234-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzuki N, Kunishima S, Ikejiri M, Maruyama S, Sone M, Takagi A, Ikawa M, Okabe M, Kojima T, Saito H, et al. Establishment of mouse model of MYH9 disorders: heterozygous R702C mutation provokes macrothrombocytopenia with leukocyte inclusion bodies, renal glomerulosclerosis and hearing disability. PLoS One. 2013;8:e71187. doi: 10.1371/journal.pone.0071187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Conti MA, Malide D, Dong F, Wang A, Shmist YA, Liu C, Zerfas P, Daniels MP, Chan CC, et al. Mouse models of MYH9-related disease: mutations in nonmuscle myosin II-A. Blood. 2012;119:238–250. doi: 10.1182/blood-2011-06-358853. [DOI] [PMC free article] [PubMed] [Google Scholar]


