Abstract
Meningiomas account for one-third of all adult central nervous system tumours and are divided into three WHO grades. In contrast to the relatively well characterized genetic alterations, our current understanding of epigenetic modifications involved in the meningioma-genesis and progression is rather incomplete. Contrary to genetic alterations, epigenetic changes do not alter the primary DNA sequence and their reversible nature serves as an excellent basis for prevention and development of novel personalised tumour therapies. Indeed, growing body of evidence suggests that disturbed epigenetic regulation plays a key role in the pathogenesis of meningiomas. Altered DNA methylation, microRNA expression, histone, and chromatin modifications are frequently noted in meningiomas bearing prognostic and therapeutic relevance. In this review we provide an overview on recently identified epigenetic alterations in meningiomas and discuss their role in tumour initiation, progression, and recurrence.
1. Introduction
Meningiomas originate from arachnoidal cap cells of the leptomeninges and account for around 30% of all central nervous system (CNS) tumours in adults. Their incidence increases with age with a female : male ratio of 2 : 1. Furthermore, autopsy and imaging studies indicate that the prevalence of subclinical meningiomas is roughly 3% in the population [1]. According to the World Health Organization (WHO) meningiomas can be divided into three histological grades: benign (grade 1), atypical (grade 2), and anaplastic meningiomas (grade 3). Besides this classification several other subtypes and variants exist (Table 1) [2, 3]. Although, 90% of meningiomas are typically benign slow-growing tumours, grade 2 and 3 tumours exhibit aggressive clinical phenotype with an increased risk of recurrence and invasive growth pattern [4]. The main histopathological characteristics of the different grades and subtypes were thoroughly reviewed by Riemenschneider et al. [5].
Table 1.
The histological subtypes of meningiomas.
| Histological subtypes | |
|---|---|
| Benign meningioma (WHO grade 1) |
Meningothelial, fibrous or fibroblastic, transitional or mixed, psammomatous, angiomatous, microcystic, secretory, lymphoplasmacyte-rich, and metaplastic meningioma |
|
| |
| Atypical meningioma (WHO grade 2) |
Chordoid, clear cell, atypical, and brain invasive meningioma |
|
| |
| Anaplastic meningioma (WHO grade 3) |
Papillary, rhabdoid, and anaplastic or malignant meningioma |
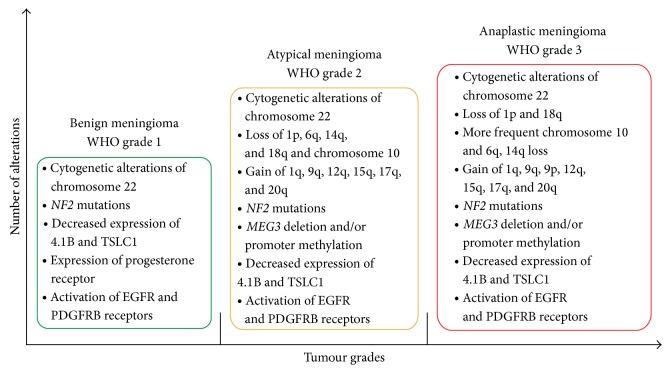
However, most meningiomas are sporadic; they are rarely associated with familial tumour syndromes such as Li-Fraumeni, Turcot, Gardener, von Hippel-Lindau, Cowden, Gorlin, and multiple endocrine neoplasia type I (MEN1) [6]. The aetiology of meningiomas is still unclear; ionizing radiation, head trauma, hormone-replacement therapy, and advanced age are the established risk factors though [7]. While the cytogenetic landscape of tumours is well known, the molecular mechanisms underlying progression and recurrence are not well defined. Cytogenetic alterations of chromosome 22 and NF2 gene are characteristic genetic alterations in early tumorigenesis and are frequently noted in higher grade tumours. In contrast to benign meningiomas, grade 2 and 3 tumours display more complex cytogenetic and molecular background with activation of oncogenes, inactivation of tumour suppressor genes, and alterations in other genes involved in several cellular pathways (Figure 1) [8]. Despite the identification of many potential molecular targets current treatment strategies are still limited to conventional forms of tumour therapy [9].
Figure 1.
Characteristic cytogenetic and molecular alterations of the different WHO grades of meningiomas.
During the last decade, disruption of normal epigenetic regulation has been recognized as a novel hallmark of cancer [10]. Deeper understanding of epigenetic modifications in meningiomas may increase our knowledge regarding tumorigenesis, progression, and recurrence. The goal of our review is to provide an overview of advances in the field of meningioma epigenetics with focus on possible biomarkers which may open the door to novel and more effective molecular diagnosis and therapies.
2. Epigenetic Profile of Meningiomas
In the past cancer was viewed as a disease initiated by the accumulation of genetic alterations causing neoplastic transformation in the respective cell type of origin. However, in the new millennium, it has become evident that also epigenetic regulation plays a prominent role in tumorigenesis. In principle, epigenetic modifications are heritable changes that influence gene expression without altering the primary sequence of DNA [11]. Besides cancer, disruption of the normal epigenetic regulation may also contribute to the pathogenesis of inflammatory, autoimmune, metabolic, neurological, and blood disorders [12]. Since the basics of epigenetics in cancer had been established it has become one of the most promising research fields of neurooncology which may reveal potential targets for drug development and therapy [13]. However, the epigenetic landscape of meningiomas remains still incomplete with altered DNA methylation, aberrant microRNA expression, and mutant epigenetic modifiers (EMGs) involved in histone and chromatin modifications being potential epigenetic markers of progression and recurrence (Table 2).
Table 2.
Epigenetic changes and their supposed role in meningiomas.
| Epigenetic alterations | Affected genes | Distribution between WHO grades | Possible effects in meningioma |
|---|---|---|---|
| Promoter methylation | TIMP3 | Grades 1 < 2 < 3 | Associated with tumour progression and aggressiveness parameters |
| HOXA7, HOXA9, and HOXA10 | Grades 1 < 2 < 3 | Associated with tumour progression and aggressiveness parameters | |
| RASSF1A | Grades 1 < 2 < 3 | Associated with the malignant transformation of a meningioma | |
| TP73 | Grades 1 < 2 < 3 | Associated with the malignant transformation of a meningioma | |
| NDRG2 | Grades 1 < 2 < 3 | Associated with malignant progression and predisposition to recurrence | |
| MAL2 | Grade 1 | Unknown | |
| IGF2BP1 | Grade 2 | Increases the malignant potential of tumours | |
| PDCD1 | Grade 2 | Increases the malignant potential of tumours | |
|
| |||
| Disturbed chromatin regulation | KDM5C | Grades 1 and 3 | Disturbed chromatin regulation |
| KDM6A | Grade 2 | Disturbed chromatin regulation | |
| SMARCB1 | Grade 1 | Abnormal chromatin remodelling | |
|
| |||
| Abnormal microRNAs expression | miR-29c-3p | Grades 1 > 2 > 3 | Associated with advanced clinical stages |
| miR-219-5p | Grades 1 > 2 > 3 | Associated with advanced clinical stages | |
| miR-190a | Grades 1 < 2 < 3 | Associated with advanced clinical stages | |
| miR-200a | Unknown | Functions as a multifunctional tumour suppressor miRNA | |
| miR-145 | Grades 1 > 2; 3 | Has an important antimigratory and antiproliferative function | |
2.1. Aberrant DNA Methylation in Meningiomas
Altered DNA methylation was the first epigenetic mark shown to be associated with cancer caused by both global DNA hypomethylation and/or promoter hypermethylation of certain genes. De novo methylation of DNA is catalysed by the enzymes DNMT3A and DNMT3B whereas maintenance of methylation is mediated by DNMT1 (Figure 2) [14, 15].
Figure 2.

Schematic representation of unmethylated (a) and methylated (b) genes. The most CpG islands in the promoter region of normal genes are unmethylated (a) [11]. DNA methylation refers to the addition of a methyl (CH3) group to the fifth carbon atom of the cytosine residues resulting in the formation of 5-methylcytosine (b). The process is mediated by DNA methyltransferase enzymes. DNA methylation occurs mainly at cytosine-guanosine dinucleotides (CpGs) which are concentrated in promoter CpG islands. CpG islands are short DNA sequences (<200 bp) with greater than 50% GC content. Methylation of CpGs in promoter regions plays an important role in both chromatin structure control and gene expression [14].
Intriguingly, NF2 (neurofibromatosis-2; a gene known to be frequently involved in development of meningiomas) promoter methylation does not play a key role in meningioma development [16]. Nevertheless, emerging evidence supports the involvement of DNA methylation in meningioma progression [13]. In contrast to NF2, the promoter methylation of tissue inhibitor of metalloproteinase 3 (TIMP3), located quite close to the NF2 gene, is inactivated in meningiomas [17]. Hypermethylation of TIMP3 is present in 67% of anaplastic meningiomas, but only in 22% of atypical and 17% of benign meningiomas. Thus, inactivation of TIMP3 by methylation may be involved in meningioma progression and can be a potential marker of an aggressive, high-grade meningioma phenotype [18]. Similar to TIMP3, repression of HOXA7, HOXA9, and HOXA10 in meningioma is also associated with clinically aggressive behaviour. DNA methylation levels of HOXA7, HOXA9, and HOXA10 were reported to be significantly higher in atypical and anaplastic meningiomas than in the benign form [19]. The methylation status of these three genes was lower in newly diagnosed grade 1 meningiomas in contrast to their recurrent counterpart and multiplex meningiomas presented with significantly higher HOXA10 methylation as compared to solitary meningiomas [20]. Promoter methylation of RASSF1A, TP73, and NDRG2 is more frequent in higher grade tumours than in benign meningioma [21, 22]. Repressed O6-methylguanine-DNA methyltransferase (MGMT) by promoter methylation is a prognostic biomarker in glioblastoma (GBM) [23]. The alkylating chemotherapeutic agent temozolomide (TMZ) increases the overall survival in GBM patients where the MGMT gene is methylated [24]. Unlike the vast majority of gliomas, testing the MGMT methylation status is not essential because the gene is unmethylated in the majority of meningiomas [25]. On the other hand, Larijani et al. found an increased but statistically not significant MGMT methylation with higher tumour grade, it was more frequent in males [26].
High-throughput techniques have made the genome-wide methylation analysis of human tumours including meningiomas possible. Evidence suggests that anaplastic meningioma could be distinguished from atypical and benign tumours according to DNA methylation patterns [19]. In addition, unlike in benign meningiomas, grade 2 and 3 tumours demonstrate increased global DNA hypomethylation. Interestingly, the majority of hypermethylated genes are suppressed in all tumours, but MAL2 is highly expressed in grade 1 and silenced in grade 3 meningiomas [27]. Another study identified nine differentially methylated genes by whole genome methylation analysis of benign and atypical meningiomas. The largest difference in methylation status was observed in IGF2BP1 and PDCD1 [28]. IGF2BP1 encodes the IGF2 mRNA binding protein 1 (IGF2BP1) which mediates the cytoplasmic fate of specific target mRNAs including ACTB and CD44. IGF2BP1 is a potent oncogenic factor that regulates the adhesion, migration, and invasiveness of tumour cells by modulating intracellular signalling [29].
Using genome-wide methylation analysis, grade 1 and 2 meningiomas can be divided into three subgroups. Based on this result, a simplified scoring system with five hypermethylated genes (HOXA6, HOXA9, PENK, UPK3A, and IGF2BP1) was proposed. This classification correlates well with recurrence and progression but there was no association with WHO histological or Simpson neurosurgical grades [30].
2.2. Mutations Related to Histone Modifications
Histone modifications are disturbed in many diseases including cancer. Histone octamer cores are formed by two copies of each histone protein (H2A, H2B, H3, and H4) and one H1 linker histone [31, 32]. Besides DNA condensation, histone proteins are also involved in regulation of gene expression by posttranslational modifications which mainly occur along their N-terminal tail. Histones can be modified by acetylation, methylation, phosphorylation, sumoylation, and ubiquitination. These modifications can result in an open (euchromatin) or closed (heterochromatin) state of the chromatin [31, 33]. Hence, methylated H3K9, H3K27, and H4K20 can result in a closed chromatin conformation, while open chromatin structure can be caused by methylated H3K4, H3K36, and H3K79 [34].
Not much is known about the precise role of these modifications in the initiation and progression of cancer. Histone modification pattern such as histone H3 lysine 9 trimethylation (H3K9me3), which has a prognostic relevance in glioblastoma, has not been detected in meningiomas yet [35]. On the other hand, current genome-wide studies reported mutations of epigenetic modifier genes encoding proteins that regulate the chromatin structure of cells. Among the affected EMGs KDM5C and KDM6A are histone demethylases, while SMARCB1 and SMARCE1 are members of the SWI/SNF chromatin-remodelling complex [36, 37].
2.3. Role of MicroRNAs in Meningiomas
MicroRNAs (miRNAs) are single-stranded noncoding RNAs, composed of 19 to 24 nucleotides, and play regulatory roles in important biological processes, such as cell cycle, proliferation, differentiation, migration, and apoptosis [38]. They cause posttranslational gene expression silencing by binding to complementary sites on their target mRNAs and initiate either degradation or inhibition of translation. Dysregulated miRNAs expression was described in a number of human diseases, including cardiovascular, autoimmune, inflammatory, neurodevelopmental diseases and cancer [39, 40]. Altered miRNA expression is associated with both genetic and epigenetic mechanisms. miRNAs usually have reduced levels in tumour cells and probably function as oncogenes or as tumour suppressors [41]. In cancer, several miRNAs correlate well with clinicopathological features, such as metastasis, recurrence, and length of survival [42].
MicroRNA expression profiling of meningiomas revealed downregulation of miR-29c-3p and miR-219-5p, which were associated with advanced clinical stages. High expression of miR-190a and low expression of miR-29c-3p and miR-219-5p are correlated with significantly higher recurrence rates in meningioma patients. Importantly, miR-190a expression level is a prognostic predictor of postsurgical outcomes [43]. The mRNA of β-catenin is a target for miR-200a; consequently β-catenin translation and Wnt signalling are suppressed by overexpressed miR-200a [44], and miR-200a may act as a multifunctional tumour suppressor miRNA. A direct correlation was found between the downregulation of miR-200a and the upregulation of β-catenin in meningiomas. Reduced miR-200a causes decreased expression of the ZEB1 and SIP1 transcription factors resulting in the downregulation of E-cadherin [45]. Moreover, it provokes an increased β-catenin and cyclin D1 expression and activates the Wnt signalling pathway in meningiomas [46]. miR-145 may have an important antimigratory and antiproliferative function in cancer [47]. Contrary to grade 1 meningiomas, significantly decreased miR-145 levels were detected in grade 2 and 3 tumours. Increased levels of miR-145 may result in downregulated collagen type V alpha (COL5A1) expression. Accordingly, COL5A1 expression is upregulated in atypical and anaplastic meningiomas [48].
3. Conclusion
In the past decade it has become evident that epigenetic factors are involved in tumour pathogenesis. Contrary to genetic alterations, epigenetic changes do not alter the primary DNA sequence and their reversible nature serves as an excellent basis for prevention and development of novel and personalised cancer therapies. Similar to other tumours, disturbed epigenetic regulation plays a key role in the pathogenesis of meningiomas. DNA methylation, microRNA expression, histone, and chromatin modifications can be altered in meningiomas with a prognostic relevance and may become a potential therapeutic target in the future.
Acknowledgments
Recent review was supported by the National Brain Research Program, Hungary, Grants nos. KTIA_13_NAP-A-II/7 (Tibor Hortobágyi) and KTIA_13_NAP-A-V/3 (Álmos Klekner) and by the Janos Bolyai Scholarship of the Hungarian Academy of Sciences (Álmos Klekner).
Disclosure
Both Tibor Hortobágyi and Álmos Klekner are “last/senior authors” of this paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Wiemels J., Wrensch M., Claus E. B. Epidemiology and etiology of meningioma. Journal of Neuro-Oncology. 2010;99(3):307–314. doi: 10.1007/s11060-010-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis D. N., Ohgaki H., Wiestler O. D., et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathologica. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csonka T., Murnyák B., Szepesi R., Kurucz A., Klekner Á., Hortobágyi T. Poly(ADP-ribose) polymerase-1 (PARP1) and p53 labelling index correlates with tumour grade in meningiomas. Folia Neuropathologica. 2014;52(2):111–120. doi: 10.5114/fn.2014.43782. [DOI] [PubMed] [Google Scholar]
- 4.Klekner Á., Röhn G., Schillinger G., Schröder R., Klug N., Ernestus R.-I. ODC mRNA as a prognostic factor for predicting recurrence in meningiomas. Journal of Neuro-Oncology. 2001;53(1):67–75. doi: 10.1023/A:1011878928318. [DOI] [PubMed] [Google Scholar]
- 5.Riemenschneider M. J., Perry A., Reifenberger G. Histological classification and molecular genetics of meningiomas. The Lancet Neurology. 2006;5(12):1045–1054. doi: 10.1016/s1474-4422(06)70625-1. [DOI] [PubMed] [Google Scholar]
- 6.Mawrin C., Perry A. Pathological classification and molecular genetics of meningiomas. Journal of Neuro-Oncology. 2010;99(3):379–391. doi: 10.1007/s11060-010-0342-2. [DOI] [PubMed] [Google Scholar]
- 7.Murnyák B., Csonka T., Hortobágyi T. Molecular pathology of meningiomas. doi: 10.18071/isz.68.0292. Ideggyogy Sz. In press. [DOI] [PubMed] [Google Scholar]
- 8.Suvà M. L., Louis D. N. Next-generation molecular genetics of brain tumours. Current Opinion in Neurology. 2013;26(6):681–687. doi: 10.1097/WCO.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 9.Choy W. C., Kim W., Nagasawa D., et al. The molecular genetics and tumor pathogenesis of meningiomas and the future directions of meningioma treatments. Neurosurgical Focus. 2011;30(5, article E6) doi: 10.3171/2011.2.focus1116. [DOI] [PubMed] [Google Scholar]
- 10.Azad N., Zahnow C. A., Rudin C. M., Baylin S. B. The future of epigenetic therapy in solid tumours—lessons from the past. Nature Reviews Clinical Oncology. 2013;10(5):256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandoval J., Esteller M. Cancer epigenomics: beyond genomics. Current Opinion in Genetics and Development. 2012;22(1):50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 12.DeWoskin V. A., Million R. P. The epigenetics pipeline. Nature Reviews Drug Discovery. 2013;12(9):661–662. doi: 10.1038/nrd4091. [DOI] [PubMed] [Google Scholar]
- 13.Rodríguez-Paredes M., Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Medicine. 2011;17(3):330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 14.Tsai H.-C., Baylin S. B. Cancer epigenetics: linking basic biology to clinical medicine. Cell Research. 2011;21(3):502–517. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M. Cancer epigenetics for the 21st century: what's next? Genes and Cancer. 2011;2(6):604–606. doi: 10.1177/1947601911423096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansson C. M., Buckley P. G., Grigelioniene G., et al. Comprehensive genetic and epigenetic analysis of sporadic meningioma for macro-mutations on 22q and micro-mutations within the NF2 locus. BMC Genomics. 2007;8, article 16 doi: 10.1186/1471-2164-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linsler S., Kraemer D., Driess C., et al. Molecular biological determinations of meningioma progression and recurrence. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0094987.e94987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski D., Wolter M., Reifenberger G., Riemenschneider M. J. Hypermethylation and transcriptional downregulation of the TIMP3 gene is associated with allelic loss on 22q12.3 and malignancy in meningiomas. Brain Pathology. 2010;20(3):623–631. doi: 10.1111/j.1750-3639.2009.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He S., Pham M. H., Pease M., et al. A review of epigenetic and gene expression alterations associated with intracranial meningiomas. Neurosurgical Focus. 2013;35(6, article E5) doi: 10.3171/2013.10.focus13360. [DOI] [PubMed] [Google Scholar]
- 20.Di Vinci A., Brigati C., Casciano I., et al. HOXA7, 9, and 10 are methylation targets associated with aggressive behavior in meningiomas. Translational Research. 2012;160(5):355–362. doi: 10.1016/j.trsl.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Nakane Y., Natsume A., Wakabayashi T., et al. Malignant transformation-related genes in meningiomas: allelic loss on 1p36 and methylation status of p73 and RASSF1A . Journal of Neurosurgery. 2007;107(2):398–404. doi: 10.3171/jns-07/08/0398. [DOI] [PubMed] [Google Scholar]
- 22.Lusis E. A., Watson M. A., Chicoine M. R., et al. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Research. 2005;65(16):7121–7126. doi: 10.1158/0008-5472.can-05-0043. [DOI] [PubMed] [Google Scholar]
- 23.Rivera A. L., Pelloski C. E., Gilbert M. R., et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro-Oncology. 2010;12(2):116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegi M. E., Diserens A.-C., Gorlia T., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England Journal of Medicine. 2005;352(10):997–1003. doi: 10.1056/nejmoa043331. [DOI] [PubMed] [Google Scholar]
- 25.de Robles P., McIntyre J., Kalra S., et al. Methylation status of MGMT gene promoter in meningiomas. Cancer Genetics and Cytogenetics. 2008;187(1):25–27. doi: 10.1016/j.cancergencyto.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Larijani L., Madjd Z., Samadikuchaksaraei A., et al. Methylation of O6-methyl guanine methyltransferase gene promoter in meningiomas—comparison between tumor grades I, II, and III. Asian Pacific Journal of Cancer Prevention. 2014;15(1):33–38. doi: 10.7314/apjcp.2014.15.1.33. [DOI] [PubMed] [Google Scholar]
- 27.Gao F., Shi L., Russin J., et al. DNA methylation in the malignant transformation of meningiomas. PLoS ONE. 2013;8(1) doi: 10.1371/journal.pone.0054114.e54114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vengoechea J., Sloan A. E., Chen Y., et al. Methylation markers of malignant potential in meningiomas. Journal of Neurosurgery. 2013;119(4):899–906. doi: 10.3171/2013.7.JNS13311. [DOI] [PubMed] [Google Scholar]
- 29.Stöhr N., Hüttelmaier S. IGF2BP1: a post-transcriptional “driver” of tumor cell migration. Cell Adhesion & Migration. 2012;6(4):312–318. doi: 10.4161/cam.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishida Y., Natsume A., Kondo Y., et al. Epigenetic subclassification of meningiomas based on genome-wide DNA methylation analyses. Carcinogenesis. 2012;33(2):436–441. doi: 10.1093/carcin/bgr260. [DOI] [PubMed] [Google Scholar]
- 31.Waldmann T., Schneider R. Targeting histone modifications—epigenetics in cancer. Current Opinion in Cell Biology. 2013;25(2):184–189. doi: 10.1016/j.ceb.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Chervona Y., Costa M. Histone modifications and cancer: biomarkers of prognosis? American Journal of Cancer Research. 2012;2(5):589–597. [PMC free article] [PubMed] [Google Scholar]
- 33.Spyropoulou A., Piperi C., Adamopoulos C., Papavassiliou A. G. Deregulated chromatin remodeling in the pathobiology of brain tumors. NeuroMolecular Medicine. 2013;15(1):1–24. doi: 10.1007/s12017-012-8205-y. [DOI] [PubMed] [Google Scholar]
- 34.Kim Y. Z. Altered histone modifications in gliomas. Brain Tumor Research and Treatment. 2014;2(1):7–21. doi: 10.14791/btrt.2014.2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venneti S., Felicella M. M., Coyne T., et al. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. Journal of Neuropathology and Experimental Neurology. 2013;72(4):298–306. doi: 10.1097/nen.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith M. J., O'Sullivan J., Bhaskar S. S., et al. Loss-of-function mutations in SMARCE1 cause an inherited disorder of multiple spinal meningiomas. Nature Genetics. 2013;45(3):295–298. doi: 10.1038/ng.2552. [DOI] [PubMed] [Google Scholar]
- 37.Brastianos P. K., Horowitz P. M., Santagata S., et al. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nature Genetics. 2013;45(3):285–289. doi: 10.1038/ng.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Q., Zhang C., Huang B., et al. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. European Journal of Gastroenterology and Hepatology. 2013;25(8):953–957. doi: 10.1097/MEG.0b013e32835ed691. [DOI] [PubMed] [Google Scholar]
- 39.Ardekani A. M., Naeini M. M. The role of microRNAs in human diseases. Avicenna Journal of Medical Biotechnology. 2010;2(4):161–179. [PMC free article] [PubMed] [Google Scholar]
- 40.Arunachalam G., Upadhyay R., Ding H., Triggle C. R. MicroRNA signature and cardiovascular dysfunction. Journal of Cardiovascular Pharmacology. 2014 doi: 10.1097/fjc.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 41.Melo S. A., Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Letters. 2011;585(13):2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 42.Monroig P. D. C., Calin G. A. MicroRNA and epigenetics: diagnostic and therapeutic opportunities. Current Pathobiology Reports. 2013;1(1):43–52. doi: 10.1007/s40139-013-0008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhi F., Zhou G., Wang S., et al. A microRNA expression signature predicts meningioma recurrence. International Journal of Cancer. 2013;132(1):128–136. doi: 10.1002/ijc.27658. [DOI] [PubMed] [Google Scholar]
- 44.Huang K., Zhang J.-X., Han L., et al. MicroRNA roles in beta-catenin pathway. Molecular Cancer. 2010;9, article 252 doi: 10.1186/1476-4598-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory P. A., Bert A. G., Paterson E. L., et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 46.Saydam O., Shen Y., Würdinger T., et al. Downregulated microRNA-200a in meningiomas promotes tumor growth by reducing E-cadherin and activating the Wnt/β-catenin signaling pathway. Molecular and Cellular Biology. 2009;29(21):5923–5940. doi: 10.1128/mcb.00332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dynoodt P., Speeckaert R., de Wever O., et al. miR-145 overexpression suppresses the migration and invasion of metastatic melanoma cells. International Journal of Oncology. 2013;42(4):1443–1451. doi: 10.3892/ijo.2013.1823. [DOI] [PubMed] [Google Scholar]
- 48.Kliese N., Gobrecht P., Pachow D., et al. MiRNA-145 is downregulated in atypical and anaplastic meningiomas and negatively regulates motility and proliferation of meningioma cells. Oncogene. 2013;32(39):4712–4720. doi: 10.1038/onc.2012.468. [DOI] [PubMed] [Google Scholar]